Abstract
Human artificial chromosomes (HACs), which carry a fully functional centromere and are maintained as a single-copy episome, are not associated with random mutagenesis and offer greater control over expression of ectopic genes on the HAC. Recently, we generated a HAC with a conditional centromere, which includes the tetracycline operator (tet-O) sequence embedded in the alphoid DNA array. This conditional centromere can be inactivated, loss of the alphoidtet-O (tet-O HAC) by expression of tet-repressor fusion proteins. In this report, we describe adaptation of the tet-O HAC vector for gene delivery and gene expression in human cells. A loxP cassette was inserted into the tet-O HAC by homologous recombination in chicken DT40 cells following a microcell-mediated chromosome transfer (MMCT). The tet-O HAC with the loxP cassette was then transferred into Chinese hamster ovary cells, and EGFP transgene was efficiently and accurately incorporated into the tet-O HAC vector. The EGFP transgene was stably expressed in human cells after transfer via MMCT. Because the transgenes inserted on the tet-O HAC can be eliminated from cells by HAC loss due to centromere inactivation, this HAC vector system provides important novel features and has potential applications for gene expression studies and gene therapy.
Keywords: human artificial chromosome, conditional centromere, gene delivery
1. Introduction
Human artificial chromosomes (HACs) are an important alternative to conventional gene-transfer techniques, such as viral, plasmid, P1 phage-derived artificial chromosomes, bacterial artificial chromosomes (BACs) and yeast artificial chromosomes, because they offer greater control over their location and function in recipient human cells. Advantages of HAC vectors include their stability as episomes in human cells, a low level of associated insertional mutagenesis, a low potential for causing cell transformation compared with the conventional vectors1,2 and capacity to carry and facilitate expression of the largest mammalian genes from their native gene regulatory elements.
Two strategies have been used to construct HAC vectors3,4: natural human chromosomes have been modified and reduced in size to generate minichromosomes that maintain the essential functional properties of a eukaryotic chromosome and cloning vector (top-down), or de novo artificial chromosomes have been constructed from synthetic or cloned DNA that provide such essential functions (i.e. alphoid DNA for kinetochore function) in human and other mammalian cells (bottom-up). Top-down HACs have been generated from human chromosomes by telomere-seeding and homologous recombination in chicken DT40 cells.5,6 For example, our group recently constructed a human chromosome 14- and 21-derived HAC vector that replicates and segregates properly during mitotic divisions in vitro and is stably maintained in mice.4–8 The chromosome 21-derived HAC is a powerful gene delivery vector capable of carrying large genes such as the dystrophin gene (2.4 Mb) and may be useful for gene and cell therapy.4,9–11
Bottom-up HACs have been constructed from arrays of synthetic or natural alphoid DNA and BAC vector sequences required for HAC assembly and propagation.12–14 Recently, our group constructed a bottom-up HAC in which ∼6000 copies of a 42-bp tetracycline operator (tet-O) sequence are inserted into a mega-base size synthetic alphoid DNA array. Because tet-O is bound with very high affinity and specificity by the tet repressor (tet-R), the tet-O sequences in the HAC can be targeted with tet-R fusion proteins that inactivate the centromere and induce HAC loss. Thus, the alphoidtet-O HAC hereafter, tet-O HAC has a conditional centromere, that can be turned on or off by co-expression of tet-O binding proteins. This feature of the tet-O HAC was used to target chromatin-modifying proteins into the HAC centromere and alter the balance between open and condensed chromatin, which is critical for the kinetochore function.15,16 In this study, we describe adaptation of the tet-O HAC for gene expression studies.
2. Materials and methods
2.1. Cell culture
HT1080 cells were cultured in Dulbecco's modified Eagle's medium (DMEM, Sigma, St Louis, MD, USA) supplemented with 10% fetal bovine serum (FBS; Biowest, Nuaille, France) with 4 μg/ml of Blasticidin S (BS; Funakoshi, Tokyo, Japan) at 37°C in 5% CO2. The HT1080 and A9 hybrid cells were maintained in DMEM (Sigma) containing 10% FBS (Biowest) and the appropriate antibiotics. The hypoxanthine phosphoribosyltransferase (HPRT)-deficient Chinese hamster ovary (CHO) cells (JCRB0218) were maintained in Ham's F-12 nutrient mixture (Invitrogen, USA) plus 10% FBS with 8 µg/ml of BS (Funakoshi). After introduction of the EGFP gene, the CHO cells retaining the tet-O-EGFP HAC were cultured with 1× HAT medium. Chicken DT40 cells were maintained at 37°C in RPMI 1640 medium supplemented with 10% FBS, 1% chicken serum and 50 µM 2-mercaptoethanol.
