Abstract
Background
Mutations in FLT3 result in activated tyrosine kinase activity, cell growth stimulation, and a poor prognosis among various subtypes of leukemia. The causes and timing of the mutations are not currently known. We evaluated the prevalence and timing of origin of FLT3 mutations in a population series of childhood leukemia patients from Northern California.
Methods
We screened and sequenced FLT3 mutations (point mutations and internal tandem duplications, ITDs) among 517 childhood leukemia patients, and assessed whether these mutations occurred before or after birth using sensitive "backtracking" methods.
Results
We determined a mutation prevalence of 9 of 73 acute myeloid leukemias (AMLs, 12%) and 9 of 441 acute lymphocytic leukemias (ALLs, 2%). Among AMLs, FLT3 mutations were more common in older patients, and among ALLs, FLT3 mutations were more common in patients with high hyperdiploidy (3.7%) than those without this cytogenetic feature (1.4%). Five FLT3 ITDs, one deletion mutation, and 3 point mutations were assessed for their presence in neonatal Guthrie spots using sensitive real-time PCR techniques, and no patients were found to harbor FLT3 mutations at birth.
Conclusions
FLT3 mutations were not common in our population-based patient series in California, and patients who harbor FLT3 mutations most likely acquire them after they are born.
Background
Certain, but not all, chromosome translocations in childhood leukemia are known to be present at birth. This phenomenon of prenatal origin was initially presumed from twin studies where it was observed that mono-amniotic twins always harbored the same translocations (reviewed in [1]). In addition, several studies have shown that specific mutations found at diagnosis in children with leukemia were present at birth, that is, "backtracked" to neonatal Guthrie Cards (blood spots used for newborn screens) (reviewed in [2]). The MLL rearrangement in infant ALL "backtracks" in nearly all cases and TEL-AML1 is found on 75% of Guthrie cards matched to leukemia cases with the translocation [3-5]. The E2A-PBX1 fusion generated by the t(1;19) translocation is a likely exception, with a postnatal origin [6], along with possibly others. These results collectively support a "two hit" model of leukemia, with one hit early in life or in utero, and another at a later date in temporal proximity to leukemia diagnosis.
The FLT3 gene is located on chromosome 13q12 and encodes a Type III membrane receptor kinase that regulates normal hematopoiesis. Mutations in FLT3 in AML occur in approximately 5-15% in children and 25-35% in adults, and account for the most common single gene defect in AML (reviewed in [7]). Several studies have indicated that children and adults with AML and the FLT3 mutation have a very poor prognosis. FLT3 mutations have also been documented in adult and pediatric ALL. An initial report demonstrated a 14% frequency of FLT3 mutations among childhood ALL overall, with mutations concentrated among the cytogenetic subgroups high hyperdiploidy (> 50 chromosomes in diagnostic karyotype) and MLL-translocation [8]; more recent studies have indicated a lower overall frequency in childhood ALL (in the 1-8% range) while consistently demonstrating a higher incidence among those with MLL rearrangement and high hyperdiploidy [9-13]. The absolute incidence of FLT3 mutations in pediatric leukemia is of interest in part because of the existence of several promising FLT3 inhibitors currently under development (reviewed in [14]) such inhibitors are more effective in the presence of FLT3 activation.
Internal Tandem Duplications (ITDs) and activation loop mutations are two unique FLT3 mutations that have been characterized. ITDs, which occur on exon 14, are insertions of repeated base pairs that range from 3-400 base pairs each. Activating loop mutations occur on exon 20; they are most commonly missense point mutations that occur at codon 835/836. Point mutations at codon 840, 841 and 842 have been described as well as insertion of base pairs between codons 841 and 842. In adult AML, ITDs comprise the majority of mutations, while in pediatric AML, ITD mutations are less common. It has been proposed that FLT3 mutation may be a late event in leukemia, given that FLT3 mutation status are often changed between paired diagnosis and relapse of adult and childhood patients with AML [15-17] or ALL [18]. The present study was conducted to formally assess this timing. We screened a large population case series of pediatric leukemia for FLT3 mutations of both types and determined whether FLT3 ITD mutations were present at birth by examining DNA on the corresponding Guthrie cards.
Methods
The research presented here was reviewed and approved by the UCSF Committee on Human Research, protocol # H10806-17300-12, and all study personnel completed appropriate human subjects training courses. Research material was derived from the Northern California Childhood Leukemia Study (NCCLS), and epidemiology study based at UC Berkeley.
