Abstract
The evolution of the floral homeotic genes has been characterized using phylogenetic and functional studies. It is possible to enhance these studies by comparing gene content and order between species to determine the evolutionary history of the regulatory genes. Here, we use a synteny-based approach to trace the evolution of the floral B- and C-function genes that are required for specification of the reproductive organs. Consistent with previous phylogenetic studies, we show that the euAP3–TM6 split occurred after the monocots and dicots diverged. The Arabidopsis TM6 and papaya euAP3 genes are absent from the respective genomes, and we have detected loci from which these genes were lost. These data indicate that either the TM6 or the euAP3 lineage genes can be lost without detriment to flower development. In contrast, PI is essential for male reproductive organ development; yet, contrary to predictions, complex genomic rearrangements have resulted in almost complete breakdown of synteny at the PI locus. In addition to showing the evolution of B-function genes through the prediction of ancestral loci, similar reconstructions reveal the origins of the C-function AG and PLE lineages in dicots, and show the shared ancestry with the monocot C-function genes. During our studies, we found that transposable elements (TEs) present in sequenced Antirrhinum genomic clones limited comparative studies. A pilot survey of the Antirrhinum data revealed that gene-rich regions contain an unusually high degree of TEs of very varied types, which will be an important consideration for future genome sequencing efforts.
Keywords: synteny, B-function, C-function, comparative genomics, evolution
Introduction
In nature, flowers can be seen in an enormous variety of forms. However, the mechanisms controlling flower development are largely conserved. A good example of this is the genetic functions that control the identity of the floral organs. Model flowers are made up of four concentric whorls of organs that are specified by three gene functions (A, B, and C) (Coen and Meyerowitz 1991). Flower development begins with and is maintained by the expression of A-function genes that confer floral identity to emerging tissues. As a result, leaf-like sepals develop in the first whorl. Development of second-whorl organs coincides with the expression of B-function genes that, in combination with the A function, specify petal identity. The development of the floral reproductive organs requires the activity of B- and C-function genes, the expression domains of which are partly controlled by the A function. Male reproductive organs, the stamens, result from the co-expression of B- and C-function genes in the third whorl, whereas the female organs (carpels) are specified by expression of the C function alone in the fourth whorl (Litt 2007; Causier et al. 2010).
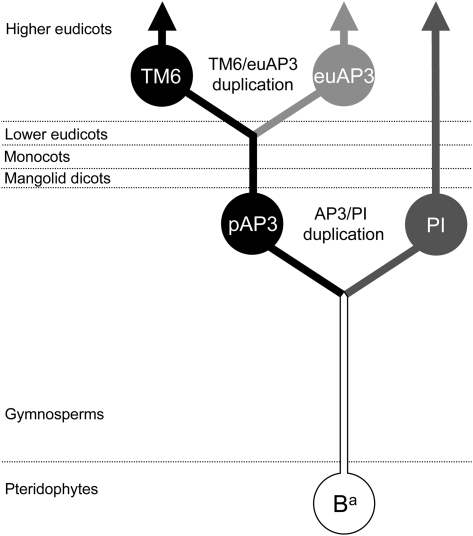
The B- and C-function genes encode transcription factor proteins belonging to the MADS-box family (Schwarz-Sommer et al. 1990; Weigel and Meyerowitz 1994). In plants, extensive gene and genome duplications have resulted in large families of MADS-box genes, leading to the evolution of diverse functions (Martínez-Castilla and Alvarez-Buylla 2003; Parenicová et al. 2003). Duplication of the ancestral B-function gene, before the emergence of the angiosperms, resulted in the paralogous paleoAPETALA3 (paleoAP3) and PISTILLATA (PI) gene lineages (Purugganan et al. 1995; Kramer et al. 1998; Kim et al. 2004, fig. 1). In angiosperms, genes from both lineages are required for a fully active B function. For example, in Arabidopsis, loss of either the AP3 or PI lineage gene functions results in homeotic changes to floral organs, including loss of stamen identity (Jack et al. 1992; Goto and Meyerowitz 1994). Similarly, null alleles of the Antirrhinum genes DEFICIENS (DEF; AP3 lineage) or GLOBOSA (GLO; PI lineage) are characterized by conversion of stamens to female structures (Sommer et al. 1990; Tröbner et al. 1992). A later duplication of paleoAP3 at the base of the core eudicots produced the euAP3 and TM6 (named after tomato MADS-box gene 6) lineages (Kramer et al. 1998, fig. 1). The TM6 lineage proteins share a C-terminal protein motif with paleoAP3 proteins found in monocots, lower dicots, and magnolid dicots. In euAP3 proteins, a frameshift mutation replaced the paleoAP3 motif with a new C-terminal motif, the euAP3 motif (Kramer et al. 1998; Vandenbussche et al. 2003). Thus, the euAP3 lineage, which is unique to the higher eudicots, represents a divergent paralogous group (Irish 2006). The TM6 lineage protein has been lost from Arabidopsis (Rijpkema, Gerats, et al. 2006), but studies in Petunia and tomato have suggested that TM6 lineage genes function predominantly in stamen development (de Martino et al. 2006; Rijpkema, Royaert, et al. 2006). The emergence of the euAP3 lineage coincided with the radiation of the higher eudicots and the evolution of a petal-specific AP3 function (Kramer et al. 1998).
FIG. 1.
Evolution of the B-function gene lineages. The ancestral B-function gene (Ba) duplicated prior to the origin of the angiosperms, to create the paleoAP3 (pAP3) and PI lineages. A later duplication of the paleoAP3 gene, at the base of the higher eudicots, resulted in the TM6 and divergent euAP3 gene lineages (after Kramer et al. 1998).
In the dicots, duplication of the ancestral C-function led to the AGAMOUS (AG) and PLENA (PLE) lineages. Independent evolution of the paralogues following speciation resulted in the two lineages adopting different functions in different species. For example, in Arabidopsis the AG lineage gene retained the C-function activity (specification of the reproductive organs), whereas the function of the PLE lineage genes, SHATTERPROOF (SHP) 1 and 2, became limited to carpel and fruit development. Conversely, in Antirrhinum, C-function activity was retained by the PLE lineage gene, whereas the AG lineage gene FARINELLI (FAR) functions only in pollen development (Davies et al. 1999; Liljegren et al. 2000; Pinyopich et al. 2003; Kramer et al. 2004; Causier et al. 2005).
Phylogenetic studies, in combination with functional data, have been instrumental in elucidating the evolution of B- and C-function genes. In addition, the use of unbiased synteny-based approaches has revealed the conserved nature of the AG and PLE lineage loci between Arabidopsis and Antirrhinum. With the availability of large amounts of genome sequence data for numerous species, synteny is emerging as a useful tool in comparative genomics. In particular, it is being used to identify gene orthologies and to trace the evolution of genes over large evolutionary distances.
Using synteny-based approaches, we followed the evolution of the floral B- and C-function gene families by recreating ancestral gene maps. Consistent with previous data, we show that the origins of the AP3/TM6 lineages can be traced back to before the monocot–dicot split. Interestingly, petal and stamen development can seemingly tolerate the loss of either the TM6 or the euAP3 lineage gene. However, PI is essential for stamen identity, and contrary to predictions, we show how complex genome rearrangements have resulted in an almost complete breakdown in synteny at the PI locus. By generating similar ancestral gene maps for the C-function genes, we also show the common ancestry of the PLE and AG lineages, and their shared heritage with the monocot C function.
During the course of these studies, we found that the presence of multiple transposable elements (TEs) in the bacterial artificial chromosome (BAC)/transformation-competent artificial chromosome (TAC) genomic sequences containing the Antirrhinum floral homeotic genes limited comparative studies. Because large TEs within gene-rich genomic sequences would be an important consideration in future genome-sequencing efforts, we also present the first survey of the Antirrhinum genome at the sequence level.
Materials and Methods
Isolation of Antirrhinum BAC and TAC Genomic Clones
The Antirrhinum TAC library used in this study has been described previously (Zhou et al. 2003). Preparation of the Antirrhinum BAC library will be described elsewhere (Castillo R, Kuckenberg M, Schwarz-Sommer Z, unpublished data). Antirrhinum BAC and TAC libraries were screened by polymerase chain reaction or using standard hybridization protocols, as described (Sambrook and Russell 2001; Zhou et al. 2003).
