Abstract
Orientia tsutsugamushi (OT) is an obligate intracellular bacterium belonging to the family Rickettsiaceae and is the causative agent of scrub typhus, or Tsutsugamushi disease. The complete genome sequences of two OT strains (Boryong and Ikeda) have recently been determined. In the present study, we performed a fine genome sequence comparison of these strains. Our results indicate that although the core gene set of the family Rickettsiaceae is highly conserved between the two strains, a common set of repetitive sequences have been explosively amplified in both genomes. These amplified repetitive sequences have induced extensive genome shuffling and duplications and deletions of many genes. On the basis of the results of the genome sequence comparison, we selected 11 housekeeping genes and carried out multilocus sequence analysis of OT strains using the nucleotide sequences of these genes. This analysis revealed for the first time the phylogenetic relationships of representative OT strains. Furthermore, the results suggest the presence of an OT lineage with higher potential for virulence, which may explain the clinical and epidemiological differences between ‘classic’ and ‘new’ types of Tsutsugamushi disease in Japan.
Keywords: Orientia tsutsugamushi, genome sequence comparison, genome rearrangement, phylogenetic analysis, multilocus sequence analysis
1. Introduction
Orientia tsutsugamushi (OT), an obligate intracellular bacterium belonging to the family Rickettsiaceae of the subdivision alpha-Proteobacteria, is the causative agent of scrub typhus, or Tsutsugamushi disease. The vector and the reservoir of OT are trombiculid mites. The bacteria reside in the cytosol of mite cells in various organs and are efficiently inherited by their offspring through transovarial transmission.1–5 Reverse transfer from infected animals to mites occurs infrequently, and the bacteria transmitted in this way are not usually passed on to offspring. Thus, limited lines of mites retain the bacterium,6–8 and a correlation is observed between the species of host mite and the serotype of the colonizing OT strain.9
Strain typing of OT was first made on the basis of antigenic variation (serotyping), but a universal scheme for serotyping has not yet been established. Later, on the basis of nucleotide (nt) sequence variation in the gene encoding a major outer membrane protein called the 56-kDa Orientia type-specific antigen (TSA), Tamura and his collaborators divided OT strains into the following subtypes: Gilliam, JG (Japanese Gilliam), Karp, JP-1 (Japanese Karp type-1), JP-2 (Japanese Karp type-2), Kato, Kawasaki, Kuroki, Shimokoshi and others.10,11 This typing system is widely used for epidemiological studies in Japan. Importantly, Ohashi et al.11 determined the 50% mouse lethal doses (MLD50) for representative strains of each subtype and found that strains from subtypes Kato, JG, Gilliam and Karp exhibit high-level virulence to mice, that from JP-2 intermediate-level virulence and those from JP-1, Kawasaki, Kuroki and Shimokoshi low-level virulence. The factors responsible for the strain-to-strain (or serotype-to-serotype) difference of mouse MLD50 have not yet been identified.
The complete genome sequences of two OT strains have recently been determined. Cho et al. sequenced strain Boryong, which was isolated from a patient in Korea,12 and our group sequenced strain Ikeda, which was isolated in 1979 from a patient in Niigata prefecture, Japan.13 Ikeda is a representative strain of subtype JG and thus highly virulent in mice.11 Genomic analysis of the two OT strains revealed that extensive reductive genome evolution as well as explosive and comprehensive amplification of repetitive sequences have occurred in OT. In both strains, repetitive sequences occupy nearly half the genome. Through intensive analysis of the repetitive sequences identified in Ikeda, we categorized them into three types13: (i) an integrative and conjugative element (ICE) named ‘OT amplified genetic element’ (OtAGE); (ii) transposable elements (TEs) and (iii) short repetitive sequences of unknown origins (short repeats). TEs included five types of insertion sequence (IS), four types of miniature inverted-repeat TEs (MITEs) and a Group II (GII) intron.13 The results of our preliminary analysis suggested that extensive genome rearrangements mediated by the repetitive sequences have taken place between the two strains. However, because of the highly complex and repeat-rich feature of the OT genomes, details of the genomic differences between the two strains remain to be clarified.
In the present study, we performed fine comparison of the Ikeda and Boryong genomes to identify the common and variable genomic features among OT strains. In addition, using 11 genes that are conserved in OT and closely related Rickettsia species, we executed multilocus sequencing (MLS) analysis of 10 OT strains representing each TSA subtype to reveal the precise phylogenetic relationship of the strains and examined the distribution of strain-specific sequences identified in Ikeda or Boryong among the OT strains.
2. Materials and methods
2.1. Bacterial strains and DNA preparation
Strains used in this study are listed in Table 1. Nine strains other than Boryong represent each of the nine TSA subtypes, and their virulence in mice was analyzed in a previous study.11 Bacterial cells were inoculated onto confluent monolayers of L929 cells grown in maintenance medium (MEM medium supplemented with 1% FBS, 0.075% NaHCO3, 0.03% glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin and 0.25 μg/ml amphotericin B) and incubated at 37°C for 7 days in 5% CO2. Strain Matsuzawa was the exception in that it was incubated for 14 days because of its slow growth rate. After 7 or 14 days of cultivation, infected L929 cells were scraped from culture dishes, and the supernatant and L929 cells were separated by centrifugation at 200 g for 5 min. The cell pellet was resuspended in the maintenance medium and homogenized gently with a glass Dounce homogenizer (GPE Scientific Ltd). The cell homogenate was re-mixed with the supernatant and centrifuged at 200 g for 10 min to remove the host cell debris. Bacterial cells were collected by centrifugation at 20 000 g for 10 min and suspended in PBS for genomic DNA extraction with the DNeasy Blood and Tissue kit (Qiagen). To reconfirm the subtype of each strain, the TSA gene was amplified by polymerase chain reaction (PCR) and sequenced using primers listed in Supplementary Table S1. JP-1 and JP-2 strains were distinguished by restriction fragment length polymorphism (RFLP) of the PCR-amplified product of the TSA gene,11 and the TSA gene of strain 423H (subtype JP-2) had not been determined previously. Therefore, based on the RFLP pattern deduced from the 423H sequence, we confirmed that our strain belongs to the JP-2 subtype. The TSA gene sequence of strain Matsuzawa showed a significant number of one-base mismatches to that in the database (AF173043.1). Therefore, we resequenced the entire TSA gene of strain Matsuzawa and deposited it in the DDBJ/EMBL/GenBank databases (accession no. AB534164). We used this new sequence for our analysis.