2.2. Construction of the targeting vectors
The targeting vector 5–4–3 for introducing a 3′-HPRT-loxP-Hyg-thymidine kinase (Tk) cassette was constructed as follows. The 2.0- and 3.1-kb fragment homologous for BAC vector sequences in the tet-O HAC were amplified by PCR using primers HR1-Fi/Ri 5′-AGAGTTAACGTTACCTTCCACGAGCAAAACACGTA-3′ and 5′- AGAGTTAACCTTGTAGGCCTTTATCCATGCTGGTT-3′; HR3S-F/R 5′-ATACCGCGGGTTCTGTGTTCATTAGGTTGTTCTGT-3′ and 5′-ATACCGCGGTGAAGCGTATATAGGACGAGTAACTG-3′, digested with either HpaI or SacII and subcloned into the equivalent sites of pKO Scrambler V901 backbone vector (Lexicon Genetics, Woodlands, TX, USA), respectively, which contains the following three selection/insertion cassettes between the EcoRI and the BglII sites: a 2-kb fragment from pMC1-Tk (Stratagene, La Jolla, CA, USA) containing the HSV-Tk gene, a 2.3-kb XbaI/AscI fragment from pKO SelectHPRT V820 (Lexicon Genetics) containing a part of the human HPRT gene with the loxP site inserted into the XbaI site of intron B and a 1.3-kb XhoI fragment from the PGK/Hyg vector containing the hygromycin-resistance gene. The transgene sequences are described previously.7
2.3. Fluorescence in situ hybridization
Fluorescence in situ hybridization (FISH) analyses of CHO and DT40 cells were performed with either fixed metaphase or interphase nuclei using digoxigenin-labelled (Roche, Basel, Switzerland) human hCot1 DNA (Invitrogen) and biotin-labelled plasmid DNA, as described previously.7 For FISH analysis of HAC in HT1080 cells, tet-O alpha-satellite DNA was used as a probe.15 Chromosomal DNA was counterstained with DAPI (Sigma). The images were captured using the Argus system (Hamamatsu Photonics, Hamamatsu, Japan) and NIS elements (Nikon, Tokyo, Japan).
2.4. Microcell-mediated chromosome transfer
Microcell-mediated chromosome transfer (MMCT) was performed as described previously.7,10 The tet-O HAC containing the loxP cassette was transferred from DT40 into CHO cells by MMCT technology. Briefly, microcells were prepared by centrifugation of 1 × 109 DT40 cells attached on flasks (Nalge Nunc, Rochester, NY, USA) coated with poly-l-lysine (Sigma) and were fused to 1 × 106 CHO cells by 47% polyethylene glycol 1000 (WAKO, Osaka, Japan). CHO hybrids were selected in the presence of 4 µg/ml of BS and collected for expansion. After construction of the tet-O-EGFP HAC vector by Cre/loxP recombination in CHO cells, the HAC was transferred from CHO into HT1080 cells via MMCT. The HT1080 hybrids were selected in the presence of 4 µg/ml of BS.
2.5. PCR analyses
Genomic DNA from cell lines was extracted using a genomic extraction kit (Gentra System, Minneapolis, MN, USA). PCR analyses were carried out using standard techniques. The primer pairs for detection of the HAC were: BACpr-F/BACpr-R (4075 bp), 5′-CTCTATACACTCAGTTGGAACACGAGAC-3′ and 5′- GAGCCTGTGTAGCGTTTATAGGAAGTAG-3′. The primer pairs for confirmation of targeting of the loxP cassette into the HAC were: HRL-F/TKpAF (4.1 kb), 5′-CAGAGCCAGACAGGAAGGAATAATGTCAAG-3′ and 5′-CCGGACGAACTAAACCTGAC-3′; PGKr-3/HRR-R (3.5 kb), 5′-CCAGAGGCCACTTGTGTA-3′ and 5′-GCATCTCAATTAGTCAGCAACCATAGTCCC-3′. The primer pairs for detection of the HPRT gene reconstruction were: Trans-L1/R1 (409 bp), 5′-TGGAGGCCATAAACAAGAAGAC-3′ and 5′-CCCCTTGACCCAGAAATTCCA-3′. The primer pairs for determination of the EGFP gene were: EGFPL/R (479 bp), 5′-CCTGAAGTTCATCTGCACCA-3′ and 5′-TGCTCAGGTAGTGGTTGTCG-3′. Primers FurinF/R, 5′-ACTCAGAGATCCACTGCACCAGGATCCAAGGGAGG-3′ and 5′-CCGCTCGAGCGGCTACACCACAGACACCATTGTTGGCTACTGCTGCC-3′, were used as an internal control for CHO cells after HAC transfer into human cells via MMCT.
2.6. HAC elimination by its targeting with chromatin modifiers
HT1080 cells expressing the tTS fusion proteins were obtained using a retroviral vector as described previously.15 A retroviral expression vector was transfected into 293FT cells (Invitrogen) with Lipofectamine2000 (Invitrogen). After 4 h, the medium was changed with a fresh medium. After 48 h, the supernatant was collected by centrifugation at 1500 rpm and an aliquot (2–5 µl) was added to HT1080 cells carrying the HAC. The infected cells were cultured with 800 µg/ml of geneticin and 1 µg/ml of doxycycline (dox) for 24 h. After 7 days of culture in the presence of geneticin, an elimination assay was performed under the culture conditions: dox(−) 7 days, dox(+) 7 days and dox(+) 7 days ganciclovir(+) 5 days.
