Abstract
Cullin-RING ubiquitin ligases (CRLs) are critical regulators of multiple developmental and cellular processes in eukaryotes. CAND1 is a biochemical inhibitor of CRLs, yet has been shown to promote CRL activity in plant and mammalian cells. Here we analyze CAND1 function in the context of a developing metazoan organism. C. elegans CAND-1 is capable of binding to all of the cullins, and we show that it physically interacts with CUL-2 and CUL-4 in vivo. The covalent attachment of the ubiquitin-like protein Nedd8 is required for cullin activity in animals and plants. In cand-1 mutants, the levels of the neddylated isoforms of CUL-2 and CUL-4 are increased, indicating that CAND-1 is a negative regulator of cullin neddylation. cand-1 mutants are hypersensitive to the partial loss of cullin activity, suggesting that CAND-1 facilitates CRL functions. cand-1 mutants exhibit impenetrant phenotypes, including developmental arrest, morphological defects of the vulva and tail, and reduced fecundity. cand-1 mutants share with cul-1 and lin-23 mutants the phenotypes of supernumerary seam cell divisions, defective alae formation, and the accumulation of the SCFLIN-23 target the glutamate receptor GLR-1. The observation that cand-1 mutants have phenotypes associated with the loss of the SCFLIN-23 complex, but lack phenotypes associated with other specific CRL complexes, suggests that CAND-1 is differentially required for the activity of distinct CRL complexes.
Keywords: CAND1, CRL, cullin, Nedd8, neddylation, ubiquitin ligase, seam cells
Introduction
Regulated protein degradation controls a wide-range of cellular processes. The majority of cellular proteins are degraded by the ubiquitin proteasome system (Ciechanover et al., 1984; Rock et al., 1994). Ubiquitin is a highly-conserved 76 amino acid polypeptide that can be covalently linked to substrates as monoubiquitin or in polyubiquitin chains to affect the activity or localization of substrates, or to mark them for degradation by the 26S proteasome (Pickart and Fushman, 2004). The ubiquitylation reaction involves a multi-enzymatic pathway mediated by a ubiquitin activating enzyme (E1), a ubiquitin conjugating enzyme (E2), and a ubiquitin protein ligase (E3) (Glickman and Ciechanover, 2002). E3s provide the target specificity in this reaction by binding the substrate and bringing it in close proximity to the E2 to facilitate the transfer of ubiquitin either directly from the E2 to the substrate, or, in the case of HECT-domain E3s, indirectly via transfer of ubiquitin to the E3 and then to the substrate.
Cullin-RING ligases (CRLs) are the largest family of E3s in metazoa (Bosu and Kipreos, 2008; Petroski and Deshaies, 2005). There are five major classes of cullins in metazoa, and each forms a distinct type of CRL complex. The cullin functions as a rigid platform for the assembly of the multisubunit complex. CRL complexes that contain the cullin CUL1 are denoted SCF complexes, while other CRL complexes are named based on the cullin in the complex, e.g., CRL2 complexes contain CUL2. The C-terminus of the cullin binds a RING H2 finger protein, either Rbx1/Roc1/Hrt1 or Rbx2/Roc2, that recruits the E2. The N-termini of cullins 1, 2, 4, and 5 bind to adaptor proteins that bring substrate-recognition subunits (SRSs) to the complex. The adaptors are Skp1 (for SCF complexes); elongin C (for CRL2 and CRL5); and DDB1 (for CRL4); while the SRSs are F-box proteins (SCF); VHL-box proteins (CRL2); SOCS-box proteins (CRL5); and WDXR/DXR-proteins (CRL4). CRL3 complexes are unique in that the SRSs (BTB/POZ proteins) bind directly to CUL3, obviating the need for an adaptor. Notably, SRSs are variable components, and the binding of different SRSs to the core components forms distinct CRL complexes that target different subsets of substrates. The in vivo functions of multiple classes of CRL complexes have been extensively studied in the context of animal development, particularly in the nematode C. elegans, where CRL complexes regulate multiple aspects of development, including polarity, cell fate, cell signaling, morphogenesis, meiosis, neuronal outgrowth, and synapse formation (Kipreos, 2005).
CRLs are activated by Nedd8, which is a ubiquitin-like protein that is conjugated to cullins at a conserved C-terminal lysine residue in a process termed neddylation (Pan et al., 2004). The neddylation reaction is similar to the ubiquitylation reaction and involves the heterodimeric E1 APP-BP-1/Uba3, the E2 UBC12, and the E3s DCN1 (defective in cullin neddylation) and Rbx1 (Gong and Yeh, 1999; Kamura et al., 1999; Kurz et al., 2008; Liakopoulos et al., 1998).
Neddylation is required for the function of SCF, CRL2, and CRL3 complexes in a number of species (Morimoto et al., 2000; Ohh et al., 2002; Osaka et al., 2000; Ou et al., 2002; Pintard et al., 2003; Podust et al., 2000; Read et al., 2000; Wu et al., 2000). Nedd8 can interact directly with the E2, and Nedd8 conjugation to the cullin potentiates the recruitment of the E2 to the CRL complex (Kawakami et al., 2001; Saha and Deshaies, 2008; Sakata et al., 2007). Analysis of crystal structures indicates that Nedd8 conjugation induces a major conformational change to the C-terminal domain of the cullin that causes the RING domain of Rbx1 to escape from its binding pocket so that it is flexibly tethered to the cullin by an extended β-sheet, reminiscent of a balloon on a string (Duda et al., 2008). The extension of Rbx1 provides the flexibility to conjugate ubiquitin onto both the substrate and the elongating polyubiquitin chain (Duda et al., 2008; Saha and Deshaies, 2008). In budding yeast, the Nedd8 ortholog Rub1p is not required for essential SCF functions although it does enhance SCF activity (Lammer et al., 1998; Liakopoulos et al., 1998). Other covalent modifications of the neddylation site are possible. SCF complexes can autoubiquitylate themselves on the neddylation site in vitro to activate the SCF complex (Duda et al., 2008). The budding yeast cullin Rtt101p is modified at the neddylation site in vivo by both Rub1p and another protein with a similar mass to that of Rub1p, potentially ubiquitin or a ubiquitin-like protein (Laplaza et al., 2004).
The COP9 signalosome (CSN) contains a Nedd8 isopeptidase activity that removes Nedd8 from the cullin (deneddylation) (Cope et al., 2002; Lyapina et al., 2001); and inactivation of CSN increases the levels of neddylated cullins in vivo (Lyapina et al., 2001; Menon et al., 2007; Pintard et al., 2003; Schwechheimer et al., 2001). Counterintuitively, loss of CSN activity is observed to reduce the activity of SCF, CRL3, and CRL4 complexes in vivo (Cope et al., 2002; Doronkin et al., 2003; Groisman et al., 2003; Liu et al., 2003; Pintard et al., 2003; Schwechheimer et al., 2001; Zhou et al., 2003). The loss of CRL activity in cells lacking CSN has been attributed to reductions in the levels of SRS proteins caused by increased autoubiquitylation (Chew et al., 2007; Cope and Deshaies, 2006; He et al., 2005; Wee et al., 2005; Wu et al., 2005; Zhou et al., 2003). In C. elegans, loss of CSN causes embryonic lethality and is associated with an increased proportion of neddylated CUL-3 (Pintard et al., 2003). Interestingly, combining CSN mutations with mutations in the neddylation pathway, which reduce cullin neddylation levels, rescues the inviability (Pintard et al., 2003). This implies either that the proportion of cullin neddylation is critical for CRL function, or that a balanced cycling between neddylated and unneddylated states is essential for CRL activity.
Human CAND1 (cullin-associated and neddylation-dissociated) is a 120 kDa protein that contains multiple HEAT repeats and binds to the cullin-Rbx1 dimeric complex (Goldenberg et al., 2004). The crystal structure of CAND1 bound to CUL1-Rbx1 reveals that the CAND1 N-terminus binds to the cullin C-terminus, and the CAND1 C-terminus binds to the cullin N-terminus (Goldenberg et al., 2004). CAND1 binding prevents CUL1 from interacting with the adapter Skp1 and blocks access to the neddylation-site lysine residue of CUL1, thus preventing the formation of an active SCF complex (Liu et al., 2002; Zheng et al., 2002). CAND1 can bind to unneddylated cullins but not to neddylated cullins (Liu et al., 2002; Min et al., 2003; Oshikawa et al., 2003; Zheng et al., 2002). In certain human cell lines, CAND1 appears to preferentially associate with CUL1 (Liu et al., 2002; Oshikawa et al., 2003), but in other cell lines, it has been found to interact with all cullins (Chew and Hagen, 2007; Min et al., 2003). In C. elegans, CAND1 has been shown to bind to CUL-2 (Starostina et al., 2007), but was reported to have no detectable binding to CUL-3 (Luke-Glaser et al., 2007).
While CAND1 acts as a biochemical inhibitor of CRL complex formation, CAND1 inactivation leads to decreased SCF and CRL3 activity in Arabidopsis and human cells (Cheng et al., 2004; Chew et al., 2007; Chuang et al., 2004; Feng et al., 2004; Lo and Hannink, 2006; Zheng et al., 2002). There are at least two potential mechanisms for this phenomenon. First, similar to loss of CSN, the inactivation of human CAND1 can lead to the loss of SRSs through autoubiquitylation. Inactivation of human CAND1 is associated with reduced levels of the SRS Skp2 through autoubiquitylation, leading to a decrease in SCFSkp2 activity (Chew et al., 2007; Zheng et al., 2002). In contrast, CAND-1 inactivation leads to a reduction in the activities of the mammalian CRL3Keap1 complex and the Arabidopsis SCFTIR1 complex even though in both cases increased levels of the SRS are bound to the cullin. This suggests that CAND1 can also promote CRL activity through a mechanism that is independent of SRS stabilization (Lo and Hannink, 2006; Zhang et al., 2008).
In this study we have analyzed the C. elegans CAND1 ortholog, CAND-1. We observe that CAND-1 binds the cullins CUL-2 and CUL-4 in vivo and negatively regulates the levels of neddylated cullin isoforms. This is unlike the situation in mammals, Arabidopsis, and fission yeast, where CAND1 inactivation does not affect cullin neddylation (Chew and Hagen, 2007; Chuang et al., 2004; Feng et al., 2004; Schmidt et al., 2009; Zheng et al., 2002). We show that CAND-1 is required for a number of developmental processes in C. elegans, including vulva and tail morphogenesis, and the control of seam cell divisions. While critical CRL-dependent functions still occur in cand-1 mutants, the mutants are hypersensitive to the inactivation of cullins, suggesting that CAND-1 promotes, but is not essential for, CRL functions.
Materials and Methods
Strains and RNAi
The following C. elegans strains were used, N2: wild type, ET327: cand-1(tm1683)/unc-76(e911); in which cand-1(tm1683) was outcrossed six times, ET342: him-8(e1489); ekEx19[Pcul-2∷CUL-2∷FLAG∷cul-2 3′UTR plus the pRF4 plasmid containing rol-6(su1006)], ET361: unc-119(ed3); ekIs9[Pcul-4∷CUL-4∷FLAG∷cul-4 3′UTR], EU626: rfl-1(or198), ET271: ekEx13[Pwrt-2∷CDC-6∷tdTomato∷unc-54 3′UTR plus pRF4]; enjIs1[Phis-24∷HIS-24∷GFP∷unc-54 3′UTR], ET363: cand-1(tm1683); ekEx13, ET113: unc-119(ed3); ekIs2[Ppie-1∷GFP∷CYB-1∷pie-1 3′UTR + unc-119(+)], ET364: cand-1(tm1683); ekIs2, ET438: zyg-11(ax491); ekIs2, WS2072: unc-119(ed3); opIs76[Pcyb-1∷CYB-1∷YFP + unc-119(+)], ET446: nuIs24[Pglr-1∷GLR-1∷GFP] (Dreier et al., 2005), ET447: lin-23(e1883)/mnC1; nuIs24, ET448: cand-1(tm1683); nuIs24, ET450: cand-1(tm1683); cul-2(ek4) unc-64(e246)/bli-5(e518), VC1033: cul-4(gk434)/mIn1[mIs14 dpy-10(e128)], JR667: unc-119(e2498(x02237)Tc1); wIs51[scm∷GFP∷unc-54 3′UTR + unc-119(+)] (Koh and Rothman, 2001), ET396: him-5(e1467); wIs51, ET365: cand-1(tm1683); wIs51, ET350: cul-1(e1756)/unc-69(e587); him-5(e1467); wIs51, ET283: cul-4(gk434)/mIn1; cand-1(tm1683), ET427: ekIs20[Plin-23∷LIN-23∷GFP∷unc-54 3′UTR + rol-6(su1006)]; ekIs20 was made by gamma irradiation-mediated integration of the extrachromosomal array otEx840, which contains a functionally-rescuing LIN-23∷GFP (Mehta et al., 2004), ET449: cand-1(tm1683); ekIs20, and ET335: cand-1(tm1683) allele outcrossed eight times into the Hawaiian CB4856 background with Hawaiian SNPs detected on all chromosomes (with at least 8 SNPs per chromosome) except for chromosome V region +7.5 to +11.5, which contains the cand-1 gene and retains N2 sequence.
