Abstract
Huntington disease (HD) is a fatal neurodegenerative disorder caused by an expanded CAG repeat. Its length can be used to estimate the time of clinical diagnosis, which is defined by overt motor symptoms. Non-motor symptoms begin before motor onset, and involve changes in hypothalamus-regulated functions such as sleep, emotion and metabolism. Therefore we hypothesized that hypothalamic changes occur already prior to the clinical diagnosis. We performed voxel-based morphometry and logistic regression analyses of cross-sectional MR images from 220 HD gene carriers and 75 controls in the Predict-HD study. We show that changes in the hypothalamic region are detectable before clinical diagnosis and that its grey matter contents alone is sufficient to distinguish HD gene carriers from control cases. In conclusion, our study shows, for the first time, that alterations in grey matter contents in the hypothalamic region occur at least a decade before clinical diagnosis in HD using MRI.
Keywords: Huntington, hypothalamus, imaging, basal ganglia, neuroendocrine, atrophy
Introduction
Huntington disease (HD) is a fully penetrant hereditary neurodegenerative disorder caused by an expanded CAG repeat in the HD gene (HDCRG, 1993). The length of the CAG repeat correlates negatively with the age of onset and can be used to estimate it (Langbehn et al. 2004). The clinical diagnosis of HD is currently defined by the presence of overt motor disturbances (Philips et al., 2008). Striatal atrophy is well established in clinical HD and has been linked to chorea, whereas changes in the cerebral cortex have been correlated with cognitive decline (Aylward et al., 2004; Kipps et al., 2005; 2007; Paulsen et al., 2006b; 2010; Wolf et al., 2007; Rosas et al., 2005; 2008). However, circadian rhythm changes, sleep abnormalities, psychiatric complications, as well as increased appetite and metabolic alterations are common and often precede motor symptoms by many years (Trejo et al., 2004; Morton et al., 2005; Underwood et al., 2006; Mochel et al., 2007; Julien et al., 2007; van Duijn et al., 2007; Duff et al., 2007; Arnulf et al., 2008; Videnovic et al., 2009). These disturbed functions may be caused by changes in the hypothalamus and neuroendocrine circuitries (Petersén et al., 2009). In fact, recent studies have shown that hypothalamic atrophy is present in HD patients in early disease stages as well as in transgenic HD mice using voxel based morphometry (VBM) of MR images (Kassubek et al., 2004; Douaud et al., 2006; Sawiak et al., 2009). Furthermore, microglia activation and reductions in dopamine D2 receptor levels occur in the hypothalamic region even before onset of motor symptoms in HD gene carriers (Politis et al., 2008). Finally orexin loss in the hypothalamus and alterations in the hypothalamic-pituitary-adrenal axis including increased levels of cortisol and insulin growth factor-1 have been shown in patients with HD from an early disease stage (Petersén et al., 2005; Björkqvist et al., 2006; Aziz et al., 2008; 2009; Saleh et al., 2009).
Despite intense research during the last decade, there is still no cure or satisfactory treatment for this fatal disorder. Biomarkers that define disease states before clinical diagnosis are urgently needed in the development of potential therapeutic interventions. The Predict-HD study constitutes a multi-national effort to identify such early disease related changes by longitudinally characterizing individuals carrying the HD gene but who have not yet reached the stage of clinical diagnosis (Paulsen et al., 2006a; 2008). In light of the recent findings of hypothalamic dysfunction and atrophy in early clinical stages in HD and the fact that non-motor symptoms and signs precede motor disturbances by many years, we hypothesized that changes in the hypothalamic region would be present already prior to clinically defined disease onset. In order to test this hypothesis, we first performed VBM analyses of cross-sectional MR images from the Predict-HD study. We then used a classification approach of the MR images as another means to assess whether changes were present in the hypothalamic region in prodromal HD gene carriers with different estimated time to clinical diagnosis.
