Abstract
Cutaneous squamous cell carcinoma (SCC) occurs commonly and can metastasize. Identification of specific molecular aberrations and mechanisms underlying the development and progression of cutaneous SCC may lead to better prognostic and therapeutic approaches and more effective chemoprevention strategies. To identify genetic changes associated with early stages of cutaneous SCC development, we analyzed a series of 40 archived skin tissues ranging from normal skin to invasive SCC. Using high-resolution array-based comparative genomic hybridization (aCGH) we identified deletions of a region on chromosome 10q harboring INPP5A gene in 24% of examined SCC tumors. Subsequent validation by immunohistochemistry (IHC) on an independent sample set of 71 SCC tissues demonstrated reduced INPP5A protein levels in 72% of primary SCC tumors. Decrease in INPP5A protein levels appears to be an early event in SCC development as it also is observed in 9 of 26 (35%) examined actinic keratoses (AK), the earliest stage in SCC development. Importantly, further reduction of INPP5A levels is seen in a subset of SCC patients as the tumor progresses from primary to metastatic stage. The observed frequency and pattern of loss indicate that INPP5A, a negative regulator of inositol signaling, may play a role in development and progression of cutaneous SCC tumors.
Keywords: Inositol polyphosphate-5-phosphatase (INPP5A), Squamous Cell Carcinoma (SCC), Actinic Keratosis (AK), array based Comparative Genomic Hybridization (aCGH), Immunohistochemistry (IHC)
Introduction
Over 1,000,000 non-melanoma skin cancers are diagnosed annually in the US, making these the most common type of cancer and the fifth most costly cancer type in the Medicare population (1). The vast majority of non-melanoma skin cancers are basal cell carcinoma (BCC) and squamous cell carcinoma (SCC). Unlike BCC, where distal spread is exceedingly rare, SCC can metastasize with appreciable frequency. Current clinical prognostic algorithms are suboptimal and therapeutic options for aggressive disease inadequate. Incomplete understanding of the molecular mechanisms leading to the development and progression of SCC have hindered development of accurate prognostic markers and targeted therapies as well as more effective early chemopreventive strategies.
Development of genomic technologies in recent years has provided unparalleled opportunities for rapid and detailed study of cancer on the molecular level. Use of high-resolution, genome-wide approaches such as aCGH has significantly impacted understanding of cancer and facilitated better disease classification (2) and development of novel diagnostic and therapeutic approaches (3–5). Thus, application of high-resolution aCGH has potential to expand the small but growing body of knowledge focused on genomic characterization of cutaneous SCC (6–13) and identify novel, clinically relevant molecular targets.
INPP5A belongs to a large family of inositol polyphosphate 5-phosphatases (14). This 40 kDa membrane-associated type I inositol phosphatase has preferential substrate affinity for inositol 1,4,5-trisphosphate (Ins(1,4,5)P3) and inositol 1,3,4,5-tetrakisphosphate (Ins(1,3,4,5)P4) (15–18), functioning mostly as a signal-terminating enzyme with implication for several cellular processes, including proliferation. Loss of INPP5A may be linked to cancer development and progression. Deletions in the general chromosomal region encoding INPP5A on chromosome 10q26 are associated with brain tumors (19, 20) and decreased inositol polyphosphate 5-phosphatase activity is associated with several human leukemias (21, 22). In addition, reduction of INPP5A expression using the antisense approach has lead to transformation in cell culture as well as tumor growth in mice (17), suggesting a potential tumor suppressor role for INPP5A.
Herein we perform genome-wide survey of gene copy number changes in skin tissues and identify frequent deletions of INPP5A gene in human SCC tumors. In addition, we demonstrate that marked decrease of INPP5A protein levels is observed in most cutaneous SCCs. This event occurs early in the development of SCC as it can be detected even at the stage of actinic keratosis, a common precursor to SCC. However, progressive reduction of INPP5A levels is seen in a subset of SCC patients as the tumor progresses from primary to metastatic stage. Loss of INPP5A, therefore, could play an important role in development and progression of cutaneous SCC.
Materials and Methods
Tissues
Tissue samples analyzed in this study were formalin fixed, paraffin embedded (FFPE), archived specimens obtained under the Institutional Review Board approved protocols at the Arizona Cancer Center, University of Arizona, Tucson, AZ; Southern Arizona Veterans Affairs Health Care System, Tucson, AZ; Loyola University Medical Center, Chicago, IL; and Mayo Clinic. The study was conducted according to the Declaration of Helsinki Principles.