Feeding RNAi was performed as described (Kamath and Ahringer, 2003). At least two generations of larvae were maintained on cand-1 feeding RNAi prior to analysis of cand-1 RNAi phenotypes in wild-type or cand-1(tm1683) mutant backgrounds. The comparisons of ubc-12, csn-5, and cand-1 single RNAi, and ubc-12 + cand-1 double RNAi were performed by feeding L1-stage larvae with RNAi bacteria and analyzing the next generation. The cul-1, cul-2, cul-3, and cul-4 RNAi was performed as described in the Table 3 legend. All feeding-RNAi constructs (expressed in HT115 bacteria) were from the Ahringer library (Kamath et al., 2003), with the exception of that for cul-2 RNAi, which has been previously described (Starostina et al., 2007).
Two-hybrid assay
Two-hybrid analysis was performed with full-length and truncated cand-1 genes in the pACT2 vector (Gal4 activation domain) and with full-length cullin genes in the pAS1-CYH2 vector (Gal4 DNA binding domain) (Clontech). Transformation of the S. cerevisiae strain pJ69-4A (James et al., 1996) and the liquid-based lacZ enzymatic assay were performed as described (Janssen, 1995). Both histidine- and adenine-deficient selective media were used to test for interaction in the two-hybrid system.
Antibody production and immunofluorescence
Antisera to CAND-1 was produced in rabbits by immunization with a fusion protein expressed in E. coli comprising the C-terminal 374 amino acids of CAND-1 linked to a His tag in the pET15b vector (Novagen). The His-CAND-1 fusion protein was isolated under denaturing conditions using Ni-NTA agarose (Qiagen) according to the manufacturer's instructions. Anti-CAND-1 antibodies were affinity purified against recombinant CAND-1 protein linked to PVDF membrane, as described (Harlow and Lane, 1988); and used at a concentration of 1:500 for immunofluorescence. Anti-rabbit Alexa Fluor 488 (Molecular Probes) (1:400) and anti-mouse rhodamine (Cappel) (1:50) were used as secondary antibodies. DNA was stained with 1 μg/ml Hoechst 33258 dye. Immunofluorescence was performed on animals fixed using the “freeze-crack” method as described (Miller and Shakes, 1995).
Microscopy
Animals were observed by differential interference contrast (DIC) and immunofluorescence microscopy using a Zeiss Axioskop microscope. Images were taken with a Hamamatsu ORCA-ER digital camera with Openlab 4.0.2 software (Improvision). Images were processed with Adobe Photoshop 7.0. Matched images were taken with the same exposure time and processed identically.
Co-Immunoprecipitation, western blots, and mass spectrometry
Mixed-stage C. elegans were lysed with NP-40 buffer containing 10 mM HEPES (pH 7.2), 150 mM NaCl, 1% NP-40, 2 mM EDTA, complete protease inhibitors cocktail (Roche), and 50 μM N-acetyl-L-leucinyl-L-leucinal-L-norleucinal (LLnL; Sigma-Aldrich). Immunoprecipitation was performed with M2 anti-FLAG agarose (Sigma; 40 μl/ml of lysate). The primary antibodies used in western blot analysis were monoclonal anti-Flag (M2; Sigma; 1:2000), rabbit polyclonal anti-CUL-2 (1:2000) (Zhong et al., 2003), anti-CAND-1 (1:10,000), anti-NEDD-8 (Zymed; 1:300), and anti-tubulin (Sigma; 1:4000). Anti-rabbit-HRP (1:4000) and anti-mouse-HRP (Pierce; 1:10,000) were used as secondary antibodies for western blots that were processed with the Advanced ECL chemiluminescence system (GE healthcare). Quantitation of western blots was performed with non-saturated images; background signals were subtracted, and the signals of the bands of interest were normalized with the signals from either α-tubulin or non-specific anti-FLAG proteins in the same lysate. Statistical significance was determined with the Student's T-test; error bars are standard error of the mean (SEM). In-gel tryptic digestion and tandem mass spectrometry were carried out by the University of Georgia Proteomics Center.
RT-PCR
RT-PCR (reverse transcription-polymerase chain reaction) was used to isolate cand-1(tm1683) cDNA. Total RNA was isolated from whole-worm lysate using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. RNA was reverse transcribed into cDNA using the RT-PCR kit from Promega; the resulting cDNAs were used as templates for PCR amplification using the primer pair: gtccatgggtAGTGCTTATCATGTCGGGC and tcaggatccTTATGCAGTTTCCATTGGAGT. The PCR product was sequenced using the following cand-1 primers: gtagatcacttgtattcattccgt; atcactttcaacgatcctttg; atcggtccagtagtgattgga; cgtgagctgtcggcttgtggt; ctattgcacgcgttgaaagag; and gacatcactcaattctcgtca.
Results
CAND-1 can interact with all cullins and associates with CUL-2 and CUL-4 in vivo
The C. elegans cand-1 gene includes 12 exons that cover 11.7 kb of genomic DNA, and is predicted to encode a 1274 amino acid polypeptide (Fig. 1D). cand-1 is the only CAND1 homolog in C. elegans, and has significant levels of sequence identity with orthologous CAND1 proteins: 37% protein identity with H. sapiens CAND1; 37% for X. laevis; 36% for D. melanogaster; and 22% for S. pombe CAND1.
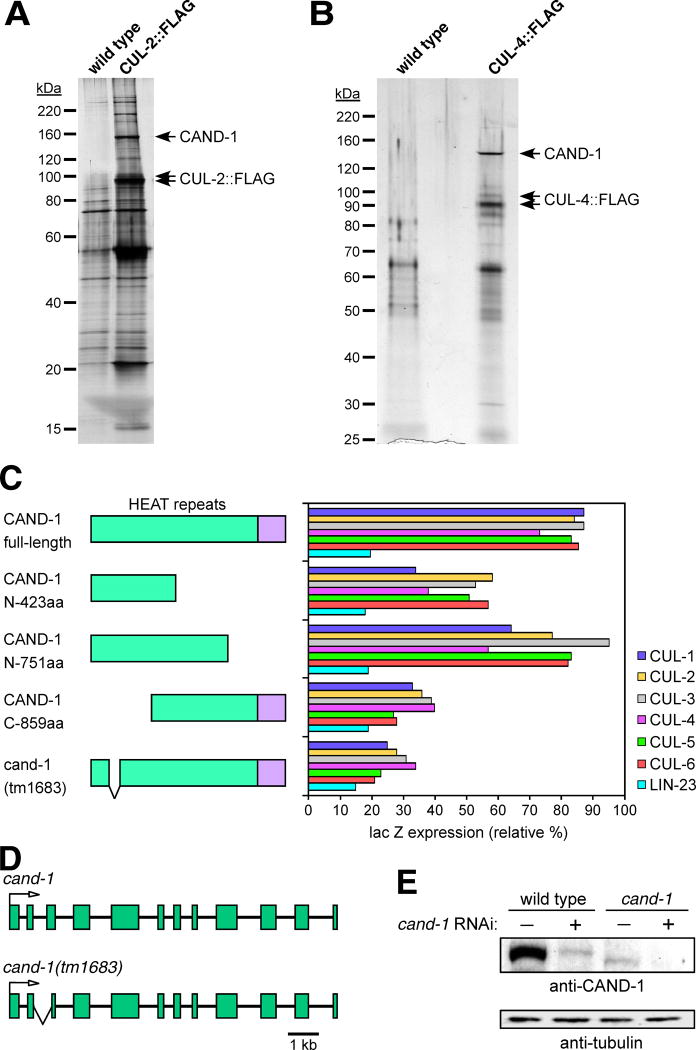
Figure 1. Interaction of CAND-1 with C. elegans cullins.
CAND-1 co-immunoprecipitates with CUL-2∷FLAG (A) and CUL-4∷FLAG (B). Silver-stained SDS-PAGE gels are shown for anti-FLAG affinity purifications from strains containing CUL-2∷FLAG or CUL-4∷FLAG, and from control wild-type animals. The CAND1 protein band (labeled) was identified by mass spectrometry. (C) Two-hybrid analysis of interaction between CAND-1 and the C. elegans cullins. On the left are diagrams of full-length, truncated, and tm1683 mutant CAND-1 proteins (the names reflect the number of N- or C-terminal amino acids remaining in the truncations; the HEAT-repeat region is in green). On the right is a graph of quantitation of interactions between the six cullin proteins and the CAND-1 proteins using a two-hybrid lacZ expression assay. CAND-1 was expressed from the pACT2 vector (fused to the Gal4 activation domain), and cullins or the negative control LIN-23 (Kipreos et al., 2000) are in the pAS1-CYH2 vector (fused to the Gal4 DNA binding domain). The scale derives from the level of the positive control (interaction between pACT2/SKR-1 and pAS1-CYH2/CUL-1; not shown), which is set at 100%. (D) Schematic of the cand-1 genomic region on chromosome V for wild type and the tm1683 deletion mutant. Exons are represented as boxes and lines represent introns. An arrow indicates the translational start point. The region deleted in the tm1683 mutant allele is encompassed by a ‘V-shaped’ lower line. (E) Effect of RNAi on CAND-1 protein levels in wild type and cand-1 mutants. cand-1 RNAi depletion is denoted by a plus above the lanes. Note that cand-1 mutants have lower CAND-1 levels than wild type, and that cand-1 RNAi further reduces CAND-1 levels.
CAND1 was identified by mass spectrometry as a protein that co-immunoprecipitates with CUL-2∷FLAG and CUL-4∷FLAG at high stoichiometry (Fig.1A,B). To determine if CAND-1 can physically associate with all C. elegans cullins, we tested interactions with the two-hybrid system using full-length and truncated forms of CAND1 (Fig. 1C; Suppl. Fig. 1). We observed that full-length CAND-1 interacts with all six C. elegans cullins in the two-hybrid system. Deletion of the N-terminal 415 amino acids of CAND-1 (C-859 truncation) decreased interaction significantly, while deletion of the C-terminal 523 amino acids (N-751 truncation) had less effect on cullin binding (Fig.1C), indicating that the N-terminal region is more critical for interaction.
To study the function of CAND-1, we obtained a cand-1 deletion allele, tm1683, from the National Bioresource Project for C. elegans (Japan) (Gengyo-Ando and Mitani, 2000). The tm1683 deletion removes 513 base pairs of intron 2 (leaving 21 base pairs at the 5′ end) and 141 bps of exon 3 (Fig.1D). We isolated the cand-1(tm1683) cDNA using RT-PCR, and the sequence indicates that the deletion produces an in-frame fusion that is predicted to encode a protein of 132 kDa, slightly smaller than the wild-type protein (141 kDa). The tm1683 deletion removes part of the N-terminal region that makes direct contact with cullins (Goldenberg et al., 2004). In the two-hybrid system, the CAND-1(tm1683) mutant protein has significantly decreased ability to bind cullins, similar to a deletion of the entire N-terminal region (Fig.1C).
We generated affinity-purified anti-CAND-1 antibody against the C-terminal 374 amino acids of CAND-1. Western blot analysis of whole-animal lysate using the anti-CAND-1 antibody revealed a single protein band. The CAND-1(tm1683) mutant protein migrates slightly faster than the wild-type CAND-1 protein as expected. Strikingly, the CAND-1(tm1683) protein is present at ∼12-fold lower level than the wild-type CAND-1 protein (11.9 ± 3.2; n=3), suggesting that the mutant protein is unstable (Fig.1E). Treatment of cand-1(tm1683) mutants with cand-1 RNAi reduced the level of CAND-1 protein to essentially undetectable levels (Fig.1E).
CAND-1 developmental expression pattern
We performed immunofluorescence with anti-CAND-1 antibody to determine the developmental expression pattern of CAND-1. RNAi depletion of CAND-1 significantly decreased the anti-CAND-1 signal, indicating that the staining is specific (Fig. 2M-P). In early embryos, CAND-1 is expressed in all cells, predominantly in the nucleus. Embryonic staining is strongest during early stages and is reduced in late-stage embryos (Fig. 2C-F). The observation of CAND-1 protein in the one-cell stage zygote indicates that CAND-1 is provided as maternal product by the hermaphrodite parent (Fig. 2A,B). During larval stages, CAND-1 is observed in proliferative cell lineages, including the seam cells, intestine, P-lineage cells, somatic gonad, and germline (Fig. 2G-J; data not shown). CAND-1 is also observed in a subset of non-proliferative tissues, including hypodermal cells, rectal epithelia and neuronal cells in the head and tail (Fig. 2G-H, data not shown). In older adults, anti-CAND-1 staining is restricted to the intestine and germline, with the strongest staining in oocyte nuclei (Fig. 2K-N). In younger adults, faint vulval cell staining is observed, potentially reflecting the perdurance of CAND-1 protein from the L4 larval stage (data not shown). The absence of CAND-1 expression in the majority of adult somatic tissues suggests that CAND-1 is not required for tissue homeostasis in the adult. Overall, our results indicate that CAND-1 is expressed in proliferating cells of the embryo and larvae, but also has expression in a subset of non-proliferating larval cells. This developmental expression pattern, and the predominantly nuclear localization are similar to that of core components of the CRL2 and CRL4 complexes (Feng et al., 1999; Kim and Kipreos, 2007; Zhong et al., 2003).
