Abstract
In rice, the class I small heat shock protein (sHSP-CI) genes were found to be selectively induced by L-azetidine-2-carboxylic acid (AZC) on chromosome 3 but not chromosome 1. Here it is shown that a novel cis-responsive element contributed to the differential regulation. By serial deletion and computational analysis, a 9 bp putative AZC-responsive element (AZRE), GTCCTGGAC, located between nucleotides –186 and –178 relative to the transcription initiation site of Oshsp17.3 was revealed. Deletion of this putative AZRE from the promoter abolished its ability to be induced by AZC. Moreover, electrophoretic mobility shift assay (EMSA) revealed that the AZRE interacted specifically with nuclear proteins from AZC-treated rice seedlings. Two AZRE–protein complexes were detected by EMSA, one of which could be competed out by a canonical heat shock element (HSE). Deletion of the AZRE also affected the HS response. Furthermore, transient co-expression of the heat shock factor OsHsfA4b with the AZRE in the promoter of Oshsp17.3 was effective. The requirement for the putative AZRE for AZC and HS responses in transgenic Arabidopsis was also shown. Thus, AZRE represents an alternative form of heat HSE, and its interaction with canonical HSEs through heat shock factors may be required to respond to HS and AZC.
Keywords: Amino acid analogue, azetidine-2-carboxylic acid, cycloheximide, electrophoretic mobility shift assay, small heat shock protein
Introduction
Exposure to a sudden temperature upshift invokes a conserved physiological phenomenon known as the heat shock response (HSR) in all organisms. This response is characterized by elevated synthesis of heat shock proteins (HSPs) and repressed synthesis of most normal proteins and their mRNAs (Key et al., 1981; Lin et al., 1984). Some chemical agents such as alcohol (Kuo et al., 2000), amino acid analogues (Lee et al., 1996), and ozone (Banzet et al., 1998), as well as heavy metals such as arsenite (As) or cadmium (Cd) (Lin et al., 1984; Edelman et al., 1988), can induce a physiological stress response comparable with the HSR in cells. Among HSPs, small HSPs (sHSPs), ranging from ∼10 kDa to 30 kDa and characterized by a conserved α-crystallin domain at their C-terminus, are the most abundant and diverse group of HSPs in plants. These proteins are dispersed in the cytoplasmic–nuclear compartment (classes CI, CII, and CIII), endoplasmic reticulum (class ER), mitochondria (class M), and plastids (class P) (Scharf et al., 2001). How the sHSPs act as a chaperone for thermoprotection is not fully understood, but they have been shown to function as a reservoir to bind non-native (unfolding) protein intermediates, thus preventing their aggregation and maintaining them in a state competent for ATP-dependent refolding by other chaperones (Lee and Vierling, 2000).
In plants, sHSP genes are involved in the HSR; in the responses to chilling, osmotic, oxidative, salt, wounding, and chemical stresses; and in various developmental stages, such as zygotic embryogenesis (Sun et al., 2002). The expression of the HSP genes is mainly attributed to interaction of activated HS transcription factors (HSFs) and HS elements (HSEs) under heat stress. The eukaryotic HSE consensus sequence was defined as alternating units of 5′-nGAAn-3′, with efficient binding by HSFs requiring at least three contiguous and inverted units, which results in a perfect form of 5′-nGAAnnTTCnnGAAn-3′ (Scharf et al., 2001). However, in most cases, a variant type of imperfect HSEs usually occurs, as in the Arabidopsis sHSP gene family. In addition, plants contain multiple HSFs that evolved with functional diversification and/or genetic redundancy (Nover et al., 2001). Hence, plant HSP genes could be delicately regulated by a complex HSF network. A specific combination of an HSF and an imperfect HSE was shown to be essential for the differential induction of specific members of sHSP genes in sunflower and Arabidopsis (Carranco et al., 1999; Almoguera et al., 2002; Rojas et al., 2002; Diaz-Martin et al., 2005, Nishiwaza et al., 2006; Kotak et al., 2007). The interaction between an HSE and other transcriptional binding sites [activating protein 1 (AP-1), CCAAT/enhancer-binding protein element (C/EBP), and metal regulatory element (MRE)] specifies the expression of the Arabidopsis AtHsp90-1 gene (Haralampidis et al., 2002). In a previous study, it was shown that L-azetidine-2-carboxylic acid (AZC), Cd, and Cu were able to induce the expression of sHSP-CI genes on chromosome 3 but not chromosome 1 (Guan et al., 2004). Several putative HSEs are found in promoters of all sHSP-CI genes, which suggests that cis-response elements other than an HSE may be involved in the differential induction. The bidirectional promoter located between Oshsp17.3 and Oshsp18.0 on chromosome 3 contains only 355 bp, which provides a good opportunity to find the cis-response element(s) involved in the HS-like response induced by AZC.
In the current study, serial deletion analysis of the bidirectional promoter by a rice coleoptile transient expression system and electrophoretic mobility shift assay (EMSA) were used to reveal a cis-response element involved in selective activation by AZC.
Materials and methods
Plant materials
Rice (Oryza sativa L. cv. Tainung No.67) seedlings were germinated in rolls of moist paper towels at 28 °C in a dark growth chamber as described by Lin et al. (1984). Three-day-old rice seedlings without endosperms were incubated at 28 °C in shaking buffer [1% (w/v) sucrose and 5 mM potassium phosphate buffer, pH 6.0]. AZC, As, Cd, and cycloheximide (CHX; 2 μg ml−1) were added in the shaking buffer as indicated. Samples were harvested, flash-frozen in liquid nitrogen, and stored at –80 °C for subsequent RNA or protein extraction. Arabidopsis thaliana (Columbia ecotype) was grown at 22 °C in a 16 h light growth chamber.
RNA isolation and RT-PCR
RNA preparation and RT-PCR were described previously (Guan et al., 2004). Total RNA was extracted with use of TRIZOL reagent (Invitrogen, Rockville, MD, USA) according to the manufacturer's protocol. A 16 ng aliquot of DNase I-treated total RNA was used for RT-PCR analyses with use of the Superscript one-step RT-PCR kit (Invitrogen) according to the manufacturer's protocol. For all treatments, RT-PCR was repeated three times, with three batches of total RNA samples isolated independently. DNA from 10 μl of each PCR was fractionated by electrophoresis on a 2.0% (w/v) agarose gel with 0.01% (w/v) ethidium bromide in 0.5× TRIS-borate EDTA buffer. The gels were digitally photographed with use of a FloGel-1 fluorescent gel digital imaging system (TOPBIO, Taipei, Taiwan).
Preparation of DNA constructs
The reporter constructs were prepared in a vector derived from pGN100, a pUC19 vector bearing the GUS (β-glucuronidase) gene fused with the nopaline synthase (Nos) termination sequence. The AZC-inducible Oshsp17.3 (p567) and Oshsp18.0 [p567(–)] and non-AZC-inducible Oshsp16.9A (p631) promoter regions were separately amplified by PCR and cloned into the sites between EcoRI and BamHI. The reporter constructs contained the different transcriptional fusions between the 5′ untranslated regions (UTRs) of sHSP genes and GUS. During cloning, purification of DNA from solution or gel bands involved the GFX PCR DNA and Gel Band Purification Kit (Amersham Biosciences, Piscataway, NJ, USA). The templates and oligonucleotides used in creating the promoter constructs are listed in Table 1. To prepare the constructs for the truncated promoter analysis, two fragments (F268 and F289) flanking the truncated motif were amplified and then ligated together with an EcoRI- and BamHI-digested pGN100 vector. To generate the effector constructs, pHsfA4b and pHsfB4b, the coding regions were synthesized by PCR using the primer sets 5′-AAGGATCCATGGAGGGGGGCGGG-3′/5′-GGATCCTCAGGTTTTCTCCGTTG-3′ and 5′-GGATCCATGGCTTTCTTGGTGGAGA-3′/5′-GGATCCTCAAGAAATCAAGCAGCA-3′, respectively (the BamHI restriction site is underlined). The PCR products were gel purified and subcloned into the BamHI site of a vector containing the promoter and the first intron of the maize Ubi1 gene and the Nos termination sequence (Christensen and Quail, 1996). For promoter analysis in Arabidopsis, the full-length or AZC response element (AZRE)-truncated promoter of Oshsp17.3 was separately cloned into the sites between BamHI and EcoRI of the pCAMBIA1391 binary vector to obtain Oshsp17.3Pro::GUS or Oshsp17.3ProΔAZRE::GUS. All of the constructs were verified by sequencing analysis. The resulting Oshsp17.3Pro::GUS and Oshsp17.3ProΔAZRE::GUS constructs were transferred to Agrobacterium and used to transform Arabidopsis by the floral dip technique (Weigel and Glazebrook, 2002). All of the constructs were verified by sequencing analysis.