Array CGH (aCGH)
To obtain genomic DNA for aCGH, microscopic examination by pathologist was used to select the areas for harvest and DNA extraction. In all samples only regions that showed greater than 50% lesional content were harvested. aCGH profiling was performed using a method developed by the authors (23). Briefly, DNA was extracted from FFPE tissue blocks using the DNeasy tissue kit (Qiagen, Valencia, CA). Normal pooled lymphocyte DNA (Promega, Madison, WI) was used as a reference. A total of 5 μg of sample genomic DNA and 1 μg of reference genomic DNA was fragmented using the thermolabile recombinant shrimp DNase (TS-DNase; Affymetrix, Santa Clara, CA) to achieve an average DNA fragment length of 200 – 600 bp. Fragmented sample and reference DNA was labeled with Cy 5 and Cy 3 fluorescent dUTP, respectively, using the Bioprime Array CGH Genomic Labeling System (Invitrogen, Carlsbad, CA). Hybridizations were performed on Agilent 44K feature microarrays for aCGH (Agilent Technologies, Santa Clara, CA) per manufacturers specifications and scanned on an Agilent DNA Microarray scanner, followed by image analysis with Feature Extraction software (Agilent Technologies, Santa Clara, CA) and data visualization with DNA Analytics software (Agilent Technologies, Santa Clara, CA) using the aberration calling algorithm, ADM-1 (24).
Fluorescence in situ hybridization (FISH)
FISH was carried out using a centromeric probe to chromosome 10 (Abbott Molecular, Des Plaines, IL) and INPP5A directed probes (BACs RP11-500B2 and RP11-288G11; BACPAC Resource Center) to either metaphase spreads or sections prepared from FFPE blocks. FFPE slices were prepared for hybridization using the Paraffin Pretreatment Kit II (Abbott Molecular, Des Plaines, IL). Slides were examined and photographed on a Zeiss Axiophot equipped with interference filters (Chroma, Bellows Falls, VT) and a CoolSnap HQ2 digital camera (Photometrics, Tucson, AZ). The FISH evaluation was semi-quantitative. Whenever the tissue was of sufficient size 100+ nuclei were examined. However, in cases where a lesion of interest was small (e.g. AK lesions) all available lesional nuclei (i.e., less than 100) were examined.
Immunohistochemistry (IHC)
FFPE tissue blocks were sectioned on glass slides at 5 μm thickness and baked for 60 min at 60 °C. Slides were subsequently subjected to heat induced epitope retrieval using a proprietary citrate based retrieval solution for 20 min. The tissue sections were incubated for 30 min with anti-INPP5A mouse monoclonal antibody, clone 3D8 (Novus Biologicals, Littleton, CO). The sections were visualized with the Bond Polymer Refine Detection kit (Leica Microsystems Inc., Bannockburn, IL) using diaminobenzidine chromogen as substrate.
Statistics
The two-tailed Fisher’s exact test was used to compare the staining patterns between the cohorts of primary SCC tumors that have subsequently metastasized to those that have not. P-values of <0.05 were considered statistically significant.
Results and Discussion
INPP5A gene is frequently deleted in cutaneous SCC tumors
Genomic instability in cancer commonly leads to gross DNA ploidy changes as well as focal gene copy number aberrations. These genetic events contribute to development and progression of cancer by providing inappropriate proliferative stimuli or eliminating essential growth-regulating mechanisms, as illustrated by amplification of oncogenes and deletion of tumor suppressor genes, respectively. Identification of such changes in cancer has elucidated pathogenic molecular mechanisms that could be exploited for clinical benefit (3, 25). In cutaneous SCC, a global picture of genomic aberrations is starting to emerge (6–13) and high-resolution analysis of SCC genomes for identification of focal, clinically relevant gene copy number aberrations is warranted.
To identify novel genes and molecular mechanisms associated with cutaneous SCC development and progression, we analyzed a series of archived skin tissues spanning a range from normal skin to invasive SCC. To this end we employed a high resolution, oligomer aCGH method that we recently optimized specifically for use on archived, formalin fixed, paraffin embedded (FFPE) tissues. This approach is capable of detecting gene copy number changes with sensitivity and accuracy comparable to that obtained by analysis of DNA derived from frozen tissues (23).
