Abstract
Functional divergence between homologous proteins is expected to affect amino acid sequences in two main ways, which can be considered as proxies of biochemical divergence: a “covarion-like” pattern of correlated changes in evolutionary rates, and switches in conserved residues (“conserved but different”). Although these patterns have been used in case studies, a large-scale analysis is needed to estimate their frequency and distribution. We use a phylogenomic framework of animal genes to answer three questions: 1) What is the prevalence of such patterns? 2) Can we link such patterns at the amino acid level with selection inferred at the codon level? 3) Are patterns different between paralogs and orthologs? We find that covarion-like patterns are more frequently detected than “constant but different,” but that only the latter are correlated with signal for positive selection. Finally, there is no obvious difference in patterns between orthologs and paralogs.
Keywords: covarion, positive selection, whole-genome duplication, vertebrate evolution, rate shift, heterotachy
Introduction
Gene function changes during evolution, including changes in biochemical function that are expected to be reflected in the amino acid sequence. Most evolutionary models assume that duplication plays a major role in the evolution of such changes (Ohno 1970; Semon and Wolfe 2007; Conant and Wolfe 2008). We have previously shown that positive selection affects vertebrate protein-coding genes relatively frequently, but that there is no increase in its prevalence after duplication (Studer et al. 2008). A general trend of higher divergence after duplication is in fact not so well supported as expected and needs more investigation at all levels of divergence (Studer and Robinson-Rechavi 2009b).
Positive selection is expected to be correlated with functional changes, although a direct link remains to be established (Eyre-Walker 2006). In this work, we explore the divergence between homologous proteins, this time directly in the amino acid sequences. We used two different measures, considered as proxies of biochemical divergence: the “covarion-like” pattern and the “conserved-but-different” pattern (Anisimova and Liberles 2007; Liberles 2007; Studer and Robinson-Rechavi 2009a). Covarion-like evolution means that several sites experience acceleration or deceleration in their evolutionary rate in a correlated way (i.e., in the same evolutionary period), presumably due to variation in selective pressure. Conserved but different refers to a pattern of change from one amino acid to another in a specific evolutionary period for a site that is conserved the rest of the time. It is generally assumed that both these patterns are linked to functional change and driven by positive selection. It is unclear whether this would be the same type of selective events detected by codon models because these amino acid patterns do not necessarily imply a high rate of nonsynonymous to synonymous change.
We answer three questions: 1) What is the prevalence of such patterns? 2) Can we link such patterns at the amino acid level with selection inferred at the codon level? 3) Are patterns different between paralogs and orthologs?
Materials and Methods
Data
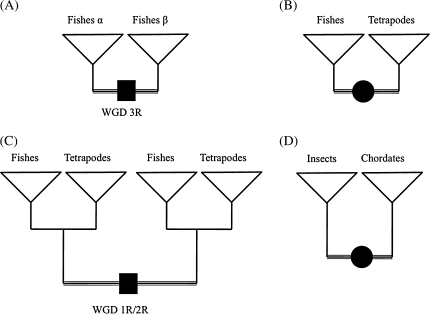
We used gene sequences and trees from the database HomolEns release 4 (Penel et al. 2009), which is based on Ensembl release 49 (March 2008; Hubbard et al. 2009) March 2008 being the date of release 49. Genes are organized in families, which include precalculated alignments and phylogenies. The main advantage of the Homolens system is the FamFetch query system and the TreePattern function (Dufayard et al. 2005). We can scan for specific topologies among the 23,155 family trees. We selected four different branches of the evolution of animals (fig. 1): 1) the whole-genome duplication at the basis of teleost fishes (3R) (549 subfamilies extracted), 2) the split between teleost fishes and tetrapods (4,024 subfamilies), 3) the whole-genome duplications at the base of the vertebrates (1R/2R) (1,014 subfamilies), and 4) the split between Protostomia (limited to insects in our data set) and Deuterostomia (vertebrates and the two chordates Ciona intestinalis and C. savignyi in our data set) (1,234 subfamilies). This represents a total of 6,821 branches to test. These events were chosen because they are important in the evolution of animals, and they contain at least four sequenced genomes on each side of the branch, which appears from pilot studies to be a minimum requirement to be able to detect shifts in a significant manner.
FIG. 1.
Tree topologies studied. Boxes represent speciation events and circles represent duplication events. The branches tested are the 3R genome duplication at the basis of teleost fishes (A), the speciation between fishes and tetrapods (B), the 1R/2R genome duplications at the origin of vertebrates (C), and the speciation between insects and chordates (D).
For the families thus recovered, we removed species with 2× genome coverage (mostly coming from the Mammalian Genome Project of the the National Institute of Health). The restricted alignments were refined with MUSCLE (Edgar 2004). Computations were then done on the new alignment after removing all columns with at least one gap and extracting the well-aligned part using GBLOCKS (Castresana 2000). Phylogenetic subtrees were extracted from the global trees, and branch lengths reestimated with PhyML release 3.0 (Guindon and Gascuel 2003). For the manipulations of sequences and trees, we combined scripts in Python, BioPython (Cock et al. 2009), Jalview (Waterhouse et al. 2009), and the R library APE (Paradis et al. 2004).
Our data set includes ten species of tetrapods: the frog Xenopus tropicalis (Hellsten et al. 2010), the chicken Gallus gallus (International Chicken Genome Sequencing Consortium 2004), and the nine mammals: Monodelphis domestica (Mikkelsen et al. 2007), Bos taurus (Elsik et al. 2009), Canis familiaris (Lindblad-Toh et al. 2005), Equus caballus (Wade et al. 2009), Mus musculus (Waterston et al. 2002), Rattus norvegicus (Rat Genome Sequencing Project Consortium 2004), Pongo pygmaeus abelii (unpublished data), Macaca mulatta (Gibbs et al. 2007), Pan troglodytes (The Chimpanzee Sequencing and Analysis Consortium 2005), and Homo sapiens (International Human Genome Sequencing Consortium 2001, 2004); five species of teleost fishes: Danio rerio (unpublished), Gasterosteus aculeatus (unpublished), Oryzias latipes (Kasahara et al. 2007), Tetraodon nigroviridis (Jaillon et al. 2004), and Takifugu rubripes (Aparicio et al. 2002); the two Ciona: C. intestinalis (Dehal et al. 2002) and C. savignyi (Hill et al. 2008); and four species of insects: Aedes aegypti (Nene et al. 2007), Anopheles gambiae (Holt et al. 2002), Apis melifera (Honeybee Genome Sequencing Consortium 2006), and Drosophila melanogaster (Celniker et al. 2002).