Table 1.
OT strains used in this study
| Strain name | Subtypea | Geographic origin | Year of isolation | Ref. |
|---|---|---|---|---|
| Ikeda | JG | Japan/Niigata pref. | 1979 | 28 |
| Boryong | not defined | Korea/Boryong pref. | 1995 | 29 |
| Gilliam | Gilliam | Myanmar | 1943 | 30 |
| Kato | Kato | Japan/Niigata pref. | 1955 | 31 |
| Karp | Karp | New Guinea | 1943 | 32 |
| Matsuzawa | JP-1 | Japan/Niigata pref. | 1984 | 10 |
| 423H | JP-2 | Japan/Niigata pref. | 1984 | 11 |
| Kuroki | Kuroki | Japan/Miyazaki pref. | 1981 | 33 |
| Kawasaki | Kawasaki | Japan/Miyazaki pref. | 1981 | 34 |
| Shimokoshi | Shimokoshi | Japan/Niigata pref. | 1980 | 28 |
aSubtypes of each strain are derived from Ohashi et al.11
2.2. Genome comparison of strains Ikeda and Boryong
An all-to-all BLASTN search14 of the protein-coding sequences (CDSs) identified in the two strains was performed to examine the conservation of protein-coding genes in each strain. CDSs annotated as pseudogenes in each genome were excluded from this analysis. CDSs that were not detected, or those with different sizes, were further examined by CLUSTALW.15
To identify repetitive sequences in the two strains with the same criteria, we constructed consensus sequences for each type of repetitive sequence that we previously identified in Ikeda, searched the two genomes for sequences homologous to each consensus using BLASTN and manually inspected each sequence. Thus, the positions of some repetitive sequences in Ikeda differ slightly from those described in the previous report.13 The repetitive sequence-free genome sequences (RSFGSs) of each strain were obtained by removing the repetitive sequences from the entire genome sequence. Regions where the linearity of the genome sequence is conserved between the two strains were identified by Harplott analysis of the RSFGSs using the GenomeMatcher ver. 1.11 software.16 These regions (>1 kb) were defined as linearity-conserved regions (LCRs) and were used to analyze genome rearrangement. Strain-specific sequences were identified by reciprocal BLASTN analysis of the RSFGSs. Sequences specific to each strain (>1 kb) were defined as Ikeda- or Boryong-specific sequences (Ike_spe1 to Ike_spe19 and Bor_spe1 to Bor_spe19, respectively). To search additional Boryong-specific repetitive sequences, a self-to-self BLASTN search was performed for all Boryong-specific sequences (>100 bp).
2.3. PCR scanning
Distribution of the Ikeda- and Boryong-specific sequences among OT strains was examined by PCR scanning analysis17 using the LA Long PCR kit (Takara) and 20 ng genomic DNA as template. Each strain-specific sequence (>1 kb) was examined by two pairs of scanning primers (F1/R1 and F2/R2) except for Ike_spe6, which was examined using six primer pairs. When only one PCR product was obtained, the second PCR using different primer combinations (F1/R2 and F2/R1) was performed. The sequences of scanning primers are listed in Supplementary Table S1.
2.4. MLS analysis
Internal segments of the following 11 housekeeping genes were amplified from each of the eight strains (Gilliam, Karp, Matsuzawa, 423H, Kato, Kawasaki, Kuroki and Shimokoshi) by PCR using the LA Long PCR kit and sequenced using the ABI3730 sequencer (Applied Biosystems): atpD (coding for ATP synthase beta-chain), clpX (ATP-dependent Clp protease ATP-binding subunit), dnaJ (DnaJ protein), dnaK (DnaK protein), fabD (malonyl CoA-acyl carrier protein transacylase), gyrB (DNA gyrase subunit B), icd (isocitrate dehydrogenase), mdh (malate dehydrogenase), nrdA (ribonucleoside-diphosphate reductase alpha-chain), sucD (succinyl-CoA synthetase alpha-chain) and ubiD (3-octaprenyl-4-hydroxybenzoate carboxy-lyase). Amplification and sequencing primers are listed in Supplementary Table S1. The nt sequences obtained from each strain were concatenated (5247 bp) and used for MLS analysis. Phylogenetic trees were constructed by the neighborhood-joining method under the Tamura–Nei model using MEGA4 in the Molecular Evolutionary Genetics Analysis software.18 All sequences used for MLS analysis have been deposited in the DDBJ/EMBL/GenBank database with the accession numbers AB537240–AB537327.
3. Results and discussion
3.1. Comparison of general genomic features between strains Ikeda and Boryong
Both strains have a single circular chromosome and possess no plasmid. The chromosomes are very similar in size (2 008 987 bp in Ikeda and 2 127 051 bp in Boryong) with almost identical average G + C contents (30.5% in both strains). The numbers of rRNA and tRNA genes are identical. The numbers of protein-coding genes and pseudogenes, the coding content (%) and the repeat content (%) were unable to be directly compared as the annotation criteria were different between the two genomes.
