Abstract
The transcriptome profile in leaves and roots of the transgenic cotton line T-34 expressing hpa1Xoo from Xanthomonas oryzae pv. oryzae was analysed using a customized 12k cotton cDNA microarray. A total of 530 cDNA transcripts involved in 34 pathways were differentially expressed in the transgenic line T-34, in which 123 differentially expressed genes were related to the cotton defence responses including the hypersensitive reaction, defence responses associated with the recognition of pathogen-derived elicitors, and defence signalling pathways mediated by salicylic acid, jasmonic acid, ethylene, auxin, abscicic acid, and Ca2+. Furthermore, transcripts encoding various leucine-rich protein kinases and mitogen-activated protein kinases were up-regulated in the transgenic line T-34 and expression of transcripts related to the energy producing and consuming pathway was also increased, which suggested that the enhanced metabolism related to the host defence response in the transgenic line T-34 imposed an increased energy demand on the transgenic plant.
Keywords: Chitin catabolism, gene expression, phenylpropanoid pathway, phosphorylation and dephosphorylation, plant hormones
Introduction
Cotton (Gossypium spp.) is one of the most important textile fibre crops worldwide. A great many fundamental and applied studies have been conducted on this crop in the areas of weed and pest management due to the large and profitable market for growers. In the past two decades, genetically modified cotton has been developed in many countries, which has reduced reliance on pesticides for the production of this crop by ∼80%, compared with the areas where conventional cotton varieties were grown (Phipps and Park, 2002). In China, genetically engineered cotton expressing δ-endotoxins (Cry proteins) from Bacillus thuringiensis (Bt) was developed in 1997 and then adopted by 95% of cotton growers in northern China (Wu et al., 2008). Although Bt cotton reduced the overall need for the insecticide spray initially, it was recently discovered that the wide application of Bt cotton in China had resulted in an increase in the mirid bug population on cotton due to the decrease of pesticide usage on this crop (Lu et al., 2010). Therefore, it is imperative to develop new sources of transgenes for genetically modified cotton that can provide a broader spectrum of resistance for pest management on cotton and increase the diversity of genetically engineered cotton in China.
Harpin is a heat-stable, glycine-rich and acidic protein secreted by Gram-negative plant pathogenic bacteria into the plant intercellular space through the type III secretion system (T3SS) (Wei et al., 1992; Perino et al., 1999). To date, a few harpins or harpin-like proteins, such as hrpNEa from Erwinia amylovora (Kim and Beer, 1998; Dong et al., 1999), hrpZPss from Pseudomonas syringae pv. syringae (He et al., 1993; Strobel et al., 1996), and hpa1Xoo from Xanthomonas oryzae pv. oryzae (Peng et al., 2004) have been characterized and their roles in the non-host interaction have been investigated.
Harpins can induce hypersensitive response (HR) and systemic acquired resistance in non-host plants through exogenous application (Wei and Beer, 1993). In addition, they are capable of inducing many other host responses such as enhanced growth and drought tolerance in tobacco and Arabidopsis (Dong et al., 1999; Dong et al., 2004; Dong et al., 2005). Furthermore, harpins regulate the plasma membrane ion channels that are considered to be putative components of the signalling pathway related to the host defence response (El-Maarouf et al., 2001). Harpins can also induce the potassium efflux and rapid inhibition of ATP synthesis that result in plasma membrane depolarization and growth medium alkalinization in various plant species (Popham et al., 1995; Hoyos et al., 1996; Xie and Chen, 2000). More recently, it was found that HrpN of E. amylovora interacted with HrpN-interacting protein from Malus (HIPM) and its orthologue, AtHIPM, of Arabidopsis. Both genes contained functional signal peptides associated with the plasma membrane (Oh and Beer, 2007). It has been hypothesized that these plant phenotypes induced by harpins are related to the change in expression of genes involved in various signalling pathways, such as salicylic acid (SA)-, jasmonate (JA)-, and ethylene (ET)-mediated signalling pathways (Somssich and Hahlbrock, 1998; Dong et al., 1999).
In addition to exogenous application, it was reported that harpins expressed in transgenic plants also induced the host defence response (Peng et al., 2004). To date, several hrp genes have been transformed into plant species, including tobacco, rice, and Arabidopsis, and enhanced resistance to various bacterial and fungal pathogens was obtained in the transgenic plants (Peng et al., 2004; Sohn et al., 2007). Several pathogenicity-related genes (pr), such as pr-1a, pr-1b, pr-2, pr-3, and Chia5, and genes related to the production of ET (e.g. NT-EFE26, NT-1A1C, DS321, NTACS1, and NT-ACS2) were up-regulated in transgenic plants expressing harpins (Peng et al., 2004; Sohn et al., 2007; Shao et al., 2008). Nevertheless, the up-regulation of npr-1 (non-expressor of pr genes) in hrp-transformed plants was variable, which suggested that expression of npr-1 differed depending on the host and the origin of the hrp gene used in the transformation (Peng et al., 2004; Sohn et al., 2007).
Although harpins induced the HR in non-host plants through exogenous application (He et al., 1993), no visible HR was found in the transgenic cotton line T-34 harbouring a constitutively expressed hpa1Xoo in our previous study (Miao et al., 2010). The hpa1Xoo-transformed cotton line T-34 showed improved resistance to Verticillum and Fusarium wilt caused by Verticillium dahliae and Fusarium oxysporum f.sp. vasinfectum, respectively. In addition, the oxidative burst and up-regulation of several key defence-related genes were observed in transgenic T-34 in response to infection caused by V. dahliae, which suggested that transformation of cotton with hpa1Xoo conferred enhanced defence response on pathogens through a priming mechanism. Since the mechanism of harpin-mediated plant responses is still largely unknown, the possibility of genome-wide modification of genetically modified plants or plants treated by harpins remains to be investigated. With the recent development of gene chip technology, it is possible to investigate the defence responses and signalling pathways involved in plants transformed with hpa1Xoo on a genome-wide scale (Kim et al., 2006; Chibucos et al., 2009). In the present study, we investigated the pattern of global gene expression in the transgenic cotton line T-34 expressing hpa1Xoo, and in the wild-type receptor (Z35) using a customized cotton 12k cDNA microarray. This study is of importance for understanding the function of hpa1Xoo in genetically modified cotton and its effects on the cotton defence signalling pathway.
Materials and methods
Plant and fungal materials
The transgenic cotton line T-34 was developed previously through the genetic transformation of the susceptible cotton variety Zhong-Mian-35 (Gossypium hirsutum L., abbreviated Z35) with hpa1Xoo derived from X. oryzae pv. oryzae using a modified Agrobacterium-mediated method (Miao et al., 2010). Homozygous transgenic lines from T1 to T6 were screened using kanamycin resistance, PCR analysis, Southern and Northern analysis as previously described (Miao et al., 2010). Three T6 progeny of the transgenic T-34 used in this study were grown in a sterile incubator (MLR-351; Sanyo Electric Co., Ltd, Japan) at 28°C with a photoperiod of 12 h under incandescent light. Cotton leaves were collected at the true leaf stage and stored in liquid nitrogen prior to RNA extraction. Wild-type cotton Z35 was used as the negative control.
V. dahliae strain (Vbps) was provided by Dr Ling Lin (Jiangsu Academy of Agricultural Science, China). V. dahliae was maintained on potato dextrose agar (PDA) at 25°C. The method of Joost et al. (1995) was used to prepare the inoculum and the V. dahliae conidia suspension was adjusted to a concentration of 1 × 107 conidia/ml. Two millilitres of conidia suspension was used to inoculate leaves of T-34 and Z35 by freshly cutting at the four to five leaf stage or dipping petioles for 3 h.
Microarray procedure and data analysis
Total RNA samples were extracted from 1 g of cotton leaves and roots using the Plant Total RNA Extract Kit (Autolab Biotech, Beijing, China) according to the manufacturer's instructions. RNA samples were collected from three biological replicates. The 12k cDNA microarray was conduced at CapitalBio Corp. (Beijing, China) using the method described by Shi et al. (2006). Briefly, PCR products from 11 236 cotton unigene expressed sequence tags (ESTs) were printed on to amino-silanized glass slides in triplicate for each PCR product. Total RNA (5 μg) was used to synthesize cDNA in an in vitro transcription reaction and fluorescently labelled using Klenow enzyme (Promega, Beijing, China). After hybridizations, arrays were scanned with a confocal LuxScan™ scanner (CapitalBio, Beijing, China) and analysed using the software LuxScanTM 3.0 (CapitalBio, Beijing, China). For extraction of data from the individual channels, faint spots with intensity <400 units after subtraction of the background were removed. A space- and intensity-dependent normalization method based on a LOWESS program (Yang et al., 2002) was used. The gene annotation and pathway identification were performed according to the method described by Shi et al. (2006).