Figure 2. CAND-1 expression pattern.
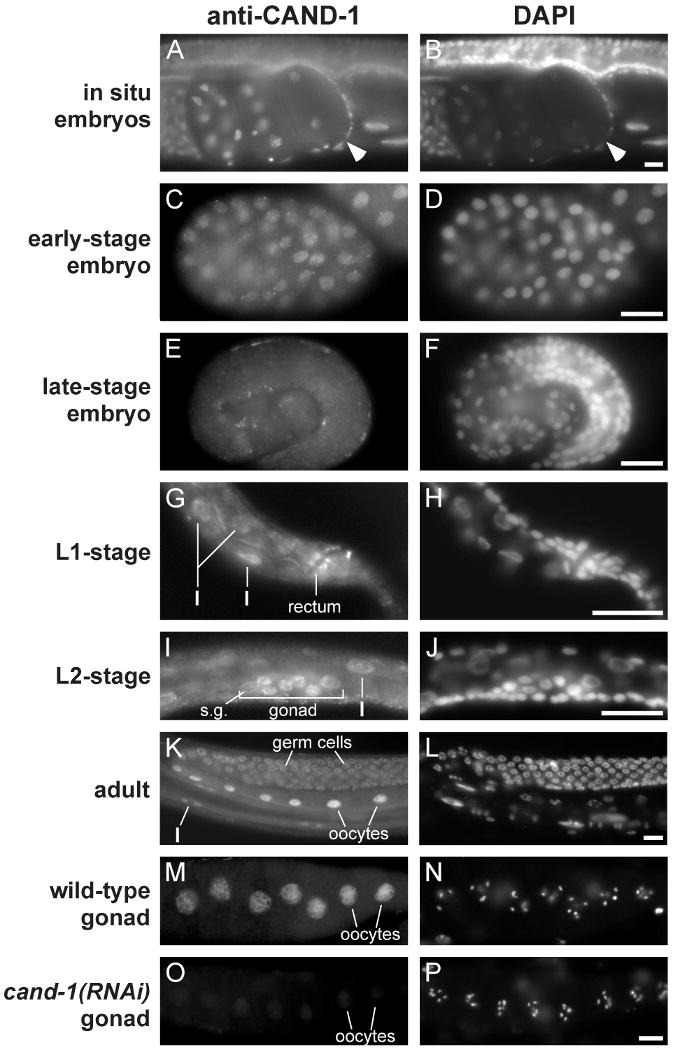
Images of wild-type animals stained with anti-CAND-1 antibody and DAPI. (A, B) in situ staining of early embryos. The one-cell stage zygote is denoted by an arrowhead, and has maternal and paternal pronuclei visible. (C, D) Early embryo at ∼100-cell stage. (E, F) A pretzel-stage embryo, which is the last of the defined embryo stages and has largely completed embryonic cell divisions and morphogenesis. Note that CAND-1 staining decreases in the older embryo. (G, H) The posterior of an L1-stage larva with CAND1 expression in the rectal epithelia and intestinal cells (I). (I, J) An L2-stage larva with CAND-1 staining in somatic gonadal cells (s.g.), germ cells, and intestine cells. (K, L) A wild-type adult with CAND-1 staining in germ cells, oocytes, and intestinal cells. (M-P) Dissected adult gonads from wild-type (M, N) and cand-1(RNAi) (O, P) hermaphrodite adults. Note that CAND-1 staining is significantly decreased in the cand-1(RNAi) gonad. Scale bars, 10 μm.
Loss of CAND-1 increases the proportion of the neddylated isoforms of CUL-2 and CUL-4
All animal cullins are modified by the conjugation of Nedd8, which promotes E3 activity (Pan et al., 2004). Endogenous CUL-2 and transgenic CUL-2∷FLAG and CUL-4∷FLAG exhibit two protein bands on SDS-PAGE gels. The slower-mobility band corresponds to the neddylated isoform, as determined by staining of immunoprecipitated protein with anti-Nedd8 antibody (Fig. 3A; data not shown). We will refer to the slower-migrating band as the ‘neddylated’ isoform as neddylated cullins migrate at that position, although, as described above, the slower-mobility band may also contain cullins that are conjugated to non-Nedd8 proteins, such as ubiquitin or ubiquitin-like proteins (Duda et al., 2008; Laplaza et al., 2004).
Figure 3. CAND-1 negatively regulates CUL-2 and CUL-4 neddylated isoform levels.
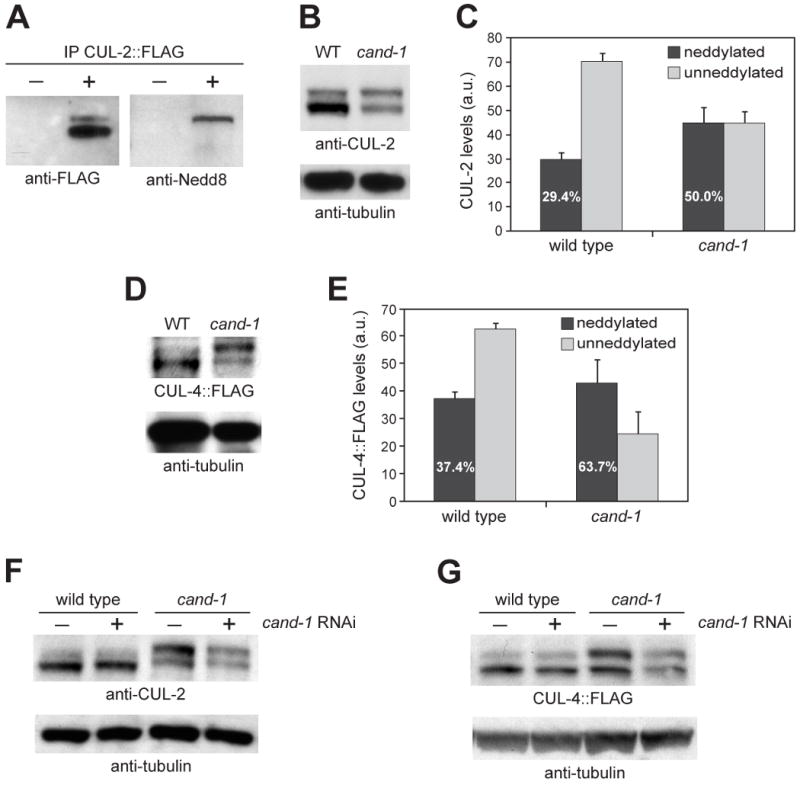
(A) Anti-FLAG immunoprecipitation from wild type (-) or animals expressing CUL-2∷FLAG (+), probed by Western blot with anti-FLAG and anti-Nedd8 antibodies. (B) Whole-worm lysates from wild type and cand-1 mutants blotted with anti-CUL-2 and anti-tubulin antibodies. (C) Graph of the average level of covalently-modified (neddylated) or unneddylated CUL-2 isoforms in wild type and cand-1 mutants from nine independent experiments. The levels of CUL-2 isoforms in cand-1 mutants are normalized to the total CUL-2 signal of wild-type animals, which is set to 100 arbitrary units (a.u.). The percentage of CUL-2 that is in the neddylated isoform relative to the total CUL-2 within a genotype is provided in white lettering within the neddylated isoform bar. This percentage is significantly different between wild type and cand-1 mutants (p < 0.0001, n=9; Student's T-test). (D) Whole-worm lysates prepared from wild type and cand-1 mutants expressing CUL-4∷FLAG and probed with anti-FLAG and anti-tubulin antibodies. (E) Graph of the average covalently-modified (neddylated) and unneddylated CUL-4∷FLAG isoforms in wild type and cand-1 mutants from seven independent experiments. Normalization and labeling are as in (C). The percentage of the neddylated CUL-4∷FLAG isoform is significantly different between wild type and cand-1 mutants (p < 0.0005, n=7). (F) Whole-worm lysates from wild type and cand-1 mutants with (+) or without (-) cand-1 RNAi, probed with anti-CUL-2 and anti-tubulin antibodies. (G) Whole worm lysates from wild type and cand-1 mutants expressing CUL-4∷FLAG with or without cand-1 RNAi, probed with anti-FLAG and anti-tubulin antibodies.
To determine the effect of loss of CAND-1 on the levels of neddylated CUL-2 and CUL-4 isoforms, we analyzed CUL-2 and CUL-4∷FLAG proteins in cand-1 mutants. The proportion of the neddylated CUL-2 isoform in cand-1 mutants (with or without cand-1 RNAi) was significantly increased relative to that observed in wild-type animals (Fig. 3B,C,F). In wild type, the neddylated CUL-2 band was on average 29% of the total CUL-2, while in cand-1 mutants without cand-1 RNAi, the neddylated band was 50% of the total, an increase of 70% (Fig. 3C). This difference was due to both a 53% increase in the level of neddylated CUL-2 in cand-1 mutants relative to wild type (45.1 ± 6.0 vs. 29.4 ± 3.0 arbitrary units; n=9; p < 0.05) and a 36% decrease in the level of unneddylated CUL-2 in cand-1 mutants (45.0 ± 4.3 vs. 70.6 ± 3.0 a.u.; p < 0.0005).
CUL-4∷FLAG also exhibited a higher proportion of the neddylated isoform in cand-1 mutants with or without cand-1 RNAi (Fig. 3D,E,G). In wild type, the neddylated CUL-4∷FLAG band was on average 37% of the total, while in cand-1 mutants without cand-1 RNAi, the neddylated band was 64% of the total, an increase of 70% (Fig. 3E). This difference was due to both a 14% increase in the level of neddylated CUL-4∷FLAG in cand-1 mutants relative to wild type (42.8 ± 8.6 vs. 37.4 ± 2.1 a.u.; n=7) and a 61% decrease in the level of the unneddylated CUL-4∷FLAG isoform in cand-1 mutants (24.4 ± 8.2 vs. 62.6 ± 2.1 a.u.; p < 0.005). These results indicate that C. elegans CAND-1 negatively regulates the formation of the active, neddylated isoform of CUL-2 and CUL-4∷FLAG.
While there was significant variability, the total levels of CUL-2 and CUL-4∷FLAG (both neddylated and unneddylated isoforms) in cand-1 mutants were on average lower than in wild type due to disproportionate reductions in the level of the unneddylated isoforms (Fig. 3B-E). This was more apparent when comparing the wild-type cullin pattern to that of cand-1 mutants with cand-1 RNAi, which had greater reductions in total cullin levels than the cand-1 mutant alone (Fig. 3F,G). These results suggest that CAND-1 promotes cullin stability in vivo.
cand-1 mutant phenotype
The cand-1(tm1683) deletion allele was outcrossed six times prior to phenotypic analysis. The cand-1(tm1683) mutant in the absence of cand-1 RNAi exhibits a number of phenotypes that reduce the viability of individual animals, although a homozygous strain can be maintained. Approximately 8% of the progeny of cand-1(tm1683) homozygous mutants arrest as embryos with hundreds of cells that generally lack overt morphogenesis; and 10% of cand-1(tm1683) progeny arrest as L2-stage larvae (Table 1). cand-1 mutant larvae have slower developmental timing to the adult stage than wild type (data not shown). Adult cand-1 mutant hermaphrodites produce lower numbers of eggs than wild type (Table 1). cand-1 mutants show impenetrant phenotypes of defective tail morphology, including tail bobs (10% of larvae), and protruding vulva (40% of adults) (Fig. 4C,D,G,H; Table 1). Defects in vulval morphology are observed at low penetrance in L4-stage cand-1 mutants (23%, n=22), but vulva cell numbers are normal (22 ± 2 cells in cand-1 vs. 22 ± 0 in wild type; n=40) (Fig. 4E,F). In an effort to ensure that unrelated mutations in the 6×-outcrossed cand-1 mutant strain were not generating these phenotypes, the cand-1(tm1683) allele was outcrossed an additional eight times into the CB4856 Hawaiian genetic background with crossovers that closely-flanked the cand-1 loci (see Methods). Similar mutant phenotypes were observed in this outcrossed Hawaiian-background strain, suggesting that the phenotypes arise from the loss of CAND-1 (Table 1).