Antirrhinum BAC/TAC Sequencing
Each BAC/TAC was sequenced commercially. The sequences of the PLE, FAR, DEF, and GLO BAC/TACs were completed by Eurofins MWG Operon. The OVATE BAC was sequenced by Qiagen, and a complete contiguous sequence was only obtained by sequencing clones from two separate shotgun libraries. The first library had an average insert size of 1.2–1.5 kb. However, this library only covered 74.9 kb of the BAC (estimated insert size of 120 kb) and resulted in an unusually high number of remaining gaps. Assuming that large fragments from the OVATE region may cause toxicity problems in the Escherichia coli strain used, a second library, with an average insert size of 0.6–0.8 kb, was prepared. After merging the sequence data from both libraries, the total contig size was 104 kb with 11 gaps, which were closed by direct primer walking on the BAC. The final BAC sequence was 111.3 kb and was generated from 2,422 reads from both libraries, with an average coverage of 14.6-fold.
The complete BAC/TAC insert sequences were submitted to GenBank with the following accession numbers: OVATE (GenBank: FJ404770), DEF (GenBank: FJ404768), GLO (GenBank: FJ404769), PLE (GenBank: AY935269), and FAR (GenBank: AY935268).
Antirrhinum Gene Identification and Bioinformatics
Putative open reading frames were identified by comparing outputs from various gene prediction algorithms, including Genscan (http://genes.mit.edu/GENSCAN.html; Burge and Karlin 1997), GeneMark.hmm (http://opal.biology.gatech.edu/GeneMark/eukhmm.cgiref; Lukashin and Borodovsky 1998; each using the Arabidopsis data set, with default settings), and FGENESH (www.softberry.com; using both Arabidopsis and tobacco data sets, with default settings). In most cases, each algorithm predicted a similar gene set, and further analyses were conducted using only the Genscan and GeneMark predictions. Basic local alignment search tool (BLAST) homology searches were performed, against the plant EST database at European Molecular Biology Laboratory (www.ebi.ac.uk/blast2, with default settings), to validate Antirrhinum gene predictions. Not all predicted genes had high-scoring homologous Antirrhinum ESTs. In some cases, this was due to poor gene prediction (none of the algorithms accurately predicted any of the previously characterized genes), but in others it was more likely that the corresponding EST was not present in the database. However, highly homologous sequences, present in collinear regions of the Arabidopsis (and tomato in the case of OVATE) genome, provided sufficient evidence for the validity of the gene predictions.
Comparisons with the Arabidopsis genome were made in a number of ways. First, each contig was compared directly with the Arabidopsis collection of BAC sequences using the WU-Blast2 algorithm at www.arabidopsis.org. Second, each predicted peptide was subjected to BLAST homology searches against both the Arabidopsis protein data set at www.arabidopsis.org (WU-Blast2 algorithm) and the Viridiplantae data set at www.ncbi.nlm.nih.gov/BLAST/ (BlastP), to determine the identity of the predicted genes (using default settings in each case).
Genome synteny between Antirrhinum, tomato, and other sequenced species was detected manually. Synteny between the available genome sequences of other species was detected using the Plant Genome Duplication Database (PGDD; http://chibba.agtec.uga.edu/duplication/index/home; Tang, Bowers, et al. 2008; Tang, Wang, et al. 2008).
TEs identified by gene prediction algorithms and BLAST homology searches were further analyzed for signature features of retroelements using the REPuter (DNA repeat identification; www.genomes.de) and InterProScan (protein domain searches; www.ebi.ac.uk/interpro) programs.
Identification of Tomato Genomic Regions Containing Putative Floral Homeotic Genes
Appropriate regions of the tomato genome were identified by BLAST homology searches against the prerelease of the tomato genome shotgun sequence (solgenomics.net), using the TAP3 (DQ674532), TM6 (DQ539419), TPI (DQ674531), TAG1 (L26295), and TAGL1 (AY098735) gene sequences. For collinearity studies, we took approximately 50 kb of sequence upstream and downstream of the homeotic gene location on the appropriate tomato scaffold sequences—TAP3: scaffold02164, nt 2821393–2921940; TM6: scaffold00090, nt 1649405–1749540; TPI; scaffold05575, nt 5347891–5450898; TAG1: scaffold00226, nt 2286871–2387868; and TAGL1: scaffold07408, nt 4054000–4154832. Gene predictions were made using Genscan (Arabidopsis settings), and gene identities by BLAST homology searches.
Results
The synteny and genome property studies described in this article involved the isolation, sequencing, and annotation of three Antirrhinum genomic BAC/TAC clones, totaling 227.4 kb of sequence data containing 27 predicted genes. Together with two previously published BAC/TAC sequences (Causier et al. 2005), we now have approximately 367 kb of Antirrhinum genome sequence. Gene predictions and homologies are presented in table 1 and Supplementary table S1 (Supplementary Material online).
Table 1.
Gene Predictions for Four Antirrhinum BAC/TAC Sequences.
| Identity |
|||||||||
| BAC | Insert Size (kb) | Gene | Gene Length (bp) | Protein Length (aa) | No. of introns | Plants | Arabidopsis | Antirrhinum majus EST | Description |
| PLE | 85.0 | P1 | 15,680 | 1,602 | 4 | AAG10812 (Arabidopsis) | At4g23160 | AJ787930 | Copia retroelement |
| P2 | 3,065 | 413 | 3 | At3g58790 (Arabidopsis) | At3g58790 | AJ559559 | Glycosyl transferase | ||
| P3 | 7,467 | 241 | 5 | AAB25101 (A. majus) | At3g58780 | AJ800374 | PLENA | ||
| P4 | 7,658 | 690 | 0 | AAG52564 (Arabidopsis) | At1g18560 | AJ791306 | hAT-like transposon | ||
| P5 | 1,959 | 115 | 3 | At2g42820 (Arabidopsis) | At2g42820 | AJ802015 | HVA22 | ||
| P6 | 8,802 | 1,053 | 3 | At3g58770 (Arabidopsis) | At3g58770 | — | Expressed protein | ||
| P7 | 6,382 | 298 | 3 | At2g37210 (Arabidopsis) | At2g37210 | AJ805299 | Expressed protein | ||
| P8 | 9,510 | 1,754 | 11 | BAB03109 (Arabidopsis) | — | AJ805299 | Gypsy retroelement | ||
| P9 | >5,789 | >1,217 | >1 | T00499 (Arabidopsis) | At4g23160 | AJ787930 | Copia retroelement | ||
| FAR | 54.3 | F1 | 14,926 | 607 | 8 | At1g29670 (Arabidopsis) | At1g29670 | AJ794332 | GDSL lipase |
| F2 | 3,952 | 285 | 4 | At1g29660 (Arabidopsis) | At1g29660 | AJ794515 | GDSL lipase | ||
| F3 | 3,564 | 350 | 5 | At1g29670 (Arabidopsis) | At1g29670 | AJ794515 | GDSL lipase | ||
| F4 | 3,072 | 321 | 4 | At1g29670 (Arabidopsis) | At1g29670 | AJ794515 | GDSL lipase | ||
| F5 | 4,549 | 303 | 4 | At1g29670 (Arabidopsis) | At1g29670 | — | GDSL lipase | ||
| F6 | 2,991 | 190 | 4 | At1g29670 (Arabidopsis) | At1g29670 | — | GDSL lipase | ||
| F7 | 5,688 | 235 | 5 | CAB42988 (A. majus) | At4g18960 | AJ568412 | FARINELLI | ||
| F8 | 2,032 | 547 | 3 | CAD12809 (A. majus) | — | — | ROSINA-like | ||
| F9 | 3,953 | 483 | 3 | AAF79374 (Arabidopsis) | — | AJ807947 | En-Spm/Tnp2 transposon | ||
| DEF | 65.1 | D1 | >5,342 | >127 | >2 | — | — | AJ568083 | — |
| D2 | 4,279 | 762 | 0 | AAZ67526 (B. rapa) | — | — | Copia retroelement | ||
| D3 | 9,409 | 1,145 | 3 | At2g21390 (Arabidopsis) | At2g21390 | AJ802033 | Coatomer α-subunit | ||
| D4 | 2,296 | ∼765 | 0 | AAT38766 (S. demissum) | — | — | Copia retroelement | ||
| D5 | 1,841 | ∼613 | 0 | AAC25101 (Arabidopsis) | — | — | Idle-like transposon/copia element | ||
| D6 | 4,630 | 184 | 4 | CAA44629 (A. majus) | At3g54340 | AJ559303 | DEFICIENS | ||
| D7 | 11,602 | 349 | 6 | NP_922593 (rice) | At1g75560 | AJ790141 | Zn finger | ||
| GLO | 51.0 | G1 | 15,284 | 214 | 3 | At3g15680 (Arabidopsis) | At3g15680 | — | Zn finger |
| G2 | 8,795 | 1,500 | 4 | AAD19773 (Arabidopsis) | — | — | Copia retroelement | ||
| G3 | 12,860 | 1,526 | 21 | At3g43920 (Arabidopsis) | At3g43920 | AJ793377 | Ribonuclease III | ||
| G4 | 5,432 | 177 | 5 | CAA48725 (A. majus) | At5g20240 | AJ801781 | GLOBOSA | ||
| G5 | 3,748 | 387 | 6 | BAD36149 (rice) | At2g26270 | AJ795996 | Expressed protein | ||
Note.—BlastP was used to identify the closest matches from the Viridiplantae (plants) and Arabidopsis data sets. TBlastN was used to identify matching Antirrhinum ESTs. Only those ESTs that gave a significant e value (<10−55) and aligned well to the predicted genes are listed. BAC accession numbers: PLE, AY935269; FAR, AY935268; DEF, FJ404768; and GLO, FJ404769.