Detection of Shifts in Evolutionary Rate (Covarion-Like)
A shift in evolutionary rate can be observed when a particular amino acid is constrained in one part of the phylogenetic tree (subtree) and is relaxed in the other subtree. This has been called “heterotachy” (Lopez et al. 2002; Philippe et al. 2003), “Type I of functional divergence” (Gu 1999, 2001), or “rate-shifting sites” (Abhiman and Sonnhammer 2005). A particular case is the “concomitantly variable codons” (covarions) process (Fitch 1971; Miyamoto and Fitch 1995; Pupko and Galtier 2002). This pattern of evolution postulates that a subset of sites shifting at the same time are more likely to be structurally linked in the protein (Pupko and Galtier 2002). For simplicity, we will use the term covarions for the rest of the paper. Although it should be noted that we do not study structural linkage, we only report cases where many sites have shifted at the same time (same branch of the tree). To detect such covarions, we used Checkcov (Galtier N, personal communication), an implementation of the algorithm described by Pupko and Galtier (2002). Checkcov performs statistical tests for each residue and can be implemented in an automatic pipeline. The method performs a likelihood ratio test between a null model with only one evolutionary rate per site [rate-among-sites (RAS) model] against an alternative model with two evolutionary rates, one for each subtree. It manages repetition test inside an alignment by using a binomial distribution B(n, P). Briefly, Checkcov assumes that if a covarion process has affected an alignment of length n, we should detect significantly more that 1% of sites at P value <0.01. We used the binomial test from R (binom.test(i, n, P)) to compute the corresponding P value.
We also used Procov to validate the results of Checkcov (Wang et al. 2009). Procov is a general method to detect covarions anywhere in a tree, whereas Checkcov focuses only on a single branch in the tree. To detect whether an alignment includes significant evidence of covarion-like evolution, Procov performs a likelihood ratio test between the RAS model and the general covarion model. For each site, Procov provides the log-likelihood value under the RAS model (1 degree of freedom) and the log-likelihood value under the COV model (3 degrees of freedom). To choose the cutoff to assign a site under a COV model, we used the Akaike information criterion. The difference in log likelihood should be higher than 2 (3 degrees of freedom for COV − 1 degree of freedom for RAS) to be significant. This is an approximation relative to the parametric bootstrap method used in the original paper of Procov (Wang et al. 2009); the parametric bootstrap necessitates a new computation for each data set, resulting in a slightly different cutoff in each case (1.62 for the data set of Wang et al.).
Detection of Change of Conservation Pattern (Constant but Different)
A change in conservation pattern can be seen when a residue is constrained in one subtree for a given property (e.g., a specific amino acid or hydrophobicity) and constrained in the other subtree for a different property (e.g., a different amino acid or polarity). This has been called “Type II of functional divergence” (Gu 2006), “conservation-shifting sites” (Abhiman and Sonnhammer 2005), or “constant but different” (CBD) (Gribaldo et al. 2003). We will use the term CBD for the rest of the paper. We focus only on radical changes in physicochemical properties, such as an acidic to a basic amino acid or a polar to a hydrophobic amino acid. We use Burst after Duplication with Ancestral Sequence Prediction (BADASP) (Edwards and Shields 2005) (release 1.3), which implements the Burst after Duplication (BAD) algorithm (Caffrey et al. 2000). BAD computes the observed differences between two subtrees by the comparison between ancestral conservation and recent conservation. The cutoff value and the minimum number of sites per family were chosen after simulations and after comparison with a positive selection data set (see Results).
Simulations
We performed simulations to test the accuracy and the power of the tests we use. For each subfamily from Homolens, we simulated five alignments of amino acid sequences. We specified the sequence length after removing gaps from the original alignment. We simulated the first 90% of the alignment under a nearly neutral process and the last 10% under a covarion process. The nearly neutral process was based on a RAS model with four categories and the alpha parameter estimated from the real data, using Evolver from PAML (Yang 2007), with a fixed tree topology. The covarion process was simulated by applying for each column a randomly chosen amino acid constraint in one subtree (100% conservation) and random amino acids in the other subtree (no conservation). Using Checkcov, we expect to find 0–1% of covarion sites in the first 90% of the simulated alignments and up to 100% of covarion sites in the last 10%.
We also used simulations derived from the 767 nonduplication trees of our positive selection data set of vertebrates (Studer et al. 2008). This was used to test the CBD under the neutral hypothesis and to define the minimal percentage of sites expected. These simulated multiple alignments followed a RAS model using Evolver.
Statistical Analysis
Over- and underrepresentation in gene ontology (GO) terms was estimated using the TopGO library (Alexa et al. 2006) from Bioconductor (Gentleman et al. 2004). We used false discovery rate to correct for repetition test for enrichment. All other statistical analyses were performed using R (R Development Core Team 2007) and the QVALUE package for test repetition (Storey and Tibshirani 2003).
Results
Comparison with Data of Positive Selection
The data from our previous study of positive selection consists in 767 families of singleton genes of vertebrates (Studer et al. 2008). The analysis has been performed by CodeML from the PAML package (Yang 2007). The model used was the branch-site model (Zhang et al. 2005), able to predict episodic positive selection, and identify amino acids position by providing a Bayes empirical Bayes (BEB) score (Yang et al. 2005).
