Abstract
Background
The cortical cytoskeleton network connects the actin cytoskeleton to various membrane proteins, influencing cell adhesion, polarity, migration and response to extracellular signals. Previous studies have suggested changes in the expression of specific components in prostate cancer, especially of 4.1 proteins (encoded by EPB41 genes) which form nodes in this network.
Methods
Expression of EPB41L1, EPB41L2, EPB41L3 (protein: 4.1B), EPB41L4B (EHM2), EPB41L5, EPB49 (dematin), VIL2 (ezrin), and DLG1 (summarized as „cortical cytoskeleton" genes) as well as ERG was measured by quantitative RT-PCR in a well-characterized set of 45 M0 prostate adenocarcinoma and 13 benign tissues. Hypermethylation of EPB41L3 and GSTP1 was compared in 93 cancer tissues by methylation-specific PCR. Expression of 4.1B was further studied by immunohistochemistry.
Results
EPB41L1 and EPB41L3 were significantly downregulated and EPB41L4B was upregulated in cancer tissues. Low EPB41L1 or high EPB41L4B expression were associated with earlier biochemical recurrence. None of the other cortical cytoskeleton genes displayed expression changes, in particular EPB49 and VIL2, despite hints from previous studies. EPB41L3 downregulation was significantly associated with hypermethylation of its promoter and strongly correlated with GSTP1 hypermethylation. Protein 4.1B was detected most strongly in the basal cells of normal prostate epithelia. Its expression in carcinoma cells was similar to the weaker one in normal luminal cells. EPB41L3 downregulation and EPB41L4B upregulation were essentially restricted to the 22 cases with ERG overexpression. Expression changes in EPB41L3 and EPB41L4B closely paralleled those previously observed for the extracellular matrix genes FBLN1 and SPOCK1, respectively.
Conclusions
Specific changes in the cortical cytoskeleton were observed during prostate cancer progression. They parallel changes in the expression of extracellular matrix components and all together appear to be associated with oncogenic ERG overexpression. We hypothesize that these alterations may contribute to the increased invasivity conferred to prostate cancer cells by ERG deregulation.
Background
The progression of epithelial tumors to invasive carcinomas involves changes in cell polarity, adhesion and motility that permit the detachment of cancer cells from the epithelial layer, their invasion into adjacent tissue layers and eventually their spread throughout the body. These processes require reorganization of the cellular cytoskeleton and altered expression of proteins that connect it to the cell membrane as well as remodelling of the extracellular matrix including changes in the composition and processing of its constituents.
The 4.1 proteins, encoded by the EPB41 (erythrocyte protein band 4.1) genes, are components of the cortical cytoskeleton underlying the cell membrane [1,2]. The family of 4.1 proteins consists of the eponymous 4.1R protein first identified in erythrocytes (gene: EPB41), 4.1N (EPB41L1), 4.1G (EPB41L2), 4.1B (EPB41L3) as well as the less closely related members NBL4 (EPB41L4A), EHM2 (EPB41L4B) and EPB41L5 (EPB41L5). They form nodes in the cell cortex connecting further components of the cortical cytoskeleton like spectrins, actin and transmembrane adhesion proteins, receptors and transporters with each other. In this fashion 4.1 proteins contribute to the organization of cell polarity, adhesion and motility, and affect transport through the membrane and responses to growth factors.
The 4.1B protein is most strongly expressed in neurons and is enriched in the basal cells of certain epithelia [2,3]. In addition to spectrins and actin, known binding partners comprise the adhesion molecule CD44 that binds hyaluronic acid in the extracellular matrix [1] and the candidate tumor suppressor disc large 1/DLG1 [4]. The 4.1B protein is downregulated in several carcinomas, including prostate cancer [5], likely by deletion or promoter hypermethylation of the EPB41L3 gene promoter [6]. Mouse models of prostate cancer progression suggest that it acts as a metastasis suppressor [7]. In contrast, EHM2 is conspicuous in tumor cells with high migratory potential, such as metastatic melanoma and fibrosarcoma cells [8,9]. In prostate cancer, EHM2 has been reported to be overexpressed [6,10] and to diminish adhesion of prostate cancer cells to collagen [10]. The most recently discovered 4.1 family member, the product of the EPB41L5 gene (also called Limulus), regulates cell adhesion during development [11], but has not yet been investigated in the context of human cancer.
The 4.1 proteins are part of a larger protein family characterized by FERM domains, of which many have related functions. For instance, the FERM domain protein ezrin, encoded by the VIL2 gene, connects CD81 at the cell membrane to the actin cytoskeleton. Ezrin has been shown to mediate invasion of prostate cancer cells [12,13], but whether it is overexpressed in prostate cancer is not known. The more distantly related protein dematin, too, interacts with the actin cytoskeleton and growth factor receptors. It is encoded by the EPB49 gene on 8p21.1, a region frequently deleted in prostate cancer. Overexpression of dematin in PC3 prostate cancer cells changed their morphology towards a more epithelial phenotype [14], but no investigations of EPB49 expression in prostate cancer tissues have been published.
The fundamental reorganization of the cytoskeleton, its attachment to the cell membrane and the extracellular matrix during cancer progression are orchestrated by transcription factors that activate cellular programs for cell migration and invasion, which are physiologically employed during embryogenesis, wound healing and tissue regeneration [15,16]. In cancer, such transcription factors, like Snail/SNAI1, Slug/SNAI2 and ZEB1, often become deregulated and promote tumor progression.