Table 1. cand-1 mutant phenotype with or without cand-1 RNAi.
| Genotype | WT | WT | cand-1 | cand-1 | Hw-cand-1 | Hw-cand-1 | CYB-1∷YFP | CYB-1∷YFP |
|---|---|---|---|---|---|---|---|---|
| cand-1 RNAi | - | + | - | + | - | + | - | + |
| Egg productiona | 357 ± 5 | 347 ± 5 | 165 ± 7 | 167 ± 4 | 258 ± 5 | 248 ± 9 | 257 ± 12 | 225 ± 8 |
| Arrested embryosb | 0% | 5% | 8% | 15% | 10% | 11% | 0.3% | 0% |
| Protruding vulvac | 0% | 2% | 40% | 56% | 42% | 52% | 2% | 9% |
| Bob-tail defectc | 0% | 0% | 9.5% | 21% | 12% | 21% | 0% | 19% |
| Sterilityc | 0% | 0% | 0% | 2.5% | 5% | 7.5% | 0% | 0% |
| L2 arrestc | 0% | 0% | 10% | 15% | 5% | 8% | 0.7% | 0% |
The percentages of protruding vulvae and sterility were among adults; and the percentages of tail defect and L2 arrest were among larvae. ‘cand-1’ denotes cand-1(tm1683) homozygotes. ‘Hw’ denotes Hawaiian genetic background. ‘CYB-1∷YFP’ denotes a strain with wild-type genetic background that expresses a Pcyb-1∷CYB-1∷unc-54 3′UTR transgene.
n=10 hermaphrodites;
n=250-300 embryos;
n=200-271 larvae or adults
Figure 4. cand-1 mutant phenotypes.
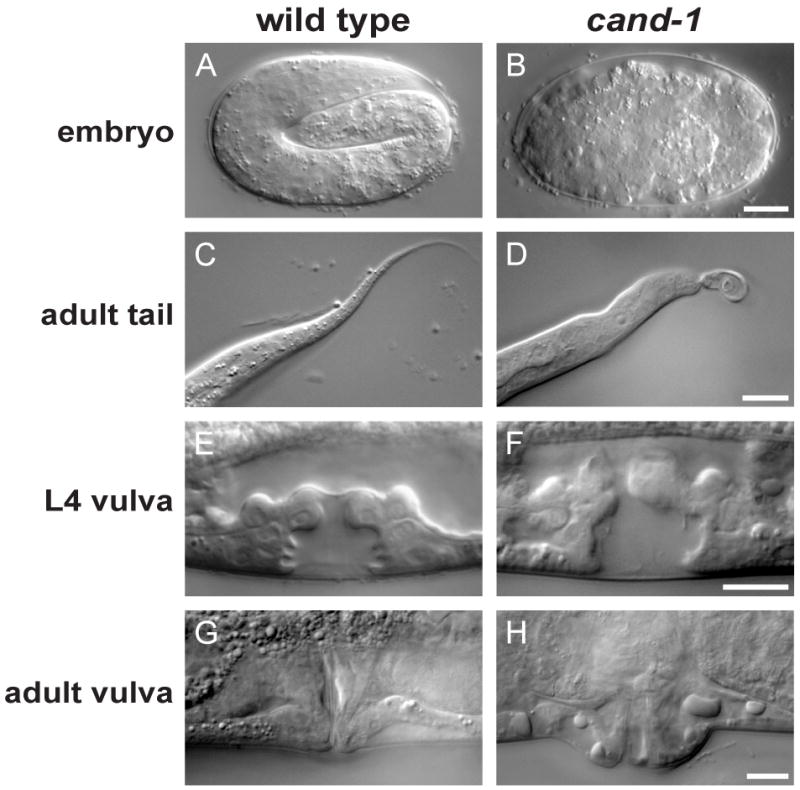
DIC images of a pretzel-stage wild-type embryo (A) and an arrested cand-1 mutant embryo (B). DIC images for wild type and cand-1 mutants of the adult tail (C, D), L4-stage vulva (E, F), and adult vulva (G, H). Scale bars, 10 μm.
The tm1683 allele appears to be a hypomorph based on the observation that the cand-1 mutant phenotypes become worse after cand-1 RNAi (Table 1). The finding that cand-1 RNAi reduces the level of CAND-1 protein in cand-1(tm1683) mutants to almost undetectable levels (Fig. 1E) suggests that combining the mutant with RNAi produces a virtually complete loss of function. In this regard, it should be noted that cand-1(tm1683); cand-1(RNAi) animals could still be maintained as a viable population, albeit with severely reduced fitness (Table 1). cand-1 RNAi in a wild-type background does not produce cand-1 mutant phenotypes except for a minor increase in protruding vulvae and embryonic arrest (Table 1). This suggests that CAND-1 protein can function at greatly reduced levels. Given this observation, we cannot rule out the possibility that a null allele of cand-1 may have more severe phenotypes than the combination of the cand-1(tm1683) allele and cand-1 RNAi.
cand-1 mutants do not exhibit the majority of cullin mutant phenotypes
Given the published observations that loss of CAND1 in Arabidopsis and mammalian cells leads to a loss of cullin function, we were interested in whether CAND-1 was important for CRL activity in C. elegans. The inactivation of each of four cullins (CUL-1, CUL-2, CUL-3, or CUL-4) causes lethality with severe cellular defects. We examined cand-1 mutants treated with cand-1 RNAi for major cullin mutant phenotypes to determine if CAND-1 is required for these cullin functions. cul-1 mutants exhibit extensive hyperplasia of multiple tissues resulting from a failure of cells to exit the cell cycle after proliferation (Kipreos et al., 1996). cand-1 mutants with cand-1 RNAi do not exhibit hyperplasia in postembryonic tissues, with the exception of modest hyperplasia in seam cells (described below). cul-2 mutants exhibit a number of distinct phenotypes including: a defect in progression through meiosis II; a cell cycle arrest of germ cells (with 2n DNA content); a failure of chromosome condensation; defective anterior-posterior polarity; and a delay in mitotic prometaphase in the early embryo (Feng et al., 1999; Liu et al., 2004; Sonneville and Gonczy, 2004). These phenotypes, which are each potentially lethal, are not observed in cand-1 mutants plus cand-1 RNAi (data not shown). Similarly, cul-3 mutant phenotypes affecting meiosis and mitoses of the early embryo (Pintard et al., 2003) are not observed in cand-1 mutants with cand-1 RNAi (data not shown). cul-4 mutants exhibit a fully-penetrant L2-stage arrest and DNA re-replication in larval blast cells (Zhong et al., 2003). cand-1 mutants with cand-1 RNAi exhibit an impenetrant L2-stage arrest, but do not exhibit DNA re-replication (Table 1 and data not shown). Inactivation of cul-4 in later larval stages produces a characteristic germ cell defect (Kim and Kipreos, 2007), but this phenotype is not observed in cand-1 mutants treated with cand-1 RNAi. Therefore, the majority of severe cullin phenotypes are not observed upon inactivating cand-1, implying that many C. elegans CRLs can perform their essential cellular functions in the absence of CAND-1.
cand-1 and cullin genetic analysis
To probe the interaction of CAND-1 and cullins further, we addressed whether the loss of one copy of the cul-2 or cul-4 genes would affect the cand-1 mutant phenotype. Both cul-2 and cul-4 null mutants are recessive and cul-2/+ or cul-4/+ heterozygotes do not exhibit obvious mutant phenotypes (Feng et al., 1999; Zhong et al., 2003). cand-1(tm1683), cul-4(gk434)/+ double mutants have increased levels of L2-stage arrest relative to cand-1 mutants alone (Table 2). cand-1(tm1683); cul-4(gk434)/+ double mutants also have increased levels of protruding vulvae relative to cand-1 mutants alone (Table 2). The L2-stage arrest is a cul-4 mutant phenotype, and protruding vulvae are associated with inactivation of either cul-4 or cul-1, although in these mutants, the vulvae have altered cell numbers (Kim and Kipreos, 2007; Kipreos et al., 1996). In a cand-1(tm1683); cul-2(ek1)/+ strain, each gonad arm has a small number of germ cells that undergo a cell cycle arrest with increased cell size and 2n DNA content (4.2 ± 0.8 cells per gonad arm) (Table 2). This phenotype is similar to, but much less penetrant than the 100% germ cell arrest observed in cul-2 homozygous mutants (Feng et al., 1999). Neither cand-1(tm1683) homozygotes nor cul-2(ek1)/+ heterozygotes by themselves exhibit arrested germ cells (Table 2).
Table 2. Phenotypes observed with heterozygous cul-2 or cul-4 and homozygous cand-1 double mutants.
| Strain | L2 arresta | Protruding vulvab | arrested germ cells (per gonad arm)c |
|---|---|---|---|
| cand-1(tm1683) | 12% | 40% | 0 ± 0 |
| cul-4(gk434)/mIn1; cand-1(tm1683) | 25% | 60% | ND |
| cul-4(gk434)/mIn1 | 0% | 0% | ND |
| mIn1; cand-1(tm1683) | 13% | 42.5% | ND |
| mIn1 | 0% | 0% | ND |
| cul-2(ek4) unc-64(e246)/bli-5(e518); cand-1(tm1683) | ND | ND | 4.2 ± 0.8 |
| cul-2(ek4) unc-64(e246)/bli-5(e518) | ND | ND | 0 ± 0 |
n=40-46 larvae;
n=40 adults;
n=10 gonad arms. ND, not done.
cand-1 mutants are hypersensitive to cul-1, cul-2, cul-3, and cul-4 RNAi treatments (Table 3). Eighty percent of cand-1 mutant L1-stage larvae on cul-1 RNAi underwent larval arrest with the remainder becoming sterile adults; in contrast, wild-type L1 larvae on cul-1 RNAi did not undergo larval arrest and became adults that were capable of producing eggs (which arrested). Similarly, on cul-3 RNAi, cand-1 L1 larvae developed to become sterile adults, while wild-type L1 larvae became adults that could produce eggs. On cul-4 RNAi, L4-stage cand-1 mutant larvae developed into adults for which 68% of their progeny arrested as embryos, while L4-stage wild-type larvae produced only 5% arrested embryos. In general, the enhancement of cullin phenotypes suggests that loss of CAND-1 reduces cullin effectiveness. We analyzed two proteins that are regulated by cullins: cyclin B1, whose protein level is negatively regulated by CUL-2 (Liu et al., 2004); and CDC-6, whose nuclear export requires CUL-4 activity (Kim et al., 2007). In cand-1 mutants with cand-1 RNAi, these proteins behaved similar to wild type: cyclin B1 levels did not perdure in zygotes, and CDC-6 remained cytoplasmic in seam cells after entry into S phase (180-210 minutes post hatch) (data not shown). We conclude that CAND-1 is required for optimal cullin function, but that many CRL complexes can maintain their essential functions in the absence of CAND-1.
Table 3. The effect of the cand-1 mutation on cullin RNAi phenotypes.
| Strain | no RNAi | cul-1 RNAi | cul-2 RNAi | cul-3 RNAi | cul-4 RNAi |
|---|---|---|---|---|---|
| wild type (RNAi on L4) |
0% Emb, 0% Lva (n=100) | 84% Emb (n=278) | 88% Emb (n=298) | 74% Emb (n=258) | 5% Emb, 95% Lva (n=262) |
|
cand-1(tm1683) (RNAi on L4) |
22% Emb 12% Lva (n=180) | 100% Emb (n=160) | 96% Emb (n=156) | 97% Emb (n=105) | 68% Emb, 32% Lva (n=115) |
| wild type (RNAi on L1) (n=20) |
100% became adults with 0% Emb, Lva progeny | 100% became adults with 100% Emb progeny | 100% became adults with 100% Emb progeny | 100% became adults with 100% Emb progeny | 100% became adults with 100% Emb progeny |
|
cand-1(tm1683) (RNAi on L1) (n=20) |
100% became adults with 22% Emb 12% Lva (n=180) | 80% larval arrest, 20% sterile adults | 100% became adults with 100% Emb progeny | 100% became sterile adults | 100% became adults with 100% Emb progeny |
L4- or L1-stage larvae were placed on cullin RNAi plates, n=20 each. L4 larvae placed on RNAi all developed to the adult stage, and the F1 progeny were scored for phenotype. L1 larvae placed on RNAi were scored for their own phenotypes. Phenotypes: Lva, larval arrest; and Emb, embryonic lethal. Note that a sterile adult is a more severe defect than an adult that is not sterile but produces Emb progeny.
cand-1 mutants have lin-23 mutant phenotypes and accumulate LIN-23 substrates
We observed that adult cand-1 mutants have a modest but significant increase in seam cell numbers (18.7 ± 0.7 seam cells per lateral side for cand-1 mutant; 18.7 ± 0.6 for cand-1 mutant + cand-1 RNAi vs. 16.0 ± 0 for wild type; n=20 for each, p < 1×10-4 for comparisons to wild type) (Fig. 5A). Inactivation of cul-1 or lin-23, which encode components of the SCFLIN-23 complex, results in hyperplasia of multiple tissues, including seam cells (Kipreos et al., 2000; Kipreos et al., 1996). In adults, differentiated seam cells produce alae, which are lateral cuticular ridges that run the length of the nematode (Sulston and Horvitz, 1977). We observed that ∼65% of cand-1 mutants show gaps in alae, as well as irregular alae patterns (n=20) (Fig. 5B). These phenotypes are similar to the disrupted alae observed in lin-23 mutants that are associated with extra seam cells (Fig. 5B, data not shown). CAND-1 therefore appears to negatively regulate seam cell divisions with secondary effects on alae formation.