Real-time RT-PCR for gene expression
Three hours after inoculation, the complete laminae of cotton leaves at the V4 stage (four-leaf stage) were harvested from T-34 and Z35 and frozen in liquid nitrogen until the RNA extraction. RNA was extracted from cotton leaves using the RNAiso kit for polysaccharide-rich plant tissue (TaKaRa® Biotechnology, Dalian, China). The concentration of RNA was quantified using a biophotometer (Eppendorf AG, Hamburg, Germany). cDNA was synthesized using a PrimeScriptTM RT-PCR Kit (TaKaRa® Biotechnology, Dalian, China). Two-step real-time RT-PCR was performed on an ABI PRISM 7000 (ABI, Foster City, CA, USA) according to the procedure optimized for the SYBR Premix Ex TaqTM kit (TaKaRa® Biotechnology, Dalian, China). EF-1α, a conserved plant housekeeping gene (Peng et al., 2004), was used as the internal control to normalize the level of expression. All real-time RT-PCRs were performed in duplicate and three biological replicates were included. The expression data were normalized to EF-1α using the ΔΔCT method described by Livak and Schmittgen (2001).
Results
The selection of plant materials for microarray analysis
Three plants from T6 progeny of the transgenic T-34 were randomly selected. The presence and the constitutive expression of hpa1Xoo in the selected T-34 were verified using PCR, Southern and Northern blot analysis. In all three plants, bands representing hpa1Xoo insert, the 35s promoter, and the NOS terminator (420, 310, and 180 bp in length, respectively) were found in the PCR analysis with hpa1Xoo-specific primers. In Southern blot analysis, three positive bands (4, 6, and 10 kb in length) were detected in all three T-34 progeny indicating the insertion of hpa1Xoo at multiple chromosomal locations. The constitutive expression of hpa1Xoo in selected T-34 plants was observed in the Western blot using the polyclonal antibody raised against purified hpa1Xoo (data not shown).
The construction and verification of the cotton cDNA microarray analysis
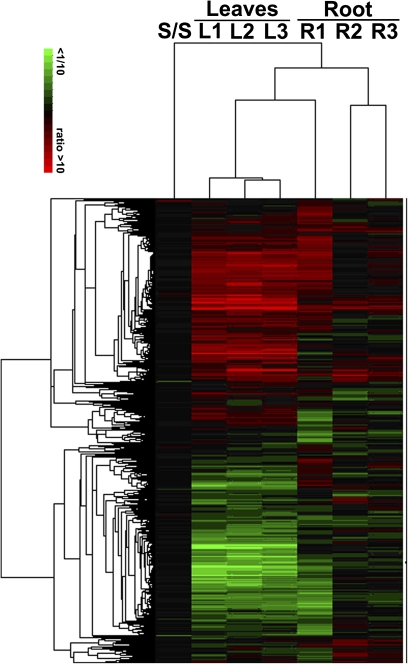
Of 12 233 unigene ESTs initially selected, 11 236 ESTs were successfully amplified through PCR amplifications and printed onto aminosilane slides. These ESTs were used to construct a cDNA microarray for the identification of genes specifically or preferentially expressed in transgenic T-34. Differentially expressed genes (DEGs) that showed up-regulation (2-fold increase in expression compared with expression in wild-type Z35) or down-regulation (2-fold decrease in expression compared with expression in wild-type Z35) in all three replicates of transgenic T-34 (corrected for the false discovery rate (FDR), P < 0.01) were considered as related to the transformation of hpa1Xoo (Figure. 1).
Fig. 1.
Hierarchical clustering of DEGs in leaves and roots of transgenic cotton T-34 compared with wild-type Z35 in the microarray analysis. Each column represents a single biological replicate and each row represents a differentially expressed probe set. L1, L2, and L3 represent biological replicates from leaves and R1, R2, and R3 represent biological replicates from roots. S/S represented self-to-self of Z35. The signal ratios were shown in a red–green colour scale, where red indicated up-regulation and green indicated down-regulation.
The quality of microarray data was verified based on the value of correlation coefficient (r values) and the swap-dye experiment. The correlation coefficient, which measured the biological reproducibility, was calculated from each replicate (Table 1). The value from the swap-dye experiment measured the technical reproducibility. After the self-hybridization of Cy3- and Cy5-labelled probes prepared using the same RNA sample prepared from leaves of wild-type Z35 (V4 stage), all but 13 data points were scattered inside the 2-fold line, which indicated that the microarray was precisely executed (data not shown). In addition, we applied a space- and intensity-dependent normalization based on a LOWESS program (Yang et al., 2002). Forty internal controls were deployed to obtain evenly distributed signal intensities (see Supplementary Table 1, at JXB Online).
Table 1.
Correlation coefficients of microarray hybridization with total RNA from leaves of transgenic cotton T-34 and wild-type Z35
| Microarray samples | Biological coefficients |
||
| (1,2)a | (2,3) | (1,3) | |
| Leaf | 0.9402 | 0.9546 | 0.9502 |
| Root | 0.9632 | 0.9306 | 0.9294 |
1, 2, and 3 represent three biological replicates from leaves or roots.
To validate results from the microarray analysis, nine genes representing a wide range of relative fluorescence-signal intensity and different functions were selected from 530 DEGs found in leaves of T-34 and subjected to real-time RT-PCR analysis (Table 2). The expression of eight genes analysed with real-time RT-PCR was consistent with the data obtained from the microarray analysis (Table 2).
Table 2.
Characteristics of PCR primers used in real-time RT-PCR analysis
| cDNA/A. thaliana orthologue | Oligonucleotide primers | Gene function or GenBank accession No. | Fold change |
||
| Microarray | Real-time RT-PCR | Real-time RT-PCR (challenged with V. dahliae) | |||
| CM011H11/ AT3G12500 | F: 5′-AGCGACAACAACAATGGC-3′ R: 5′-TAGCAAACAGGTCCTCAAA-3′ |
Pathogenesis and PCD-related protein | 0.44 ± 0.034 | 11.63 ± 3.183 | 273.32 ± 69.88 |
| CM021D04/ AT3G17390 | F: 5′-TATTTACTTCCGAGTCTGTCGTT-3′ R: 5′-TTTGCTTTCTGGGTCTTGTT-3′ |
SA biosynthesis and ET biosynthesis from methionine | 0.47 ± 0.089 | 0.52 ± 0.410 | 385.72 ± 24.64 |
| CM025C08/ AT2G41410 | F: 5′-CTCCAAAGCCATCATAGAATC-3′ R: 5′-AAAATCGGACCAGTCACCT-3′ |
Calcium signalling pathway and phosphatidylinositol signalling system | 4.21 ± 0.005 | 1.43 ± 1.159 | 352.94 ± 65.99 |
| CM031A04/ AT2G07689 | F: 5′-GCACGGCTCCTAAGTGATAA-3′ R: 5′-TGCTCCTACGGAACCAAGT-3′ |
Oxidative phosphorylation | 5.76 ± 0.018 | 4.26 ± 0.521 | 135 ± 13.86 |
| CM107B04/ AT3G53260 | F: 5′-GCCAATGGTGACAATGAAA-3′ R: 5′- CAACAACCCAGTTCCAAGC-3′ |
SA biosynthesis | 3.07 ± 0.018 | 10.23 ± 0.156 | 1011.34 ± 17.91 |
| CM048G07/ AT5G06320 | F: 5′-GAACGGAGCCTATTATGGCCCTTCC-3′ R:5′-CATGTATATCAATGAACACTAAACGCCGG-3′ |
Non-race-specific disease resistance 1 | 2.06 ± 0.002 | 6.98 ± 1.595 | 470.61 ± 30.8 |
| CM111F04/ AT1G19850 | F:5′-ATGCTAAGTTTCAAAAACCTTC-3′ R:5′-TTAGTCGCGGGGTGATTTGC-3′ |
ARF5, auxin response factor 5 | 5.15 ± 2.450 | 6.62 ± 0.173 | |
| CM024A04/ AT2G46690 | F: 5′-ATGTTTGGAATAGAAAGATC-3′ R: 5′-TCACGCAAATATGCTTAAGG-3′ |
ARF7, auxin response factor 7 | 2.09 ± 0.125 | 6.05 ± 1.142 | |
| CM024E02/ AT2G28350 | F: 5′-ATGGTGGGATCACAAGGAGAG-3′ R: 5′-TCAAACCCTAAAGCAACC-3′ |
ARF10, auxin response factor 10 | 1.96 ± 0.157 | 9.58 ± 0.520 | |
| hsr203J | F: 5′-TGTACTACACTGTCTACACGC-3′ R: 5′-GATAAAAGCTATGTCCCACTCC-3′ |
Hypersensitive reaction marker gene | 5.24 ± 1.324 | ||
| EF-1α | F: 5′-AGACCACCAAGTACTACTGCAC-3′ R: 5′-CCACCAATCTTGTACACATCC-3′ |
Housekeeping gene used as internal reference | |||
Global cell expression patterns in the transgenic line T-34 and wild-type receptor Z35
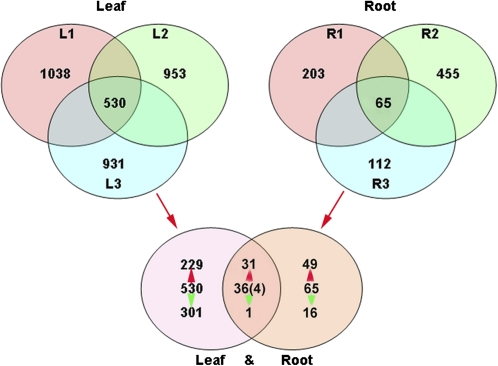
The microarray data were deposited in the GEO database of the National Center for Biotechnology (accession number GSE20446). In leaves and roots of transgenic T-34, a total of 1552 and 672 DEGs, respectively, were related to the hpa1Xoo transformation with the exhibited ratio of >2.0 (2-fold increase in expression) or <0.5 (2-fold decrease in expression) in the microarray (Fig. 2). In leaves of T-34, 530 DEGs related to the hpa1Xoo transformation were found in all three replicates, in which 229 transcripts (43.2%) were up-regulated and 301 transcripts (56.7%) were down-regulated. In comparison, 65 DEGs related to the hpa1Xoo transformation were identified in three root replicates, in which 49 (74.5%) transcripts were up-regulated and 16 (24.5%) transcripts were down-regulated. The comparison of DEGs in leaves and roots from transgenic T-34 and wild-type Z35 showed that 36 DEGs were up/down-regulated in both leaves and roots of transgenic T-34. Of these DEGs, 31 DEGs were up-regulated whereas only one DEG was down-regulated. Four DEGs were up-/down-regulated conversely in leaves and roots of transgenic T-34 (Fig. 2 and Table 3).