In prostate carcinoma, transcription factors of the ETS family are prominent candidates for oncogenes driving this facet of tumor progression. Specific members of this protein family are activated towards oncogenes by chromosomal translocations [17] placing a structural ETS transcription factor gene under the control of an androgen-responsive promoter, resulting in its deregulation and overexpression. The most common translocation, found in 30-70% of all cases, creates a fusion gene placing the androgen-responsive promoter of TMPRSS2 in control of the ERG structural gene encoding an ETS family transcription factor. This genetic aberration results in the androgen-driven overexpression of intact or amino-terminally truncated ERG proteins in prostate epithelial cells. ERG oncoproteins influence tumor cell proliferation, but exert a more pronounced effect on migration and invasion through broad changes in gene expression [18-20]. In accord with a function in promoting tumor progression, TMPRSS2-ERG translocations are observed in a significantly lower fraction of high-grade prostate intraepithelial neoplasias, a non-invasive precursor stage, than in invasive carcinomas [21].
We have previously reported downregulation of EPB41L3 encoding protein band 4.1B and upregulation of EPB41L4B encoding EHM2, respectively, in prostate cancer [6], in accord with observations by other groups [7,10]. In the present study, we have investigated the expression of further members of the family as well as selected genes encoding related or interacting proteins such as disc large 1, ezrin and dematin, and the relation of the changes to activation of oncogenic ERG.
Methods
Tissues
High quality RNA was available from 13 benign prostate tissues and 45 cancer samples collected from patients undergoing radical prostatectomy for prostate carcinoma between 1997 and 2002 in our institution as described in a prior study [Schulz et al, 2007]. TNM classification was performed according to the rules of the International Union against Cancer from 2002. Twenty cancers were staged as pT2, 23 as pT3 and two as pT4. Twenty-six cancer specimens had a Gleason score of 7, 13 < 7 and 6 > 7. At the time of surgery, no distant metastases were detectable, but 11 patients had lymph node metastases. The patients' age ranged from 59 to 74 years. Follow-up data were available for all patients with a median follow-up time of 98 months. High quality DNA was available from 93 cancer tissues essentially encompassing the 45 specimens used for RNA analysis. Of these 93 cases, 44 were staged as pT2 and 49 as pT3 or pT4. Sixteen patients had lymph node metastases, but no distant metastases were detected. Each 27 carcinomas were assigned a Gleason score > 7 or <7 and 39 a score of 7. The study was approved by the ethics committee of the Heinrich Heine University medical faculty.
DNA and RNA extraction
DNA and RNA were extracted from identical powdered tissues as described previously [22,23]. High molecular weight genomic DNA was isolated using the blood and cell culture DNA kit (Qiagen, Hilden, Germany). Total RNA was isolated using the RNeasy Mini Kit (Qiagen) following guanidinium/acid phenol/chloroform extraction (peqGOLD TriFast, peqLab, Erlangen, Germany). Quality of DNA and RNA was initially checked by spectrophotometry and subsequently by agarose gel or capillary electrophoresis, respectively. Only high quality DNA and RNA preparations were used in the present study.
Reverse transcription and quantitative RT-PCR
cDNA was prepared using SuperscriptII (Invitrogen, Karlsruhe, Germany) according to the manufacturer's protocol with a mixture of random and oligo-dT primers. Quantitative real-time RT-PCR was performed on an ABI 7900 instrument using commercially available Taqman assays specific for the respective mRNAs (Applied Biosystems, Weiterstadt, Germany), namely HS 00938024-m1 (DLG1), HS 00385004_m1 (EPB41L1), HS 00154988_m1 (EPB41L2), HS 00202360_m1 (EPB41L3), HS 00603031_m1 (EPB41L4B), HS 01554437_m1 (EPB41L5), HS 00157387_m1 (EPB49), HS 00267195_m1 (MKI67), HS 0018574_m1 (VIL2). TBP (HS 00427620_m1) was used as a reference gene throughout. The assay for ERG was chosen to cover exons common to all oncogenic transcripts [19]. All reactions were performed with cDNA corresponding to 10 ng of RNA each in at least duplicates using 40 cycles of 15 s at 95°C and 60 s at 60°C after an initial 10 min denaturation at 95°C. Each run was standardized using a dilution series of cDNA from a strongly expressing cell line or normal tissue. Experimental variation for each sample was below 10%.
DNA methylation analysis
DNA was bisulfite-treated using the EZ DNA Methylation Gold Kit (Zymo Research, Freiburg, Germany), Methylation-specific PCR for GSTP1 was performed as described [22] and accordingly for EPB41L3 using the primer pairs 5'-TTTGTGTATTGTTGTTGAGGAGTG-3' and 5'-CACAATCCCCCACTCCAAAAAACA-3' to detect unmethylated sequences or 5'-TTGCGTATCGTCGTCGTCGAGGACG-3' and 5'-CGCAATCCCCCACTCCGAAAAACG-3' to detect methylated sequences at 61°C and 64°C annealing temperature, respectively. Bisulfite sequencing of the EPB41L1 promoter was conducted by the method described previously [22] with the newly designed primers 5'-AGAGGAGATAGGTAGAGAGGA-3' and 5'-CTAAACCC(A/G)AAATTCCCCATATC-3' at 58°C.
