Abstract
Prion diseases or transmissible spongiform encephalopathies (TSEs) are fatal diseases associated with the conversion of the cellular prion protein (PrPC) to the abnormal prion protein (PrPSc). Since the molecular mechanisms in pathogenesis are widely unclear, we analyzed the global phospho-proteome and detected a differential pattern of tyrosine- and threonine phosphorylated proteins in PrPSc-replicating and pentosan polysulfate (PPS)-rescued N2a cells in two-dimensional gel electrophoresis. To quantify phosphorylated proteins, we performed a SILAC (stable isotope labeling by amino acids in cell culture) analysis and identified 105 proteins, which showed a regulated phosphorylation upon PrPSc infection. Among those proteins, we validated the dephosphorylation of stathmin and Cdc2 and the induced phosphorylation of cofilin in PrPSc-infected N2a cells in Western blot analyses. Our analysis showed for the first time a differentially regulated phospho-proteome in PrPSc infection, which could contribute to the establishment of novel protein markers and to the development of novel therapeutic intervention strategies in targeting prion-associated disease.
Findings
Transmissible spongiform encephalopathies (TSEs) are fatal neurodegenerative diseases occurring in many different host species including humans, which develop e.g. Creutzfeld Jacob disease (sCJD) [1]. The development of TSEs is associated with the self-propagating conversion of the normal host cellular prion protein (PrPC) into the abnormal protease-resistant isoform (PrPSc or PrPres) in an autocatalytic manner [2]. PrPSc plays a key role as an infectious agent in certain degenerative diseases of the central nervous system [3].
The cellular functions of PrPC and PrPSc still remain enigmatic. The cellular prion protein can be variably glycosylated at two N-glycosylation sites and is C-terminally attached to the cell surface by a glycosyl phosphatidylinositol (GPI) anchor. GPI-anchored proteins are found in lipid rafts, highly cholesterol- and glycolipid-enriched membrane domains associated with a large number of signaling molecules such as G-protein-coupled receptors and protein kinases suggesting that signaling transduction pathways might play a role in TSEs [4]. Hence, previous publications described a functional role of PrPC as a signaling molecule with major findings indicating that PrPC interacts with and activates Src family kinases [5-7]. Increased levels of active Src kinases in scrapie-infected cells then led to the activation of downstream signal transduction pathways [8]. Recently, activation of the JAK-STAT signaling pathway in astrocytes of scrapie-infected brains was observed underlining that signal transduction pathways may play pivotal roles in prion pathogenesis [9]. Interestingly, it was demonstrated that inhibition of the non-receptor tyrosine kinase c-Abl strongly activates the lysosomal degradation of PrPSc [10]. These data indicate that specific interference with cellular signaling pathways could represent a novel strategy in treatment of TSEs.
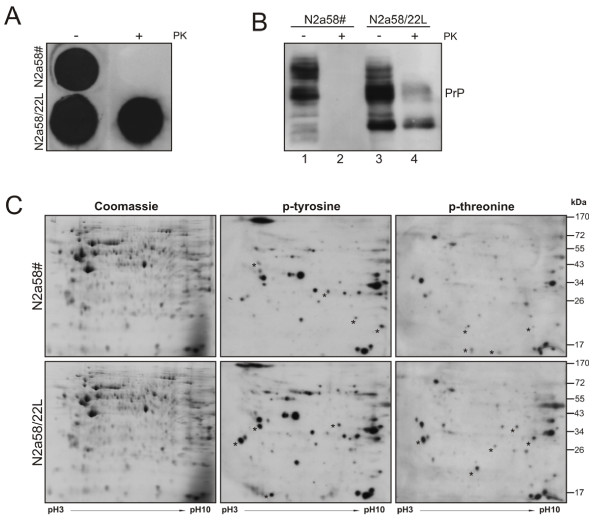
We have performed a quantitative analysis of the phospho-proteome to obtain a global insight into deregulated signal transduction pathways in scrapie-infected neuronal cells. We analyzed tyrosine- and threonine-phosphorylated proteins in the murine neuroblastoma cell line N2a58/22L, which were infected with the PrPSc strain 22L [11]. We have treated N2a58/22L cells with pentosan polysulfate (PPS), a known inhibitor of 22L PrPSc replication in N2a cells [12], resulting in the PrPSc-rescued cell line N2a58# which served as an uninfected control. Successful rescue from PrPSc was demonstrated in the colony assay as reflected by the absence of proteinase K (PK)-resistant PrPSc in N2a58# cells after PPS treatment (Figure 1A). PrPSc replication and the effect of PPS-treatment were further studied in an immunoblot. After PK digestion, PrPSc replication was only observed in N2a58/22L cells (Figure 1B, lanes 2 and 4). Compared to 22L-infected N2a58/22L cells, PPS-treated N2a58# cells showed a different glycosylation profile as expected for PrPC [13-15]. The glycosylation pattern of PrPC in N2a58# cells displayed high amounts of di- and mono-glycosylated PrPC, whereas in N2a58/22L cells predominantly mono- and non-glycosylated PrPSc was detected (Figure 1B, lanes 1 and 3). Altogether, PPS treatment of N2a58/22L cells successfully abolished PrPSc formation in N2a58# cells, which served as a non-infected control cell line in our study.
Figure 1.
Differentially phosphorylated proteins in PrPSc-positive and -negative N2a cells. (A) PrPres-positive N2a58/22L cells were treated with pentosan polysulfate (PPS) to obtain PrPres-negative N2a58# cells. Successful PPS treatment was validated in a colony assay. Cells were grown to confluence on cover slips and directly lysed on nitrocellulose. Where indicated 20 μg/ml proteinase K (PK) was added followed by the detection of PrP expression using the 6H4 monoclonal antibody. In non-treated cells (-), PrP was detected in both, cured and infected N2a cells. Upon digestion with PK (+), PrPres was only observed in N2a58/22L cells. (B) Equal amounts of protein lysates were incubated with 20 μg/ml PK or left untreated. PrP was detected with the 8H4 monoclonal antibody showing the typical migration pattern of PrP and PrPres in infected and PPS-treated N2a58# cells. In parallel, lysates were incubated with PK to visualize PK-resistant PrPres in N2a58/22L. (C) 150 μg of N2a58# or prion-infected N2a58/22L cell lysates were separated by two-dimensional gel electrophoresis followed either by Coomassie staining or immunoblotting for detection of tyrosine- and threonine-phosphorylated proteins. Black asterisks indicate changed intensities of protein phosphorylation.