Materials and Methods
MR images
This study was performed using a subset of the Predict-HD cohort. Predict-HD is a multi-national study of individuals known to be at risk for HD (Paulsen et al., 2006a; 2008) recruited at 17 sites in the USA, four sites in Canada, seven sites in Europe and three sites in Australia (Paulsen et al., 2006a). The study was approved by institutional review boards at all study and data processing sites. Participants underwent informed consent procedures and signed consents both for participation and to allow de-identified research data to be sent to collaborative institutions for analyses. Inclusion criteria for Predict-HD required to have undergone genetic testing for the presence of the CAG repeat expansion in the HD gene, and exclusion criteria included clinical evidence of unstable medical or psychiatric illness, history of other central nervous system disease or event such as head trauma or seizures as described previously (Paulsen et al., 2006a; 2008).
Data obtained from two hundred and twenty prodromal HD gene carriers, hereafter referred to as “prHD” participants, were included in the present analyses. Individuals with prHD were stratified by the predicted time to clinical diagnosis, as calculated by the method described by Langbehn et al (2004). Participants with a predicted time to clinical diagnosis less than 9 years were grouped into the “prHDnear” group (7.20 ± 1.43 years to predicted clinical diagnosis; mean ± SD), those with 9–15 years to predicted clinical diagnosis formed the “prHDmid” group (11.36 ± 1.95 years), and the “prHDfar” group consisted of participants with more than 15 years to predicted clinical diagnosis (21.04 ± 5.09 years). Demographic data of the study population are shown in Table 1. The prHD participants for the present study, as well as seventy-five control participants, were selected from the first 348 individuals in the Predict-HD study with quality controlled MRI scans, to obtain approximately equally sized age and gender matched groups. The control participants constituted members of families with HD that were found not to carry the HD gene and that were enrolled in the Predict-HD study. There were no significant age differences or differences in the gender distribution between the control group and any of the prHD groups (age differences analyzed by unpaired t-tests, gender distributions by Fisher’s exact test. Significance threshold p<0.05, uncorrected). As could be expected, the mean age of participants in the prHDnear group was significantly higher than that of the prHDfar group. Otherwise, no significant differences in age or gender distribution were found between any pair of prHD groups. We also compared the mean CAG repeat lengths between the groups with unpaired t-tests, and found significant differences in all pairwise comparisons of prHD groups (p<0.05, uncorrected).
Table 1.
Demographic data for the study population
| Control | prHDfar | prHDmid | prHDnear | Statistical difference |
|
|---|---|---|---|---|---|
| n | 75 | 75 | 75 | 70 | |
|
Predicted time to clinical diagnosis (years) |
NA | > 15 | 9 – 15 | < 9 | |
|
Predicted time to clinical diagnosis mean ± SD (years) |
NA | 21.04 ± 5.09 | 11.36 ± 1.95 | 7.20 ± 1.43 | |
|
CAG repeat length mean ± SD |
19.51 ± 2.74 | 40.75 ± 1.46 | 42.79 ± 2.18 | 44.13 ± 2.55 |
* between all pairs of prHD groups |
|
Age, mean ± SD (years) |
42.1 ± 10.7 | 39.9 ± 7.6 | 42.2 ± 9.5 | 44.2 ± 8.5 |
* between prHDfar and prHDnear |
| Age, range (years) | 19 – 64 | 22 – 59 | 26 – 63 | 26 – 64 | |
| Gender (F/M) | 47/28 | 49/26 | 45/30 | 37/33 |
= p<0.05, significant difference between the indicated groups
All participants had MRI scans at 1.5 Tesla, and 30 sites used a General Electric 1.5 T scanner (with two exceptions, both using 1.5 T Siemens scanners) (Paulsen et al., 2008). Briefly, a sagittal localizer preceded the axial 3DSPGR with the following specifications; ~1×1×1.5 mm voxels, TR = 18, TE = 3, FOV = 24, thickness = 1.5mm, 0 gap, matrix = 256 × 192 with ¾ phase FOV, NEX = 2, flip angle = 20, bandwidth = 15, 124 slices. The 3DSPGR was followed by a coronal T2/PD (proton density) image with the following specificities: ~1×1×3 mm voxels, TR = 3000, TE = 28, FOV = 26, thickness = 3.0, 0 gap, matrix = 256 × 192, NEX = 1, flip angle = 90, 64 slices. The acquired images were inhomogeneity corrected, rigidly AC-PC aligned and resampled with 1 mm isotropic voxels.