Immunohistochemistry
For immunohistochemistry paraffin sections of tissue microarrays comprising prostate cancer and corresponding benign prostatic tissue samples were treated with xylene and ethanol. Sections were rehydrated and endogenous peroxidase activity was eliminated by H2O2. After antigen retrieval using a citrate buffer in a pressure cooker, endogenous biotin was blocked. Slides were incubated with a mouse monoclonal antibody against 4.1B (Santa Cruz Biotechnology, dilution 1:10) overnight at 4°C. After a second incubation with a biotin-conjugated polyvalent antibody, slides were mixed with an avidin-biotin-peroxidase reagent (Scy Tek, Logan, USA). Reaction products were visualized by immersing slides in diaminobenzidine tetrachloride and finally counter-stained with haematoxilin. For negative controls, slides were subjected to the entire immunohistochemical procedure except for the fact that the primary antibody was omitted. Under these conditions no staining was observed.
Statistical methods
All statistical calculations were performed with SPSS 18.0. For comparisons between groups Mann-Whitney U test was employed, correlations were evaluated by Spearman's rho, nominal data were evaluated by chi-square test and follow-up data by log-rank test.
Results
Expression of EPB41 and related genes in prostate cancer tissues
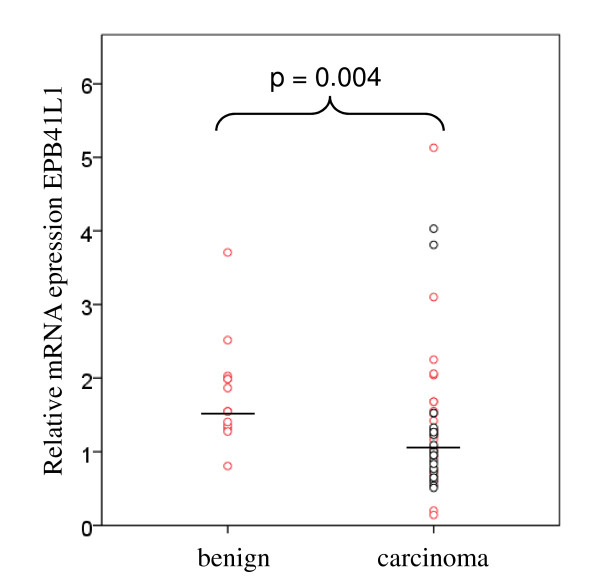
Expression of EPB41L1, EPB41L2, EPB41L3, EPB41L4B, EPB41L5, EPB49, VIL2, and DLG1 (summarized as „cortical cytoskeleton" genes) was measured by quantitative RT-PCR in a well-characterized set of 45 M0 prostate adenocarcinoma tissues and 13 benign tissues from cancer-carrying prostates. As reported previously [6], EPB41L3 mRNA was highly significantly (Mann-Whitney p < 0.001) diminished and EPB41L4B increased (p = 0.002) in these cancer tissues. The expression of EPB41L1 was decreased moderately, but significantly in cancer tissues (Fig. 1). The expression of the five other genes remained unchanged on average (Additional file 1, Figure S1).
Figure 1.
Expression of EPB41L1 in prostate cancers. Expression of EPB41L1 mRNA relative to TBP in prostate carcinoma and benign tissues. Black circles denote carcinoma specimens with ERG overexpression. Median values are indicated by horizontal lines.
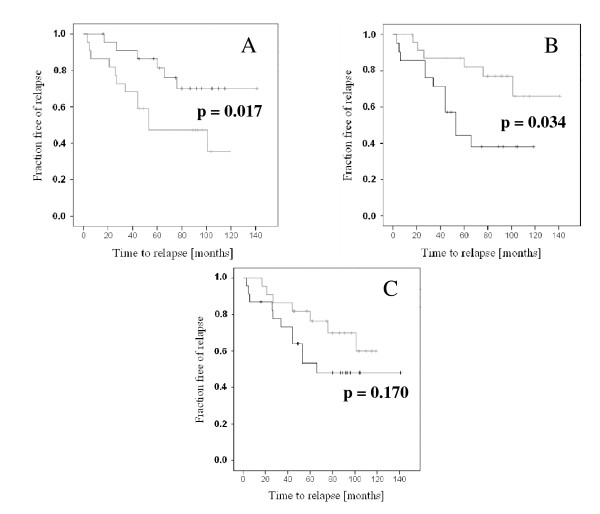
Within the cancer specimens, none of the genes showed a significant association of its expression level with tumor stage (pT2 vs. > pT2), Gleason score (< 7 vs. 7 vs. > 7) or lymph node metastasis. Below median expression of EPB41L1 was associated with significantly earlier biochemical recurrence, as was above median expression of EPB41L4B (Fig. 2). In contrast, decreased expression of EPB41L3 showed only a tendency towards earlier tumor relapse (Fig. 2).
Figure 2.
Relation of EPB41L3 and EPB41L4B expression to prostate cancer recurrence. Kaplan-Meyer analysis of biochemical recurrence in patients with below (lower line in B and C, upper line in A) or above (upper line in B and C, lower line in A) median expression of EPB41L4B (A), EPB41L1 (B) and EPB41L3 (C). The respective log rank p-values were 0.017, 0.034, and 0.170.