Table 1.
Oligonucleotides used in promoter constructs
| Construct/insert | Primer name | Sequence |
| p567 | BamHI-HTHPro-Fw | 5′-CGggatccGTCGGAATAGCTGCGAA-3′ |
| HTHPro-EcoRI-Rv | 5′-GgaattcTGTGTATTGTGTCTTGCTG-3′ | |
| p448 | BamHI-HTHPro-Fw | 5′-CGggatccGTCGGAATAGCTGCGAA-3′ |
| HTHPro-EcoRI-Rv-2 | 5′-GgaattcCTGTCGCTGCGTGGT-3′ | |
| p310, p310ΔAZRE | BamHI-HTHPro-Fw | 5′-CGggatccGTCGGAATAGCTGCGAA-3′ |
| HTHPro-EcoRI-Rv-3 | 5′-GgaattcAGGCCTGCTTATGGCC-3′ | |
| p270 | BamHI-HTHPro-Fw | 5′-CGggatccGTCGGAATAGCTGCGAA-3′ |
| HTHPro-EcoRI-Rv-5 | 5′-GgaattcGCTATGGGCTACAGC-3′ | |
| p290 | BamHI-HTHPro-Fw | 5′-CGggatccGTCGGAATAGCTGCGAA-3′ |
| HTHPro-EcoRI-Rv-6 | 5′-GgaattcTTATTCTGGAGAGTCCTGGACG-3′ | |
| F268 | BamHI-HTHPro-Fw | 5′-CGggatccGTCGGAATAGCTGCGAA-3′ |
| HTHPro-Rv-7 | 5′-CTATGGGCTACAGCCCAC-3′ | |
| F289 | HTHPro-Fw-8 | 5′-TCTCCAGAATAATCTAGGCC-3′ |
| HTHPro-EcoRI-Rv | 5′-GgaattcTGTGTATTGTGTCTTGCTG-3′ | |
| p567(–), p567(–)ΔAZRE | EcoRI-HTHPro-Fw-1 | 5′-CGgaattcGTCGGAATAGCTGCGAA-3′ |
| HTHPro-BamHI-Rv-1 | 5′-CGggatccTGTGTATTGTGTCTTGTG-3′ | |
| p265 | EcoRI-HTHPro-Fw-4 | 5′-CGgaattcGCAGGCCTAGAACAAT-3′ |
| HTHPro-BamHI-Rv-1 | 5′-CGggatccTGTGTATTGTGTCTTGTG-3′ |
The recognition site of the restriction enzyme is shown by lowercase letters.
Analysis of GUS activity in Arabidopsis
Four-week-old transgenic plants were treated as indicated and incubated in the fixation solution (0.3% formaldehyde, 0.1% Triton X-100, 0.1% β-mercaptoethanol, 100 mM sodium phosphate buffer, pH 7.0) for 15 min, vacuum-infiltrated for 15 min in GUS staining buffer (1 mM X-Gluc, 0.5 mM ferricyanide, 0.5 mM ferrocyanide, 0.1% Triton X-100, 10 mM EDTA, 100 mM sodium phosphate buffer, pH 7.0), and then incubated at 37 °C for 24 h. After staining, the reaction was stopped, and chlorophyll was cleared from plants with several washes with 70% ethanol (v/v). Plants were photographed to record deposition of the transgene product.
Particle bombardment and transient expression assay
Particle bombardment and transient expression assay were conducted as described (Guan et al., 2004). Briefly, the mixture (in a 1:1 molar ratio) of a test DNA construct and a maize ubiquitin–luciferase internal control construct were coated onto gold particles (1.0 μm particle size, 30 mg) (Bio-Rad, Hercules, CA, USA). The coleoptile of a rice seedling was cut from the embryonic root and positioned in the middle of a 10 cm Petri dish containing Murashige and Skoog (MS) salts supplemented with 0.6% (w/v) agar and 3% (w/v) sucrose. A helium biolistic particle delivery system (model PDS-1000, Bio-Rad) was used for particle bombardment. After bombardment, the Petri dishes were incubated at 28 °C in the dark for at least 6 h and then subjected to treatments as indicated. The luciferase activity was assayed by use of the Bright-Glo™ luciferase assay system (Promega, Madison, WI, USA) according to the technical manual. For the GUS activity assay, the fluorescence was measured in a Fluoroskan Ascent FL fluorometer (Labsystems, Helsinki, Finland). Normalized GUS activity was calculated by dividing GUS activity by luciferase activity for each sample. For each independent experiment, at least four samples placed on MS plates were bombarded. After bombardment, the samples from each plate were separated into two batches, one used as a control and the other treated with AZC or HS. Thus, for each construct assay of one particular condition, the total sample size was 12 (3 experiments×4 plates=12). Data analysis involved analysis of variance (ANOVA) with Microsoft Excel software. A P-value <0.05 was considered statistically significant.
Preparation of nuclear extracts from rice seedlings
The nuclear isolation procedure was modified from the method developed for wheat embryos (Luthe and Quatrano, 1980). All procedures were performed in a 4 °C cold room. Rice seedlings (5–10 g) were ground in liquid nitrogen to a fine powder with use of a mortar and pestle. The powder was transferred into 35 ml of Honda buffer [0.44 M sucrose, 2.5% (w/v) Ficoll, 5.0% (w/v) dextran 40, 25 mM TRIS-HCl (pH 7.5), 10 mM MgCl2, 10 mM β-mercaptoethanol, 0.5% (v/v) Triton X-100] with 2 mM spermine. The homogenate was filtered through four layers of gauze and two layers of miracloth (Calbiochem-Novabiochem, La Jolla, CA, USA). The filtrate was centrifuged at 5850 g (Sorvall RC-5C, SS-34 rotor) for 5 min and the supernatant was discarded. The pellet was suspended in 10 ml of Honda buffer and centrifuged again, then washed twice in 10 ml of nuclear washing buffer (25 mM TRIS-HCl, 10 mM MgCl2, 10 mM β-mercaptoethanol, 20% glycerol). The crude nuclear pellet was gently suspended in a small amount of freshly prepared ice-cold nuclear resuspension buffer [10 mM HEPES (pH 8.0), 50 mM NaCl, 0.5 M sucrose, 0.1 mM EDTA (pH 8.0), 5 mM MgCl2, 1 mM dithiothreitol (DTT), 0.5% Triton X-100]. For isolation of nuclear extract, the crude nuclei were perforated in a solution containing 5 mM spermidine and 0.5 M NaCl. After perforation in ice for 30–45 min, the lysate was centrifuged at maximal speed for 10 min in a 4 °C microcentrifuge. The supernatant was dialysed for 5 h in 200 ml of dialysis buffer [10 mM HEPES (pH 8.0), 50 mM NaCl, 1 mM MgCl2, 1 mM DTT, 50% glycerol, 0.8 mM phenylmethylsulphonyl fluoride (PMSF)]. After dialysis, the supernatant was centrifuged again and then transferred to a new microcentrifuge tube. The concentration of nuclear extract was quantified according to the Bradford method with Dye Reagent concentrate (Bio-rad). An aliquot of the extract was snap-frozen by use of liquid nitrogen and kept in a –80 °C refrigerator.