Voxel-based morphometry
VBM is an automated technique for statistical comparisons of tissue composition between groups of individuals (Ashburner and Friston, 2000; Wright et al., 1995). T1-weighted MR images are first normalized to the same stereotactic space and segmented into grey matter, white matter and cerebrospinal fluid. The grey matter segment is then extracted and further processed. After the normalization and segmentation, the intensity of each voxel in this segment represents the relative grey matter content of the voxel. During the normalization step, some brain regions may be compressed while others may be expanded. A modulation step, in which the intensity of each voxel is scaled to compensate for such volume changes, may be introduced to preserve the total amount of grey matter in the image. Hence, after the modulation each voxel value is a measure of the actual grey matter content of the voxel. Finally, the images are smoothed, so that the value in each voxel is a measure of the local average grey matter content. The smoothing compensates for some imperfections in the normalization procedure and also renders the voxel values more normally distributed. To yield statistical parametric maps, the local average grey matter content of each voxel is compared between two or more groups.
We performed VBM using the SPM5 software (Wellcome Department of Cognitive Neurology, London) in MATLAB v. 7.5.0 (R2007b; Mathworks). The T1-weighted images from all participants were normalized to the tissue templates supplied with SPM5 by means of the unified segmentation algorithm described in Ashburner and Friston, 2005. After normalization, the voxel size was 2 mm × 2 mm × 2 mm. A modulation step was performed to ascertain that the intensity of each voxel reflected the actual grey matter content of that voxel. All images were subsequently smoothed with an isotropic gaussian kernel with FWHM = 4 mm, which was chosen based on the estimated size of the hypothalamic region and according to Ridgway et al., 2008. Voxel-wise comparisons of the local average grey matter content were performed within the general linear model framework, contrasting the control group and each of the prHD groups, using age and gender as co-variates and hence removing the confounding effects of those on the result. As noted by Ridgway et al (2009), performing the statistical tests only for voxels within a properly specified mask may clarify the interpretation of the VBM results. They proposed an automated method for creating such a mask, by finding the binary image which has the highest correlation with the mean grey matter image. We applied this method to the mean grey matter image from the control group, and the statistical tests were performed for all voxels within the resulting mask. We applied two-sided t-tests to find brain regions with a difference in grey matter content between the compared groups. The resulting statistical parametric maps were corrected for multiple comparisons using the false discovery rate (FDR). Values with q<0.01 were considered significant. The MNI Space Utility (http://www.ihb.spb.ru/~pet_lab/MSU/MSUMain.html) was used to transform the MNI coordinates of the significant voxels to the Talairach space and infer the corresponding brain regions.
Validation of the normalization process for the hypothalamic region
Interpretation of VBM results requires that the images are correctly aligned to each other to ensure that a certain voxel corresponds to the same physical space in all images. Since the hypothalamic region is the main region of interest in this study, and as a normalization algorithm can not be expected to perform equally well throughout the brain, we assessed the performance of the applied normalization algorithm in this region only. The aim was to investigate whether voxels in the hypothalamic region were systematically shifted in the different groups after the normalization, which could potentially influence the results from the VBM. First, six points of reference were marked in the native space T1-weighted images of all participants by an individual who was blinded to the genotype. The points were chosen to provide a boundary for the hypothalamus. They were placed in a coronal plane 2.7 mm posterior to the anterior commissure according to a human brain atlas (Mai et al., 2007). In each hemisphere, the landmarks were placed in the medial and ventral edge of the fornix, the medial and ventral edge of the optic tract, and in the inferior and medial edge of the hypothalamus. The individual transformations estimated by the unified segmentation in SPM5 were then used to map these points to the MNI space, and the location of the points in this space was compared across groups and individuals.