Expression of 4.1B protein in normal and cancerous prostate tissue
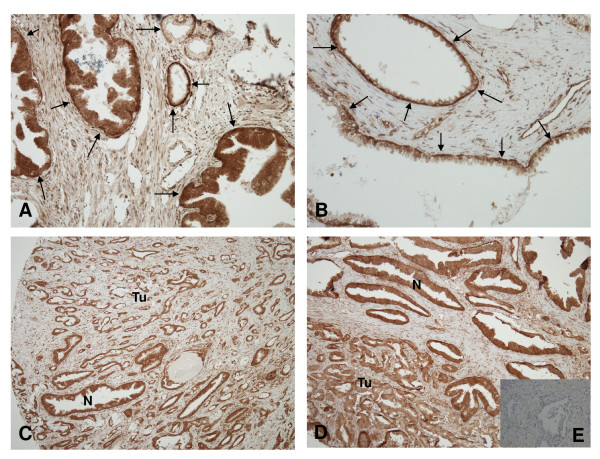
Immunohistochemical investigation (Fig. 3) revealed that in normal prostate glands, 4.1B expression was usually much stronger in basal cells than in secretory epithelial cells. Of note, although all glands stained positive, 4.1B expression was quite variable between individual glands (Fig. 3A, B). In cancerous glands, 4.1B protein remained detectable. In most cases staining intensity in tumor glands was similar to that of corresponding benign secretory epithelial cells within the same section (Fig. 3C). In some cases, 4.1B expression in carcinoma cells was decreased as compared to secretory epithelial cells of normal glands (Fig. 3D), but the opposite tendency was also observed in a few specimens.
Figure 3.
Immunohistochemical analysis of protein 4.1B expression in prostate cancer and benign prostatic glands. A and B: Heterogeneous expression of protein 4.1B in benign prostatic glands. Note stronger expression levels in basal cells (arrows) than in secretory epithelial cells. C-D: Comparison of 4.1B expression levels in tumor glands (TU) and corresponding benign prostatic glands (N): C and D: equivalent and slightly decreased, respectively, 4.1B expression in cancer cells compared to secretory cells of normal glands. Original magnifications: A, D, x200; B, x400; C, x100. E: negative control without primary antibody.
DNA methylation analysis of EPB41L3 and EPB41L1
Decreased expression of EPB41L3 was previously shown to be associated with methylation of its gene promoter in prostate cancer cell lines and a smaller series of prostate cancer tissues [6]. To more precisely measure its frequency and relationship to clinical parameters, the previously analyzed series was extended to 93 cancer specimens that were investigated by MS-PCR as described (a typical result is shown in Additional File 2, Figure S2). Of these, 61 (65%) cases displayed EPB41L3 hypermethylation. Hypermethylation was more frequent in higher stage cases, i.e. detectable in 25/44 (57%) of pT2 vs. 35/49 (71%) of higher stage specimens and 12/16 (75%) lymph-node positive vs. 49/78 (63%) lymph-node negative cancers, but the difference was not statistically significant (χ2 > 0.05). Average expression of EPB41L3 mRNA was significantly lower in carcinomas with hypermethylation of the gene than in those without (1.79 ± 0.50 vs. 1.33 ± 0.61; p = 0.025). In the same tissue series, 71 (76%) carcinomas displayed hypermethylation of GSTP1. Hypermethylation of EPB41L3 was significantly (p = 0.002) more frequent in tumors with GSTP1 hypermethylation than in those without.
To investigate methylation as a potential cause of diminished EPB41L1 expression in cancer, 47 CpG sites around the transcriptional start site (from bp -208 to +157) were analyzed by bisulfite sequencing in the prostate cancer cell lines PC3 and LNCaP, in two prostate cancer tissues with lowered expression, one benign prostate sample and in leucocytes. None of them displayed any methylation, whereas an artificially methylated DNA yielded the expected fully methylated sequence (data not shown).
Relation to changes in expression of other genes
Next, the expression of all cortical cytoskeleton genes in the cancer tissues was compared to that of MKI67, encoding the well established marker of cell proliferation in cancer, Ki67. Expression of EPB41L4B was positively (Spearman rho = 0.535, p < 0.001) and expression of EPB41L3 negatively (ς = - 0.411, p < 0.003) correlated with the proliferation marker.
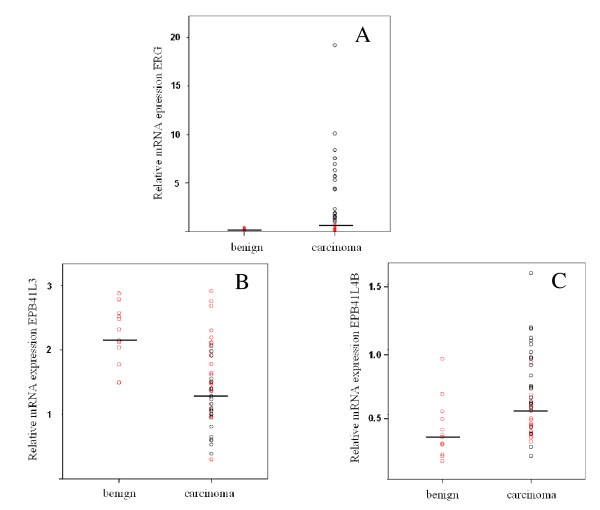
In various studies, approximately half of all prostate cancers have been observed to overexpress the ETS family gene ERG, predominantly as a consequence of different chromosomal translocations [24]. To cover all instances of ERG overexpression, we determined the expression of an mRNA segment common to all normal and oncogene transcripts by qRT-PCR and defined overexpression of ERG as a level exceeding twice that of the maximum of benign tissues. By that definition in the present series 22 of 45 cancers overexpressed ERG, by up to two orders of magnitude (Fig. 4A). Across all cases, ERG expression was significantly different between cancer and benign tissues (Mann-Whitney-U p = 0.029).