Electrophoretic mobility shift assay (EMSA)
The synthetic oligonucleotides with sequences from positions –192 to –170 of the Oshsp17.3 promoter relative to the transcription start site (AZRE-sense: 5′-CCATAGCGTCCAGGACTCTCCAG-3′ and AZRE-antisense: 5′-CTGGAGAGTCCTGGACGCTATGG-3′) were heated at 100 °C for 5 min and then allowed to anneal to double-stranded oligonucleotides for 12 h at room temperature. The annealed oligonucleotides were isotopically labelled by use of T4 polynucleotide kinase (New England Biolabs, Beverly, MA, USA) together with [γ-32P]ATP (10 mCi ml−1) (NEN, Boston, MA, USA). Binding reactions were carried out in a total volume of 20 μl of a solution that contained 25 mM HEPES-KOH (pH 7.5), 100 mM KCI, 0.1 mM EDTA, 17% glycerol, 1 mM DTT, 4 μg of poly(dI–dC)/poly(dI–dC), 10 μg of protein in a crude nuclear extract, labelled probe (∼106 cpm, 1 pmol), and competitor DNA (0, 0.5, 1, 10, 100, or 200 pmol). The assay mixtures were incubated for 20 min at room temperature. The reaction mixtures were layered on 4% acrylamide gels containing 0.5× TBE buffer and 3.6% glycerol. After a pre-run in the 0.5× TBE buffer for 1 h at 100 V at room temperature, the sample was electrophoresed for another 1 h under the same conditions. The gel was blotted on Whatman 3 MM paper, dried, subjected to a storage phosphor screen for 12 h at room temperature, and imaged by use of a Typhoon 9400 scanner (Amersham Biosciences). In competition experiments, the reaction conditions were the same as for the standard binding reaction, and specific competitor oligonucleotides were added. For the specific competitor oligonucleotides, non-isotopically labelled double-stranded oligonucleotides were used. In cross-competition experiments, the synthetic double-stranded oligonucleotides with a perfect HSE motif were used as the non-specific competitors. The sequences of oligomers, designated HSE-sense and HSA-antisense, were 5′-AATAAGCTTCTAGAAACTTCCAG-3′ and 5′-CTGGAAGTTTCTAGAAGCTTATT-3′, respectively. The sequence locates at positions –87 to –65 of the Oshsp17.3 promoter relative to the transcription start site.
Bioinformatics
The sHSP-CI gene promoter sequences were searched for motifs identical or similar to the previously reported cis-element motifs in the PLACE database by ‘Signal Scan Search’ or by ‘Homology Search’ (http://www.dna.affrc.go.jp/PLACE/) and in the PlantCARE database by ‘Search for CARE’ (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Lescot et al., 2002). To find cis-acting regulatory elements from the co-regulated sHSP-CI gene promoters that respond to chemical inducers, the Motif Sampler program was used (Thijs et al., 2001). The ‘Plants (EPD)’ was chosen as a background model, and the other parameters were the default.
Results
The promoter of Oshsp17.3 contains nine putative HSEs but no other known stress-related elements
It was previously shown that a bidirectional promoter controlled the expression of both Oshsp17.3 (corresponding to Os03g1602000 in Nipponbare) and Oshsp18.0 (Os03g1603000) by treatment with AZC or other chemicals (Guan et al., 2004). The nucleotide sequence of the intergenic region of Oshsp17.3 and Oshsp18.0 is shown in Fig. 1A. In addition to an HSE, only a putative C/EBP was related to the HSR (Rieping and Schöffl, 1992). Nine putative HSEs were found with the canonical HS consensus of at least three core units, with the repeating pentanucleotide sequence 5′-nGAAn-3′ arranged in alternate orientation. Three putative perfect core units of the HSE (gGAAgtTTCtaGAAg) were found 39 nt upstream of the putative TATA box of Oshp17.3; the other imperfect HSEs consisted of only one perfect core unit of the pentanucleotide and two imperfect units in the distal one, gTTCgtGAtggTcCa. A putative perfect CCAAT box is also present at position –240 relative to the transcription start site of Oshsp17.3. The CCAAT box sequences were shown to act cooperatively with HSEs to increase promoter activity (Rieping and Schöffl, 1992; Haralampidis et al., 2002). In addition, three putative CAAT box sequences are present at positions –222, –330, and –406. Although the intergenic region between Oshsp17.3 and Oshp18.0 could also respond to heavy metal treatments (Guan et al., 2004), the stress-response elements AP-1 or MRE were not found.
Fig. 1.
Localization of the L-azetidine-2-carboxylic acid response element (AZRE) by 5′ deletion analysis of the Oshsp17.3 promoter. (A) Nucleotide sequence of the intergenic region between Oshsp17.3 and Oshsp18.0. The transcription start site is in bold. The direction of transcription is shown by an arrow with a vertical line. The putative TATA box is indicated by a thin solid underline. The putative heat shock elements (HSEs) are underlined in bold with dots to designate matches to the core consensus (GAA/TTC) of the canonical HSE. The putative CAAT/CCAAT box is indicated by bold italic letters. The downward-pointing arrowhead shows the position used to generate the promoter–GUS-deleted construct in the Oshsp17.3 direction, and the upward-pointing arrowhead shows the same construct in the Oshsp18.0 direction. The putative AZRE is indicated by a dashed line. (B) Schematic representation of the 5′ deletion and internal deletion derivatives of the Oshsp17.3 promoter fused to the GUS reporter gene shown on the left. The number indicates the distance from the transcription start site of Oshsp17.3. The TATA box is shown as a vertical black rectangle. The putative perfect HSE is represented by a grey ellipse, and the putative imperfect HSE is represented by a hollow ellipse. The putative 9 bp AZRE is indicated by a hatched box. The thin black angled lines in the p567ΔAZRE and p310ΔAZRE constructs represent the truncation of the 9 bp AZRE in the Oshsp17.3 promoter. The bombarded coleoptiles were incubated at 28 °C in the dark for at least 6 h and then incubated with 5 mM AZC in phosphate buffer for 4 h. Then, the samples were incubated in fresh phosphate buffer at 28 °C in the dark for at least 12 h. The relative GUS activity was determined as shown in the right-hand panel. The fold induction relative to the control (28 °C) is indicated to the right of the scale bar. Each experiment was repeated at least three times. Relative GUS activites with ±SE are from at least 12 independent bombardments. Constructs with <2-fold induction by AZC and their P-values are shown at the right of the scale bar. All P-values are >0.05, which suggests no significant difference in induction.
A 51 bp fragment in the promoter of Oshsp17.3 is important for the AZC response
To define promoter regions that confer the AZC-responsive expression of Oshsp17.3, a series of constructs was designed with 5' deletions of the Oshsp17.3 promoter fused to the GUS reporter gene for transient expression assay (Fig. 1B). These constructs were introduced by particle bombardment into 3- to 4-d-old rice coleoptiles and then treated with AZC. By comparing the relative GUS activity of the control and AZC-treated coleoptiles, it was found that the full-length promoter of Oshsp17.3 showed a 15.1-fold induction after AZC treatment, which is similar to a previous result (Guan et al., 2004). To identifiy a more specific region, three constructs (p448, p310, and p270) were designed, with a deletion in each from the 5′ end of the Oshsp17.3 promoter. When the promoter was deleted to position –355 (p448), the induction by AZC was 11.7-fold that of the control, which was 23% less induction than that of the full-length promoter. Oshsp17.3 and Oshsp18.0 are arranged in a head-to-head configuration (Fig. 1A), so the deleted region belongs to the 5′ UTR of Oshsp18.0 and contains one CAAT box. Deletion of the promoter to position –217 (p310) decreased the induction substantially to 5.2-fold (∼34% of that of the full-length construct). This deleted region contains five putative HSEs and three putative CAAT/CCAAT boxes. Further deletion of the promoter to position –167 (p270), which contains two putative HSEs (Fig. 1A), abolished the induction by AZC. Serial deletion analyses revealed that the 51 bp fragment between nucleotides –217 and –167 of the Oshsp17.3 promoter is the functional region for AZC response.
