Abstract
Background
Published data regarding the associations between genetic variants and asthma risk in Chinese population were inconclusive. The aim of this study was to investigate asthma susceptible genes in Chinese population.
Methods
The authors conducted 18 meta-analyzes for 18 polymorphisms in 13 genes from eighty-two publications.
Results
Seven polymorphisms were found being associated with risk of asthma, namely: A Disintegrin and Metalloprotease 33 (ADAM33) T1-C/T (odds ratio [OR] = 6.07, 95% confidence interval [CI]: 2.69-13.73), Angiotensin-Converting Enzyme (ACE) D/I (OR = 3.85, 95%CI: 2.49-5.94), High-affinity IgE receptor β chain (FcεRIβ) -6843G/A (OR = 1.49, 95%CI: 1.01-2.22), Interleukin 13(IL-13) -1923C/T (OR = 2.99, 95%CI: 2.12-4.24), IL-13 -2044A/G (OR = 1.49, 95%CI: 1.07-2.08), Regulated upon Activation, Normal T cell Expressed and Secreted (RANTES) -28C/G (OR = 1.64, 95%CI: 1.09-2.46), Tumor Necrosis Factor-α (TNF-α) -308G/A(OR = 1.42, 95%CI: 1.09, 1.85). After subgroup analysis by age, the ACE D/I, β2-Adrenergic Receptor (β2-AR) -79G/C, TNF-α -308G/A, Interleukin 4 receptor(IL-4R) -1902G/A and IL-13 -1923C/T polymorphisms were found significantly associated with asthma risk in Chinese children. In addition, the ACE D/I, FcεRIβ -6843G/A, TNF-α -308G/A, IL-13 -1923C/T and IL-13 -2044A/G polymorphisms were associated with asthma risk in Chinese adults.
Conclusion
ADAM33, FcεRIβ, RANTES, TNF-α, ACE, β2-AR, IL-4R and IL-13 genes could be proposed as asthma susceptible genes in Chinese population. Given the limited number of studies, more data are required to validate these associations.
Introduction
Asthma is one of the most common chronic respiratory diseases, affecting about 300 millions of children and adults worldwide[1]. In China, more than 25 millions people are asthmatic patients, which includes almost 10 million children[2]. Compared with the western world, the preventive controls and treatments for asthma were not well established in China [3]. Only a few percent of asthma patients received proper treatment. Poverty and inadequate resources are the main hindrance to reduce the burden of disease in China especially in numerous of Chinese villagers. Therefore, the best approach to reduce asthma is primary prevention through modifying the risk factors of asthma.
It is well accepted that asthma is a complex disease and both genetic and environmental factors contribute to its inception and evolution[4,5]. Many studies regarding associations between genetic variants and asthma risk have been published and many genes were proposed as asthma susceptible genes[6-9]. However, the conclusions obtained from different populations were often different or even controversial. Possible roles may be that different genetic backgrounds and environment exposures in different ethnic population that may affect the pathogenesis of asthma. Thus, asthma susceptible genes in different population may not be the same.
In recent years, host genetic susceptibility to asthma has been a research focus in scientific community in China. Many genes were suggested as asthma risk factors for Chinese population; however, many of the studies drew incompatible or even contradictory results. Considering a small number of sample size may be lack of power to reveal the reliable conclusion, we carried out a meta-analysis to assess the susceptible genes for asthma in Chinese population. To our knowledge, this is the first comprehensive and largest genetic meta-analysis conducted in people of Chinese descent for any respiratory diseases.
Materials and methods
Literature search
We conducted a literature search by using the electronic database Medline (Ovid), Pubmed, Embase, ScienceDirect, Springer, CNKI, Wanfang database, Weipu database and CBM database to identify articles that evaluated the association between genetic variants and the risk of asthma in Chinese population (Last search was updated on May 13, 2010). The search terms were used as follows: 'asthma or asthmatic', in combination with 'polymorphism or variant or mutation' and 'Chinese or China' for Medline (Ovid), Pubmed, Embase, ScienceDirect, Springer database; 'asthma or asthmatic', in combination with 'polymorphism or variant or mutation' for CNKI database, Wanfang database, Weipu database and CBM database. All languages were included. The following criteria were used for selecting literatures in the meta-analysis: (1) the study should evaluate the association between genetic variants and risk of asthma in Chinese population from either mainland, overseas or both, (2) the study should be a case-control design published in a journal (3) genotype distributions in both cases and controls were available for estimating an odds ratio with 95% confidence interval (CI) and P value, (4)genotype distributions of control population must be consistent with Hardy-Weinberg equilibrium(HWE), P > 0.05 (5) the polymorphism for data synthesis should be studied in at least three case-control studies, (6) polymorphisms for data synthesis should be characterized as -A/B, with the following genotypes: AA, AB and BB. Accordingly, the following exclusion criteria were used: (1) abstracts and reviews, (2)genotype frequency not reported, (3) repeated or overlapping publications (4) polymorphisms with data less than three case-control studies (5) genotype distributions of control population not consistent with HWE, (6)genetic variants not characterized as -A/B. For duplication or overlapping publications, the studies with larger number of cases and controls or been published latest were included.
Data extraction
Two independent authors (Xiaobo Li and Yonggang Zhang) checked all potentially relevant studies and reached a consensus on all items. In case of disagreement, a third author(Jie Zhang) would assess these articles. The following data were collected from each study: first author, year of publication, location of the people, ages, genotype frequencies in cases and controls.
Statistical Analysis
For each case-control study, we first examined whether the genotype distribution in control group was according to Hardy-Weinberg equilibrium by Pearson's X2 test http://ihg2.helmholtz-muenchen.de/cgi-bin/hw/hwa1.pl.
Any polymorphism that had been studied in at least three case-control studies was included in the meta-analysis. The strength of the associations between asthma risk and genetic variants were estimated by ORs and 95% CIs. The statistical significance of summary ORs were assessed by Z-test. The evaluated genetic models for each study were based mostly on those used in primary studies. Heterogeneity was evaluated by a X2-based Q statistic and was considered statistical significant at P value < 0.10. I2 was used to measure the percentage of variability in point estimated that due to heterogeneity rather than sampling error. When the P-value is > 0.10, the pooled OR was calculated by the fixed-effects model, otherwise, a random-effects model was used. To evaluate the age-specific effects, subgroup analyses were performed by age for polymorphisms which were investigated in a sufficient number of studies(data were available from at least three case-control studies for at least one subgroup). Publication bias was examined by using the funnel plots, Begg's test and Egger's test[4]. The funnel plot is asymmetrical when there is evidence of publication bias. All statistical tests were performed by using REVMAN 4.2 software and STATA 10.0.
Results
Candidate asthma-genes in Chinese Population
The selection process is shown in Figure 1. Briefly, 2489 search results were identified from Medline (Ovid), Pubmed, Embase, ScienceDirect, Springer, CNKI database, Wanfang database, Weipu database and CBM database in the initial search. After reading the titles and abstracts, 2159 articles were excluded for abstracts, reviews, duplicated search results or not being relevant to genetic variants and asthma risk in Chinese population. By reading through the full texts of the remaining 330 articles, 7 articles were excluded for not being relevant to polymorphisms and asthma risk. The remaining 323 articles were used for data extraction. A total of 539 case-control studies were extracted from 248 articles, and 75 articles were excluded because of the absence of the usable data or not a case-control design. In meta-analysis, a small number of studies weaken the conclusions; therefore, only polymorphisms which had been investigated in at least three case-control studies were included for data synthesis. Thus, we excluded all these polymorphisms which were studied in less than three case-control studies(a total of 260 case-control studies were excluded). Hence, a total of 279 case-control studies were left. In addition, genotype frequencies for control population in 53 case-control studies were not consistent with HWE and these case-control studies were all excluded. In the remaining 226 case-control studies, data in 45 case-control studies were overlapped or duplicated with other studies and these case-control studies were all excluded. Thus, 181 case-control studies were left. Among the 181 case-control studies, some polymorphisms were studied in less than three case-control studies, and these polymorphisms were also excluded(a total of 62 case-control studies were excluded). Finally, a total of 18 polymorphisms in 13 genes in 119 case-control studies concerning genetic variants and asthma risk in Chinese population met the inclusion criteria, were identified for data synthesis (Table 1). The characteristics of each polymorphism are listed in Table 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18 and 19. The genetic models for pooling data are also listed in Table 1.
Figure 1.
Flow diagram of included/excluded studies.
Table 1.
Genes identified from individual studies
| Gene | Chromosome location of gene | Polymorphism | Aminoacid change | Genetic model | Genotypes Evaluated | Other genotypes | Cases | Controls |
|---|---|---|---|---|---|---|---|---|
| β2-AR | 5q31-32 | -46G/A | Arg16Gly | Recessive | GG | GA+AA | 1796 | 1589 |
| -79G/C | Gln27Glu | Recessive | GG | GC+CC | 823 | 692 | ||
| IL-4R | 16p11.2-12.1 | -1902G/A | Q576R | Dominant | GG+GA | AA | 2308 | 1971 |
| -223G/A | Ile/Val | Recessive | GG | GA+AA | 1623 | 1304 | ||
| IL-4 | 5q31 | -589C/T | Dominant | CC+CT | TT | 1724 | 1656 | |
| TNF-α | 6p21.1-21.3 | -308A/G | Dominant | AA+AG | GG | 1428 | 1511 | |
| FcεRIβ | 11q13 | -6843G/A | Glu237Gly | Dominant | GG+GA | AA | 1434 | 1276 |
| -109C/T | Recessive | CC | CT+TT | 428 | 371 | |||
| ACE | 17q23 | D/I | Recessive | DD | DI+II | 385 | 335 | |
| IL-13 | 5q31 | -2044A/G | Gln130Arg | Dominant | AA+AG | GG | 1512 | 1351 |
| -1923C/T | Recessive | TT | CC+CT | 645 | 588 | |||
| IL-1β | 2q12-21 | -511C/T | Dominant | TT+TC | CC | 333 | 255 | |
| LT-α | 6q21.3 | +252A/G | Dominant | GG+GA | AA | 674 | 896 | |
| TGF-β1 | 19q13 | -509C/T | Dominant | TT+TC | CC | 406 | 390 | |
| CD14 | 5q31.1 | -159C/T | Dominant | TT+TC | CC | 1381 | 1219 | |
| ADAM33 | 20p13 | T1 | Met764Thr | Recessive | CC | TT+TC | 569 | 512 |
| RANTES | 17q11.2-12 | -28G/C | Dominant | GG+GC | CC | 314 | 229 | |
| -403A/G | Dominant | AA+AG | GG | 343 | 205 |
Table 2.