Figure 4.
Expression of ERG and its relation to EPB41L3 and EPB41L4B in prostate cancers. Expression of ERG, EPB41L3 and EPB41L4B mRNAs relative to TBP in prostate carcinoma and benign tissues. Black circles denote carcinoma specimens with ERG overexpression. Median values are indicated by horizontal lines.
Subsequently, expression of the cortical cytoskeleton genes was compared between the groups with elevated (ERG-high) and close to normal (ERG-low) ERG expression (marked in black and red, respectively in Figs. 1 and 4). Expression of EPB41L4B was significantly stronger (p = 0.01) and EPB41L3 significantly lower (Mann-Whitney U p = 0.01) in ERG-high specimens (Fig. 4B, C). It is evident from the figure that the majority of ERG-high cancers displayed decreased expression of EPB41L3 and increased expression of EPB41L4B, whereas most of the ERG-low carcinomas had expression levels of the two genes within the range of benign tissues. In a similar fashion, EPB41L1 expression was on average lower in ERG-high than ERG-low cancers (Fig. 1), but in this case the difference was not statistically significant. None of the other cortical cytoskeleton genes differed significantly with respect to its expression between ERG-high and ERG-low cancer tissues.
When comparing these results to previous data from our group, we noted a close correlation (Spearman rho = 0.807, p < 0.001) between the expression of EPB41L4B and SPOCK1, the gene encoding the extracellular matrix protein testican 1. SPOCK1 expression is upregulated in prostate cancer and associated with diminished expression of FBLN1 encoding the basement membrane component fibulin 1 [25]. Accordingly, expression of FBLN1 was correlated inversely with that of EPB41L4B (rho = - 0.739, p < 0.001), but correlated well positively with that of EPB41L3 (rho = 0.684, p < 0.001). Moreover, SPOCK1 expression was significantly elevated (p = 0.002) in ERG-high cancers and that of FBLN1 was significantly diminished (p = 0.047). The expression of other cortical cytoskeleton genes was not related to either SPOCK1 or FBLN1 expression.
Discussion
Previous studies have concurrently reported changes in the expression of EPB41L3/4.1B and EPB41L4B/EHM2 in many prostate cancers. The other members of the 4.1 family had not been studied yet in prostate cancer, but previous investigations have indicated potential functions for the related proteins ezrin and dematin [12-14,26,27]. Despite these hints, we did not observe significant changes in ezrin/VIL2 or dematin/EPB49 mRNA expression in prostate cancer compared to benign tissues. Instead, the present investigation indicated that the expression changes of EPB41L3 and EPB41L4B are quite specific among cortical cytoskeleton genes, with the exception of EPB41L1. Our findings therefore call for a detailed investigation of 4.1N expression and function in prostatic cells in future studies.
Investigation of EPB41L3 methylation in a larger series of samples confirmed the presumed association between its downregulation and hypermethylation. EPB41L3 hypermethylation tended to be more frequent in higher stage tumors and downregulation of gene expression tended to be associated with earlier recurrence. The two changes did not correlate significantly with the same clinical indicators of tumor progression, which may be due the limited number of samples studied, since the tendencies were the same. In any case, these findings concur with the idea from functional studies in mouse and cell line models that 4.1B may act as a metastasis suppressor in prostate cancer [7]. Likewise, although we did not observe significant associations of EPB41L4B expression with cancer stage or grade, increased mRNA expression was associated with earlier recurrence, in accord with the immunohistochemical study of Wang et al. [10].
Moreover, the extended EPB41L3 methylation analysis allowed a comparison with the well-studied hypermethylation of GSTP1. Hypermethylation of GSTP1 can be detected in a significant fraction of HG-PINs [28], but more consistently after progression towards invasive carcinomas. It is thought to occur as part of an „epigenetic catastrophe" [29] that involves hypermethylation of further genes, e.g. APC, RARB2 and RASSF1A. In our tissue series, accordingly, cases with hypermethylation of GSTP1 have a very high likelihood of displaying hypermethylation of these genes as well [22]. EPB41L3 hypermethylation appears to occur slightly less frequently and was notably essentially restricted to cases with GSTP1 hypermethylation. A straightforward interpretation of this finding is that EPB41L3 hypermethylation occurs in many prostate cancers in conjunction with or following the hypermethylation of GSTP1.
In the present study, we observed a good correlation between the presence of EPB41L3 and EPB41L4B expression changes on one hand and ERG overexpression on the other hand. Simply stated, carcinoma tissues without ERG overexpression showed no differences in expression of the 4.1 genes towards normal tissues (Fig. 4). Our data therefore suggest that the changes in the expression of the two cortical cytoskeleton genes might be a consequence of ERG overexpression. This hypothesis fits with the presumed sequence of events during prostate cancer development. Fusion genes causing ERG overexpression can be detected in some pre-neoplastic HG-PINs, but more consistently in invasive carcinomas [21] supporting a role of ERG in promoting prostate cancer invasion and progression. As mentioned above, this transition is also associated with consistent hypermethylation of GSTP1, which may coincide with or precede hypermethylation of EPB41L3.