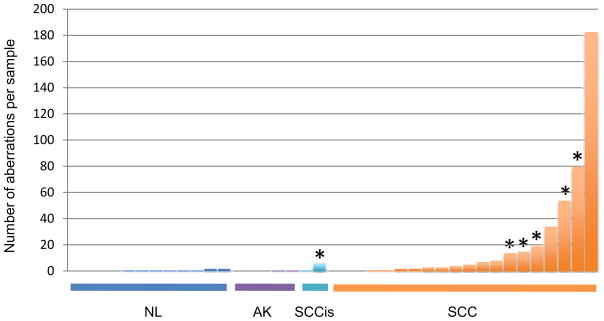
aCGH was performed using the DNA from a spectrum of 40 FFPE skin tissues including normal skin (n=12), precancerous lesions of actinic keratosis (AK; n=5), in situ SCC lesions (SCCis; n=2) and invasive SCCs (SCC; n=21). A total of 458 copy number aberrations were identified in the examined samples, 267 (58%) of which were amplifications and 191 (42%) deletions. We observed an increase in the overall frequency of gene copy number aberrations per sample in proportion to the increasing malignant characteristics of the examined tissue, with invasive SCCs harboring, on average, the highest number of aberrations per genome (Figure 1). Detailed genomic distribution of aberrations as well as their overall percent penetrance in SCC lesions is illustrated in the supplemental figure S1.
Figure 1. Distribution of gene copy number aberrations in examined tissues.
aCGH was performed and a total number of genomic aberrations was calculated for each examined sample. The bars represent individual samples, which are clustered according to the tissue type, from left to right, including normal skin (NL), actinic keratoses (AK), SCC in situ (SCCis) and invasive SCC (SCC). Within each cluster, individual samples are sorted from left to right, according to the number of gene copy aberrations per sample. Asterisk (*) designates samples harboring INPP5A deletions.
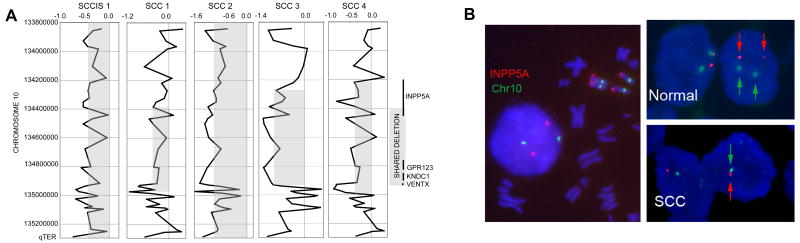
Examination of the genomic regions characterized by recurrent copy number changes across samples, as detected by aCGH, identified several previously reported regions of aberrations including amplifications of 1p, 3q, 8q, 14q and 20q as well as deletions on 3p and 9p (Figure S1) (9, 11, 12, 26). However, the most prevalent copy number aberration was deletion of the q-ter region of chromosome 10, an area harboring the INPP5A gene (Figure 2a). aCGH detected loss of INPP5A gene in 1 of 2 examined SCCis lesions and 5 of 21 (24%) examined invasive SCC tumors, but in none of the examined AK lesions or normal skin (Table 1). To verify the accuracy of aCGH calls, we performed FISH for INPP5A in 2/5 samples that demonstrated INPP5A deletions by aCGH. Both cases showed clear INPP5A loss, while control tissues showed no detectable loss of FISH signal (Figure 2b).
Figure 2. Identification of INPP5A deletions by aCGH and validation by FISH.
(a) Recurrent deletions involving chromosome 10q26.3 were identified in SCC tissues by aCGH. Log2 scale provided above each panel depicts DNA copy number change (“0” indicates no net change; negative values indicate deletions), with gray boxes depicting deletions. The gray box on the right indicates the area of overlap, shared among the five samples harboring deletions in this region. Black bars on the right depict individual genes, including INPP5A. (b) A representative FISH assay is shown with INPP5A signal in red and centromeric chromosome 10 signal in green. Two copies of INPP5A are seen in normal keratinocytes (right, upper panel) but only one copy in the SCC (right lower panel). Normal metaphase spread (left panel) is provided as a reference.
Table 1.