Patients
The patient population consisted of 517 consecutive leukemia patients enrolled in the 9 hospitals participating in the NCCLS during the years 1995 to 2002. Intensive cytogenetic, morphologic, and flow cytometry review [19], parental interviews [20,21] and biologic and environmental sampling [22] were performed, as well as characterization of NRAS and KRAS mutations [23]. A detailed description of this approximately population-based study design can be found elsewhere [24]. Parental demographic characteristics were provided by the case mother (97.5%) or father (2.5%) through in-person interviews in the home of the parents. A full cytopathological review to distinguish immunophenotype and cytogenetic characteristics was instituted as previously described [19,23]. We employed fluorescence in situ hybridization (FISH) to further assess status of two major but cryptic cytogenetic subtypes of childhood ALL, TEL-AML1 translocation and high hyperdiploidy, using combined gene-loci specific FISH probes for chromosomes 12 (TEL) and 21 (AML1), as well as centromere probe for chromosome X. Because > 96% of high hyperdiploid patients have extra copies of both chromosomes 21 and X in the same cell [25,26], cases with both extra X and 21 were classified as high hyperdiploidy in the current study. For comparisons with healthy children, controls were individually matched by birthdate, gender, race and ethnicity to cases, and utilized the same questionnaire.
Mutation Screening
Each sample was amplified by PCR at the two most common sites for the FLT3 mutation using Optimase DNA polymerase (Transgenomics) with standard methods. The following primers were used to amplify exon 14-15 where the ITD mutation occurs and exon 20 where missense point mutations commonly occur: ITD-F TATCTGCAGAACTGCCTATTCC, and ITD-R CTTTCAGCATTTTGACGGCAACC; MUT-F CTCCTACTGAAGTTGAGTGTAG, and MUT-R CAGTGAGTGCAGTTGTTTACCA, respectively. Each PCR product was analyzed on a 3% metaphor agarose gel to confirm the presence or absence of the predicted amplicon (large ITDs are indicated by an extra band of larger than expected size). Samples that exhibited ITD mutation (for exon 14 samples) on agarose gel were subsequently sequenced. RFLP analysis was used to screen for missense FLT3 mutations (exon 20). EcoRV digests wild type FLT3 DNA, but does not digest a point mutation at the most common mutation site. Twenty-five μl of the PCR product was digested with 10 U of EcoRV for 2 hours at 37°C, followed by incubation at 80°C for 20 minutes to inactivate the enzyme. Digestion products were visualized by electrophoresis on a 3% agarose gel. Positive samples were gel extracted with the Qiagen QIAquick Gel Extraction Kit and confirmed with sequencing. All PCR products were also analyzed on denaturing high pressure liquid chromatography (DHPLC) using a Varian Wave machine with a Transgenomics column at 57.5C; and suspect mutations indicated by aberrant wave patterns were sequenced. This permitted the discovery of mutations outside of codon 835.
Backtracking
ITD-junction primers that would bind the mutant and would not bind wild type DNA were specifically designed for each mutant. Patient- and mutant-specific primers were designed for each FLT3 ITD mutation. The specificity of each mutant specific primer was tested by performing PCR amplifying diluted mutant DNA from a patient in limited dilutions (1:10) within wild type DNA (from peripheral blood cells); mutant specific primers will not bind wild type DNA with optimized assays. This dilution series was used to assess the sensitivity of the assay each time the assay was performed. SYBR green PCR was performed using the mutant specific primer pair designed for each patient to amplify DNA on the diagnostic DNA dilution series and the corresponding Guthrie card for each patient sample where a FLT3 ITD mutation was identified.
For point mutation backtracking, we used methods modified from our previous experiments with KRAS2 mutation backtracking [27]. Dilution series of patient DNA diluted into wild-type DNA (from peripheral blood cells) was used to assess assay sensitivity each time the assay was performed. Guthrie card DNA (240 ng) and dilution series of diagnostic DNA from each patient diluted into normal DNA were digested with 5 units EcoRV for 16 hours in 10 ul. An additional 5 units of EcoRV was added for 2 more hours (18 hours total). The sample was split into six for subsequent PCR analysis (40 ng per reaction). PCR primers D835-BTF (ACATCACAGTAAATAACACTCTGGTG) and D835-BTR (GACACAACACAAAATAGCCGTAT) were used for the SYBR Green PCR backtracking. Nontemplate controls as well as wild-type DNA controls were run alongside patient dilution series and Guthrie DNAs.