In conventional GC skew analysis, no clear skew in leading and lagging strands was observed in either genome. Therefore, we reassigned the origin of replication (oriC) using Ori-Finder.19 The oriC region was found to lie between the rho and hemE genes, which are located from nt 539552 to nt 541039 in Ikeda and nt 996058 to nt 997543 in Boryong.
3.2. Comparison of repetitive sequences between strains Ikeda and Boryong
Because the large number of repetitive sequences in both strains makes comparison of their genome sequences difficult, we first identified all repetitive sequences for the two strains under the same criteria, established on the basis of the results of our previous analysis.13 We found that Boryong contains all types of repetitive sequence identified in Ikeda (Table 2). No additional repetitive sequence was found in Boryong. Total lengths of repetitive sequences are 851.6 kb (42.4% of the total genome) in Ikeda and 911.9 kb (42.9%) in Boryong. In both strains, the repetitive sequences are scattered throughout the genome (Fig. 1A). OtAGEs and TEs, the main components of the repetitive sequences, often form small clusters in which one element is inserted into another. Because of such complexity, we did not determine the exact copy number for each type of repetitive sequence in Boryong.
Table 2.
Repeated sequences of OT strains IKEDA and Boryong.a
| Strain Ikeda | Strain Boryong | |
|---|---|---|
| OtAGEs | 578 383 (28.79) | 597 432 (28.09) |
| Transposable elementsb | ||
| ISOts1 | 47 090 (2.34) | 91 682 (4.31) |
| mISOt1 | 8415 (0.42) | 15 468 (0.73) |
| ISOts2 | 9621 (0.48) | 9848 (0.46) |
| mISOt2a | 4455 (0.22) | 5921 (0.28) |
| mISOt2b | 6043 (0.3) | 4364 (0.21) |
| ISOts3 | 25 509 (1.27) | 3944 (0.19) |
| ISOts4 | 43 077 (2.14) | 46 343 (2.18) |
| mISOt4 | 11 442 (0.57) | 17 842 (0.84) |
| ISOts5 | 45 824 (2.28) | 20 328 (0.96) |
| GIIOt1 | 55 506 (2.76) | 79 494 (3.74) |
| Total | 256 982 (12.79) | 295 234 (13.88) |
| Short Repeats | ||
| SROt1 | 10 613 (0.53) | 13 890 (0.65) |
| SROt2 | 1006 (0.05) | 1046 (0.05) |
| SROt3 | 1432 (0.07) | 1429 (0.07) |
| SROt4 | 1246 (0.06) | 1080 (0.05) |
| SROt5 | 693 (0.03) | 495 (0.02) |
| SROt6 | 875 (0.04) | 837 (0.04) |
| SROt7 | 366 (0.02) | 409 (0.02) |
| Total | 16 231 (0.81) | 19 186 (0.9) |
| Total | 851 596 (42.39) | 911 852 (42.87) |
aValues given in parentheses are the ratio to the total genome length (%).
bNames of the IS elements have been changed from ISOt1 ∼ 5 to ISOts1 ∼ 5 according to the suggestions from LMGE (the organizer of ISFinder: http://www-is.biotoul.fr/).
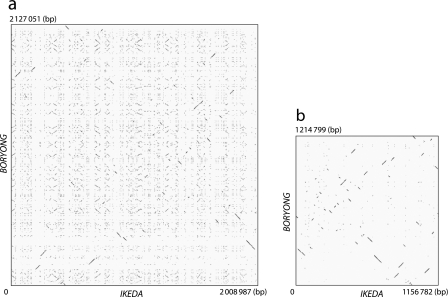
Figure 1.
Extensive shuffling of OT genomes induced by explosively amplified repetitive sequences. (a) Dot plot representation of DNA sequence homology between Ikeda and Boryong chromosomes. Explosively amplified repetitive sequences are scattered throughout the genomes in both strains, and it is therefore difficult to identify regions with colinearity between the two genomes. (b) Dot plot representation of DNA sequence homology for the repetitive sequence-free genomic sequences (RSFGSs) of the two strains. The RSFGSs were constructed by removing all repetitive sequences from each genome. By comparing the RSFGSs, extensive rearrangements of the core genome of OT can be clearly visualized. Each dot represents a high-scoring segment pair of E-value <1E−200, which was identified by BLASTN. The GenomeMatcher ver. 1.11 software16 was used for BLASTN analysis and visualization of the results.
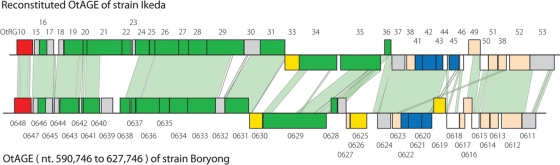
As in Ikeda, OtAGEs have been extensively amplified in Boryong (Table 2). The total length of OtAGE is 578.4 kb in Ikeda (28.8% of the total genome) and 597.4 kb in Boryong (28.1%). In Ikeda, various degrees of decay were observed in all OtAGEs, and a large number of fragmented OtAGEs have been scattered throughout the genome. We could, however, reconstitute a possible genetic organization of original OtAGE by comparing fragmented elements.13 Although extensive decay has also occurred in the OtAGEs of Boryong, we found one copy of OtAGE very similar to the reconstituted OtAGE of strain Ikeda (Fig. 2).13 However, duplication of the attachment sequence, which must have been generated upon integration, was not detected on the element, suggesting that some deletion has occurred at either or both ends of the element. Several IS insertions and genomic deletions were also observed in the element. Thus, this element of Boryong appears to be no more functional as an ICE.
Figure 2.