2.7. FACS analysis
Mitotic stability of the HAC carrying the EGFP gene was determined by FACS as described previously.17
2.8. Southern blot analysis
Southern blot hybridization was performed as described previously with a 32P-labelled probe.18 Genomic DNA from cells containing the tet-O HAC was digested by SpeI, CHEF separated, and blot-hybridized with a 201-bp probe specific to a sequence corresponding to the BAC vector region that is repeated 47 times in the HAC. The probe was amplified by PCR using the following 4243/4443 primers 5′-GGGCAATTTGTCACAGGG-3′ and 5′-ATCCACTTATCCACGGGGAT-3′. Blots were incubated for 2 h at 65°C in pre-hybridization solution (0.5 M Na-phosphate buffer containing 7% SDS and 100 mg/ml of unlabelled salmon sperm carrier DNA). Labelled probe (20 ml) was heat denatured in boiling water for 5 min and snap cooled on ice. The probe was added to the hybridization buffer and allowed to hybridize overnight at 65°C. Blots were washed twice in 2× SSC (300 mM NaCl, 30 mM sodium citrate, pH 7.0), 0.1% SDS for 30 min at room temperature, then three times in 0.1× SSC, 0.1% SDS for 30 min at 65°C. Blots were exposed to X-ray film for 24–72 h at −70°C.
3. Results and discussion
3.1. Retrofitting the tet-O HAC with a loxP cassette in DT40 cells
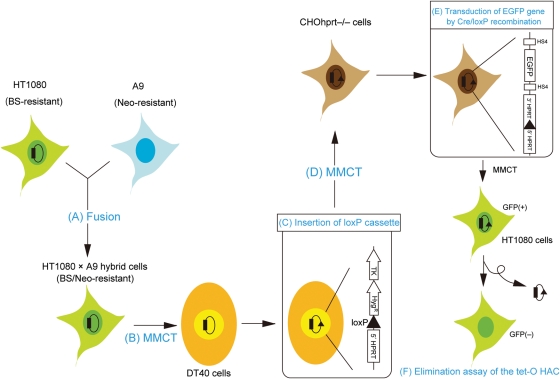
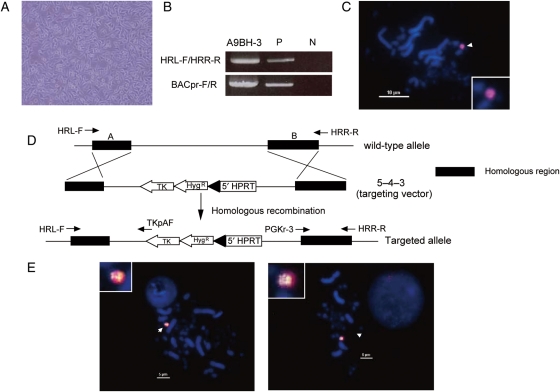
The tet-O HAC was constructed in human fibrosarcoma HT1080 cells,15 because these are the only cells that support efficient formation of HACs from alphoid DNA arrays. Subsequently, the tet-O HAC was transferred into chicken DT40 cells, the standard host for homologous targeting of HACs and mammalian chromosomes by homologous recombination. In addition, DT40 cells form microcells, which facilitate transfer of modified HACs to other mammalian cells. The overall strategy used to modify the tet-O HAC is shown in Fig. 1. First, HT1080 cells carrying the tet-O HAC were fused to high-microcell forming mouse A9 cells, and hybrid A9BH-3 cells were isolated under selection for resistance to BS and G418. The morphology of A9BH-3 cells was similar to that of A9 cells (Fig. 2A), but different from the morphology of HT1080 cells. HAC-specific PCR and FISH was used to confirm the presence of the tet-O HAC in A9BH-3 hybrid cells (Fig. 2B). The hybrid cells form microcells in response to colcemid treatment (data not shown). Second, the tet-O HAC was transferred from hybrid cells into chicken DT40 cells by MMCT.7 In DT40 cells, the tet-O HAC was maintained autonomously without integration into chromosomes (Fig. 2C). The loxP-targeting cassette containing the drug-resistance marker hygromycin, the 5′-end of the HPRT gene, and the Tk gene was introduced into the tet-O HAC by homologous recombination in DT40 cells as shown in Fig. 2D. More than 30 clones were obtained under selection for hygromycin. FISH analysis showed that the loxP cassette recombined into the HAC but not into host chromosomes, and the modified HAC remained stable as an episome (Fig. 2E and Supplementary Table S1) with no detectable change in HAC structure, as detected by Southern blot hybridization. These data show that MMCT successfully transferred the HAC to recipient DT40 cells and that HAC modification by homologous recombination proceeded with high fidelity (Supplementary Fig. S1).
Figure 1.