For both ITD and point mutations, we utilized SYBR green PCR on an ABI 7900HT with 384 well block: Reactions were performed with 1× buffer (SYBR Green PCR Core Reagents, ABI), 1× AccuPrime PCR buffer, 300 nM of each primer, the appropriate patient-specific forward primer and ITD-R2 (for the ITD reactions) and 3% DMSO, 0.4 U AccuPrime Taq DNA polymerase (Invitrogen), 40 ng template DNA, and water to yield a 10 ul reaction volume. Reactions were performed at 95°C for 5 min, followed by 50 cycles of 94°C for 15 sec, and 60°C for 15 sec. Backtracking PCR reactions were set-up in a different room of the research facility with separately stored PCR reagents from the laboratory where patient DNA was stored and processed for mutation screening, to prevent back-contamination. Test reactions with primers to ACTB (beta-actin) were routinely used to confirm the suitability of Guthrie DNA for PCR amplification.
Results
Eighteen FLT3 mutations were discovered among 517 acute leukemia cases for an overall frequency of 3.5%. Nine of 73 were found in AML (12.3% frequency) and 9/441 among ALL patients (2.0%). FLT3 mutations were as common in AML-M2 subtype (3 of 20 AML-M2 patients) as in other subtypes (6 of 40 non-AML-M2 patients). Regarding mutation type, insertion/deletion (indel) mutations were more common among AML patients (6 of the 9 indel mutations) than ALL patients (3 of the 9 indels, Table 1). All patients were concurrently assessed for NRAS and KRAS codon 12 and 13 mutations; one ALL patient had a concurrent KRAS mutation with FLT3 but no other patients were concurrent for these mutations (Table 1). AML patients with FLT3 mutations were older than those without mutations (average 11.5 yrs vs. 7.3 yrs, P = 0.01 by t-test); ALL patients with FLT3 were slightly but not significantly older (6.4 vs. 5.5 years average, P = 0.4, t-test). Our observation that indel mutations were more common among AMLs while point mutations were more common among ALLs, suggests a potential complementation or selection of mutation type with cell lineage. Five high hyperdiploid patients had FLT3 mutations among 132 in our cases series (3.7%), and four FLT3 mutations among 309 non-high hyperdiploid ALLs (1.3%) indicating some bias towards high hyperdiploidy though not significantly (P = 0.13, Fisher's exact test).
Table 1.
FLT3 mutations among 517 acute leukemia subjects from the Northern California Childhood Leukemia Study
| Patient ID | ITD or MUT* | MUT | Age | Cytogenetics | FAB (lineage) | Backtrack result† |
|---|---|---|---|---|---|---|
| 0004 | ITD | 6.1 | 46, XY [10/20]; 46, XY, del(9)(p13) [9/20]; 47, XY, +?22 [1/20] | ALL | neg | |
| 0087 | ITD | 10.5 | 46, XY [21/24] | AML-M1 | neg | |
| 0104 | ITD | 9.1 | 46, XY [19/20]; 44, XY, -14, -22 [1/20] | ALL-L1 (T-cell) | neg | |
| 0126 | ITD | 14.9 | 46, XX [20/20] | AML-M2 | ||
| 0201 | MUT | GAT→GAA D835E | 13.2 | 46, XY [20/20] | AML-M2 | |
| 0261 | ITD | 13.7 | 46, XX, t(6;9)(p23;q34) [23/24] | AML-M2 | neg | |
| 0544 ‡ | MUT | GAT→GTT D835V | 5.3 | 46, XX [20/20]; nuc ish 12p13(TEL×2), 21q22(AML1×4) [149/207]/12p13(TEL×2), 21q22(AML×2) [31/207]/12p13(TEL×2), 21q22(AML1×3) [25/207] | ALL | neg |
| 0678 | ITD | 14.5 | 46, XX [20] | AML-M2 | ||
| 0738 | MUT | TAT→TGT Y842C | 5.0 | 45, XY, -7, del(13)(q13q21) [12/20]; 46, XY [8/20] | ALL-L1 | |
| 0745 | MUT | GAT→TAT D835Y | 12.7 | 46, XY [21/21]; nuc ish 4cen(CEP4×2), 10cen(CEP10×2), 12p13(TEL×2), 21q22(AML1×2) |
ALL-L1/L2 | |
| 0796 ‡ | MUT | GAT→TAT D835Y | 1.8 | 46, XY [3/3]; nuc ish 12p13 (TEL×3), 21q22(AML1×4) [90/100], 12p13(TEL×2), 21q22(AML1×2) [10/100] FISH: +12++21/++X (presumed cryptic high hyperdiploidy) | ALL-L1 | neg |
| 0803 | MUT | GAT→TAT D835Y and GAT→CAT D835H | 0.3 | 46, XY [20/20]; nuc ish 11q23 (MLL5'x2, MLL3'x2) [200/200] | AML-M5 | |
| 0945 ‡ | MUT | GAT→TAT D835Y | 7.9 | 46, XY [21/21] | ALL | neg |
| 0999 | ITD | 8.3 | 46, XX [20] | AML | ||
| 1043 | MUT | GAT→CAT D835H | 14.0 | 46, XY, inv(16)(p12q22) [12/12] | AML | |
| 1073 ‡ | DEL | 5.9 | 46, XY [30]; nuc ish 9q34(ABL×2), 22q11.2(BCR×2) | ALL | neg | |
| 1107 ‡§ | MUT | GAT→GCT D835A | 3.5 | 56~58, XY, dup(1)(q21q32),+4,+5,+6,+10,+14,+18,+18,+19,+21, +22,+2mar [5/23]; 46, XY [18/23] | ALL-L1 | |
| 1148 | ITD | 14.5 | 47, XX, +14 [16] | AML | neg |
ITD, internal tandem duplication; MUT, point mutation; DEL, deletion.