Genetic organizations of the reconstituted OtAGE of strain Ikeda and the largest OtAGE found in strain Boryong. Both strains contain a large number of OtAGEs, but extensive decay has occurred in all OtAGE copies. A possible original structure of Ikeda OtAGE was reconstituted by comparing larger fragments in our previous report.13 The Ikeda OtAGE shown in this figure was constructed by combining OtAGE_178 and OtAGE_40. Genes for conjugal transfer are depicted by green boxes, those for integrase by red, those for transposase by yellow, those for ATPase domain-containing proteins by blue, conserved hypothetical genes by white and OT-specific hypothetical genes by gray. All genes in the OtAGEs are drawn to the scale. Numbers above the reconstituted Ikeda OtAGE indicate family numbers of OtRG families. Numbers below the Boryong OtAGE indicate CDS numbers in Boryong. Homologous regions indicated by light green shading were identified by the BLASTN program (E-value threshold: ≤1E−10). Note that the Boryong OtAGE contains several deletions and IS insertions, suggesting that this largest OtAGE copy is also defective.
Although Ikeda and Boryong share the same sets of TEs and the total lengths of the TEs are nearly equivalent (257.0 kb and 12.8% of the total genome in Ikeda and 295.2 kb and 13.9% in Boryong), amounts of each type of TE differ significantly between the strains (Table 2). This result suggests that intensive amplification and decay of these elements have occurred in each strain after their divergence.
All types of short repeats identified in Ikeda are also present in Boryong. However, in contrast to the TEs, total lengths of the short repeats and amounts of each type of short repeat are very similar in the two strains (Table 2).
3.3. Extensive repetitive sequence-mediated genome shuffling
By removing all repetitive sequences from each genome, we constructed RSFGSs of the two strains. The lengths of the RSFGSs of Ikeda and Boryong are 1156.8 and 1214.8 kb, respectively. The result of Harplott analysis of the RSFGSs clearly indicated that an extremely high degree of genome shuffling has occurred between the OT strains (Fig. 1). This genome shuffling was obviously induced by extensively amplified repetitive sequences. By comparing the RSFGSs of the two strains, we identified 66 LCRs of >1 kb in size, where the linearity of the genome sequence is conserved between the two strains (positions of all LCRs in each genome are listed in Supplementary Table S2). The average length of these LCRs is 16.0 kb, with the longest being ∼46 kb.
3.4. Strain-specific sequences
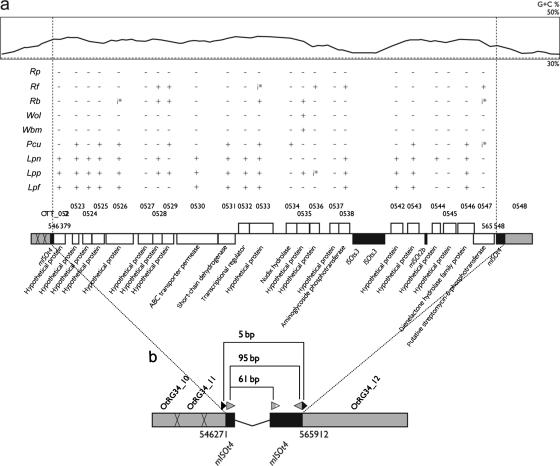
In the RSFGSs, we identified 19 Ikeda-specific (Ike_spe1 to Ike_spe19) and 19 Boryong-specific (Bor_spe1 to Bor_spe19) sequences of >1 kb in length (Supplementary Table S3 and Fig. 3). These sequences were not found in other bacteria of the family Rickettsiaceae. Several sequences have been duplicated in each genome. Their duplications appear to have been induced by repetitive sequences that flank these sequences (Supplementary Fig. S1). Total lengths of the Ikeda- and Boryong-specific sequences are 52.4 (average 2.8) and 40.8 (average 2.1) kb, respectively. Among these sequences, Ike_spe6 is particularly interesting. At 19.2 kb, this is the longest of the strain-specific sequences, and it shows a higher G + C content compared with other parts of the Ikeda genome and is flanked by ISOts4 and mISOts4, forming a composite transposon-like structure with 95-bp-long terminal inverted repeats (Fig. 4). A direct repeat of 5 bp, which was most likely created upon insertion (target sequence duplication), was also identified. Many of the 26 genes encoded in Ike_spe6 have the closest homologs in Legionella spp. and Parachlamydia spp. A few genes have homologs in Rickettsia species, but their sequence similarity to the Ike_spe6 genes was significantly lower than to homologs in Legionella and Parachlamydia, suggesting that Ikeda may have acquired this sequence by horizontal gene transfer from related bacterial species. Intriguingly, both Legionella and Parachlamydia are intracellular microorganisms, and their main niches in natural environments are thought to be amoebas. There are some articles suggesting the relationship between the order Rickettsiales and amoebae.20–22 The presence of Ike_spe6 may provide the new evidence showing that an amoeba may be involved in the life cycle of OT as well.
Figure 3.
Distribution of Ikeda- and Boryong-specific sequences among other OT strains. The presence of each strain-specific region (>1 kb) was examined by PCR scanning using two pairs of PCR primers, except for Ike_spe6, which was examined using six pairs of primers. Regions present in each strain are indicated by black cells with white plus signs. Regions where only one primer pair yielded a PCR product are indicated by gray cells with black plus signs. Regions yielding PCR products with different sizes compared with those of the reference strain (Ikeda or Boryong) are also indicated by gray cells. In Ike_spe6 of strain Kato, five primer pairs yielded PCR products. Phylogenetic relationships of the OT strains examined are also indicated (Fig. 5).
Figure 4.