Fig. 2.
Venn diagram of DEGs in leaves and roots of transgenic cotton T-34. L1, L2, and L3 represent biological replicates from leaves of T-34 and R1, R2, and R3 were biological replicates from roots of T-34. The expression level of selected genes was based on the relative ratio >2.0 or <0.5 in T-34 versus Z35. Red arrows indicate the number of up-regulated genes and green arrows indicate the number of down-regulated genes. Numbers in parentheses indicate the number of genes that were conversely regulated in leaves and roots.
Table 3.
Thirty-six genes that were differentially expressed in both leaves and roots of transgenic T-34 compared with wild-type Z35
| Array ID | A. thaliana orthologue | Fold change |
Description | Putative biological functions | |
| Leaf | Root | ||||
| CM105C09 | – | 9.37 | 2.93 | Unknown | Unknown |
| CM114B09 | – | 9.84 | 3.13 | Unknown | Unknown |
| CM102E05 | – | 9.05 | 4.85 | Unknown | Unknown |
| CM089F02 | – | 6.07 | 2.55 | Unknown | Unknown |
| CM105E06 | – | 4.91 | 2.27 | Unknown | Unknown |
| CM104E05 | – | 4.84 | 2.01 | Unknown | Unknown |
| CM104C04 | – | 4.7 | 2.39 | Unknown | Unknown |
| CM101G02 | – | 4.51 | 2.01 | Unknown | Unknown |
| CM084B08 | – | 4.01 | 2.18 | Unknown | Unknown |
| CM098D08 | – | 3.33 | 2.05 | Unknown | Unknown |
| CM104A08 | – | 3.06 | 2.05 | Unknown | Unknown |
| CM090B01 | – | 2.78 | 2.1 | Unknown | Unknown |
| CM079A04 | – | 2.64 | 2.17 | Unknown | Unknown |
| CM105C02 | AT3G57170 | 8.85 | 2.66 | N-acetylglucosaminyl transferase | Protein N-glycosylation related to biotic/abitotic stress |
| CM105H03 | AT4G22760 | 8.51 | 2.66 | Pentatricopeptide repeat-containing protein | RNA binding related to plastid ribosome biogenesis |
| CM105E03 | AT5G45140 | 8 | 2.63 | DNA-directed RNA polymerase II | DNA repair and transcription |
| CM104G02 | AT3G54220 | 7.47 | 2.05 | Scarecrow transcription factor | Transcription regulation |
| CM027H01 | AT3G22840 | 7.36 | 2.65 | Chlorophyll A–B binding protein | Receptor for energy transfer |
| CM105A02 | AT1G69440 | 7.02 | 2.11 | A member of the ARGONAUTE family | RNA-mediated gene silencing |
| CM104G01 | AT5G51850 | 4.89 | 4.82 | Expressed protein | Unknown |
| CM088H01 | AT5G49740 | 4.52 | 2.79 | Ferric chelate reductase. | Iron chelate transport related to oxidation–reduction |
| CM105C04 | AT1G30700 | 4.44 | 2.38 | FAD-binding domain-containing protein | Oxidoreductase related to oxidation–reduction |
| CM105F08 | AT3G13700 | 4.13 | 2.17 | RNA-binding protein | RNA binding |
| CM103A05 | AT5G07400 | 3.59 | 2.22 | Forkhead-associated domain-containing protein | RNA-mediated gene silencing |
| CM105E10 | AT2G29125 | 3.34 | 3.77 | Expressed protein | Unknown |
| CM105B08 | AT2G26330 | 3.32 | 3.13 | LRR protein kinase | ATP-binding protein response to biotic stress |
| CM105F10 | AT5G46370 | 3.31 | 2.06 | Outward rectifying potassium channel | Potassium transport related to abiotic stress |
| CM105G10 | AT5G43270 | 3.29 | 3.15 | Squamosa promoter-binding protein | Transcription regulation |
| CM101H09 | AT2G07689 | 3.27 | 2.31 | NADH-ubiquinone oxidoreductase | Electron transport response to oxidative stress |
| CM113A07 | AT4G30710 | 3.3 | 2.52 | Expressed protein | Unknown |
| CM101F11 | AT5G16650 | 2.42 | 2.16 | DNAJ heat shock N-terminal domain-containing protein | Heat shock protein binding response to stress |
| CM049G04 | AT1G17860 | 0.12 | 2.44 | Trypsin and protease inhibitor | Protease inhibitor response to biotic stress |
| CM110G08 | AT2G20870 | 0.08 | 3.62 | Cell wall protein precursor | Cell wall morphogenesis |
| CM112F08 | AT5G23960 | 3.94 | 0.43 | Sesquiterpene synthase | Sesquiterpenoid biosynthetic process |
| CM111E02 | AT4G20820 | 3.28 | 0.24 | FAD-binding domain-containing protein | Oxidoreductase related to oxidation–reduction |
| CM022B06 | AT1G27730 | 0.36 | 0.48 | Zinc finger protein | Metal-binding protein response to the salt tolerance |
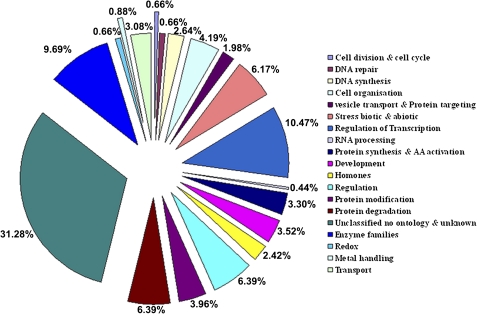
Five hundred and thirty DEGs found in leaves of transgenic T-34 were annotated through the blast against Kegg (www.genome.ad.jp/kegg), Biocyc (www.biocyc.org), and Kobas (kobas.cbi.pku.edu.cn:8080) databases (Fig. 3). The largest class of annotated DEGs was transcription factors (10.47%) followed by DEGs encoding various enzymes (9.69, and proteins involved in the protein degradation process (6.39%). In addition, 6.17% of DEGs were related to responses induced by biotic or abiotic stress and 2.42% of DEGs were involved in hormone biosynthesis. Among 36 DEGs that were differentially expressed in leaves and roots of transgenic T-34, five DEGs (CM105C02, CM105B08, CM105F10, CM101F11, and CM049G04) were related to responses induced by biotic or abiotic stress and two DEGs (CM104G02 and CM105G10) were transcription factors (Table 3).
Fig. 3.
The functional annotation of 530 DEGs in leaves of transgenic T-34 identified in the microarray analysis (P <0.001).
Twenty-one DEGs were involved in cytological and biochemical processes related to the defences response, such as programmed cell death (PCD), phenylpropanoid and flavonoid biosynthesis, lignin biosynthesis, pathogenesis-related proteins, and NAD/NADH phosphorylation and dephosphorylation (Table 4). In addition, a large group of DEGs involved in the signalling transduction cascade mediated by protein kinases, G-proteins, and calmodulin-like proteins were also identified (Table 5).