Schematic diagram of construction of the tet-O HAC-based vector for regulated expression of genes. (A) The tet-O HAC was constructed in human HT1080 cells. HT1080 carrying the tet-O HAC and mouse A9 cells were fused. (B) Hybrid cells carrying the tet-O HAC were transferred into chicken DT40 cells by MMCT. (C) The loxP/HPRT cassette was inserted into the tet-O HAC by homologous recombination in DT40 cells. (D) The tet-O HAC vector was transferred into CHO cells deficient in HPRT. (E) The EGFP/HPRT cassette was introduced into the HAC by Cre/loxP recombination in CHO cells. (F) The tet-O-EGFP HAC was transferred into human HT1080 cells by MMCT. The tet-O HAC was destabilized by expression of a chromatin modifier gene fused with tet-R.
Figure 2.
Insertion of the loxP cassette into the tet-O HAC by homologous recombination in DT40 cells. (A) Morphology of A9BH-3 hybrid cells. (B) Genomic DNA from A9BH-3 hybrid cells was analysed by PCR using HAC-specific primers, HRL-F/R and BACpr-F/R. (C) FISH of DT40/BH-1 cells using the hCot1 probe. The tet-O HAC is indicated by an arrowhead. (D) Insertion of the loxP/HPRT cassette into the tet-O HAC using the targeting vector 5–4–3, which carries the regions of homology (black boxes) to BAC vector sequences in the HAC. (E) FISH analysis of DT40/BHI 1–38 (left) and DT40/BHI 2–2 (right) cells. FISH analysis was performed using a biotin-labelled probe for the loxP cassette (green) and digoxigenin-labelled human hCot1 (red).
The tet-O HAC carries ∼47 copies of the BAC vector used for assembly and propagation of synthetic alphoid DNA arrays.15 The loxP cassette shares homology with the BAC sequences on the tet-O HAC and is targeted into these sequences at one or more sites by homologous recombination in DT40 cells. The copy number of the loxP cassette in the tet-O HAC was 1–32, as determined by a real-time PCR (Supplementary Fig. S2). Two clones, DT40/BHI 1–38 and DT40/BHI 2–2, with 1 and ∼20 copies of the loxP cassette, respectively, were chosen for further analysis.
To investigate whether the tet-O HAC with the loxP cassette retained a conditional centromere, a tet-R-transcriptional transactivator fusion protein was expressed in DT40 cells using the pTet-on advanced vector (TAKARA). The pTet-on expression construct containing the reverse transcriptional transactivator (rtTA) and the Neo gene was introduced by electroporation into DT40/BHI 1–38 and DT40/BHI 2–2 cells, and HAC retention was measured by FISH analysis. Using this ‘tet-on’ expression system, the rtTA fusion binds to tet-O sequences in the presence of doxycycline, causing centromere inactivation and HAC destabilization. HAC retention was measured by FISH analysis in two randomly selected DT40/BHI 1–38 and DT40/BHI 2–2 cultures maintained for 7 days in the presence or the absence of doxycycline (when the rtTA fusion protein does not bind to the centromere; Supplementary Fig. S3). The results indicated that retention of the tet-O HAC decreased dramatically when DT40 cells were grown in the presence of doxycycline. Thus, the tet-O HAC retains a conditional centromere after MMCT into DT40 cells and insertion of the loxP cassette by targeted recombination.
3.2. EGFP transgene insertion into tet-O HAC by Cre-loxP recombination in CHO cells
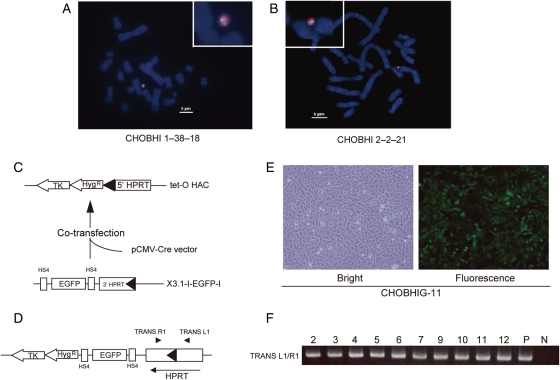
The tet-O HAC containing 1 and 20 copies of the loxP site, BHI 1–38 and BHI 2–2 (Supplementary Fig. S2), were transferred from DT40 cells to HPRT-deficient CHO cells via MMCT. More than 20 BS-resistant clones were obtained for each HAC. PCR analysis with a set of HAC-specific primers that bind to a region proximal to the loxP cassette showed that roughly half of the BS-resistant clones maintained the tet-O HAC (Supplementary Table S1). FISH analysis showed that the tet-O HAC propagated autonomously (Fig. 3A and B). No detectable changes in the HAC structure were detected by Southern blot (Supplementary Fig. S1).
Figure 3.