† neg: 240 ng of patient Guthrie card was tested and was determined to be negative. The rest of the patients were not tested.
‡ Patients exhibiting high hyperdiploidy by FISH assay (see Materials and Methods)
§Patient 1107 has a KRAS mutation, which was also negative in backtracking experiment (ref #23)
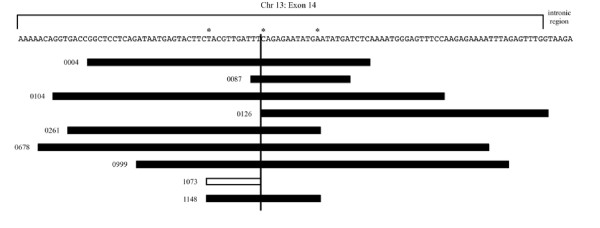
Eight indel mutations were identified consisting of 8 internal tandem duplications and 1 unconventional deletion. All indel mutants occurred in a 102 base pair region within exon 14 of the FLT3 gene, except for one case (0126) whose duplication included one base in the intronic region between exon 14 and 15 (Figure 1 and Additional File 1, Figure S1). Indels ranged from a 9 base pair deletion to a 90 base pair insertion and all preserved the open reading frame. Of the 9 indel mutations found, 6 Guthrie cards were available for backtracking. Each of these 6 Guthrie cards was used with their respective mutant specific primers (Table 2) in an attempt to amplify the DNA and determine if the mutation was present in the birth blood. None of the DNA from the 6 Guthrie cards DNA was amplified using the mutant specific primers at a sensitivity of 1 cell per 6,700 tested in 40 ng DNA (1.5 × 10-4) suggesting that the indel mutations were either not present in the DNA from the Guthrie cards, or were present but at a lower frequency than detectable by our assay.
Figure 1.
Relative location and sizes of FLT3 ITD mutations and deletion in 9 patients from the Northern California Childhood Leukemia Study. Black boxes - regions of duplication; white box - deletion of FLT3 sequence in one patient.
* - positions where multiple breaks occurred.
Table 2.
PCR Primers used for FLT3 ITD Backtracking on Guthrie card DNA: Northern California Childhood Leukemia Study
| Primer Name | Primer Sequences‡ (5'(3') |
|---|---|
| ITD-R2 | AGACAAATGGTGAGTACGTGCA |
| ITD-4-F | ATATGATCTCtccgagggcc |
| ITD-87-F | TATGAATATGAgTTCAGAGAATATG |
| ITD-104-F | GAGTTTCCcctcgggaag |
| ITD-261-F | AGAGAATATGAGACCGGCTC |
| ITD-1073-F* | GAGTACTTCccCAGAGAATATG |
| ITD-1148-F | AGAGAATATGAgTACGTTGATTTC |
‡ bold, WT sequence; underlined, ITD; lower case, N nucleotides
*1073 had an 11 bp deletion with a 2 bp insertion, giving an overall deletion of 9 bp.
ITD-R2 was used as the reverse primer for all the backtracking reactions.
Nine point mutations in the FLT3 gene were identified with 8 located at codon 835 and 1 located at codon 842. The latter was discovered using Denaturing High Pressure Liquid Chromatography (DHPLC) while the remaining by PCR-RFLP; all were sequenced for confirmation. One patient exhibited two mutations in codon 835 (patient #0803, Table 1). We cloned the PCR product using TA cloning kit (Invitrogen) and sequenced 8 clones - the mutations were present on opposite alleles meaning that two oncogenic mutations existed in this patient. Guthrie cards were available for 4 of these patients, and backtracking experiments were successfully performed on 3 patients at a similar sensitivity as the indel mutations (1 cell in 40 ng DNA), and the three patients were negative for mutant sequence. The requisite reaction sensitivity for one of the patient samples with available cards was not obtained. In sum, between indel and point mutation samples, 9 cards were tested and all were negative for presence of FLT3 mutation.
