Abstract
Epigenetic information includes heritable signals that modulate gene expression but are not encoded in the primary nucleotide sequence. We have studied natural epigenetic variation in three allotetraploid sibling orchid species (Dactylorhiza majalis s.str, D. traunsteineri s.l., and D. ebudensis) that differ radically in geography/ecology. The epigenetic variation released by genome doubling has been restructured in species-specific patterns that reflect their recent evolutionary history and have an impact on their ecology and evolution, hundreds of generations after their formation. Using two contrasting approaches that yielded largely congruent results, epigenome scans pinpointed epiloci under divergent selection that correlate with eco-environmental variables, mainly related to water availability and temperature. The stable epigenetic divergence in this group is largely responsible for persistent ecological differences, which then set the stage for species-specific genetic patterns to accumulate in response to further selection and/or drift. Our results strongly suggest a need to expand our current evolutionary framework to encompass a complementary epigenetic dimension when seeking to understand population processes that drive phenotypic evolution and adaptation.
Keywords: adaptation, epigenetics, evolution, hybridization, polyploidy, selection
Introduction
Without altering the underlying DNA sequence, epigenetic information influences the identity of cells and their response to the environment by modulating gene expression. Mechanisms providing epigenetic signals include DNA (cytosine) methylation, histone modifications, and small RNAs; these processes are at least partly interrelated (Lister et al. 2008; Zhang 2008). Laboratory-based mechanistic understanding of epigenetics in model organisms is expanding rapidly. These new findings indicate that, in addition to genetic information, epigenetic alleles (i.e., epialleles) influence phenotypic outcomes (Jablonka and Raz 2009; Johannes et al. 2009) and even have the potential to result in fitness differences subject to natural selection in identical genetic backgrounds (e.g., monozygotic twins: Fraga et al. 2005).
However, the prevalence of alternative epialleles in wild organisms and their significance to phenotypic variation, ecological interactions, and selection in real-world contexts remain largely unexplored (Rapp and Wendel 2005; Bossdorf et al. 2008; Turner 2009). Therefore, the likely importance of epigenetic information in evolution and adaptation seems underrated. This neglect has been reinforced by a general perception of epigenetic information as being short-lived, typically being reset every generation in mammals. However, several examples are known in which epialleles have been stably inherited across generations and demonstrably affect key phenotypic characters, such as floral shape, vegetative and seed pigmentation, pathogen resistance, and general development in angiosperms (Richards 2006; Jablonka and Raz 2009). Probably the best-known example in plants, hypermethylation and silencing of the gene cycloidea (Lcyc), resulted in a peloric variant of Linaria flowers (Cubas et al. 1999), which is inferred to have originated more than 250 years ago at its locus classicus and has been inherited ever since. Fewer cases, but perhaps better known, in which the environment affects biological properties of subsequent generations exist also in mammals. For example, altering the diet of a pregnant rodent can change coat color of pups (Waterland and Jirtle 2003) or lead to male fertility defects in subsequent generations (Anway et al. 2005). Jablonka and Raz (2009) recently reviewed over 100 cases of inherited epigenetic variations in bacteria, protists, fungi, plants, and animals. Most examples of spontaneous heritable epialleles are, however, found in flowering plants, reflecting frequent epigenetic meiotic persistence and the characteristically late partitioning of reproductive and vegetative cell lineages in higher plants (Richards 2008).
Heritable epialleles may mimic random genetic mutations in formation pathway and implications for evolution. However, what makes epigenetic processes fundamentally different from genetic mechanisms is that they can be directly disrupted by the environment, which gives them a neoLamarckian flavor (Richards 2006), potentially permitting heritability of acquired characteristics (Turner 2009). Also, epigenetic variation may be generated at a much higher rate than equivalent genetic variation, especially in rapidly changing environmental conditions, when organisms must respond by producing alternative phenotypes (Angers et al. 2010). Other important (yet still theoretical) evolutionary implications of epigenetics concern the relaxation of the link between natural selection and recombination: Environmental disruption can simultaneously induce several new potentially advantageous epimutations in the same individual, in marked contrast with genetic changes. Therefore, natural epigenetic processes partly underpin phenotypic variation and can play a significant role in adaptation and natural selection.
Epigenetic alterations are especially prevalent after genomic stress, such as that caused by hybridization and/or polyploidization—phenomena of global importance in angiosperm evolution and diversification (De Bodt et al. 2005; Paun et al. 2009; Soltis et al. 2009; Van de Peer et al. 2009; Wood et al. 2009). By activating mobile elements (Parisod et al. 2009) and silencing redundant genes, epigenetic effects constitute an effective and flexible mechanism for stabilizing cellular processes immediately after genome doubling (Chen 2007; Paun et al. 2007; Doyle et al. 2008). Once stabilized, polyploidy offers a duplicated genetic background on which epigenetic novelties may have higher chances to become adaptive. Little information exists, however, regarding the stability of the epigenetic changes in later-generation polyploids and their long-term adaptive and evolutionary implications.
We have studied genome-wide natural epigenetic variation in three closely related allotetraploid (2n = 80) species of the Dactylorhiza majalis complex (Orchidaceae). This complex has arisen by multiple, independent origins from the same diploid parental lineages (D. fuchsii and D. incarnata; both 2n = 40). Dactylorhiza majalis s.str., D. traunsteineri s.l. (fig. 1A), and the putative narrow endemic D. ebudensis have been formed at different times during the last part of the quaternary (Pillon et al. 2007). The three allotetraploids differ substantially in geography and ecology (see below). We demonstrate here that the widespread epigenetic alterations triggered by genome doubling within these sibling allopolyploids have been molded by divergent selection under environmental induction. This results in species-specific epigenetic patterns that have a direct impact on the ecology, distribution, and evolution of these lineages, potentially hundreds of generations after their formation.
FIG. 1.