To analyze differentially phosphorylated proteins in N2a58/22L cells in comparison to N2a58# cells, we separated equal protein amounts by two-dimensional gel electrophoresis. Gels were stained with Coomassie Blue to demonstrate equal protein amounts in N2a58/22L and N2a58# cells (Figure 1C, left panels). In parallel, gels were blotted onto membranes and incubated with phospho-specific antibodies to detect tyrosine- (Figure 1C, middle panels) or threonine-phosphorylated proteins (Figure 1C, right panels). Interestingly, considerable differences in phosphorylation patterns were observed (Figure 1C, asterisks), while other phosphorylated proteins were not changed in N2a58/22L and N2a58# cells (Figure 1C). These data imply differentially regulated phosphoproteins in response to 22L infection of neuronal cells.
Generally, global detection of phosphorylated proteins is still challenging, as antisera often recognize phosphorylated residues dependent on the surrounding sequence. For a general detection of proteins post-translationally phosphorylated at those sites, we performed a SILAC analysis allowing the identification and relative quantification of differential phosphoprotein regulation. Therefore, N2a58# cells were grown in light isotope containing and N2a58/22L cells in heavy isotope containing medium. Equal amounts of protein lysates were mixed, separated by gel electrophoresis, trypsinized and followed by enrichment of phosphoproteins, which were then analyzed by mass spectrometry. We identified 109 different phosphoproteins of which 105 were also quantified (Tables 1 and 2). We observed 75 proteins with a ratio of identified peptides in N2a58/22L versus N2a58# cells ranging from 0.46 to 0.99 (Table 1). Conversely, 30 phosphoproteins showed a ratio between 1.01 and 1.79 (Table 2). We defined proteins exhibiting a ratio < 0.70 as dephosphorylated proteins and proteins with ratios between 0.70 and 1.40 as proteins, whose phosphorylation was not altered in 22L-infected N2a58/22L cells. Ratios > 1.40 were considered as proteins whose phosphorylation increased upon Scrapie infection.
Table 1.
Proteins exhibiting decreased phosphorylation in N2a58/22L cells.
| No. | Uniprot | Protein Names | Ratioa | Peptb | sequence coverage [%] |
PEPc | Biological Process |
|---|---|---|---|---|---|---|---|
| 1 | P43276 | Histone H1.5 | 0.46237 | 1 | 13.9 | 5.61E-16 | nucleosome assembly |
| 2 | P30681 | High mobility group protein B2 | 0.48683 | 2 | 11 | 3.33E-02 | genome maintenance; differentiation |
| 3 | P11440 | Cell division control protein 2 homolog | 0.49428 | 5 | 7.7 | 3.03E-03 | cell cycles; protein phosphorylation |
| 4 | P97310 | DNA replication licensing factor MCM2 | 0.54657 | 1 | 2.4 | 1.65E-05 | cell cycle; nucleosome assembly; transcription |
| 5 | P43275 | Histone H1.1 | 0.56637 | 1 | 19.2 | 8.42E-04 | nucleosome assembly |
| 6 | P43274 | Histone H1.4 | 0.58367 | 4 | 24.2 | 1.04E-14 | nucleosome assembly |
| 7 | Q9Z2X1-1 | Heterogeneous nuclear ribonucleoprotein F | 0.60054 | 2 | 6.5 | 5.45E-22 | RNA processing |
| 8 | P70670 | Nascent polypeptide-associated complex subunit alpha, muscle-specific form | 0.60501 | 2 | 1.2 | 5.07E-11 | protein transport; transcription |
| 9 | P60843 | Eukaryotic initiation factor 4A-I | 0.65781 | 5 | 4.9 | 5.39E-03 | translation |
| 10 | P27659 | 60S ribosomal protein L3 | 0.65906 | 12 | 7.7 | 1.49E-03 | translation |
| 11 | P28656 | Nucleosome assembly protein 1-like 1 | 0.70087 | 1 | 7.2 | 1.16E-05 | nucleosome assembly |
| 12 | Q62167 | ATP-dependent RNA helicase DDX3X | 0.70816 | 3 | 3.3 | 3.12E-19 | putative helicase activity |
| 13 | P68040 | Guanine nucleotide-binding protein subunit beta-2-like1 | 0.71371 | 1 | 7.9 | 5.15E-08 | unknown |
| 14 | P47911 | 60S ribosomal protein L6 | 0.71887 | 7 | 12 | 7.26E-07 | translation |
| 15 | Q61937 | Nucleophosmin | 0.72062 | 7 | 29.8 | 4.38E-07 | cell cycle; nuclear export |
| 16 | P15532 | Nucleoside diphosphate kinase A | 0.72108 | 2 | 17.8 | 7.93E-03 | NTP biosynthesis; nervous system development |
| 17 | O70251 | Elongation factor 1-beta | 0.73137 | 1 | 24 | 2.20E-18 | translation |
| 18 | Q61656 | Probable ATP-dependent RNA helicase DDX5 | 0.73832 | 4 | 3.7 | 1.41E-03 | RNA processing; transcription |
| 19 | Q9ERK4 | Exportin-2 | 0.74333 | 1 | 2.1 | 8.97E-05 | cell proliferation; protein transport |
| 20 | P09411 | Phosphoglycerate kinase 1 | 0.74902 | 2 | 6.7 | 3.40E-03 | glycolysis; phosphorylation |
| 21 | P48962 | ADP/ATP translocase 1 | 0.75149 | 6 | 18.8 | 3.87E-36 | transmembrane transport |
| 22 | Q9D8N0 | Elongation factor 1-gamma | 0.76191 | 3 | 7.8 | 1.45E-10 | translation |