A putative 9 bp AZRE was found in the AZC-inducible sHSP-CI gene promoters
A previous study showed that the sHSP-CI genes Oshsp17.3, Oshsp17.7, Oshsp17.9A, and Oshsp18.0 on chromosome 3 were induced by AZC (Guan et al., 2004). To search for potential common motifs in the promoter regions of these co-regulated genes, ∼600 bp promoter regions from these sHSP-CI genes were analysed by use of the MotifSampler program (Thijs et al., 2001). In addition to HSEs, a partially conserved motif was found in all four AZC-inducible putative promoter regions (Fig. 2). In contrast, no similar motif was found in the non-AZC-inducible promoters of sHSP-CI genes on chromosome 1. Of interest, this motif is located between nucleotides –350 and –250 upstream of the translation start sites in all four AZC-inducible promoters. As expected, this motif is located inside the 51 bp functional region of the Oshsp17.3 promoter described above, which suggests that this motif is the putative AZRE. In the bidirectional promoter of Oshsp17.3 and Oshsp18.0, the putative 9 bp AZREs overlapping each other in opposite directions (shown in Fig. 1A) are located between nucleotides –185 and –177 (5′-GTCCTGGAC-3′) relative to the transcription start site of Oshsp17.3 (Fig. 1A). Of interest, this 9 bp putative AZRE contains contiguous inverted repeats of GTCC separated by the nucleotide ‘T’ and forms an odd palindromic configuration. The consensus nucleotide sequence of the putative AZREs is GTyCwGGAm (where y is C or T, w is A or T, and m is A or C) (Fig. 2). This putative AZRE in the Oshsp17.9A or Oshsp17.7 promoters overlapped with a putative HSE (underlined in Fig. 2). However, the conserved ‘G’ base at positions 1 and 6 in the AZRE is not the same as a typical HSE (nTTCnnGAAnnTTCn). None of the known elements found in the PLACE database is identical to this putative AZRE; thus, it is considered to be a new cis-response element in plants.
Fig. 2.
Position and nucleotide sequence alignment of the four putative AZREs in AZC-inducible sHSP-CI gene promoters. The numbers indicate the number of nucleotides relative to the translation start codon (ATG). The putative AZREs are in bold uppercase letters. The putative HSEs in the promoters of Oshsp17.9A and Oshsp17.7 are underlined.
The putative 9 bp AZRE is essential for responsiveness to AZC
To test whether the putative 9 bp AZRE is necessary for the AZC response, it was deleted from p567 and the construct was designated p567ΔAZRE. As expected, the p567ΔAZRE construct did not respond to AZC treatment (Fig. 1B). Deletion of the Oshsp18.0 promoter to position –146 (p265) or deletion of the 9 bp AZRE from the Oshsp18.0 promoter [p567(–)ΔAZRE] also lost AZC inducibility (Fig. 1B). p265 contains several HSEs but without the AZRE in the Oshsp18.0 transcription direction. These deletion analyses provide strong evidence that the putative 9 bp AZRE is essential for the AZC responsiveness of Oshsp17.3 and Oshsp18.0.
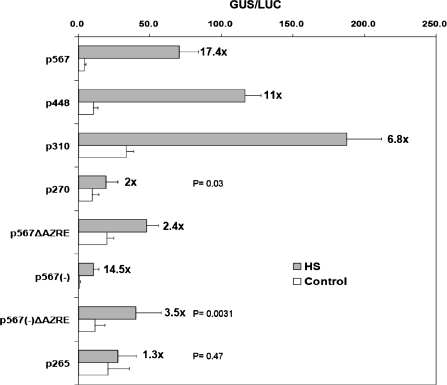
The putative 9 bp AZRE is indispensable for HS responsiveness
Because Oshsp17.3 is induced by HS and by chemicals such as As, it was decided to investigate whether the putative 9 bp AZRE is involved in these responses. The full-length promoter (p567) and its deletant (p310) separately showed 17.4-fold and 6.8-fold induction after HS treatment (Fig. 3). Interestingly, the amount of induction was diminished to 2.4-fold in the construct without the AZRE (p567ΔAZRE), which was ∼ 86% less induction than that of p567. The same reduction effect was shown in the p567(–)ΔAZRE or p265 construct (75% and 91% reduction, respectively) (Fig. 3). Thus, the putative AZRE is also necessary for the HS responsiveness of Oshsp17.3 and Oshsp18.0. In addition, it was found that the p567ΔAZRE construct lost its response to As treatment, whereas the p567 construct showed 5.3-fold induction (Supplementary Fig. S1 available at JXB online).
Fig. 3.
Effect of the 9 bp AZRE deletion on response to HS. The bombarded coleoptiles were incubated at 28 °C in the dark for at least 6 h and then at 41 °C for 2 h. Then, the samples were incubated in fresh incubation buffer at 28 °C in the dark for at least 12 h. The fold induction relative to the control (28 °C) is indicated next to the bars. Each experiment was repeated at least three times. Mean ±SE GUS activites are from at least 12 independent bombardments. The P-values for p270, p567(–)ΔAZRE, and p265 are shown. p270 and p567(–)ΔAZRE show a significant difference from the control with HS treatment (P ≤0.05), and p265 shows no significant difference (P=0.47).
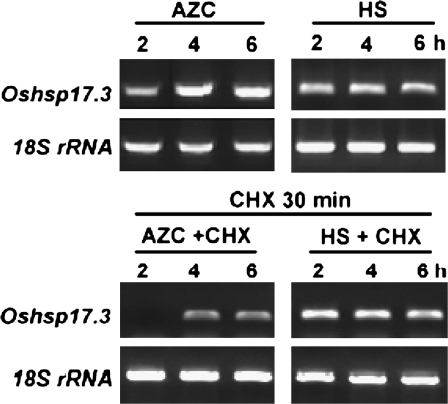
Induction of Oshsp17.3 transcripts by AZC is delayed by inhibition of protein synthesis
In soybean, the induction of a subset of sHSP genes by AZC at 28 °C was delayed in the presence of the protein synthesis inhibitor CHX (Lee et al., 1996). In the present study, CHX (2 μg ml−1) was added to the incubation medium 30 min before AZC or HS treatment. The induction of Oshsp17.3 was detected with a delay of 2 h with AZC in the presence of CHX relative to AZC alone (Fig. 4). In contrast, CHX had no significant effect on the induction of Oshsp17.3 transcripts with HS treatment, as was shown previously in soybean. CHX treatment alone did not affect the induction of Oshsp17.3 transcripts at 28 °C (data not shown). The results suggest that the induction mechanisms of the AZC response and the HSR are distinct but may share some components in pathways.
Fig. 4.
Effect of a protein synthesis inhibitor on accumulation of Oshsp17.3 transcript induced by AZC and HS. Before 5 mM AZC or HS (41°C) treatment, 3-d-old rice seedlings were treated or not with cycloheximide (CHX) (2 μg ml−1) for 30 min at 28 C. A 16 ng aliquot of DNase I-treated total RNA was used for RT-PCR. The RT-PCR products of Oshsp17.3 are shown by ethidium bromide staining, and the RT-PCR product of the 18S rRNA was used as an internal PCR control.