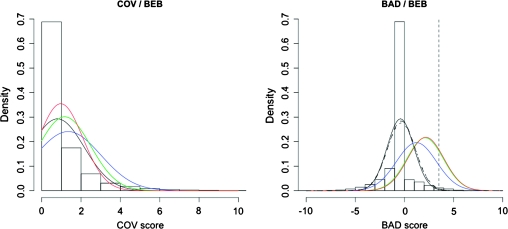
First, we focused on sites. The BEB scores per site were retrieved for the test on the branch between tetrapods and teleost fishes. We assigned a value of 0 for sites with a BEB < 50% and used the real BEB value otherwise. We then associated to these sites the corresponding chi-square value from Checkcov (COV) and the BAD score from BADASP. We have data for a total of 330,067 sites. We found a weak positive correlation between BEB and COV (Pearson’s rxy = 0.07, P < 2.2 × 10−16), a higher positive correlation between BEB and BAD (Pearson’s rxy = 0.26, P < 2.2 × 10−16), and, as expected, a weak negative correlation between COV and BAD (Pearson’s rxy = −0.13, P < 2.2 × 10−16). To verify that results are not influenced by potential saturation of synonymous changes, we repeated the analysis excluding branches with dS > 1. All trends are similar (BEB–COV rxy = 0.04, BEB–BAD rxy = 0.19, and COV–BAD rxy = −0.16) consistent with the lack of saturation reported in Studer et al. (2008). In a second step, we made four different classes of sites (BEB < 50%, 50% ≤ BEB < 95%, 95% ≤ BEB < 99%, and BEB ≥ 99%), and we plotted the distribution of covarion and CBD scores among these classes. These distributions show that the correlation between covarions and positive selection is not biologically significant (fig. 2A) because sites have the same distribution of covarion scores whatever their posterior probability of positive selection. On the other hand, there is a clear shift in CBD scores between the different classes of positive selection (fig. 2B). Of note, these results are consistent with the detection of a “bona fide” signal by the test for positive selection.
FIG. 2.
Correlation of sites under positive selection with covarions and CBDs. (A) The histogram presents the values of the chi-square score per site for covarions. The curves are the chi-square values for different BEB intervals (posterior probability for a site to be under positive selection): black for BEB < 50%, blue for BEB < 95%, green for 99% < BEB, and red for BEB ≥ 99%. (B) The histogram represents the values of BAD scores per site for CBD. The curves are the BAD values for different BEB intervals: black for BEB < 50%, blue for BEB < 95%, green for 99% < BEB, and red for BEB ≥ 99%. The dashed curve represents BAD scores of nearly neutral simulated sequences. The dashed line at 3.5 is the 99th percentile of BAD scores for BEB < 50%.
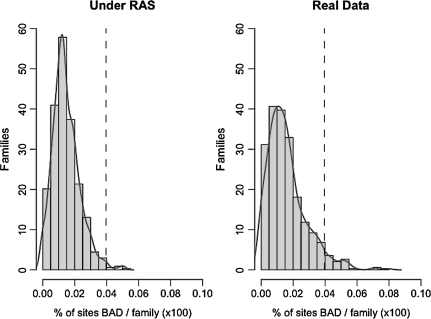
Because BAD scores are not associated to a statistical test but are associated to positive selection, we used BEB values to identify a cutoff for BAD scores. We identified the 99th percentile for the distribution of sites in the class of BEB < 50%. We found a BAD score value of 3.50, which will be defined as our cutoff to detect CBD. This is consistent with the cutoff value of 3+ recommended by the authors (BADASP manual, p. 23). It is also consistent with the distribution of BAD scores for sequences simulated under nearly neutral evolution (dashed curve in fig. 2). To fix the expected proportion of CBD sites (BAD score ≥3.5) in an alignment under the null hypothesis, we used these simulated alignments and selected the 99th percentile of the distribution (fig. 3). We obtain a limit of 3.9% (rounded to 4% for further analyses). These cutoff values (BAD ≥ 3.5; sites >4%) provide us with a stringent test of CBD for further analyses.
FIG. 3.
Percentage of CBDs per families under RAS model and real data. Histograms of proportion of CBD sites with BAD score >3.5 per subtree in data simulated under a neutral model of RAS (A) and in real data (B). The dashed line at 3.9% is the 99th percentile under the neutral model.
Covarion-Like Sites
We first performed tests on simulated data (table 1). For fish specific duplicates (3R), we recovered 75% of covarion sites. This power is not very high, but the accuracy is more than 99.6%. In other branches, we recovered 94–99% of covarion sites without losing accuracy, which is always above 99.2%.
Table 1.
Evaluation of the Accuracy and Power of the Test for Covarions.
| Event Studied | Number of Familiesa | Average Number of Taxa | Average Sequences Length | Accuracy under Neutral Evolution | Power under Covarion Process |
| (=1 − Percentage Covarion) (%) | (=Percentage Covarion) (%) | ||||
| Duplication 3R | 2,745 | 9.1 | 390.7 | 99.6 | 75.2 |
| Speciation tetrapods–fishes | 20,120 | 16.2 | 353.8 | 99.4 | 94.5 |
| Duplication 2R | 5,070 | 31.3 | 229.5 | 99.4 | 99.2 |
| Speciation chordates–insects | 6,170 | 28.0 | 254.9 | 99.2 | 94.5 |
Five simulations per set of parameters derived from one family of real data.
On real data, various percentages of families under covarion processes were found, above the 1% threshold (table 2). In the most recent event tested, the 3R duplication, only two families (0.4%) are significant, and we suspect misalignment for one case (ENSDARP00000012602; Dup3R/HBG059441-2 in our Web supdata: http://bioinfo.unil.ch/supdata/). The 3R is the event with the lowest number of sequences per family (nine in average), and this could provide less power, as seen in simulations. For the speciation between tetrapods and fishes, there are twice as many genes per family (16 on average), and we find 33 families (0.8%). This result is coherent with a preliminary study (Studer and Robinson-Rechavi 2009a), where we found a similar trend using another tool (ShiftFinder; http://sites.univ-provence.fr/evol/phylogenomics-lab/PageWeb/SHIFT-FINDER.htm) to detect shifts in evolutionary rates.
Table 2.
Results of the Detection of Covarions.
| Event Studied | Number of Families | Average Number of Sequences | Average Number of Sites | Families without Significanta Signal for Covarions |
Families with Significanta Signal for Covarions |
|||
| Number of Families (%) | Mean Branch Lengthb | Number of Families (%) | Percentage of Covarions Sites (%) | Mean Branch Length | ||||
| Duplication 3R | 549 | 9.1 | 391.8 | 547 (99.6) | 0.134 | 2 (0.4) | 3.2 | 0.201 |
| Speciation tetrapods–fishes | 4,024 | 16.2 | 355.2 | 3991 (99.2) | 0.247 | 33 (0.8) | 3.5 | 0.445 |
| Duplication 2R | 1,014 | 31.3 | 231.0 | 928 (91.5) | 0.290 | 86 (8.5) | 4.8 | 0.522 |
| Speciation chordates–insects | 1,234 | 28.0 | 256.2 | 1161 (94.1) | 0.426 | 73 (5.9) | 4.4 | 0.667 |
P value = 1% and Q value threshold at 10%.