| 23 | P49312-2 | Heterogeneous nuclear ribonucleoprotein A1 | 0.77303 | 5 | 12.1 | 7.20E-05 | alternative splicing; nuclear export/import |
| 24 | Q9CZM2 | 60S ribosomal protein L15 | 0.77591 | 9 | 10.3 | 2.04E-15 | translation |
| 25 | P97855 | Ras GTPase-activating protein-binding protein 1 | 0.78021 | 2 | 7.3 | 1.11E-15 | protein transport |
| 26 | Q9EQU5-1 | Protein SET | 0.79357 | 7 | 7.9 | 5.77E-12 | nucleosome assembly |
| 27 | Q7TPV4 | Myb-binding protein 1A | 0.79496 | 2 | 1.5 | 2.39E-11 | cytoplasmic transport; transcription |
| 28 | P80318 | T-complex protein 1 subunit gamma | 0.79504 | 1 | 6.6 | 2.89E-05 | protein folding |
| 29 | P25444 | 40S ribosomal protein S2 | 0.79516 | 20 | 12.3 | 2.99E-04 | translation |
| 30 | P10126 | Elongation factor 1-alpha 1 | 0.80203 | 5 | 18.6 | 1.06E-33 | translational elongation |
| 31 | P61979-2 | Heterogeneous nuclear ribonucleoprotein K | 0.80502 | 6 | 11.9 | 1.17E-15 | RNA processing |
| 32 | P07901 | Heat shock protein HSP 90-alpha | 0.80637 | 2 | 32.7 | 1.38E-94 | CD8 T-cell differentiation; chaperone activity |
| 33 | Q61598-1 | Rab GDP dissociation inhibitor beta | 0.81242 | 3 | 5.6 | 1.93E-05 | protein transport; regulation of GTPase activity |
| 34 | P54775 | 26S protease regulatory subunit 6B | 0.81795 | 2 | 7.2 | 1.59E-09 | blastocyst development; protein catabolism |
| 35 | Q20BD0 | Heterogeneous nuclear ribonucleoprotein A/B | 0.82349 | 3 | 19.3 | 3.26E-11 | nucleotide binding |
| 36 | P14206 | 40S ribosomal protein SA;Laminin receptor 1 | 0.8261 | 7 | 20.7 | 2.57E-17 | translation |
| 37 | P68134 | Actin, alpha skeletal muscle | 0.83457 | 10 | 27.3 | 4.38E-26 | cytoskeleton |
| 38 | P80314 | T-complex protein 1 subunit beta | 0.83579 | 2 | 11.6 | 1.05E-21 | protein folding |
| 39 | P50580 | Proliferation-associated protein 2G4 | 0.84048 | 2 | 8.1 | 1.53E-03 | rRNA processing; transcription; translation |
| 40 | P11983-1 | T-complex protein 1 subunit alpha B | 0.84687 | 4 | 8.8 | 5.73E-23 | protein folding |
| 41 | P35564 | Calnexin | 0.85134 | 1 | 6.3 | 2.08E-06 | protein folding |
| 42 | Q8BUP7 | Putative uncharacterized protein;26S protease regulatory subunit 6A | 0.85207 | 3 | 7.3 | 9.72E-03 | blastocyst development; protein catabolism |
| 43 | P63017 | Heat shock cognate 71 kDa protein | 0.85947 | 9 | 32.2 | 2.16E-96 | response to stress |
| 44 | Q01768 | Nucleoside diphosphate kinase B | 0.86107 | 4 | 17.8 | 1.35E-04 | NTP biosynthesis |
| 45 | P62082 | 40S ribosomal protein S7 | 0.86306 | 3 | 10.3 | 8.39E-03 | translation |
| 46 | P80315 | T-complex protein 1 subunit delta | 0.86423 | 1 | 6.9 | 3.81E-12 | protein folding |
| 47 | Q71LX8 | Heat shock protein 84b | 0.8654 | 4 | 32 | 6.13E-136 | protein folding; stress response |
| 48 | P58252 | Elongation factor 2 | 0.86672 | 2 | 7.9 | 3.57E-25 | translation |
| 49 | P08249 | Malate dehydrogenase, mitochondrial | 0.86743 | 1 | 9.2 | 2.49E-35 | glycolysis |
| 50 | P70168 | Importin subunit beta-1 | 0.87257 | 2 | 2.7 | 3.47E-13 | nuclear import |
| 51 | P51859 | Hepatoma-derived growth factor | 0.87395 | 2 | 12.7 | 2.95E-07 | transcription |
| 52 | P14152 | Malate dehydrogenase, cytoplasmic | 0.87522 | 1 | 7.8 | 1.09E-03 | glycolysis |
| 53 | P80313 | T-complex protein 1 subunit eta | 0.89251 | 1 | 8.6 | 1.22E-39 | protein folding |
| 54 | P62827 | GTP-binding nuclear protein Ran | 0.89794 | 3 | 22.7 | 2.27E-09 | cell cycle; nuclear import; signal transduction |
| 55 | P20029 | 78 kDa glucose-regulated protein | 0.89996 | 1 | 7 | 4.16E-04 | cerebellar Purkinje cell development/organization |
| 56 | P56480 | ATP synthase subunit beta, mitochondrial | 0.90399 | 1 | 18.5 | 1.85E-42 | proton transport; lipid metabolism |
| 57 | P17742 | Peptidyl-prolyl cis-trans isomerase A | 0.90719 | 10 | 20.4 | 4.97E-20 | neuron differentiation; protein folding |
| 58 | P20152 | Vimentin | 0.90814 | 10 | 26.2 | 2.22E-34 | cytoskeleton |
| 59 | P09103 | Protein disulfide-isomerase | 0.91019 | 2 | 4.7 | 8.84E-02 | redox homeostasis |
| 60 | P80317 | T-complex protein 1 subunit zeta | 0.91579 | 1 | 10.2 | 1.49E-02 | protein folding |
| 61 | Q569Z6 | Thyroid hormone receptor-associated protein 3 | 0.93507 | 3 | 3.7 | 4.27E-04 | transcription |
| 62 | P09405 | Nucleolin | 0.93545 | 1 | 12.7 | 8.06E-34 | nucleotide binding |
| 63 | Q9D6F9 | Tubulin beta-4 chain | 0.93722 | 4 | 23.2 | 9.43E-32 | cytoskeleton |
| 64 | Q8C2Q7 | Heterogeneous nuclear ribonucleoprotein H1 | 0.93795 | 3 | 5.7 | 2.30E-06 | nucleotide binding |
| 65 | Q8K019-1 | Bcl-2-associated transcription factor 1 | 0.94132 | 4 | 5.3 | 2.22E-05 | transcription |