Epigenetic patterns in sibling polyploid Dactylorhiza. (A) The allotetraploid Dactylorhiza traunsteineri at a natural site in Yorkshire, England. (B) PCoA (goodness of fit 0.87 at P = 0.001) of methylation status of allotetraploids D. majalis s. str. (blue symbols), D. traunsteineri s.l. (dark purple symbols), and D. ebudensis (pink symbols). Geographical provenance is indicated by symbol shapes: squares, Pyrenees; diamonds, Britain; triangles, Scandinavia; circles, Alps. The dotted line encloses samples from Yorkshire, England, and the dashed line D. traunsteineri s.l. samples from northwestern Scotland.
Materials and Methods
Plant Material
The allotetraploid D. majalis complex has evolved polytopically from unidirectional hybridization between the diploids D. fuchsii (in all known cases the maternal parent) and D. incarnata (Pillon et al. 2007). Different polyploid species can occur sympatrically but less often at exactly the same site; their ecological requirements and distributions are generally distinct (fig. 2). The evolutionary history of the complex has been inferred from data on the distribution of plastid haplotypes among allotetraploids and their parentals (Pillon et al. 2007; Hedrén et al. 2008), nuclear microsatellite and amplified fragment length polymorphism (AFLP) markers (Hedrén et al. 2001, Hedrén M, Nordström S, Bateman RM, unpublished data), the degree of concerted evolution in ITS alleles (Pillon et al. 2007), and in agreement with the patterns of morphology and ecological preferences. Dactylorhiza majalis is genetically homogeneous but comparatively derived, including a wide variety of plastid haplotypes that are no longer encountered in the maternal lineage. Its transcriptome, studied with cDNA-AFLP, proved to be also slightly more derived than that of D. traunsteineri, including, for example, more novel transcripts (Paun et al. forthcoming 2010). Dactylorhiza majalis is accordingly viewed as the oldest of the three allotetraploids; it has most probably been molded by glacially induced bottlenecks in southern Europe. Today, it has a fairly wide ecological tolerance of soil moisture and occurs in damp meadows and fens in western and central Europe, the Baltic region, and northwestern Russia. By contrast, D. traunsteineri s.l. is a more recently evolved set of allotetraploids that are more heterogeneous and still maintain both parental ITS alleles in most accessions (Pillon et al. 2007). It probably originated postglacially and at present exhibits a more localized and disjunct distribution in northwestern and central Europe. It has narrow tolerances of both soil moisture and pH and grows in calcareous fens and marshes. The third allotetraploid studied here, D. ebudensis, is a narrow endemic forming at present a single near-contiguous population on the island of North Uist, in northwestern Scotland. Most probably it has only a single relatively recent origin, being as young as, or more probably younger than, D. traunsteineri (cf. Pillon et al. 2007; Hedrén M, Nordström S, Bateman RM, unpublished data). The eustatic–epistatic history of the regions means that its distinctive habitat is unlikely to have formed more than 2,500 years ago. The coastal dune slack habitat occupied by D. ebudensis indicates relatively narrow tolerances of both soil moisture and pH. All individual plants included in the present study (supplementary table S1, Supplementary Material online) were first genotyped using various nuclear and plastid markers to be certain that they conformed to type.
FIG. 2.
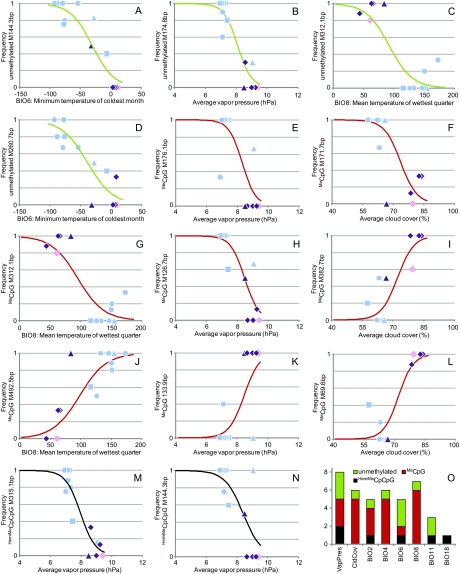
Loci under selection as indicated by SAM (Joost et al. 2007). (A–N) Graphs of the logistic sigmoid functions, symbolized with lines, corresponding to relevant pairs of epigenetic markers and their most significantly associated environmental variable (table 1). Symbols indicate the observed within-population frequency of the given marker for the corresponding value of the investigated ecoclimatic parameter. The shape and color of the symbols follow figure 1. (O) Histogram showing the environmental variables that significantly explain patterns of alternative epialleles. Some methylation markers are associated with more than one ecological variable (table 1).
The MSAP Technique
The methylation-sensitive amplified polymorphism (MSAP) approach is similar to standard AFLP (Vos et al. 1995) but uses two methylation-sensitive isoschizomers (MspI and HpaII) as frequent cutters, each in combination with the same rare cutter (EcoRI) in parallel batches (Baurens et al. 2003). The two isoschizomers recognize the same sequence (5′-CCGG) but differ in their sensitivity to DNA methylation. Comparison of the two profiles for each individual allowed assessment of the methylation state of the restriction sites. MeCpG sites (maintained during cell division by MET1 DNA methyltransferase; Finnegan and Kovac 2000) are recognized by MspI only, whereas plant-specific HemiMeCpCpG sites (established by the CMT3 and DRM methyltransferases; Finnegan and Kovac 2000) are recognized by HpaII only (e.g., REBASE or Salmon et al. 2005). Sites that are hypermethylated (i.e., both at the internal and external Cs), and sites that are fully methylated at the external Cs (i.e., on both strands) are not cut by either enzyme, whereas sites that are free from methylation are recognized by both.