In amino acid substitutions.
The highest number of significant families (8.5%) is found for the genes associated to the 2R duplication at the base of vertebrates. This is also the data set with the most sequences per family, 31 on average. The oldest event tested is the speciation event between insects and chordates, where we found 5.9% of families with an average of 28 sequences per family.
A different approach to the detection of covarion-like sites is implemented in Procov (Wang et al. 2009). Procov detects shifts that occurred on any branch of the tree and expectedly detects more shifts than the targeted approach of Checkcov. In our data set, Checkcov identifies 3% of significant families, whereas Procov identifies 66% of significant families. Among the 194 families identified by Checkcov, Procov also detected 97% of them. Of the 17,787 sites detected by Checkcov at the 1% threshold, 95% present some signal in Procov (ln(L_cov) − ln(L_RAS) > 0). Moreover, there is a strong correlation of Checkcov and Procov scores, when comparing sites that are significant under Checkcov (Spearman’s ρ = 0.41, P < 2.2 × 10−16). Finally, using very conservative thresholds (ten for Checkcov and two for Procov), more than 50% of the 2,961 sites from Checkcov are recovered by Procov, whereas detecting exact covarion sites is a difficult problem.
CBD Sites
We found no significant CBD sites in 3R duplicates and very few in other events (table 3): tetrapode–teleost speciation (0.5%), 2R duplication (0.4%), and chordate–insect speciation (1.4%). This is probably due to our stringent cutoff and control for false discovery rate. Interestingly, the average of CBD sites in significant families is quite high (≈10%), supporting the relevance of those shifts that we do detect.
Table 3.
Results of the Detection of CBD.
| Event Studied | Number of Families | Average Number of Sequences | Average Number of Sites | Families without Significanta Signal for CBD |
Families with significanta signal for CBD |
|||
| Number of Families (%) | Mean Branch Lengthb | Number of Families (%) | Percentage of BADASP Sites (%) | Mean Branch Length | ||||
| Duplication 3R | 549 | 9.1 | 391.8 | 549 (100) | 0.134 | 0 (0) | NA | NA |
| Speciation tetrapods–fishes | 4,024 | 16.2 | 355.2 | 4003 (99.5) | 0.245 | 21 (0.5) | 9.7 | 0.900 |
| Duplication 2R | 1,014 | 31.3 | 231.0 | 1010 (99.6) | 0.305 | 4 (0.4) | 12.7 | 1.592 |
| Speciation chordates–insects | 1,234 | 28.0 | 256.2 | 1217 (98.6) | 0.430 | 17 (1.4) | 10.3 | 1.149 |
Cutoff at 4%, based on simulation data, and Q value threshold at 10%.
In amino acid substitutions.
Global Trends
A potential confounding effect in the analysis of 2R duplicate genes is the evolutionary rate of fish genes, which evolve faster than tetrapods (Brunet et al. 2006; Steinke et al. 2006). We recomputed the 2R families after removing all fish sequences. The first observation, as expected, is a global increase of the mean length in the tested branch, from 0.257 amino acid substitutions per site to 0.439, because we measure the branch between the 2R event and the amphibian–amniote speciation, instead of the branch between the 2R event and the earlier teleost–tetrapode speciation. More interesting, there is a decrease in the number of families with significant covarion patterns (from 8.5% to 4.4%) and inversely a large increase in the number of families significant for CBD (from 0.4% to 16.8%). The fast-evolving fish genes probably increased the global heterogeneity in amino acid alignments, decreasing the signal for paralog-specific conservation of amino acids (CBD).
GO enrichment shows that our data set is biased toward slow-evolving genes, as in Studer et al. (2008), for similar reasons of stringent data collection. Significant results in covarions revealed only a few categories, typical of fast-evolving proteins; no GO categories were enriched in CBD genes. With these results, we can only say that it seems that covarions and CBDs can be found among most categories of genes.
Finally, we evaluated possible confounding factors by computing a linear model (analysis of variance) testing the effect of different parameters describing gene families (tables 4 and 5). The number of sequences and the branch length are correlated with the percentage of significant sites for both tests, whereas the number of sites analyzed has no impact. It should be noted that the branch length explains up to half of the variance in the CBD analyses. However, at least 72% of the variance is explained by none of these parameters for covarions and at least 44% of the variance for CBD sites. Presumably, most of this remaining variance is due to shifts in function and selection.
Table 4.
Effect of Potential Confounding Factors on the Detection of Covarions.
| Variable | Duplication 3R |
Speciation Tetrapods–Fishes |
Duplication 2R |
Speciation Vertebrates–Insects |
||||
| Variance Explained (%) | P Valuea | Variance Explained (%) | P Valuea | Variance Explained (%) | P Valuea | Variance Explained (%) | P Valuea | |
| Number of genes | 2 | 3.8 × 10−04 | 2 | 2.2 × 10−16 | 2 | 1.5 × 10−07 | 3 | 5.5 × 10−12 |
| Number of sites | 0 | 1.5 × 10−01 | 0 | 2.1 × 10−01 | 0 | 8.8 × 10−01 | 0 | 7.2 × 10−01 |
| Branch length separating subtrees | 2 | 2.0 × 10−04 | 12 | 2.2 × 10−16 | 16 | 2.2 × 10−16 | 6 | 2.2 × 10−16 |
| Number of branches in subtree alpha | 2 | 1.1 × 10−04 | 0 | 8.6 × 10−04 | 0 | 8.3 × 10−01 | 0 | 2.9 × 10−01 |
| Sum of branch lengths in subtree alpha | 8 | 2.9 × 10−13 | 3 | 2.2 × 10−16 | 1 | 5.6 × 10−05 | 1 | 1.7 × 10−03 |
| Number of branches in subtree betab | NA | NA | 0 | 6.1 × 10−01 | 0 | 3.7 × 10−01 | NA | NA |
| Sum of branch lengths in subtree beta | 10 | 2.2 × 10−16 | 5 | 2.2 × 10−16 | 1 | 8.5 × 10−04 | 1 | 3.3 × 10−04 |
| Median_diffc | 2 | 2.8 × 10−05 | 4 | 2.2 × 10−16 | 0 | 1.3 × 10−02 | 5 | 2.2 × 10−16 |
| Residuals | 74 | 72 | 79 | 84 | ||||
NOTE.—NA, non-available.