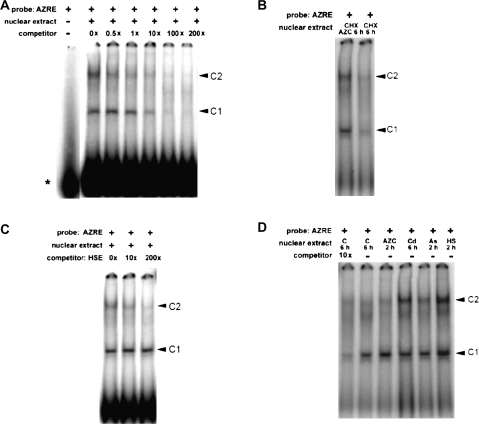
Specificity of nuclear proteins interacting with the 9 bp AZRE in vitro by EMSA
To characterize the putative AZRE further, EMSA was used to determine whether it can interact with trans-acting factor(s) present in nuclear extracts. A 23 bp synthetic oligonucleotide encompassing the AZRE was used as a probe and was labelled with 32P by T4 polynucleotide kinase. Then, the probe was incubated with nuclear extracts from AZC-treated rice seedlings. Two DNA-bound protein bands (C1 and C2) were detected on the gel (Fig. 5A). The intensity of C1 appeared to be stronger than that of C2, and C1 could be further competed out by a 200-fold excess of AZRE probe without a 32P label. In addition, nuclear extracts from seedlings treated with AZC plus CHX for 6 h or CHX alone did not affect the levels of C1 and C2 interacting with the AZRE (Fig. 5B), which suggests that de novo synthesis of proteins is not required for the interaction. Because the 9 bp AZRE is involved in the HSR, it was also tested whether the 9 bp AZRE could be cross-competed by an HSE. This 23 bp HSE probe encompasses a perfect HSE (CTTCTAGAAACTTCC). Interestingly, the C2 but not the C1 band could be competed out by a 200-fold excess of HSE without being 32P labelled (Fig. 5C). Nevertheless, these results show that the AZRE can be specifically recognized by the nuclear proteins from AZC-treated rice seedlings.
Fig. 5.
Electrophoretic mobility shift assay results for the AZRE. (A) Sequence-specific interaction of nuclear proteins with the AZRE. The 23 bp AZRE containing a fragment between nucleotides –190 and –168 relative to the transcription start site of Oshsp17.3 was radiolabelled, and 1 ng of the probe was incubated with 10 μg of the nuclear proteins prepared from rice etiolated seedlings under AZC treatment for 6 h. The positions of the DNA–protein complexes are shown as C1 and C2 by arrows. The free probe is indicated with an asterisk (*). (B) Effect of CHX on the interaction of nuclear proteins with the AZRE. Three-day-old rice seedlings were treated with CHX (2 μg ml−1) and with (left) or without (right) AZC for 6 h. (C) Cross-competition analysis of the HSE. The 23 bp HSE-containing fragment located at –87 to –65 relative to the transcription start site of Oshsp17.3 was used as a competitor. (D) Stress-dependent interaction of nuclear proteins with the AZRE. Nuclear extracts were prepared from seedlings treated with AZC, As, or HS for 2 h and by 28 °C [control (C)] or Cd for 6 h.
Because the AZRE was required for the response to HS and AZC, EMSA was used to examine whether nuclear extracts from rice seedlings treated with different inducers can bind to the AZRE. In nuclear extracts from seedlings treated with As (2 h), Cd (6 h), HS (2 h), or AZC (2 h), the two DNA–protein complexes C1 and C2 were also detected in all extracts, including control extracts (Fig. 5D). Additionally, the C1 band in the control was competed out by a 10-fold amount of the AZRE (Fig. 5D, first left lane). The intensity of the C1 complex with AZC treatment was found to be ∼5-fold higher than that of the control by measuring the integrated density with Scion image software. However, the intensity of the C2 band in 2 h AZC-treated and control nuclear extracts was lower than that in extracts receiving 6 h AZC, As, Cd, and HS treatments. Nevertheless, the results demonstrate that nuclear proteins from rice seedlings bind to the AZRE in a stress-dependent manner and depend on the duration of AZC treatment.
Deletion of the AZRE on the promoter of Oshsp17.3 can reduce OsHsfA4b-modulated activation
According to the indispensable role of the AZRE for HS responsiveness (Fig. 3) and the results of EMSA (Fig. 5C), it was speculated that the AZRE can be specifically recognized by HSFs. Thus, tests were conducted to determine whether the AZRE was necessary for the interaction between OsHsfs and the promoter of Oshsp17.3. According to the expression analysis of 23 rice HSF genes, OsHsfA4b (Os01g54550) and OsHsfB4b (Os07g44690) were found to be induced by AZC treatment (data not shown). Two effector constructs, pHsfA4b and pHsfB4b, under the control of the maize constitutive Ubi1 promoter, were made (Fig. 6A). GUS activity in coleoptiles co-transformed with one reporter (p567 or p567ΔAZRE) and one effector (pHsfA4b or pHsfB4b) construct at a ratio of 1:1 was compared. As shown in Fig. 6B, when co-transformed with pHsfA4b, p567 achieved 2.8-fold induction in relative GUS activity but p567ΔAZRE achieved only a 1.8-fold induction. However, co-transformation of pHsfB4b was not significantly induced on p567 or p567ΔAZRE. To confirm these results further, the p631 reporter construct was also co-bombarded with pHsfA4b or pHsfB4b and no effect on the non-AZC-inducible Oshsp16.9A promoter was found (1.3- and 1.5-fold induction, respectively) (Fig. 6B). Also, transient expression of another highly AZC-inducible HSF gene, OsHsfC1b (Os01g53220), had no effect on the activation of Oshsp17.3 and Oshsp16.9A (data not shown). Thus, HSFs are possibly an integral part of the AZRE-dependent induction pathway of sHSP-CIs.
Fig. 6.
Effect of the 9 bp AZRE on activation of HSFs on the Oshsp17.3 promoter. (A) Schemes of gene constructs. Arrowheads indicate the orientation of each gene. (B) The heat-inducible reporter construct was co-bombarded with the Ubi-Empty vector (–) or the effector construct (+) at a 1:1 ratio. The bombarded coleoptiles were incubated in fresh shaking buffer at 28 °C in the dark for at least 12 h, and then their relative GUS activity was determined. Each experiment was repeated at least three times. Mean ±SE relative GUS activites are from at least 12 independent bombardments. The P-values of p567+pHsfA4b and p567+pHsfB4b are shown on the right. (This figure is available in colour at JXB online.)