Frequency of INPP5A gene deletions as detected by CGH.
| Tissue type | INPP5A deletion |
|---|---|
| Normal | 0/12 |
| AK | 0/5 |
| SCCis | 1/2 |
| SCC | 5/21 (24%) |
AK, actinic keratosis; SCCis, SCC in situ; SCC, invasive SCC
Observed deletions of INPP5A represent a highly selected, non-random genetic event in SCC. Most of 191 deletions identified among SCC samples occur only once, while a smaller proportion are observed as recurrent deletions, affecting more than one sample. The region of INPP5A is the single most frequently deleted segment in the interrogated SCC genomes, as well as the only recurrent deletion detected in 5 independent SCC samples. The core INPP5A deletion, characterized as the smallest area of overlap among the aberrations harboring INPP5A deletions (Figure 2a), covers a genetic segment containing 587,219 bp, of which 91,861 bp are in the INPP5A gene itself. In addition to INPP5A, this segment contains three genes, GPR123, KNDC1 and VENTX. However, INPP5A is the only gene in this cluster repeatedly affected by the copy number transition (the edge of the aberration), being affected in 3 of 5 samples harboring deletion of this region. Taken together, this data strongly suggests that INPP5A gene deletions are highly selected genetic events, rather than nonspecific bystander events in the context of the overall genomic instability of the SCC genome.
INPP5A protein level is frequently reduced in primary SCC tissues
Genomic aberrations detected by aCGH, such as gene deletions, can indicate a “tip of the iceberg” phenomenon, where loss of a gene on the DNA level is seen in a subset of tumors, while in remaining cases the implicated gene may be deregulated by other mechanisms, including those affecting its mRNA and protein products. To evaluate whether the genetic loss of INPP5A observed with aCGH might be similarly indicative of a more general phenomenon of INPP5A loss in SCC, we examined INPP5A protein levels in an independent cohort of FFPE skin tissues by IHC using a monoclonal antibody to INPP5A. We evaluated a total of 71 archived SCC tumors and compared them to the matched normal skin from the same patient using the histologically normal epidermis, immediately adjacent to the SCC tumor as control. Stained slides were evaluated using a standard scoring system based on the intensity of staining (0–3) with score of 0 representing no staining and score of 3 as intense staining. If a relative difference in signal was observed between tissues being compared, it was recorded as a change in INPP5A protein level.
Detection of INPP5A by IHC demonstrated mainly diffuse cytoplasmic signal. A comparison of INPP5A staining intensity between SCC tissues and matched normal epidermis identified three general staining patterns. The most prevalent pattern of expression, observed in 51/71 (72%) of examined tissues, manifested as a relative reduction of INPP5A in SCC tissues when compared to matched normal skin (Figure 3a). Only 20/71 (28%) of examined tissues showed no difference in INPP5A staining between the SCC and matched normal skin. Importantly, no single case was observed where INPP5A staining was more intense in SCC tumor than in matched normal skin, further highlighting the specificity of the observed pattern (Table 2, top; Figure 3b). Notably, 10 SCC lesions examined in this cohort were classified by pathology as SCC in situ (SCCis). 6/10 of these SCCis lesions demonstrated reduced INPP5A IHC signal, indicating no significant difference to the frequency detected in primary SCC in general. The observed reduction of INPP5A signal in SCC tissues is tumor-specific, as the history of sun exposure and the extent of sun damage is comparable between the SCC lesion and the examined, adjacent normal epithelium used as a control. Taken together, a significantly higher frequency of INPP5A loss at the protein level compared to loss at the DNA level indicates that gene deletions may represent only one mechanisms of INPP5A suppression, while a sizable proportion of SCC tumors likely achieve the same effect through deregulation of INPP5A by other mechanisms.
Figure 3. Detection of INPP5A protein loss in primary SCC tissues relative to the matched, normal epidermis.