B-function
In angiosperms, petal and stamen development are specified by the action of genes belonging to the AP3 and PI lineages. To trace the evolution of the various B-gene lineages, we compared gene content, order, and orientation for B-gene-containing loci from a diverse range of species.
APETALA3 Lineages
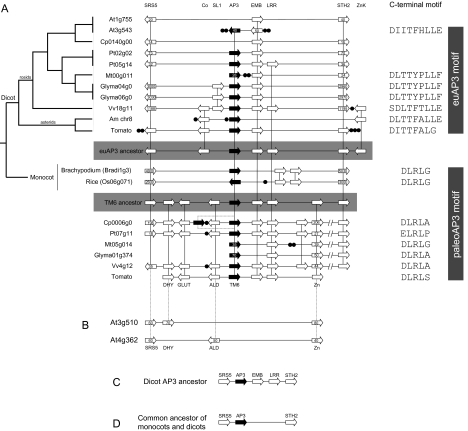
In the basal eudicots, duplication at the paleoAP3 locus resulted in the euAP3 and TM6 gene lineages (Kramer et al. 1998, fig. 1). In Arabidopsis and Antirrhinum, the TM6 ortholog has apparently been lost (Rijpkema, Gerats, et al. 2006). To trace the evolution of the AP3 gene lineages, we examined the extent of gene collinearity at the euAP3- and TM6-containing loci from a range of species both manually and using the PGDD (chibba.agtec.uga.edu/duplication/; Tang, Bowers, et al. 2008; Tang, Wang, et al. 2008). Such analyses reveal that both the euAP3 and the TM6 loci have a number of genes in common, indicating their common ancestry. In addition, the euAP3 locus also contains a number of genes that are distinct from the TM6 locus, and vice versa (fig. 2A).
FIG. 2.
Relationships between the AP3 and TM6 loci. (A) Synteny at the euAP3 and TM6 loci from Arabidopsis (At), papaya (Cp), poplar (Pt), Medicago (Mt), soybean (Glyma), grape (Vv), Antirrhinum (Am), tomato, rice, and Brachypodium (shown to the left of each genomic segment with the chromosome designations; where appropriate, the identifiers for the genes at either end of the displayed segment are shown). The phylogenetic tree to the left shows the relationships between the species. The euAP3 lineage locus from the dicots is shown at the top of the panel, AP3 locus from the monocots is shown in the center, with dicot TM6 locus at the bottom of the panel. Reconstructed ancestral euAP3 and TM6 are shown, highlighted in gray. The C-terminal sequence for each AP3/TM6 protein is shown on the right. Current gene predictions do not identify the euAP3 motif for the Populus euAP3 orthologs. (B) Two Arabidopsis genomic segments are shown that are syntenic with the TM6 locus but that are missing the TM6, EMB, and LRR orthologous genes. Genes syntenic with the TM6 lineage are connected to part (A) by dashed lines. (C) A reconstructed gene map for the dicot AP3 ancestor. (D) A reconstructed gene map for the last common ancestor of monocots and dicots. Gene repertoire and orientation are shown by block arrows for each species, and vertical lines link syntenic genes. Genes highlighted in black are the AP3/TM6 orthologs. Black circles represent nonsyntenic genes. Landmark genes discussed in the text (SRS5, Co, SL1, AP3/TM6, EMB, LRR, STH2, ZnK, DHY, GLUT, ALD, and Zn) are indicated.
Although gene content and order are well conserved at the euAP3 locus for many of the rosids (Populus trichocarpa, Medicago truncatula, Glycine max, and Vitis vinifera), the same is not true for the Arabidopsis AP3 locus on chromosome 3. Indeed, this locus only shares two genes in common with the euAP3-containing loci of other rosid species: AP3 and EMB1967 (fig. 2A). Furthermore, the orientation of these genes relative to one another is unique in Arabidopsis, suggesting that the contemporary Arabidopsis AP3 locus is the product of gene duplication, gene loss, and genomic rearrangement events. A region of Arabidopsis chromosome 1 that contains several genes syntenic with the euAP3 locus of other rosids, but that is missing an AP3-like gene, may represent the other product of a duplication event (fig. 2A). Comparative studies also revealed a region of the papaya (Carica papaya) genome syntenic with rosid euAP3 locus. However, this genomic segment does not contain an euAP3-like gene (fig. 2A). Furthermore, BLAST homology searches of the papaya genome sequence failed to identify an euAP3 gene (data not shown). Two TM6 lineage genes, which appear to be the result of a linked duplication (fig. 2A), have been identified in papaya (Ackerman et al. 2008), but our data indicate that the euAP3 gene has been completely lost from its genome.
The euAP3 locus of the asterid Antirrhinum shares several genes in common with the rosid euAP3 locus. However, comparisons involving this Antirrhinum genomic region are limited by the presence of several TEs (table 1). To further the comparisons between the rosids and the asterids, we annotated a region of the tomato (Solanum lycopersicum) genome containing the TAP3 gene. This showed that gene content and order were as equally well conserved between asterids and rosids as they are among the rosid loci (fig. 2A). Together, these data provide the opportunity to predict the ancestral euAP3 locus. A locus containing SRS5–Co–euAP3–EMB–LRR–STH2–ZnK may have been the progenitor of modern-day euAP3 locus that existed at the base of the core eudicots.
Collinearity between the rosid and the asterid TM6 loci was also well conserved, suggesting that the ancestral dicot TM6 locus may have had the following gene content and order: SRS5–DHY–GLUT–ALD–TM6–EMB–LRR–Zn–STH2. However, the TM6 ortholog was lost from the Arabidopsis and Antirrhinum genomes. Among the asterids, TM6 was maintained in the Solanales but has been lost from Antirrhinum and Mimulus guttatus (from homology searches of the draft Mimulus genome at www.phytozome.net; data not shown), which belong to the closely related Lamiales. One possibility is that TM6 was lost at the base of the Lamiales radiation, or later after Antirrhinum and Mimulus split from the other Lamiales.
Using the ancestral TM6 locus, we attempted to identify the region of the Arabidopsis genome from which the TM6 ortholog was lost. Two segments, on chromosomes 3 and 4, were identified that shared gene order with the ancestral TM6 locus. Interestingly, in both segments the sequence containing the TM6, EMB, and LRR genes is missing (fig. 2B).
A number of genes are common to the reconstructed euAP3 and TM6 ancestral loci (fig. 2A), revealing the common ancestry of the two lineages. This comparison suggests that the ancestral dicot AP3 locus that pre-dated the euAP3–TM6 split was likely to include the SRS5, EMB, LRR, and STH2 genes adjacent to the AP3 gene (fig. 2C).
The euAP3 and TM6 lineages were the product of a duplication in the dicots after divergence from the monocots. To trace the evolution of these B-function genes, we compared the AP3 loci of rice (Oryza sativa) and Brachypodium distachyon to each dicot AP3 lineage. Over the interval examined, the monocot loci shared only three genes with the dicot loci, and these genes were common to both the euAP3 and TM6 lineages (fig. 2A). This confirms that both the euAP3 and the TM6 lineages are orthologous to the monocot AP3 genes. Based on these data, we are able to predict that the SRS5, AP3, and STH2 genes were all present on the ancestral AP3 loci that pre-dated the monocot–dicot split (fig. 2D). It is interesting to note that the SRS5 gene is in opposite orientations at the dicot TM6 and euAP3 loci (fig. 2). Furthermore, the orientation of SRS5 at the TM6 (paleoAP3) locus is conserved in the monocots, suggesting that SRS5 was flipped during the evolution of the euAP3 locus.