The genetic organization of Ikeda_spe6 and distribution of genes homologous to CDSs on Ike_spe6. Ike_spe6 is the longest strain-specific sequence found in the Ikeda genome. In (a), the G + C content of this region (top part; the average G + C content of the Ikeda genome is indicated by a broken line), distribution of homologs for Ike_spe6-encoded genes (middle part) and the genetic organizations of Ike_spe6 (bottom part) are shown. White, gray and black boxes indicate singleton genes, repeated genes and IS elements (or MITEs), respectively. Species and strain designations are as follows: Rp, Rickettsia prowazekii strain Madrid E; Rf, Rickettsia felis strain URRWXCal2; Rb, Rickettsia bellii strain RML369-C; Wol, Wolbachia pipientis strain wMel; Wbm, Wolbachia sp. strain TRS; Pcu, Candidatus Protochlamydia amoebophila strain UWE25; Lpn, Legionella pneumophila strain Philadelphia-1; Lpp, Legionella pneumophila strain Paris; Lpf, Legionella pneumophila strain Lens. The presence of pseudogenes is indicated by ‘i*’. In (b), the genetic organization of the insertion site for Ike_spe6 is shown. Ike_spe6 is flanked by two mISOts4 elements (ISOts4-related MITE), thus displaying a composite transposon (Tn)-like structure with terminal inverted repeats of 95 bp. This Tn-like element is integrated into OtRG34, creating a target site duplication of 5 bp (black arrowheads).
3.5. Comparison of gene content between strains Ikeda and Boryong
3.5.1. Repeated genes
In our previous analysis of the Ikeda genome,13 we defined repeated gene families as the genes whose products exhibited at least 90% amino acid sequence identity over 60% of the alignment length. According to this definition, we found 1196 repeated genes that were classified into 85 OT repeated gene (OtRG) families. OtRG1–OtRG9 are genes in TEs, and OtRG10–OtRG54 are those in OtAGEs. Thus, these families are also conserved in Boryong. Other repeated genes (OtRG55–OtRG85) are not components of repetitive elements, but their frequent association with repetitive sequences suggested that they have been duplicated or multiplexed in conjunction with amplification of repetitive sequences.13 In support of this hypothesis, copy numbers for many of these OtRGs differ significantly between the two strains (Supplementary Table S4). At least six genes have duplicated in Ikeda but not in Boryong. In addition, several repeated gene families are present in only one strain (three families specific to Ikeda and two families specific to Boryong). Extensive gene decay has also taken place in many Boryong-repeated genes as in those of Ikeda.
3.5.2. Singleton genes
We previously found that, among the 1967 CDSs of Ikeda, 771 are singleton genes and 543 of the 771 genes are shared with R. prowazekii. Because most OT/R. prowazekii shared genes (520 genes) are conserved in all five sequenced Rickettsia species, they appear to represent the core gene set of the family Rickettsiaceae. As expected, 541 of the 543 shared genes are conserved in Boryong (Table 3). No other Boryong singleton gene was conserved in R. prowazekii. Genomic locations of these 541 genes have been extensively shuffled between the two OT genomes, but the average nt sequence identity of these genes is 97.5%.
Table 3.
Conservation of Ikeda singleton genes in strain Boryong
| Ikeda singleton gene | Strain Boryong |
||
|---|---|---|---|
| Present | Absent | ||
| Ikeda (+)/R. prowazekii (+) | 543a (84)b | 541 (83) | 2 (1) |
| Present in all the five Rickettsia sp.c | 520 (80) | 519 (80) | 1 (0) |
| Variably present in the five Rickettsia sp. | 23 (4) | 22 (3) | 1 (1) |
| Ikeda (+)/R. prowazekii (−) | 195 (149) | 124 (91) | 71 (58) |
| Absent in all the five Rickettsia sp. | 161 (131) | 96 (77) | 65 (54) |
| Variably present in the five Rickettsia sp. | 34 (18) | 28 (14) | 6 (4) |
aNumber of shared genes has been changed from 542 to 543, because OTT_0717 was added to this gene set after re-inspection.
bNumbers of pseudogenes are indicated in parentheses.
cThe five Rickettsia species examined here include R. prowazekii, R. conorii, R. felis, R. typhi and R. bellii.
Two of the 543 OT/R. prowazekii shared genes are not conserved in Boryong: the atpF gene encoding the beta-subunit of ATP synthase F0 sector (OTT_0385 in Ikeda) and a gene encoding a hypothetical protein containing ankyrin repeats (OTT_1381). The atpF gene of Boryong contains a frameshift mutation, and thus the N-terminal 47 amino acids are missing. This mutation may be due to a sequence error. OTT_1481 is located in an Ikeda strain-specific sequence (Ike_spe13) that is flanked by OtAGEs (Supplementary Fig. S2). Boryong may have lost this gene by the OtAGE-mediated deletion of Ike_spe13. Conversely, one of the 543 OT/R. prowazekii shared genes (the tryptophanyl-tRNA synthetase gene) has been duplicated in Boryong (OTBS_0762 and OTBS_1824). ISOts1 and GIIOt1 flanking these genes suggest that this gene duplication has also been induced by TEs.
In addition to the 543 shared genes, Ikeda possesses 195 singleton genes. Among these, 124 are present in Boryong but 71 are absent (Supplementary Table S5). Of the former 124 genes, OTT_0440 (encoding a repeat-containing protein) and OTT_0622 (encoding a hypothetical protein) are duplicated in Boryong. OtAGEs and/or TEs are also located at both the ends of these CDSs (Supplementary Fig. S2), suggesting that their duplications have also been induced by repetitive sequences. Although many Ikeda-specific singleton genes (58 out of 71) are of unknown function, these genes may have introduced phenotypic differences between the two strains.
Boryong also possesses 74 singleton genes that are not present in Ikeda. Sequences corresponding to 49 of these genes are also present in Ikeda, but they were not annotated as CDSs under our annotation criteria. The remaining 25 Boryong-specific genes are of unknown function.
Another interesting finding is that, among 27 disrupted Ikeda singleton genes, 23 have also been disrupted and 4 were not identified in the Boryong genome (Supplementary Table S6). This finding may suggest that inactivation of these genes took place before the divergence of the two strains.