Main data of all studies included in the meta-analysis for the -46G/A (Arg16Gly) polymorphism in β2-AR gene
| Case | Control | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Study | Population location | Year | Age | AA | AG | GG | AA | AG | GG | OR | 95%CI |
| Chan, I H[16] | Hong Kong | 2008 | 10.4 ± 3.7 | 101 | 135 | 59 | 51 | 89 | 33 | 1.06 | 0.66-1.70 |
| Cui, LY(Han)[17] | Neimenggu | 2007 | 21-62 | 6 | 34 | 2 | 6 | 20 | 4 | 0.33 | 0.06-1.90 |
| Cui, LY(Meng)[17] | Neimenggu | 2007 | 26-69 | 3 | 21 | 6 | 6 | 19 | 5 | 1.25 | 0.34-4.64 |
| Gao, J M[18] | Beijing | 2004 | 38.7 ± 13.8 | 38 | 59 | 28 | 35 | 53 | 8 | 3.18 | 1.37-7.33 |
| Li, H[19] | Shanghai | 2009 | 3-12 | 86 | 76 | 30 | 46 | 100 | 46 | 0.59 | 0.35-0.98 |
| Liao, W[20] | Chongqing | 2001 | 5.8 ± 4.3 | 12 | 27 | 11 | 14 | 28 | 8 | 1.48 | 0.54-4.06 |
| Qiu, Y Y(2008)[21] | Jiangsu | 2008 | 63.2 ± 5.6 | 25 | 31 | 14 | 34 | 55 | 23 | 0.97 | 0.46-2.04 |
| Shi, X H[22] | Jiangsu | 2008 | 34(14-66) | 22 | 19 | 7 | 10 | 25 | 13 | 0.46 | 0.17-1.28 |
| Wang, Z[23] | Anhui | 2001 | 30.6 ± 16.2 | 52 | 54 | 22 | 38 | 64 | 34 | 0.62 | 0.34-1.14 |
| Xie, Y[24] | Shanghai | 2008 | 4.98 ± 2.78 | 14 | 37 | 6 | 21 | 34 | 7 | 0.92 | 0.29-2.93 |
| Xing, J[25] | Beijing | 2001 | 20-66 | 9 | 62 | 29 | 29 | 55 | 16 | 2.14 | 1.08-4.26 |
| Zhang, X Y[26] | Chongqing | 2008 | 1.08-17 | 81 | 111 | 25 | 19 | 23 | 8 | 0.68 | 0.29-1.62 |
| Wang, J Y[27] | Taiwan | 2009 | 7.82 ± 3.81 | 138 | 207 | 97 | 173 | 250 | 87 | 1.37 | 0.99-1.89 |
Table 3.
Main data of all studies included in the meta-analysis for the -79G/C (Gln27Glu) polymorphism in β2-AR gene
| Case | Control | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Study | Population location | Year | Age | CC | CG | GG | CC | CG | GG | OR | 95%CI |
| Cui, LY(Han)[17] | Neimenggu | 2007 | 21-62 | 32 | 6 | 4 | 26 | 3 | 1 | 3.05 | 0.32-28.79 |
| Gao, G K[28] | Beijing | 2002 | 4-56 | 20 | 32 | 6 | 32 | 49 | 8 | 1.17 | 0.38-3.56 |
| Liao, W[20] | Chongqing | 2001 | 5.8 ± 4.3 | 26 | 20 | 4 | 20 | 27 | 3 | 1.36 | 0.29-6.43 |
| Lin, Y C[29] | Taiwan | 2003 | 13.9 ± 0.07 | 65 | 15 | 0 | 54 | 14 | 1 | 0.28 | 0.01-7.08 |
| Pan, Y P[30] | Jiangxi | 2005 | - | 15 | 24 | 6 | 17 | 23 | 5 | 1.23 | 0.35-4.37 |
| Qiu, Y Y(2000)[31] | Jiangsu | 2000 | 42 ± 5 | 23 | 30 | 6 | 29 | 36 | 7 | 1.05 | 0.33-3.32 |
| Qiu, Y Y(2008)[21] | Jiangsu | 2008 | 63.2 ± 5.6 | 56 | 13 | 1 | 90 | 20 | 2 | 0.80 | 0.07-8.96 |
| Wang, Z[23] | Anhui | 2001 | 30.6 ± 16.2 | 108 | 19 | 1 | 113 | 22 | 1 | 1.06 | 0.07-17.18 |
| Ye, X W[32] | Guizhou | 2003 | 42.68 ± 10.55 | 25 | 39 | 10 | 15 | 20 | 4 | 1.37 | 0.40-4.68 |
| Zhang, X Y[26] | Chongqing | 2008 | 1.08-17 | 54 | 119 | 44 | 8 | 24 | 18 | 0.45 | 0.23-0.88 |
Table 4.
Main data of all studies included in the meta-analysis for the -1902G/A (Q576R) polymorphism in IL-4R gene
| Case | Control | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Study | Population location | Year | Age | AA | AG | GG | AA | AG | GG | OR | 95%CI |
| Cui, T P[33] | Hubei | 2003 | 3-68 | 129 | 89 | 23 | 130 | 41 | 4 | 2.51 | 1.64-3.83 |
| Deng, R Q[34] | Guangdong | 2006 | 8-75 | 26 | 42 | 32 | 15 | 38 | 47 | 0.50 | 0.25-1.02 |
| Gui, Q[35] | Chongqing | 2006 | 49(28-72) | 33 | 15 | 2 | 34 | 14 | 2 | 1.09 | 0.48-2.52 |
| Hu, S Y[36] | Guangdong | 2005 | 2-16 | 90 | 66 | 19 | 130 | 41 | 4 | 2.73 | 1.74-4.28 |
| Liu, L N[37] | Henan | 2005 | 3-15 | 46 | 27 | 3 | 47 | 12 | 1 | 2.36 | 1.09-5.08 |
| Mak, J C[38] | Hong Kong | 2007 | 42.4 ± 16.1 | 200 | 81 | 4 | 191 | 91 | 9 | 0.81 | 0.57-1.15 |
| Sun, J[39] | Heilongjiang | 2010 | 3-14 | 67 | 24 | 0 | 33 | 9 | 0 | 1.31 | 0.55-3.14 |
| Wu, X H[40] | Hubei | 2010 | 8.8 | 183 | 61 | 8 | 168 | 55 | 4 | 1.07 | 0.72-1.61 |
| Zhang, A M[41] | Hunan | 2005 | 3-14 | 55 | 39 | 0 | 57 | 11 | 0 | 3.67 | 1.71-7.89 |
| Zhang, H[42] | Shanghai | 2007 | - | 257 | 87 | 8 | 87 | 27 | 0 | 1.19 | 0.73-1.95 |
| Zhang, W[43] | Singapore | 2007 | - | 115 | 30 | 0 | 115 | 38 | 4 | 0.71 | 0.42-1.22 |
| Wang, J Y[27] | Taiwan | 2009 | 7.82 ± 3.81 | 326 | 112 | 9 | 360 | 140 | 12 | 0.88 | 0.66-1.17 |
Table 5.
Main data of all studies included in the meta-analysis for the -223G/A (Ile/Val) polymorphism in IL-4R gene
| Case | Control | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Study | Population location | Year | Age | AA | AG | GG | AA | AG | GG | OR | 95%CI |
| Chan, I H [16] | Hong Kong | 2008 | 10.4 ± 3.7 | 79 | 159 | 57 | 49 | 80 | 38 | 0.81 | 0.51-1.29 |
| Deng, R Q[44] | Guangdong | 2006 | 8-75 | 24 | 47 | 29 | 9 | 33 | 58 | 0.30 | 0.16-0.53 |
| Yang, Q[45] | Jiangxi | 2004 | 18-71 | 6 | 21 | 7 | 8 | 16 | 5 | 1.24 | 0.35-4.44 |
| Zhang, H[42] | Shanghai | 2007 | - | 106 | 168 | 78 | 44 | 53 | 17 | 1.62 | 0.92-2.88 |
| Zhang, W[43] | Singapore | 2007 | - | 32 | 84 | 29 | 42 | 76 | 39 | 0.76 | 0.44-1.30 |
| Wang, J Y[27] | Taiwan | 2009 | 7.82 ± 3.81 | 105 | 201 | 139 | 124 | 250 | 136 | 1.25 | 0.94-1.65 |
| Wu, X H[40] | Hubei | 2010 | 8.8 | 46 | 131 | 75 | 59 | 110 | 58 | 1.23 | 0.83-1.85 |
Table 6.
Main data of all studies included in the meta-analysis for the -589 C/T polymorphism in IL-4 gene
| Case | Control | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Study | Population location | Year | Age | TT | CT | CC | TT | CT | CC | OR | 95%CI |
| Cui, T P[33] | Hubei | 2003 | 3-68 | 141 | 89 | 11 | 114 | 52 | 9 | 1.33 | 0.89-1.98 |
| Hu, S Y[36] | Guangdong | 2005 | 2-16 | 108 | 59 | 8 | 114 | 52 | 9 | 1.16 | 0.75-1.79 |
| Liu, L N[37] | Henan | 2005 | 3-15 | 45 | 29 | 2 | 34 | 23 | 3 | 0.90 | 0.45-1.79 |
| Mak, J C[38] | Hong Kong | 2007 | 42.4 ± 16.1 | 179 | 95 | 15 | 186 | 87 | 19 | 1.08 | 0.77-1.51 |
| Wang, W[46] | Xinjiang | 2004 | 39 ± 8 | 22 | 42 | 29 | 15 | 26 | 21 | 1.03 | 0.49-2.19 |
| Wu, X H[40] | Hubei | 2010 | 8.8 | 163 | 83 | 6 | 132 | 84 | 11 | 0.76 | 0.52-1.10 |
| Zhang, W D[47] | Singapore | 2005 | - | 101 | 47 | 4 | 109 | 45 | 3 | 1.15 | 0.71-1.85 |
| Wang, J Y[27] | Taiwan | 2009 | 7.82 ± 3.81 | 279 | 145 | 22 | 309 | 183 | 16 | 0.93 | 0.72-1.21 |
Table 7.
Main data of all studies included in the meta-analysis for the -308A/G polymorphism in TNF-α gene
| Case | Control | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Study | Population location | Year | Age | GG | GA | AA | GG | GA | AA | OR | 95%CI |
| Gao, J M[48] | Beijing | 2003 | 38.7 ± 13.8 | 47 | 52 | 26 | 44 | 41 | 11 | 1.40 | 0.82-2.41 |
| Guo, Y L[49] | Jiangxi | 2004 | - | 4 | 28 | 16 | 7 | 11 | 3 | 5.50 | 1.40-21.60 |
| Li, Z F[50] | Guangdong | 2003 | 2-12 | 9 | 16 | 5 | 14 | 10 | 2 | 2.72 | 0.91-8.16 |
| Liu, R M[51] | Hubei | 2004 | 2-15 | 98 | 15 | 0 | 104 | 22 | 0 | 0.72 | 0.36-1.47 |
| Mak, J C[38] | Hong Kong | 2007 | 42.4 ± 16.1 | 244 | 47 | 1 | 250 | 40 | 2 | 1.17 | 0.75-1.84 |
| Tan, E C[52] | Singapore | 1999 | - | 49 | 18 | 0 | 115 | 36 | 0 | 1.17 | 0.61-2.26 |
| Wang, T N[53] | Taiwan | 2004 | 5-18 | 140 | 49 | 2 | 111 | 18 | 0 | 2.25 | 1.24-4.06 |
| Zhai, F Z[54] | Shandong | 2004 | 35.80 ± 10.18 | 44 | 14 | 6 | 67 | 12 | 1 | 2.34 | 1.06-5.19 |
| Zhao, H J[55] | Jilin | 2005 | - | 45 | 5 | 0 | 71 | 9 | 0 | 0.88 | 0.28-2.78 |
| Wang, J Y[27] | Taiwan | 2009 | 7.82 ± 3.81 | 345 | 100 | 3 | 409 | 94 | 7 | 1.21 | 0.89-1.65 |
Table 8.