| 66 | P62908 | 40S ribosomal protein S3 | 0.94247 | 2 | 9.1 | 1.74E-06 | translation |
| 67 | P99024 | Tubulin beta-5 chain | 0.9446 | 3 | 36.3 | 1.15E-54 | cytoskeleton |
| 68 | P15331-2 | Peripherin | 0.95087 | 3 | 17.2 | 3.41E-40 | cytoskeleton |
| 69 | P63038-1 | 60 kDa heat shock protein, mitochondrial | 0.96089 | 6 | 13.8 | 9.17E-37 | T cell activation; interferon production |
| 70 | P27773 | Protein disulfide-isomerase A3 | 0.96765 | 1 | 7.9 | 1.26E-03 | redox homeostasis; apoptosis |
| 71 | P16858 | Glyceraldehyde-3-phosphate dehydrogenase | 0.96943 | 58 | 23.4 | 5.69E-66 | glycolysis |
| 72 | Q3TED3 | Putative uncharacterized protein; ATP-citrate synthase | 0.97174 | 2 | 4.3 | 4.63E-03 | acetyl-CoA biosynthesis |
| 73 | Q03265 | ATP synthase subunit alpha, mitochondrial | 0.98925 | 2 | 13.6 | 1.81E-22 | proton transport; lipid metabolism |
| 74 | Q9ERD7 | Tubulin beta-3 chain | 0.99071 | 2 | 27.3 | 5.05E-33 | cytoskeleton |
| 75 | P17751 | Triosephosphate isomerase | 0.9967 | 1 | 11.2 | 1.52E-04 | gluconeogenesis; glycolysis |
a. Ratio of N2a58/22L vs. N2a58# cells
b. Number of identified peptides
c. posterior error probability (PEP) estimates the probability of wrong assignment of a spectrum to a peptide sequence
Table 2.
Proteins exhibiting increased phosphorylation in N2a58/22L cells.
| No. | Uniprot | Protein Names | Ratioa | Pept.b | Sequence Coverage [%] |
PEPc | Biological Process |
|---|---|---|---|---|---|---|---|
| 76 | P80316 | T-complex protein 1 subunit epsilon | 1.017 | 1 | 6.1 | 8.63E-18 | protein folding |
| 77 | Q9CX22 | Putative uncharacterized protein; Cofilin-1 | 1.0173 | 5 | 29.7 | 3.72E-41 | cytoskeleton; protein phosphorylation |
| 78 | P32067 | Lupus La protein homolog | 1.02 | 2 | 7.5 | 1.68E-02 | RNA processing |
| 79 | A6ZI44 | Fructose-bisphosphate aldolase | 1.0328 | 8 | 12.2 | 2.75E-25 | glycolysis |
| 80 | P35700 | Peroxiredoxin-1 | 1.0342 | 6 | 6.7 | 2.44E-10 | proliferation; redox homeostasis; stress response |
| 81 | P05202 | Aspartate aminotransferase, mitochondrial | 1.0344 | 1 | 12.3 | 1.04E-11 | aspartate biosynthesis; oxaloacetate metabolism |
| 82 | Q3TFD0 | Serine hydroxymethyltransferase | 1.0425 | 3 | 6.5 | 2.95E-03 | carbon metabolism |
| 83 | P63101 | 14-3-3 protein zeta/delta | 1.0478 | 19 | 19.2 | 7.09E-27 | protein binding |
| 84 | Q9CZ30-1 | Obg-like ATPase 1 | 1.0606 | 3 | 7.1 | 2.25E-16 | ATP/GTP binding; hydrolase activity |
| 85 | P06745 | Glucose-6-phosphate isomerase | 1.0652 | 3 | 7.7 | 3.34E-15 | angiogenesis; gluconeogenesis; glycolysis |
| 86 | Q71H74 | Collapsin response mediator protein 4A | 1.0688 | 5 | 6.3 | 6.94E-12 | nervous system development |
| 87 | Q6P5F9 | Exportin-1 | 1.0703 | 2 | 3.6 | 2.74E-20 | nuclear export; centrosome duplication |
| 88 | Q01853 | Transitional endoplasmic reticulum ATPase | 1.0708 | 2 | 3.7 | 2.97E-05 | apoptosis; retrograde protein transport |
| 89 | Q3TCI7 | L-lactate dehydrogenase | 1.0746 | 4 | 18.3 | 1.43E-32 | glycolysis |
| 90 | P08113 | Endoplasmin | 1.08 | 1 | 2.6 | 1.05E-05 | protein folding |
| 91 | Q8VC46 | Ubc protein;Ubiquitin | 1.0848 | 17 | 30.7 | 2.18E-06 | protein binding |
| 92 | A0PJ96 | Mtap1b protein | 1.0871 | 2 | 3.2 | 2.72E-13 | cytoskeleton |
| 93 | Q61171 | Peroxiredoxin-2 | 1.0934 | 2 | 9.1 | 4.16E-04 | signal transduction; redox homeostasis |
| 94 | Q9WVA4 | Transgelin-2 | 1.106 | 2 | 11.8 | 9.37E-05 | muscle organ development |
| 95 | P52480-1 | Pyruvate kinase isozymes M1/M2 | 1.1081 | 3 | 22.2 | 6.30E-39 | glycolysis |
| 96 | O08709 | Peroxiredoxin-6 | 1.1204 | 5 | 13.8 | 1.10E-04 | redox homeostasis |
| 97 | P17182 | Alpha-enolase | 1.128 | 11 | 41.5 | 6.35E-26 | glycolysis |
| 98 | P54227 | Stathmin | 1.1422 | 4 | 18.1 | 1.94E-02 | cell cycle; cytoskeleton |
| 99 | Q922F4 | Tubulin beta-6 chain | 1.1427 | 1 | 19.2 | 1.01E-25 | cytoskeleton |
| 100 | Q60864 | Stress-induced-phosphoprotein 1 | 1.1499 | 1 | 3.5 | 5.12E-05 | stress response |
| 101 | P52480-2 | Pyruvate kinase isozymes M1/M2 | 1.2958 | 1 | 19.6 | 3.40E-33 | glycolysis |
| 102 | Q920E5 | Farnesyl pyrophosphate synthetase | 1.3534 | 1 | 8.2 | 8.44E-16 | cholesterol/isoprenoid biosynthesis |
| 103 | P38647 | Stress-70 protein, mitochondrial | 1.357 | 2 | 4.6 | 6.57E-08 | nuclear export; protein folding |
| 104 | P14824 | Annexin A6 | 1.6252 | 4 | 7.9 | 2.47E-08 | Ca2+ transport; muscle contraction |
| 105 | Q61753 | D-3-phosphoglycerate dehydrogenase | 1.7927 | 3 | 8.1 | 6.28E-15 | cell cycle; neural development; serine biosynthesis |
a. Ratio of N2a58/22L vs. N2a58# cells
b. Number of identified peptides