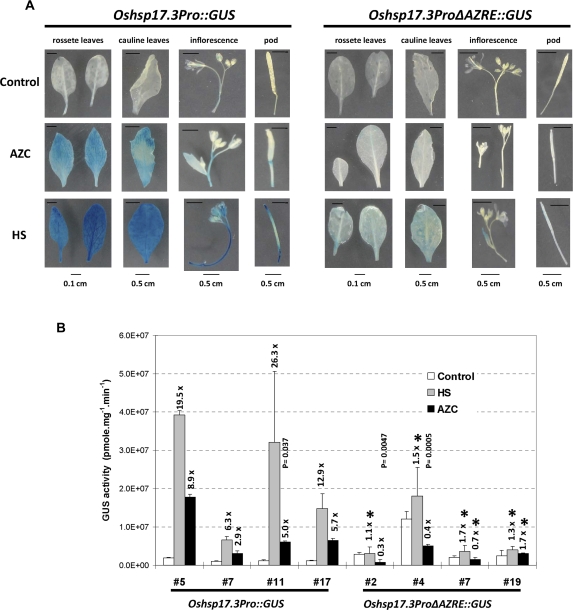
The putative 9 bp AZRE is essential for AZC and HS responsiveness of the Oshsp17.3 promoter in transgenic Arabidopsis
To gain better insight into the function of this new cis-response element, Oshsp17.3Pro::GUS and Oshsp17.3ProΔAZRE::GUS with or without the 9 bp putative AZRE were constructed and introduced into Arabidopsis by Agrobacterium-mediated transformation. T1 seedlings from independent T0 transformants were analysed by Southern blot analysis (data not shown), and seedlings containing one or two copies of the T-DNA insertion were collected for generation of T2. Four independent 20-d-old T2 transgenic lines were separately treated with AZC or HS. Compared with untreated seedlings, which showed weak GUS staining, Oshsp17.3Pro::GUS transgenic plants showed GUS expression induced by AZC, whereas no significant GUS signal was detected in Oshsp17.3ProΔAZRE::GUS transgenic plants treated with AZC (Fig. 7A). Oshsp17.3Pro::GUS and Oshsp17.3ProΔAZRE::GUS transgenic plants showed weak GUS expression with HS treatment (Fig. 7A). The reduction in AZC and HS responsiveness was further confirmed quantitatively by measuring GUS activity. As shown in Fig. 7B, the full-length promoter of Oshsp17.3 (Oshsp17.3Pro::GUS) conferred 2.9- to 8.9-fold induction with AZC treatment and 6.3- to 26.3-fold induction with HS treatment in Arabidopsis seedlings. In contrast, deletion of the AZRE (Oshsp17.3ProΔAZRE::GUS) had no significant effect on the GUS activity induced by AZC or HS (Fig. 7B). As shown by the P-values, most Oshsp17.3ProΔAZRE::GUS transgenic plants have values >0.17 under HS and AZC treatment. The lower GUS activity in trasgenic plants #2 and #4 produced by AZC treatment (P=0.0047 and 0.0005, respectively) may have resulted from the effect of prolines replaced by AZCs, which interfere with the GUS enzyme activity. Nevertheless, these stable transgenic results provide strong evidence that the putative 9 bp AZRE is also indispensible for the AZC response and the HSR in Arabidopsis.
Fig. 7.
Effect of AZRE truncation in transgenic Arabidopsis. A GUS reporter gene was fused to the full-length Oshsp17.3 promoter (Oshsp17.3Pro::GUS) and the AZRE-truncated Oshsp17.3 promoter (Oshsp17.3ProΔAZRE::GUS). (A) Seedlings from independent transgenic lines (four lines for each contruct) underwent AZC treatment for 4 h at 23 °C or HS treatment (39 °C) for 150 min and then 18 h of no treatment at 23 °C. Untreated seedlings were used as the control (23 °C). (B) The relative GUS activity of seedlings. The magnitude of induction relative to the control is indicated at the top of the bars. Mean GUS activites ±SE are from at least three independent experiments. For Oshsp17.3ProΔAZRE::GUS, the asterisks indicate no significant difference from control in induction with HS or AZC treatment (P ≥0.7). The P-values for transgenic plants #11 of Oshsp17.3Pro::GUS and #2 and #4 of Oshsp17.3ProΔAZRE::GUS are shown at the top of the scale bar. (This figure is available in colour at JXB online.)
Discussion
Because AZC and HS have a common property in causing the accumulation of abnormal proteins in the cytosol, the AZC response can reflect the ability of the cytosol to sense proteome stability in the presence of misfolded proteins (Sugio et al., 2009). AZC can act as a competent inducing signal in the HS-like response and selectively activate a subset of HSP genes (Trotter et al., 2002; Guan et al., 2004); however, the mechanism remains unclear. This work provides evidence of a novel cis-element required for the selective expression of a subset of sHSPs.
The interaction between trans-regulatory proteins and a specific cis-acting element is the initial step in the regulation of gene expression. Under normal growth conditions and various stress conditions, the binding of HSFs to the HSE of target genes is necessary for activation of transcription (Nover et al., 2001; Rojas et al., 2002). In previous work it was shown that the bidirectional promoter of Oshsp17.3 and Oshsp18.0 is AZC inducible and the Oshsp16.9A promoter is not AZC inducible, but both promoters are HS inducible (Guan et al., 2004). Several canonical HSEs were found in these promoters, which suggests that canonical HSEs may not be involved in the differential induction of the bidirectional promoter by AZC. Evidence shows that interactions between HSEs and other stress response elements contribute to differential induction of AtHsp90-1 by heavy metals (Haralampidis et al., 2002). Analysis of the bidirectional promoter sequence revealed the presence of several HSEs but no other stress response cis-element such as MRE. Therefore, other unknown cis-acting element(s) may be necessary for the specific responsiveness to AZC (Fig. 1A). Promoter deletion analyses first indicated that a 51 bp fragment (from –217 to –167) contains a putative cis-acting element for AZC-specific induction (Fig. 1B). Computational analysis revealed a common motif present on all four AZC-inducible sHSP-CI promoters on chromosome 3 but not on the non-AZC-inducible sHSP-CI promoters on chromosome 1 in this study. This agrees with a previous expression anaylsis of sHSP-CI in rice etiolated seedlings (Guan et al., 2004). Deletion of this motif confirmed the existence of an AZRE. Additionally, site-directed mutagenesis and gain-of-function analysis of the putative AZRE support its role in the AZC response (Supplementary Figs S2, S3 at JXB online). In the transient expression assays, the relative GUS activity of some deletion constructs such as p448 and p310 was increased under both control and AZC or HS treatment (Figs 1B, 3). This made the results not so convincing, even though the fold induction was drastically decreased. These experiments were repeated several times; however, very similar results were obtained. Thus, it is thought that the decreased fold induction may have resulted from the promoter per se. Since p567 contains the 5′ UTR of Oshsp18.0 and Oshsp17.3, the increase in relative GUS activities of p448 and p310 could have resulted from the basic transcription apparatus contributed mostly by the expression of Oshsp17.3, rather than by both genes. Because of the fluctuating and unstable properties of the transient assay system, stable transformation was used to confirm further the existence of the putative AZRE. GUS reporter assays in transgenic Arabidopsis provided further evidence to support the conclusions from transient assays (Fig. 7). Furthemore, they also revealed that the AZC response may function by a similar mechanism via the AZRE in both monocot rice and dicot Arabidopsis.
The differential transcription of sHSP genes may have resulted from the action of different kinds of HSFs or additional transcription factors responding to environmental stresses (Rojas et al., 2002). Two DNA–protein complexes (C1 and C2) were observed by EMSA. Both of the interaction complexes can be competed out by cold AZRE, which indicates that the AZRE–protein interactions are sequence specific and may involve two distinct trans-acting proteins (Fig. 5A). Also, nuclear extracts from untreated control rice seedlings could interact specifically with the AZRE (Fig. 5D), and the level of bound proteins was not affected by CHX (Fig. 5B). These data show that de novo protein synthesis was not required for the trans-acting factors recognizing the AZRE, and thus these factors constitutively exist in the cells. However, nuclear extracts prepared from seedlings treated for 6 h with AZC, Cd, As, or HS showed a higher intensity of C1 and C2 than those from control seedlings (Fig. 5D), which suggests that the AZRE may interact with nuclear proteins in a stress-dependent manner. However, whether trans-acting factor(s) directly sense AZC or are only indirectly activated by AZC needs further study. Interestingly, it was also found that the AZRE was indispensable for HS activation of Oshsp17.3 and Oshsp18.0 in the transient expression system (Fig. 3), that the C2 band can be competed out by the HSE (Fig. 5C), and that HS strongly boosted both C1 and C2 signals (Fig. 5D). These data are in agreement with the observation of Sugio et al. (2009), who showed HSFs to be involved in the AZC response in Arabidopsis. Among the HSFs identified in rice, OsHsfA4b, OsHsfB4b, and OsHsfC1b show high AZC responsiveness (data not shown). Co-expression of OsHsfA4b in rice coleoptiles could activate both p567 and p567ΔAZRE constructs in the absence of AZC (Fig. 6B), but the induction level in p567 was 67% higher than that in p567ΔAZRE. In contrast, no significant activation was observed for the p631 construct, which contains a non-AZC-inducible promoter. Consistent with the in vitro binding assay results, these analyses support HSFs being an integral part of the AZRE-dependent induction pathway. They also indicate that the activation of sHSP-CI genes depends on specific HSFs under HS stress.