Figure 5. cand-1 mutant has more seam cells and defective alae.
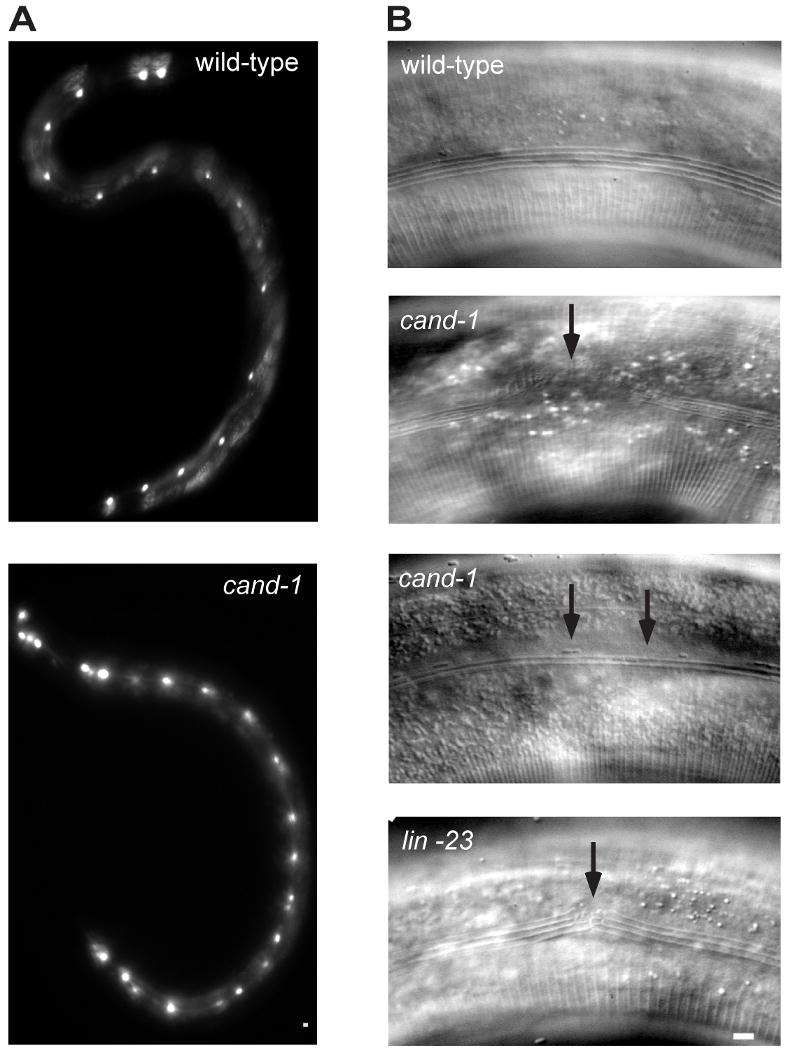
(A) Epifluorescence images of seam cell marker scm∷GFP signal in seam cells from wild type (top) and cand-1 mutant (bottom). In these images, the wild-type adult has 16 seam cells on its lateral side, and the cand-1 adult has 22 seam cells. (B) DIC images of adult alae in wild-type (top), cand-1 mutant (two middle images), and lin-23 mutant (bottom). Note that wild-type alae have four ridges. In cand-1 mutants, alae are often missing from sections (second panel) or have defective morphology (third panel). The lin-23 mutant, which also exhibits excessive seam cell numbers, has similar defects in alae formation (bottom panel and data not shown). Scale bars, 10 μm.
The observation that cand-1 shows excessive seam cell numbers similar to cul-1 and lin-23 mutants made us suspect that the cand-1 seam cell phenotype resulted from a decrease in SCFLIN-23 activity. We observed that cand-1 RNAi, while ineffective in altering seam cell numbers in a wild-type background (16.1 ± 0.2 seam cells, n=20), increased the number of seam cells in cul-1 heterozygotes (17.8 ± 0.4 for cul-1/+ with cand-1 RNAi vs. 16.1 ± 0.2 for cul-1/+ alone; n=20 for each; p < 5×10-4), suggesting genetic interaction. If CAND-1 is required for full SCFLIN-23 activity, we would expect that inactivation of CAND-1 would negatively affect the ability SCFLIN-23 to degrade its substrates. The SCFLIN-23 substrate(s) that are targeted to restrain seam cell divisions are not known, however SCFLIN-23 has been shown to target the degradation of the glutamate receptor GLR-1 in the ventral nerve cord (Dreier et al., 2005). Significantly, we observed that cand-1 mutants accumulate GLR-1∷GFP in the ventral nerve cord to an extent similar to that of lin-23 mutants, consistent with CAND-1 being required for full SCFLIN-23 activity (Fig. 6A,B). Notably, the levels of LIN-23∷GFP in cand-1 mutants were unchanged relative to wild type, indicating that CAND-1 is not required to maintain LIN-23 protein levels, and that the GLR-1 accumulation did not result from a reduction in the level of the SRS for the SCFLIN-23 complex (Fig. 6C).
Figure 6. The glutamate receptor GLR-1 accumulates in cand-1 mutants.
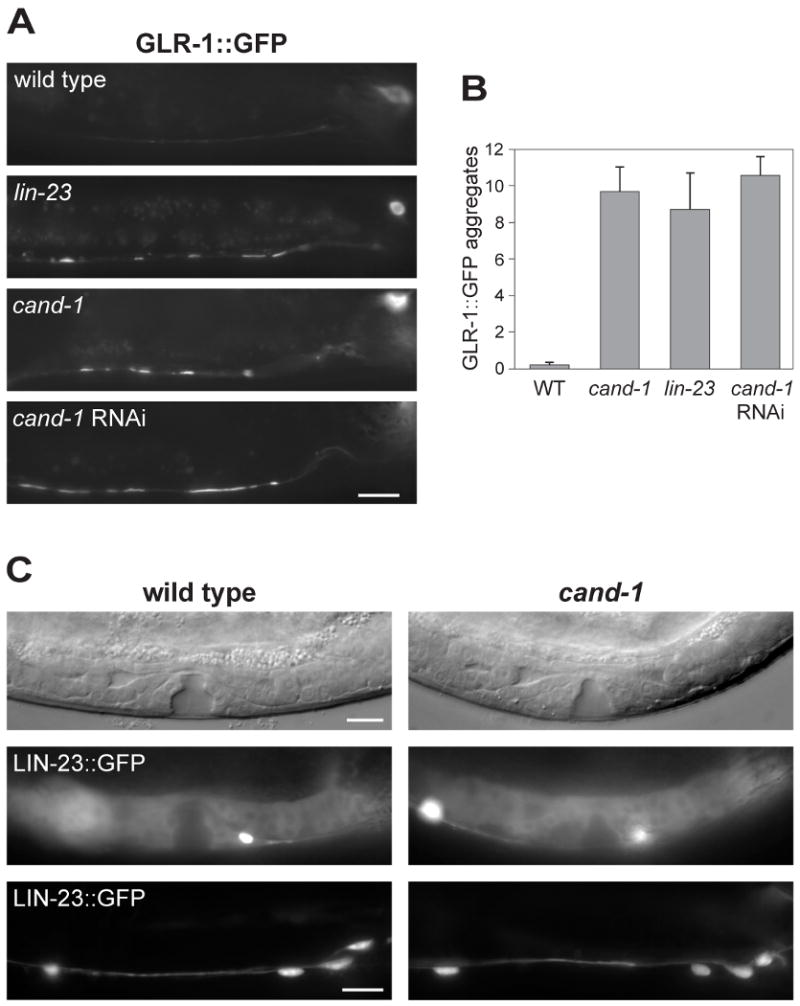
(A) Epifluorescence images of GLR-1∷GFP signal in the posterior ventral nerve cord of wild-type, lin-23(e1883) mutant, or cand-1 (tm1683) mutant young adults. Note that the signal from nerve cell bodies (upper right) is out of focus in several of the images. (B) A graph of the number of GLR-1∷GFP aggregates per ventral nerve cord that are ≥ 2 μm in length, n=10 animals. (C) Analysis of LIN-23∷GFP levels in wild type and cand-1(tm1683) mutants. The vulva and gonadal regions of early L4-stage stage animals are shown as DIC images (top) or LIN-23∷GFP epifluorescence images (middle). LIN-23∷GFP epifluorescence in the posterior ventral nerve cord of young adults are shown in the bottom panels. Posterior is to the right in all images. Scale bars, 10 μm.
While our data suggests that CAND-1 promotes SCFLIN-23 activity, CAND-1 does not appear to significantly contribute to CRL2ZYG-11-mediated degradation of cyclin B1. CRL2ZYG-11 is required for the degradation of CYB-1∷GFP during the meiotic divisions of the newly-fertilized zygote (Liu et al., 2004; Vasudevan et al., 2007). In a homozygous zyg-11(ax491) temperature-sensitive mutant, we found that CYB-1∷GFP was not degraded in the meiotic divisions at 25°C, while at 24°C, CYB-1∷GFP was partially degraded, and at 20°C, the CYB-1 degradation rate was similar to that in wild type. Significantly, at all three temperatures, the level of CYB-1∷GFP in zyg-11(ax491) mutants did not change in response to cand-1 RNAi (data not shown). While cand-1 RNAi does not produce many cand-1 mutant phenotypes in a wild-type background, in the CYB-1∷GFP strain, cand-1 RNAi produced a 21% bob-tail defect (which is associated with the cand-1 mutant). To determine if the overexpression of CYB-1 causes an enhancement of cand-1 RNAi, we tested a different strain overexpressing a CYB-1∷YFP transgenic protein from its own promoter. The Pcyb-1∷CYB-1∷YFP-expressing strain exhibited protruding vulva (9%) and bob-tail morphology (19%) defects upon cand-1 RNAi, suggesting that the overexpression of cyclin B1 produces a sensitized background for cand-1 RNAi (Table 1). It is currently unknown how increased cyclin B1 levels enhance cand-1 RNAi.
Taken as a whole, CAND-1 appears to be required for SCFLIN-23-dependent functions in both regulating seam cell divisions and the degradation of the glutamate receptor GLR-1, yet loss of CAND-1 does not markedly affect a number of other CRL-dependent functions, including the ability of CRL2ZYG-11 to regulate CYB-1, even in a sensitized background with reduced ZYG-11 activity. These results imply that CAND-1 is important for the activity of only a subset of CRL complexes.
CAND-1 is required beyond the regulation of the levels of cullin neddylated isoforms
UBC-12 (the Nedd8 E2) is required for C. elegans cullin neddylation and CSN-5 (the COP9 Signalosome deneddylase) is required for the deneddylation of cullins (Jones and Candido, 2000; Kurz et al., 2002; Pintard et al., 2003). We determined the effect of inactivating either ubc-12 or csn-5 on the levels of the neddylated isoforms in wild type or cand-1 mutants. RNAi depletion of ubc-12 reduces CUL-2 neddylated isoforms, while csn-5 RNAi increases the level of neddylated isoforms (Fig. 7A,B). Notably, ubc-12 RNAi reduced the proportion of the neddylated CUL-2 isoform in cand-1 mutants to a level that was similar to that of wild type. While csn-5 RNAi increased the level of the neddylated isoform in wild type, it had only a modest effect on the level of CUL-2 neddylation in cand-1 mutants, less than would be expected for additive effects (Fig. 7A,B). This suggests either that CSN-5 and CAND-1 function in the same pathway or that only a limited percentage of CUL-2 is available to undergo neddylation in vivo (and therefore there is a cap on the extent to which neddylation can be altered by loss of CSN-5 or CAND-1). In regard to the hypothesis that CSN-5 and CAND-1 may function in the same pathway, it has been observed that the presence of mammalian CAND1 increases the rate of CSN-mediated deneddylation of CUL1 in vitro, although the mechanism for this effect is not understood (Min et al., 2005).
Figure 7. Co-inactivation of CAND-1 and neddylation pathway components.