Main data of all studies included in the meta-analysis for the -6843G/A polymorphism in FcεRI β gene
| Case | Control | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Study | Population location | Year | Age | AA | AG | GG | AA | AG | GG | OR | 95%CI |
| Chan, I H[16] | Hong Kong | 2008 | 10.4 ± 3.7 | 267 | 23 | 1 | 154 | 13 | 0 | 1.06 | 0.53-2.15 |
| Cui, T P[56] | Hubei | 2004 | 40.37 ± 15.09 | 60 | 40 | 6 | 78 | 26 | 2 | 2.14 | 1.20-3.81 |
| Liu, T[57] | Shandong | 2006 | 36.5 | 45 | 14 | 1 | 39 | 10 | 1 | 1.18 | 0.49-2.87 |
| Tang, Y[58] | Guangdong | 2003 | 39.5(12-67) | 49 | 11 | 0 | 61 | 4 | 0 | 3.42 | 1.03-11.42 |
| Wang, L[59] | Hubei | 2003 | 2-16 | 65 | 40 | 5 | 70 | 20 | 2 | 2.20 | 1.20-4.06 |
| Zeng, L X[60] | Jiangxi | 2001 | 37(14-63) | 61 | 5 | 3 | 27 | 1 | 0 | 3.54 | 0.42-29.73 |
| Zhang, X Z[61] | Singapore | 2004 | 52 ± 16 | 81 | 57 | 3 | 108 | 42 | 7 | 1.63 | 1.02-2.62 |
| Zhao, K S[62] | Jilin | 2004 | 1.5-14 | 126 | 23 | 2 | 92 | 13 | 0 | 1.40 | 0.68-2.89 |
| Wang, J Y[27] | Taiwan | 2009 | 7.82 ± 3.81 | 309 | 121 | 16 | 314 | 165 | 27 | 0.73 | 0.55-0.95 |
Table 9.
Main data of all studies included in the meta-analysis for the -109C/T polymorphism in FcεRI β gene
Table 10.
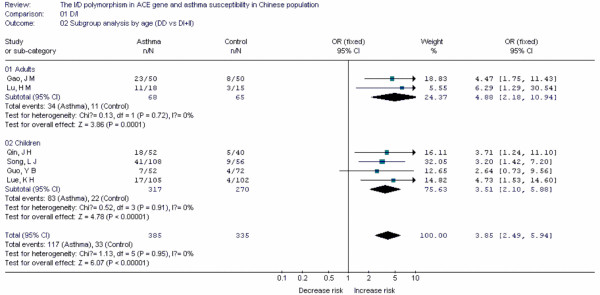
Main data of all studies included in the meta-analysis for the D/I polymorphism in ACE gene
| Case | Control | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Study | Population location | Year | Age(year) | II | DI | DD | II | DI | DD | OR | 95%CI |
| Gao, J M[64] | Beijing | 1999 | 39(16-69) | 12 | 15 | 23 | 16 | 26 | 8 | 4.47 | 1.75-11.43 |
| Guo, Y B[65] | Guangdong | 2006 | 0.33-3 | 27 | 18 | 7 | 36 | 32 | 4 | 2.64 | 0.73-9.56 |
| Lu, H M[66] | Tianjin | 2004 | 37(18-52) | 3 | 4 | 11 | 5 | 7 | 3 | 6.29 | 1.29-30.54 |
| Lue, K H[67] | Taiwan | 2006 | 9.91 ± 1.62 | 48 | 40 | 17 | 56 | 42 | 4 | 4.73 | 1.53-14.60 |
| Qin, J H[68] | Liaoning | 2000 | 6.9 ± 2.7 | 24 | 10 | 18 | 21 | 14 | 5 | 3.71 | 1.24-11.10 |
| Song, L J[69] | Jilin | 2001 | 1-14 | 22 | 45 | 41 | 18 | 29 | 9 | 3.20 | 1.42-7.20 |
Table 11.
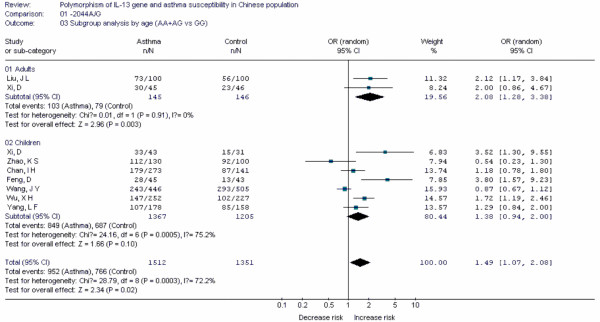
Main data of all studies included in the meta-analysis for the -2044A/G polymorphism in IL-13 gene
| Case | Control | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Study | Population location | Year | Age | GG | AG | AA | GG | AG | AA | OR | 95%CI |
| Chan, I H[16] | Hong Kong | 2008 | 10.4 ± 3.7 | 94 | 136 | 43 | 54 | 70 | 17 | 1.18 | 0.78-1.80 |
| Feng, D[70] | Heilongjiang | 2009 | 3-16 | 17 | 18 | 10 | 30 | 10 | 3 | 3.80 | 1.57-9.23 |
| Liu, J L[71] | Guangdong | 2004 | 14-67 | 27 | 54 | 19 | 44 | 46 | 10 | 2.12 | 1.17-3.84 |
| Wu, X H[40] | Hubei | 2010 | 8.8 | 105 | 111 | 36 | 125 | 84 | 18 | 1.72 | 1.19-2.46 |
| Yang, L F[72] | Gansu | 2010 | 8 ± 4 | 71 | 60 | 47 | 73 | 66 | 19 | 1.29 | 0.84-2.00 |
| Zhao, K S[73] | Jilin | 2005 | 1.5-14 | 18 | 60 | 52 | 8 | 42 | 50 | 0.54 | 0.23-1.30 |
| Wang, J Y[27] | Taiwan | 2009 | 7.82 ± 3.81 | 203 | 194 | 49 | 212 | 234 | 59 | 0.87 | 0.67-1.12 |
| Xi, D[74] | Hubei | 2004 | ≥20 | 15 | 24 | 6 | 23 | 20 | 3 | 2.08 | 1.28-3.38 |
| Xi, D[74] | Hubei | 2004 | ≥4 | 10 | 25 | 8 | 16 | 13 | 2 | 3.52 | 1.30-9.55 |
Table 12.
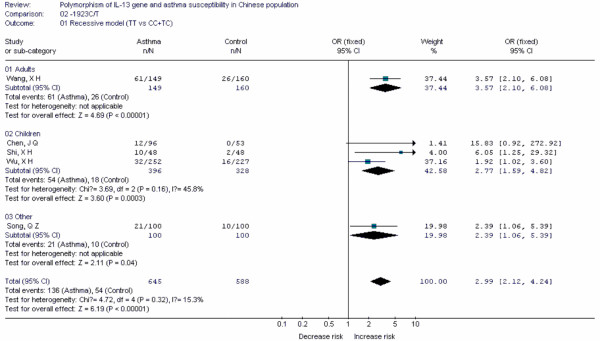
Main data of all studies included in the meta-analysis for the -1923C/T polymorphism in IL-13 gene
| Case | Control | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Study | Population location | Year | Age | CC | CT | TT | CC | CT | TT | OR | 95%CI |
| Song, Q Z[75] | Guangdong | 2005 | 14-67 | 24 | 55 | 21 | 43 | 47 | 10 | 2.39 | 1.06-5.39 |
| Shi, X H[22] | Jiangsu | 2008 | 34(14-66) | 12 | 26 | 10 | 30 | 16 | 2 | 6.05 | 1.25-29.32 |
| Chen, J Q[76] | Jiangsu | 2004 | 2.59 ± 1.45 | 41 | 43 | 12 | 39 | 14 | 0 | 15.83 | 0.92-272.92 |
| Wang, X H[77] | Shandong | 2009 | 39 ± 11 | 31 | 57 | 61 | 66 | 68 | 26 | 3.57 | 2.10-6.08 |
| Wu, X H[40] | Hubei | 2010 | 8.8 | 106 | 114 | 32 | 126 | 85 | 16 | 1.92 | 1.02-3.60 |
Table 13.
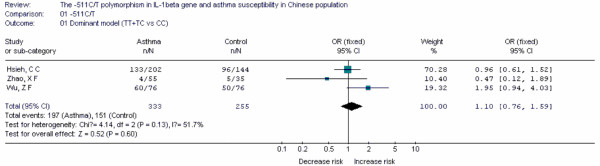
Main data of all studies included in the meta-analysis for the -511C/T polymorphism in IL-1β gene
Table 14.
Main data of all studies included in the meta-analysis for the +252A/G polymorphism in LT-α gene
| Case | Control | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Study | Population location | Year | Age | AA | AG | GG | AA | AG | GG | OR | 95%CI |
| Gao, J M[81] | Beijing | 2003 | 38.7 ± 13.8 | 13 | 63 | 49 | 14 | 46 | 36 | 1.47 | 0.66-3.30 |
| Ma, W C[82] | Guangdong | 2005 | 1.8-9 | 8 | 14 | 10 | 26 | 46 | 28 | 1.05 | 0.42-2.64 |
| Mak, J C[38] | Hong Kong | 2007 | 42.4 ± 16.1 | 70 | 146 | 69 | 79 | 134 | 76 | 1.16 | 0.80-1.68 |
| Tan, E C[52] | Singapore | 1999 | - | 13 | 38 | 15 | 30 | 84 | 39 | 0.99 | 0.48-2.06 |
| Xu, X[83] | Guangdong | 2003 | 18-69 | 12 | 21 | 19 | 26 | 47 | 30 | 1.13 | 0.51-2.46 |
| Huang, S C[84] | Taiwan | 2008 | 9.9 ± 4.1 | 20 | 69 | 25 | 45 | 69 | 41 | 1.62 | 0.98-2.66 |
Table 15.
Main data of all studies included in the meta-analysis for the -509C/T polymorphism in TGF-β1 gene
Table 16.
Main data of all studies included in the meta-analysis for the -159C/T polymorphism in CD14 gene
| Case | Control | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Study | Population location | Year | Age | CC | CT | TT | CC | CT | TT | OR | 95%CI |
| Chan, I H[16] | Hong Kong | 2008 | 10.4 ± 3.7 | 55 | 134 | 80 | 26 | 77 | 38 | 0.88 | 0.52-1.48 |
| Chen, M[88] | Guangdong | 2009 | 14-71 | 63 | 62 | 25 | 40 | 68 | 42 | 0.50 | 0.31-0.82 |
| Cui, T P[89] | Hubei | 2003 | 2-16 | 27 | 67 | 49 | 10 | 42 | 20 | 0.69 | 0.32-1.52 |
| Tan, C Y[90] | Taiwan | 2006 | - | 17 | 56 | 47 | 24 | 55 | 41 | 1.51 | 0.77-2.99 |
| Wu, X H[40] | Hubei | 2010 | 8.8 | 54 | 117 | 81 | 31 | 121 | 75 | 0.58 | 0.36-0.94 |
| Wang, J Y[27] | Taiwan | 2009 | 7.82 ± 3.81 | 160 | 230 | 57 | 177 | 236 | 96 | 0.96 | 0.73-1.25 |
Table 17.