To confirm that collinearity correctly predicts euAP3 and TM6 lineage genes, we examined the C-terminal sequence of the available predicted proteins. As expected, all those loci predicted to contain euAP3 genes encoded proteins with an euAP3 motif, whereas those B-function genes at the TM6 locus, and at the monocot AP3 locus, encoded proteins with the paleoAP3 motif (fig. 2A).
In summary, we have used synteny to trace back the origins of the AP3/TM6 lineages to before the monocot–dicot split. In addition, our comparative study has revealed the complex genome duplications, rearrangements, and gene loss that have degraded synteny at the Arabidopsis AP3 locus and that resulted in the loss of its TM6 ortholog.
PISTILLATA Lineage
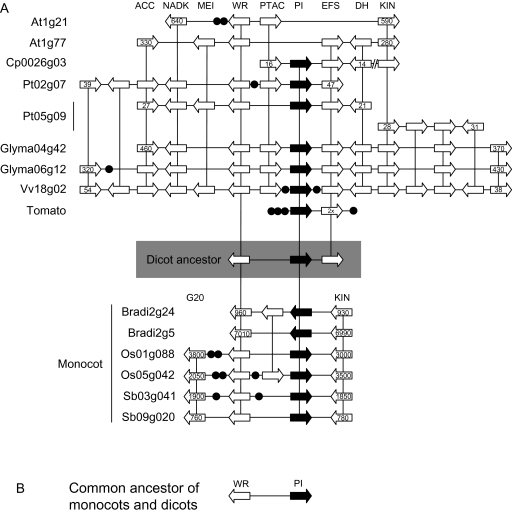
With the exception of the Arabidopsis PI locus on chromosome 5, the PI-containing locus is well conserved between the majority of the rosid species examined (fig. 3). Using the PGDD, no syntenic blocks were identified between the Arabidopsis PI locus and the genomes of any other species. However, two Arabidopsis chromosomal regions, both on separate parts of chromosome 1 and both missing a PI-like gene, were found to be syntenic with PI loci from other rosid species (fig. 3). Together, the findings would suggest that, like the Arabidopsis AP3 locus, the PI locus has diverged significantly from those of closely related species.
FIG. 3.
Synteny between the PI loci. (A) Synteny at the PI loci from Arabidopsis (At), papaya (Cp), poplar (Pt), soybean (Glyma), grape (Vv), tomato, rice (Os), Brachypodium (Bradi), and Sorghum (Sb) (shown to the left of each genomic segment with the chromosome designations; where appropriate, the identifiers for the genes at either end of the displayed segment are shown). PI lineage locus from the dicots is shown in the top section of the figure, and that of the monocots is shown at the bottom. A reconstructed ancestral dicot PI locus is shown, highlighted in gray. (B) A reconstructed gene map for the last common ancestor of the monocots and dicots. Gene repertoire and orientation are shown by block arrows for each species, and vertical lines link syntenic genes. Genes highlighted in black are the PI orthologs. Black circles represent nonsyntenic genes. Landmark genes discussed in the text (ACC, NADK, MEI, WR, PTAC, PI, EFS, DH, KIN, and G20) are indicated. Note that the kinase (KIN) genes shown on the dicot and monocot loci encode unrelated proteins.
The 51-kb Antirrhinum BAC clone containing the PI ortholog GLO is predicted to contain only five genes, including a TE (table 1). None of the genes predicted on the GLO-containing BAC were syntenic with genes at PI loci in other species, perhaps suggesting that as in Arabidopsis, the GLO locus has diverged significantly from other PI loci. Because a comparison between rosid and asterid PI loci was not possible using the Antirrhinum data, we annotated a 100-kb segment of the tomato genome containing the PI gene. This genomic fragment contained seven genes, including a direct duplication of the EFS gene. Comparison with the rosid data revealed that only PI and the EFS genes were shared with tomato (fig. 3). In addition, study of the dicot and monocot loci (see below) indicated that the WR, PI, and EFS genes were present on the dicot ancestor of the PI locus (fig. 3).
The PI lineage is common to both monocots and dicots. We compared the PI-containing loci of Brachypodium, rice, and Sorghum bicolour with the dicot data, which revealed that they shared the WR and PI genes in common. This close association between PI and WR suggests that they were together in the PI locus of the last common ancestor of the monocots and dicots.
In summary, the PI locus of Arabidopsis is poorly conserved in terms of its gene complement with PI loci from other rosids. Interestingly, this locus is also poorly conserved in the asterid species examined. As a consequence, only a limited ancestral PI locus was constructed for the common ancestor of the rosids and asterids. However, the V. vinifera locus contains all the genes found in other rosid PI loci and may represent a good model for the ancestral rosid PI locus.
C-function
We had previously revealed the evolutionary relationships between the Antirrhinum C-function genes PLE and FAR and the AG and SHP genes from Arabidopsis using a collinearity-based approach (Causier et al. 2005). However, this study was constrained by the available data and by the limited amount of Antirrhinum genome sequence for the appropriate loci. To understand better the evolution of the C-function genes, we compared C-function gene loci from a number of species, including representatives of the dicots and the monocots. Our aim was to establish the ancestral locus of the dicot PLE and AG lineages, the dicot C-function gene locus prior to the divergence of the PLE and AG lineages, and finally the locus for the common ancestor of the monocots and dicots.
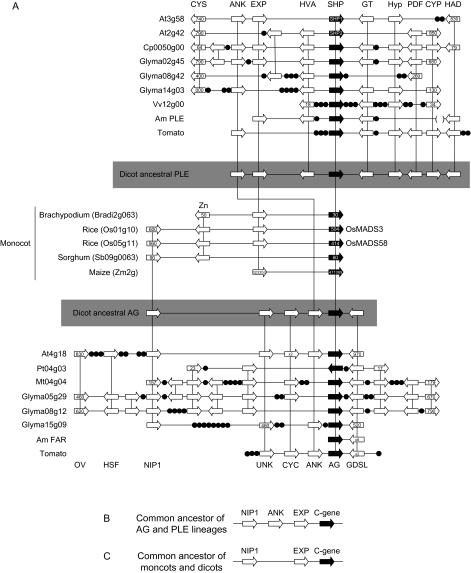
To these ends, we identified blocks of synteny between the different genomes. Previously, we showed that the Antirrhinum PLE locus was syntenic with the SHP1 and SHP2 loci of Arabidopsis. This collinearity extends to loci in the genomes of papaya, G. max, V. vinifera, and tomato, allowing for the unambiguous identification of the orthologs of PLE/SHP in these species (fig. 4A). The data reveal that collinearity at this locus is extensive between the rosids, and with the addition of the tomato locus (containing the TAGL1 gene), a high degree of collinearity between the asterids and the rosids is also revealed (fig. 4A). Together, the rosid and asterid data allow prediction of the ancestral dicot PLE/SHP locus with the following gene order: ANK–EXP–HVA–PLE/SHP–GT–Hyp–PDF–CYP–HAD (fig. 4A).
FIG. 4.
Conserved gene order at C-function loci. (A) Synteny at the PLE and AG loci from Arabidopsis (At), papaya (Cp), poplar (Pt), Medicago (Mt), soybean (Glyma), grape (Vv), Antirrhinum (Am), tomato, rice Brachypodium, Sorghum, and maize (shown to the left of each genomic segment with the chromosome designations; where appropriate, the identifier for the genes at either end of the displayed segment are shown). PLE lineage locus from the dicots is shown at the top of the panel, C-gene locus from the monocots is shown in the center, with dicot AG locus at the bottom of the panel. Reconstructed ancestral dicot PLE and AG loci are shown, highlighted in gray. (B) A reconstructed gene map for the common dicot ancestor of the PLE and AG lineages. (C) A reconstructed gene map for the last common ancestor of the monocots and dicots. Gene repertoire and orientation are shown by block arrows for each species, and vertical lines link syntenic genes. Genes highlighted in black are the PLE or AG orthologs. Black circles represent nonsyntenic genes. Landmark genes discussed in the text (CYS, ANK, EXP, HVA, PLE, GT, Hyp, PDF, CYP, HAD, OV, HSF, NIP1, UNK, CYC, AG, and GDSL) are indicated. The rice loci corresponding to the characterized C-function genes OsMADS3 and OsMADS58 are also indicated.