Insertion of the EGFP transgene into tet-O HAC in CHO cells. (A and B) FISH analysis of CHO metaphase spreads. FISH was performed using a biotin-labelled probe for the loxP cassette (green) and digoxigenin-labelled human hCot1 (red). (C) Insertion of the EGFP cassette into the tet-O HAC by Cre/loxP recombination. The pCMV-Cre expression vector and pCAG-EGFP 3′-HPRT-loxP-targeting vector were co-transfected into the CHO cells containing the tet-O HAC. (D) Map of the tet-O-EGFP HAC vector. (E) Bright and fluorescent images of CHOBHIG-11 cells carrying the tet-O-EGFP HAC. (F) PCR analysis confirming a site-specific recombination resulting in restoration of a functional HPRT. Primers TRANS-L1/TRANS-R1 were used to detect Cre/loxP recombination.
The loxP cassette in the modified tet-O HAC is designed for site-specific gene targeting using Cre recombinase. To test this feature and determine the efficiency of gene targeting into the modified tet-O HAC, an EGFP/HPRT transgene construct and a Cre-recombinase expression vector were co-transfected into CHO cells carrying the modified tet-O HAC with 1 and 20 copies of the loxP cassette (Fig. 3C and D). Recombinant clones were selected by growth in HAT medium for 12 days. PCR analysis with TRANS-L1/TRANS-R1 primers showed that the HPRT gene was reconstituted in all drug-resistant clones (Fig. 3F). All the transfectants expressed the EGFP transgene. A fluorescent image of one representative clone, CHOBHIG-11, with the HAC that was transferred from DT40/BHI 2–2 is shown in Fig. 3E. The efficiency of targeting into both HACs was ∼2 × 10−4, indicating that the number of loxP cassettes in the HAC was not a critical determinant of gene-targeting efficiency. The same efficiency of HACs with 1-loxP copy and 20-loxP copies might be explained by different accessibility of loxP sites in the HAC with 20 copies, i.e. some loxP sites may be ‘masked’ by heterochromatin. Expression of the EGFP transgene was detected by fluorescence microscopy and was stable for at least 12 weeks under selective conditions (data not shown). The tet-O-EGFP HAC also exhibited relatively high stability in CHO cells (up to 50% HAC retention after 2 months of growth under non-selective conditions). The tet-O-EGFP HAC was inactivated and destabilized when rtTA was expressed in these cells, as shown above in DT40 cells (Supplementary Fig. S3). Thus, the tet-O-EGFP HAC retains a conditional centromere in CHO cells.
3.3. Propagation of the retrofitted tet-O HAC in human HT1080 cells and expression of the reporter gene
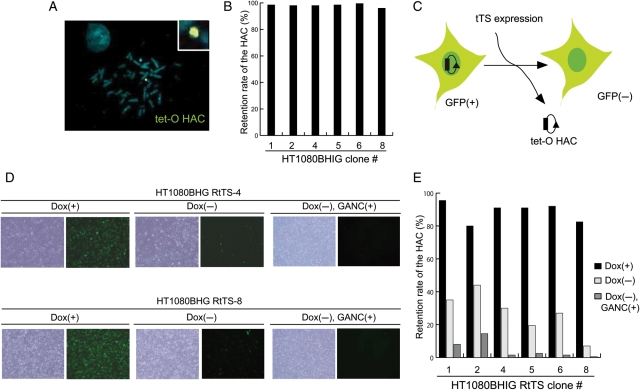
The properties of the tet-O-EGFP HAC were re-examined after transfer from CHOBHIG-11 into human HT1080 cells via MMCT. Under selection for BS, 15 drug-resistant clones that expressed the EGFP transgene were isolated (Fig. 4A). PCR analyses with a set of HAC-specific primers showed that all the 15 clones contain the HAC. FISH analysis showed that the HAC was maintained autonomously in six out of six randomly selected clones (Fig. 4B and Supplementary Table S1).
Figure 4.
Analysis of the tet-O-EGFP HAC after its transferring into HT1080 cells. (A) The tet-O-EGFP HAC was hybridized with biotin-labelled EGFP-targeting vector (green). (B) FISH analysis was performed on HT1080BHIG interphase nuclei as shown in (A). Two hundred interphase nuclei were counted and a rate of the tet-O HAC retention was calculated (as described in the ‘Materials and methods’ section) for six independent clones. (C) HT1080 cells were infected with a retroviral vector expressing the tet-R-tTS-EYFP fusion. (D) Bright and fluorescent images of HT1080 cells. After infection, the cells were cultured under three conditions: dox(+) 7 days, dox(−) 7 days and dox(−) 7 days plus GANC(+) 5 days. Two hundred interphase nuclei were counted and a rate of the tet-O HAC retention was calculated. (E) Tet-O HAC retention was calculated as described in (B).