Total genomic DNA was extracted from similar amounts of silica gel-dried flower tissue using a CTAB method (Doyle JJ and Doyle JL 1987). Care was taken to collect flowers in the field at the same developmental stage so that any developmentally related variation in methylation would not confound our ability to determine genotype-specific variation in methylation patterns. Genomic DNA (ca. 0.5 μg) was digested and ligated to double-stranded adapters in one step at 37 °C overnight. The reaction mix (final volume 11 μl) contained 1.1 μl T4 DNA ligase buffer (Promega), 0.55-μl bovine serum albumin (1 mg/ml; New England Biolabs), 1.1 μl 0.5 M NaCl, 10 U MspI or HpaII (both Fermentas) in parallel reactions, 10 U EcoRI (Promega), 1 U T4 DNA ligase (Promega), 1 μl 50 μM MspI/HpaII-adapters, 1 μl 5 μM EcoRI adapters, and approximately 0.5 μg template DNA. The sequence of the HpaII/MspI adapters and primers used followed Salmon et al. (2005). Ligated DNA fragments were diluted 10-fold. Negative control samples were included in all steps to test for contamination (Bonin et al. 2004).
Both preselective and selective amplifications were performed in a 10-μl volume in a thermocycler (GeneAmp PCR System 9700, Applied Biosystems). Polymerase chain reaction (PCR) protocols followed Vos et al. (1995). The reaction mix for the preselective amplification contained 1.14 μl 10× RedTaq PCR Reaction buffer (Sigma), 0.2 U RedTaq, 0.22 μl dinucleotide triphosphates (dNTPs) (10 mM; Applied Biosystems), 0.58 μl preselective primers (5 μM), and 2 μl diluted product of the restriction/ligation. The PCR product was diluted 10-fold. The selective primers chosen after a primer trial were EcoRI ACA (6-FAM) in combination with HpaII/MspI CTG, and EcoRI AGG (JOE) and EcoRI AGC (NED), both in combination with HpaII/MspI CAT. The reaction mix for the selective amplification contained 1 μl 10× RedTaq PCR reaction buffer (Sigma), 0.2 U RedTaq, 0.22 μl dNTPs (10 mM; Applied Biosystems), 0.54 μl of each selective primer (MspI/HpaII-primer: 5 μM; EcoRI-primer: 1 μM), and 2 μl diluted product of the preselective amplification. The selective PCR product was purified using Sephadex G-50 Superfine (GE Healthcare BioSciences) applied to a Multi Screen-HV plate (Millipore) in three steps of 200 μl each and packed at 600 g for 1, 1, and 5 min, respectively. The same rotation was used for centrifugation of the samples (5 μl of each selective PCR product), again for 5 min. Then, 1.5 μl of the elution product was combined with 10-μl formamide and 0.2 μl GeneScan ROX (Applied Biosystems) internal size standard and run on a capillary sequencer ABI 3100 (Applied Biosystems). Fragments in the range 50–500 bp were aligned using ABI PRISM GeneScan 2.1 analysis software (Applied Biosystems) and visualized, scored, and exported as binary presence/absence matrix using Genographer 1.6 (available from http://hordeum.msu.montana.edu/genographer/). Scoring was performed for all samples in the same batch, including both HpaII and MspI profiles. Each AFLP fragment was scored using the “thumbnail” option, which allows comparison of the signal of each fragment (present or absent) over all profiles and samples. Twelve samples (20% of the total) were genotyped twice to test the reproducibility of MSAP fingerprinting (Bonin et al. 2004), identifying an error rate of 1.1%. The MSAP data are deposited in the Dryad Digital Repository and can be found at http://hdl.handle.net/10255/dryad.1522.
cDNA-AFLP Technique
Being based on mRNA, this method comparatively examines changes in the transcribed regions of the genome and can compare gene expression in multiple individuals. Transcript polymorphism as identified by cDNA-AFLP may not necessarily represent expression differences: apart from gene silencing, physical loss, and nonsynonymous polymorphism (indels, substitutions, and rearrangements), synonymous substitutions may also be visible to this method (Paun et al. 2007). However, due to their relatively recent origin and similar genetic background (see above), most of the variation depicted with cDNA-AFLPs should reflect expression differentiation rather than polymorphism at the nucleotide level in the exons.
The standard AFLP procedure (Vos et al. 1995) was performed on a pool of cDNAs (Bachem et al. 1996; Paun et al. 2007) generated from leaves sampled from Dactylorhiza plants grown in uniform conditions at RBG Kew. In brief, total RNA was isolated with the SV Total RNA Isolation System (Promega), following the manufacturer’s protocol from tissue material fixed in RNALater (Sigma) and stored at −20 °C. Then, cDNA was synthesized from mRNA with SuperScriptTM Double-Stranded cDNA Synthesis Kit (Invitrogen), following the manufacturer’s protocol and using an oligo (dT)12-18 primer. The next steps generally followed the protocol of AFLP Plant Mapping Kit (Applied Biosystems) but used only half of the recommended reaction volumes. Double-stranded cDNA was digested with 1U MseI (New England Biolabs) and 5U EcoRI (Promega) and ligated (with 1U of T4 DNA Ligase; Promega) to double-stranded adapters for 2 h at 37 °C. Blind samples and two replicated samples (18% of the total) were included in all steps to test for contamination and reproducibility (Bonin et al. 2004). Preselective amplification was performed using primer pairs with one selective nucleotide. The products of 27 primer combinations (of the general type EcoRI AX [+fluorescent dye]-MseI CYZ, where X, Y, and Z are different selective nucleotides) were separated and scored as described for MSAP profiles. In total, 109 unambiguous polymorphic fragments were obtained. All individuals analyzed showed distinct transcript profiles.
Standard AFLP Technique
Genome-wide, neutral data were generated following Vos et al. (1995) and the MSAP procedure described above with minor modifications: genomic DNA (ca. 500 ng) was digested with 1 U MseI (New England BioLabs) and 5 U EcoRI (Promega) for 2 h at 37 °C. Three selective primer combinations were chosen after a primer trial (fluorescent dye in brackets): EcoRI AGG (VIC)-MseI CAG; EcoRI ACA (6-FAM)-MseI CTG; and EcoRI AGC (NED)-MseI CAG. In total, 114 polymorphic fragments were scored. Blind samples and replicates were included to test for contamination and reproducibility (Bonin et al. 2004).