Main data of all studies included in the meta-analysis for the T1-C/T polymorphism in ADAM33 gene
Table 18.
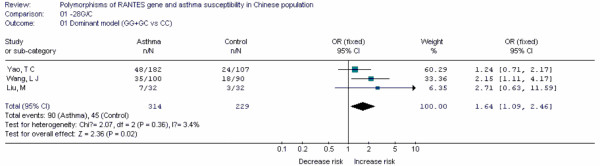
Main data of all studies included in the meta-analysis for the -28G/C polymorphism in RANTES gene
Table 19.
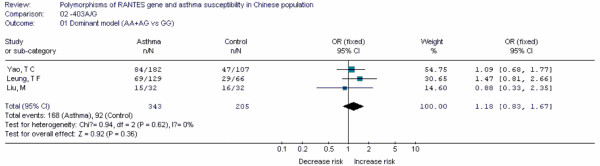
Main data of all studies included in the meta-analysis for the -403A/G polymorphism in RANTES gene
Summary results of Meta-analyzes
For each polymorphism, heterogeneity was analyzed by a X2-based Q statistic and was considered statistical significant at P-value <0.10. When the P-value is less than 0.10, the pooled OR of each meta-analysis was calculated by the fixed-effects model; otherwise, a random-effects model was used. The chosen models to synthesize the data for each polymorphism can be seen in Table 20.
Table 20.
Summary results of the meta-analysis and publications bias
| Pubilication bias (Begg's test) | |||||||
|---|---|---|---|---|---|---|---|
| Gene | Polymorphism | Genotype investigated | Studies Number | Effect Model | OR(95%CI) | t | P |
| β2-AR | -46G/A | GG | 13 | Random | 1.02(0.75, 1.38) | -0.66 | 0.525 |
| -79G/C | GG | 10 | Fixed | 0.86(0.58, 1.29) | 1.60 | 0.148 | |
| IL-4R | -1902G/A | GG+GA | 12 | Random | 1.30(0.94, 1.80) | 0.92 | 0.377 |
| -223G/A | GG | 7 | Random | 0.92(0.63, 1.35) | -0.81 | 0.453 | |
| IL-4 | -589C/T | CC+CT | 8 | Fixed | 1.01(0.88, 1.16) | 0.53 | 0.615 |
| TNF-α | -308A/G | AA+AG | 10 | Random | 1.42(1.09, 1.85) | 1.38 | 0.205 |
| FcεRIβ | -6843G/A | GG+GA | 9 | Random | 1.49(1.01, 2.22) | 2.82 | 0.026 |
| -109C/T | CC | 3 | Fixed | 0.96(0.62, 1.48) | -1.10 | 0.471 | |
| ACE | D/I | DD | 6 | Fixed | 3.85(2.49, 5.94) | 0.88 | 0.429 |
| IL-13 | -2044A/G | AA+AG | 9 | Random | 1.49(1.07, 2.08) | 1.93 | 0.095 |
| -1923C/T | TT | 5 | Fixed | 2.99(2.12, 4.24) | 1.19 | 0.320 | |
| IL-1β | -511C/T | TT+TC | 3 | Fixed | 1.10(0.76, 1.59_ | -0.16 | 0.896 |
| LT-α | +252A/G | GG+GA | 6 | Fixed | 1.26(0.98, 1.62) | -0.02 | 0.985 |
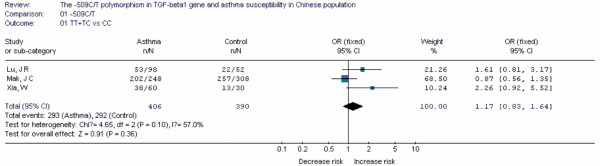
| TGF-β1 | -509C/T | TT+TC | 3 | Fixed | 1.17(0.83, 1.64) | 8.57 | 0.074 |
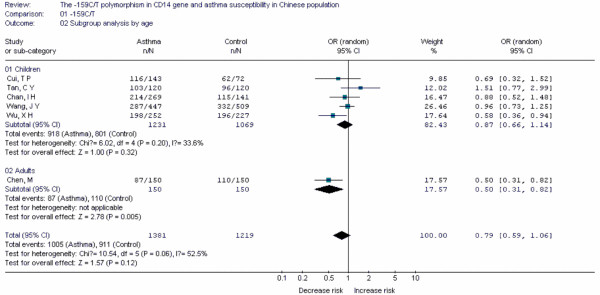
| CD14 | -159C/T | TT+TC | 6 | Random | 0.79(0.59, 1.06) | -0.41 | 0.700 |
| ADAM33 | T1-C/T | CC | 3 | Fixed | 6.07(2.69, 13.73) | -8.22 | 0.077 |
| RANTES | -28G/C | GG+GC | 3 | Fixed | 1.64(1.09, 2.46) | 0.87 | 0.544 |
| -403A/G | AA+AG | 3 | Fixed | 1.18(0.83, 1.67) | -0.37 | 0.777 | |
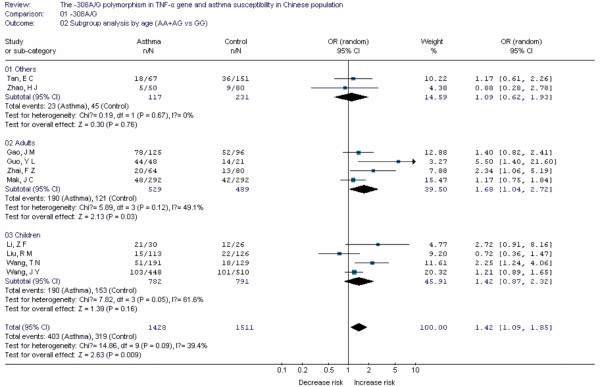
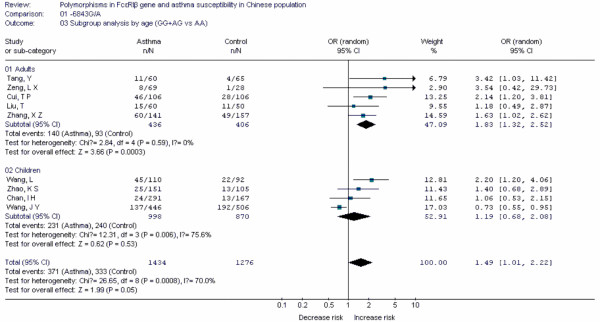
Forest plots of each polymorphism can be seen in Figure 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18 and 19. In summary, we abstained significant results for seven polymorphisms: ADAM33 T1-C/T (OR = 6.07, 95%CI: 2.69-13.73, Z = 4.33, P < 0.0001), ACE D/I(OR = 3.85, 95%CI: 2.49-5.94, Z = 6.07, P < 0.00001), FcεRIβ -6843G/A (OR = 1.49, 95%CI: 1.01-2.22, Z = 1.99, P = 0.05), IL-13 -1923C/T(OR = 2.99, 95%CI: 2.12-4.24, Z = 6.19, P< 0.00001), IL-13 -2044A/G(OR = 1.49, 95%CI: 1.07-2.08, Z = 2.34, P = 0.02), RANTES -28C/G (OR = 1.64, 95%CI: 1.09-2.46, Z = 2.36, P = 0.02), TNF-α -308G/A (OR = 1.42, 95%CI: 1.09-1.85, Z = 2.63, P = 0.009). These results indicated that these polymorphisms were significant associated with asthma risk in Chinese population. All results for all 18 meta-analyzes are summarized in table 20.
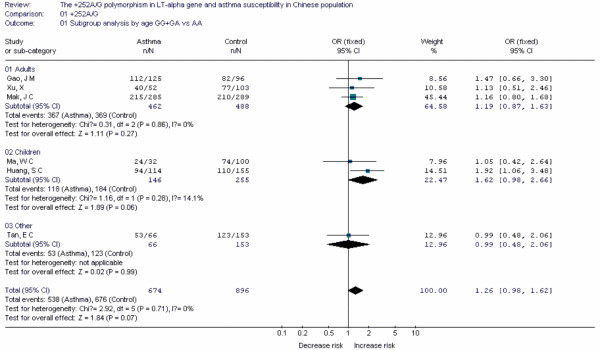
Figure 2.
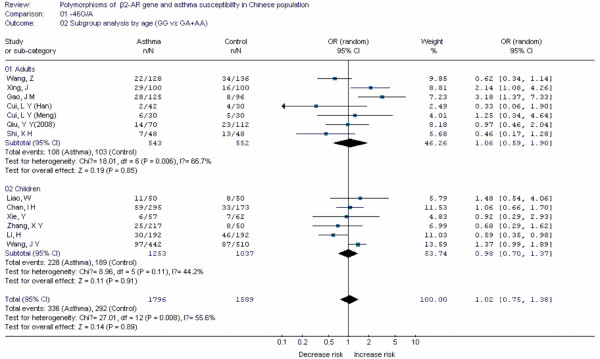
Forest plot of asthma risk associated with β2-AR -46G/A in Chinese population. Subgroup analysis by age.
Figure 3.
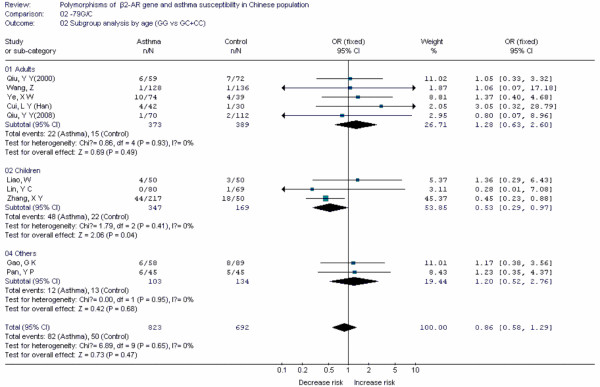
Forest plot of asthma risk associated with β2-AR -79G/C in Chinese population. Subgroup analysis by age.
Figure 4.
Forest plot of asthma risk associated with IL-4R -1902G/A in Chinese population. Subgroup analysis by age.
Figure 5.
Forest plot of asthma risk associated with IL-4R -223G/A in Chinese population.
Figure 6.
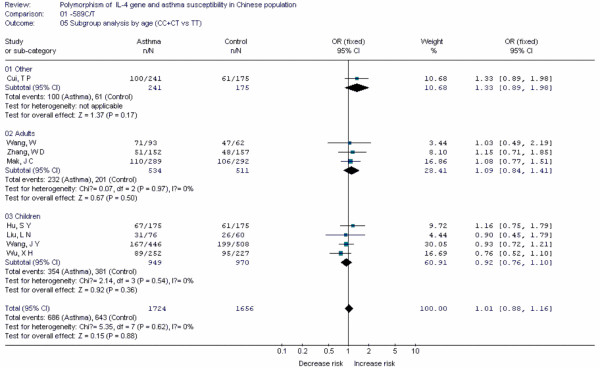
Forest plot of asthma risk associated with IL-4 -589C/T in Chinese population. Subgroup analysis by age.
Figure 7.
Forest plot of asthma risk associated with TNF-α -308A/G in Chinese population. Subgroup analysis by age.
Figure 8.
Forest plot of asthma risk associated with FcεRIβ -6843G/A in Chinese population. Subgroup analysis by age.
Figure 9.