Discussion
Using NCCLS bone marrow samples, we have screened for FLT3 mutations in the largest sample of pediatric leukemias yet reported. Our results confirm the previously reported occurrence of FLT3 mutations in both pediatric ALL and AML although the incidence was lower than that of reported for some previous patient series. The highest rates of FLT3 mutation were recorded in population series of leukemias in Japan (9% of 162 ALL patients) [28] and Sweden, with 8% of ALL and 21% of AMLs (in children up to age 17 yrs) [9]. Lower frequencies were found in other population series in Greece (2.3% among 86 ALL patients), the UK (3.5% of 86 ALL patients), and Japan (1% of ALL 95 patients) [12]. Our study is the largest FLT3 screen in a pediatric population, and was performed in a population-based series of 517 cases. The rate of FLT3 among AML patients was 12.3%, which may be lower than some pediatric series since our study includes younger children only (< 15 years). The ALL rate of 2.0% found in here is comparable with the larger more recent reports. It is unlikely that we have missed pathogenetic FLT3 mutations in exons 11 and 15 since we incorporated a DHPLC screen of mutations. An additional notable difference between the current study and other studies is the apparent lack of a significant association between high hyperdiploidy and FLT3 mutation among our ALL patients, although this lack of significance may be due to small numbers. The association between RAS mutations and high hyperdiploidy is extremely strong in our patient sample set [29] and therefore we can confirm a RAS pathway association. It is unclear whether a lower prevalence of FLT3 mutations among hyperdiploid cases in California (compared to, for instance Paulsson et al., [11]) is due to differences in etiology of this subtype based on genetic or environmental characteristics of patients in California.
Our results suggesting that the FLT3 mutation is not present at birth is consistent with the hypothesis that FLT3 mutations are a second stage mutation. However, we cannot rule out that the mutations were present in some children but at a level beneath the sensitivity of the assay, or sequestered in the bone marrow and not in blood circulation. Our results corroborate that of Burjanivova, et al., who did not find evidence of prenatal origin of FLT3 in two AML patients [30]. The "two hit" model of leukemogenesis related to FLT3 proposed by Gilliland and Griffin, hypothesizes that two specific types of mutations are required in leukemia: one mutation that promotes proliferation and another mutation that stops differentiation [31]. Mutation of FLT3, like other tyrosine kinases is associated with proliferation and may complement mutations that impair cell differentiation such as the deletion of B-cell transcription factors or translocation-associated fusion genes. NRAS mutations and FLT3 both promote myeloproliferation, so their presence together is not necessary for leukemogenesis. The MLL gene rearrangement blocks differentiation and its association with FLT3 supports this model as well. None of our FLT3-mutant patients exhibited an MLL, TEL-AML1, AML1-ETO or other common translocation, so we were unable to examine a translocation concurrently with FLT3 in a backtracking experiment. However, strong prior evidence that high hyperdiploidy is a prenatal event [32-35], combined with no evidence of prenatal origin of FLT3 mutation in five high hyperdiploid patients here, helps to place FLT3 mutation postnatally.
Conclusions
In conclusion, FLT3 mutations are not common in our California childhood leukemia population, where RAS mutations are far more common [29]. Our investigation provided no evidence that FLT3 mutation occurs before birth, compatible with a hypothesis of postnatal origin for these mutations.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
All authors have read and approved the final manuscript. PC performed FLT3 screening of patients and performed initial backtracking experiments, wrote a first draft of the manuscript, and obtained funding for the study. MK reconfirmed FLT3 mutations and performed most of the backtracking experiments. AX assisted in additional point mutation backtracking experiments. JC managed epidemiologic data and performed analysis. JF assisted in the laboratory experiments and interpretation. PB recruited patients and epidemiologic information. JW performed additional backtracking experiments, assisted in the manuscript writing, and also obtained funding.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Supplementary Material
Figure S1. FLT3 ITD Sequences in the Northern California Childhood Leukemia Study. Exact sequences of internal tandem duplications found in patients from the Northern California Childhood Leukemia Study.
Contributor Information
Patrick Chang, Email: patrickchang72@yahoo.com.
Michelle Kang, Email: KangM@peds.ucsf.edu.
Anny Xiao, Email: annyxiao2007@u.northwestern.edu.
Jeffrey Chang, Email: jeffreychang@nhri.org.tw.