3.6. Lineage analysis of OT strains by MLS analysis
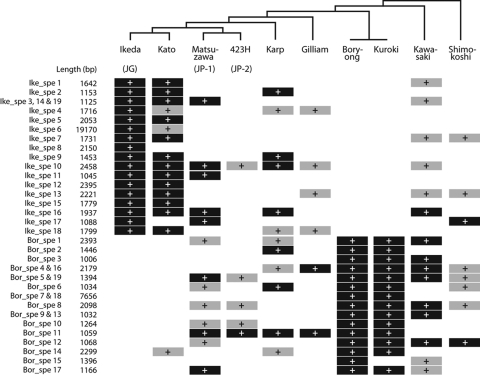
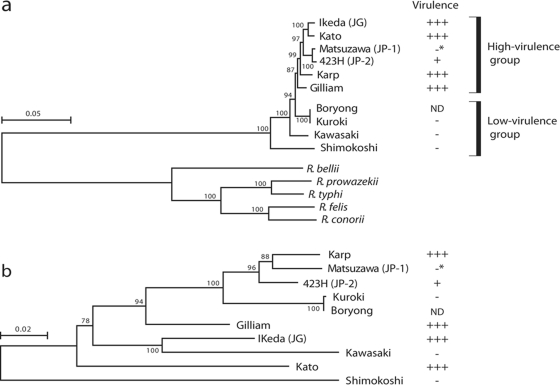
Partial nt sequences of the TSA gene were used for lineage analysis of OT strains.10 The TSA gene encodes a major surface protein. It exhibits remarkable strain-to-strain sequence variation and contains four variable domains. Phylogenetic trees constructed using each domain sequence are not consistent between the domains.23 Thus, the sequence of this gene is not suitable for phylogenetic analysis of OT strains. Therefore, we selected 11 genes among the core gene set of the family Rickettsiaceae and performed MLS analysis of various OT strains using concatenated sequences of these 11 genes. Strains examined in this analysis included nine OT strains (Ikeda, Gilliam, Kato, Karp, Matsuzawa, 423H, Kuroki, Kawasaki and Shimokoshi) representing each of the known nine TSA subtypes. Strain Boryong was also included in this analysis. The phylogenetic tree obtained by this MLS analysis differed considerably from that generated using TSA gene sequences and probably represents the true phylogenetic relationship among OT strains (Fig. 5). Strain Boryong, which can be classified as the Kuroki subtype with TSA-based subtyping, has the same MLS type as strain Kuroki.
Figure 5.
Phylogenetic relationships of OT strains representing each known TSA subtype. (a) Phylogenetic tree constructed using the concatenated nt sequences of 11 housekeeping genes. The 11 genes used were selected from the core genes of five Rickettsia species: R. prowazekii, R. conorii, R. felis, R. typhi and R. bellii. The reliability of the internal branches was assessed by bootstrapping with 250 pseudoreplicates. Strains with higher virulence to mice clustered together in this phylogram. Virulence of each strain to mice was categorized according to their 50% mice lethal doses (MLD50), which were reported by Ohashi et al.11: +++, MLD50 (log10) > 2.0; ++, 2.0 > MLD50 > 1.0; +, 1.0 > MLD50 > 0; -, all alive. The strain Matsuzawa does not show higher virulence to mice (indicated by asterisks), but this may be due to the slow growth of this strain. The virulence of strain Boryong to mice has not been determined. (b) Phylogenetic tree constructed using the sequences of TSA genes, which encode an OT-specific surface protein of 56 kDa. The scale bar represents the number of substitutions per site.
All OT strains used in this MLS analysis (except for Boryong) were previously examined for virulence in mice by Ohashi et al.11 In the new phylogram, strains that showed higher virulence to mice (strains Ikeda, Kato, Karp, 423H and Gilliam) clustered together. The sole exception is strain Matsuzawa belonging to the JP-1 type, which may be due to its slow growth in mammalian cells. This result suggests the presence of an OT lineage with higher virulence to mice.
Although the natural hosts of each OT strains are not fully understood, hosts for the OT strains of higher virulence are mainly Leptotrombidium akamushi and L. pallidum, whereas the hosts for the other strains are L. scutellare and some other Leptotrombidium spp.11,24–27 This host specificity may be linked to the generation of such sublineages of OT. In Japan, many severe scrub typhus cases previously occurred only in the several local areas where L. akamushi is predominantly distributed, and the causative agents of these diseases (so-called classic-type Tsutsugamushi diseases) were the strains of serotype Kato. Recently, many scrub typhus cases (several hundred cases per year) are occurring in other areas of Japan, where L. scutellare and L. pallidum are densely distributed. Most of these new-type Tsutsugamushi diseases are caused by strains of serotypes Kawasaki (the natural host is L. scutellare) and Kuroki (unknown), and clinical symptoms of these diseases are generally milder than that of the classic type.27 Our findings may at least partly explain these clinical and epidemiological differences between the classic- and new-type Tsutsugamushi diseases.
Because strain Boryong has the same MLS type as that of strain Kuroki which belongs to the lower virulence group,11 it is possible that some of the Ikeda-specific sequences (or genes on the sequences) could be responsible for the virulence difference between the two groups. Therefore, we examined the distribution of 19 Ikeda- and 19 Boryong-specific sequences among the 10 OT strains by PCR scanning (Fig. 3). The result of this analysis indicates that the distribution patterns of these sequences largely followed the phylogeny of each strain (Fig. 3). For example, an intact copy of the above-mentioned Ike_spe6 was found only in strain Kato, which is most closely related to Ikeda. The insertion site of Ike_spe6 in strain Kato is the same as that in Ikeda (data not shown). However, none of the Ikeda-specific sequences showed a higher virulence group-specific distribution (shared by all strains of the higher virulence group but absent in the lower virulence group). Thus, the factors responsible for the virulence difference between the two groups still remain to be elucidated.