After being incorporated into proteins by replacing proline residues, which results in the formation of misfolded proteins, AZC acts as a competent inducing signal in an HS-like response and induces a profile of HSP synthesis similar to the HSR (Lee et al., 1996; Hoshikawa et al., 2003). The regulatory mechanism for the AZC response remains unclear. It was found that AZC responsiveness is through existing transcription factor-dependent pathways. The induction of Oshsp17.3 transcripts was delayed by 2 h with AZC in the presence of CHX but had no similar effect with HS treatment (Fig. 4). The AZC response and HSR may be via distinct pathways to activate HSP genes through variable HSFs (Fig. 6B). Furthermore, mitogen-activated protein kinase signalling pathways are activated in response to environmental stresses in eukaryotes (Kyriakis and Avruch, 1996). In rice suspension cells and seedlings, OsMAPK2 was induced by heavy metals such as Cu and Cd but not by HS (Yeh et al., 2004). Similar activation effects were also found on OsMAPK2 transcripts and its kinase activity after AZC treatment (data not shown). However, there is no evidence to show the activation mechanism by AZC. One possibility is through post-translational modifications such as phosphorylation.
HSFs were shown to function in differential regulation of HSP genes under various environmental stresses and in various developmental stages (Mathew et al., 2001; Trinklein et al., 2004). In Arabidopsis, HsfA2 was identified as the major HSF for an AZC-induced response (Sugio et al., 2009). In addition, the diverse configuration of HSEs is important for the differentional regulation of HSP genes under various stresses (Ahn et al., 2001; Sakurai and Takemori, 2007). In this study, it is shown that a new cis-element, the AZRE, contributes to the differential induction of sHSP-CI genes by AZC. On EMSA, the C2 band was competed out not only by the AZRE but also by HSEs (Fig. 5A, B), which suggests that HSFs may play an important role in the interaction. In addition, OshsfA4b was shown to have some effect on the induction of the Oshsp17.3 promoter (Fig. 6B). Thus, the putative AZRE may be an alternative form of HSE. Actually, in the promoters of Oshsp17.7 and Oshsp17.9A, the location of the predicted AZRE overlapped with an HSE (Fig. 2). The interaction between the HSE and other transcriptional binding sites such as AP-1, C/EBP, and MRE has been shown to specify the expression of Arabidopsis AtHsp90-1 in response to various chemical inducers (Haralampidis et al., 2002). Deletion of the 9 bp AZRE from the Oshsp17.3 and Oshsp18.0 promoters resulted in ∼80% loss of induction by HS, although the level of all predicted putative HSEs remained unchanged in the deleted construct (Fig. 3). Additionally, the deleted constructs without AZRE sequences (p270 and p265) were almost unable to be induced by HS (Fig. 3), even though several canonical HSEs were found in the remaining region (Fig. 1B). Furthermore, transgenic Arabidopsis plants with the AZRE-truncated Oshsp17.3 promoter were barely induced by HS treatment (Fig. 7). Therefore, a possible association of the AZRE and canonical HSEs in the Oshsp17.3 promoter is suggested to specify the expression of Oshsp17.3 in response to not only AZC but also HS.
In conclusion, the present results suggest a new pathway for differential regulation of sHSP-CI genes under AZC stress, through an AZRE-dependent pathway. In addition, the AZRE may associate with HSEs to regulate the HSR via specific HSF(s) such as OsHsfA4b. However, further experiments are needed to verify the trans-acting factor(s) specific for the AZC response and the possible interaction between the AZRE and HSEs.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Effect of AZRE deletion on As responsiveness. The bombarded coleoptiles were incubated at 28 °C in the dark for at least 6 h and then treated with 250 μM As for 2 h. After that, the samples were incubated in a fresh incubation buffer at 28 °C in the dark for at least 12 h. The fold induction relative to the control (28 °C) is indicated at the top of the scale bar.
Figure S2. Effect of addition of the AZRE to the Oshsp16.9A promoter or the –46 CaMV 35S minimal promoter on AZC inducibility. (A) The 631 bp Oshsp16.9A promoter was transcriptionally fused to the GUS reporter gene as indicated. The AZRE was added to the 5' end of the Oshsp16.9A promoter or the –46 CaMV 35S minimal promoter as indicated. The AZRE is indicated by a hatched box. All other symbols are as given in Figure 1B. (B) The level of relative GUS activity was determined. The fold induction relative to the control (at 28 °C) is indicated at the top of the scale bar. Each experiment was repeated at least twice. Mean ±SE relative GUS activities are from at least four independent transformations. The P-values are also shown at the top of the scale bar.
Figure S3. Effect of base substitution of the AZRE on AZC and HS responsiveness. (A) The Oshsp17.3 promoter with base substitution was transcriptionally fused to the GUS reporter gene as indicated. Substituted nucleotides in the AZRE of the Oshsp17.3 promoter are indicated by asterisks. The nucleotide sequence of the AZRE is shown in bold. Constructs M1–M3 contain base substitutions as indicated by italic lowercase letters. (B) The relative GUS activity was determined as indicated. The magnitude of induction relative to the control (at 28 °C) is indicated at the top of the scale bar. Each experiment was repeated at least twice. Mean ±SE relative GUS activities are from at least four independent transformations. The P-value of M1 induced by AZC is indicated on the top of the scale bar.
Supplementary Material
Acknowledgments
We are grateful to Dr Anthony Huang for comments and suggestions on the manuscript. This work was supported by the National Science Council, Taiwan, ROC (grants NSC92-2311-B-002-005 and NSC93-2311-B-002-023 to CYL, and NSC97-2313-B-0080001-MY3 to CHY).