James Feusner, Email: JFeusner@mail.cho.org.
Patricia Buffler, Email: pab@berkeley.edu.
Joseph Wiemels, Email: joe.wiemels@ucsf.edu.
Acknowledgements
This analysis was funded by Hope Street Kids Foundation (PC, JLW), and the NCI R01CA89032 (JLW), and the Children with Leukaemia Foundation (JLW). Additional study funding provided by NIH P42ES0470 and R01ES09137 (PAB). We thank the children and their families for their participation in the Northern California Childhood Leukemia Study, and clinical investigators at the collaborating hospitals for help in recruiting patients: University of California-Davis Medical Center (J. Ducore), University of California-San Francisco (M. Loh), Children's Hospital of Central California (V. Crouse), Lucile Packard Children's Hospital (G. Dahl), Children's Hospital Oakland (J. Feusner), Kaiser Permanente Northern California (V. Kiley, C. Russo, A. Wong, K. Leung, S. Month). We also acknowledge the entire Northern California Childhood Leukemia Study staff and the Survey Research Center for their effort and dedication.
References
- Greaves MF, Maia AT, Wiemels JL, Ford AM. Leukemia in twins: lessons in natural history. Blood. 2003;102(7):2321–2333. doi: 10.1182/blood-2002-12-3817. [DOI] [PubMed] [Google Scholar]
- Wiemels J. Chromosomal translocations in childhood leukemia: natural history, mechanisms, and epidemiology. J Natl Cancer Inst Monogr. 2008. pp. 87–90. [DOI] [PubMed]
- Gale KB, Ford AM, Repp R, Borkhardt A, Keller C, Eden OB, Greaves MF. Backtracking leukemia to birth: identification of clonotypic gene fusion sequences in neonatal blood spots. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(25):13950–13954. doi: 10.1073/pnas.94.25.13950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHale CM, Wiemels JL, Zhang L, Ma X, Buffler PA, Guo W, Loh ML, Smith MT. Prenatal origin of TEL-AML1-positive acute lymphoblastic leukemia in children born in California. Genes Chromosomes Cancer. 2003;37(1):36–43. doi: 10.1002/gcc.10199. [DOI] [PubMed] [Google Scholar]
- Wiemels JL, Cazzaniga G, Daniotti M, Eden OB, Addison GM, Masera G, Saha V, Biondi A, Greaves MF. Prenatal origin of acute lymphoblastic leukaemia in children. Lancet. 1999;354(9189):1499–1503. doi: 10.1016/S0140-6736(99)09403-9. [DOI] [PubMed] [Google Scholar]
- Wiemels JL, Leonard BC, Wang Y, Segal MR, Hunger SP, Smith MT, Crouse V, Ma X, Buffler PA, Pine SR. Site-specific translocation and evidence of postnatal origin of the t(1;19) E2A-PBX1 fusion in childhood acute lymphoblastic leukemia. Proc Natl Acad Sci USA. 2002;99(23):15101–15106. doi: 10.1073/pnas.222481199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirewalt DL, Radich JP. The role of FLT3 in haematopoietic malignancies. Nat Rev Cancer. 2003;3(9):650–665. doi: 10.1038/nrc1169. [DOI] [PubMed] [Google Scholar]
- Armstrong SA, Mabon ME, Silverman LB, Li A, Gribben JG, Fox EA, Sallan SE, Korsmeyer SJ. FLT3 mutations in childhood acute lymphoblastic leukemia. Blood. 2004;103(9):3544–3546. doi: 10.1182/blood-2003-07-2441. [DOI] [PubMed] [Google Scholar]
- Andersson A, Paulsson K, Lilljebjorn H, Lassen C, Strombeck B, Heldrup J, Behrendtz M, Johansson B, Fioretos T. FLT3 mutations in a 10 year consecutive series of 177 childhood acute leukemias and their impact on global gene expression patterns. Genes Chromosomes Cancer. 2008;47(1):64–70. doi: 10.1002/gcc.20508. [DOI] [PubMed] [Google Scholar]
- Case M, Matheson E, Minto L, Hassan R, Harrison CJ, Bown N, Bailey S, Vormoor J, Hall AG, Irving JA. Mutation of genes affecting the RAS pathway is common in childhood acute lymphoblastic leukemia. Cancer Res. 2008;68(16):6803–6809. doi: 10.1158/0008-5472.CAN-08-0101. [DOI] [PubMed] [Google Scholar]