A relationship between ERG and the cortical cytoskeleton proteins is also plausible on functional grounds. Overexpression of ERG is thought to result in increased proliferation, invasiveness and motility of prostate cancer cells [18-20,30], whereas protein 4.1B has been shown to oppose invasiveness and metastasis of prostate cancer cells [5,7] and EHM2 to modulate adhesion of prostate cancer cells [10]. Our data invite the interpretation that the changes in EPB41 gene expression are part of a presumed invasion program directed by ERG and serve to implement the necessary changes in the cortical cytoskeleton. Whether cortical cytoskeleton genes are indeed - directly or indirectly - regulated by ERG, must now be investigated in cell line and animal models.
Along the same line of argument, we have previously observed changes in the expression of ECM genes encoding fibulin 1 (FBLN1) and testican 1 (SPOCK1). The causes of these changes are so far unclear, since we did not observe altered gene copy numbers in the majority of cases or obtain evidence for FBLN1 hypermethylation [25]. We have now become aware that upregulation of SPOCK1 parallels that of EPB41L4B and downregulation of FBLN1 parallels that of EPB41L3. Accordingly, the changes in the expression of the ECM genes were also associated with ERG overexpression. They may therefore represent changes in the composition of the extracellular matrix brought about as a consequence of ERG oncogenic activation. The observation that EHM2 influences cell adhesion to basement membrane collagen is intriguing in that respect [10]. Of note, the strikingly close correlation between EPB41L4B and SPOCK1 expression in the tissue samples may be enhanced by their common regulation by androgens [10,31]. It remains to be determined whether the ECM genes are indeed ERG targets.
In this study, we also report the first immunohistochemical investigation of 4.1B protein in prostatic tissues. Not unexpectedly [2,3], we observed the most prominent expression in the basal cells of the prostatic glands, while luminal cells contained less protein. Loss of the basal cell population is a well-established characteristic of prostate cancers. The downregulation of EPB41L3 expression may therefore largely reflect the loss of basal cells during prostate cancer development. Basal cells in the prostate epithelium express GSTP1, in contrast to luminal cells [32]. Accordingly, loss of expression and hypermethylation of GSTP1 may be related to the loss of the basal cell compartment [33]. Consequently, it is tempting to speculate that the hypermethylation of EPB41L3 in prostate cancer, too, might be associated with the loss of the basal cell compartment. Unlike GSTP1, however, 4.1B protein is expressed also in luminal cells of normal glands and the expression in cancerous prostatic glands remained overall comparable to that in the luminal cells, despite the presence of significant hypermethylation in a majority of the cases. Indeed, in prostate cancer cell lines with pronounced hypermethylation of the EPB41L3 gene and low levels of mRNA [6], a small amount of 4.1B protein remains detectable by blot techniques (Schulz et al., unpublished results). The residual expression of the gene in cancer cells could be due to a low level of transcription despite hypermethylation of the main promoter or from an alternative weaker promoter suspected in the gene [34].
Conclusions
Our findings suggest that highly specific changes in the cortical cytoskeleton occur during the progression of prostate cancers which parallel changes in the expression of extracellular matrix genes. Since these changes are found especially in cases with ERG overexpression, we hypothesize that they may constitute a part of the invasion program induced by oncogenic ERG.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
WAS designed the study. ECD and CH performed RNA and DNA analyses, RE performed and evaluated immunohistochemistry. MI and JR conducted statistical analyses. WAS, MI, JR and RE interpreted the data. WAS, MI and RE wrote the manuscript. All authors have given final approval to the manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Supplementary Material
Expression of EPB41L2, EPB41L5, EPB49, VIL2, and DLG1 mRNA in prostate tissues. Box plot representation of expression of the indicated genes relative to TBP in prostate cancer and benign tissues as measured by qRT-PCR.
MS-PCR for EPB41L3 methylation. Photograph of ethidium-bromide stained agarose gel with PCR products from bisulfite-treated DNA from numbered tissue samples, leukocytes (Leu) as unmethylated control, Du145 (Du) as methylated control, or water (W), using primers specific for the unmethylated (U) or methylated (M) EPB41L3 promoter. The last lane (MW) contained the size marker.
Contributor Information
Wolfgang A Schulz, Email: wolfgang.schulz@uni-duesseldorf.de.
Marc Ingenwerth, Email: marc.ingenwerth@uni-duesseldorf.de.
Carolle E Djuidje, Email: kroll16@yahoo.fr.
Christiane Hader, Email: hadersche@web.de.
Jörg Rahnenführer, Email: rahnenfuehrer@statistik.uni-dortmund.de.
Rainer Engers, Email: engers@uni-duesseldorf.de.
Acknowledgements
This study was supported by the Deutsche Krebshilfe.