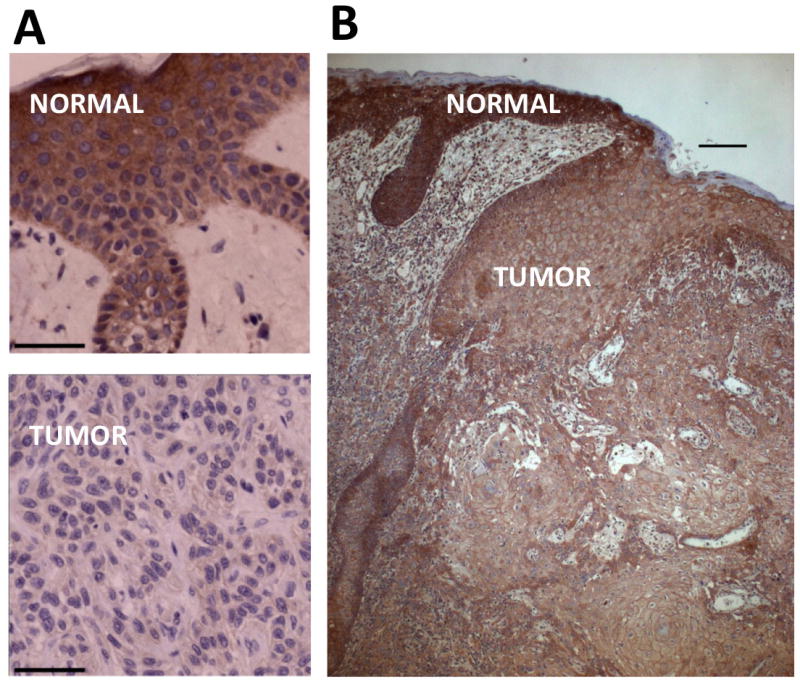
Immunohistochemistry (IHC) for INPP5A was performed on FFPE tissues and relative intensity of INPP5A staining compared between the SCC tissue and the adjacent, histologically normal epidermis. a) Primary skin SCC tissue with low level of INPP5A staining is shown in the lower panel and matched, adjacent normal epidermis from the same patient in the upper panel. b) A representative case is shown to further illustrate a relative difference in INPP5A staining between the primary SCC lesion and the adjacent normal epidermis. Bar = 50 μm.
Table 2.
Detection of INPP5A protein levels at successive stages of SCC progression.
| INPP5A staining intensity | Frequency |
|---|---|
| Primary SCC compared to matched normal skin | |
| Normal skin > Primary SCC | 51/71 (72%) |
| Normal skin = Primary SCC | 20/71 (28%) |
| Normal skin < Primary SCC | 0/71 (0%) |
| AK compared to matched normal skin | |
| Normal skin >AK | 9/26 (35%) |
| Normal skin = AK | 17/26 (65%) |
| Normal skin < AK | 0/26 (0%) |
| Primary SCC compared to matched metastatic SCC tissues | |
| Primary SCC > Met | 6/17 (35%) |
| Primary SCC = Met | 11/17 (65%) |
| Primary SCC < Met | 0/17 (0%) |
INPP5A loss is an early event in SCC development
To more precisely evaluate the timing of the reduction of INPP5A level in the development of cutaneous SCC we examined a series of 26 AKs, the earliest step in SCC development. Using IHC, as described above, we compared INPP5A protein levels between the AK lesions and adjacent normal epidermis. A relative reduction of INPP5A in AK lesions was seen in 9/26 (35%) of examined tissues, whereas 17/26 (65%) of examined tissues showed no difference in INPP5A levels between the AK and normal epidermis (Table 2, middle).
We next asked whether the observed reduction of INPP5A protein in AKs is caused by genetic loss at the DNA level, such as seen in a subset of SCC tumors. To this end we carried out FISH analysis and detected no INPP5A gene deletion in any of the examined cases (data not shown). Although a small lesion size and limited number of lesional nuclei available for analysis in some of the studied AKs calls for cautious interpretation of these results, it is important to note that no single lesion showed evidence of a clonal population with uniform loss of INPP5A FISH signal, even in cases where such clonal loss was suggested by IHC data. This absence of perturbations on DNA level in AKs is not surprising given the relative paucity of gene copy number aberrations at early stages of disease detected by aCGH (Figure 1) and likely indicates that deregulation of INPP5A expression in these precursor lesions occurs mainly on mRNA or protein level.
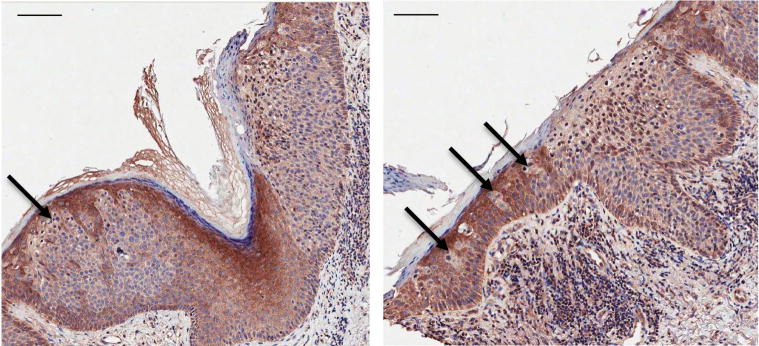
The less frequent reduction of INPP5A levels in AK than in SCC lesions (35% vs. 72%) is also informative and may reflect selection that favors progression of AK lesions with low INPP5A to the next stage of disease. Interestingly, a pattern of INPP5A loss in a subset of AKs, occurring in the form of strikingly demarcated regions of low INPP5A signal, is suggestive of clonally expanding populations of affected cells within epidermis (Figure 4). As SCCs in situ often arise within the preexisting lesions of AKs, this focal loss of INPP5A in AKs might represent an early step towards a full oncogenic transformation along the spectrum of evolving epidermal neoplasia.