Table 4.
Differentially expressed defence-related genes in leaves of transgenic T-34
| Array ID | A. thaliana orthologue | Fold change | P-value | Putative functions | |
| Apoptosis and PCD | CM105A03 | AT1G29340 | 6.74 | 0.00662 | E3 ubiquitin ligase related to PCD |
| CM103E04 | AT5G02190 | 2.45 | 0.00095 | Aspartic protease related to PCD | |
| CM025G05 | AT5G17540 | 2.09 | 0.00157 | PCD-related protein with transferase activity | |
| CM009E10 | AT4G19500 | 2.07 | 0.00378 | Disease resistance protein with a signature TIR-NBS-LRR domain | |
| CM048G07 | AT5G06320 | 2.06 | 0.00246 | Hrp-induced HR-related proteins in tobacco and Arabidopsis | |
| CM099D07 | AT1G61190 | 0.43 | 0.02809 | Disease resistance protein with a signature CC-NBS-LRR domain | |
| Phenylpropanoid biosynthesis | CM107B04 | AT3G53260 | 2.63 | 0.01784 | Phenylalanine ammonia lyase (PAL-2) related to phenylpropanoid and salicylic biosynthesis |
| CM048C05 | AT5G48930 | 2.48 | 0.00212 | Quinate hydroxycinnamoyltransferase in the phenylpropanoid pathway | |
| CM091F11 | AT5G53970 | 1.96 | 0.00312 | Tyrosine aminotransferase related to ET and phenylalanine biosynthesis | |
| CM092H07 | AT1G76490 | 0.48 | 0.05042 | 3-Hydroxy-3-methylglutaryl-CoA reductase related to isopropanoid biosynthesis | |
| CM099F12 | AT3G07630 | 0.43 | 0.01476 | Prephenate dehydratase related to phenylalanine biosynthesis | |
| Flavonoid biosynthesis | CM087F05 | AT5G13930 | 3.63 | 0.05291 | Chalcone synthase |
| CM100D01 | AT5G13930 | 2.09 | 0.23097 | Chalcone synthase | |
| CM113E06 | AT5G13930 | 2.52 | 0.08218 | Chalcone synthase | |
| CM092E03 | AT3G51240 | 2.44 | 0.04194 | Naringenin 3-dioxygenase | |
| Lignin biosynthesis | CM072F03 | AT2G23910 | 5.21 | 0.04636 | Cinnamoyl-CoA reductase |
| CM089D11 | AT2G23910 | 2.39 | 0.08661 | Cinnamoyl-CoA reductase | |
| CM078C09 | AT5G42800 | 2.9 | 0.00135 | Dihydroflavonol 4-reductase | |
| Pathogenesis-related proteins | CM011H11 | AT3G12500 | 0.44 | 0.0339 | Basic endochitinase (PR-3) |
| CM113A02 | AT3G12500 | 0.29 | 0.00453 | Basic endochitinase (PR-3) | |
| CM072H01 | AT4G21960 | 0.28 | 0.03461 | Peroxidase (PR-9) |
aP-value (t-test) was calculated from three replicates.
bThe function annotation was based on the consensus of multiple blasts.
cFold change of expression was based on the comparison between expression in leaves of T-34 versus Z35.
Table 5.
Differentially expressed genes related to the signal transduction in leaves of transgenic T-34
| Array ID | A. thaliana orthologue | Fold change | P-value | Description | |
| Kinase-mediated signalling | CM079E09 | AT4G32300 | 3.09 | 0.00721 | Lectin protein kinase |
| CM113G02 | AT4G08850 | 3 | 0.03893 | LRR protein with kinase domain | |
| CM030A02 | AT5G48740 | 2.34 | 0.00938 | LRR protein with kinase domain | |
| CM057G05 | AT5G10530 | 2.3 | 0.01311 | Lectin protein kinase | |
| CM023F07 | AT5G49760 | 2.17 | 0.00131 | LRR protein with kinase domain | |
| CM019F07 | AT2G24230 | 2.08 | 0.01277 | LRR transmembrane protein kinase | |
| CM089A03 | AT4G08850 | 2.06 | 0.03666 | LRR protein with kinase domain | |
| CM076F10 | AT1G75820 | 0.48 | 0.01157 | Receptor kinase with an extracellular leucine-rich domain | |
| CM093F12 | AT5G54380 | 0.47 | 0.01072 | Protein kinase | |
| CM081B01 | AT2G18170 | 0.41 | 0.00271 | Mitogen-activated protein kinase | |
| CM105D02 | AT1G14390 | 0.37 | 0.00242 | LRR transmembrane protein kinase | |
| ET-mediated signalling | CM105E12 | AT5G61600 | 8.83 | 0.00375 | ET responsive ERF/AP2 transcription factor |
| CM038C12 | AT5G47230 | 2.16 | 0.20507 | ET responsive ERF/AP2 transcription factor | |
| CM058G10 | AT3G16770 | 2.12 | 0.00402 | ET responsive ERF/AP2 transcription factor | |
| Auxin-mediated signalling | CM111F04 | AT1G19850 | 3.5 | 0.02526 | Auxin-responsive transcription factor |
| CM024E02 | AT2G46690 | 2.09 | 0.01753 | Auxin-responsive transcription factor | |
| CM024A04 | AT2G28350 | 2 | 0.03526 | Auxin-responsive transcription factor | |
| ABA-mediated signalling | CM105G04 | AT4G28950 | 2.56 | 0.02261 | ROP GTPase gene family related to ABA signalling |
| CM040B08 | AT3G09600 | 2.04 | 0.03666 | myb transcription factor response to SA/ABA/JA | |
| CM032D01 | AT3G55730 | 2 | 0.00482 | myb transcription factor response to SA/ABA/JA | |
| GTP-binding protein-mediated signalling | CM019F05 | AT1G52280 | 2.69 | 0.00204 | Ras-related GTP-binding protein |
| CM105C07 | AT3G18820 | 0.52 | 0.00052 | Ras-related GTP-binding protein | |
| CM028G08 | AT2G44610 | 0.46 | 0.00538 | Ras-related GTP-binding protein | |
| Jasmonic acid-mediated signalling | CM104D06 | AT1G13280 | 3.18 | 0.01822 | Allene oxide cyclase related to jasmonic acid biosynthesis |
| CM077D03 | AT3G25770 | 0.45 | 0.00544 | Allene oxide cyclase related to jasmonic acid biosynthesis | |
| Transcripts involved in the intracellular signalling cascade | CM025C08 | AT2G41410 | 3.71 | 0.01343 | Calcium-binding protein |
| CM006A06 | AT4G20780 | 2.52 | 0.01252 | Calcium-binding protein | |
| CM033E11 | AT3G08510 | 0.42 | 0.02265 | Phosphoinositide-specific phospholipase | |
| CM052F01 | AT3G56800 | 0.39 | 0.03461 | Calcium-binding protein | |
| CM092E09 | AT3G43810 | 0.45 | 0.057 | Calcium-binding protein |
Differentially expressed transcripts related to the defence response in leaves of transgenic T-34
Five up-regulated DEGs were related to PCD. The up-regulation of CM105A03 was the most significant followed by CM103E04 and CM025G05, which were increased by 6.74-, 2.45- and 2.09-fold, respectively. CM105A03 encoded an E3 ligase, an inhibitor of PCD in plants, and CM103E04 had sequence similarity to an aspartic protease gene involved in PCD by hydrolysing important cell components and/or activating other proteinases. CM025G05 was similar to the hypersensitivity-related gene hsr201 found in Nicotiana tabacum (Table 4).
Among five DEGs related to phenylpropanoid biosynthesis, the up-regulation of CM107B04, which encoded a phenylalanine ammonia lyase (PAL) catalysing the initial reaction of the phenylpropanoid biosynthetic pathway, was the most significant (2.63-fold) (Table 4). In addition, expression of CM048C05 was increased by 2.48-fold in transgenic T-34. CM048C05 was nearly identical to a quinate hydroxycinnamoyl transferase (HCT), which synthesized hydroxycinnamoylesters in the phenylpropanoid pathway. In comparison, two DEGs (CM092H07 and CM099F12) were down-regulated in leaves of transgenic T-34. CM092H07 was similar to a 3-hydroxy-3-methylglutaryl-CoA reductase, which was the rate-limiting enzyme in the mevalonate pathway related to isoprenoid biosynthesis. CM099F12 encoded a prephenate dehydratase participating in phenylalanine, tyrosine, and tryptophan biosynthesis.