3.7. Conclusion
Fine genome sequence comparison of two recently sequenced OT strains revealed that they contain the same set of repetitive sequences and that these repetitive sequences have been amplified explosively in both strains and have induced extensive genome shuffling. In addition, the core gene set of family Rickettsiaceae is highly conserved between the two strains. On the basis of the results of MLS analysis, we present a phylogram that represents a true phylogenetic relationship among OT strains. This analysis further revealed the presence of an OT sublineage with higher potential virulence, which might explain the clinical and epidemiological differences between the classic and new types of Tsutsugamushi disease in Japan.
Supplementary Data
Supplementary data are available at www.dnaresearch.oxfordjournals.org.
Funding
This work was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan and the Ministry of Health, Labor and Welfare of Japan, and partly by a grant from Miyazaki Prefecture.
Supplementary Material
Acknowledgements
We thank Yoshiro Terawaki and Akira Tamura for their encouragement and Akemi Yoshida, Yumiko Takeshita, Noriko Kanemaru, Miki Shinbara, and Nobuko Fujii for their technical assistance.
Footnotes
Edited by Katsumi Isono
References
- 1.Rai J., Bandopadhyay D. Vertical transmission in chigger borne rickettsiosis. Indian J. Med. Res. 1978;68:31–8. [PubMed] [Google Scholar]
- 2.Rapmund G., Dohany A.L., Manikumaran C., Chan T.C. Transovarial transmission of Rickettsia tsutsugamushi in Leptotrombidium (Leptotrombidium) arenicola Traub (Acarina: Trombiculidae) J. Med. Entomol. 1972;9:71–2. doi: 10.1093/jmedent/9.1.71. [DOI] [PubMed] [Google Scholar]
- 3.Rapmund G., Upham R.W., Jr, Kundin W.D., Manikumaran C., Chan T.C. Transovarial development of scrub typhus rickettsiae in a colony of vector mites. Trans. R. Soc. Trop. Med. Hyg. 1969;63:251–8. doi: 10.1016/0035-9203(69)90155-2. doi:10.1016/0035-9203(69)90155-2. [DOI] [PubMed] [Google Scholar]
- 4.Takahashi M., Murata M., Nogami S., Hori E., Kawamura A., Tanaka H. Transovarial transmission of Rickettsia tsutsugamushi in Leptotrombidium pallidum successively reared in the laboratory. Jpn J. Exp. Med. 1988;58:213–8. [PubMed] [Google Scholar]
- 5.Urakami H., Takahasi M., Hori E., Tamura A. An ultrastructural study of vertical transmission of Rickettsia tsutsugamushi during oogenesis and spermatogenesis in Leptotrombidium pallidum. Am. J. Trop. Med. Hyg. 1994;50:219–28. doi: 10.4269/ajtmh.1994.50.219. [DOI] [PubMed] [Google Scholar]
- 6.Takahashi M., Murata M., Misumi H., Hori E., Kawamura A., Tanaka H. Failed vertical transmission of Rickettsia tsutsugamushi (Rickettsiales: Rickettsiaceae) acquired from rickettsemic mice by Leptotrombidium pallidum (Acari: Trombiculidae) J. Med. Entomol. 1994;31:212–6. doi: 10.1093/jmedent/31.2.212. [DOI] [PubMed] [Google Scholar]
- 7.Walker J.S., Chan C.T., Manikumaran C., Elisberg B.L. Attempts to infect and demonstrate transovarial transmission of R. tsutsugamushi in three species of Leptotrombidium mites. Ann. N Y Acad. Sci. 1975;266:80–90. doi: 10.1111/j.1749-6632.1975.tb35090.x. doi:10.1111/j.1749-6632.1975.tb35090.x. [DOI] [PubMed] [Google Scholar]
- 8.Traub R., Wisseman C.L., Jones M.R., O'Keefe J.J. The acquisition of Rickettsia tsutsugamushi by chiggers (trombiculid mites) during the feeding process. Ann. N Y Acad. Sci. 1975;266:91–114. doi: 10.1111/j.1749-6632.1975.tb35091.x. doi:10.1111/j.1749-6632.1975.tb35091.x. [DOI] [PubMed] [Google Scholar]
- 9.Kelly D.J., Fuerst P.A., Ching W.M., Richards A.L. Scrub typhus: the geographic distribution of phenotypic and genotypic variants of Orientia tsutsugamushi. Clin. Infect. Dis. 2009;48:S203–30. doi: 10.1086/596576. doi:10.1086/596576. [DOI] [PubMed] [Google Scholar]
- 10.Enatsu T., Urakami H., Tamura A. Phylogenetic analysis of Orientia tsutsugamushi strains based on the sequence homologies of 56-kDa type-specific antigen genes. FEMS Microbiol. Lett. 1999;15:163–9. doi: 10.1111/j.1574-6968.1999.tb08791.x. [DOI] [PubMed] [Google Scholar]
- 11.Ohashi N., Koyama Y., Urakami H., et al. Demonstration of antigenic and genotypic variation in Orientia tsutsugamushi which were isolated in Japan, and their classification into type and subtype. Microbiol. Immunol. 1996;40:627–38. doi: 10.1111/j.1348-0421.1996.tb01120.x. [DOI] [PubMed] [Google Scholar]