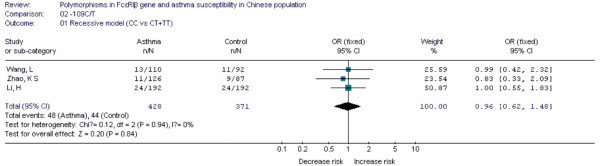
Forest plot of asthma risk associated with FcεRIβ -109C/T in Chinese population.
Figure 10.
Forest plot of asthma risk associated with ACE D/I in Chinese population. Subgroup analysis by age.
Figure 11.
Forest plot of asthma risk associated with IL-13 -2044A/G in Chinese population. Subgroup analysis by age.
Figure 12.
Forest plot of asthma risk associated with IL-13 -1923C/T in Chinese population.
Figure 13.
Forest plot of asthma risk associated with IL-1β-511C/T in Chinese population.
Figure 14.
Forest plot of asthma risk associated with LT-α +252A/G in Chinese population.
Figure 15.
Forest plot of asthma risk associated with TGF-β1 -509C/T in Chinese population.
Figure 16.
Forest plot of asthma risk associated with CD14 -159C/T in Chinese population.
Figure 17.
Forest plot of asthma risk associated with ADAM33 T1-C/T in Chinese population.
Figure 18.
Forest plot of asthma risk associated with RANTES -28G/C in Chinese population.
Figure 19.
Forest plot of asthma risk associated with RANTES -403A/G in Chinese population.
To evaluate the age-specific effects, subgroup analyses were performed by age for polymorphisms which were investigated in a sufficient number of studies(data were available from at least three case-control studies for at least one subgroup). Three subgroups were used: adults, children, others(ages in these case-control studies were not mentioned or mixed with adults and children). Briefly, we obtained significant results from five polymorphisms(ACE D/I, β2-AR -79G/C, TNF-α -308G/A, IL-4R -1902G/A and IL-13 -1923C/T) in children and five polymorphisms(ACE D/I, FcεRIβ -6843G/A, TNF-α -308G/A, IL-13 -1923C/T, IL-13 -2044A/G) in adults.
Publication bias
The Begg's funnel plots and Egger's tests were performed to assess the potential publication bias (Begg's funnel plots can be seen in Additional File 1). The results did not suggest evidence of publication bias except for the FcεRIβ -6843G/A polymorphism. Statistical results of Begg's test are summarized in Table 20.
Discussion
The aim of meta-analysis is to combine results from studies on the same topic and to produce more precise results. The current study is to reveal the roles of genetic variants and their associations with risk of asthma in Chinese population. In summary, we finally identified 18 polymorphisms in 13 genes. Among them, seven polymorphisms (ADAM33 T1-C/T, ACE D/I, FcεRIβ -6843G/A, IL-13 -1923C/T, IL-13 -2044A/G, RANTES -28C/G and TNF-α -308G/A) were statistically associated with increased risk of asthma. In order to analysis the age-specific associations, subgroup analysis were performed by age. The ACE D/I, β2-AR -79G/C, TNF-α -308G/A, IL-4R -1902G/A and IL-13 -1923C/T polymorphisms were found being associated with asthma risk in Chinese children, while the ACE D/I, FcεRIβ -6843G/A, TNF-α -308G/A, IL-13 -1923C/T, IL-13 -2044A/G polymorphisms were associated with asthma risk in Chinese adults. Given that the data for each polymorphism were from at least three case-control studies, the obtained results could be more precise than results obtained form any individual study.
The β2-AR gene is a critical gene in the pathogenesis of asthma. β2-ARs are present on many airway cells, especially in smooth muscle cells which are hyperreactive in asthmatic patients. At present, β2-AR agonists were major methods for treating asthmatic patients. In this meta-analysis, ten case-control studies for β2-AR -79G/C and eleven for -46G/A polymorphism were identified. The results indicated the two polymorphisms were not associated with asthma risk in Chinese population. After subgroup analysis by age, the -79G/C polymorphism was associated with decreased risk of asthma in Chinese children. Up to now, three meta-analyses had been performed to investigate the association between polymorphism of β2-AR gene and risk of asthma [10-12]. Thakkinstian A[12] found that the heterozygote in -79G/C was associated with decreased risk of asthma in both adults and children. However, we didn't find these associations in Chinese adults, which suggested different roles of this polymorphism may exist in the pathogenesis of asthma in difference age groups. Previous study indicated that the -46G allele enhanced agonist-induced down regulation of the receptor, and the -79G allele might enhance resistance to down regulation. In combination with our results, personalized therapy of asthma patients in different age population with different genetic backgrounds in Chinese population should also be carried out in clinical practices.
The TNF-α gene, encodes a key proinflammatory cytokine in airway, is located on an asthma susceptible region-chromosome 6p. The TNF-α protein plays a central role in inflammation and involves in pathogenesis of asthma. Several polymorphisms have been identified in this gene, such as -308A/G, -238A/G. The -308A/G polymorphism in the promoter may affect the expression of this cytokine, which may affect the occurrence of asthma. In the meta-analysis performed by Gao and colleagues[13], they found the A allele was significant with increased risk of asthma (OR = 1.37, 95%CI = 1.02-1.84 for A vs. G). Consistently, we found the TNF-α-308A/G polymorphism was significantly associated with increased risk of asthma (OR = 1.36, 95%CT = 1.13-1.63 for AA+AG vs. GG) in Chinese population. For A vs G, the pooled OR is 1.26 with 95%CI: 1.08-1.47 in this study, which suggested a weaker association between this polymorphism and asthma risk in Chinese population.
IL-4 gene is located on chromosome 5q31, it was suggested to be associated with asthma risk, including elevated serum IgE levels and airway hypersensitiveness. A few studies indicated the -589C/T polymorphism in the promoter as a risk factor for asthma, but with inconclusive results. Li and colleagues performed a meta-analysis and found the T allele was associated with decrease risk of asthma(T vs C: OR = 0.86, 95%CI = 0.78-0.94)[14]. However, our results didn't reveal a positive association between this polymorphism and risk of asthma in Chinese. Compared with Li's study, the total number of studies concerning the Chinese population was smaller, which suggested more studies should be carried out to reveal these associations.
IL-4 and IL-13 signal through binding to a receptor complex comprised of the IL-13Rα1 and IL-4Rα with subsequent phosphorylation of JAKs and STAT6[15]. IL-4 receptor plays its role in inflammation through IL-4 and IL-13. The IL-4 receptor gene is located on chromosome 16 p12.1-p11.2. Some polymorphisms had been identified as risk factors for asthma, such as -1902G/A and -223G/A. Our results indicated the -1902G/A polymorphism was associated with increased risk of asthma in Chinese children, but not in Chinese adults. The results also indicated the -223G/A polymorphism was not associated with risk of asthma in Chinese population.
The FcεRIβ gene is a major candidate gene, involving in the pathogenesis of asthma. It is located on the chromosome 11q13. The -6843G/A polymorphism, leading change in an amino acid sequence at residue 237 from glutamic acid to glycine, is associated with increased IgE levels in atopic asthmatic children. In Chinese population, the -6843G/A polymorphism is the most extensively studied polymorphism in FcεRIβ gene. Our study revealed this polymorphism as a risk factor of asthma in Chinese population. Chinese who carry the GG or GA genotype have an 49% increased risk of asthma than AA carriers. Our results also demonstrated the -109C/T polymorphism in this gene was not associated with increased risk of asthma in Chinese population.
Up to date, we first found that ADAM33 T1-C/T, ACE D/I, IL-13 -1923C/T, RANTES -28C/G and IL-13 -2044A/G polymorphisms were associated with risk of asthma in Chinese population by using meta-analyzes. Some results are similar to other studies performed in other ethnic- groups and some are not. In future, more published results should be included to update and validate these associations in Chinese population.
In this study, the rigorous inclusive criteria made the results more precise. Any study in which genotype distribution of control group divorced from HWE was excluded. In this meta-analysis, 11 polymorphisms were synthesized by using the fixed-effect model, 7 used random-effects model. Because the fixed-effect model is more precise than random effect model, the strength of evidence of ADAM33 T1-C/T, ACE D/I, IL-13 -1923C/T, RANTES -28C/G, as risk factors for asthma was greater than that of FcεRIβ -6843G/A, IL-13 -2044A/G and TNF-α -308G/A.
The heterogeneity of clinical information among studies should also be mentioned. Heterogeneity is an important issue when interpreting the results of meta-analysis. Significant heterogeneity existed in overall comparisons in a few meta-analyses, such as FcεRIβ -6843G/A. After subgroup analyses by age, the heterogeneity was effectively decreased or removed in adults. Possible explanation may be that differences in etiology may exist in difference age groups. Another important factor contributing to heterogeneity was that homogeneity in either the case and control groups was uncertain. Ideally, all cases and controls in this meta-analysis should be matched for age, sex, atopic status and environmental exposures. However, these issues could not all be explained precisely because of insufficient clinical information for individual person. In addition, because this study is based on population of Chinese descent with the same genetic background, so the similarity of these studies might be very good, despite most studies were conducted in different areas of China.
Some limitations of this meta-analysis should be acknowledged when explaining our results. First, only published articles in the selected electronic databases were included in this study, it may be possible that some studies were not included in those databases or some unpublished studies which had null results, which might bias the results. Second, due to lack of sufficient data, the homogeneity in either the case and control groups was uncertain and data were not stratified by other factors such as atopic status or sex. The tests for gene-environment interactions were not carried out either. Third, publication bias may affect the results. Although P values of Begg's test were more than 0.05 in 18 meta-analyses, we could not rule out this possibility, because for some polymorphisms, the included number of studies were relatively small. Third, this study didn't included some polymorphisms with lack of number of studies, or polymorphisms which were not characterized as -A/B for lack of quality analysis for HWE, some polymorphism, such as GSTM1-P/N, or HLA DR1 alleles and MHC alleles were not included, future studies should performed to analysis the effect of these polymorphism in Chinese population.
To our knowledge, this is the first and most comprehensive genetic meta-analysis to date conducted in Chinese descent for any respiratory diseases. In conclusion, this meta-analysis indicated the T1-C/T polymorphism in ADAM33 gene, the D/I polymorphism in ACE gene, the -6843G/A polymorphism in FcεRIβ gene, the -1923C/T polymorphism in IL-13 gene, the -2044A/G polymorphism in IL-13 gene, the -28C/G polymorphism in RANTES gene and the -308G/A polymorphism in TNF-α gene are associated with asthma risk in Chinese population. And these results may also implicate in personalized therapy for asthma in Chinese population. In future, more studies should be conducted to investigate the gene-gene and gene-environment interactions between these polymorphisms in Chinese population.
Abbreviations
ADAM33: A Disintegrin and Metalloprotease 33; FcεRIβ: High-affinity IgE receptor β chain; ACE: Angiotensin-Converting Enzyme; β2-AR: β2-Adrenergic Receptor; IL-4: Interleukin 4; IL-13: Interleukin 13; IL-1β: Interleukin 1β; LT-α: Lymphotoxin-α; RANTES: Regulated upon Activation, Normal T cell Expressed and Secreted; TNF-α: Tumor Necrosis Factor-α; TGF-β1: Transforming Growth Factor β1.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
HF designed the study, provided resources, coordinated the study and directed its implementation; XBL, YGZ and JZ searched the publications, extracted the data and wrote the materials and methods, results; YLX wrote the discussion and checked all data, JH was responsible for data synthesis, CT and CH helped designed the study's analytic strategy, YD edited the manuscript, YYY wrote the introduction. All authors read and approved the final manuscript.