Four DEGs related to flavonoid biosynthesis were also up-regulated in leaves of transgenic T-34 (2.09- to 3.63-fold increase in expression). Three DEGs (CM087F05, CM100D01, and CM113E06) encoded products similar to chalcone synthase. Chalcone synthase is important in the production of chalcones, a class of organic compounds commonly found in plants related to natural defence mechanisms. CM092E03 encoded a flavanone 3-dioxygenase. Furthermore, expression of three DEGs (CM072F03, CM089D11, and CM078C09) related to the biosynthesis of lignin was also increased in leaves of transgenic T-34 (5.21-, 2.39-, and 2.9-fold, respectivley). CM072F03 and CM089D11 were both similar to the gene encoding cinnamoyl-CoA reductase whereas CM078C09 had similarity to a gene encoding dihydroflavonol 4-reductase. Both of these two genes are very important for lignin biosynthesis in plants (Table 4).
Surprisingly, three DEGs with similarities to previously described pathogenesis-related (pr) genes were down-regulated in transgenic T-34. CM011H11 and CM113A02, which encoded products similar to a basic endochitinase (PR-3), were down-regulated by 0.44- and 0.29-fold, respectively. A decrease in expression was also found for CM072H01, which encodes a putative peroxidase (PR-9) (Table 4).
Differentially expressed transcripts related to signal transduction in leaves of transgenic T-34
Receptor protein kinases play a fundamental signalling role in plant growth, plant development and host responses to biotic and abiotic stress. In the present study, 11 DEGs with similarities to various receptor protein kinases were identified in transgenic T-34 in which the leucine-rich repeat receptor kinases represented the largest group (CM113G02, CM030A02, CM057G05, CM023F07, CM019F07, CM089A03, CM076F10, CM093F12, CM081B01, and CM105D02). Additionally, expression of two DEGs (CM079E09 and CM057G05) similar to lectin protein kinases was also up-regulated 3.09- and 2.3-fold, respectively. In comparison, CM081B01, which encoded a mitogen-activated protein kinase (MAPK), was down-regulated (0.41-fold) (Table 5). It has been reported that MAPKs relay signals from receptors and activate downstream defence responses in the host defence response (Desikan et al., 1999; Dievart and Clark, 2004).
Ethylene (ET), auxin, and abscisic acid (ABA) are three major plant hormones with important roles in signalling related to plant growth and the defence response. Three DEGs (CM105E12, CM038C12, and CM058G10), which encoded products similar to ET-responsive ERF/AP2 transcription factors, were up-regulated in transgenic T-34 (Table 5). The increase in expression of CM105E12 was the highest (8.83-fold) followed by CM038C12 and CM058G10, which were increased by 2.16- and 2.12-fold, respectively. CM111F04, CM024E02, and CM024A04 were all annotated as auxin-responsive transcription factors and their expression was increased 2.0- to 3.5-fold in leaves of transgenic T-34. In addition, one DEG (CM105G04) with similarity to a ROP GTPase related to an ABA-mediated signalling pathway and two DEGs similar to myb transcription factors were also up-regulated in transgenic T-34 (Table 5). Myb family transcription factors play regulatory roles in plant developmental processes and host responses to one or multiple types of hormone and stress treatment.
Small GTP-binding proteins are another class of signalling molecules that play a critical role in the regulation of a range of cellular processes including growth, differentiation, and intracellular transportation. In our study, three DEGs (CM019F05, CM105C07, and CM028G08) were identified as Ras-related GTP-binding proteins (Table 5). Although expression of CM019F05 was increased in transgenic T-34 (2.69-fold), CM105C07 and CM028G08 were down-regulated. Five DEGs involved in the intracellular signalling cascade were also differentially regulated in transgenic plants. CM025C08, CM006A06, CM052F01, and CM092E09 encoded calcium-binding proteins and CM033E11 encoded a phosphoinositide-specific phospholipase.
Differentially expressed transcripts related to energy production in leaves of transgenic T-34
Since plant defence responses are often energy consuming, up- or down-regulation of DEGs related to the defence response, the biosynthesis of secondary metabolites, transcription factors and cellular signalling molecules may alter the state of the energy production in the transgenic plant. In our study, several DEGs related to various energy-producing pathways were up-regulated (>2.0-fold increase in expression) (Table 6). CM103E07 (phosphopyruvate hydratase) and CM019F08 (peroxisomal biogenesis factor) were related to glycolysis. CM110G09 (aconitate hydratase) and CM024A06 (isocitrate dehydrogenase) were involved in the tricarboxylic acid (TCA) cycle. CM102G09, CM006G10, and CM032E11, which were annotated as acetyl-CoA carboxylase, biotin carboxyl carrier protein, and the malic enzyme, were important for fatty acid biosynthesis. Furthermore, three DEGs (CM031A04, CM101H09, and CM097C03), related to NAD/NADH phosphorylation and dephosphorylation, were also up-regulated in transgenic T-34.
Table 6.
Up-regulated genes related to the energy production and consumption in transgenic T-34
| Array ID | A. thaliana orthologue | Fold change | P-value | Description |
| CM103E07 | AT2G36530 | 3.27 | 0.0124 | Phosphopyruvate hydratase related to glycolysis |
| CM019F08 | AT3G61070 | 2.1 | 0.0114 | Peroxisomal biogenesis factor related to glycolysis |
| CM102G09 | AT1G36160 | 2.22 | 0.1386 | Acetyl-CoA carboxylase related to fatty acid biosynthesis |
| CM006G10 | AT5G16390 | 2.18 | 0.0027 | Biotin carboxyl carrier protein related to fatty acid biosynthesis |
| CM032E11 | AT1G79750 | 2.1 | 0.0147 | Malic enzyme related to fatty acid biosynthesis |
| CM110G09 | AT2G05710 | 2.84 | 0.0024 | Aconitate hydratase related to the TCA cycle |
| CM024A06 | AT5G14590 | 2.12 | 0.0107 | isocitrate dehydrogenase related to the TCA cycle |
| CM031A04 | AT2G07689 | 4.2 | 0.1226 | NADH-ubiquinone oxidoreductase related to NAD/NADH phosphorylation and dephosphorylation |
| CM101H09 | AT2G07689 | 3.27 | 0.01533 | NADH-ubiquinone oxidoreductase related to NAD/NADH phosphorylation and dephosphorylation |
| CM097C03 | AT3G21070 | 3.53 | 0.01809 | NAD(H) kinase related to nitrogen compound metabolism |
Enhanced expression of defence-related genes from T-34 after challenge by V. dahliae
Based on the result of microarray, six DEGs including CM011H11 (basic chitinase), CM048G (harpin-induced family protein), CM021D04 (S-adenosylmethionine synthetase), CM031A04 (NADH-ubiquinone oxidoreductase), CM107B04 (PAL2), and CM025C08 (calmodulin-like protein) were selected from different pathways and their expression in transgenic T-34 in response to V. dahliae infection was analysed using real-time RT-PCR analysis (Fig. 4 and Table 3). All six DEGs were significantly up-regulated in transgenic T-34, a >100 to 1000-fold increase in expression, after the transgenic T-34 plants were challenged with V. dahliae.
Discussion
Microarray is a useful technology to measure the transcriptome profile, which has been used successfully in various plant species to examine the host response to abiotic and biotic stress (Clarke and Zhu, 2006; Miyama and Tada, 2008; Van Hoewyk et al., 2008). In the present study, we used the microarray to investigate the transcriptome profile in leaves and roots of transgenic T-34 and wild-type Z35. Among 11 236 unigene ESTs included in the 12k cDNA microarray, 4.7% and 0.57% of ESTs were differentially expressed in leaves and roots of transgenic T-34, respectively. The ratios of DEGs to total ESTs included in the microarray chip are similar to that found in transgenic Arabidopsis overexpressing a stress response brassinosteroid receptor gene (BR1) (Kim et al., 2010) but significantly higher than that found in transgenic rice transformed with choline oxidase (0.032%) (Su et al., 2006), and with alanine aminotransferase (0.11%) (Shrawat et al., 2008). Although it is possible that this difference is merely due to the relatively larger number of transcripts included in gene chips used in previous studies, the higher ratios of DEGs found in transgenic T-34 could indicate that a larger portion of genes was differentially regulated in transgenic T-34.
Five hundred and thirty and 65 DEGs related to hpa1Xoo transformation were found in leaves and roots, respectively, of transgenic T-34, which indicateded that the transcriptome profile was altered not only in leaves but also in roots of transgenic T-34. Since no tissue specific promoter was added in front of the hpa1Xoo transgene in the transformation and our expression analysis indicated that hpa1Xoo was expressed in both leaves and roots of T-34 (data not shown), it is unlikely that the change in the transcriptome profile in different tissues of T-34 was due to the systemic effect of hpa1Xoo. Although 36 common DEGs were found in leaves and roots of transgenic T-34, which indicated that cells from different tissues of T-34 had a similar reaction to the transformation of hpa1Xoo, some differences were found between DEGs from leaves and roots of T-34. For examples, transcripts CM049G04, CM110G08, CM111E02, and CM022B06 were transversely regulated in leaves and roots of transgenic T-34. Some of DEGs found in leaves were not present in roots of T-34 (data not shown). These differences suggested the presence of certain tissue specificity in cotton in response to the hpa1Xoo transformation.