References
- Sun CX, Robb VA, Gutmann DH. Protein 4.1 tumor suppressors: getting a FERM grip on growth regulation. J Cell Sci. 2002;115:3991–4000. doi: 10.1242/jcs.00094. [DOI] [PubMed] [Google Scholar]
- Calinisan V, Gravem D, Chen RPH, Brittin S, Mohandas N, Lecomte MC, Gascard P. New insights into potential functions for the protein 4.1 superfamily of proteins in kidney epithelium. Front Biosci. 2006;11:1646–66. doi: 10.2741/1911. [DOI] [PubMed] [Google Scholar]
- Parra M, Gascard P, Walensky LD, Gimm JA, Blackshaw S, Chan N, Takakuwa Y, Berger T, Lee G, Chasis JA, Snyder SH, Mohandas N, Conboy JG. Molecular and functional characterization of protein 4.1B, a novel member of the protein 4.1 family with high level, focal expression in brain. J Biol Chem. 2000;275:3247–55. doi: 10.1074/jbc.275.5.3247. [DOI] [PubMed] [Google Scholar]
- Hanada T, Takeuchi A, Sondarva G, Chishti AH. Protein 4.1-mediated membrane targeting of human discs large in epithelial cells. J Biol Chem. 2003;278:34445–50. doi: 10.1074/jbc.M305209200. [DOI] [PubMed] [Google Scholar]
- Bernkopf DB, Williams ED. Potential role of EPB41L3 (protein 4.1B/Dal-1) as a target for treatment of advanced prostate cancer. Expert Opin Ther Targets. 2008;12:845–53. doi: 10.1517/14728222.12.7.845. [DOI] [PubMed] [Google Scholar]
- Schulz WA, Alexa A, Jung V, Hader C, Hoffmann MJ, Yamanaka M, Fritzsche S, Wlazlinski A, Müller M, Lengauer T, Engers R, Florl AR, Wullich B, Rahnenführer J. Factor interaction analysis for chromosome 8 and DNA methylation alterations highlights innate immune response suppression and cytoskeletal changes in prostate cancer. Mol Cancer. 2007;6:14. doi: 10.1186/1476-4598-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong SY, Haack H, Kissil JL, Barry M, Bronson RT, Shen SS, Whittaker CA, Crowley D, Hynes RO. Protein 4.1B suppresses prostate cancer progression and metastasis. Proc Natl Acad Sci USA. 2007;104:12784–9. doi: 10.1073/pnas.0705499104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu K, Nagamachi Y, Tani M, Kimura K, Shiroishi T, Wakana S, Yokota J. Molecular cloning of a novel NF2/ERM/4.1 superfamily gene, ehm2, that is expressed in high-metastatic K1735 murine melanoma cells. Genomics. 2000;65:113–20. doi: 10.1006/geno.2000.6154. [DOI] [PubMed] [Google Scholar]
- Chauhan S, Pandey R, Way JF, Sroka TC, Demetriou MC, Kunz S, Cress AE, Mount DW, Miesfeld RL. Androgen regulation of the human FERM domain encoding gene EHM2 in a cell model of steroid-induced differentiation. Biochem Biophys Res Commun. 2003;310:421–32. doi: 10.1016/j.bbrc.2003.08.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Cai Y, Penland R, Chauhan S, Miesfeld RL, Ittmann M. Increased expression of the metastasis-associated gene Ehm2 in prostate cancer. Prostate. 2006;66:1641–52. doi: 10.1002/pros.20474. [DOI] [PubMed] [Google Scholar]
- Laprise P, Lau KM, Harris KP, Silva-Gagliardi NF, Paul SM, Beronja S, Beitel GJ, McGlade CJ, Tepass U. Yurt, Coracle, Neurexin IV and the Na(+), K(+)-ATPase form a novel group of epithelial polarity proteins. Nature. 2009;459:1141–5. doi: 10.1038/nature08067. [DOI] [PubMed] [Google Scholar]
- Chuan YC, Pang ST, Cedazo-Minguez A, Norstedt G, Pousette A, Flores-Morales A. Androgen induction of prostate cancer cell invasion is mediated by ezrin. J Biol Chem. 2006;281:29938–48. doi: 10.1074/jbc.M602237200. [DOI] [PubMed] [Google Scholar]
- Chuan YC, Iglesias-Gato D, Fernandez-Perez L, Cedazo-Minguez A, Pang ST, Norstedt G, Pousette A, Flores-Morales A. Ezrin mediates c-Myc actions in prostate cancer cell invasion. Oncogene. 2010;29:1531–42. doi: 10.1038/onc.2009.442. [DOI] [PubMed] [Google Scholar]
- Lutchman M, Pack S, Kim AC, Azim A, Emmert-Buck M, van Huffel C, Zhuang Z, Chishti AH. Loss of heterozygosity on 8p in prostate cancer implicates a role for dematin in tumor progression. Cancer Genet Cytogenet. 1999;115:65–9. doi: 10.1016/S0165-4608(99)00081-3. [DOI] [PubMed] [Google Scholar]
- Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9:265–73. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–90. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Kumar-Sinha C, Tomlins SA, Chinnaiyan AM. Recurrent gene fusions in prostate cancer. Nat Rev Cancer. 2008;8:497–511. doi: 10.1038/nrc2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermans KG, Bressers AA, van der Korput HA, Dits NF, Jenster G, Trapman J. Two unique novel prostate-specific and androgen-regulated fusion partners of ETV4 in prostate cancer. Cancer Res. 2008;68:3094–8. doi: 10.1158/0008-5472.CAN-08-0198. [DOI] [PubMed] [Google Scholar]