Supplementary Material
Begg's funnel plots for publication bias in selection of studies on asthma susceptibility genes in Chinese. Figure S1 Begg's funnel plots for publication bias in selection of studies on β2-AR -46G/A polymorphism. Figure S2 Begg's funnel plots for publication bias in selection of studies on β2-AR -79G/C polymorphism. Figure S3 Begg's funnel plots for publication bias in selection of studies on IL-4R -1902G/A polymorphism. Figure S4 Begg's funnel plots for publication bias in selection of studies on IL-4R -223G/A polymorphism. Figure S5 Begg's funnel plots for publication bias in selection of studies on IL-4 -589C/T polymorphism. Figure S6 Begg's funnel plots for publication bias in selection of studies on TNF-α -308A/G polymorphism. Figure S7 Begg's funnel plots for publication bias in selection of studies on FcεRIβ -6843G/A polymorphism. Figure S8 Begg's funnel plots for publication bias in selection of studies on FcεRIβ -109C/T polymorphism. Figure S9 Begg's funnel plots for publication bias in selection of studies on ACE D/I polymorphism. Figure S10 Begg's funnel plots for publication bias in selection of studies on IL-13 -2044A/G polymorphism. Figure S11 Begg's funnel plots for publication bias in selection of studies on IL-13 -1923C/T polymorphism. Figure S12 Begg's funnel plots for publication bias in selection of studies on IL-1β-511C/T polymorphism. Figure S13 Begg's funnel plots for publication bias in selection of studies on LT-α +252A/G polymorphism. Figure S14 Begg's funnel plots for publication bias in selection of studies on TGF-β1 -509C/T polymorphism. Figure S15 Begg's funnel plots for publication bias in selection of studies on CD14 -159C/T polymorphism. Figure S16 Begg's funnel plots for publication bias in selection of studies on ADAM33 T1-C/T polymorphism. Figure S17 Begg's funnel plots for publication bias in selection of studies on RANTES -28G/C polymorphism. Figure S18 Begg's funnel plots for publication bias in selection of studies on RANTES -403A/G polymorphism
Contributor Information
Xiaobo Li, Email: litanhua01@yahoo.com.cn.
Yonggang Zhang, Email: yonggang-zhang@sohu.com.
Jie Zhang, Email: jiezhangwzyx@sohu.com.
Yuling Xiao, Email: xylpq@163.com.
Jin Huang, Email: huang.jin2010@sohu.com.
Can Tian, Email: doudouniao24@163.com.
Chao He, Email: xiaoqiao123@126.com.
Yao Deng, Email: day861023@163.com.
Yingying Yang, Email: gll0098@yahoo.cn.
Hong Fan, Email: z85445417@yahoo.cn.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (30470761 and 30871117).
References
- Lima JJ, Mohapatra S, Feng H, Lockey R, Jena PK, Castro M, Irvin C, Johnson JA, Wang J, Sylvester JE. A polymorphism in the NPPA gene associates with asthma. Clin Exp Allergy. 2008;38(7):1117–1123. doi: 10.1111/j.1365-2222.2008.02955.x. [DOI] [PubMed] [Google Scholar]
- Chen YZ. Recent status of prevention and treatment of asthma in children in China (Chinese) Zhonghua Er Ke Za Zhi. 2004;42(2):81–82. [PubMed] [Google Scholar]
- Chen X, Lin JT. The current prevention and treatment situation of asthma in China (Chinese) Journal of Internal Intensive Medicine. 2008;14(5):225–226. [Google Scholar]
- Zhang Y, Zhang J, Huang J, Li X, He C, Tian C, Peng C, Guo L, Xiao Y, Fan H. Polymorphisms in the transforming growth factor-beta1 gene and the risk of asthma: A meta-analysis. Respirology. 2010;15(5):643–650. doi: 10.1111/j.1440-1843.2010.01748.x. [DOI] [PubMed] [Google Scholar]
- Denham S, Koppelman GH, Blakey J, Wjst M, Ferreira MA, Hall IP, Sayers I. Meta-analysis of genome-wide linkage studies of asthma and related traits. Respir Res. 2008;9:38. doi: 10.1186/1465-9921-9-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss ST, Raby BA, Rogers A. Asthma genetics and genomics 2009. Curr Opin Genet Dev. 2009;19(3):279–282. doi: 10.1016/j.gde.2009.05.001. [DOI] [PubMed] [Google Scholar]
- Pinto LA, Depner M, Klopp N, Illig T, Vogelberg C, von Mutius E, Kabesch M. MMP-9 gene variants increase the risk for non-atopic asthma in children. Respir Res. 2010;11:23. doi: 10.1186/1465-9921-11-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litonjua AA, Tantisira KG, Lake S, Lazarus R, Richter BG, Gabriel S, Silverman ES, Weiss ST. Polymorphisms in signal transducer and activator of transcription 3 and lung function in asthma. Respir Res. 2005;6:52. doi: 10.1186/1465-9921-6-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossé Y, Lemire M, Poon AH, Daley D, He JQ, Sandford A, White JH, James AL, Musk AW, Palmer LJ, Raby BA, Weiss ST, Kozyrskyj AL, Becker A, Hudson TJ, Laprise C. Asthma and genes encoding components of the vitamin D pathway. Respir Res. 2009;10:98. doi: 10.1186/1465-9921-10-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migita O, Noguchi E, Jian Z, Shibasaki M, Migita T, Ichikawa K, Matsui A, Arinami T. ADRB2 polymorphisms and asthma susceptibility: transmission disequilibrium test and meta-analysis. Int Arch Allergy Immunol. 2004;134(2):150–157. doi: 10.1159/000078648. [DOI] [PubMed] [Google Scholar]
- Contopoulos-Ioannidis DG, Manoli EN, Ioannidis JP. Meta-analysis of the association of beta2-adrenergic receptor polymorphisms with asthma phenotypes. J Allergy Clin Immunol. 2005;115(5):963–972. doi: 10.1016/j.jaci.2004.12.1119. [DOI] [PubMed] [Google Scholar]
- Thakkinstian A, McEvoy M, Minelli C, Gibson P, Hancox B, Duffy D, Thompson J, Hall I, Kaufman J, Leung TF, Helms PJ, Hakonarson H, Halpi E, Navon R, Attia J. Systematic review and meta-analysis of the association between {beta}2-adrenoceptor polymorphisms and asthma: a HuGE review. Am J Epidemiol. 2005;162(3):201–211. doi: 10.1093/aje/kwi184. [DOI] [PubMed] [Google Scholar]
- Gao J, Shan G, Sun B, Thompson PJ, Gao X. Association between polymorphism of tumour necrosis factor alpha-308 gene promoter and asthma: a meta-analysis. Thorax. 2006;61(6):466–471. doi: 10.1136/thx.2005.051284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Guo B, Zhang L, Han J, Wu B, Xiong H. Association between C-589T polymorphisms of interleukin-4 gene promoter and asthma: a meta-analysis. Respir Med. 2008;102(7):984–992. doi: 10.1016/j.rmed.2008.02.008. [DOI] [PubMed] [Google Scholar]
- Moynihan BJ, Tolloczko B, El Bassam S, Ferraro P, Michoud MC, Martin JG, Laberge S. IFN-gamma, IL-4 and IL-13 modulate responsiveness of human airway smooth muscle cells to IL-13. Respir Res. 2008;9:84. doi: 10.1186/1465-9921-9-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan IH, Tang NL, Leung TF, Huang W, Lam YY, Li CY, Wong CK, Wong GW, Lam CW. Study of gene-gene interactions for endophenotypic quantitative traits in Chinese asthmatic children. Allergy. 2008;63:1031–1039. doi: 10.1111/j.1398-9995.2008.01639.x. [DOI] [PubMed] [Google Scholar]
- Cui LY, Liu XH, Gao LX, Fan DS. Study on the association between β2-adrenergic receptor genetic polymorphisms and asthma in the population of Inner Mongolia (Chinese) Zhong Guo Lin Chuang Yi Xue. 2007;14(4):477–481. [Google Scholar]
- Gao JM, Lin YG, Qiu CC, Liu YW, Ma Y, Liu Y. β2-adrenergic receptor gene polymorphism in Chinese northern asthmatics. Chinese Medical Sciences Journal. 2004;19(3):164–169. [PubMed] [Google Scholar]
- Li H, Xiaoyan D, Quanhua L, Jie L, Yixiao B. Single-nucleotide polymorphisms in genes predisposing to asthma in children of Chinese Han nationality. J Investig Allergol Clin Immunol. 2009;19(5):391–395. [PubMed] [Google Scholar]
- Liao W, Li WM, Zhao CM, Guang LX, Yin XJ, Ai YP, Xi M. Preliminary study on the realtionship between β2-adrenergic receptors genetic polymorphisms and asthma in children of Han nationality of Chongqing (Chinese) Di San Jun Yi Da Xue Xue Bao. 2001;23(8):968–971. [Google Scholar]
- Qiu YY, Yin KS. Relationship between beta 2-adrenergic receptor haplotype/polymorphisms and bronchial asthma in the elderly (Chinese) Shi Yong Lao Nian Yi Xue. 2008;22(2):105–107. [Google Scholar]
- Shi XH, Zhou JP. Relationship between polymorphisms of IL-13 gene and β2-AR gene and ashma (Chinese) Shan Dong Yi Yao. 2008;48(32):119–121. [Google Scholar]
- Wang Z, Chen C, Niu T, Wu D, Yang J, Wang B, Fang Z, Yandava CN, Drazen JM, Weiss ST, Xu X. Association of asthma with beta(2)-adrenergic receptor gene polymorphism and cigarette smoking. Am J Respir Crit Care Med. 2001;163(6):1404–1409. doi: 10.1164/ajrccm.163.6.2001101. [DOI] [PubMed] [Google Scholar]
- Xie Y, Yang ZZ, Chai BC. Relationship of genetic polymorphisms of β2-adrenergic receptor and asthma in children in Shanghai area (Chinese) Shi Yong Er Ke Lin Chuang Za Zhi. 2008;23(4):272–273. 303. [Google Scholar]
- Xing J, Wang C, Liu JZ, Yan M, Huang KW, Xiao B. Study on the beta2-AR gene and asthma risk in Chinese northern asthma patients (Chinese) Zhong Hua Nei Ke Za Zhi. 2001;40(5):340–342. [Google Scholar]
- Zhang XY, Zhao WL, Gui Q, He NH. Relationship between genetic polymorphisms of β2-adrenergic receptor and childhood asthma (Chinese) Lin Chuang Er Ke Za Zhi. 2008;26(5):399–402. 408. [Google Scholar]