Italic values indicate significant after a Bonferroni correction (α = 0.05/4 = 0.0125).
The values NA were removed from the analysis of variance because the number of branches in the subtree beta is identical to the number of branches in the subtree alpha.
Median_diff represents the difference between the medians of all branch lengths for both trees.
Table 5.
Effect of Potential Confounding Factors on the Detection of CBD Sites.
| Variable | Duplication 3R |
Speciation Tetrapods–Fishes |
Duplication 2R |
Speciation Vertebrates–Insects |
||||
| Variance Explained (%) | P Valuea | Variance Explained (%) | P Valuea | Variance Explained (%) | P Valuea | Variance Explained (%) | P Valuea | |
| Number of genes | 3 | 2.2 × 10−09 | 4 | 2.2 × 10−16 | 1 | 1.5 × 10−06 | 4 | 2.2 × 10−16 |
| Number of sites | 0 | 5.3 × 10−01 | 0 | 2.1 × 10−02 | 0 | 5.2 × 10−01 | 0 | 4.6 × 10−01 |
| Branch length separating subtrees | 51 | 2.2 × 10−16 | 52 | 2.2 × 10−16 | 39 | 2.2 × 10−16 | 39 | 2.2 × 10−16 |
| Number of branches in subtree alpha | 0 | 9.6 × 10−01 | 0 | 8.0 × 10−01 | 0 | 5.7 × 10−01 | 0 | 2.4 × 10−01 |
| Sum of branch lengths in subtree alpha | 0 | 2.3 × 10−01 | 0 | 6.6 × 10−02 | 0 | 2.6 × 10−02 | 0 | 5.7 × 10−01 |
| Number of branchesin subtree betab | NA | NA | 0 | 6.8 × 10−01 | 0 | 2.1 × 10−01 | NA | NA |
| Sum of branch lengths in subtree beta | 1 | 1.8 × 10−04 | 0 | 9.0 × 10−02 | 0 | 2.4 × 10−01 | 0 | 9.1 × 10−04 |
| Median_diffc | 0 | 5.2 × 10−01 | 1 | 6.9 × 10−16 | 1 | 5.2 × 10−04 | 2 | 8.1 × 10−13 |
| Residuals | 44 | 44 | 59 | 55 | ||||
NOTE.—NA, non-available.
Italic values indicates significant after a Bonferroni correction (α = 0.05/4 = 0.0125).
The values NA were removed from the ANOVA because the number of branches in the subtree beta is identical to the number of branches in the subtree alpha.
Median_diff represents the difference between the medians of all branch lengths for both trees.
Discussion
Covarion-Like and CBD patterns as Proxies of Functional Divergence
We found a variable proportion of protein families to be significantly under covarion process, depending of the branch tested, from 0.8% to 8.5% (table 2), and very few families with a CBD pattern. The proportion of 2% found by Gruenheit et al. (2008) in their balanced data set between two monophyletic groups is within the same range. This suggests that covarion-like patterns may reflect relatively frequent small refinements in function or even compensatory mutations without change in function. Importantly, most cases of covarion that we detect with Checkcov are also detected by the more recent Procov method (Wang et al. 2009). The rarity of CBD is consistent with more radical functional changes. It is also possible that CBD sites are more difficult to discriminate from other slow-evolving sites, resulting in rare detection under stringent criteria. Interestingly, the proportion of sites found when a branch is significant for a family is quite high for both patterns, and highest for CBD, which is consistent with the detection of radical functional shifts in protein function in at least some cases. Of note, removing fast-evolving fish genes increased our capacity to detect CBD patterns between tetrapode paralogs. Thus, we should be careful to take into account evolutionary rate differences between species in the study of paralog divergence.
We have tried to apply strict cutoffs for both covarion and CBD, but it is difficult to estimate the true level of changes in amino acid evolution that will affect function in a biologically meaningful way (Levasseur et al. 2007). Case studies present both examples of the evolution of a new function (Braasch et al. 2006) and of more subtle optimization of the original function (Christin et al. 2008). In general, minor sequence changes can affect structural properties that are directly linked to the biochemical function (Tokuriki and Tawfik 2009). At the extreme, the bacterial melamine deaminase shares 98% of identity with the atrazine chlorohydrolase (Seffernick et al. 2001). A correlation of covarions as detected in sequences with structural and functional divergence has been found in several case studies. This is the case for bacterial and eukaryotic elongation factors, which differ in function (Gaucher et al. 2001), and for which covarions were confirmed by crystal structure (Gaucher, Das, et al. 2002). It is also the case in caspases (Wang and Gu 2001). In vertebrate hemoglobin α and β, CBD sites seem to be a more reliable predictor of function divergence than covarions (Gribaldo et al. 2003). Other case studies have highlighted the importance of studying covarion or CBD patterns in correlation with protein function (Gaucher, Gu, et al. 2002; Philippe et al. 2003; Liberles 2007). A difficulty in comparing results from various studies is the use of different methods, with no common standard, and notably differences in the treatment of radical versus conservative changes (Liberles 2007).
Both covarions and CBD sites can result from either gain or loss of function. A covarion pattern may indicate a site, which was kept functionally constrained in one subtree but lost this constraint in the other, or the recruitment of a site (newly constrained) for a novel function. And a CBD pattern may indicate a site conserved in the ancestor which changed by positive selection to get a new function, or a partition of the ancestral protein into different functions, especially through escape from adaptive conflict (Conant and Wolfe 2008).