Volumetric analysis of the third ventricle
The hypothalamic region has no clear anatomical landmarks that can be used to accurately delineate it in MR images obtained from a 1.5 T scanner. We therefore assessed the volume of the third ventricle in the slices where the hypothalamic region is present as an indirect measurement to determine whether hypothalamic atrophy had taken place. This was performed in native space T1-weighted images. The third ventricle was semi-manually delineated based on its tissue-CSF borders in all slices from where the anterior commissure is present ending with the slice where the posterior commissure is first present using the MRIcro software (http://www.cabiatl.com/mricro/)(Tisserand et al., 2000). The intracranial volume (ICV) was measured with a three-step procedure using FSL (www.fmrib.ox.ac.uk/fsl/). First, a 3D box that covers the brain was used to mask out partial skull and neck regions. Second, a brain extraction tool (BET) was used to extract only the brain tissue after a manually adjusted intensity threshold to remove high-intensity areas of the eyes and low-intensity non-brain tissue. Finally, a morphological operation was used to fill the holes inside the brain with the original intensity. Normalization for ICV was performed using MATLAB version 7.9.0 (R2009b) (Tisserand et al., 2000). We then compared the mean value of the third ventricle volume between the groups of participants using a one-way ANCOVA, including age as a covariate. The statistical analyses were performed using the statistical package R (version 2.10.1, R Development Core Team, 2009) and PASW Statistics 18 (Release Version 18.0.0, SPSS, Inc., 2009).
Estimation of the classification accuracy of different brain regions
Next, we investigated whether it was possible to use information extracted from the structural MR images to discriminate between participants with different predicted time to disease onset. The rationale behind this approach was that the potential of a region to discriminate between different groups of participants should be an indication of the extent of disease-related changes occurring in this region. The normalized, segmented and smoothed images created by SPM5, i.e. the same images which were employed for the VBM, were used to train an L2-regularized logistic regression model, with each variable representing a voxel (see also supplementary methods). Regularized logistic regression can be applied to large binary classification problems with many variables. An efficient implementation is provided by the LIBLINEAR software (Fan et al., 2008, Lin et al., 2008) which together with its Python interface (http://public.procoders.net/liblinear2scipy/src/dist/) was used in this study. Before entered into the classifier, each variable was linearly scaled to the interval [−1,1], to avoid numerical problems and possibly improve classification performance. To evaluate the discrimination power of different brain regions, five regions of interest (ROIs) were extracted from the grey matter images. These were placed around the hypothalamus, the caudate and the insula, as well as in the cerebral cortex and cerebellum. The effects from the VBM appeared to be symmetric, and hence the results from the left hemisphere can be expected to parallel those that would be found in the right hemisphere. Therefore we considered only the left insula and caudate. All ROIs measured 22 mm × 16 mm × 18 mm (792 voxels), and all contained a CSF-tissue border. Furthermore, all ROIs contained voxels with a significantly altered grey matter content in the prHDnear group compared to the control group, according to the VBM analysis. In the “whole brain” region we included voxels where the value in the smoothed grey matter image exceeded 0.0001 in all participants. This yielded in total 252,682 voxels. The discriminative power of each of the ROIs as well as that of the whole brain was estimated by 5-fold cross-validation, hence leaving out a group of 29–30 participants at a time. Finally, 95% confidence intervals for the classification accuracies were calculated as described by Wilson (1927), with correction for continuity, by means of the statistical package R (Version 2.10.1, R Development Core Team, 2009).