Data Analyses
Any monomorphic fragments and fragments present/absent in all but one individual were removed from all data sets to avoid biased parameter estimates (Bonin et al. 2004). To visualize the pattern of population differentiation, we constructed a principal coordinates analysis (PCoA) with NTSYS-pc 2.02h following the procedure detailed in Paun et al. (2008). Furthermore, using Mantel tests as implemented in NTSYS-pc (module “MxComp”) and 1,000 permutations, we compared the cDNA-AFLP matrix with a reduced MSAP data set (based on 11 individuals and 176 polymorphic fragments), with the genomic AFLP matrix and, respectively, with a combined matrix of AFLP and MSAP data. All comparisons were computed between the same individuals using the Nei and Li (1979) similarity. The Nei–Li algorithm, which does not treat shared band absence as homologous, was chosen because absence of MSAP fragments can result from either a full methylation of cytosines on both strands or from genetic clues (e.g., absence of restriction site in the DNA sequence).
To identify candidate adaptive epiloci that are selected by native environmental conditions and may therefore play a role favoring the presence of the individuals in a given landscape, we performed multiple univariate logistic regressions in spatial analysis method (SAM) (Joost et al. 2007, 2008), testing for association between methylation markers and environmental variables. As SAM takes the individual as the reference unit, the analysis functions independently of any notion of population and is largely assumption free (Joost et al. 2008). The analysis used 21 GIS-based ecoclimatic variables, including 19 bioclimatic parameters from Worldclim (http://www.worldclim.org/bioclim.htm) (Hijmans et al. 2005), plus yearly averages for vapor pressure (in hPa) and percentage cloud cover from IWMI Climate and Water Atlas (http://dw.iwmi.org).
Epiloci showing an atypical pattern of variability compared with the rest of the epigenome and thus inferred to be potentially under selection were detected with BayeScan (Foll and Gaggiotti 2008) using a Bayesian outlier locus approach. BayeScan relies on estimates of epigenetic structure among sampling plots, with the rationale that epiloci influenced by divergent selection will show greater genetic differentiation than neutral epiloci, and epiloci under purifying selection will exhibit the opposite trend (Foll and Gaggiotti 2008). Descriptive statistical measures used in population genetics, such as FST, are equally transferable to describe population differentiation at epigenetic level (Bossdorf et al. 2008). Moreover, simulation studies demonstrated the robustness of BayeScan to a wide range of biological scenarios (Foll and Gaggiotti 2008). BayeScan was run on the methylation data for 1,250,000 iterations, with the first 250,000 excluded as burn-in. An additional burn-in was represented by 10 pilot runs that were performed before starting the sampling; these were used to better estimate the mean and variance of alpha.
Results
Methylation Data
Analyses were performed on a data set of estimated presence/absence of methylation status obtained by comparing the paired MSAP profiles for each individual and removing any redundant information in the paired profiles. As a result, the 251 scored fragments provided information for a total of 332 loci, out of which 133 unmethylated markers (when present), 110 loci methylated at CpG sites, and 89 markers that were interpreted as being hemimethylated at CpCpG sites. The genome-wide methylation information clearly separates the three species (fig. 1B) and partly discriminates geographic provenance at the population level. Within-species clustering was most pronounced for D. traunsteineri, where individuals from northeastern England, western Scotland, and Scandinavia were clearly distinguished by similar distances to those separating them from D. ebudensis. The situation was different for D. majalis, where only the Pyrenean samples were slightly separated from the intermingled Alpine and Scandinavian individuals.
Comparisons of Methylation, Neutral Genetic, and Gene Expression Patterns
Gene expression differences between the allopolyploid species are reflected better by methylation information, as estimated with MSAP, than by neutral genome-wide genetic data (r = 0.81 vs. r = 0.67, both significant at P < 0.01). A combined data set of genetic and epigenetic information correlates marginally more strongly with gene expression patterns (r = 0.84, P < 0.01).
Detection of Loci Under Selection
To detect signatures of selection within epigenetic information and pinpoint candidate epiloci, we used two complementary genome-wide scan approaches. After Bonferroni correction of the significance level for multiple comparisons (set to 7.17 × 10−06, corresponding to 95% confidence), univariate logistic regression models inferred with SAM indicated that 87 methylation markers (26%) are correlated with at least one environmental parameter using the G statistical test, although only 14 markers were confirmed as significantly associated with ecoclimatic data via the more stringent Wald test (Joost et al. 2007) (table 1 and fig. 2A–N). Eight of these markers were affected by methylation at the internal C positions (MeCpG), 4 of the candidate loci were unmethylated (when present), and only 2 were hemimethylated at the external C of the cutting site (HemiMeCpCpg). Most of the markers were correlated with vapor pressure and/or mean temperature of the wettest quarter, BIO8 (fig. 2O). Partial correlation between environmental parameters meant that most of the candidate epiloci correlated significantly with more than one ecoclimatic variable (table 1).
Table 1.