The covarion model has been recently extended to detect shifts in any branch in a phylogenetic tree (Penn et al. 2008). However, this method needs dozens of sequences, which is not yet applicable to the comparison of completely sequenced animal genomes. It would be interesting in the future to develop methods to analyze such patterns between three subtrees: the two lineages of interest and an outgroup, which will help to discriminate between gain and loss of function (Studer and Robinson-Rechavi 2009b).
Can We Link These Shifts at the Amino Acid Level with Positive Selection Inferred at the Codon Level?
Although the models of amino acid changes are not able to discriminate between a relaxation of purifying selection and positive selection, this is the main focus of codon models (Anisimova and Liberles 2007). But an advantage of amino acid patterns is the potential for comparisons between sequences for which synonymous substitutions are saturated. Thus, a correlation between amino acid shifts and the detection of positive selection would be interesting. We found only a weak positive correlation between scores of positive selection and of covarions. Among sites identified as under positive selection on a specific branch, there are more sites under purifying selection on background branches (type K2a of CodeML; Yang 2007) (∼6.5% in Studer et al. 2008), than sites under neutral evolution on background branches (type K2b) (∼1% in Studer et al. 2008). The type K2a of codon model with its slow evolutionary rate except on one branch is closest to a CBD pattern. The covarion pattern may be more similar to the type K2b but is not quite the same. CodeML K2b sites are expected to be variable in all background branches, whereas covarion sites are only variable in one subtree. In fact, the covarion pattern is most similar to the clade model of CodeML (Bielawski and Yang 2004), which has not been used in any large scan to our knowledge.
In any case, it appears that by detecting CBD sites, we do detect a signal of positive selection similar to that inferred by the branch-site model. This opens the possibility to scan for such shifts in very ancient events, where synonymous substitutions are clearly saturated.
Is the Incidence of Covarion-Like and CBD Patterns Different between Paralogs and Orthologs?
In a study of vertebrate hemoglobin α and β, metrics of protein divergence were compared in their capacity to distinguish paralog divergence from ortholog divergence (Gribaldo et al. 2003). But on a larger data set, there is no obvious excess of either covarion-like or CBD sites on duplication branches relative to speciation branches (tables 2 and 3). The main explanatory variables for test results are branch length and the number of genes analyzed (tables 4 and 5). If anything, there seems to be a slight deficit of CBD sites after duplication, but this could be due to other factors, such as the evolutionary rate acceleration in teleost fishes. This is consistent with a previous report of similar levels of amino acid variability in orthologs and paralogs on a smaller sample (Conant et al. 2007). It has been suggested that the divergence of paralogs owes more to expression patterns than to biochemical function (Wapinski et al. 2007), although a stronger divergence of expression between paralogs than between orthologs remains to be established (Studer and Robinson-Rechavi 2009b). At the protein level, in any case, it appears that functional shifts could be frequent not only between paralogs but also between orthologs and that we should be careful when applying orthology to define the function of new genes (Lynch 2009; Studer and Robinson-Rechavi 2009b).
Conclusions
In our study, we found that the CBD pattern correlates well with sites predicted by a branch-site model. This pattern could be used as a proxy of positive selection in very ancient evolutionary events, avoiding the classical problem of saturation of synonymous sites. Positive selection is expected to be involved in strong changes that affect the protein structure and function, which could be causing the CBD pattern. Most evolutionary models predict a burst of functional change after duplication (Force et al. 1999; Conant and Wolfe 2008). We find that changes at the amino acid level, while not infrequent, affect both orthologs and paralogs similarly. Thus, it seems that the same pattern of functional divergence affects both paralogs and orthologs genes, and this might be, rather than an exception, a general trend (Studer and Robinson-Rechavi 2009b). If there are any differences in gene evolution after duplication, it is possible that they affect expression patterns rather than protein biochemical function as reflected in sequences (Wapinski et al. 2007).
Acknowledgments
We acknowledge funding from Etat de Vaud and Swiss National Science Foundation (grant 116798). The computations were performed at the Vital-IT (http://www.vital-it.ch) center for High Performance Computing of the Swiss Institute of Bioinformatics. We thank Nicolas Galtier, Pierre Pontarotti, Huaichun Wang, Tal Pupko, Adi Stern, and anonymous reviewers for helpful discussions.
References
- Abhiman S, Sonnhammer EL. Large-scale prediction of function shift in protein families with a focus on enzymatic function. Proteins. 2005;60:758–768. doi: 10.1002/prot.20550. [DOI] [PubMed] [Google Scholar]
- Alexa A, Rahnenfuhrer J, Lengauer T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics. 2006;22:1600–1607. doi: 10.1093/bioinformatics/btl140. [DOI] [PubMed] [Google Scholar]
- Anisimova M, Liberles DA. The quest for natural selection in the age of comparative genomics. Heredity. 2007;99:567–579. doi: 10.1038/sj.hdy.6801052. [DOI] [PubMed] [Google Scholar]
- Aparicio S, Chapman J, Stupka E, et al. (41 co-authors) Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science. 2002;297:1301–1310. doi: 10.1126/science.1072104. [DOI] [PubMed] [Google Scholar]
- Bielawski JP, Yang Z. A maximum likelihood method for detecting functional divergence at individual codon sites, with application to gene family evolution. J Mol Evol. 2004;59:121–132. doi: 10.1007/s00239-004-2597-8. [DOI] [PubMed] [Google Scholar]
- Braasch I, Salzburger W, Meyer A. Asymmetric evolution in two fish-specifically duplicated receptor tyrosine kinase paralogons involved in teleost coloration. Mol Biol Evol. 2006;23:1192–1202. doi: 10.1093/molbev/msk003. [DOI] [PubMed] [Google Scholar]