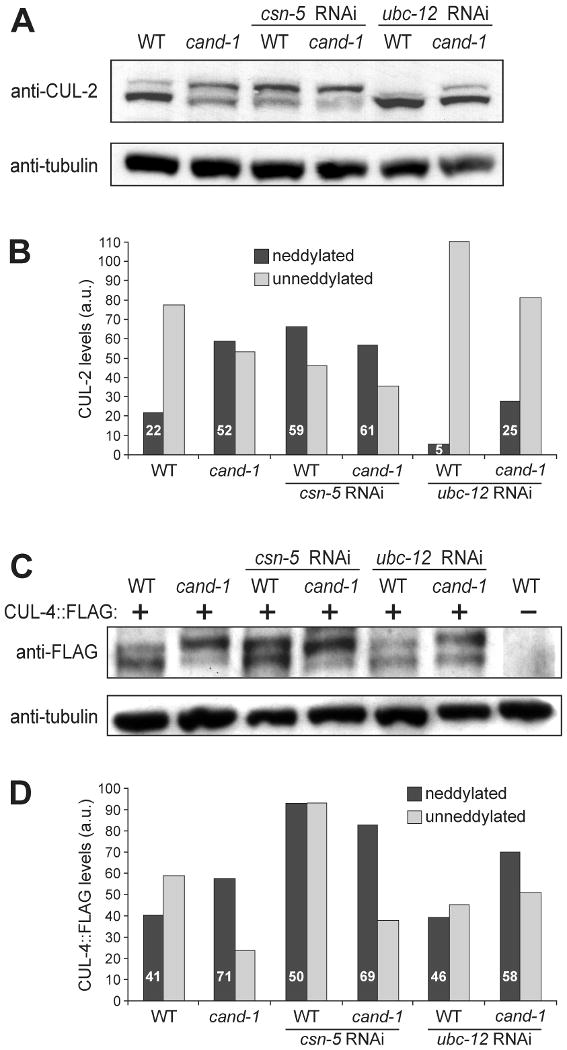
(A) Whole-worm lysates from wild type and cand-1 mutants with or without RNAi depletion of csn-5 (COP9 Signalosome deneddylase) or ubc-12 (Nedd8 E2), and probed with anti-CUL-2 and anti-tubulin antibodies. (B) Quantitation of the neddylated and unneddylated CUL-2 isoform levels in (A) relative to tubulin levels. The levels of CUL-2 isoforms in cand-1 mutants are normalized to the total CUL-2 signal of wild-type animals (set to 100 a.u.); and the percentage of neddylated isoform within a genotype is provided in white lettering. (C) Total-worm lysates prepared from wild type and cand-1 mutants expressing CUL-4∷FLAG with or without csn-5 or ubc-12 RNAi, and probed with anti-FLAG or anti-tubulin antibodies. (D) Quantitation of slower-migrating covalently-modified and unmodified CUL-4∷FLAG isoforms in (C). Normalization and labeling are as in (B).
The level of the slower-migrating isoform of CUL-4∷FLAG was not significantly affected by ubc-12 RNAi in a wild-type background, while the proportion, but not absolute level, was modestly reduced in a cand-1 mutant background relative to the cand-1 mutant without RNAi (Fig. 7C,D). RNAi depletion of csn-5 modestly increased the levels of the slower-migrating isoform in wild type and cand-1 mutants. The observation that ubc-12 and csn-5 RNAi had more limited effects on the level of the slower migrating CUL-4∷FLAG band (relative to their effect on CUL-2) may reflect the presence of a non-Nedd8 modification of CUL-4∷FLAG. In this regard, we have observed that the anti-Nedd8 signal of the slower-migrating band of immunoprecipitated CUL-4∷FLAG is over ten times lower than that of the slower-migrating CUL-2∷FLAG band, suggesting that a significant proportion of the CUL-4∷FLAG slower-migrating band is modified by a mechanism other than neddylation, such as conjugation to ubiquitin or ubiquitin-like proteins as has been proposed for other animal and yeast cullins (Duda et al., 2008; Laplaza et al., 2004) (data not shown).
Interestingly, despite ubc-12 RNAi reducing the proportion of the neddylated cullin isoforms in cand-1 mutants so that they were more similar to that of wild type, ubc-12 RNAi did not rescue the cand-1 mutant or ubc-12 RNAi phenotypes. In fact, the co-inactivation of ubc-12 and cand-1 produced an enhanced mutant phenotype when compared to either single inactivation: the double inactivation produced 100% sterile F1 adults (n=100) versus ubc-12(RNAi) F1 non-sterile adults that laid eggs of which 65% arrested (n=50), and cand-1 mutant adults that laid 22% arrested eggs (n=180). An analogous result was observed when feeding wild-type hermaphrodites 1:1 mixes of feeding RNAi bacteria for ubc-12 + cand-1 (36% arrested embryos) vs. ubc-12 + empty vector (19% arrested), or cand-1 + empty vector (4% arrested) (n=100 for each; chi-squares p < 5×10-4 for all comparisons). Therefore, simultaneously inactivating ubc-12 and cand-1 produced a more severe defect despite levels of cullin neddylated isoforms that were more similar to wild type.
Discussion
In this study, we report the expression pattern and mutant phenotype of C. elegans CAND-1, and describe its role in regulating the levels of neddylated isoforms of C. elegans cullins. CAND-1 is expressed in germ cells and mitotically-dividing postembryonic blast cells, including the intestine, seam cells, P lineage, and somatic gonad. CAND-1 protein is provided as maternal product to early embryos, and CAND-1 expression is higher in early embryonic stages, which have the highest rates of proliferation (Sulston, 1983). CAND-1 is also expressed in a subset of non-proliferating tissues, including the rectal epithelia and a few neuronal cells in the head and tail regions, implying non-cell cycle-related functions for CAND-1. CAND-1 shares with core CUL-2 and CUL-4 complex components a similar expression pattern in proliferating larval tissues and a lack of expression in adult somatic tissues, which matches the expectation that CAND-1 functions in cells that express CRL complexes (Feng et al., 1999; Kim and Kipreos, 2007; Zhong et al., 2003).
CAND-1 is required to maintain the proper levels of neddylated cullin isoforms
The covalent modification of cullins by Nedd8 is associated with the activation of the E3 complexes. Neddylation and deneddylation activities are crucial for proper CRL activities in vivo (Jones and Candido, 2000; Ohh et al., 2002; Osaka et al., 2000; Ou et al., 2002; Pintard et al., 2003; Read et al., 2000; Wu et al., 2005). In mammals, Arabidopsis, and fission yeast, loss of CAND1 does not alter the level of neddylated or unneddylated CUL1 isoforms (Chew and Hagen, 2007; Chuang et al., 2004; Feng et al., 2004; Schmidt et al., 2009; Zheng et al., 2002). Mammals have a second CAND1 paralog, CAND2 (Shiraishi et al., 2007), and it is possible that redundancy between the two paralogs obscures effects on cullin neddylation upon inactivation of CAND1. In fission yeast, the CAND1 ortholog Knd1p interacts stably with only a small fraction of Cul1p and does not appear to interact with Cul3p (Schmidt et al., 2009), which is consistent with its having negligible effects on neddylation levels. We provide the first in vivo example of a CAND1 ortholog that regulates the level of neddylation (or alternate covalent modification) of cullins. In C. elegans, the loss of cand-1 activity significantly increases the ratio of neddylated to unneddylated isoforms of CUL-2 and CUL-4∷FLAG. CAND-1 may regulate neddylation levels either by sequestering cullins to prevent their interaction with the neddylation machinery or by increasing the rate of deneddylation. The latter hypothesis is supported by the observation that mammalian CAND1 enhances the rate of CUL1 deneddylation in vitro (Min et al., 2005).
In Drosophila and Neurospora, increased neddylation of CUL1 and CUL3 induces their degradation (He et al., 2005; Wu et al., 2005). In contrast, mammalian CUL1, CUL3, and CUL4 are not destabilized by increased neddylation, and CUL2 exhibits only a minor decrease in overall level upon increased neddylation (Cope and Deshaies, 2006). The total levels of endogenous CUL-2 and CUL-4∷FLAG levels were moderately lower in cand-1 mutants with cand-1 RNAi relative to wild type, suggesting that CAND-1 promotes cullin stability in C. elegans.
Cullin-dependent cellular functions are largely unaffected in cand-1 mutants
cand-1(tm1683) mutant homozygotes are viable but exhibit a range of impenetrant phenotypes, including: arrested late-stage embryos and L2-stage larvae, protruding vulva, slower postembryonic development, reduced numbers of progeny, and defective tail and vulva morphogenesis. The CAND-1(tm1683) mutant protein has an in-frame deletion in the N-terminal region that reduces interactions with cullins in the two-hybrid system to almost background levels. Additionally, the level of CAND-1 protein in cand-1(tm1683) mutants is greatly reduced. Treating cand-1(tm1683) mutants with cand-1 RNAi reduces CAND1 protein levels to practically undetectable levels. These animals, which lack detectable CAND-1 can still be maintained as a viable population, although the penetrance of mutant phenotypes increases along with the inviability of individual animals in the population. As there are no other CAND1 homologs in C. elegans, our results suggest that the CAND1 regulatory system is not essential for viability in the nematode, although it markedly increases fitness.
cand-1 mutants in combination with cand-1 RNAi exhibit only limited phenotypic overlap with cullin mutants. cand-1 mutants share an L2-stage arrest with cul-4 mutants, although the cand-1 mutant arrest occurs at lower penetrance, and it is not known if the arrests arise from the same underlying defect. cand-1 mutants show limited hyperplasia of seam cells, which is similar to cul-1 and lin-23 mutants; although the cand-1 mutant phenotype is impenetrant and is restricted to seam cells, while cul-1 or lin-23 mutants have robust hyperplasia in all somatic blast cell lineages (Kipreos et al., 1996). cand-1 mutants do not exhibit a number of distinct embryonic and germ cell phenotypes that are associated with cul-2 and cul-3 mutants (Feng et al., 1999; Liu et al., 2004; Pintard et al., 2003; Sonneville and Gonczy, 2004) or the DNA replication and germ cell defects associated with cul-4 mutants (Kim and Kipreos, 2007; Zhong et al., 2003). As loss of CAND-1 does not show many of the severe cellular defects that are associated with loss of CUL-1, CUL-2, CUL-3, or CUL-4, we conclude that C. elegans CAND-1 is not essential for many CRL functions.
CAND-1 is a positive regulator of CRL activity in C. elegans
The biochemical function of CAND-1 is to inhibit CRL complex formation, yet in humans and plants, loss of CAND1 leads to a loss of CRL activities (Cheng et al., 2004; Chew and Hagen, 2007; Chuang et al., 2004; Feng et al., 2004; Lo and Hannink, 2006; Zheng et al., 2002). Loss of the CAND1 ortholog in fission yeast does not produce an overall loss of CRL activities but shifts the proportion of different SRSs that are bound to SCF complexes (Schmidt et al., 2009). To address whether C. elegans CAND-1 negatively or positively regulates CRL functions in vivo, we asked what effect loss of CAND-1 had on partial cullin inactivations. If CAND-1 negatively regulates CRLs in vivo, then the loss of CAND-1 should suppress hypomorphic cullin mutant phenotypes. Conversely, if CAND-1 positively regulates CRLs in vivo, then the loss of CAND-1 should enhance hypomorphic cullin phenotypes. We observed that double mutants of homozygous cand-1 and heterozygous cul-2 or cul-4 alleles exhibited impenetrant phenotypes associated with cul-2 or cul-4 homozygous mutants. Additionally, cand-1 mutants were hypersensitive to cul-1, cul-2, and cul-4 RNAi relative to wild-type animals. These results suggest that CAND-1 is required for optimal CRL functions in C. elegans.
Our data suggests that the SCFLIN-23 complex is particularly reliant on the presence of CAND-1 for full activity relative to other CRL complexes. The seam cell hyperplasia observed in cand-1 mutants is similar to the hyperplasia observed upon loss of the SCFLIN-23 complex. Further evidence that the hyperplasia in cand-1 mutants arises from a loss of SCFLIN-23 activity is that cand-1 RNAi (which does not produce hyperplasia in a wild-type background) induces seam cell hyperplasia in cul-1/+ heterozygotes (which also normally do not exhibit hyperplasia). The SCFLIN-23 substrate GLR-1, a glutamate receptor, accumulates in both cand-1 mutants and cand-1(RNAi) animals, providing further evidence that SCFLIN-23 activity is compromised if CAND-1 is inactivated. In contrast, cand-1 RNAi had no effect on the level of maternal cyclin B1, a downstream target of CRL4ZYG-11, even in a highly sensitized background with partially-inactive ZYG-11. This suggests that CAND-1 is required for the full activity of SCFLIN-23 but has no appreciable effect on the ability of CRL4ZYG-11 to promote the degradation of cyclin B1.