- Wang JY, Liou YH, Wu YJ, Hsiao YH, Wu LS. An association study of 13 SNPs from seven candidate genes with pediatric asthma and a preliminary study for genetic testing by multiple variants in Taiwanese population. J Clin Immunol. 2009;29(2):205–209. doi: 10.1007/s10875-008-9256-6. [DOI] [PubMed] [Google Scholar]
- Gao GK, Wang SW, Zhang JC. Study on β2 adrenergic receptor genetic polymorphisms in asthmatics in the people of the Han nationality of northern China (Chinese) Zhong Hua Jie He He Hu Xi Za Zhi. 2000;23(2):93–97. [PubMed] [Google Scholar]
- Lin YC, Lu CC, Shen CY, Lei HY, Guo YL, Su HJ. Roles of genotypes of β2-adrenergic receptor in the relationship between eosinophil counts and lung function in Taiwanese adolescents. J Asthma. 2003;40(3):265–272. doi: 10.1081/JAS-120018323. [DOI] [PubMed] [Google Scholar]
- Pan YP, Zhou SL, Kuang JL, Rao WH. Study on the relationship between the genetic polymorphisms of β2-adrenergic receptor gene and asthma (Chinese) Jiang Xi Yi Xue Yuan Xue Bao. 2005;45(4):44–47. [Google Scholar]
- Qiu YY, Yin KS. Study on polymorphism of β2-AR and asthma risk in China (Chinese) Zhong Hua Jie He He Hu Xi Za Zhi. 2000;23(7):435–436. [Google Scholar]
- Ye XW, Feng DX, Wen XP, Zhang XY, Yu H, Diao XY, Zhang XR, Luo RR. Study on β2-adrenergic receptor genetic polymorphisms in asthmatics in the people of the Han nationality of Guizhou (Chinese) Gui Zhou Yi Yao. 2003;27(10):878–880. [Google Scholar]
- Cui TP, Wu JM, Pan SX, Xie JG. Polymorphisms in the IL-4 and IL-4R[α] genes and allergic asthma. Clin Chem Lab Med. 2003;41(7):888–892. doi: 10.1515/CCLM.2003.134. [DOI] [PubMed] [Google Scholar]
- Deng RQ, Wu B, He XL, Chen M, Xie SQ. Correlation between IL-4R α Arg551Gln gene polymorphism and asthma (Chinese) Lin Chuang Fei Ke Za Zhi. 2006;11(2):164–165. [Google Scholar]
- Gui Q, Qian GS, Zhao ZQ, Li SP. Study on association between IL-4R gene mutation and asthmatic patients of Han nationality of Chongqing in China (Chinese) Chong Qing Yi Xue. 2006;35(22):2055–2057. [Google Scholar]
- Hu SY, Yang XG, Li P, Yu ZD. Relation of polymorphism of IL-4 and IL-4R to allergic asthma in children (Chinese) Zhong Hua Yi Xue Jian Yan Za Zhi. 2005;6(6):460–462. [Google Scholar]
- Liu LN, Zhang YW. Study on relationship between asthma and polymorphisms of interleukin-4 receptor and interleukin-4 (Chinese) Yi Yao Lun Tan Za Zhi. 2005;26(19):38–40. [Google Scholar]
- Mak JC, Ko FW, Chu CM, Leung HC, Chan HW, Cheung AH, Ip MS, Chan-Yeung M. Polymorphisms in the IL-4, IL-4 receptor α chain, TNF-α, and lymphotoxin-α genes and risk of asthma in Hong Kong Chinese adults. Int Arch Allery Immunol. 2007;144(2):114–122. doi: 10.1159/000103222. [DOI] [PubMed] [Google Scholar]
- Sun J, Yu XH, Chen Y, Zhao HL, Yu JB, Zhou Y, Yi LY, Zhang YQ. Relationship between polymorphisms of interleukin-4 receptor gene and childhood asthma in Harbin (Chinese) Lin Chuang Er Ke Za Zhi. 2010;28(2):138–141. [Google Scholar]
- Wu XH, Li Y, Chen Q, Chen F, Cai P, Wang L, Hu L. Association and gene-gene interactions of eight common single-nucleotide polymorphisms with pediatric asthma in middle china. J Asthma. 2010;47(3):238–244. doi: 10.3109/02770900903509099. [DOI] [PubMed] [Google Scholar]
- Zhang AM, Li HL, Hao P, Chen YH, Li JM, Mo YX, Dai M. Association of Q576R polymorphism in the interleukin-4 receptor gene with serum IgE levels in children with asthma (Chinese) Zhong Guo Dang Dai Er Ke Za Zhi. 2006;8(2):109–112. [PubMed] [Google Scholar]
- Zhang H, Zhang Q, Wang L, Chen H, Li Y, Cui T, Huang W, Zhang L, Yan F, Wang L, Xu Y, Hu L, Kong X. Association of IL4R gene polymorphisms with asthma in Chinese populations. Hum Mutat. 2007;28(10):1046. doi: 10.1002/humu.9508. [DOI] [PubMed] [Google Scholar]
- Zhang W, Zhang X, Qiu D, Sandford A, Tan WC. IL-4 receptor genetic polymorphisms and asthma in Asian populations. Respir Med. 2007;101(1):186–190. doi: 10.1016/j.rmed.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Deng RQ, Wu B, Yan SF, Chen M. Association between IL-4R gene polymorphism and level of sIL-4R and TIgE in patients with asthma (Chinese) Gan Nan Yi Xue Yuan Xue Bao. 2006;26(3):321–323. [Google Scholar]
- Yang Q, Zou YQ, Kuang JL. A study on the relationship between interleukin-4 receptor polymorphism and asthma (Chinese) Jiang Xi Yi Xue Yuan Xue Bao. 2004;44(1):37–39. [Google Scholar]
- Wang W, Hamulati WFE, Yilihamujiang SBT, Xiang YB, Abulikemu ABL. A study on the relationship between interleukin-4 promoter polymorphism and asthma in a Xinjiang Uyger population (Chinese) Zhong Hua Jie He He Hu Xi Za Zhi. 2004;27(7):460–464. [PubMed] [Google Scholar]
- Zhang WD, Zhang XZ, Qiu DW, Tan WC. Relation between IL-4 promoter gene polymorphisms and asthma in Chinese, Malay and Indian (Chinese) Yi Xue Lin Chuang Yan Jiu. 2005;22(3):293–296. [Google Scholar]
- Gao JM, Lin YG, Qiu CC, Liu YW, Ma Y, Liu Y. The association between tumor necrosis factor-alpha gene polymorphism and asthma. Chinese Medical Sciences Journal. 2003;18(4):248–253. [Google Scholar]
- Guo YL, Zhou SL. Investigation of the association between tumour necrosis factor alpha promotor polymorphism and asthma (Chinese) Jiang Xi Yi Xue Yuan Xue Bao. 2004;44(5):28–30. 33. [Google Scholar]
- Li ZF, Li JR, Sun XF, Liao BP. Lack of association between childhood asthma and the tumor necrosis factor α gene-308 polymorphism (Chinese) Xin Yi Xue. 2003;34(4):217–218. [Google Scholar]
- Liu RM, Wu JM, Liu DF, Cui TP. Polymerase chain reaction analysis for the tumor factor alpha-308(G-A) gene polymorphism in relation to susceptibility of asthma in infants (Chinese) Hua Zhong Yi Xue Za Zhi. 2004;28(3):201–202. 154. [Google Scholar]
- Tan EC, Lee BW, Tay AW, Chew FT, Tay AH. Asthma and TNF variants in Chinese and Malays. Allergy. 1999;54(4):402–403. doi: 10.1034/j.1398-9995.1999.00082.x. [DOI] [PubMed] [Google Scholar]
- Wang TN, Chen WY, Wang TH, Chen CJ, Huang LY, Ko YC. Gene-gene synergistic effect on atopic asthma: tumour necrosis factor-α-308 and lymphotoxin-α-NcoI in Taiwan's children. Clin Exp Allergy. 2004;34(2):184–188. doi: 10.1111/j.1365-2222.2004.01867.x. [DOI] [PubMed] [Google Scholar]
- Zhai FZ, Li Y. Association between polymorphism of tumor necrosis factor-α promoter gene and asthma (Chinese) Shan Dong Yi Yao. 2004;44(25):4–6. [Google Scholar]
- Zhao HJ, Ding YC, Liu Y, Shi JP, Liu HF, Zhang J, Cheng HJ, Cui YN, Hou SP. Association between polymorphism of tumor necrosis factor promoter gene and asthma (Chinese) Ji Lin Da Xue Xue Bao (Yi Xue Ban) 2005;31(3):449–451. [Google Scholar]
- Cui TP, Jiang WC, Wang L, Xie JG, Wu JM. The association analysis of FcεRIβ with allergic asthma in a Hubei Han adults population (Chinese) Zhong Guo Bing Li Sheng Li Za Zhi. 2004;20(11):2049–2052. [Google Scholar]
- Liu T, Teng L, Guan LX, Wu LP, Sun KY. Study on the E237G polymorphism of the FcepsilonR Ι beta gene with asthma (Chinese) Zhong Guo Shi Yong Nei Ke Za Zhi. 2006;26(19):1520–1522. [Google Scholar]
- Tang Y, Wu XQ, Liu XY, Zeng Y, Li YQ, Wu Q, Zhou TH. Study on mutations of β-chain of high affinity IgE receptor gene in people of Han nationality in the southern China (Chinese) Zhong Guo Xian Dai Yi Xue Za Zhi. 2003;13(9):6–10. [Google Scholar]
- Wang L, Cui TP. Relationship between FcεRI β gene polymorphism and juvenile allergic asthma in Hubei area (Chinese) Hua Zhong Ke Ji Da Xue Xue Bao (Yi Xue Ban) 2003;32(3):332–335. [Google Scholar]
- Zeng LX, Zhou SL, Kuang JL, Rao WH. Study on mutations of β-chain of high affinity IgE receptor gene in asthmatic patients (Chinese) Jiang Xi Yi Xue Yuan Xue Bao. 2001;41(5):43–45. [Google Scholar]
- Zhang XZ, Zhang WD, Qiu DW, Andrew S, Cheng TW. The E237G polymorphism of the high-affinity IgE receptor β chain and asthma. Ann Allergy Asthma Immunol. 2004;93(5):499–503. doi: 10.1016/S1081-1206(10)61419-6. [DOI] [PubMed] [Google Scholar]
- Zhao KS, Cheng HJ, Qiao HM, Zhuo FX, Sun MY, Fu WY. Analysis of gene mutation for high affinity immunoglobulin E receptor chain in asthmatic children (Chinese) Lin Chuang Er Ke Za Zhi. 2004;22(12):794–797. [Google Scholar]
- Zhao KS, Lu JR, Wang ZH, Guo Y, Yu LY, Fu WY. Association between FcεRI-β gene promoter polymorphism and total serum IgE levels of asthma in children (Chinese) Zhong Guo Shi Yong Er Ke Za Zhi. 2004;19(12):744–746. [Google Scholar]
- Gao JM, Lin YG, Xiao Y, Xu KF, Xu WB, Ma Y. Polymorphism of angiotensin-converting enzyme gene and susceptibility to asthma with familial aggregation (Chinese) Zhong Hua Jie He He Hu Xi Za Zhi. 1999;22(11):669–672. [PubMed] [Google Scholar]
- Guo YB, Lu Y, Cai HW, Chen YH, Cheng YS, Ye XF. Genetic polymorphism of angiotensin converting enzyme (ACE) gene in kidney-deficiency asthma from Guangdong population (Chinese) Zhong Guo You Sheng Yu Yi Chuan Za Zhi. 2006;14(8):20–22. [Google Scholar]
- Lu HM, Li LY. Polymorphism of angiotensin-converting enzyme gene and susceptibility to patients of asthma in Tianjin (Chinese) Shan Xi Yi Yao Za Zhi. 2004;33(12):1016–1017. [Google Scholar]