Figure 4. Detection of INPP5A loss in actinic keratoses (AK).
Immunohistochemistry (IHC) for INPP5A was performed on FFPE tissues. Two representative lesions are shown in left and right panel, each containing an area of SCCis, (evident by full epidermal thickness neoplasia; right half of each image), arising in association with an AK (partial epidermal thickness neoplastic change, consistent with AK; left half of each image). Arrows highlight populations of cells demonstrating low level of INPP5A staining within the AK lesions. Bar = 100 μm.
Loss of INPP5A in association with progression to metastatic disease
The above data implicates deregulation of INPP5A levels as an early event in the development of keratinocyte neoplasia, which may provide a selective advantage in progression from AK to SCC. To assess a potential role of INPP5A loss in the process of tumor maintenance and progressions, we queried whether reduction of INPP5A level is associated with the subsequent biological step in SCC progression, development of metastatic disease. To this end, we evaluated INPP5A protein levels in a selected cohort of 17 patients with cutaneous SCC tumors that have subsequently metastasized, and where both primary tumor tissue and matched regional metastatic tissue were available for examination. IHC analysis of these paired tissues detected further reduction of INPP5A levels in the transition from primary to metastatic SCC in 6/17 (35%) of examined tissue pairs (Table 2, bottom). Though the remaining 11/17 (65%) of studied pairs show no further loss of INPP5A levels in transition from primary to metastatic disease, it is important to note that no single case was identified where INPP5A staining was stronger in the metastatic tissue than in the primary SCC tumor, further highlighting the specificity of the observed INPP5A loss in SCC progression. These data suggest that reduction of INPP5A levels, although an early event in development of SCC, may also play a role in progression of SCC from primary to metastatic disease in a significant subset of aggressive primary SCC tumors. Future exploration of this question on a larger number of tissue specimens is warranted.
As we evaluated INPP5A staining in this cohort of 17 patients with metastatic SCCs, we noted the presence of normal epidermis immediately adjacent to the primary SCC tissue in 13/17 examined primary SCCs. It is interesting to note that a relative difference in INPP5A staining between the SCC tumors and adjacent normal epidermis in these 13 patients demonstrate strikingly high frequency of INPP5A loss in SCC tumors. 12/13 (92%) of these aggressive primary SCC tumors demonstrated loss of INPP5A staining when compared to the adjacent, normal epidermis. This is in contrast to the above described pattern of INPP5A loss observed in randomly selected primary SCC tumors where 51/71 (72%) of SCC tumors demonstrated reduction of INPP5A protein levels by IHC (Figure 3a and Table 2). Thus, higher frequency of INPP5A loss in primary SCC tumors that have demonstrated an aggressive clinical course (i.e., subsequent development of metastases) may indicate more aggressive primary disease and suggest a potential prognostic value of INPP5A levels in assessing the risk of progression in primary SCC tumors. Although this is an exploratory study and the sample sets are not powered to provide robust statistical quantification (comparison by Fisher’s exact test did not reach statistical significance) the observed patterns strongly highlight a trend that merits further exploration on a larger sets of matched, clinically annotated specimens.
In summary, we identify loss of INPP5A as an early event in development of cutaneous SCC. The gene itself is deleted in a significant proportion of SCC tumors, and its protein levels are reduced in the majority of SCC tumors. More frequent reduction of INPP5A levels in aggressive primary SCC tumors, as well as further reduction in metastatic disease, point to a potential role of INPP5A in the development and progression of cutaneous SCC. Our findings support the previously reported observations that implicate INPP5A as a novel tumor suppressor in other human cancers (17). Understanding the precise mechanism(s) of INPP5A loss in SCC and exploring the connection between INPP5A and uncontrolled cellular proliferation in skin cancer may provide novel insights into relevant mechanisms of epithelial carcinogenesis and facilitate development of clinically applicable prognostic markers, therapeutic strategies as well as novel chemopreventive approaches.