It has been proposed that the cycling of CRLs between active and inactive states via neddylation is required for full ubiquitin ligase activity (Cope and Deshaies, 2003; Pintard et al., 2003; Wolf et al., 2003). In particular, Pintard et al. proposed that a cycle of neddylation and deneddylation is required for the addition of each ubiquitin in a polyubiquitin chain. This hypothesis was based on the observation that C. elegans CUL-3 is inactive if either neddylation or deneddylation activity is separately reduced, but co-reducing both processes restores CUL-3 activity, suggesting that CUL-3 requires a balance between neddylation and deneddylation (Pintard et al., 2003). This data is also consistent with the hypothesis that the most important function of neddylation and deneddylation regulatory enzymes is to ensure the proper level of neddylated to unneddylated cullin. Based on our observations, it would be reasonable to consider that the major role of CAND-1 is to maintain the proper level of the active neddylated cullin isoform in vivo. However, inactivating the Nedd8 E2 UBC-12 in cand-1 mutants produced levels of the CUL-2 and CUL-4 neddylated isoforms that were more similar to those in wild type, but did not suppress the cand-1 or ubc-12 mutant phenotypes. On the contrary, depleting ubc-12 in cand-1 mutants produced a more severe phenotype than was observed with inactivation of either cand-1 or ubc-12 alone. This implies that modulating cullin neddylation levels is not the sole critical function of CAND-1.
In humans and Arabidopsis, loss of CAND1 has been shown to decrease the activity of SCF complexes because of reductions in the levels of the SRS. In particular, this has been shown for the SRSs UFO in Arabidopsis, and Skp2 in human cells (Chew et al., 2007; Feng et al., 2004; Zheng et al., 2002). The reduction in SRS levels appears to be due to increased autoubiquitylation (Chew et al., 2007), presumably resulting from the overactivity of E3 complexes in the absence of CAND1. In fission yeast, loss of the CAND1 ortholog does not affect SRS levels, but has been reported to alter the relative levels of specific SRSs interacting with the core CRL complex (Schmidt et al., 2009). However, in Arabidopsis, inactivation of CAND1 leads to a decrease in SCFTIR1 activity despite an increase in the level of assembled SCFTIR1 complexes (Zhang et al., 2008). Similar results were observed with mammalian CRL3Keap1, which had increased levels of Keap1 associated with CUL-3 in CAND1 knockdown cells, yet had lower activity (Lo and Hannink, 2006). These results suggest that CAND1 can promote CRL activity independently of SRS stability. In C. elegans, we found that CAND1 promotes SCFLIN-23 activity, but does not affect LIN-23 levels, suggesting that CAND1 regulates SCFLIN-23 independently of SRS stabilization.
Our results suggest that CAND-1 is a positive regulator of cullins in C. elegans, whose activity is required for optimal viability and proper development of the seam cells, vulva, and tail. CAND-1 has a major role in regulating the levels of cullin neddylated isoforms in vivo, and is required for full activity of at least a subset of CRL complexes in vivo. C. elegans provides a useful genetic system for addressing the many unresolved issues in how CAND-1 regulates CRL functions, particularly as the C. elegans cand-1 mutant is viable. Defects in CRLs are linked to multiple cancers (Pagano and Benmaamar, 2003) and CAND1 levels are downregulated in high-grade neuroendocrine lung tumors (Salon et al., 2007). Therefore, understanding how CAND-1 functions is likely to provide translational implications in addition to illuminating the regulation of numerous CRL-dependent developmental processes.
Supplementary Material
Acknowledgments
We thank the National Bioresource Project for the Experimental Animal Nematode C. elegans for providing the cand-1(tm1683) allele; and Josh Kaplan and the Caenorhabditis Genetics Center for providing strains. This work was supported by grants from the NIH National Institute of General Medical Sciences (R01 GM055297 and R01 GM074212) to ETK, and from the Korean Research Foundation (KRF-2008-357-C00125) to KM.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bosu DR, Kipreos ET. Cullin-RING ubiquitin ligases: global regulation and activation cycles. Cell Div. 2008;3 doi: 10.1186/1747-1028-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y. AtCAND1, a HEAT-repeat protein that participates in auxin signaling in Arabidopsis. Plant Physiol. 2004;135:1020–6. doi: 10.1104/pp.104.044495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew EH, Hagen T. Substrate-mediated regulation of cullin neddylation. J Biol Chem. 2007;282:17032–40. doi: 10.1074/jbc.M701153200. [DOI] [PubMed] [Google Scholar]
- Chew EH, Poobalasingam T, Hawkey CJ, Hagen T. Characterization of cullin-based E3 ubiquitin ligases in intact mammalian cells--evidence for cullin dimerization. Cell Signal. 2007;19:1071–80. doi: 10.1016/j.cellsig.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Chuang HW, Zhang W, Gray WM. Arabidopsis ETA2, an apparent ortholog of the human cullin-interacting protein CAND1, is required for auxin responses mediated by the SCF(TIR1) ubiquitin ligase. Plant Cell. 2004;16:1883–97. doi: 10.1105/tpc.021923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A, Finley D, Varshavsky A. Ubiquitin dependence of selective protein degradation demonstrated in the mammalian cell cycle mutant ts85. Cell. 1984;37:57–66. doi: 10.1016/0092-8674(84)90300-3. [DOI] [PubMed] [Google Scholar]
- Cope GA, Deshaies RJ. COP9 signalosome: a multifunctional regulator of SCF and other cullin-based ubiquitin ligases. Cell. 2003;114:663–71. doi: 10.1016/s0092-8674(03)00722-0. [DOI] [PubMed] [Google Scholar]
- Cope GA, Deshaies RJ. Targeted silencing of Jab1/Csn5 in human cells downregulates SCF activity through reduction of F-box protein levels. BMC Biochem. 2006;7:1. doi: 10.1186/1471-2091-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cope GA, Suh GS, Aravind L, Schwarz SE, Zipursky SL, Koonin EV, Deshaies RJ. Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science. 2002;298:608–11. doi: 10.1126/science.1075901. [DOI] [PubMed] [Google Scholar]
- Doronkin S, Djagaeva I, Beckendorf SK. The COP9 signalosome promotes degradation of Cyclin E during early Drosophila oogenesis. Dev Cell. 2003;4:699–710. doi: 10.1016/s1534-5807(03)00121-7. [DOI] [PubMed] [Google Scholar]
- Dreier L, Burbea M, Kaplan JM. LIN-23-mediated degradation of beta-catenin regulates the abundance of GLR-1 glutamate receptors in the ventral nerve cord of C. elegans. Neuron. 2005;46:51–64. doi: 10.1016/j.neuron.2004.12.058. [DOI] [PubMed] [Google Scholar]
- Duda DM, Borg LA, Scott DC, Hunt HW, Hammel M, Schulman BA. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell. 2008;134:995–1006. doi: 10.1016/j.cell.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H, Zhong W, Punkosdy G, Gu S, Zhou L, Seabolt EK, Kipreos ET. CUL-2 is required for the G1-to-S-phase transition and mitotic chromosome condensation in Caenorhabditis elegans. Nat Cell Biol. 1999;1:486–92. doi: 10.1038/70272. [DOI] [PubMed] [Google Scholar]
- Feng S, Shen Y, Sullivan JA, Rubio V, Xiong Y, Sun TP, Deng XW. Arabidopsis CAND1, an unmodified CUL1-interacting protein, is involved in multiple developmental pathways controlled by ubiquitin/proteasome-mediated protein. Degradation Plant Cell. 2004;16:1870–82. doi: 10.1105/tpc.021949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gengyo-Ando K, Mitani S. Characterization of mutations induced by ethyl methanesulfonate, UV, and trimethylpsoralen in the nematode Caenorhabditis elegans. Biochem Biophys Res Commun. 2000;269:64–9. doi: 10.1006/bbrc.2000.2260. [DOI] [PubMed] [Google Scholar]
- Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- Goldenberg SJ, Cascio TC, Shumway SD, Garbutt KC, Liu J, Xiong Y, Zheng N. Structure of the Cand1-Cul1-Roc1 complex reveals regulatory mechanisms for the assembly of the multisubunit cullin-dependent ubiquitin ligases. Cell. 2004;119:517–28. doi: 10.1016/j.cell.2004.10.019. [DOI] [PubMed] [Google Scholar]
- Gong L, Yeh ET. Identification of the activating and conjugating enzymes of the NEDD8 conjugation pathway. J Biol Chem. 1999;274:12036–42. doi: 10.1074/jbc.274.17.12036. [DOI] [PubMed] [Google Scholar]
- Groisman R, Polanowska J, Kuraoka I, Sawada J, Saijo M, Drapkin R, Kisselev AF, Tanaka K, Nakatani Y. The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell. 2003;113:357–67. doi: 10.1016/s0092-8674(03)00316-7. [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies (A Laboratory Manual) Cold Spring Harbor Laboratories; Cold Spring Harbor, N.Y: 1988. [Google Scholar]
- He Q, Cheng P, He Q, Liu Y. The COP9 signalosome regulates the Neurospora circadian clock by controlling the stability of the SCFFWD-1 complex. Genes Dev. 2005;19:1518–31. doi: 10.1101/gad.1322205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two- hybrid selection in yeast. Genetics. 1996;144:1425–36. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen K, editor. Current Protocols in Molecular Biology. John Wiley & Sons; Boston, MA: 1995. [Google Scholar]
- Jones D, Candido EP. The NED-8 conjugating system in Caenorhabditis elegans is required for embryogenesis and terminal differentiation of the hypodermis. Dev Biol. 2000;226:152–65. doi: 10.1006/dbio.2000.9847. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Ahringer J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 2003;30:313–21. doi: 10.1016/s1046-2023(03)00050-1. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, Welchman DP, Zipperlen P, Ahringer J. Systematic functional analysis of the Caenorhabditis elegans genome using. RNAi Nature. 2003;421:231–7. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Kamura T, Conrad MN, Yan Q, Conaway RC, Conaway JW. The Rbx1 subunit of SCF and VHL E3 ubiquitin ligase activates Rub1 modification of cullins Cdc53 and Cul2. Genes Dev. 1999;13:2928–33. doi: 10.1101/gad.13.22.2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami T, Chiba T, Suzuki T, Iwai K, Yamanaka K, Minato N, Suzuki H, Shimbara N, Hidaka Y, Osaka F, Omata M, Tanaka K. NEDD8 recruits E2-ubiquitin to SCF E3 ligase. EMBO J. 2001;20:4003–12. doi: 10.1093/emboj/20.15.4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Feng H, Kipreos ET. Curr Biol. Vol. 17. 2007. C elegans CUL-4 prevents rereplication by promoting the nuclear export of CDC-6 via a CKI-1-dependent pathway; pp. 966–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Kipreos ET. The Caenorhabditis elegans replication licensing factor CDT-1 is targeted for degradation by the CUL-4/DDB-1 complex. Mol Cell Biol. 2007;27:1394–406. doi: 10.1128/MCB.00736-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipreos ET. Ubiquitin-mediated pathways in C. elegans. WormBook. 2005:1–24. doi: 10.1895/wormbook.1.36.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipreos ET, Gohel SP, Hedgecock EM. The C. elegans F-box/WD-repeat protein LIN-23 functions to limit cell division during development. Development. 2000;127:5071–82. doi: 10.1242/dev.127.23.5071. [DOI] [PubMed] [Google Scholar]
- Kipreos ET, Lander LE, Wing JP, He WW, Hedgecock EM. cul-1 is required for cell cycle exit in C. elegans and identifies a novel gene family. Cell. 1996;85:829–39. doi: 10.1016/s0092-8674(00)81267-2. [DOI] [PubMed] [Google Scholar]
- Koh K, Rothman JH. ELT-5 and ELT-6 are required continuously to regulate epidermal seam cell differentiation and cell fusion in C. elegans. Development. 2001;128:2867–80. doi: 10.1242/dev.128.15.2867. [DOI] [PubMed] [Google Scholar]