In our study, several DEGs (e.g. CM025G05, CM009E10, and CM048G07) with similarities to genes encoding HR-related proteins (HSR203 and proteins with signature TIR-NBS-LRR domains) were up-regulated in transgenic T-34. The up-regulation of these HR-related genes in transgenic T-34 is in agreement with our previous finding that micro-HRs were observed in leaves of transgenic T-34 in the absence of the pathogen (Miao et al., 2010). Up to date, three types of hrp-induced HR have been described including the visible HRs in response to foliar infiltration (He et al., 1993), microscopic HRs induced by foliar spray (Dong et al., 2004), and up-regulated HR-related marker genes in the absence of visible HR phenotypes in transgenic plants expressing harpins (Peng et al., 2004). This difference in HR phenotypes suggests that hrp-mediated HRs in hrp-transformed transgenic plants differ from plants treated exogenously with harpins. It is interesting that CM103E04, which encoded an aspartic protease, was up-regulated in transgenic T-34 2.45-fold, compared with its expression in the wild-type Z35. Ge et al. (2005) previously reported that the aspartic protease functioned as an anti-cell death component in the reproduction and embryogenesis of Arabidopsis. Similarly, a rice nucellin gene (OsAsp1), which encoded an aspartic protease, was found to play an important role in PCD of the rice nucleolus (Bi et al., 2005). It is possible that the up-regulation of DEGs encoding anti-cell death inhibitor-like products in transgenic T-34 indicates the counter-response of the hpa1Xoo-transformed plants to reduce the excess rate of the cell death since hpa1Xoo is an effector protein inducing hypertensive reaction in non-host plants. Nevertheless we cannot rule out the possibility that the absence of visible HR phenotypes in transgenic T-34 is due merely to the dosage effect of hpa1Xoo expressed in the transgenic plant (Miao et al., 2010).
In addition to DEGs related to HRs, DEGs involved in the biosynthesis of secondary metabolites were also up-regulated in transgenic T-34. For example, CM107B04, which encoded a phenylalanine ammonia-lyase (pal), was up-regulated in transgenic T-34. The expression of three transcripts (CM087F05, CM100D01, and CM113E06) similar to a chalcone synthase (Chs) was also increased in transgenic T-34. Both Pal and Chs are considered as the classical marker genes for the defence response since end products of phenylpropanoid and flavonoid biosynthesis, such as flavonoid phytoalexins and lignin, often play diverse roles in host responses to biotic or abiotic stimuli (Sewalt et al., 1997). Accordingly, DEGs related to flavonoid and lignin biosynthesis were up-regulated in transgenic T-34. The up-regulation of DEGs related to the biosynthesis of defence-related secondary metabolites suggested the constitutive activation of the host defence response in transgenic T-34. This result is in line with our previous finding that transgenic T-34 has enhanced resistance to a range of pathogens such as F. oxysporium and V. dahliae (Miao et al., 2010).
It is surprising that several DEGs with similarities to basic endochitinase (PR-3) and peroxidase (PR-9) were down-regulated in transgenic T-34. PR proteins can either promote the degradation of fungal cell wall or release endogenous elicitors from the plant cell wall, which further stimulate the defence response (Baron and Zambryski, 1995). It was unlikely that the down-regulation of DEGs similar to pr proteins in transgenic T-34 was due to the suppression of the host defence response, since DEGs related to HR, flavonoid, and lignin biosynthesis were up-regulated in transgenic T-34. In addition, our previous study showed that several key defence-related genes were also up-regulated in transgenic T-34 in response to inoculation with V. dahliae (Miao et al., 2010). Since only specific isoforms of basic chitinases and peroxidases exhibited antifungal activity (Sela-Buurlage et al., 1993), these pr-like DEGs may be not be related to the host defence response. For example, basic endochitinases are also expressed in tobacco and tomato during flower formation. Furthermore, several DEGs with similarity to endo-β-1,4-glucanases and glucan 1,3-β-glucosidases were also down-regulated in transgenic T-34 (data not shown). It is possible that the down-regulation of these DEGs is the result of a shift in carbohydrate metabolism in the transgenic plant due to the increase in secondary metabolite biosynthesis and energy production.
In addition to the up-regulation of DEGs related to the defence response, a number of DEGs involved in different signalling pathways were also up-regulated in transgenic T-34. For example, CM105A03, which encodes an E3 ubiquitin ligase, was up-regulated by 6.74-fold in transgenic T-34. There is emerging evidence that E3 ligase is important in the signalling of the plant immune system. For example, Yang et al. (2006) reported that E3 ligases were positive regulators of cell death and the host defence response across the Solanaceae and Brassicaceae families. Similarly, Abramovitch et al. (2006) found that the ubiquitin liagse activity of bacterial TypIII effector AvrPtoB suppressed plant cell death and immunity. To date, the regulation of defence gene expression by ET and JA has been linked to ubiquitination, which supports the hypothesis that multiple pathogen perception systems converge on common ubiquitination-based signalling pathways (Devoto et al., 2003). The up-regulation of CM105A03 in transgenic T-34 suggests that the ubiquitination-based signalling pathway is important for hpa1Xoo-mediated signalling in transgenic cotton T-34.
It has been reported that various plant hormones are involved in hrp-regulated plant PCD (Dong, 1998). For example, the ET-mediated signalling pathway was associated with pathogen and insect resistance in Arabidopsis treated with harpinEa from E. amylovora (Dong et al., 2004). In our study, three DEGs with similarities to the ET-responsive ERF/AP2 transcription factor were up-regulated in transgenic T-34. The ERF/AP2 transcription factor binds specifically to the GCC box present in many pr genes, to modulate their expression and participate in disease-resistance signalling pathways (Brown et al., 2003). The up-regulation of ERF/AP2 transcription factors has been previously reported in the defence response of Arabidopsis to infection caused by P. syringae pv. tomato DC3000 (avrRpt2) (Onate-Sanchez et al., 2007). Sasaki et al. (2007) reported that overexpression of ERF genes resulted in broad-spectrum resistance in transgenic plants. Dong et al. (2004) found that harpin activated ET signalling in Arabidopsis through EIN2 and EIN5, which conferred insect resistance and plant root growth in Arabidopsis. Nevertheless, two ET-responsive proteins, EIN2 and EIN5, were not differentially expressed in transgenic T-34 (data not shown). This discrepancy suggests that the EIN gene family may be not involved in the signalling pathway in transgenic T-34 in response to hpa1Xoo transformation.
The plant hormone auxin regulates diverse aspects of plant reactions, including plant growth and development, and HRs (Dharmasiri and Estelle, 2004; Badescu and Napier, 2006). Gopalan (2008) reported that PCD initiated by harpinEa was reversed by auxins without affecting the expression of marker genes related to local and systemic immunity. So, an interesting question is whether enhanced expression of auxin can be observed in genetically modified cotton. In our study, several auxin-related DEGs were up-regulated in transgenic T-34 which supported the hypothesis that the auxin-mediated signalling cascade played an important role in the hpa1Xoo-mediated response in transgenic T-34.
ABA, as an essential signal molecule modulating the plant response to abiotic and biotic stress, has divergent effects on defence responses (Asselbergh et al., 2004; Adie et al., 2007). The results of microarray analysis herein provided further evidence of the complexity of hormonally controlled signalling networks. For example, DEGs encoding products similar to myb transcription factors and GTPases responsive to ABA stimulus were up-regulated in transgenic T-34. These genes have been reported to be also responsive to other plant hormones such as SA and JA (Dong et al., 2005). Additionally, DEGs with similarities to GTP- and calucium-binding proteins related to the intercellular signalling cascade were also up-regulated in transgenic T-34. Although the mechanism for the up-regulation of these signalling-related products in transgenic T-34 is still not clear, our previous study showed that the transformation of hpa1Xoo conferred enhanced resistance in transgenic cotton through a priming mechanism (Miao et al., 2010). It is possible that the up-regulation of these signalling-related transcripts in hpa1Xoo-transformed cotton increased the sensitivity of the transgenic plant to pathogen infection. This hypothesis is supported by the observation that multiple DEGs with similarities to leucine-rich repeat (LRR) protein were up-regulated in transgenic T-34. It is well known that LRR proteins are not only essential for the activation of defence genes after recognition between the plant and the product of an avirulence gene from the pathogen but also play important roles in various actions after ligand recognition. Nevertheless, further study is required to better understand the role and interplay of multiple hormones and other signalling molecules involved in pathways regulated by hpa1Xoo in the transgenic plant.