The following experiments examine whether the tet-O-EGFP HAC retains a conditional centromere and determines the conditions that induce loss of this HAC from HT1080 cells. In our previous study,15 we demonstrated that retro virus-induced tTS expression is highly efficient in tet-O HAC elimination in HT1080 cells but not efficient from the plasmid. Therefore, a retroviral vector expressing tTS-EYFP was introduced into HAC containing HT1080 cells as described previously.15 This vector expresses a fusion of tet-R and the Kruppel-associated box (KRAB)-AB silencing a domain of the Kid-1 protein,19,20 which binds to tet-O in the absence of doxycycline. After transfection with the tTS fusion construct, neomycin-resistant HT1080 clones were selected in the presence of doxycycline, conditions that support proper HAC segregation and HAC stability. The HAC elimination assay was then performed by transfer of the stable transfectants to media lacking doxycycline. FISH analysis revealed dramatic HAC loss after removal of doxycycline (Fig. 4E), which allows the tTS fusion protein to bind to tet-O in the HAC centromere. Thus, after three rounds of MMCT, the HAC retained a conditional centromere. It is worth noting that no detectable cell death was observed during inactivation of kinetochore in the HAC, as reported previously.15
The loxP cassette in the tet-O HAC also carries the HSV-Tk gene, which can be selected against, because it encodes a gene product that causes cell death in a medium containing ganciclovir (Fig. 3C). Consistent with this, FISH and FACS analyses showed that the cells without HAC were selected when cells were grown in the presence of ganciclovir (data not shown). A fraction of tet-O HAC-less cells was even higher when ganciclovir selection was combined with tTS expression (Fig. 4E). Additional experiments demonstrated that EGFP was stably expressed in HT1080 cells carrying the tet-O-EGFP HAC for at least 4 weeks (data not shown).
Together, these data show that the tet-O HAC with a conditional centromere retains all its functional features through three rounds of MMCT and two cycles of targeted engineering. Furthermore, the engineered features of the tet-O HAC and its derivatives, including ability to stably express cloned transgenes, function well in chicken DT40, hamster CHO and human HT1080 cells. This suggests that the host cell environment does not significantly influence the functionality of the tet-O HAC.
3.4. Potential of the tet-O HAC vector
In our previous studies, we developed the tet-O HAC, whose centromere can be inactivated in HT1080 cells by targeting with tet-R fusions with eukaryotic chromatin modifiers.15,16 The tet-O HAC is a unique system for the study of epigenetic modifications in the human kinetochore, but it cannot be used for gene delivery and gene expression studies. In this work, the tet-O HAC was further engineered by insertion of a loxP-5′ HPRT-Hyg-Tk cassette by homologous recombination in chicken DT40 cells. The modified HAC reported here has features that allow it to be a useful vector for gene cloning, delivery and expression in mammalian cells.
A transgene of ∼20 kb in size was efficiently inserted into the loxP site in the modified tet-O HAC by Cre-mediated recombination in CHO cells. Insertion of human genes >60 kb in length was recently also achieved (V.L., unpublished data). Because CHO cells form microcells at a high frequency, the HAC can be easily moved from donor CHO cells into any recipient human or mouse cell lines via MMCT for additional functional analysis.
Importantly, the centromere in the HAC can be inactivated in all three animal cell types tested in this study (human, hamster and chicken). This observation indicates that no significant primary sequence or gross structural changes were introduced into the HAC during three cycles of MMCT and that the functionality of the conditional centromere is relatively insensitive to host cell factors. Indeed, Southern blot analysis of the tet-O HAC confirmed identical profiles in all three host cells. In addition, an EGFP transgene inserted into the loxP cassette could be stably expressed for at least 12 weeks, despite the presence of adjacent functional centromeric DNA. This suggests that the functional kinetochore co-exists with permanently open chromatin domains in mammalian cells. However, additional studies are needed to determine whether a transgene in the loxP cassettes on the tet-O HAC requires specific insulator sequences, which prevent gene silencing due to spreading of pericentromeric heterochromatin during cell propagation.
It is worth noting that this HAC cloning system is compatible with the TAR cloning technology allowing selective isolation of full-length genes from complex genomes by homologous recombination in yeast.21 In particular, a newly developed TAR cloning vector that contains the loxP/HPRT cassette can be used to insert any TAR-isolated gene into the tet-O HAC with a conditional centromere directly in CHO cells using Cre/loxP recombination (V.L., unpublished data).
The tet-O HAC vector has significant advantages over other expression/cloning systems, because it provides a mechanism to compare the phenotype of a mammalian cell with or without a functional copy of any cloned gene of interest. Thus, a rigorous negative control for phenotypic changes attributed to expression of the cloned gene can be conducted easily in any population of dividing cells by simply inactivating the tet-O HAC centromere. Such controls are required for proper interpretation of gene function studies. In addition, the modified tet-O HAC vector can be used for experiments that require transient expression of a cloned gene of interest. For example, iPS cells are generated by transient expression of specific cellular factors, including OCT4, SOX2, KLF4, cMYC and LIN28. In this case, HAC elimination and removal of the stem cell-inducing factors could provide a strategy to avoid insertional mutagenesis and cell transformation, complications that are frequently observed during cell re-programming.22
The capacity for transient expression of cloned genes is also important for other translational research studies. For example, somatic mesenchymal stem cells (MSCs) are mortal, so it is difficult to propagate and expand MSC cultures for transplantation experiments. This problem can be overcome by transient expression of human telomerase reverse transcriptase (hTERT), which extends MSC life span by preventing telomere shortening.23,24 However, our study has shown that constitutive hTERT expression in MSCs prevents their differentiation into muscle cells, possibly because hTERT is normally down-regulated during MSC differentiation (unpublished data). Reversible immortalization by transient expression of hTERT from a viral vector has already been reported in human fibroblasts.25 However, even if the gene is eliminated after transient expression from a viral vector, use of the viral vector is associated with risk of insertional mutagenesis leading to cancer or other cellular dysfunction.2 So, complete elimination of hTERT and vector sequences from MSCs after reversible immortalization is preferred. The tet-O HAC cloning system described in this work provides an important tool for engineering differentiated cells that can safely be used for cell transplantation.