- Kurz T, Chou YC, Willems AR, Meyer-Schaller N, Hecht ML, Tyers M, Peter M, Sicheri F. Dcn1 functions as a scaffold-type E3 ligase for cullin neddylation. Mol Cell. 2008;29:23–35. doi: 10.1016/j.molcel.2007.12.012. [DOI] [PubMed] [Google Scholar]
- Kurz T, Pintard L, Willis JH, Hamill DR, Gonczy P, Peter M, Bowerman B. Cytoskeletal regulation by the Nedd8 ubiquitin-like protein modification pathway. Science. 2002;295:1294–8. doi: 10.1126/science.1067765. [DOI] [PubMed] [Google Scholar]
- Lammer D, Mathias N, Laplaza JM, Jiang W, Liu Y, Callis J, Goebl M, Estelle M. Modification of yeast Cdc53p by the ubiquitin-related protein rub1p affects function of the SCFCdc4 complex. Genes Dev. 1998;12:914–26. doi: 10.1101/gad.12.7.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplaza JM, Bostick M, Scholes DT, Curcio MJ, Callis J. Saccharomyces cerevisiae ubiquitin-like protein Rub1 conjugates to cullin proteins Rtt101 and Cul3 in vivo. Biochem J. 2004;377:459–67. doi: 10.1042/BJ20030755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liakopoulos D, Doenges G, Matuschewski K, Jentsch S. A novel protein modification pathway related to the ubiquitin system. Embo J. 1998;17:2208–14. doi: 10.1093/emboj/17.8.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Powell KA, Mundt K, Wu L, Carr AM, Caspari T. Cop9/signalosome subunits and Pcu4 regulate ribonucleotide reductase by both checkpoint-dependent and -independent mechanisms. Genes Dev. 2003;17:1130–40. doi: 10.1101/gad.1090803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Furukawa M, Matsumoto T, Xiong Y. NEDD8 modification of CUL1 dissociates p120(CAND1), an inhibitor of CUL1-SKP1 binding and SCF ligases. Mol Cell. 2002;10:1511–8. doi: 10.1016/s1097-2765(02)00783-9. [DOI] [PubMed] [Google Scholar]
- Liu J, Vasudevan S, Kipreos ET. CUL-2 and ZYG-11 promote meiotic anaphase II and the proper placement of the anterior-posterior axis in C. elegans. Development. 2004;131:3513–25. doi: 10.1242/dev.01245. [DOI] [PubMed] [Google Scholar]
- Lo SC, Hannink M. CAND1-mediated substrate adaptor recycling is required for efficient repression of Nrf2 by Keap1. Mol Cell Biol. 2006;26:1235–44. doi: 10.1128/MCB.26.4.1235-1244.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke-Glaser S, Roy M, Larsen B, Le Bihan T, Metalnikov P, Tyers M, Peter M, Pintard L. CIF-1, a shared subunit of the COP9/signalosome and eukaryotic initiation factor 3 complexes, regulates MEL-26 levels in the Caenorhabditis elegans embryo. Mol Cell Biol. 2007;27:4526–40. doi: 10.1128/MCB.01724-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyapina S, Cope G, Shevchenko A, Serino G, Tsuge T, Zhou C, Wolf DA, Wei N, Shevchenko A, Deshaies RJ. Promotion of NEDD-CUL1 conjugate cleavage by COP9 signalosome. Science. 2001;292:1382–5. doi: 10.1126/science.1059780. [DOI] [PubMed] [Google Scholar]
- Mehta N, Loria PM, Hobert O. A genetic screen for neurite outgrowth mutants in Caenorhabditis elegans reveals a new function for the F-box ubiquitin ligase component LIN-23. Genetics. 2004;166:1253–67. doi: 10.1534/genetics.166.3.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon S, Chi H, Zhang H, Deng XW, Flavell RA, Wei N. COP9 signalosome subunit 8 is essential for peripheral T cell homeostasis and antigen receptor-induced entry into the cell cycle from quiescence. Nat Immunol. 2007;8:1236–45. doi: 10.1038/ni1514. [DOI] [PubMed] [Google Scholar]
- Miller DM, Shakes DC. Immunofluorescence microscopy. Methods Cell Biol. 1995;48:365–94. [PubMed] [Google Scholar]
- Min KW, Hwang JW, Lee JS, Park Y, Tamura TA, Yoon JB. TIP120A associates with cullins and modulates ubiquitin ligase activity. J Biol Chem. 2003;278:15905–10. doi: 10.1074/jbc.M213070200. [DOI] [PubMed] [Google Scholar]
- Min KW, Kwon MJ, Park HS, Park Y, Yoon SK, Yoon JB. CAND1 enhances deneddylation of CUL1 by COP9 signalosome. Biochem Biophys Res Commun. 2005;334:867–74. doi: 10.1016/j.bbrc.2005.06.188. [DOI] [PubMed] [Google Scholar]
- Morimoto M, Nishida T, Honda R, Yasuda H. Modification of cullin-1 by ubiquitin-like protein Nedd8 enhances the activity of SCF(skp2) toward p27(kip1) Biochem Biophys Res Commun. 2000;270:1093–6. doi: 10.1006/bbrc.2000.2576. [DOI] [PubMed] [Google Scholar]
- Ohh M, Kim WY, Moslehi JJ, Chen Y, Chau V, Read MA, Kaelin WG., Jr An intact NEDD8 pathway is required for Cullin-dependent ubiquitylation in mammalian cells. EMBO Rep. 2002;3:177–82. doi: 10.1093/embo-reports/kvf028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osaka F, Saeki M, Katayama S, Aida N, Toh EA, Kominami K, Toda T, Suzuki T, Chiba T, Tanaka K, Kato S. Covalent modifier NEDD8 is essential for SCF ubiquitin-ligase in fission yeast. Embo J. 2000;19:3475–84. doi: 10.1093/emboj/19.13.3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshikawa K, Matsumoto M, Yada M, Kamura T, Hatakeyama S, Nakayama KI. Preferential interaction of TIP120A with Cul1 that is not modified by NEDD8 and not associated with Skp1. Biochem Biophys Res Commun. 2003;303:1209–16. doi: 10.1016/s0006-291x(03)00501-1. [DOI] [PubMed] [Google Scholar]
- Ou CY, Lin YF, Chen YJ, Chien CT. Distinct protein degradation mechanisms mediated by Cul1 and Cul3 controlling Ci stability in Drosophila eye development. Genes Dev. 2002;16:2403–14. doi: 10.1101/gad.1011402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagano M, Benmaamar R. When protein destruction runs amok, malignancy is on the loose. Cancer Cell. 2003;4:251–6. doi: 10.1016/s1535-6108(03)00243-5. [DOI] [PubMed] [Google Scholar]
- Pan ZQ, Kentsis A, Dias DC, Yamoah K, Wu K. Nedd8 on cullin: building an expressway to protein destruction. Oncogene. 2004;23:1985–97. doi: 10.1038/sj.onc.1207414. [DOI] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- Pickart CM, Fushman D. Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol. 2004;8:610–6. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- Pintard L, Kurz T, Glaser S, Willis JH, Peter M, Bowerman B. Neddylation and deneddylation of CUL-3 is required to target MEI-1/Katanin for degradation at the meiosis-to-mitosis transition in C. elegans. Curr Biol. 2003;13:911–21. doi: 10.1016/s0960-9822(03)00336-1. [DOI] [PubMed] [Google Scholar]
- Podust VN, Brownell JE, Gladysheva TB, Luo RS, Wang C, Coggins MB, Pierce JW, Lightcap ES, Chau V. A Nedd8 conjugation pathway is essential for proteolytic targeting of p27Kip1 by ubiquitination. Proc Natl Acad Sci U S A. 2000;97:4579–84. doi: 10.1073/pnas.090465597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read MA, Brownell JE, Gladysheva TB, Hottelet M, Parent LA, Coggins MB, Pierce JW, Podust VN, Luo RS, Chau V, Palombella VJ. Nedd8 modification of cul-1 activates SCF(beta(TrCP))-dependent ubiquitination of IkappaBalpha. Mol Cell Biol. 2000;20:2326–33. doi: 10.1128/mcb.20.7.2326-2333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock KL, Gramm C, Rothstein L, Clark K, Stein R, Dick L, Hwang D, Goldberg AL. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 1994;78:761–71. doi: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- Saha A, Deshaies RJ. Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Mol Cell. 2008;32:21–31. doi: 10.1016/j.molcel.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakata E, Yamaguchi Y, Miyauchi Y, Iwai K, Chiba T, Saeki Y, Matsuda N, Tanaka K, Kato K. Direct interactions between NEDD8 and ubiquitin E2 conjugating enzymes upregulate cullin-based E3 ligase activity. Nat Struct Mol Biol. 2007;14:167–8. doi: 10.1038/nsmb1191. [DOI] [PubMed] [Google Scholar]
- Salon C, Brambilla E, Brambilla C, Lantuejoul S, Gazzeri S, Eymin B. Altered pattern of Cul-1 protein expression and neddylation in human lung tumours: relationships with CAND1 and cyclin E protein levels. J Pathol. 2007;213:303–10. doi: 10.1002/path.2223. [DOI] [PubMed] [Google Scholar]
- Schmidt MW, McQuary PR, Wee S, Hofmann K, Wolf DA. F-box-directed CRL complex assembly and regulation by the CSN and CAND1. Mol Cell. 2009;35:586–97. doi: 10.1016/j.molcel.2009.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwechheimer C, Serino G, Callis J, Crosby WL, Lyapina S, Deshaies RJ, Gray WM, Estelle M, Deng XW. Interactions of the COP9 signalosome with the E3 ubiquitin ligase SCFTIRI in mediating auxin response. Science. 2001;292:1379–82. doi: 10.1126/science.1059776. [DOI] [PubMed] [Google Scholar]
- Shiraishi S, Zhou C, Aoki T, Sato N, Chiba T, Tanaka K, Yoshida S, Nabeshima Y, Nabeshima Y, Tamura TA. TBP-interacting protein 120B (TIP120B)/cullin-associated and neddylation-dissociated 2 (CAND2) inhibits SCF-dependent ubiquitination of myogenin and accelerates myogenic differentiation. J Biol Chem. 2007;282:9017–28. doi: 10.1074/jbc.M611513200. [DOI] [PubMed] [Google Scholar]
- Sonneville R, Gonczy P. Zyg-11 and cul-2 regulate progression through meiosis II and polarity establishment in C. elegans. Development. 2004;131:3527–43. doi: 10.1242/dev.01244. [DOI] [PubMed] [Google Scholar]
- Starostina NG, Lim JM, Schvarzstein M, Wells L, Spence AM, Kipreos ET. A CUL-2 ubiquitin ligase containing three FEM proteins degrades TRA-1 to regulate C. elegans sex determination. Dev Cell. 2007;13:127–39. doi: 10.1016/j.devcel.2007.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston JE. Neuronal cell lineages in the nematode Caenorhabditis elegans. Cold Spring Harb Symp Quant Biol. 1983;48:443–52. doi: 10.1101/sqb.1983.048.01.049. Pt 2. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol. 1977;56:110–56. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- Vasudevan S, Starostina NG, Kipreos ET. The Caenorhabditis elegans cell-cycle regulator ZYG-11 defines a conserved family of CUL-2 complex components. EMBO Rep. 2007;8:279–86. doi: 10.1038/sj.embor.7400895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wee S, Geyer RK, Toda T, Wolf DA. CSN facilitates Cullin-RING ubiquitin ligase function by counteracting autocatalytic adapter instability. Nat Cell Biol. 2005;7:387–91. doi: 10.1038/ncb1241. [DOI] [PubMed] [Google Scholar]
- Wolf DA, Zhou C, Wee S. The COP9 signalosome: an assembly and maintenance platform for cullin ubiquitin ligases? Nat Cell Biol. 2003;5:1029–33. doi: 10.1038/ncb1203-1029. [DOI] [PubMed] [Google Scholar]
- Wu JT, Lin HC, Hu YC, Chien CT. Neddylation and deneddylation regulate Cul1 and Cul3 protein accumulation. Nat Cell Biol. 2005;7:1014–20. doi: 10.1038/ncb1301. [DOI] [PubMed] [Google Scholar]
- Wu K, Chen A, Pan ZQ. Conjugation of Nedd8 to CUL1 enhances the ability of the ROC1-CUL1 complex to promote ubiquitin polymerization. J Biol Chem. 2000;275:32317–24. doi: 10.1074/jbc.M004847200. [DOI] [PubMed] [Google Scholar]
- Zhang W, Ito H, Quint M, Huang H, Noel LD, Gray WM. Genetic analysis of CAND1-CUL1 interactions in Arabidopsis supports a role for CAND1-mediated cycling of the SCFTIR1 complex. Proc Natl Acad Sci U S A. 2008;105:8470–5. doi: 10.1073/pnas.0804144105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Yang X, Harrell JM, Ryzhikov S, Shim EH, Lykke-Andersen K, Wei N, Sun H, Kobayashi R, Zhang H. CAND1 binds to unneddylated CUL1 and regulates the formation of SCF ubiquitin E3 ligase complex. Mol Cell. 2002;10:1519–26. doi: 10.1016/s1097-2765(02)00784-0. [DOI] [PubMed] [Google Scholar]
- Zhong W, Feng H, Santiago FE, Kipreos ET. CUL-4 ubiquitin ligase maintains genome stability by restraining DNA-replication licensing. Nature. 2003;423:885–9. doi: 10.1038/nature01747. [DOI] [PubMed] [Google Scholar]
- Zhou C, Wee S, Rhee E, Naumann M, Dubiel W, Wolf DA. Fission yeast COP9/signalosome suppresses cullin activity through recruitment of the deubiquitylating enzyme Ubp12p. Mol Cell. 2003;11:927–38. doi: 10.1016/s1097-2765(03)00136-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.