It is interesting that several DEGs related to energy usage were also up-regulated in transgenic T-34. Although the effects of the up-regulation of DEGs in relation to the energy production and consumption in transgenic cotton is still not clear, our previous study (Miao et al., 2010) showed that the hpa1Xoo-transformed transgenic line T-34 shared similar phenotypic characteristics, such as leaf morphology and fibre quality. The height of the T-34 line was lower than wild-type Z35 before the flowering stage. Nevertheless, no significant difference was found between the height of T-34 and Z35 after the flowering stage. Further study will be required to investigate whether the gain of cotton disease resistance resulting from the transformation by hpa1Xoo will outweigh the possible decreased yield due to the change in the defence energy cost.
In summary, comprehensive information has been provided on transcriptome analysis of the hpa1Xoo-transformed transgenic cotton T-34. Transcripts related to the defence response, secondary metabolite biosynthesis, and various signalling pathways were differentially expressed in the transgenic plant in response to hpa1Xoo transformation. In addition, transcripts related to the energy producing pathways were also up-regulated in transgenic plants, which suggested that a high energy demand was imposed on transgenic cotton to support the cellular energy consumption resulting from the induction of multiple metabolic and host defence responses. This is somewhat similar to race non-specific resistance mediated by Lr34 as reported by Bolton et al. (2008).
Supplementary data
Supplementary data can be found at JXB online.
Supplementary Table S1. Expression ratio of 40 internal control genes in leaves and roots of transgenic T-34 cotton versus wild-type Z35 cotton
Supplementary Material
Acknowledgments
This research was supported by grants of the National Key Basic Research Plan of China (No. 2006CB101902, 2003CB114204), National Key Science Plan (No. 2004BA901A36), and the National High Technology Research and Development Program of China (863 Program) (No. 2006AA02Z180).
References
- Abramovitch RB, Janjusevic R, Stebbins CE, Martin GB. Type III effector AvrPtoB requires intrinsic E3 ubiquitin ligase activity to suppress plant cell death and immunity. Proceedings of the National Academy of Sciences, USA. 2006;103:2851–2856. doi: 10.1073/pnas.0507892103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adie BAT, Perez-Perez J, Perez-Perez MM, Godoy M, Sanchez-Serrano JJ, Schmelz EA, Solano R. ABA is an essential signal for plant resistance to pathogens affecting JA biosynthesis and the activation of defenses in Arabidopsis. Plant Cell. 2007;19:1665–1681. doi: 10.1105/tpc.106.048041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asselbergh B, Vleesschauwer DD, Höfte M. Global switches and fine-tuning—ABA modulates plant pathogen defense. Molecular Plant-Microbe Interactions. 2004;21:709–719. doi: 10.1094/MPMI-21-6-0709. [DOI] [PubMed] [Google Scholar]
- Badescu GO, Napier RM. Receptors for auxin: will it all end in TIRs? Trends in Plant Science. 2006;11:217–223. doi: 10.1016/j.tplants.2006.03.001. [DOI] [PubMed] [Google Scholar]
- Baron C, Zambryski PC. The plant response in pathogenesis, symbiosis, and wounding: variations on a common theme? Annual Review of Genetics. 1995;29:107–129. doi: 10.1146/annurev.ge.29.120195.000543. [DOI] [PubMed] [Google Scholar]
- Bi X, Khush GS, Bennett J. The rice nucellin gene ortholog OsAsp1 encodes an active aspartic protease without a plant-specific insert and is strongly expressed in early embryo. Plant and Cell Physiology. 2005;46:87–98. doi: 10.1093/pcp/pci002. [DOI] [PubMed] [Google Scholar]
- Bolton MD, Kolmer JA, Xu W, Garvin DF. Lr34-mediated leaf rust resistance in wheat: transcript profiling reveals a high energetic demand supported by transient recruitment of multiple metabolic pathways. Molecular Plant-Microbe Interactions. 2008;21:1515–1527. doi: 10.1094/MPMI-21-12-1515. [DOI] [PubMed] [Google Scholar]
- Brown RL, Kazan K, McGrath KC, Maclean DJ, Manners JM. A role for the GCC-box in jasmonate-mediated activation of the PDF1.2 gene of Arabidopsis. Plant Physiology. 2003;132:1020–1032. doi: 10.1104/pp.102.017814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chibucos MC, Collmer CW, Trudy TA, Michelle GG, Magdalen L, Li D, Tyler BM. Programmed cell death in host-symbiont associations, viewed through the gene ontology. BMC Microbiology. 2009;9(Suppl 1):S5. doi: 10.1186/1471-2180-9-S1-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke JD, Zhu T. Microarray analysis of the transcriptome as a stepping stone towards understanding biological systems: practical considerations and perspectives. The Plant Journal. 2006;45:630–650. doi: 10.1111/j.1365-313X.2006.02668.x. [DOI] [PubMed] [Google Scholar]
- Desikan R, Clarke A, Atherfold P, Hancock JT, Neill SJ. Harpin induces mitogen-activated protein kinase activity during defence responses in Arabidopsis thaliana suspension cultures. Planta. 1999;210:97–103. doi: 10.1007/s004250050658. [DOI] [PubMed] [Google Scholar]
- Devoto A, Muskettz PR, Shirasu K. Role of ubiquitination in the regulation of plant defence against pathogens. Current Opinion in Plant Biology. 2003;6:307–311. doi: 10.1016/s1369-5266(03)00060-8. [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Estelle M. Auxin signaling and regulated protein degradation. Trends in Plant Science. 2004;9:302–308. doi: 10.1016/j.tplants.2004.04.003. [DOI] [PubMed] [Google Scholar]
- Dievart A, Clark SE. LRR-containing receptors regulating plant development and defense. Development. 2004;131:251–261. doi: 10.1242/dev.00998. [DOI] [PubMed] [Google Scholar]
- Dong H, Delaney TP, Bauer DW, Beer SV. Harpin induces disease resistance in Arabidopsis through the systemic acquired resistance pathway mediated by salicylic acid and the NIM1 gene. The Plant Journal. 1999;20:207–215. doi: 10.1046/j.1365-313x.1999.00595.x. [DOI] [PubMed] [Google Scholar]
- Dong HP, Peng J, Bao Z, Meng X, Bonasera JM, Chen G, Beer SV, Dong H. Downstream divergence of the ethylene signaling pathway for harpin-stimulated Arabidopsis growth and insect defense. Plant Physiology. 2004;136:3628–3638. doi: 10.1104/pp.104.048900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong HP, Yu H, Bao Z, Guo X, Peng J, Yao Z, Chen G, Qu S, Dong H. The ABI2-dependent abscisic acid signalling controls HrpN-induced drought tolerance in Arabidopsis. Planta. 2005;221:313–327. doi: 10.1007/s00425-004-1444-x. [DOI] [PubMed] [Google Scholar]
- Dong X. SA, JA, ethylene, and disease resistance in plants. Current Opinion in Plant Biology. 1998;1:316–323. doi: 10.1016/1369-5266(88)80053-0. [DOI] [PubMed] [Google Scholar]
- El-Maarouf H, Barny MA, Rona JP, Bouteau F. Harpin, a hypersensitive response elicitor from Erwinia amylovora, regulates ion channel activities in Arabidopsis thaliana suspension cells. FEBS Letters. 2001;497:82–84. doi: 10.1016/s0014-5793(01)02441-3. [DOI] [PubMed] [Google Scholar]
- Ge X, Dietrich C, Matsuno M, Li G, Berg H, Xia Y. An Arabidopsis aspartic protease functions as an anti-cell-death component in reproduction and embryogenesis. EMBO Reports. 2005;6:282–288. doi: 10.1038/sj.embor.7400357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalan S. Reversal of an immunity associated plant cell death program by the growth regulator auxin. BMC Research Notes. 2008;1:126. doi: 10.1186/1756-0500-1-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He SY, Huang HC, Collmer A. Pseudomonas syringae pv. syringae HarpinPss: a protein that is secreted via the Hrp pathway and elicits the hypertensive response in plants. Cellular Microbiology. 1993;73:1255–1266. doi: 10.1016/0092-8674(93)90354-s. [DOI] [PubMed] [Google Scholar]
- Hoyos AE, Stanley CM, He SY, Pike S, Pu XA, Novacky A. The interaction of harpinPss with plant cell walls. Molecular Plant-Microbe Interactions. 1996;9:608–616. [Google Scholar]