c. posterior error probability (PEP) estimates the probability of wrong assignment of a spectrum to a peptide sequence
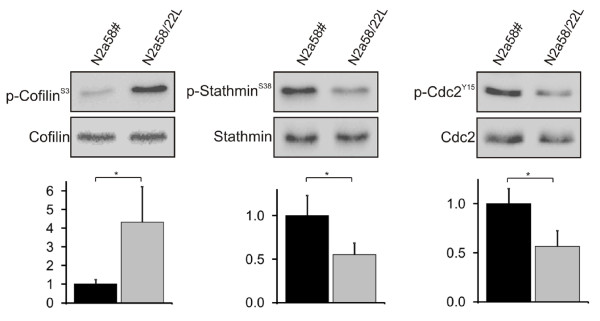
Among quantified phosphoproteins, we then considered specific phosphosites in selected target proteins, such as Cdc2, stathmin, and cofilin as analyzed by mass-spectrometry (Table 3). An increase of cofilinS3 phosphorylation in N2a58/22L cells was suggested by a ratio 1.63, while the amount of the two tyrosine phosphorylation sites (Y15, Y160) in Cdc2 were decreased upon 22L infection. Stathmin phosphopeptides containing serine 38 were increased, whereas the amount of stathmin phosphopeptides harboring serine 25 in N2a58/22L cells was significantly lower (Table 3).
Table 3.
Identified phosphorylation sites
| Protein names | Ratio (total)a |
Phosphosite | Ratio (specific phospho-site)b |
|---|---|---|---|
| Cdc2 | 0.49428 | Y15 | 0.43086 |
| Y160 | 0.64359 | ||
| stathmin | 1.1422 | S25 | 1.2155 |
| S38 | 0.45601 | ||
| cofilin | 1.0173 | S3 | 1.6328 |
a. Ratio of phosphorylation N2a58/22L vs. N2a58# cells
b. Ratio of phospho sites in N2a58/22L and N2a58# cells
To validate the results obtained in the SILAC phosphoproteomic analysis we performed Western blots for cofilin 1, Cdc2, and stathmin using antibodies for the detection of specific phosphosites. As predicted by the SILAC analysis, cofilin 1 phosphorylation was significantly induced in Scrapie-infected N2a58/22L cells compared to PPS-treated N2a58# cells (Figure 2, left panels). Cofilin represents a potent regulator of the actin filaments, which is controlled by phosphorylation of serine 3 mediated through the LIM-kinase 1 (LIMK-1) in vitro and in vivo [16]. These data support previous studies indicating a direct interaction of PrPSc with cofilin [17]. Together with our finding that phosphorylation of cofilin is induced in PrPSc-infected neuronal cells; the results indicate a significant role for the protein in neurodegeneration processes. Stathmin acts as an important regulatory protein of microtubule dynamics, which can be directly targeted by Cdc2 [18]. In our analysis, we showed that stathminS38 phosphorylation was decreased (Figure 2, middle panels), which correlates with the inactivation of Cdc2 in N2a58/22L cells (Figure 2, right panels) implying that there is a functional interaction. Cdc2 is a crucial kinase in starting M phase events during the cell cycle progression and regulates important mitotic structure changes, including nuclear envelope breakdown and spindle assembly [19]. Dephosphorylation of stathminS38 led to an inhibition of cells at G2/M phase, lack of spindle assembly, and growth inhibition [20,21]. Together with the finding that the prion gene is transcriptionally activated in the G1 phase in confluent and terminally differentiated cells [22], we assume that control of the cell cycle might be important in prion diseases.
Figure 2.
Specific regulation of cofilin, Cdc2, and stathmin phosphorylation in scrapie-infected neuronal cells. Cell lysates of N2a58# and 22L-infected N2a58/22L cells were analyzed by Western blot using phospho-specific antibodies to detect p-cofilinS3, p-cdc2Y15, and p-stathminS38 (left panels). As loading controls, equal amounts of cofilin, Cdc2 and stathmin were shown. Quantification of intensities of phosphorylation signal was performed by normalizing the corresponding loading control (* p < 0.05) (right panels).
Aberrant signal transduction pathways are implicated in many diseases. However, perturbations in phosphorylation-based signaling networks are typically studied in a hypothesis-driven approach. In this study, we performed the first global analysis of the phosphoproteome of scrapie-infected neuronal cells, since the knowledge of PrP-dependent deregulation of the signalling network is poor. SILAC provides a powerful and accurate technique for relative proteome-wide quantification by mass-spectrometry. Its versatility has been demonstrated by a wide range of applications, especially for intracellular signal transduction pathways [23-25]. Since we applied SILAC for the quantitative detection of the phosphoproteome in scrapie-infected neuroblastoma cells, we found 105 different phosphoproteins. Among identified proteins, we validated the regulated phosphorylation of cofilin, stathmin and Cdc2 indicating that the identification of phosphoproteins in scrapie-infected neuronal cells by SILAC is reliable. Future work is necessary to determine whether the identified novel phosphoproteins are involved in prion diseases and if they probably represent sensitive and specific biomarkers for diagnosis or therapeutic intervention strategies.