The Arabidopsis genomic locus containing the C-function gene AG has been shown to be partially syntenic with the Antirrhinum FAR locus (Causier et al. 2005). Local duplication of the GDSL gene together with the presence of TEs at the FAR locus (table 1) provided only limited data on which to examine synteny between these species. To understand evolution of the AG/FAR locus, we used PGDD data, together with gene predictions for the tomato locus containing the TAG1 gene and our previous data, to examine synteny across a broad range of species. As we found with PLE/SHP, collinearity was extensive between the rosids, but only with the addition of the tomato data was it possible to see that gene content and order were also conserved between the rosids and the asterids (fig. 4A). Interestingly, as at the FAR locus, the tomato GDSL gene has duplicated, whereas in the rosid lineages this has not occurred, suggesting that the duplication occurred in a common ancestor of Antirrhinum and tomato. The asterid and rosid data provide sufficient information to predict the ancestral dicot AG locus, containing the following genes: NIP1–UNK–ANK–AG–GDSL.
The ANK gene is present on both the ancestral PLE/SHP and AG loci, suggesting that it pre-dates the divergence of the PLE and AG lineages. These lineages arose in the dicots following the split from the monocots. This was confirmed when we examined the rice locus containing the C-function gene OsMADS3 and found that it shared genes with each of the Arabidopsis AG, SHP1, and SHP2 loci (Causier et al. 2005, fig. 4A). To determine the extent of collinearity between C-function gene loci of the monocots, and between monocots and dicots, we expanded these initial studies. We found that the C-function gene loci of rice, Brachypodium, Sorghum, and maize (Zea mays) were partially syntenic (fig. 4A). In addition, these loci were also syntenic with the ancestral dicot AG and PLE loci (fig. 4A).
Together, the data from the C-function gene loci of the monocots and dicots allow us to predict the ancestral dicot C-function gene locus that pre-dates the divergence of the AG and PLE gene lineages: NIP1–ANK–EXP–C (fig. 4B). In addition, we can also predict the C-function gene locus of the common ancestor of the monocots and dicots: NIP1–EXP–C (fig. 4C). This would suggest that the ANK gene either translocated to the dicot C-function gene locus or was lost from the monocot locus, after the monocot–dicot split. Another notable feature of the C-function gene loci revealed by our comparative approach, which was not seen for the B function, is a translocation of genes to the loci in some species (e.g., G. max, M. trunculata, and V. vinifera). Although significant, these gene additions did not affect our comparative analyses.
In summary, we have traced the evolution of the dicot C-function gene lineages using genome synteny. We have reconstructed the ancestral dicot C-function gene locus that pre-dates the AG–PLE split, which reveals the common ancestry with the monocot C-function genes.
Properties of the Antirrhinum Genome
In general, the Antirrhinum floral homeotic BAC/TAC sequences showed conserved microsynteny, along the full length of the clones, with genomic regions from diverse species. However, in some cases the presence of large TEs, which resulted in lower than anticipated true-gene numbers within the available sequence, limited these comparisons. Because the presence of these large mobile elements not only complicates comparative studies but must also be a major consideration in genome sequencing efforts, we thought it prudent to make some predictions about the Antirrhinum genome that may benefit future studies. To facilitate this, and to avoid potential bias from studying only floral homeotic MADS loci, we also sequenced an additional genomic BAC of 111.3 kb, containing an OVATE-like gene (table S1). The OVATE BAC shows conserved microsynteny, along its entire length, with the previously published tomato OVATE BAC and syntenic Arabidopsis genomic segments (Ku et al. 2000) and to genomic regions from various other species (Supplementary fig. S1, Supplementary Material online).
The five BAC/TACs described in this study map to different chromosomes, and total approximately 367 kb of genomic sequence, which represents approximately 0.1% of the 360-Mb haploid Antirrhinum genome (Bennett et al. 2000). Gene prediction algorithms identified 45 genes in the five BAC/TAC clones, including 11 putative TEs (table 1 and Supplementary table S1, Supplementary Material online). Non-TE gene densities range from 6.8 to 17 kb/gene over the five separate genomic regions, averaging 10.8 kb/gene. Consequently, we can predict that the Antirrhinum genome contains in the order of 33,000 non-TE genes. The G + C content for the five BAC/TACs was approximately 34%. This value is lower than the average reported for tomato (37%), which has one of the lowest G + C contents of any plant species (Messeguer et al. 1991; Wang et al. 2005).
Importantly, our data indicate that approximately 24% of the genes encoded by the Antirrhinum genome are TEs. Of the 11 putative TEs, 7 were similar to the long terminal repeat (LTR) class of retroelements (based on homology searches, identification of repeated sequence elements, and protein motif scans; Supplementary fig. S2, Supplementary Material online). LTR retroelements are characterized by long terminal repeats (LTRs; Kidwell 2002) and are found in distinct groups that include the Ty1-copia and the Ty3-gypsy elements. Both copia and gypsy elements contain two major genes, gag and pol, that encode proteins required for the life cycle of the retroelement (Feschotte et al. 2002; Casacuberta and Santiago 2003). The order of the coding units within pol differs between the copia and the gypsy groups (Supplementary fig. S2, Supplementary Material online). Of the LTR elements found in the Antirrhinum sequences, six appear to belong to the copia family (PLE P1 and P9, DEF D2 and D4, GLO G2, OVATE Ov10; table 1 and Supplementary table S1, Supplementary Material online), and one to the gypsy-like family (PLE P8; Supplementary fig. S2, Supplementary Material online). Analysis of the structure of the predicted retroelements suggests that many have become degraded over time (Supplementary fig. S2, Supplementary Material online). Combined approaches, including analysis of DNA repeats (REPuter, www.genomes.de) and protein domain searches (InterProScan, www.ebi.ac.uk/interpro), indicate that at least two of the elements are likely to be complete (GLO G2 and PLE P8). The remainder appear to have suffered degradation of signature features.
Four DNA transposon-like elements were also identified. OVATE Ov3 and PLE P4 show similarity to the hAT family of transposons, whereas FAR F9 appears to belong to the En-Spm/Tnp2 family (table 1 and Supplementary table S1, Supplementary Material online). DEF D5 shows similarity to the newly identified Idle group of transposons (Castillo R, Schwarz-Sommer Z, unpublished data), although the prediction of the D5 gene is complicated by the presence of Ty1-copia sequences (data not shown). Idle transposons belong to a growing list of short TEs and may be missed by gene prediction algorithms. To identify small elements within the BAC/TAC inserts, BLAST homology searches were performed using each contig sequence against the Viridiplantae nucleotide sequence data set (at www.ncbi.nlm.nih.gov/BLAST/). For each BAC/TAC at least two Idle transposon sequences were revealed (data not shown), suggesting that transposon numbers in the Antirrhinum genome are somewhat higher than indicated by gene prediction. With the exception of the Idle elements, none of the putative transposons identified by gene prediction software in this study share absolute homology with database sequences, suggesting that these may represent novel Antirrhinum mobile elements.
In summary, our combined large-scale analysis provides some preliminary data about the arrangement of the Antirrhinum genome. Critically, it reveals that gene-rich regions also contain a high degree of TEs, of varied types.
Discussion
Evolution of the Floral Homeotic Genes Can Be Traced by Genome Synteny
Extensive functional and phylogenetic studies have established the evolutionary relationships between floral homeotic genes from diverse species. In most cases, phylogeny is sufficient to reconstruct evolutionary histories. However, occasionally phylogenetic studies are complicated by incorrect gene models or high levels of substitution. Therefore, approaches that can determine gene relationships without functional constraints and that are largely insensitive to incorrectly predicted genes might be more useful in these cases. In a previous study, we used genome synteny to show the relationships between the duplicate C-function genes in Arabidopsis and Antirrhinum (Causier et al. 2005). Genome synteny can be efficiently used to unambiguously identify orthologous genes, to infer ancestral gene loci, and to trace the evolutionary heritage of a gene. However, in cases such as Arabidopsis PI, where loss of synteny at a particular locus would limit its use in determining gene relationships, phylogeny remains the best approach.