Significant Associations between MSAP Loci and Ecoclimatic Parameters, as Indicated by SAM Analysis (see also fig. 2).
| Marker Type | Marker |
Environmental Variables—SAM Results |
BayeScan |
|||||||
| I (P) |
II (P) |
III (P) |
log10(BF) | PP | ||||||
| Unmethylated | a1 | 144.3 bp | BIO6 | 295 × 10−8 | BIO11 | 313 × 10−8 | VapPres | 664 × 10−8 | 1.18 | 0.9482 |
| a2 | 174.8 bp | VapPres | 62 × 10−8 | BIO2 | 268 × 10−8 | BIO6 | 520 × 10−8 | 1.17 | 0.9372 | |
| a3 | 260.7 bp | BIO6 | 238 × 10−8 | BIO11 | 390 × 10−8 | VapPres | 406 × 10−8 | 0.67 | 0.8254 | |
| a4 | 312.1 bp | BIO8 | 202 × 10−8 | CldCov | 218 × 10−8 | BIO4 | 262 × 10−8 | 1.42 | 0.9632 | |
| MeCpG | b1 | 69.6 bp | CldCov | 197 × 10−8 | BIO8 | 228 × 10−8 | — | — | 1.47 | 0.9672 |
| b2 | 126.7 bp | VapPres | 169 × 10−8 | BIO8 | 232 × 10−8 | BIO2 | 436 × 10−8 | 1.59 | 0.9752 | |
| b3 | 133.9 bp | VapPres | 206 × 10−8 | BIO2 | 471 × 10−8 | BIO4 | 631 × 10−8 | 3.7 | 0.9998 | |
| b4 | 171.7 bp | CldCov | 158 × 10−8 | BIO4 | 323 × 10−8 | BIO8 | 557 × 10−8 | 1.67 | 0.9790 | |
| b5 | 176.1 bp | VapPres | 59 × 10−8 | BIO2 | 243 × 10−8 | BIO6 | 651 × 10−8 | 1,000 | 1.0000 | |
| b6 | 308.8 bp | — | — | — | — | — | — | 2.4 | 0.9960 | |
| b7 | 312.1 bp | BIO8 | 240 × 10−8 | CldCov | 341 × 10−8 | BIO4 | 411 × 10−8 | 0.96 | 0.9006 | |
| b8 | 382.7 bp | CldCov | 211 × 10−8 | BIO4 | 243 × 10−8 | BIO8 | 246 × 10−8 | 1.05 | 0.9176 | |
| b9 | 492.5 bp | BIO8 | 282 × 10−8 | BIO4 | 437 × 10−8 | — | — | 0.58 | 0.7902 | |
| HemiMeCpCpG | c1 | 144.3 bp | VapPres | 487 × 10−8 | — | — | — | — | 2.25 | 0.9944 |
| c2 | 315.1 bp | VapPres | 185 × 10−8 | BIO6 | 262 × 10−8 | BIO18 | 396 × 10−8 | 0.63 | 0.6937 | |
| c3 | 367.4 bp | — | — | — | — | — | — | 2.92 | 0.9988 | |
NOTE.—Most MSAP markers were significantly associated with more than one ecoclimatic parameter (only the first three most significant are shown here). Abbreviations: VapPres, vapor pressure (hPa); CldCov, cloud coverage (%); BIO2, mean diurnal range; BIO4, temperature seasonality; BIO6, minimum temperature of coldest month; BIO8, mean temperature of wettest quarter; BIO18, precipitation of warmest quarter; P, significance. Selection type for some of the markers was inferred with BayeScan (see fig. 3).
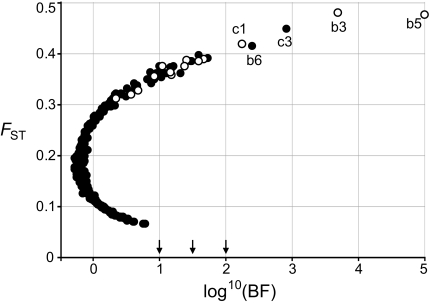
To confirm these results, a Bayesian outlier locus approach was used to estimate the posterior probability (PP) of each epilocus being under selection and differentiate the type of selection each marker was subjected to (divergent vs. purifying). BayeScan analyses (fig. 3) identified 23 MSAP candidate markers under selection with “strong evidence” (log10(BF) > 1, PP > 0.91) on the Jeffrey’s scale (Foll and Gaggiotti 2008). Nine of these markers were “very strongly” indicated to be under selection (log10(BF) > 1.5, PP > 0.97), and evidence for five of them was judged “decisive” (log10(BF) > 2, PP > 0.99). All these epiloci are characterized by FST values significantly higher than the average (0.192) and are therefore interpreted as having been affected by divergent (i.e., bidirectional) selection. Ten methylation markers were confirmed by both SAM and BayeScan as being under selection (table 1).
FIG. 3.
Scan for epiloci under selection performed using BayeScan (Foll and Gaggiotti 2008). The PP for a locus to be under natural selection is shown on a log scale on the x axis (see also table 1). The open symbols indicate adaptive markers indentified with SAM. The three arrows on the x axis from left to right show the minimum threshold for strong, very strong, and decisive evidence for selection on Jeffrey’s scale (Foll and Gaggiotti 2008). Note that for epilocus b5, log10(BF) = 1,000, but the program places it in the graph at the value 5 because of space limitations.
Discussion
For much of the last century, our understanding of evolution has been based on the Modern Synthesis, assuming that natural selection is acting solely on the amount and structure of “chance” genetic variation, of which the ultimate origin is “random” mutation. However, natural selection directly targets phenotypic variation. The neo-Darwinian evolutionary foundation has recently been deemed incomplete by the discovery that novel, variable, and heritable gene expression could be achieved via a suite of epigenetic changes “under environmental influence” (Richards 2006; Turner 2009), even in the complete absence of genetic variability. Here, we emphasize this ostensibly heterodox idea and provide new corroborating evidence. By analyzing established alloplyploid Dactylorhiza species that share a highly similar genetic heritage, we demonstrate that ecological divergence in sibling allopolyploids is largely the result of adaptation achieved by epigenetic effects that modulate gene expression under environmental influence. Such a process may occur commonly after genome doubling in frequently observed polyploid complexes (Soltis et al. 2009).