Methods
Cell culture
N2a58/22L cells have been described previously [11] and were kindly provided by Prof. Schätzl (LMU, Munich). Cells were cultured in DMEM containing 10% FCS and 4 mM L-glutamine at 37°C. Cells were treated with 5 μg/ml pentosan polysulfate (Cartrophen Vet, A. Albrecht GmbH + Co. KG, Germany) for two passages, resulting in a stable rescued cell line for more than 15 passages (N2a58# cells). Cell lysates were prepared by scraping cells in lysis buffer containing 150 mM NaCl, 0.5% Triton X-100, 0.5% DOC, 50 mM Tris pH 7.5, 1 mM Na-vanadate, 1 mM Na-molybdate, 20 mM NaF, 10 mM NaPP, 20 mM β-glycerophosphat, 1× protease inhibitor cocktail (Roche, Mannheim, Germany). For digestion with proteinase K (PK) 80 μg protein were treated with 20 μg/ml PK for 30 min at 37°C. PK digestion was stopped by addition of laemmli sample buffer and protein denaturation at 95°C for 7 min.
Colony assay
The colony assay was performed as previously described with minor modifications [26]. In brief, cells were grown on glass cover slips to confluence using a 24 well plate. The cell layer was soaked in lysis buffer (150 mM NaCl, 0.5% Triton X-100, 0.5% DOC, 50 mM Tris pH 7.5) on a nitrocellulose membrane. After drying for 30 min at room temperature, the membrane was incubated in lysis buffer containing 5 μg/ml proteinase K (PK) for 90 min at 37°C, rinsed twice with water, and incubated in 2 mM PMSF for 10 min. The membrane was shaken in 3 M guanidinium thiocyanate, 10 mM Tris-HCl (pH 8.0) for 10 min, followed by rinsing five times with water. 5% nonfat dry milk in TBS-T was used for blocking for 1 h at room temperature. PrP was detected using an anti-PrP antibody 6H4 (Prionics) and a HRP-conjugated sheep anti-mouse antibody (GE Healthcare).
SDS-PAGE and Western Blot
Proteins were separated by 12% SDS-PAGE and transferred to polyvinylidene difluoride membranes (PVDF, Millipore) by semidry blotting. PrP was detected using the PrP-specific mouse mAb 8H4 (Alicon AG). For validation of phosphorylated proteins anti-phospho-stathmin (Ser38) (#3426, Cell Signaling Technology), anti-phospho-cdc2 (Tyr15) (#4539, Cell Signaling Technology), and anti-phospho-cofilin (Ser3) antibodies (#3313, Cell Signaling Technology) were used. Antibodies recognizing stathmin (#3352), cdc2 (#9112) and cofilin (#3312) were also obtained from Cell Signaling Technology.
Two dimensional gel electrophoresis
For 2D electrophoresis 150 μg protein of cell lysates were purified by trichloroacetic acid precipitation and re-suspended in DeStreak Rehydration Solution (Amersham Biosciences) containing 0.5% Bio-Lyte pH3-10 (Bio-Rad Laboratories GmbH, München). The isoelectric focusing was run on IPG strips with a non-linear pH range of 3-10 and a length of 7 cm (Bio-Rad) using the ZOOM® IPGRunner™system from Invitrogen. After focussing strips were equilibrated in 50 mM Tris, 1 mM Urea, 30% Glycerin, 2% SDS, 1% DTT for 25 min and in 50 mM Tris, 1 mM Urea, 30% Glycerin, 2% SDS, 5% Iodacetamid for 25 min. Strips were then separated in 10% SDS-PAGE gels in the second dimension and analyzed by Coomassie staining or immunoblotting using an anti-phospho-tyrosine (sc-7020, Santa Cruz) or an anti-phospho-threonine antibody (#9381, Cell Signaling Technology).
SILAC phosphoproteomics analysis
SILAC ready-to-use cell culture media and dialyzed FBS were obtained from Dundee Cell Products Ltd, UK. While N2a58# cells were cultured in control SILAC DMEM media containing unlabelled arginine and lysine amino acids (R0K0), N2a58/22L cells were cultured in ready-to-use SILAC DMEM medium containing 13C labeled arginine and lysine amino acids (R6K6) for seven cell division cycles. After preparation of cell lysates and measurement of protein concentration, lysates of N2a58# and N2a58/22L cells were mixed in a ratio 1:1. Each sample was reduced in SDS PAGE loading buffer containing 10 mM DTT and alkylated in 50 mM iodoacetamide prior to separation by one-dimensional SDS-PAGE (4-12% Bis-Tris Novex mini-gel, Invitrogen) and visualization by colloidal Coomassie staining (Novex, Invitrogen). The entire protein gel lane was excised and cut into 10 gel slices each. Every gel slice was subjected to in-gel digestion with trypsin [27]. The resulting tryptic peptides were extracted by 1% formic acid, acetonitrile, lyophilized in a speedvac (Helena Biosciences).
Phosphopeptide enrichment
The lyophilized peptides above were resuspended in 5% acetic acid (binding buffer) and phosphopeptide enrichment was carried out using immobilized metal ion affinity chromatography (IMAC). Immobilized gallium in the Pierce Ga-IDA Phosphopeptide Enrichment Kit was used to enrich for phosphopeptides prior to MS/MS analysis according to the manufacturer's instructions (Thermo Scientific).
LC-MS/MS
Trypsin digested peptides were separated using an Ultimate U3000 (Dionex Corporation) nanoflow LC-system consisting of a solvent degasser, micro and nanoflow pumps, flow control module, UV detector and a thermostated autosampler. 10 μl of sample (a total of 2 μg) was loaded with a constant flow of 20 μl/min onto a PepMap C18 trap column (0.3 mm id × 5 mm, Dionex Corporation). After trap enrichment peptides were eluted off onto a PepMap C18 nano column (75 μm × 15 cm, Dionex Corporation) with a linear gradient of 5-35% solvent B (90% acetonitrile with 0.1% formic acid) over 65 minutes with a constant flow of 300 nl/min. The HPLC system was coupled to a LTQ Orbitrap XL (Thermo Fisher Scientific Inc) via a nano ES ion source (Proxeon Biosystems). The spray voltage was set to 1.2 kV and the temperature of the heated capillary was set to 200°C. Full scan MS survey spectra (m/z 335-1800) in profile mode were acquired in the Orbitrap with a resolution of 60,000 after accumulation of 500,000 ions. The five most intense peptide ions from the preview scan in the Orbitrap were fragmented by collision induced dissociation (normalised collision energy 35%, activation Q 0.250 and activation time 30 ms) in the LTQ after the accumulation of 10,000 ions. Maximal filling times were 1,000 ms for the full scans and 150 ms for the MS/MS scans. Precursor ion charge state screening was enabled and all unassigned charge states as well as singly charged species were rejected. The dynamic exclusion list was restricted to a maximum of 500 entries with a maximum retention period of 90 seconds and a relative mass window of 10 ppm. The lock mass option was enabled for survey scans to improve mass accuracy [28]. Data were acquired using the Xcalibur software.