- Lue KH, Ku MS, Li C, Sun HL, Lee HS, Chou MC. ACE gene polymorphism might disclose why some Taiwanese children with allergic rhinitis develop asthma symptoms but others do not. Pediatr Allergy Immunol. 2006;17(7):508–513. doi: 10.1111/j.1399-3038.2006.00452.x. [DOI] [PubMed] [Google Scholar]
- Qin JH, Wang LS. DD genotype of angiotensin-converting enzyme may be a risk factor for development of asthma in children (Chinese) Zhong Hua Er Ke Za Zhi. 2000;38(8):487–489. [Google Scholar]
- Song LJ, Quan CS, Peng L, Fu WY. Correlation between asthma and polymorpilsm of anglotensin-converting enzyme gene with insertion or deletion in 108 Chinese northern children with asthma (Chinese) Lin Chuang Er Ke Za Zhi. 2001;19(6):364–365. [Google Scholar]
- Feng D. Research on the polymorphism of gene IL-13 in asthma and their first degree relatives. Hei Long Jiang Yi Xue. 2009;33(7):481–485. [Google Scholar]
- Liu JL, Wu B, Chen HJ, He CW, Liu ZH, Xie JX. Relationship among IL-13 gene polymorphism, asthma and plasma cytokine levels. Lin Chuang Fei Ke Za Zhi. 2004;9(2):122–124. [Google Scholar]
- Yang LF, Zhang Y, Liu QL. Genetic Arg144Gln polymorphism of interleukin-13 and asthma in children (Chinese) Zhong Guo Xian Dai Yi Yao Za Zhi. 2010;12(3):46–47. [Google Scholar]
- Zhao KS, Lu JR, Li SY, Wang ZH, Fu WY, Sun MY. Correlationship between interleukin-13 genotype and phenotype in children with bronchial asthma (Chinese) Lin Chuang Er Ke Za Zhi. 2005;23(5):312–314. 330. [Google Scholar]
- Xi D, Pan S, Cui T, Wu J. Association between IL-13 gene polymorphism and asthma in Han nationality in Hubei Chinese population. J Huazhong Univ Sci Technolog Med Sci. 2004;24(3):219–222. doi: 10.1007/BF02831994. [DOI] [PubMed] [Google Scholar]
- Song QZ, Wu B, Li W, Liu JL, Zhang WZ, Zhang YL. Association between IL-13 gene polymorphism and level of IL-13 and TIgE in patients with asthma (Chinese) Zhong Guo Mian Yi Xue Za Zhi. 2005;21(6):469–471. [Google Scholar]
- Chen JQ, Sun HP, Guo XR, Chen RH. Effect of IL-13 gene polymorphism on the levels of serum IL-13 and total IgE in asthmatic children (Chinese) Zhong Guo Shi Yong Er Ke Za Zhi. 2004;19(4):209–211. [Google Scholar]
- Wang XH, Zhao W, Liu SG, Feng XP. Correlation of IL-4 and IL-13 gene polymorphisms with asthma and total serum IgE levels. Zhong Hua Jie He He Hu Xi Za Zhi. 2009;32(3):161–164. [PubMed] [Google Scholar]
- Hsieh CC, Tsai FJ, Chow WC, Wu CR, Kobayashi H. There is no evidence of difference in polymorphisms in the IL-1 beta-511 promoter and IL-1Ra gene between asthmatic and healthy groups. Pediatr Asthma Allergy Immunol. 2004;17(1):53–57. doi: 10.1089/088318704322994949. [DOI] [Google Scholar]
- Wu ZF, Yang H, Liu YL, Chen XW, Cui XM, Liang ZH. Relationship of interleukin-1beta and interleukin-1 receptor antagonist gene polymorphisms with asthma (Chinese) Mian Yi Xue Za Zhi. 2007;23(6):699–700. [Google Scholar]
- Zhao XF, Li HL, Huang YK. Study on association between interleukin-1 beta gene polymorphism and childhood asthma (Chinese) Shi Yong Er Ke Lin Chuang Za Zhi. 2006;21(16):1074–1075. [Google Scholar]
- Gao JM, Lin YG, Qiu CC, Liu YW, Ma Y, Liu Y, Zhu YY. TNFa/LTa genes polymorphism and bronchial asthma susceptibility (Chinese) Ji Chu Yi Xue Yu Lin Chuang. 2003;23(5):512–516. [Google Scholar]
- Ma WC, Zhu MH. Polymorphism of TNF-beta gene and asthma risk in children (Chinese) Xian Dai Lin Chuang Yi Xue Sheng Wu Gong Cheng Xue Za Zhi. 2005;11(3):204–206. [Google Scholar]
- Xu X, Chen SQ, Liu LD, Sun BQ, Chen SC. HLA-DRB, LMP, TNFbeta alleles polymorphism in susceptibility to asthma in Guangdong Chinese (Chinese) Xian Dai Lin Chuang Yi Xue Sheng Wu Gong Cheng Xue Za Zhi. 2003;9(3):188–190. [Google Scholar]
- Huang SC, Wu WJ, Sun HL, Lue KH, Hsu CH, Liao PF, Ku MS. Association of a lymphotoxin-alpha gene polymorphism and atopic asthma in Taiwanese children. Pediatr Neonatol. 2008;49(2):30–34. doi: 10.1016/S1875-9572(08)60008-X. [DOI] [PubMed] [Google Scholar]
- Lu JR, Liu WD, Zhao KS, Sun MY, Fu WY. Study on TGFβ1 polymorphism and asthma susceptibility (Chinese) Lin Chuang Er Ke Za Zhi. 2004;22(4):212–215. [Google Scholar]
- Mak JC, Leung HC, Ho SP, Law BK, Ho AS, Lam WK, Ip MS, Chan-Yeung MM. Analysis of TGF-beta(1) gene polymorphisms in Hong Kong Chinese patients with asthma. J Allergy Clin Immunol. 2006;117(1):92–96. doi: 10.1016/j.jaci.2005.08.049. [DOI] [PubMed] [Google Scholar]
- Xia W, Zhou SL, Xu P, Li P, Wang FX. Study on TGF-β1 promoter polymorphism in asthmatics (Chinese) Jiang Xi Yi Xue Yuan Xue Bao. 2006;46(6):102–103. 106. [Google Scholar]
- Chen M, Wu B, Li W. Influence of CD14 gene-159C/T polymorphism on IL-5 level in patients with asthma (Chinese) Shang Dong Yi Yao. 2009;49(5):13–15. [Google Scholar]
- Cui TP, Jiang WC, Wu JM. Genetic polymorphism of CD14 and allergic asthma in children (Chinese) Hua Zhong Yi Xue Za Zhi. 2003;27(5):235–236. [Google Scholar]
- Tan CY, Chen YL, Wu LS, Liu CF, Chang WT, Wang JY. Association of CD14 promoter polymorphisms and soluble CD14 levels in mite allergen sensitization of children in Taiwan. J Hum Genet. 2006;51(1):59–67. doi: 10.1007/s10038-005-0323-z. [DOI] [PubMed] [Google Scholar]
- Su DJ, Zhang XM, Sui H, Lu FZ, Jin LH, Zhang J. Association of ADAM33 gene polymorphisms with adult allergic asthma and rhinitis in a Chinese Han population. BMC Med Genet. 2008;9:82. doi: 10.1186/1471-2350-9-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Liu QJ, Li JS, Li HC, Wei CH, Guo CH, Gong YQ. Lack of association between ADAM33 gene and asthma in a Chinese population. Int J Immunogenet. 2006;33(4):303–306. doi: 10.1111/j.1744-313X.2006.00617.x. [DOI] [PubMed] [Google Scholar]
- Xiong JY, He QQ, Jiang ZQ, Li JF. Association of polymorphism of T1 locus allele in ADAM33 gene with bronchial athma (Chinese) Shi Yong Er Ke Lin Chuang Za Zhi. 2009;24(16):1241–1243. [Google Scholar]
- Liu M, Li HL, Huang YK, Chen YH, Liu H, Jin P. The SNPs of chemokine RANTES promoter in children with asthma (Chinese) Zhong Guo You Sheng Yu Yi Chuan Za Zhi. 2005;13(11):20–23. [Google Scholar]
- Wang LJ, Li YR, Chen JH, Cui TP, Wu JM. Polymorphism of regulated upon activation, normal T cell expressed and secreted promoter region-28 position in Chinese allergic asthmaic children (Chinese) Zhong Hua Jie He He Hu Xi Za Zhi. 2004;27(6):394–397. [PubMed] [Google Scholar]
- Yao TC, Kuo ML, See LC, Chen LC, Yan DC, Ou LS, Shaw CK, Huang JL. The RANTES promoter polymorphism: a genetic risk factor for near-fatal asthma in Chinese children. J Allergy Clin Immunol. 2003;111(6):1285–1292. doi: 10.1067/mai.2003.1506. [DOI] [PubMed] [Google Scholar]
- Leung TF, Tang NL, Lam CW, Li AM, Fung SL, Chan IH, Wong GW. RANTES G-401A polymorphism is associated with allergen sensitization and FEV1 in Chinese children. Respir Med. 2005;99(2):216–219. doi: 10.1016/j.rmed.2004.06.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Begg's funnel plots for publication bias in selection of studies on asthma susceptibility genes in Chinese. Figure S1 Begg's funnel plots for publication bias in selection of studies on β2-AR -46G/A polymorphism. Figure S2 Begg's funnel plots for publication bias in selection of studies on β2-AR -79G/C polymorphism. Figure S3 Begg's funnel plots for publication bias in selection of studies on IL-4R -1902G/A polymorphism. Figure S4 Begg's funnel plots for publication bias in selection of studies on IL-4R -223G/A polymorphism. Figure S5 Begg's funnel plots for publication bias in selection of studies on IL-4 -589C/T polymorphism. Figure S6 Begg's funnel plots for publication bias in selection of studies on TNF-α -308A/G polymorphism. Figure S7 Begg's funnel plots for publication bias in selection of studies on FcεRIβ -6843G/A polymorphism. Figure S8 Begg's funnel plots for publication bias in selection of studies on FcεRIβ -109C/T polymorphism. Figure S9 Begg's funnel plots for publication bias in selection of studies on ACE D/I polymorphism. Figure S10 Begg's funnel plots for publication bias in selection of studies on IL-13 -2044A/G polymorphism. Figure S11 Begg's funnel plots for publication bias in selection of studies on IL-13 -1923C/T polymorphism. Figure S12 Begg's funnel plots for publication bias in selection of studies on IL-1β-511C/T polymorphism. Figure S13 Begg's funnel plots for publication bias in selection of studies on LT-α +252A/G polymorphism. Figure S14 Begg's funnel plots for publication bias in selection of studies on TGF-β1 -509C/T polymorphism. Figure S15 Begg's funnel plots for publication bias in selection of studies on CD14 -159C/T polymorphism. Figure S16 Begg's funnel plots for publication bias in selection of studies on ADAM33 T1-C/T polymorphism. Figure S17 Begg's funnel plots for publication bias in selection of studies on RANTES -28G/C polymorphism. Figure S18 Begg's funnel plots for publication bias in selection of studies on RANTES -403A/G polymorphism