Glossary
Abbreviations
- As
arsenite
- AZC
L-azetidine-2-carboxylic acid
- AZRE
AZC response element
- EMSA
electrophoretic mobility shift assay
- ER
endoplasmic reticulum
- GUS
β-glucuronidase
- HSE
heat shock element
- HSF
heat-shock transcription factor
- sHSP-CI
class I small heat shock protein
References
- Ahn S-G, Liu PCC, Klyachko K, Morimoto R, Thiele D. The loop domain of heat shock transcription factor 1 dictates DNA-binding specificity and responses to heat stress. Genes and Development. 2001;15:2134–2145. doi: 10.1101/gad.894801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almoguera C, Rojas A, Diaz-Martin J, Prieto-Dapena P, Carranco R, Jordano J. A seed-specific heat-shock transcription factor involved in developmental regulation during embryogenesis in sunflower. Journal of Biological Chemistry. 2002;277:43866–43872. doi: 10.1074/jbc.M207330200. [DOI] [PubMed] [Google Scholar]
- Banzet N, Richaud C, Deveaux Y, Kazmaier M, Gagnon J, Triantaphylidès C. Accumulation of small heat shock proteins, including mitochondrial HSP22, induced by oxidative stress and adaptive response in tomato cells. The Plant Journal. 1998;13:519–527. doi: 10.1046/j.1365-313x.1998.00056.x. [DOI] [PubMed] [Google Scholar]
- Carranco R, Almoguera C, Jordano J. An imperfect heat shock element and different upstream sequences are required for the seed-specific expression of a small heat shock protein gene. Plant Physiology. 1999;121:723–730. doi: 10.1104/pp.121.3.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen AH, Quail PH. Ubiquitin promoter-based vectors for high-level expression of selectable and/or screenable marker genes in monocotyledonous plants. Transgenic Research. 1996;5:213–218. doi: 10.1007/BF01969712. [DOI] [PubMed] [Google Scholar]
- Diaz-Martin J, Almoguera C, Prieto-Dapena P, Espinosa JM, Jordano J. Functional interaction between two transcription factors involved in the developmental regulation of a small heat stress protein gene promoter. Plant Physiology. 2005;139:1483–1494. doi: 10.1104/pp.105.069963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelman L, Czarnecka E, Key JL. Induction and accumulation of heat shock-specific poly(A+) RNAs and proteins in soybean seedlings during arsenite and cadmium treatments. Plant Physiology. 1988;86:1048–1056. doi: 10.1104/pp.86.4.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan JC, Jinn TL, Yeh CH, Feng SP, Chen YM, Lin CY. Characterization of the genomic structures and selective expression profiles of nine class I small heat shock protein genes clustered on two chromosomes in rice (Oryza sativa L.) Plant Molecular Biology. 2004;56:795–809. doi: 10.1007/s11103-004-5182-z. [DOI] [PubMed] [Google Scholar]
- Haralampidis K, Milioni D, Rigas S, Hatzopoulos P. Combinatorial interaction of cis elements specifies the expression of the Arabidopsis AtHsp90-1 gene. Plant Physiology. 2002;129:1138–1149. doi: 10.1104/pp.004044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshikawa C, Shichiri M, Nakamori S, Takagi H. A nonconserved Ala401 in the yeast Rsp5 ubiquitin ligase is involved in degradation of Gapl permease and stress-induced abnormal proteins. Proceedings of the National Academy of Sciences, USA. 2003;100:11505–11510. doi: 10.1073/pnas.1933153100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Key JL, Lin CY, Chen YM. Heat shock proteins of higher plants. Proceedings of the National Academy of Sciences, USA. 1981;78:3526–3530. doi: 10.1073/pnas.78.6.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotak S, Vierling E, Bäumlein H, von Koskull-Döring P. A novel transcriptional cascade regulating expression of heat stress proteins during seed development of Arabidopsis. The Plant Cell. 2007;19:182–195. doi: 10.1105/tpc.106.048165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo HF, Tsai YF, Young LS, Lin CY. Ethanol treatment triggers a heat shock-like response but no thermotolerance in soybean (Glycine max cv. Kaohsiung No.8) seedlings. Plant, Cell and Environment. 2000;23:1099–1108. [Google Scholar]
- Kyriakis JM, Avruch J. Sounding the alarm: protein kinase cascades activated by stress and inflammation. Journal of Biological Chemistry. 1996;271:24313–24316. doi: 10.1074/jbc.271.40.24313. [DOI] [PubMed] [Google Scholar]
- Lee GJ, Vierling E. A small heat shock protein cooperates with heat shock protein 70 systems to reactivate a heat-denatured protein. Plant Physiology. 2000;122:189–197. doi: 10.1104/pp.122.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YRJ, Nagao RT, Lin CY, Key JL. Induction and regulation of heat-shock gene expression by an amino acid analog in soybean seedlings. Plant Physiology. 1996;110:241–248. doi: 10.1104/pp.110.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescot M, Déhais P, Moreau Y, De Moor B, Rouzé P, Rombauts S. PlantCARE: a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Research. 2002;30:325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CY, Roberts JK, Key JL. Acquisition of thermotolerance in soybean seedlings 1. Synthesis and accumulation of heat shock proteins and their cellular localization. Plant Physiology. 1984;74:152–160. doi: 10.1104/pp.74.1.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luthe DS, Quatrano RS. Transcription in isolated wheat nuclei: I. Isolation of nuclei and elimination of endogenous ribonuclease activity. Plant Physiology. 1980;65:305–308. doi: 10.1104/pp.65.2.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew A, Mathur SK, Jolly C, Fox SG, Kim S, Morimoto RI. Stress-specific activation and repression of heat shock factor 1 and 2. Molecular and Cellular Biology. 2001;21:7163–7171. doi: 10.1128/MCB.21.21.7163-7171.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiwaza A, Yabuta Y, Yoshida E, Maruta T, Yoshimura K, Shigeoka S. Arabidopsis heat shock transcription factor as a key regulator in response to several types of environmental stress. The Plant Journal. 2006;48:535–547. doi: 10.1111/j.1365-313X.2006.02889.x. [DOI] [PubMed] [Google Scholar]
- Nover L, Bharti K, Doring P, Mishra SK, Ganguli A, Scharf KD. Arabidopsis and the heat stress transcription factor world: how many heat stress transcription factors do we need? Cell Stress and Chaperones. 2001;6:177–189. doi: 10.1379/1466-1268(2001)006<0177:aathst>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieping M, Schöffl F. Synergistic effect of upstream sequences, CCAAT box elements, and HSE sequences for enhanced expression of chimaeric heat shock genes in transgenic tobacco. Molecular and General Genetics. 1992;231:226–232. doi: 10.1007/BF00279795. [DOI] [PubMed] [Google Scholar]
- Rojas A, Almoguera C, Carranco R, Scharf KD, Jordano J. Selective activation of the developmentally regulated Hahsp17.6 G1 promoter by heat stress transcription factors. Plant Physiology. 2002;129:1207–1215. doi: 10.1104/pp.010927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai H, Takemori Y. Interaction between heat shock transcription factor (HSFs) and divergent binding sequences. Journal of Biological Chemistry. 2007;282:13334–13341. doi: 10.1074/jbc.M611801200. [DOI] [PubMed] [Google Scholar]
- Scharf KD, Siddique M, Vierling E. The expanding family of Arabidopsis thaliana small heat stress proteins and a new family of proteins containing α-crystallin domains (Acd proteins) Cell Stress and Chaperones. 2001;6:225–237. doi: 10.1379/1466-1268(2001)006<0225:tefoat>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugio A, Dreos R, Aparicio F, Maule AJ. The cytosolic protein response as a subcomponent of the wider heat shock response in Arabidopsis. The Plant Cell. 2009;21:642–645. doi: 10.1105/tpc.108.062596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, van Montagu M, Verbruggen N. Small heat shock proteins and stress tolerance in plants. Biochimica et Biophysica Acta. 2002;1577:1–9. doi: 10.1016/s0167-4781(02)00417-7. [DOI] [PubMed] [Google Scholar]
- Thijs G, Lescot M, Marchal K, Rombauts S, De Moor B, Rouze P, Moreau Y. A higher-order background model improves the detection of promoter regulatory elements by Gibbs sampling. Bioinformatics. 2001;17:1113–1122. doi: 10.1093/bioinformatics/17.12.1113. [DOI] [PubMed] [Google Scholar]
- Trinklein ND, Chen WC, Kingston RE, Myers RM. Transcriptional regulation and binding of heat shock factor 1 and heat shock factor 2 to 32 human heat shock genes during thermal stress and differentiation. Cell Stress and Chaperones. 2004;9:21–28. doi: 10.1379/481.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotter EW, Kao CM, Berenfeld L, Botstein D, Petsko GA, Gray JV. Misfolded proteins are competent to mediate a subset of the responses to heat shock in Saccharomyces cerevisiae. Journal of Biological Chemistry. 2002;277:44817–44825. doi: 10.1074/jbc.M204686200. [DOI] [PubMed] [Google Scholar]
- Weigel D, Glazebrook J. Arabidopsis: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2002. [Google Scholar]
- Yeh CM, Hsiao LJ, Huang HJ. Cadmium activates a mitogen-activated protein kinase gene and MBP kinases in rice. Plant and Cell Physiology. 2004;45:1306–1312. doi: 10.1093/pcp/pch135. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.