Quantification and Bioinformatic Analysis
Quantification was performed with MaxQuant version 1.0.7.4 [29], and was based on two-dimensional centroid of the isotope clusters within each SILAC pair. To minimize the effect of outliers, protein ratios were calculated as the median of all SILAC pair ratios that belonged to peptides contained in the protein. The percentage variability of the quantitation was defined as the standard deviation of the natural logarithm of all ratios used for obtaining the protein ratio multiplied by a constant factor 100.
The generation of peak list, SILAC- and extracted ion current-based quantitation, calculated posterior error probability, and false discovery rate based on search engine results, peptide to protein group assembly, and data filtration and presentation was carried out using MaxQuant. The derived peak list was searched with the Mascot search engine (version 2.1.04; Matrix Science, London, UK) against a concatenated database combining 80,412 proteins from International Protein Index (IPI) human protein database version 3.6 (forward database), and the reversed sequences of all proteins (reverse database). Alternatively, database searches were done using Mascot (Matrix Science) as the database search engine and the results saved as a peptide summary before quantification using MSQuant http://msquant.sourceforge.net/. Parameters allowed included up to three missed cleavages and three labeled amino acids (arginine and lysine). Initial mass deviation of precursor ion and fragment ions were up to 7 ppm and 0.5 Da, respectively. The minimum required peptide length was set to 6 amino acids. To pass statistical evaluation, posterior error probability (PEP) for peptide identification (MS/MS spectra) should be below or equal to 0.1. The required false positive rate (FPR) was set to 5% at the peptide level. False positive rates or PEP for peptides were calculated by recording the Mascot score and peptide sequence length-dependent histograms of forward and reverse hits separately and then using Bayes' theorem in deriving the probability of a false identification for a given top scoring peptide. At the protein level, the false discovery rate (FDR) was calculated as the product of the PEP of a protein's peptides where only peptides with distinct sequences were taken into account. If a group of identified peptide sequences belong to multiple proteins and these proteins cannot be distinguished, with no unique peptide reported, these proteins are reported as a protein group in MaxQuant. Proteins were quantified if at least one MaxQuant-quantifiable SILAC pair was present. Identification was set to a false discovery rate of 1% with a minimum of two quantifiable peptides. The set value for FPR/PEP at the peptide level ensures that the worst identified peptide has a probability of 0.05 of being false; and proteins are sorted by the product of the false positive rates of their peptides where only peptides with distinct sequences are recognized. During the search, proteins are successively included starting with the best-identified ones until a false discovery rate of 1% is reached; an estimation based on the fraction of reverse protein hits.
Enzyme specificity was set to trypsin allowing for cleavage of N-terminal to proline and between aspartic acid and proline. Carbamidomethylation of cysteine was searched as a fixed modification, whereas N-acetyl protein, oxidation of methionine and phosphorylation of serine, threonine and tyrosine were searched as variable modifications.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
WW carried out the experimental work, drafted and wrote the manuscript. PA performed and interpreted the SILAC analysis. JL participated in the design of the study. SW conceived of the study, and participated in its design and coordination and wrote the manuscript. All authors read and approved the final manuscript.
Contributor Information
Wibke Wagner, Email: wagwi@pei.de.
Paul Ajuh, Email: pajuh@dundeecellproducts.com.
Johannes Löwer, Email: loejo@pei.de.
Silja Wessler, Email: silja.wessler@sbg.ac.at.
Acknowledgements
We thank Prof. Schätzl from the LMU in Munich for providing N2a58/22L cells.
References
- Prusiner SB. Prions. Proc Natl Acad Sci USA. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusiner SB. Novel proteinaceous infectious particles cause scrapie. Science. 1982;216:136–144. doi: 10.1126/science.6801762. [DOI] [PubMed] [Google Scholar]
- Aguzzi A, Polymenidou M. Mammalian prion biology: one century of evolving concepts. Cell. 2004;116:313–327. doi: 10.1016/S0092-8674(03)01031-6. [DOI] [PubMed] [Google Scholar]
- Taylor DR, Hooper NM. The prion protein and lipid rafts. Mol Membr Biol. 2006;23:89–99. doi: 10.1080/09687860500449994. [DOI] [PubMed] [Google Scholar]
- Mattei V, Garofalo T, Misasi R, Circella A, Manganelli V, Lucania G, Pavan A, Sorice M. Prion protein is a component of the multimolecular signaling complex involved in T cell activation. FEBS Lett. 2004;560:14–18. doi: 10.1016/S0014-5793(04)00029-8. [DOI] [PubMed] [Google Scholar]
- Hugel B, Martinez MC, Kunzelmann C, Blattler T, Aguzzi A, Freyssinet JM. Modulation of signal transduction through the cellular prion protein is linked to its incorporation in lipid rafts. Cell Mol Life Sci. 2004;61:2998–3007. doi: 10.1007/s00018-004-4318-2. [DOI] [PubMed] [Google Scholar]
- Mouillet-Richard S, Ermonval M, Chebassier C, Laplanche JL, Lehmann S, Launay JM, Kellermann O. Signal transduction through prion protein. Science. 2000;289:1925–1928. doi: 10.1126/science.289.5486.1925. [DOI] [PubMed] [Google Scholar]