Supplementary Material
Acknowledgments
We are grateful to Dr. Rafael Fonseca, Scott Van Wier, Daniel Johnson and Christopher Gooden for their input and assistance with FISH experiments; to Dr. Mark Dahl for helpful discussions and comments; and to Ms. Susan Wing for administrative assistance with manuscript preparation. This work was supported by National Institute of Health grant 5P01CA027502-27.
Footnotes
Conflict of Interest: The authors state no conflict of interest.
References
- 1.Housman TS, Feldman SR, Williford PM, et al. Skin cancer is among the most costly of all cancers to treat for the Medicare population. J Am Acad Dermatol. 2003 Mar;48(3):425–9. doi: 10.1067/mjd.2003.186. [DOI] [PubMed] [Google Scholar]
- 2.Curtin JA, Fridlyand J, Kageshita T, et al. Distinct sets of genetic alterations in melanoma. N Engl J Med. 2005 Nov 17;353(20):2135–47. doi: 10.1056/NEJMoa050092. [DOI] [PubMed] [Google Scholar]
- 3.Gerami P, Jewell SS, Morrison LE, et al. Fluorescence in situ hybridization (FISH) as an ancillary diagnostic tool in the diagnosis of melanoma. Am J Surg Pathol. 2009 Aug;33(8):1146–56. doi: 10.1097/PAS.0b013e3181a1ef36. [DOI] [PubMed] [Google Scholar]
- 4.Curtin JA, Busam K, Pinkel D, Bastian BC. Somatic activation of KIT in distinct subtypes of melanoma. J Clin Oncol. 2006 Sep 10;24(26):4340–6. doi: 10.1200/JCO.2006.06.2984. [DOI] [PubMed] [Google Scholar]
- 5.Garrido MC, Bastian BC. KIT as a therapeutic target in melanoma. J Invest Dermatol. Jan;130(1):20–7. doi: 10.1038/jid.2009.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Quinn AG, Sikkink S, Rees JL. Basal cell carcinomas and squamous cell carcinomas of human skin show distinct patterns of chromosome loss. Cancer Res. 1994 Sep 1;54(17):4756–9. [PubMed] [Google Scholar]
- 7.Dobler M, Schuh J, Kiesewetter F, Schell H, Liehr T, Gebhart E. Deletion monitoring in skin tumors by interphase-FISH using band-specific DNA probes. Int J Oncol. 1999 Mar;14(3):571–6. doi: 10.3892/ijo.14.3.571. [DOI] [PubMed] [Google Scholar]
- 8.Popp S, Waltering S, Holtgreve-Grez H, et al. Genetic characterization of a human skin carcinoma progression model: from primary tumor to metastasis. J Invest Dermatol. 2000 Dec;115(6):1095–103. doi: 10.1046/j.1523-1747.2000.00173.x. [DOI] [PubMed] [Google Scholar]
- 9.Ashton KJ, Weinstein SR, Maguire DJ, Griffiths LR. Chromosomal aberrations in squamous cell carcinoma and solar keratoses revealed by comparative genomic hybridization. Arch Dermatol. 2003 Jul;139(7):876–82. doi: 10.1001/archderm.139.7.876. [DOI] [PubMed] [Google Scholar]
- 10.Ashton KJ, Carless MA, Griffiths LR. Cytogenetic alterations in nonmelanoma skin cancer: a review. Genes Chromosomes Cancer. 2005 Jul;43(3):239–48. doi: 10.1002/gcc.20183. [DOI] [PubMed] [Google Scholar]
- 11.Clausen OP, Aass HC, Beigi M, et al. Are keratoacanthomas variants of squamous cell carcinomas? A comparison of chromosomal aberrations by comparative genomic hybridization. J Invest Dermatol. 2006 Oct;126(10):2308–15. doi: 10.1038/sj.jid.5700375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salgado R, Toll A, Espinet B, et al. [Analysis of cytogenetic abnormalities in squamous cell carcinoma by array comparative genomic hybridization] Actas Dermosifiliogr. 2008 Apr;99(3):199–206. doi: 10.1016/s1578-2190(08)70232-4. [DOI] [PubMed] [Google Scholar]