Dactylorhiza has long been recognized as an evolutionarily complex genus with a history of recurrent hybridization and polyploidization (Heslop-Harrison 1968; Hedrén 1996; Hedrén et al. 2001, 2008; Pillon et al. 2007). Reticulate evolution has resulted in significant but often subtle morphological/ecological variation that challenges species delimitation. In addition, neutral genetic differentiation between Dactylorhiza allopolyploids is rarely detectable (Hedrén et al. 2001; Pillon et al. 2007), which is typical in such polyploid complexes. In stark contrast, genome-wide epigenetic variation, studied using methylation-sensitive enzymes, clearly separates the three allopolyploid species studied here (fig. 1B) and confirms previous hypotheses of their evolutionary history (Heslop-Harrison 1968). For example, both D. majalis and D. traunsteineri clearly have polytopic origins, but the former is likely to have formed around the last glacial maximum (Hedrén et al. 2001; Pillon et al. 2007), presumably responding to profound climate change by migrating alongside its progenitors. This migration-induced bottleneck resulted in decreased genetic variation (Hedrén et al. 2001; Pillon et al. 2007), but it seems also to have reshaped within-species epigenetic differentiation, as D. majalis now exhibits a relatively homogeneous methylation pattern (fig. 1B). By contrast, as a result of its postglacial formation and present disjunct distribution, the genome-wide methylation patterns of D. traunsteineri are more heterogeneous and correlate more clearly with geography. Genetic bottlenecks have been hypothesized on several occasions to result in an immediate release of epigenetic variation (e.g., Rapp and Wendel 2005). This is at present a poorly explored arena; however, in D. majalis, only parts of this novel epigenetic variation seem to have achieved fixation in response to subsequent selection.
The fact that recent evolutionary history is discernable in the patterns of epigenetic variation indicates that the pace of resetting at a significant number of loci may be much less rapid than was previously thought (Reik and Dean 2002). It is noteworthy that, in this group, the release of epigenetic variation triggered in first-generation allopolyploids has been found to provide lineage-specific patterns and that some of this variation seems to persist stably over many generations. Although containing information from both coding and noncoding DNA regions, the methylation patterns reflect better gene-expression differences in these species than neutral genome-wide genetic data. Therefore, physical (genetic) diversification per se may be less relevant for the three allopolyploids studied. Divergence between them may instead reside in quantitative partitioning of expression patterns via epigenetic changes at individual genes (see also King and Wilson 1975). Indeed, expression levels of a gene alone can determine phenotypic variation, contributing substantially to the natural variation on which selection can act (Bossdorf et al. 2008). Modeling studies suggest that epigenetic variation can facilitate jumps between fitness slopes by reducing genetic/ecological barriers represented by valleys in an adaptive landscape (Pal and Miklos 1999); these potentially lethal valleys have received far less attention than peaks in evolutionary modeling (Bateman and DiMichele 2002; Gavrilets 2004).
Identifying loci under selection that play a role in adaptation to different environments is a long-standing ambition of evolutionary biologists (Nielsen 2005; Foll and Gaggiotti 2008; Nosil et al. 2009). Our results pinpoint several methylation markers that correlate significantly with environmental parameters (fig. 2 and table 1) and are probably invoked by native ecological conditions to maximize the fitness of individuals in that environment. This indicates that the environment shapes methylation patterns in independently formed allopolyploids to create similar races. Indeed, the epigenetic constitution of an individual or species is sensitive to its environment (Richards 2006; Bossdorf et al. 2008; Angers et al. 2010). The relevant environmental factors (table 1) are hypothesized to exert divergent selection pressures responsible for the presence of a particular individual/population/species in a given portion of the ecological landscape (Joost et al. 2007). Water availability in combination with temperature (i.e., as indicated by vapor pressure and mean temperature of the wettest quarter) appears to be a key factor causing environmental allopatry in Dactylorhiza, being identified both at the epigenetic level (table 1 and fig. 2O) and by transcriptome profiling (Paun et al. forthcoming 2010). If maintained over evolutionary timescales, environmental allopatry, even when associated with limited specialization to only subtle differences in ecological conditions, may effectively limit dispersal between populations, thus promoting divergence via the stochastic effects of genetic drift (Nosil et al. 2009) and further directional selection.
Therefore, stable epigenetic divergence between genetically similar Dactylorhiza species may be largely responsible for lasting ecological differences. It remains uncertain whether the visibly cohesive epigenetic patterns of the allopolyploid lineages studied here are a result of past single environmental inductions, followed by long-term meiotic inheritance and selective fixation or instead originate from repeated and ongoing disruptions under environmental pressure. However, repeated environmental induction is unlikely to explain the entire range of epigenetic variation described here because the oldest allopolyploid (i.e., D. majalis) exhibits less epigenetic variation than D. traunsteineri but occupies a larger distribution area which may be ecologically more diverse. Aside from being extensively involved in developmental controls and parent-of-origin imprinted gene expression (Steimer et al. 2004; Henderson and Jacobsen 2007), epigenetic processes are clearly playing a key role in incipient adaptation and evolution by influencing primary phenotypic diversity at the interface between genetics and the environment. Adaptation through selection of heritable epialleles implies a need to expand the gene-centered view that still dominates evolutionary thinking about variation, heritability, and evolution (Jablonka and Lamb 2005; Richards 2006; Bossdorf et al. 2008). Accordingly, our focus in the study of evolution should shift from single genes to developmental/regulatory networks and holistic phenotypes.
Supplementary Material
Supplementary table S1 is available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Supplementary Material
Acknowledgments
We are grateful to I. Denholm, F. Horsman, G. Joseph, and A. Tribsch for help in finding/collecting plant material, to C. Ryan for maintaining Dactylorhiza plants in cultivation, and to R. Cowan, M. Frohlich, A. Leitch, I. Leitch, B. Sayers, and two anonymous reviewers for helpful comments. J. Luna assisted with genotypic plant samples at various loci prior to these analyses. J. Moat provided environmental data points from Worldclim for the sampling localities; N. Gamage granted access to the IWMI Climate and Water Atlas. Natural England and Forestry Commission England kindly provided collecting permits. This work was supported by a Marie Curie Fellowship (EU Commission, MEIF-CT-2007-040494) and by an Austrian Science Fund (FWF) project (P222260-B16), both to O.P.