- Gyllberg H, Lofgren K, Lindegren H, Bedecs K. Increased Src kinase level results in increased protein tyrosine phosphorylation in scrapie-infected neuronal cell lines. FEBS Lett. 2006;580:2603–2608. doi: 10.1016/j.febslet.2006.03.092. [DOI] [PubMed] [Google Scholar]
- Na YJ, Jin JK, Kim JI, Choi EK, Carp RI, Kim YS. JAK-STAT signaling pathway mediates astrogliosis in brains of scrapie-infected mice. J Neurochem. 2007;103:637–649. doi: 10.1111/j.1471-4159.2007.04769.x. [DOI] [PubMed] [Google Scholar]
- Ertmer A, Gilch S, Yun SW, Flechsig E, Klebl B, Stein-Gerlach M, Klein MA, Schatzl HM. The tyrosine kinase inhibitor STI571 induces cellular clearance of PrPSc in prion-infected cells. J Biol Chem. 2004;279:41918–41927. doi: 10.1074/jbc.M405652200. [DOI] [PubMed] [Google Scholar]
- Nishida N, Harris DA, Vilette D, Laude H, Frobert Y, Grassi J, Casanova D, Milhavet O, Lehmann S. Successful transmission of three mouse-adapted scrapie strains to murine neuroblastoma cell lines overexpressing wild-type mouse prion protein. J Virol. 2000;74:320–325. doi: 10.1128/JVI.74.1.320-325.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocisko DA, Engel AL, Harbuck K, Arnold KM, Olsen EA, Raymond LD, Vilette D, Caughey B. Comparison of protease-resistant prion protein inhibitors in cell cultures infected with two strains of mouse and sheep scrapie. Neurosci Lett. 2005;388:106–111. doi: 10.1016/j.neulet.2005.06.053. [DOI] [PubMed] [Google Scholar]
- Kascsak RJ, Rubenstein R, Merz PA, Carp RI, Wisniewski HM, Diringer H. Biochemical differences among scrapie-associated fibrils support the biological diversity of scrapie agents. J Gen Virol. 1985;66(Pt 8):1715–1722. doi: 10.1099/0022-1317-66-8-1715. [DOI] [PubMed] [Google Scholar]
- Collinge J, Sidle KC, Meads J, Ironside J, Hill AF. Molecular analysis of prion strain variation and the aetiology of 'new variant' CJD. Nature. 1996;383:685–690. doi: 10.1038/383685a0. [DOI] [PubMed] [Google Scholar]
- Somerville RA, Chong A, Mulqueen OU, Birkett CR, Wood SC, Hope J. Biochemical typing of scrapie strains. Nature. 1997;386:564. doi: 10.1038/386564a0. [DOI] [PubMed] [Google Scholar]
- Yang N, Higuchi O, Ohashi K, Nagata K, Wada A, Kangawa K, Nishida E, Mizuno K. Cofilin phosphorylation by LIM-kinase 1 and its role in Rac-mediated actin reorganization. Nature. 1998;393:809–812. doi: 10.1038/31735. [DOI] [PubMed] [Google Scholar]
- Giorgi A, Di Francesco L, Principe S, Mignogna G, Sennels L, Mancone C, Alonzi T, Sbriccoli M, De Pascalis A, Rappsilber J. et al. Proteomic profiling of PrP27-30-enriched preparations extracted from the brain of hamsters with experimental scrapie. Proteomics. 2009;9:3802–3814. doi: 10.1002/pmic.200900085. [DOI] [PubMed] [Google Scholar]
- Beretta L, Dobransky T, Sobel A. Multiple phosphorylation of stathmin. Identification of four sites phosphorylated in intact cells and in vitro by cyclic AMP-dependent protein kinase and p34cdc2. J Biol Chem. 1993;268:20076–20084. [PubMed] [Google Scholar]
- Satyanarayana A, Kaldis P. Mammalian cell-cycle regulation: several Cdks, numerous cyclins and diverse compensatory mechanisms. Oncogene. 2009;28:2925–2939. doi: 10.1038/onc.2009.170. [DOI] [PubMed] [Google Scholar]
- Brattsand G, Marklund U, Nylander K, Roos G, Gullberg M. Cell-cycle-regulated phosphorylation of oncoprotein 18 on Ser16, Ser25 and Ser38. Eur J Biochem. 1994;220:359–368. doi: 10.1111/j.1432-1033.1994.tb18632.x. [DOI] [PubMed] [Google Scholar]
- Marklund U, Osterman O, Melander H, Bergh A, Gullberg M. The phenotype of a "Cdc2 kinase target site-deficient" mutant of oncoprotein 18 reveals a role of this protein in cell cycle control. J Biol Chem. 1994;269:30626–30635. [PubMed] [Google Scholar]
- Gougoumas DD, Vizirianakis IS, Tsiftsoglou AS. Transcriptional activation of prion protein gene in growth-arrested and differentiated mouse erythroleukemia and human neoplastic cells. Exp Cell Res. 2001;264:408–417. doi: 10.1006/excr.2001.5151. [DOI] [PubMed] [Google Scholar]
- Zhang G, Neubert TA. Use of stable isotope labeling by amino acids in cell culture (SILAC) for phosphotyrosine protein identification and quantitation. Methods Mol Biol. 2009;527:79–92. doi: 10.1007/978-1-60327-834-8_7. xi. full_text. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amanchy R, Kalume DE, Iwahori A, Zhong J, Pandey A. Phosphoproteome analysis of HeLa cells using stable isotope labeling with amino acids in cell culture (SILAC) J Proteome Res. 2005;4:1661–1671. doi: 10.1021/pr050134h. [DOI] [PubMed] [Google Scholar]
- Kruger M, Kratchmarova I, Blagoev B, Tseng YH, Kahn CR, Mann M. Dissection of the insulin signaling pathway via quantitative phosphoproteomics. Proc Natl Acad Sci USA. 2008;105:2451–2456. doi: 10.1073/pnas.0711713105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klohn PC, Stoltze L, Flechsig E, Enari M, Weissmann C. A quantitative, highly sensitive cell-based infectivity assay for mouse scrapie prions. Proc Natl Acad Sci USA. 2003;100:11666–11671. doi: 10.1073/pnas.1834432100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- Olsen JV, de Godoy LM, Li G, Macek B, Mortensen P, Pesch R, Makarov A, Lange O, Horning S, Mann M. Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol Cell Proteomics. 2005;4:2010–2021. doi: 10.1074/mcp.T500030-MCP200. [DOI] [PubMed] [Google Scholar]
- Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]