In summary, the modified tet-O HAC described here is a unique HAC-based cloning system for gene expression and gene function studies. Because the tet-O HAC is generated from synthetic alphoid DNA arrays and vector DNA whose DNA sequence is completely defined, this mammalian cell cloning system may have significant potential for gene therapy.
Supplementary Data
Supplementary Data are available at www.dnaresearch.oxfordjournals.org.
Funding
This study was supported by the 21st Century COE program from the Ministry of Education, Culture, Sports, Science and Technology of Japan (M.O.), the Grand-in-Aid for Scientific Research from Ministry of Education, Culture, Sports, Science and Technology of Japan (M.O. and H.M.). Work in the WCE lab was supported by The Wellcome Trust. This research was also supported by the intramural research program of the NIH, National Cancer Institute, Center for Cancer Research.
Supplementary Material
Acknowledgements
We thank Dr Jun-ichirou Ohzeki for his help with a quantitative PCR analysis; Satoshi Abe, Hajime Kurosaki, Hiroyuki Kugoh, Masaharu Hiratsuka, Tetsuya Ohbayashi, Motonobu Katoh, Toshiaki Inoue, Akihiro Kurimasa and Masako Tada for valuable discussions.
Footnotes
Edited by Osamu Ohara
References
- 1.O'Connor T.P., Crystal R.G. Genetic medicines: treatment strategies for hereditary disorders. Nat. Rev. Genet. 2006;7:261–76. doi: 10.1038/nrg1829. doi:10.1038/nrg1829. [DOI] [PubMed] [Google Scholar]
- 2.Hacein-Bey-Abina S., Von Kalle C., Schmidt M., et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–9. doi: 10.1126/science.1088547. doi:10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 3.Larin Z., Mejia J.E. Advances in human artificial chromosome technology. Trends Genet. 2002;18:313–9. doi: 10.1016/S0168-9525(02)02679-3. doi:10.1016/S0168-9525(02)02679-3. [DOI] [PubMed] [Google Scholar]
- 4.Oshimura M., Katoh M. Transfer of human artificial chromosome vectors into stem cells. Reprod. Biomed. Online. 2008;16:57–69. doi: 10.1016/s1472-6483(10)60557-3. doi:10.1016/S1472-6483(10)60557-3. [DOI] [PubMed] [Google Scholar]
- 5.Kuroiwa Y., Tomizuka K., Shinohara T., et al. Manipulation of human minichromosomes to carry greater than megabase-sized chromosome inserts. Nat. Biotechnol. 2000;18:1086–90. doi: 10.1038/80287. [DOI] [PubMed] [Google Scholar]
- 6.Katoh M., Ayabe F., Norikane S., et al. Construction of a novel human artificial chromosome vector for gene delivery. Biochem. Biophys. Res. Commun. 2004;321:280–90. doi: 10.1016/j.bbrc.2004.06.145. doi:10.1016/j.bbrc.2004.06.145. [DOI] [PubMed] [Google Scholar]
- 7.Tomizuka K., Yoshida H., Uejima H., et al. Functional expression and germline transmission of a human chromosome fragment in chimaeric mice. Nat. Genet. 1997;16:133–43. doi: 10.1038/ng0697-133. doi:10.1038/ng0697-133. [DOI] [PubMed] [Google Scholar]
- 8.Suzuki N., Kazuhiro N., Okazaki T., Ikeno M. Human artificial chromosomes constructed using the bottom-up strategy are stably maintained in mitosis and efficiently transmissible to progeny mice. J. Biol. Chem. 2006;281:26615–23. doi: 10.1074/jbc.M603053200. doi:10.1074/jbc.M603053200. [DOI] [PubMed] [Google Scholar]
- 9.Hoshiya H., Kazuki Y., Abe S., et al. A highly stable and nonintegrated human artificial chromosome (HAC) containing the 2.4 Mb entire human dystrophin gene. Mol. Ther. 2009;17:309–17. doi: 10.1038/mt.2008.253. doi:10.1038/mt.2008.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kazuki Y., Hiratsuka M., Takiguchi M., et al. Complete genetic correction of iPS cells from Duchenne muscular dystrophy. Mol. Ther. 2009;18:386–93. doi: 10.1038/mt.2009.274. doi:10.1038/mt.2009.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kazuki Y., Hoshiya H., Kai Y., et al. Correction of a genetic defect in multipotent germline stem cells using a human artificial chromosome. Gene Ther. 2008;15:617–24. doi: 10.1038/sj.gt.3303091. doi:10.1038/sj.gt.3303091. [DOI] [PubMed] [Google Scholar]