References
- Angers B, Castonguay E, Massicotte R. Environmentally induced phenotypes and DNA methylation: how to deal with unpredictable conditions until the next generation and after. Mol Ecol. 2010;19:1283–1295. doi: 10.1111/j.1365-294X.2010.04580.x. [DOI] [PubMed] [Google Scholar]
- Anway MD, Cupp AS, Uzumcu M, Skinner MK. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science. 2005;308:1466–1469. doi: 10.1126/science.1108190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachem CWB, van der Hoeven RS, de Bruijn SM, Vreugdenhil D, Zabeau M, Visser RGF. Visualization of differential gene expression using a novel method of RNA fingerprinting based on AFLP: analysis of gene expression during potato tuber development. Plant J. 1996;9:745–753. doi: 10.1046/j.1365-313x.1996.9050745.x. [DOI] [PubMed] [Google Scholar]
- Bateman RM, DiMichele WA. Generating and filtering major phenotypic novelties: neoGoldschmidtian saltation revisited. In: Cronk QCB, Bateman RM, Hawkins JA, editors. Developmental genetics and plant evolution. London: Taylor & Francis; 2002. pp. 109–159. [Google Scholar]
- Baurens F-C, Bonnot F, Bienvenu D, Causse S, Legavre T. Using SD-AFLP and MSAP to assess CCGG methylation in the banana genome. Plant Mol Biol Rep. 2003;21:339–348. [Google Scholar]
- Bonin A, Bellemain E, Bronken Eidesen P, Pompanon F, Brochmann C, Taberlet P. How to track and assess genotyping errors in population genetic studies. Mol Ecol. 2004;13:3261–3273. doi: 10.1111/j.1365-294X.2004.02346.x. [DOI] [PubMed] [Google Scholar]
- Bossdorf O, Richards CL, Pigliucci M. Epigenetics for ecologists. Ecol Lett. 2008;11:106–115. doi: 10.1111/j.1461-0248.2007.01130.x. [DOI] [PubMed] [Google Scholar]
- Chen ZJ. Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annu Rev Plant Biol. 2007;58:377–406. doi: 10.1146/annurev.arplant.58.032806.103835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubas P, Vincent C, Coen E. An epigenetic mutation responsible for natural variation in floral symmetry. Nature. 1999;401:157–161. doi: 10.1038/43657. [DOI] [PubMed] [Google Scholar]
- De Bodt S, Maere S, Van de Peer Y. Genome duplication and the origin of angiosperms. Trends Ecol Evol. 2005;20:591–597. doi: 10.1016/j.tree.2005.07.008. [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 1987;19:11–15. [Google Scholar]
- Doyle JJ, Flagel LE, Paterson AH, Rapp RA, Soltis DE, Soltis PS, Wendel JF. Evolutionary genetics of genome merger and doubling in plants. Annu Rev Genet. 2008;42:443–461. doi: 10.1146/annurev.genet.42.110807.091524. [DOI] [PubMed] [Google Scholar]
- Finnegan EJ, Kovac KA. Plant DNA methyltransferases. Plant Mol Biol. 2000;43:189–201. doi: 10.1023/a:1006427226972. [DOI] [PubMed] [Google Scholar]
- Foll M, Gaggiotti O. A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective. Genetics. 2008;180:977–993. doi: 10.1534/genetics.108.092221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraga MF, Ballestar E, Paz MF, et al. (21 co-authors) Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci U S A. 2005;102:10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrilets S. Fitness landscapes and the origin of species. Princeton (NJ): Princeton University Press; 2004. [Google Scholar]
- Hedrén M. Genetic differentiation, polyploidization and hybridization in northern European Dactylorhiza (Orchidaceae): evidence from allozymes markers. Plant Syst Evol. 1996;201:31–55. [Google Scholar]
- Hedrén M, Fay MF, Chase MW. Amplified fragment length polymorphisms (AFLP) reveal details of polyploidy evolution in Dactylorhiza (Orchidaceae) Am J Bot. 2001;88:1868–1880. [PubMed] [Google Scholar]
- Hedrén M, Nordström S, Ståhlberg D. Polyploid evolution and plastid DNA variation in the Dactylorhiza incarnata/maculata complex (Orchidaceae) in Scandinavia. Mol Ecol. 2008;17:5075–5091. doi: 10.1111/j.1365-294X.2008.03965.x. [DOI] [PubMed] [Google Scholar]
- Henderson IR, Jacobsen SE. Epigenetic inheritance in plants. Nature. 2007;447:418–424. doi: 10.1038/nature05917. [DOI] [PubMed] [Google Scholar]
- Heslop-Harrison J. Genetic system and ecological habit as factors in dactylorchid variation. Jber Naturw Ver Wuppertal. 1968;21+22:20–27. [Google Scholar]
- Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A. Very high resolution interpolated climate surfaces for global land areas. Int J Climatol. 2005;25:1965–1978. [Google Scholar]
- Jablonka E, Lamb MJ. Evolution in four dimensions: genetic, epigenetic, behavioral, and symbolic variation in the history of life. Cambridge (MA): The MIT Press; 2005. [Google Scholar]
- Jablonka E, Raz G. Transgenerational epigenetic inheritance: prevalence, mechanisms, and implications for the study of heredity and evolution. Q Rev Biol. 2009;84:131–176. doi: 10.1086/598822. [DOI] [PubMed] [Google Scholar]
- Johannes F, Porcher E, Teixeira FK, et al. (15 co-authors) Assessing the impact of transgenerational epigenetic variation on complex traits. PLoS Genet. 2009;5:1–11. doi: 10.1371/journal.pgen.1000530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joost S, Bonin A, Bruford MW, Conord C, Erhardt G, Taberlet P. A spatial analysis method (SAM) to detect candidate loci for selection: towards a landscape genomics approach to adaptation. Mol Ecol. 2007;16:3955–3969. doi: 10.1111/j.1365-294X.2007.03442.x. [DOI] [PubMed] [Google Scholar]