- Brunet FG, Crollius HR, Paris M, Aury JM, Gibert P, Jaillon O, Laudet V, Robinson-Rechavi M. Gene loss and evolutionary rates following whole-genome duplication in teleost fishes. Mol Biol Evol. 2006;23:1808–1816. doi: 10.1093/molbev/msl049. [DOI] [PubMed] [Google Scholar]
- Caffrey DR, O’Neill LA, Shields DC. A method to predict residues conferring functional differences between related proteins: application to MAP kinase pathways. Protein Sci. 2000;9:655–670. doi: 10.1110/ps.9.4.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Celniker SE, Wheeler DA, Kronmiller B, et al. (32 co-authors) Finishing a whole-genome shotgun: release 3 of the Drosophila melanogaster euchromatic genome sequence. Genome Biol. 2002 doi: 10.1186/gb-2002-3-12-research0079. doi:10.1186/gb-2002-3-12-research0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christin PA, Salamin N, Muasya AM, Roalson EH, Russier F, Besnard G. Evolutionary switch and genetic convergence on rbcL following the evolution of C4 photosynthesis. Mol Biol Evol. 2008;25:2361–2368. doi: 10.1093/molbev/msn178. [DOI] [PubMed] [Google Scholar]
- Cock PJ, Antao T, Chang JT, et al. (11 co-authors) Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009;25:1422–1423. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conant GC, Wagner GP, Stadler PF. Modeling amino acid substitution patterns in orthologous and paralogous genes. Mol Phylogenet Evol. 2007;42:298–307. doi: 10.1016/j.ympev.2006.07.006. [DOI] [PubMed] [Google Scholar]
- Conant GC, Wolfe KH. Turning a hobby into a job: how duplicated genes find new functions. Nat Rev Genet. 2008;9:938–950. doi: 10.1038/nrg2482. [DOI] [PubMed] [Google Scholar]
- Dehal P, Satou Y, Campbell RK, et al. (87 co-authors) The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science. 2002;298:2157–2167. doi: 10.1126/science.1080049. [DOI] [PubMed] [Google Scholar]
- Dufayard JF, Duret L, Penel S, Gouy M, Rechenmann F, Perriere G. Tree pattern matching in phylogenetic trees: automatic search for orthologs or paralogs in homologous gene sequence databases. Bioinformatics. 2005;21:2596–2603. doi: 10.1093/bioinformatics/bti325. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards RJ, Shields DC. BADASP: predicting functional specificity in protein families using ancestral sequences. Bioinformatics. 2005;21:4190–4191. doi: 10.1093/bioinformatics/bti678. [DOI] [PubMed] [Google Scholar]
- Elsik CG, Tellam RL, Worley KC, et al. The genome sequence of taurine cattle: a window to ruminant biology and evolution. Science. 2009;324:522–528. doi: 10.1126/science.1169588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker A. The genomic rate of adaptive evolution. Trends Ecol Evol. 2006;21:569–575. doi: 10.1016/j.tree.2006.06.015. [DOI] [PubMed] [Google Scholar]
- Fitch WM. Rate of change of concomitantly variable codons. J Mol Evol. 1971;1:84–96. doi: 10.1007/BF01659396. [DOI] [PubMed] [Google Scholar]
- Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaucher EA, Das UK, Miyamoto MM, Benner SA. The crystal structure of eEF1A refines the functional predictions of an evolutionary analysis of rate changes among elongation factors. Mol Biol Evol. 2002;19:569–573. doi: 10.1093/oxfordjournals.molbev.a004113. [DOI] [PubMed] [Google Scholar]
- Gaucher EA, Gu X, Miyamoto MM, Benner SA. Predicting functional divergence in protein evolution by site-specific rate shifts. Trends Biochem Sci. 2002;27:315–321. doi: 10.1016/s0968-0004(02)02094-7. [DOI] [PubMed] [Google Scholar]
- Gaucher EA, Miyamoto MM, Benner SA. Function-structure analysis of proteins using covarion-based evolutionary approaches: elongation factors. Proc Natl Acad Sci U S A. 2001;98:548–552. doi: 10.1073/pnas.98.2.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, et al. (25 co-authors) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs RA, Rogers J, Katze MG, et al. (176 co-authors) Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- Gribaldo S, Casane D, Lopez P, Philippe H. Functional divergence prediction from evolutionary analysis: a case study of vertebrate hemoglobin. Mol Biol Evol. 2003;20:1754–1759. doi: 10.1093/molbev/msg171. [DOI] [PubMed] [Google Scholar]
- Gruenheit N, Lockhart PJ, Steel M, Martin W. Difficulties in testing for covarion-like properties of sequences under the confounding influence of changing proportions of variable sites. Mol Biol Evol. 2008;25:1512–1520. doi: 10.1093/molbev/msn098. [DOI] [PubMed] [Google Scholar]
- Gu X. Statistical methods for testing functional divergence after gene duplication. Mol Biol Evol. 1999;16:1664–1674. doi: 10.1093/oxfordjournals.molbev.a026080. [DOI] [PubMed] [Google Scholar]
- Gu X. Maximum-likelihood approach for gene family evolution under functional divergence. Mol Biol Evol. 2001;18:453–464. doi: 10.1093/oxfordjournals.molbev.a003824. [DOI] [PubMed] [Google Scholar]
- Gu X. A simple statistical method for estimating type-II (cluster-specific) functional divergence of protein sequences. Mol Biol Evol. 2006;23:1937–1945. doi: 10.1093/molbev/msl056. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Hellsten U, Harland RM, Gilchrist MJ, et al. (48 co-authors) The genome of the Western clawed frog Xenopus tropicalis. Science. 2010;328:633–636. doi: 10.1126/science.1183670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill MM, Broman KW, Stupka E, Smith WC, Jiang D, Sidow A. The C. savignyi genetic map and its integration with the reference sequence facilitates insights into chordate genome evolution. Genome Res. 2008;18:1369–1379. doi: 10.1101/gr.078576.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt RA, Subramanian GM, Halpern A, et al. (123 co-authors) The genome sequence of the malaria mosquito Anopheles gambiae. Science. 2002;298:129–149. doi: 10.1126/science.1076181. [DOI] [PubMed] [Google Scholar]
- Honeybee Genome Sequencing Consortium. Insights into social insects from the genome of the honeybee Apis mellifera. Nature. 2006;443:931–949. doi: 10.1038/nature05260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard TJ, Aken BL, Ayling S, et al. (58 co-authors) Ensembl 2009. Nucleic Acids Res. 2009;37:D690–D697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Chicken Genome Sequencing Consortium. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- Jaillon O, Aury JM, Brunet F, et al. (61 co-authors) Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature. 2004;431:946–957. doi: 10.1038/nature03025. [DOI] [PubMed] [Google Scholar]