- 13.Purdie KJ, Harwood CA, Gulati A, et al. Single nucleotide polymorphism array analysis defines a specific genetic fingerprint for well-differentiated cutaneous SCCs. J Invest Dermatol. 2009 Jun;129(6):1562–8. doi: 10.1038/jid.2008.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Majerus PW. Inositols do it all. Genes Dev. 1996 May 1;10(9):1051–3. doi: 10.1101/gad.10.9.1051. [DOI] [PubMed] [Google Scholar]
- 15.Mitchell CA, Speed CJ, Nicholl J, Sutherland GR. Chromosomal mapping of the gene (INPP5A) encoding the 43-kDa membrane-associated inositol polyphosphate 5-phosphatase to 10q26.3 by fluorescence in situ hybridization. Genomics. 1996 Jan 1;31(1):139–40. doi: 10.1006/geno.1996.0023. [DOI] [PubMed] [Google Scholar]
- 16.Connolly TM, Bansal VS, Bross TE, Irvine RF, Majerus PW. The metabolism of tris- and tetraphosphates of inositol by 5-phosphomonoesterase and 3-kinase enzymes. J Biol Chem. 1987 Feb 15;262(5):2146–9. [PubMed] [Google Scholar]
- 17.Speed CJ, Little PJ, Hayman JA, Mitchell CA. Underexpression of the 43 kDa inositol polyphosphate 5-phosphatase is associated with cellular transformation. EMBO J. 1996 Sep 16;15(18):4852–61. [PMC free article] [PubMed] [Google Scholar]
- 18.Laxminarayan KM, Chan BK, Tetaz T, Bird PI, Mitchell CA. Characterization of a cDNA encoding the 43-kDa membrane-associated inositol-polyphosphate 5-phosphatase. J Biol Chem. 1994 Jun 24;269(25):17305–10. [PubMed] [Google Scholar]
- 19.Fults D, Pedone C. Deletion mapping of the long arm of chromosome 10 in glioblastoma multiforme. Genes Chromosomes Cancer. 1993 Jul;7(3):173–7. doi: 10.1002/gcc.2870070311. [DOI] [PubMed] [Google Scholar]
- 20.Lee SH, Davison JA, Vidal SM, Belouchi A. Cloning, expression and chromosomal location of NKX6B TO 10Q26, a region frequently deleted in brain tumors. Mamm Genome. 2001 Feb;12(2):157–62. doi: 10.1007/s003350010247. [DOI] [PubMed] [Google Scholar]
- 21.Nye KE, Riley GA, Poulter LW, Porfiri E, Hoffbrand AV, Wickremasinghe RG. Impaired degradation of Ca(2+)-regulating second messengers in myeloid leukemia cells. Implications for the regulation of leukemia cell proliferation. Leukemia. 1992 Aug;6(8):801–5. [PubMed] [Google Scholar]
- 22.Mengubas K, Jabbar SA, Nye KE, Wilkes S, Hoffbrand AV, Wickremasinghe RG. Inactivation of calcium ion-regulating inositol polyphosphate second messengers is impaired in subpopulations of human leukemia cells. Leukemia. 1994 Oct;8(10):1718–25. [PubMed] [Google Scholar]
- 23.Hostetter G, Kim SY, Savage S, et al. Random DNA fragmentation allows detection of single-copy, single-exon alterations of copy number by oligonucleotide array CGH in clinical FFPE samples. Nucleic Acids Res. 2009 Oct 29; doi: 10.1093/nar/gkp881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lipson D, Aumann Y, Ben-Dor A, Linial N, Yakhini Z. Efficient calculation of interval scores for DNA copy number data analysis. J Comput Biol. 2006 Mar;13(2):215–28. doi: 10.1089/cmb.2006.13.215. [DOI] [PubMed] [Google Scholar]
- 25.Carter CA, Kelly RJ, Giaccone G. Small-molecule inhibitors of the human epidermal receptor family. Expert Opin Investig Drugs. 2009 Dec;18(12):1829–42. doi: 10.1517/13543780903373343. [DOI] [PubMed] [Google Scholar]
- 26.Purdie KJ, Lambert SR, Teh MT, et al. Allelic imbalances and microdeletions affecting the PTPRD gene in cutaneous squamous cell carcinomas detected using single nucleotide polymorphism microarray analysis. Genes Chromosomes Cancer. 2007 Jul;46(7):661–9. doi: 10.1002/gcc.20447. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.