Evolution of the B-Function AP3 Lineages
At the base of the core eudicots, duplication of the ancestral paleoAP3 gene produced the TM6 and euAP3 lineages (fig. 1), which can be distinguished by particular sequence motifs. TM6 genes encode proteins with a C-terminal sequence referred to as the paleoAP3 motif (DLRLA*), whereas in euAP3 proteins a frameshift replaced the paleoAP3 motif with the euAP3 motif (DLTTFALLE*; Kramer et al. 1998; Vandenbussche et al. 2003). We examined whether the euAP3 and TM6 lineages could also be distinguished based on conservation of gene content and order at the gene loci, using data from rosid and asterid genomes. Reconstructed ancestral genetic maps revealed gene additions and deletions that distinguished the two lineages in the dicots. The presence of DHY, GLUT, ALD, and Zn adjacent to AP3 is diagnostic for a TM6 locus, whereas the complete absence of these genes would be indicative of an euAP3 locus (fig. 2A). In cases where AP3 genes are incorrectly annotated, these features may be important identifiers of orthology. For example, database predictions for the protein encoded by the AP3-like gene annotated on poplar chromosome 2 do not include a lineage-defining C-terminal motif (Pt02g0272 in the PGDD; Pt_APETALA3.1 in the poplar genome database at www.phytozome.net). However, comparison of the gene complement of the Pt_APETALA3.1 locus with other AP3/TM6 loci places the gene in the euAP3 group (fig. 2A). The Arabidopsis genome does not have a TM6 ortholog, and the papaya genome appears to lack an euAP3 gene. However, by searching for the defining features of AP3 lineage loci, we identified genomic segments in each species from which these B-function genes were possibly lost (fig. 2).
The paleoAP3 motif is found throughout the angiosperms, whereas the euAP3 motif is unique to the higher eudicots (Kramer et al. 1998; Vandenbussche et al. 2003). Consistent with previous studies, which suggest that the duplication resulting in the TM6 and euAP3 lineages occurred before the diversification of the major higher dicots (Kramer et al. 1998), synteny between AP3 loci from the monocots and dicots reveals that the euAP3–TM6 split was specific to the dicots.
TM6 and euAP3 diverged from a common paleoAP3 ancestor, the ortholog of which is present in the monocots. Interestingly, although the monocots and dicots diverged approximately 140 My (Chaw et al. 2004), it was possible to trace the common heritage of the AP3 lineages from these groups of plants in the synteny data (fig. 2A).
Evolution of the B-Function PI Lineage
Comparison of the PI loci from a range of rosid and asterid species reveals that the genomic regions are generally syntenic (fig. 3). However, closer inspection reveals that collinearity between rosid and asterid loci is poor. Indeed, the Antirrhinum GLO locus shows no collinearity with any other PI locus, and the tomato PI genomic region shows very limited shared gene content. Interestingly, the Arabidopsis genome segment containing the PI gene also shows no collinearity with any of the rosid or asterid locus. Thus, it appears that conservation of gene order at the PI locus is restricted to a small group of rosids. AP3 and PI lineage gene activities are essential for male reproductive organ development. In contrast to higher eudicot AP3 lineages, where loss of either the TM6 or the euAP3 duplicate has no impact on stamen development (see below), loss of PI results in the complete absence of stamens. One would predict that, as shown for yeast (Pál and Hurst 2003), essential genes would be in regions of low recombination. However, in the case of PI, it appears that the loci have diverged significantly following speciation. One possibility for this is that the PI regulatory region is compact (Chen et al., 2000), suggesting that genomic rearrangements were less likely to disrupt the locus.
Complex Genomic Rearrangements and the Evolution of the Arabidopsis B-function
The Arabidopsis genome has undergone one or more large-scale gene or whole-genome duplications. In addition, considerable genomic rearrangements and gene loss are also evident, which reduce collinearity and complicate comparative studies (Blanc et al. 2000; Simillion et al. 2002; Raes et al. 2003; Tang, Bowers, et al. 2008). Although the ortholog of TM6 has been lost from the Arabidopsis genome, our synteny-based approach has identified two genomic segments that are syntenic with TM6 loci in other species. The loss of TM6 may have coincided with the deletion of a small region of the TM6 locus, which also included the EMB and LRR genes. In species where both lineages are maintained, aspects of the B-function appear to be partitioned between the TM6 and euAP3 genes (Poupin et al. 2007; Ackerman et al. 2008, and references therein). In contrast, in Arabidopsis and Antirrhinum, where the euAP3 genes are indispensable for stamen development, loss of TM6 genes may reflect a reduced selective pressure to maintain these genes. The role of TM6 and euAP3 genes in stamen development is likely to be the ancestral function because stamens evolved only once (Kramer et al. 1998). In contrast, evolution of petals at the base of the higher eudicots correlates with the emergence of the divergent euAP3 lineage. However, petal-like structures have evolved several times and are found in flowers of the monocots and lower dicots, suggesting that each time these arose, the paleoAP3 genes adopted new functions (Kramer et al. 1998). In the higher eudicot papaya, the euAP3 ortholog has been lost from the genome (Ackerman et al. 2008, fig. 2A); yet, the flowers produce five petals (Yu et al. 2008). Together with the Arabidopsis and Antirrhinum data, this suggests that euAP3 and TM6 lineage genes can be equally co-opted into petal and stamen developmental pathways and that only one duplicate, from either the euAP3 or the TM6 lineage, needs to be maintained for normal flower development. A clear example of this is the rescue of both petal and stamen identity in the Arabidopsis ap3-3 mutant expressing the maize paleoAP3 gene Silky1 (Whipple et al. 2004).
Complex gene/genome duplications, genome rearrangements, and gene loss processes have resulted in a breakdown of synteny at both the Arabidopsis AP3 and the PI loci. Loss-of-function ap3 and pi mutants were critical for defining the B-function (Coen and Meyerowitz 1991). However, it seems that although these genes are good models for the functionality of the B function, the genomic loci are poor templates for comparative genomics and gene synteny studies.
Evolution of the C-Function Lineages
C-function gene activity is essential for both male and female reproductive organ development in flowering plants. In dicots, the C-function is performed by a single gene, loss of which results in the conversion of reproductive organs to perianth organs and in total sterility. In a common ancestor of the dicots, this essential gene duplicated, resulting in the modern-day AG and PLE lineages. However, following speciation, the critical C-function activity was retained by a different duplicate in Arabidopsis (AG) and Antirrhinum (PLE; Kramer et al. 2004; Causier et al. 2005; Irish and Litt 2005). Comparison of the reconstructed ancestral AG and PLE gene loci reveals traces of the common ancestry of the two lineages. However, because the duplication that produced these two lineages occurred uniquely in the dicots, PLE and AG are both orthologous to monocot C-function genes. In rice and maize, the C-function duplicated independently to that in dicots and resulted in a partitioning of C-function activity between the two copies (Mena et al. 1996; Yamaguchi et al. 2006). Comparison of the loci containing the maize C-function genes ZAG1 and ZMM2 (Mena et al. 1996) failed to identify syntenic segments in any of the other monocot or dicot genomes. Interestingly, a third maize locus on chromosome 3 shared two genes in common with monocot C-function loci. The first is a MADS-box gene that is a recent duplicate of ZMM2 (ZMM23; Münster et al. 2002) and the second a gene with weak homology to the At3g58770 (EXP) gene from Arabidopsis (fig. 4A). Together, this suggests that complex genome rearrangements have occurred during the evolution of the C-function in maize. The duplicate rice C-function genes OsMADS3 and OsMADS58 (Yamaguchi et al. 2006) lie in regions syntenic with genomic segments from the monocot species Sorghum and Brachypodium, which are all syntenic with both the PLE and the AG loci (fig. 4A).
Although comparison of the reconstructed ancestral AG and PLE loci reveals traces of the common evolutionary histories of these lineages, inclusion of the monocot loci provides a better template from which ancestry can be studied. What emerges is a reconstructed locus for the last common ancestor of the AG and PLE lineages (fig. 4B) that is remarkably similar to the predicted ancestor of the monocots and dicots. Together, this suggests that the arrangement of the C-function loci has remained relatively constant over large evolutionary distances.