- Kasahara M, Naruse K, Sasaki S, et al. The medaka draft genome and insights into vertebrate genome evolution. Nature. 2007;447:714–719. doi: 10.1038/nature05846. [DOI] [PubMed] [Google Scholar]
- Levasseur A, Orlando L, Bailly X, Milinkovitch MC, Danchin EG, Pontarotti P. Conceptual bases for quantifying the role of the environment on gene evolution: the participation of positive selection and neutral evolution. Biol Rev Camb Philos Soc. 2007;82:551–572. doi: 10.1111/j.1469-185X.2007.00024.x. [DOI] [PubMed] [Google Scholar]
- Liberles DA. Ancestral sequence reconstruction. New York: Oxford University Press; 2007. [Google Scholar]
- Lindblad-Toh K, Wade CM, Mikkelsen TS, et al. (234 co-authors) Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- Lopez P, Casane D, Philippe H. Heterotachy, an important process of protein evolution. Mol Biol Evol. 2002;19:1–7. doi: 10.1093/oxfordjournals.molbev.a003973. [DOI] [PubMed] [Google Scholar]
- Lynch VJ. Use with caution: developmental systems divergence and potential pitfalls of animal models. Yale J Biol Med. 2009;82:53–66. [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Wakefield MJ, Aken B, et al. Genome of the marsupial Monodelphis domestica reveals innovation in non-coding sequences. Nature. 2007;447:167–177. doi: 10.1038/nature05805. [DOI] [PubMed] [Google Scholar]
- Miyamoto MM, Fitch WM. Testing the covarion hypothesis of molecular evolution. Mol Biol Evol. 1995;12:503–513. doi: 10.1093/oxfordjournals.molbev.a040224. [DOI] [PubMed] [Google Scholar]
- Nene V, Wortman JR, Lawson D, et al. Genome sequence of Aedes aegypti, a major arbovirus vector. Science. 2007;316:1718–1723. doi: 10.1126/science.1138878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. Evolution by gene duplication. Heidelberg: Springer-Verlag; 1970. [Google Scholar]
- Paradis E, Claude J, Strimmer K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- Penel S, Arigon AM, Dufayard JF, Sertier AS, Daubin V, Duret L, Gouy M, Perriere G. Databases of homologous gene families for comparative genomics. BMC Bioinformatics. 2009;10(Suppl 6):S3. doi: 10.1186/1471-2105-10-S6-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn O, Stern A, Rubinstein ND, Dutheil J, Bacharach E, Galtier N, Pupko T. Evolutionary modeling of rate shifts reveals specificity determinants in HIV-1 subtypes. PLoS Comput Biol. 2008;4:e1000214. doi: 10.1371/journal.pcbi.1000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe H, Casane D, Gribaldo S, Lopez P, Meunier J. Heterotachy and functional shift in protein evolution. IUBMB Life. 2003;55:257–265. doi: 10.1080/1521654031000123330. [DOI] [PubMed] [Google Scholar]
- Pupko T, Galtier N. A covarion-based method for detecting molecular adaptation: application to the evolution of primate mitochondrial genomes. Proc Biol Sci. 2002;269:1313–1316. doi: 10.1098/rspb.2002.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: a language and environment for statistical computing. Vienna (Austria): Foundation for Statistical Computing. 2007. [Google Scholar]
- Rat Genome Sequencing Project Consortium. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- Seffernick JL, de Souza ML, Sadowsky MJ, Wackett LP. Melamine deaminase and atrazine chlorohydrolase: 98 percent identical but functionally different. J Bacteriol. 2001;183:2405–2410. doi: 10.1128/JB.183.8.2405-2410.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semon M, Wolfe KH. Consequences of genome duplication. Curr Opin Genet Dev. 2007;17:505–512. doi: 10.1016/j.gde.2007.09.007. [DOI] [PubMed] [Google Scholar]
- Steinke D, Salzburger W, Braasch I, Meyer A. Many genes in fish have species-specific asymmetric rates of molecular evolution. BMC Genomics. 2006;7:20. doi: 10.1186/1471-2164-7-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer RA, Penel S, Duret L, Robinson-Rechavi M. Pervasive positive selection on duplicated and nonduplicated vertebrate protein coding genes. Genome Res. 2008;18:1393–1402. doi: 10.1101/gr.076992.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer RA, Robinson-Rechavi M. Evidence for an episodic model of protein sequence evolution. Biochem Soc Trans. 2009a;37:783–786. doi: 10.1042/BST0370783. [DOI] [PubMed] [Google Scholar]
- Studer RA, Robinson-Rechavi M. How confident can we be that orthologs are similar, but paralogs differ? Trends Genet. 2009b;25:210–216. doi: 10.1016/j.tig.2009.03.004. [DOI] [PubMed] [Google Scholar]
- The Chimpanzee Sequencing and Analysis Consortium. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- Tokuriki N, Tawfik DS. Protein dynamism and evolvability. Science. 2009;324:203–207. doi: 10.1126/science.1169375. [DOI] [PubMed] [Google Scholar]
- Wade CM, Giulotto E, Sigurdsson S, et al. Genome sequence, comparative analysis, and population genetics of the domestic horse. Science. 2009;326:865–867. doi: 10.1126/science.1178158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HC, Susko E, Roger AJ. PROCOV: maximum likelihood estimation of protein phylogeny under covarion models and site-specific covarion pattern analysis. BMC Evol Biol. 2009;9:225. doi: 10.1186/1471-2148-9-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Gu X. Functional divergence in the caspase gene family and altered functional constraints: statistical analysis and prediction. Genetics. 2001;158:1311–1320. doi: 10.1093/genetics/158.3.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wapinski I, Pfeffer A, Friedman N, Regev A. Natural history and evolutionary principles of gene duplication in fungi. Nature. 2007;449:54–61. doi: 10.1038/nature06107. [DOI] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E, et al. (222 co-authors) Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yang Z, Wong WS, Nielsen R. Bayes empirical Bayes inference of amino acid sites under positive selection. Mol Biol Evol. 2005;22:1107–1118. doi: 10.1093/molbev/msi097. [DOI] [PubMed] [Google Scholar]
- Zhang J, Nielsen R, Yang Z. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol. 2005;22:2472–2479. doi: 10.1093/molbev/msi237. [DOI] [PubMed] [Google Scholar]