- Joost S, Kalbermatten M, Bonin A. Spatial analysis method (SAM): a software tool combining molecular and environmental data to identify candidate loci for selection. Mol Ecol Res. 2008;8:957–960. doi: 10.1111/j.1755-0998.2008.02162.x. [DOI] [PubMed] [Google Scholar]
- King M-C, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- Lister R, O'Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Excker JR. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:1–14. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Li WH. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci U S A. 1979;76:5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R. Molecular signatures of natural selection. Annu Rev Genet. 2005;39:197–218. doi: 10.1146/annurev.genet.39.073003.112420. [DOI] [PubMed] [Google Scholar]
- Nosil P, Funk DJ, Ortiz-Barrientos D. Divergent selection and heterogeneous genomic divergence. Mol Ecol. 2009;18:375–402. doi: 10.1111/j.1365-294X.2008.03946.x. [DOI] [PubMed] [Google Scholar]
- Pal C, Miklos I. Epigenetic inheritance, genetic assimilation and speciation. J Theor Biol. 1999;200:19–37. doi: 10.1006/jtbi.1999.0974. [DOI] [PubMed] [Google Scholar]
- Parisod C, Salmon A, Zerjal T, Tenaillon M, Grandbastien M-A, Ainouche M. Rapid structural and epigenetic reorganization near transposable elements in hybrid and allopolyploid genomes in Spartina. New Phytol. 2009;184:1003–1015. doi: 10.1111/j.1469-8137.2009.03029.x. [DOI] [PubMed] [Google Scholar]
- Paun O, Fay MF, Soltis DE, Chase MW. Genetic and epigenetic alterations after hybridization and genome doubling. Taxon. 2007;56:649–656. [PMC free article] [PubMed] [Google Scholar]
- Paun O, Forest F, Fay MF, Chase MW. Hybrid speciation in angiosperms: parental divergence drives ploidy. New Phytol. 2009;182:507–518. doi: 10.1111/j.1469-8137.2009.02767.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paun O, Luna JA, Fay MF, Bateman RM, Chase MW. Genomic responses drive adaptation in allotetraploid species of Dactylorhiza (Orchidaceae; Orchidinae) In: Seberg O, Petersen G, Barfod AS, Davis JI, editors. Diversity, phylogeny, and evolution in the monocotyledons. Århus: Aarhus University Press; 2010. pp. 169–192. [Google Scholar]
- Paun O, Schönswetter P, Winkler M, Consortium I, Tribsch A. Historical divergence vs. contemporary gene flow: evolutionary history of the calcicole Ranunculus alpestris group (Ranunculaceae) in the European Alps and the Carpathians. Mol Ecol. 2008;17:4263–4275. doi: 10.1111/j.1365-294x.2008.03908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillon Y, Fay MF, Hedrén M, Bateman RM, Devey DS, Shipunov AB, van der Bank M, Chase MW. Evolution and temporal diversification of western European polyploid species complexes in Dactylorhiza (Orchidaceae) Taxon. 2007;56:1185–1208. [Google Scholar]
- Rapp RA, Wendel JF. Epigenetics and plant evolution. New Phytol. 2005;168:81–91. doi: 10.1111/j.1469-8137.2005.01491.x. [DOI] [PubMed] [Google Scholar]
- Reik W, Dean W. Epigenetic reprogramming: back to the beginning. Nature. 2002;420:127. doi: 10.1038/420127a. [DOI] [PubMed] [Google Scholar]
- Richards EJ. Inherited epigenetic variation—revisiting soft inheritance. Nat Rev Genet. 2006;7:395–402. doi: 10.1038/nrg1834. [DOI] [PubMed] [Google Scholar]
- Richards EJ. Population epigenetics. Curr Opin Genet Dev. 2008;18:221–226. doi: 10.1016/j.gde.2008.01.014. [DOI] [PubMed] [Google Scholar]
- Salmon A, Ainouche ML, Wendel JF. Genetic and epigenetic consequences of recent hybridization and polyploidy in Spartina (Poaceae) Mol Ecol. 2005;14:1163–1175. doi: 10.1111/j.1365-294X.2005.02488.x. [DOI] [PubMed] [Google Scholar]
- Soltis DE, Albert VA, Leebens-Mack J, Bell CD, Paterson AH, Zheng C, Sankoff D, dePamphilis CW, Wall PK, Soltis PS. Polyploidy and angiosperm diversification. Am J Bot. 2009;96:336–348. doi: 10.3732/ajb.0800079. [DOI] [PubMed] [Google Scholar]
- Steimer A, Schöb H, Grossniklaus U. Epigenetic control of plant development: new layers of complexity. Curr Opin Plant Biol. 2004;7:11–19. doi: 10.1016/j.pbi.2003.11.008. [DOI] [PubMed] [Google Scholar]
- Turner BM. Epigenetic responses to environmental change and their evolutionary implications. Philos Trans R Soc Lond B Biol Sci. 2009;364:3403–3418. doi: 10.1098/rstb.2009.0125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Peer Y, Fawcett JA, Proost S, Sterck L, Vandepoele K. The flowering world: a tale of duplications. Trends Plant Sci. 2009;14:680–688. doi: 10.1016/j.tplants.2009.09.001. [DOI] [PubMed] [Google Scholar]
- Vos P, Hogers R, Bleeker M, et al. (11 co-authors) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterland RA, Jirtle RL. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol. 2003;23:5293–5300. doi: 10.1128/MCB.23.15.5293-5300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood TE, Takebayashi N, Barker MS, Mayrose I, Greenspoon PB, Rieseberg LH. The frequency of polyploid speciation in vascular plants. Proc Natl Acad Sci U S A. 2009;106:13875–13879. doi: 10.1073/pnas.0811575106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. The epigenetic landscape of plants. Science. 2008;320:489–492. doi: 10.1126/science.1153996. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.