The Antirrhinum Genome Is Unusually Rich in TEs
All the Antirrhinum genomic clones sequenced as part of this investigation contained TEs. We found that the presence of these elements limited our comparative studies, which suggested that they might also be a problem for future Antirrhinum genome sequencing and assembly projects. The total sequence presented in this article, isolated from different chromosomes, represents approximately 0.1% of the Antirrhinum genome, allowing us to perform a pilot survey of the genome. Application of the gene prediction data to the whole genome suggests that it contains about 48,000 genes (both TE and non-TE), at a density of one gene every 8.1 kb. Gene number in eukaryotes is constant, ranging from 25,000 to 43,000 (Caldwell et al. 2004). However, plant genomes are rich in TEs and gene number is often overestimated in plants due to poor annotation of these elements (Kumar and Bennetzen 1999; Fedoroff 2000; Casacuberta and Santiago 2003; Bennetzen et al. 2004). Identification of TEs is complicated by the fact that the majority identified in plant genomes are no longer functional, and most copies are nonautonomous or are fragments of full-length elements (Kidwell 2002). Of the 45 genes predicted in the 367 kb of Antirrhinum genome sequenced, 11 had strong homology to TEs (table 1 and Supplementary table S1, Supplementary Material online). Further study of these elements revealed that the majority were incomplete and presumably nonfunctional (Supplementary fig. S2, Supplementary Material online). Taking this into account, we predict that, similar to other eukaryote genomes, the Antirrhinum euchromatin contains approximately 33,000 non-TE genes, at a density of one gene every 10.8 kb.
However, our data suggest that the Antirrhinum genome has a much higher density of TEs (24% of predicted genes are TEs) than that of other dicot species, including tomato (12–96% TEs of which are retroelements; Wang et al. 2005), Arabidopsis (mobile elements constitute only 4–8% of the genome; Casacuberta and Santiago 2003), and chickpea (8.6%; Rajesh et al. 2008). Many of the TEs identified here are unlike TEs previously characterized in Antirrhinum. TEs have proved invaluable in identifying the functions of Antirrhinum genes (Schwarz-Sommer et al. 2003), and the discovery of new elements will facilitate reverse genetics screens in the future.
A number of short elements were also identified in the BAC/TAC nucleotide sequences. These short elements belong to a novel group of nonautonomous class II DNA transposons identified in Antirrhinum, Misopates, and Linaria, termed Idle (Castillo and Schwarz-Sommer, unpublished data). In Antirrhinum, 105 unique Idle transposon sequences have been identified to date (accession numbers AM422777 and FM992410-FM992514). Consequently, the presence of multiple Idle transposons in the BAC/TAC sequences suggests that the prediction of TE numbers in the Antirrhinum genome may be a significant underestimation.
Supplementary Material
Supplementary figures S1 and S2, and supplementary table S1 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Supplementary Material
Acknowledgments
This work was funded by grants from the Biotechnology and Biological Sciences Research Council to B.D. and B.C. and from the British Council to B.D. and Z.S.-S. We gratefully acknowledge the excellent technical assistance provided by Richard Ingram and Markus Kuckenberg. We would also like to thank Chiara Airoldi for comments made during the preparation of the manuscript, and the anonymous reviewers for their insightful suggestions for improving this article.
The authors wish to dedicate this article to the memory of Zsuzsanna Schwarz-Sommer (1946–2009).
References
- Ackerman CM, Yu Q, Kim S, Paull RE, Moore PH, Ming R. B-class MADS-box genes in trioecious papaya: two paleoAP3 paralogs, CpTM6-1 and CpTM6-2, and a PI ortholog CpPI. Planta. 2008;227:741–753. doi: 10.1007/s00425-007-0653-5. [DOI] [PubMed] [Google Scholar]
- Bennett MD, Bhandol P, Leitch IJ. Nuclear DNA amounts in angiosperms and their modern uses—807 new estimates. Ann Bot. 2000;86:859–909. [Google Scholar]
- Bennetzen JL, Coleman C, Liu R, Ma J, Ramakrishna W. Consistent over-estimation of gene number in complex plant genomes. Curr Opin Plant Biol. 2004;7:732–736. doi: 10.1016/j.pbi.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Blanc G, Barakat A, Guyot R, Cooke R, Delseny M. Extensive duplication and reshuffling in the Arabidopsis genome. Plant Cell. 2000;12:1093–1101. doi: 10.1105/tpc.12.7.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J Mol Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- Caldwell KS, Langridge P, Powell W. Comparative sequence analysis of the region harboring the hardness locus in barley and its colinear region in rice. Plant Physiol. 2004;136:3177–3190. doi: 10.1104/pp.104.044081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casacuberta JM, Santiago N. Plant LTR-retrotransposons and MITEs: control of transposition and impact on the evolution of plant genes and genomes. Gene. 2003;311:1–11. doi: 10.1016/s0378-1119(03)00557-2. [DOI] [PubMed] [Google Scholar]
- Causier B, Castillo R, Zhou J, Ingram R, Xue Y, Schwarz-Sommer Z, Davies B. Evolution in action: following function in duplicated floral homeotic genes. Curr Biol. 2005;15:1508–1512. doi: 10.1016/j.cub.2005.07.063. [DOI] [PubMed] [Google Scholar]
- Causier B, Schwarz-Sommer Z, Davies B. Floral organ identity: 20 years of ABCs. Semin Cell Dev Biol. 2010;21:73–79. doi: 10.1016/j.semcdb.2009.10.005. [DOI] [PubMed] [Google Scholar]
- Chaw SM, Chang CC, Chen HL, Li WH. Dating the monocot-dicot divergence and the origin of core eudicots using whole chloroplast genomes. J Mol Evol. 2004;58:424–441. doi: 10.1007/s00239-003-2564-9. [DOI] [PubMed] [Google Scholar]
- Chen X, Riechmann JL, Jia D, Meyerowitz E. Minimal regions in the Arabidopsis PISTILLATA promoter responsive to the APETALA3/PISTILLATA feedback control do not contain a CArG box. Sex Plant Reprod. 2000;13:85–94. [Google Scholar]
- Coen ES, Meyerowitz EM. The war of the whorls: genetic interactions controlling flower development. Nature. 1991;353:31–37. doi: 10.1038/353031a0. [DOI] [PubMed] [Google Scholar]
- Davies B, Motte P, Keck E, Saedler H, Sommer H, Schwarz-Sommer Z. PLENA and FARINELLI: redundancy and regulatory interactions between two Antirrhinum MADS-box factors controlling flower development. EMBO J. 1999;18:4023–4034. doi: 10.1093/emboj/18.14.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Martino G, Pan I, Emmanuel E, Levy A, Irish VF. Functional analyses of two tomato APETALA3 genes demonstrate diversification in their roles in regulating floral development. Plant Cell. 2006;18:1833–1845. doi: 10.1105/tpc.106.042978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedoroff N. Transposons and genome evolution in plants. Proc Natl Acad Sci U S A. 2000;97:7002–7007. doi: 10.1073/pnas.97.13.7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C, Jiang N, Wessler SR. Plant transposable elements: where genetics meets genomics. Nat Rev Genet. 2002;3:329–341. doi: 10.1038/nrg793. [DOI] [PubMed] [Google Scholar]
- Goto K, Meyerowitz EM. Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Dev. 1994;8:1548–1560. doi: 10.1101/gad.8.13.1548. [DOI] [PubMed] [Google Scholar]
- Irish VF. Duplication, diversification, and comparative genetics of angiosperm MADS-box genes. Adv Bot Res. 2006;44:129–161. [Google Scholar]
- Irish VF, Litt A. Flower development and evolution: gene duplication, diversification and redeployment. Curr Opin Genet Dev. 2005;15:454–460. doi: 10.1016/j.gde.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Jack T, Brockman LL, Meyerowitz EM. The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell. 1992;68:683–697. doi: 10.1016/0092-8674(92)90144-2. [DOI] [PubMed] [Google Scholar]
- Kidwell MG. Transposable elements and the evolution of genome size in eukaryotes. Genetica. 2002;115:49–63. doi: 10.1023/a:1016072014259. [DOI] [PubMed] [Google Scholar]
- Kim S, Yoo M, Albert VA, Farris JS, Soltis PS, Soltis DE. Phylogeny and diversification of B-function genes in angiosperms: evolutionary and functional implications of a 260-million year old duplication. Am J Bot. 2004;91:2102–2118. doi: 10.3732/ajb.91.12.2102. [DOI] [PubMed] [Google Scholar]