- 12.Grimes B.R., Schindelhauer D., McGill N.I., Ross A., Ebersole T.A., Cooke H.J. Stable gene expression from a mammalian artificial chromosome. EMBO Rep. 2001;2:910–4. doi: 10.1093/embo-reports/kve187. doi:10.1093/embo-reports/kve187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ikeno M., Grimes B., Okazaki T., et al. Construction of YAC-based mammalian artificial chromosomes. Nat. Biotechnol. 1998;16:431–9. doi: 10.1038/nbt0598-431. doi:10.1038/nbt0598-431. [DOI] [PubMed] [Google Scholar]
- 14.Grimes B.R., Monaco Z.L. Artificial and engineered chromosomes: developments and prospects for gene therapy. Chromosoma. 2005;114:230–41. doi: 10.1007/s00412-005-0017-5. doi:10.1007/s00412-005-0017-5. [DOI] [PubMed] [Google Scholar]
- 15.Nakano M., Cardinale S., Noskov V.N., et al. Inactivation of a human kinetochore by specific targeting of chromatin modifiers. Dev. Cell. 2008;14:507–22. doi: 10.1016/j.devcel.2008.02.001. doi:10.1016/j.devcel.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cardinale S., Bergmann J.H., Nakano M., et al. Hierarchical inactivation of a synthetic human kinetochore by a chromatin modifier. Mol. Biol. Cell. 2009;19:4194–204. doi: 10.1091/mbc.E09-06-0489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim J-H., Ebersole T., Kouprina N., et al. Human pericentromeric gamma-satellite DNA maintains open chromatin structure and protects a transgene from epigenetic silencing at an ectopic site. Genome Res. 2009;19:533–44. doi: 10.1101/gr.086496.108. doi:10.1101/gr.086496.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kouprina N., Ebersole T., Koriabine M., et al. Cloning of human centromeres by transformation-associated recombination in yeast and generation of functional human artificial chromosomes. Nucleic Acids Res. 2003;31:922–34. doi: 10.1093/nar/gkg182. doi:10.1093/nar/gkg182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Witzgall R., O'Leary E., Leaf A., Onaldi D., Bonventre J.V. The Kruppel-associated box-A domain of zinc finger proteins mediates transcriptional repression. Proc. Natl Acad. Sci. USA. 1994;91:4514–8. doi: 10.1073/pnas.91.10.4514. doi:10.1073/pnas.91.10.4514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Freundlieb S., Schirra-Müller C., Bujard H. A tetracycline controlled activation/repression system with increased potential for gene transfer into mammalian cells. J. Gene Med. 1999;1:4–12. doi: 10.1002/(SICI)1521-2254(199901/02)1:1<4::AID-JGM4>3.0.CO;2-Y. doi:10.1002/(SICI)1521-2254(199901/02)1:1<4::AID-JGM4>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 21.Kouprina N., Larionov V. TAR cloning: insights into gene function, long-range haplotypes, and genome structure and evolution. Nat. Rev. Genet. 2006;7:805–12. doi: 10.1038/nrg1943. doi:10.1038/nrg1943. [DOI] [PubMed] [Google Scholar]
- 22.Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–76. doi: 10.1016/j.cell.2006.07.024. doi:10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 23.Mori T., Kiyono T., Imabayashi H., et al. Combination of hTERT and bmi-1, E6, or E7 induces prolongation of the life span of bone marrow stromal cells from an elderly donor without affecting their neurogenic potential. Mol. Cell Biol. 2005;25:5183–95. doi: 10.1128/MCB.25.12.5183-5195.2005. doi:10.1128/MCB.25.12.5183-5195.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shitara S., Kakeda M., Nagata K., et al. Telomerase-mediated life-span extension of human primary fibroblasts by human artificial chromosome (HAC) vector. Biochem. Biophys. Res. Commun. 2008;3:807–11. doi: 10.1016/j.bbrc.2008.02.119. [DOI] [PubMed] [Google Scholar]
- 25.Narushima M., Kobayashi N., Okitsu T., et al. A human beta-cell line for transplantation therapy to control type 1 diabetes. Nat. Biotechnol. 2005;10:1274–82. doi: 10.1038/nbt1145. [DOI] [PubMed] [Google Scholar]
- 26.Ebersole T., Okamoto Y., Noskov V.N., et al. Rapid generation of long synthetic tandem repeats and its application for analysis in human artificial chromosome formation. Nucleic Acids Res. 2005;33:e130. doi: 10.1093/nar/gni129. doi:10.1093/nar/gni129. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.