- 12.Cho N.H., Kim H.R., Lee J.H., et al. The Orientia tsutsugamushi genome reveals massive proliferation of conjugative type IV secretion system and host-cell interaction genes. Proc. Natl Acad. Sci. USA. 2007;8:7981–6. doi: 10.1073/pnas.0611553104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakayama K., Yamashita A., Kurokawa K., et al. The whole-genome sequencing of the obligate intracellular bacterium Orientia tsutsugamushi revealed massive gene amplification during reductive genome evolution. DNA Res. 2008;15:185–99. doi: 10.1093/dnares/dsn011. doi:10.1093/dnares/dsn011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Altschul S.F., Madden T.L., Schaffer A.A., et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. doi:10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pearson W.R., Lipman D.J. Improved tools for biological sequence comparison. Proc. Natl Acad. Sci. USA. 1988;85:2444–8. doi: 10.1073/pnas.85.8.2444. doi:10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ohtsubo Y., Ikeda-Ohtsubo W., Nagata Y., Tsuda M. GenomeMatcher: a graphical user interface for DNA sequence comparison. BMC Bioinformatics. 2008;16:376. doi: 10.1186/1471-2105-9-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ohnishi M., Terajima J., Kurokawa K., et al. Genomic diversity of enterohemorrhagic Escherichia coli O157 revealed by whole genome PCR scanning. Proc. Natl Acad. Sci. USA. 2002;99:17043–8. doi: 10.1073/pnas.262441699. doi:10.1073/pnas.262441699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar S., Nei M., Dudley J., Tamura K. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform. 2008;9:299–306. doi: 10.1093/bib/bbn017. doi:10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao F., Zhang C.T. Ori-Finder: a web-based system for finding oriCs in unannotated bacterial genomes. BMC Bioinformatics. 2008;1:79. doi: 10.1186/1471-2105-9-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fritsche T.R., Horn M., Seyedirashti S., et al. In situ detection of novel bacterial endosymbionts of Acanthamoeba spp. phylogenetically related to members of the order Rickettsiales. Appl. Environ. Microbiol. 1999;65:206–12. doi: 10.1128/aem.65.1.206-212.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ogata H., La Scola B., Audic S., et al. Genome sequence of Rickettsia bellii illuminates the role of amoebae in gene exchanges between intracellular pathogens. PLoS Genet. 2006;2:733–44. doi: 10.1371/journal.pgen.0020076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmitz-Esser S., Tischler P., Arnold R., et al. The genome of the amoeba symbiont ‘Candidatus Amoebophilus asiaticus’ reveals common mechanisms for host cell interaction among amoeba-associated bacteria. J. Bacteriol. 2010;194:1045–57. doi: 10.1128/JB.01379-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohashi N., Nashimoto H., Ikeda H., Tamura A. Diversity of immunodominant 56-kDa type-specific antigen (TSA) of Rickettsia tsutsugamushi. Sequence and comparative analyses of the genes encoding TSA homologues from four antigenic variants. J. Biol. Chem. 1992;267:12728–35. [PubMed] [Google Scholar]
- 24.Kawamori F., Akiyama M., Sugieda M., et al. Epidemiology of Tsutsugamushi disease in relation to the serotypes of Rickettsia tsutsugamushi isolated from patients, field mice, and unfed chiggers on the eastern slope of Mount Fuji, Shizuoka Prefecture, Japan. J. Clin. Microbiol. 1992;30:2842–6. doi: 10.1128/jcm.30.11.2842-2846.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kitaoka M., Okubo K., Asanuma K. Epidemiological survey by means of complement fixation test on scrub typhus in Japan. Acta Med. Biol. 1967;15(Suppl):69–85. [PubMed] [Google Scholar]
- 26.Shishido A. Strain variation of Rickettsia orientalis in the complement fixation test. Jpn J. Med. Sci. Biol. 1964;17:59–72. doi: 10.7883/yoken1952.17.59. [DOI] [PubMed] [Google Scholar]
- 27.Tamura A., Yamamoto N., Koyama S., et al. Epidemiological survey of Orientia tsutsugamushi distribution in field rodents in Saitama Prefecture, Japan, and discovery of a new type. Microbiol. Immunol. 2001;45:439–46. doi: 10.1111/j.1348-0421.2001.tb02643.x. [DOI] [PubMed] [Google Scholar]
- 28.Tamura A., Takahashi K., Tsuruhara T., et al. Isolation of Rickettsia tsutsugamushi antigenically different from Kato, Karp, and Gilliam strains from patients. Microbiol. Immunol. 1984;28:873–882. doi: 10.1111/j.1348-0421.1984.tb00743.x. [DOI] [PubMed] [Google Scholar]
- 29.Chang W.H., Kang J.S., Lee W.K., et al. Serological classification by monoclonal antibodies of Rickettsia tsutsugamushi isolated in Korea. J. Clin. Microbiol. 1990;28:685–688. doi: 10.1128/jcm.28.4.685-688.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bengston I.A. Apparent serological heterogeneity among strains of tsutsugamushi disease (scrub typhus) Public Health Rep. 1945;60:1483–1488. [PubMed] [Google Scholar]
- 31.Shishido A., Ohtawara M., Tateno S., et al. The nature of immunity against scrub typhus in mice. I. The resistance of mice, surviving subcutaneous infection of scrub typhus rickettsia, to intraperitoneal reinfection of the same agent. Jpn. J. Med. Sci. Biol. 1958;11:383–399. [Google Scholar]
- 32.Derrick E.H., Brown H.E. Isolation of the Karp strain of rickettsia tsutsugamushi. Lancet. 1949;2:150–151. doi: 10.1016/s0140-6736(49)90996-4. [DOI] [PubMed] [Google Scholar]
- 33.Ohashi N., Tamura A., Sakurai H., Yamamoto S. Characterization of a new antigenic type, Kuroki, of Rickettsia tsutsugamushi isolated from a patient in Japan. J. Clin. Microbiol. 1990;28:2111–2113. doi: 10.1128/jcm.28.9.2111-2113.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamamoto S., Kawabata N., Tamura A., et al. Immunological properties of Rickettsia tsutsugamushi, Kawasaki strain, isolated from a patient in Kyushu. Microbiol. Immunol. 1986;30:611–620. doi: 10.1111/j.1348-0421.1986.tb02988.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.