- Kramer EM, Dorit RL, Irish VF. Molecular evolution of genes controlling petal and stamen development: duplication and divergence within the APETALA3 and PISTILLATA MADS-box gene lineages. Genetics. 1998;149:765–783. doi: 10.1093/genetics/149.2.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer EM, Jaramillo MA, Di Stilio VS. Patterns of gene duplication and functional evolution during the diversification of the AGAMOUS subfamily of MADS box genes in angiosperms. Genetics. 2004;166:1011–1023. doi: 10.1534/genetics.166.2.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku HM, Vision T, Liu J, Tanksley SD. Comparing sequenced segments of the tomato and Arabidopsis genomes: large-scale duplication followed by selective gene loss creates a network of synteny. Proc Natl Acad Sci U S A. 2000;97:9121–9126. doi: 10.1073/pnas.160271297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Bennetzen JL. Plant retrotransposons. Annu Rev Genet. 1999;33:479–532. doi: 10.1146/annurev.genet.33.1.479. [DOI] [PubMed] [Google Scholar]
- Liljegren SJ, Ditta GS, Eshed Y, Savidge B, Bowman JL, Yanofsky MF. SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature. 2000;404:766–770. doi: 10.1038/35008089. [DOI] [PubMed] [Google Scholar]
- Litt A. An evaluation of the A-function: evidence from the APETALA1 and APETALA2 gene lineages. Int J Plant Sci. 2007;168:73–91. [Google Scholar]
- Lukashin AV, Borodovsky M. GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res. 1998;26:1107–1115. doi: 10.1093/nar/26.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Castilla LP, Alvarez-Buylla ER. Adaptive evolution in the Arabidopsis MADS-box gene family inferred from its complete resolved phylogeny. Proc Natl Acad Sci U S A. 2003;100:13407–13412. doi: 10.1073/pnas.1835864100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mena M, Ambrose BA, Meeley RB, Briggs SP, Yanofsky MF, Schmidt RJ. Diversification of C-function activity in maize flower development. Science. 1996;274:1537–1540. doi: 10.1126/science.274.5292.1537. [DOI] [PubMed] [Google Scholar]
- Messeguer R, Ganal MW, Steffens JC, Tanksley SD. Characterization of the level, target sites and inheritance of cytosine methylation in tomato nuclear DNA. Plant Mol Biol. 1991;16:753–770. doi: 10.1007/BF00015069. [DOI] [PubMed] [Google Scholar]
- Münster T, Deleu W, Wingen LU, Ouzunova M, Cacharrón J, Faigl W, Werth S, Kim JTT, Saedler H, Theissen G. Maize MADS-box genes galore. Maydica. 2002;47:287–301. [Google Scholar]
- Pál C, Hurst LD. Evidence for co-evolution of gene order and recombination rate. Nat Genet. 2003;33:392–395. doi: 10.1038/ng1111. [DOI] [PubMed] [Google Scholar]
- Parenicová L, de Folter S, Kieffer M, et al. (12 co-authors) Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: new openings to the MADS world. Plant Cell. 2003;15:1538–1551. doi: 10.1105/tpc.011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinyopich A, Ditta GS, Savidge B, Liljegren SJ, Baumann E, Wisman E, Yanofsky MF. Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature. 2003;424:85–88. doi: 10.1038/nature01741. [DOI] [PubMed] [Google Scholar]
- Poupin MJ, Federici F, Medina C, Matus JT, Timmermann T, Arce-Johnson P. Isolation of the three grape sub-lineages of B-class MADS-box TM6, PISTILLATA and APETALA3 genes which are differentially expressed during flower and fruit development. Gene. 2007;404:10–24. doi: 10.1016/j.gene.2007.08.005. [DOI] [PubMed] [Google Scholar]
- Purugganan MD, Rounsley SD, Schmidt RJ, Yanofsky MF. Molecular evolution of flower development: diversification of the plant MADS-box regulatory gene family. Genetics. 1995;140:345–356. doi: 10.1093/genetics/140.1.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raes J, Vandepoele K, Simillion C, Saeys Y, Van de Peer Y. Investigating ancient duplication events in the Arabidopsis genome. J Struct Funct Genomics. 2003;3:117–129. [PubMed] [Google Scholar]
- Rajesh PN, O'Bleness M, Roe BA, Muehlbauer FJ. Analysis of genome organization, composition and microsynteny using 500 kb BAC sequences in chickpea. Theor Appl Genet. 2008;117:449–458. doi: 10.1007/s00122-008-0789-0. [DOI] [PubMed] [Google Scholar]
- Rijpkema A, Gerats T, Vandenbussche M. Genetics of floral development in Petunia. Adv Bot Res. 2006;44:237–278. [Google Scholar]
- Rijpkema AS, Royaert S, Zethof J, van der Weerden G, Gerats T, Vandenbussche M. Analysis of the Petunia TM6 MADS box gene reveals functional divergence within the DEF/AP3 lineage. Plant Cell. 2006;18:1819–1832. doi: 10.1105/tpc.106.042937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning: a laboratory manual. New York: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Schwarz-Sommer Z, Davies B, Hudson A. An everlasting pioneer: the story of Antirrhinum research. Nat Rev Genet. 2003;4:657–666. doi: 10.1038/nrg1127. [DOI] [PubMed] [Google Scholar]
- Schwarz-Sommer Z, Huijser P, Nacken W, Saedler H, Sommer H. Genetic control of flower development by homeotic genes in Antirrhinum majus. Science. 1990;250:931–936. doi: 10.1126/science.250.4983.931. [DOI] [PubMed] [Google Scholar]
- Simillion C, Vandepoele K, Van Montagu MC, Zabeau M, Van de Peer Y. The hidden duplication past of Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2002;99:13627–13632. doi: 10.1073/pnas.212522399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer H, Beltrán JP, Huijser P, Pape H, Lönnig WE, Saedler H, Schwarz-Sommer Z. Deficiens, a homeotic gene involved in the control of flower morphogenesis in Antirrhinum majus: the protein shows homology to transcription factors. EMBO J. 1990;9:605–613. doi: 10.1002/j.1460-2075.1990.tb08152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Bowers JE, Wang X, Ming R, Alam M, Paterson AH. Synteny and collinearity in plant genomes. Science. 2008;320:486–488. doi: 10.1126/science.1153917. [DOI] [PubMed] [Google Scholar]
- Tang H, Wang X, Bowers JE, Ming R, Alam M, Paterson AH. Unraveling ancient hexaploidy through multiply-aligned angiosperm gene maps. Genome Res. 2008b;18:1944–1954. doi: 10.1101/gr.080978.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tröbner W, Ramirez L, Motte P, Hue I, Huijser P, Lönnig WE, Saedler H, Sommer H, Schwarz-Sommer Z. GLOBOSA: a homeotic gene which interacts with DEFICIENS in the control of Antirrhinum floral organogenesis. EMBO J. 1992;11:4693–4704. doi: 10.1002/j.1460-2075.1992.tb05574.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche M, Theissen G, Van de Peer Y, Gerats T. Structural diversification and neo-functionalization during floral MADS-box gene evolution by C-terminal frameshift mutations. Nucleic Acids Res. 2003;31:4401–4409. doi: 10.1093/nar/gkg642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, van der Hoeven RS, Nielsen R, Mueller LA, Tanksley SD. Characteristics of the tomato nuclear genome as determined by sequencing undermethylated EcoRI digested fragments. Theor Appl Genet. 2005;112:72–84. doi: 10.1007/s00122-005-0107-z. [DOI] [PubMed] [Google Scholar]
- Weigel D, Meyerowitz EM. The ABCs of floral homeotic genes. Cell. 1994;78:203–209. doi: 10.1016/0092-8674(94)90291-7. [DOI] [PubMed] [Google Scholar]
- Whipple CJ, Ciceri P, Padilla CM, Ambrose BA, Bandong SL, Schmidt RJ. Conservation of B-class floral homeotic gene function between maize and Arabidopsis. Development. 2004;131:6083–6091. doi: 10.1242/dev.01523. [DOI] [PubMed] [Google Scholar]
- Yamaguchi T, Lee DY, Miyao A, Hirochika H, An G, Hirano HY. Functional diversification of the two C-class MADS box genes OSMADS3 and OSMADS58 in Oryza sativa. Plant Cell. 2006;18:15–28. doi: 10.1105/tpc.105.037200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q, Steiger D, Kramer EM, Moore PH, Ming R. Floral MADS-box genes in trioecious papaya: characterization of AG and AP1 subfamily genes revealed a sex-type-specific gene. Trop Plant Biol. 2008;1:97–107. [Google Scholar]
- Zhou J, Wang F, Ma W, Zhang Y, Han B, Xue Y. Structural and transcriptional analysis of S-locus F-box genes in Antirrhinum. Sex Plant Reprod. 2003;16:165–177. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.