- Wang J, Cai Y, Yu W, Ren C, Spencer DM, Ittmann M. Pleiotropic biological activities of alternatively spliced TMPRSS2/ERG fusion gene transcripts. Cancer Res. 2008;68:8516–24. doi: 10.1158/0008-5472.CAN-08-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver BS, Tran J, Gopalan A, Chen Z, Shaikh S, Carracedo A, Alimonti A, Nardella C, Varmeh S, Scardino PT, Cordon-Cardo C, Gerald W, Pandolfi PP. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat Genet. 2009;41:619–24. doi: 10.1038/ng.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perner S, Mosquera JM, Demichelis F, Hofer MD, Paris PL, Simko J, Collins C, Bismar TA, Chinnaiyan AM, De Marzo AM, Rubin MA. TMPRSS2-ERG fusion prostate cancer: an early molecular event associated with invasion. Am J Surg Pathol. 2007;31:882–8. doi: 10.1097/01.pas.0000213424.38503.aa. [DOI] [PubMed] [Google Scholar]
- Florl AR, Steinhoff C, Müller M, Seifert HH, Hader C, Engers R, Ackermann R, Schulz WA. Coordinate hypermethylation at specific genes in prostate carcinoma precedes LINE-1 hypomethylation. Br J Cancer. 2004;91:985–94. doi: 10.1038/sj.bjc.6602030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann MJ, Müller M, Engers R, Schulz WA. Epigenetic control of CTCFL/BORIS and OCT4 expression in urogenital malignancies. Biochem Pharmacol. 2006;72:1577–88. doi: 10.1016/j.bcp.2006.06.020. [DOI] [PubMed] [Google Scholar]
- Tomlins SA, Bjartell A, Chinnaiyan AM, Jenster G, Nam RK, Rubin MA, Schalken JA. ETS gene fusions in prostate cancer: from discovery to daily clinical practice. Eur Urol. 2009;56:275–86. doi: 10.1016/j.eururo.2009.04.036. [DOI] [PubMed] [Google Scholar]
- Wlazlinski A, Engers R, Hoffmann MJ, Hader C, Jung V, Müller M, Schulz WA. Downregulation of several fibulin genes in prostate cancer. Prostate. 2007;67:1770–80. doi: 10.1002/pros.20667. [DOI] [PubMed] [Google Scholar]
- Pang ST, Fang X, Valdman A, Norstedt G, Pousette A, Egevad L, Ekman P. Expression of ezrin in prostatic intraepithelial neoplasia. Urology. 2004;63:609–12. doi: 10.1016/j.urology.2003.09.068. [DOI] [PubMed] [Google Scholar]
- Valdman A, Fang X, Pang ST, Nilsson B, Ekman P, Egevad L. Ezrin expression in prostate cancer and benign prostatic tissue. Eur Urol. 2005;48:852–7. doi: 10.1016/j.eururo.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Brooks JD, Weinstein M, Lin X, Sun Y, Pin SS, Bova GS, Epstein JI, Isaacs WB, Nelson WG. CG island methylation changes near the GSTP1 gene in prostatic intraepithelial neoplasia. Cancer Epidemiol Biomarkers Prev. 1998;7:531–6. [PubMed] [Google Scholar]
- Yegnasubramanian S, Kowalski J, Gonzalgo ML, Zahurak M, Piantadosi S, Walsh PC, Bova GS, De Marzo AM, Isaacs WB, Nelson WG. Hypermethylation of CpG islands in primary and metastatic human prostate cancer. Cancer Res. 2004;64:1975–86. doi: 10.1158/0008-5472.CAN-03-3972. [DOI] [PubMed] [Google Scholar]
- King JC, Xu J, Wongvipat J, Hieronymus H, Carver BS, Leung DH, Taylor BS, Sander C, Cardiff RD, Couto SS, Gerald WL, Sawyers CL. Cooperativity of TMPRSS2-ERG with PI3-kinase pathway activation in prostate oncogenesis. Nat Genet. 2009;41:524–6. doi: 10.1038/ng.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris JD, Chang CY, Wittmann BM, Kunder RS, Cui H, Fan D, Joseph JD, McDonnell DP. The homeodomain protein HOXB13 regulates the cellular response to androgens. Mol Cell. 2009;36:405–16. doi: 10.1016/j.molcel.2009.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cookson MS, Reuter VE, Linkov I, Fair WR. Glutathione S-transferase PI (GST-pi) class expression by immunohistochemistry in benign and malignant prostate tissue. J Urol. 1997;157:673–6. doi: 10.1016/S0022-5347(01)65248-0. [DOI] [PubMed] [Google Scholar]
- Schulz WA, Hatina J. Epigenetics of prostate cancer: beyond DNA methylation. J Cell Mol Med. 2006;10:100–25. doi: 10.1111/j.1582-4934.2006.tb00293.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan JS, Mohandas N, Conboy JG. Evolutionarily conserved coupling of transcription and alternative splicing in the EPB41 (protein 4.1R) and EPB41L3 (protein 4.1B) genes. Genomics. 2005;86:701–7. doi: 10.1016/j.ygeno.2005.08.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression of EPB41L2, EPB41L5, EPB49, VIL2, and DLG1 mRNA in prostate tissues. Box plot representation of expression of the indicated genes relative to TBP in prostate cancer and benign tissues as measured by qRT-PCR.
MS-PCR for EPB41L3 methylation. Photograph of ethidium-bromide stained agarose gel with PCR products from bisulfite-treated DNA from numbered tissue samples, leukocytes (Leu) as unmethylated control, Du145 (Du) as methylated control, or water (W), using primers specific for the unmethylated (U) or methylated (M) EPB41L3 promoter. The last lane (MW) contained the size marker.