- Joost O, Bianchini G, Bell AA, Benedict CR, Magill CW. Differential induction of 3-hydroxy-3-methylglutaryl CoA reductase in two cotton species following inoculation with Verticillium. Molecular Plant-Microbe Interactions. 1995;8:880–885. doi: 10.1094/mpmi-8-0880. [DOI] [PubMed] [Google Scholar]
- Kim JF, Beer SV. HrpW of Erwinia amylovora, a new harpin that contains a domain homologous to pectate lyases of a distinct class. Journal of Bacteriology. 1998;180:5203–5210. doi: 10.1128/jb.180.19.5203-5210.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Lee S, Park K, Jeong EJ, Ryu CM, Choi D, Pai HS. Comparative microarray analysis of programmed cell death induced by proteasome malfunction and hypersensitive response in plants. Biochemical and Biophysical Research Communications. 2006;342:514–521. doi: 10.1016/j.bbrc.2006.01.176. [DOI] [PubMed] [Google Scholar]
- Kim SY, Lim BH, Lim CJ, Nam CO, Hee K. Constitutive activation of stress-inducible genes in a brassinosteroid-insensitive 1 (bri1) mutant results in higher tolerance to cold. Physiologia Plantarum. 2010;138:191–204. doi: 10.1111/j.1399-3054.2009.01304.x. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lu YH, Wu KM, Jiang YY, Xia B, Li P, Feng HQ, Kris AG, Guo YY. Mirid bug outbreaks in multiple crops correlated with wide-scale adoption of Bt cotton in China. Science. 2010;328:1151–1154. doi: 10.1126/science.1187881. [DOI] [PubMed] [Google Scholar]
- Miao WG, Wang XB, Li M, Song CF, Wang Y, Hu DW, Wang JS. Genetic transformation of cotton with a harpin-encoding gene hpaXoo confers an enhanced defense response against different pathogens through a priming mechanism. BMC Plant Biology. 2010;10:67. doi: 10.1186/1471-2229-10-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyama M, Tada Y. Transcriptional and physiological study of the response of Burma mangrove (Bruguiera gymnorrhiza) to salt and osmotic stress. Plant Molecular Biology. 2008;68:119–129. doi: 10.1007/s11103-008-9356-y. [DOI] [PubMed] [Google Scholar]
- Oh CS, Beer SV. AtHIPM, an ortholog of the apple HrpN-interacting protein, is a negative regulator of plant growth and mediates the growth-enhancing effect of HrpN in Arabidopsis. Plant Physiology. 2007;145:426–436. doi: 10.1104/pp.107.103432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onate-Sanchez L, Anderson JP, Young J, Singh KB. AtERF14, a member of the ERF family of transcription factors, plays a nonredundant role in plant defense. Plant Physiology. 2007;143:400–409. doi: 10.1104/pp.106.086637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng JL, Bao ZL, Ren HY, Wang JS, Dong HS. Expression of harpin (Xoo) in transgenic tobacco induces pathogen defense in the absence of hypersensitive cell death. Phytopathology. 2004;94:1048–1055. doi: 10.1094/PHYTO.2004.94.10.1048. [DOI] [PubMed] [Google Scholar]
- Perino C, Gaudriault S, Vian B, Barny MA. Visualization of harpin secretion in planta during infection of apple seedlings by Erwinia amylovora. Cellular Microbiology. 1999;1:131–141. doi: 10.1046/j.1462-5822.1999.00013.x. [DOI] [PubMed] [Google Scholar]
- Phipps RH, Park JR. Environmental benefits of genetically modified crops: global and European perspectives on their ability to reduce pesticide use. Journal of Animal and Feed Sciences. 2002;11:1–18. [Google Scholar]
- Popham PL, Pike SM, Novacky A. The effect of harpin from Erwinia amylovora on the plasmalemma of suspension-cultured tobacco cells. Physiological and Molecular Plant Pathology. 1995;47:39–50. [Google Scholar]
- Sasaki K, Mitsuhara I, Seo S, Ito H, Matsui H, Ohashi Y. Two novel AP2/ERF domain proteins interact with cis-element VWRE for wound-induced expression of the tobacco tpoxN1 gene. The Plant Journal. 2007;50:1079–92. doi: 10.1111/j.1365-313X.2007.03111.x. [DOI] [PubMed] [Google Scholar]
- Sela-Buurlage MB, Ponstein AS, Bres-Vloemans SA, Melchers LS, Van Den Elzen PJM, Cornelissen BJC. Only specific tobacco (Nicotiana tabacum) chitinases and [beta]-1,3-glucanases exhibit antifungal activity. Plant Physiology. 1993;101:857–863. doi: 10.1104/pp.101.3.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewalt VJH, Ni W, Jung HG, Dixon RA. Lignin impact on fiber degradation: increased enzymatic digestibility of genetically engineered tobacco (Nicotiana tabacum) stems reduced in lignin content. Journal of Agricultural and Food Chemistry. 1997;45:1977–1983. [Google Scholar]
- Shao M, Wang JS, Dean RA, Lin YG, Gao XW, Hu SJ. Expression of a harpin-encoding gene in rice confers durable nonspecific resistance to Magnaporthe grisea. Plant Biotechnology Journal. 2008;6:73–81. doi: 10.1111/j.1467-7652.2007.00304.x. [DOI] [PubMed] [Google Scholar]
- Shi YH, Zhu SW, Mao XZ, Feng JX, Qin YM, Zhang L, Cheng J, Wei LP, Wang ZY, Zhu YX. Transcriptome profiling, molecular biological, and physiological studies reveal a major role for ethylene in cotton fiber cell elongation. Plant Cell. 2006;18:651–664. doi: 10.1105/tpc.105.040303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrawat AK, Carroll RT, DePauw M, Taylor GJ, Good AG. Genetic engineering of improved nitrogen use efficiency in rice by the tissue-specific expression of alanine aminotransferase. Plant Biotechnology Journal. 2008;6:722–32. doi: 10.1111/j.1467-7652.2008.00351.x. [DOI] [PubMed] [Google Scholar]
- Sohn SI, Kim YH, Kim BR, Lee SY, Lim CK, Hur JH, Lee JY. Transgenic tobacco expressing the hrpNEP gene from Erwinia pyrifoliae triggers defense responses against Botrytis cinerea. Molecular Cells. 2007;24:232–239. [PubMed] [Google Scholar]
- Somssich IE, Hahlbrock K. Pathogen defence in plants – a paradigm of biological complexity. Trends in Plant Science. 1998;3:86–90. [Google Scholar]
- Strobel NE, Ji C, Gopalan S, Kuc JA, He SY. Induction of systemic acquired resistance in cucumber by Pseudomonas syringae pv. syringae 61 HrpZPss protein. The Plant Journal. 1996;9:431–439. [Google Scholar]
- Su J, Hirji R, Zhang L, He C, Selvaraj G, Wu R. Evaluation of the stress-inducible production of choline oxidase in transgenic rice as a strategy for producing the stress-protectant glycine betaine. Journal of Experimental Botany. 2006;57:1129–1135. doi: 10.1093/jxb/erj133. [DOI] [PubMed] [Google Scholar]
- Van Hoewyk D, Garifullina GF, Ackley AR, Abdel-Ghany SE, Marcus MA, Fakra S, Ishiyama K, Inoue E, Pilon M, Takahashi H, et al. Overexpression of AtCpNifS enhances selenium tolerance and accumulation in Arabidopsis. Plant Physiology. 2005;139:1518–1528. doi: 10.1104/pp.105.068684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei ZM, Beer SV. HrpI of Erwinia amylovora functions in secretion of harpin and is a member of a new protein family. Journal of Bacteriology. 1993;175:7958–7967. doi: 10.1128/jb.175.24.7958-7967.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei ZM, Laby RJ, Zumoff CH, Bauer DW, He SY, Collmer A, Beer SV. Harpin, elicitor of the hypersensitive response produced by the plant pathogen Erwinia amylovora. Science. 1992;257:85–88. doi: 10.1126/science.1621099. [DOI] [PubMed] [Google Scholar]
- Wu KM, Lu YH, Feng HQ, Jiang YY, Zhao JZ. Suppression of cotton bollworm in multiple crops in China in areas with Bt toxin-containing cotton. Science. 2008;321:1676. doi: 10.1126/science.1160550. [DOI] [PubMed] [Google Scholar]
- Xie Z, Chen Z. Harpin-induced hypersensitive cell death is associated with altered mitochondrial functions in tobacco cells. Molecular Plant-Microbe Interactions. 2000;13:183–190. doi: 10.1094/MPMI.2000.13.2.183. [DOI] [PubMed] [Google Scholar]
- Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acid Research. 2002 doi: 10.1093/nar/30.4.e15. 30, e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CW, González-Lamothe R, Ewan RA, Rowland O, Yoshioka H, Shenton M, Ye H, O'Donnell E, Jones JDG, Sadanandom A. The E3 ubiquitin ligase activity of Arabidopsis PLANT U-BOX17 and its functional tobacco homolog ACRE276 are required for cell death and defense. The Plant Cell. 2006;18:1084–1098. doi: 10.1105/tpc.105.039198. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.