- Paulsson K, Horvat A, Strombeck B, Nilsson F, Heldrup J, Behrendtz M, Forestier E, Andersson A, Fioretos T, Johansson B. Mutations of FLT3, NRAS, KRAS, and PTPN11 are frequent and possibly mutually exclusive in high hyperdiploid childhood acute lymphoblastic leukemia. Genes Chromosomes Cancer. 2008;47(1):26–33. doi: 10.1002/gcc.20502. [DOI] [PubMed] [Google Scholar]
- Yamamoto T, Isomura M, Xu Y, Liang J, Yagasaki H, Kamachi Y, Kudo K, Kiyoi H, Naoe T, Kojma S. PTPN11, RAS and FLT3 mutations in childhood acute lymphoblastic leukemia. Leuk Res. 2006;30(9):1085–1089. doi: 10.1016/j.leukres.2006.02.004. [DOI] [PubMed] [Google Scholar]
- Braoudaki M, Karpusas M, Katsibardi K, Papathanassiou C, Karamolegou K, Tzortzatou-Stathopoulou F. Frequency of FLT3 mutations in childhood acute lymphoblastic leukemia. Med Oncol. 2009;26(4):460–462. doi: 10.1007/s12032-008-9146-z. [DOI] [PubMed] [Google Scholar]
- Weisberg E, Barrett R, Liu Q, Stone R, Gray N, Griffin JD. FLT3 inhibition and mechanisms of drug resistance in mutant FLT3-positive AML. Drug Resist Updat. 2009;12(3):81–89. doi: 10.1016/j.drup.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachas C, Schuurhuis GJ, Hollink IH, Kwidama ZJ, Goemans BF, Zwaan CM, van den Heuvel-Eibrink MM, de Bont ES, Reinhardt D, Creutzig U, de Haas V, Assaraf YG, Kaspers GJ, Cloos J. High frequency type I/II mutational shifts between diagnosis and relapse are associated with outcome in pediatric AML: implications for personalized medicine. Blood. 2010. in press . [DOI] [PubMed]
- Kottaridis PD, Gale RE, Langabeer SE, Frew ME, Bowen DT, Linch DC. Studies of FLT3 mutations in paired presentation and relapse samples from patients with acute myeloid leukemia: implications for the role of FLT3 mutations in leukemogenesis, minimal residual disease detection, and possible therapy with FLT3 inhibitors. Blood. 2002;100(7):2393–2398. doi: 10.1182/blood-2002-02-0420. [DOI] [PubMed] [Google Scholar]
- Cloos J, Goemans BF, Hess CJ, van Oostveen JW, Waisfisz Q, Corthals S, de Lange D, Boeckx N, Hahlen K, Reinhardt D, Creutzig U, Schuurhuis GJ, Zwaan Ch M, Kaspers GJ. Stability and prognostic influence of FLT3 mutations in paired initial and relapsed AML samples. Leukemia. 2006;20(7):1217–1220. doi: 10.1038/sj.leu.2404246. [DOI] [PubMed] [Google Scholar]
- Davidsson J, Paulsson K, Lindgren D, Lilljebjorn H, Chaplin T, Forestier E, Andersen MK, Nordgren A, Rosenquist R, Fioretos T, Young BD, Johansson B. Relapsed childhood high hyperdiploid acute lymphoblastic leukemia: presence of preleukemic ancestral clones and the secondary nature of microdeletions and RTK-RAS mutations. Leukemia. 2010;24(5):924–931. doi: 10.1038/leu.2010.39. [DOI] [PubMed] [Google Scholar]
- Aldrich MC, Zhang L, Wiemels JL, Ma X, Loh ML, Metayer C, Selvin S, Feusner J, Smith MT, Buffler PA. Cytogenetics of Hispanic and White children with acute lymphoblastic leukemia in California. Cancer Epidemiol Biomarkers Prev. 2006;15(3):578–581. doi: 10.1158/1055-9965.EPI-05-0833. [DOI] [PubMed] [Google Scholar]
- Ma X, Buffler PA, Selvin S, Matthay KK, Wiencke JK, Wiemels JL, Reynolds P. Daycare attendance and risk of childhood acute lymphoblastic leukaemia. Br J Cancer. 2002;86(9):1419–1424. doi: 10.1038/sj.bjc.6600274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JS, Selvin S, Metayer C, Crouse V, Golembesky A, Buffler PA. Parental smoking and the risk of childhood leukemia. Am J Epidemiol. 2006;163(12):1091–1100. doi: 10.1093/aje/kwj143. [DOI] [PubMed] [Google Scholar]
- Metayer C, Buffler PA. Residential Exposures to Pesticides and Childhood Leukaemia. Radiat Prot Dosimetry. 2009;132:212–219. doi: 10.1093/rpd/ncn266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiemels JL, Kang M, Chang JS, Zheng L, Kouyoumji C, Zhang L, Smith MT, Scelo G, Metayer C, Buffler P, Wiencke JK. Backtracking RAS mutations in high hyperdiploid childhood acute lymphoblastic leukemia. Blood Cells Mol Dis. 2010;45(3):186–91. doi: 10.1016/j.bcmd.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Buffler PA, Gunier RB, Dahl G, Smith MT, Reinier K, Reynolds P. Critical windows of exposure to household pesticides and risk of childhood leukemia. Environ Health Perspect. 2002;110(9):955–960. doi: 10.1289/ehp.02110955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorman AV, Clark R, Farrell DM, Hawkins JM, Martineau M, Secker-Walker LM. Probes for hidden hyperdiploidy in acute lymphoblastic leukaemia. Genes Chromosomes Cancer. 1996;16(1):40–45. doi: 10.1002/(SICI)1098-2264(199605)16:1<40::AID-GCC6>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Raimondi SC, Pui CH, Hancock ML, Behm FG, Filatov L, Rivera GK. Heterogeneity of hyperdiploid (51-67) childhood acute lymphoblastic leukemia. Leukemia. 1996;10(2):213–224. [PubMed] [Google Scholar]
- Wiemels J, Kang M, Greaves M. Backtracking of leukemic clones to birth. Methods Mol Biol. 2009;538:7–27. doi: 10.1007/978-1-59745-418-6_2. full_text. [DOI] [PubMed] [Google Scholar]
- Taketani T, Taki T, Sugita K, Furuichi Y, Ishii E, Hanada R, Tsuchida M, Ida K, Hayashi Y. FLT3 mutations in the activation loop of tyrosine kinase domain are frequently found in infant ALL with MLL rearrangements and pediatric ALL with hyperdiploidy. Blood. 2004;103(3):1085–1088. doi: 10.1182/blood-2003-02-0418. [DOI] [PubMed] [Google Scholar]
- Wiemels JL, Zhang Y, Chang J, Zheng S, Metayer C, Zhang L, Smith MT, Ma X, Selvin S, Buffler PA, Wiencke JK. RAS mutation is associated with hyperdiploidy and parental characteristics in pediatric acute lymphoblastic leukemia. Leukemia. 2005;19:415–419. doi: 10.1038/sj.leu.2403641. [DOI] [PubMed] [Google Scholar]
- Burjanivova T, Madzo J, Muzikova K, Meyer C, Schneider B, Votava F, Marschalek R, Stary J, Trka J, Zuna J. Prenatal origin of childhood AML occurs less frequently than in childhood ALL. BMC Cancer. 2006;6:100. doi: 10.1186/1471-2407-6-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilliland DG. Molecular genetics of human leukemias: new insights into therapy. Semin Hematol. 2002;39(4 Suppl 3):6–11. doi: 10.1053/shem.2002.36921. [DOI] [PubMed] [Google Scholar]
- Fasching K, Panzer S, Haas OA, Marschalek R, Gadner H, Panzer-Grumayer ER. Presence of clone-specific antigen receptor gene rearrangements at birth indicates an in utero origin of diverse types of early childhood acute lymphoblastic leukemia. Blood. 2000;95(8):2722–2724. [PubMed] [Google Scholar]
- Gruhn B, Taub JW, Ge Y, Beck JF, Zell R, Hafer R, Hermann FH, Debatin KM, Steinbach D. Prenatal origin of childhood acute lymphoblastic leukemia, association with birth weight and hyperdiploidy. Leukemia. 2008;22(9):1692–1697. doi: 10.1038/leu.2008.152. [DOI] [PubMed] [Google Scholar]
- Taub JW, Konrad MA, Ge Y, Naber JM, Scott JS, Matherly LH, Ravindranath Y. High frequency of leukemic clones in newborn screening blood samples of children with B-precursor acute lymphoblastic leukemia. Blood. 2002;99(8):2992–2996. doi: 10.1182/blood.V99.8.2992. [DOI] [PubMed] [Google Scholar]
- Yagi T, Hibi S, Tabata Y, Kuriyama K, Teramura T, Hashida T, Shimizu Y, Takimoto T, Todo S, Sawada T, Imashuku S. Detection of clonotypic IGH and TCR rearrangements in the neonatal blood spots of infants and children with B-cell precursor acute lymphoblastic leukemia. Blood. 2000;96(1):264–268. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. FLT3 ITD Sequences in the Northern California Childhood Leukemia Study. Exact sequences of internal tandem duplications found in patients from the Northern California Childhood Leukemia Study.