Results
VBM analysis revealed changes in MR images from prHD participants
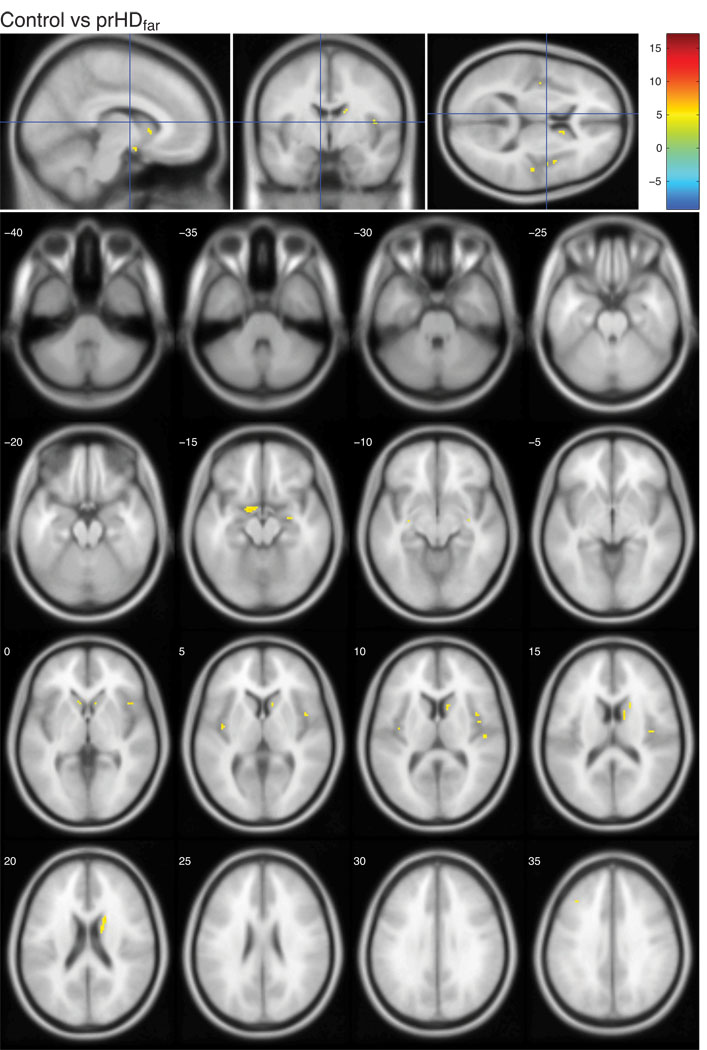
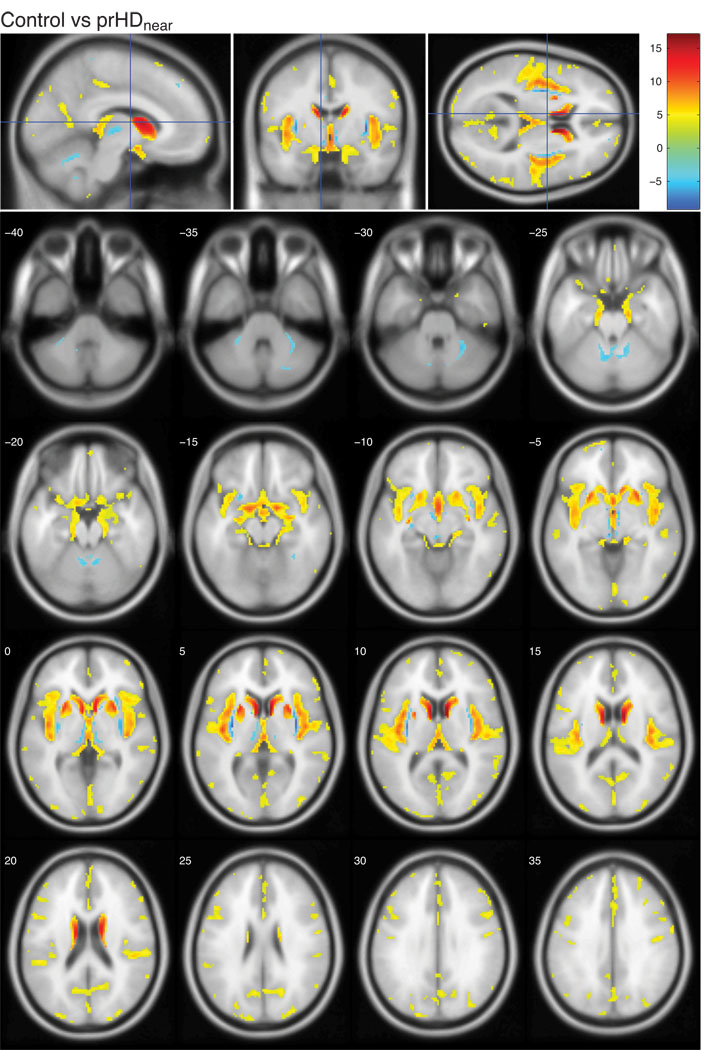
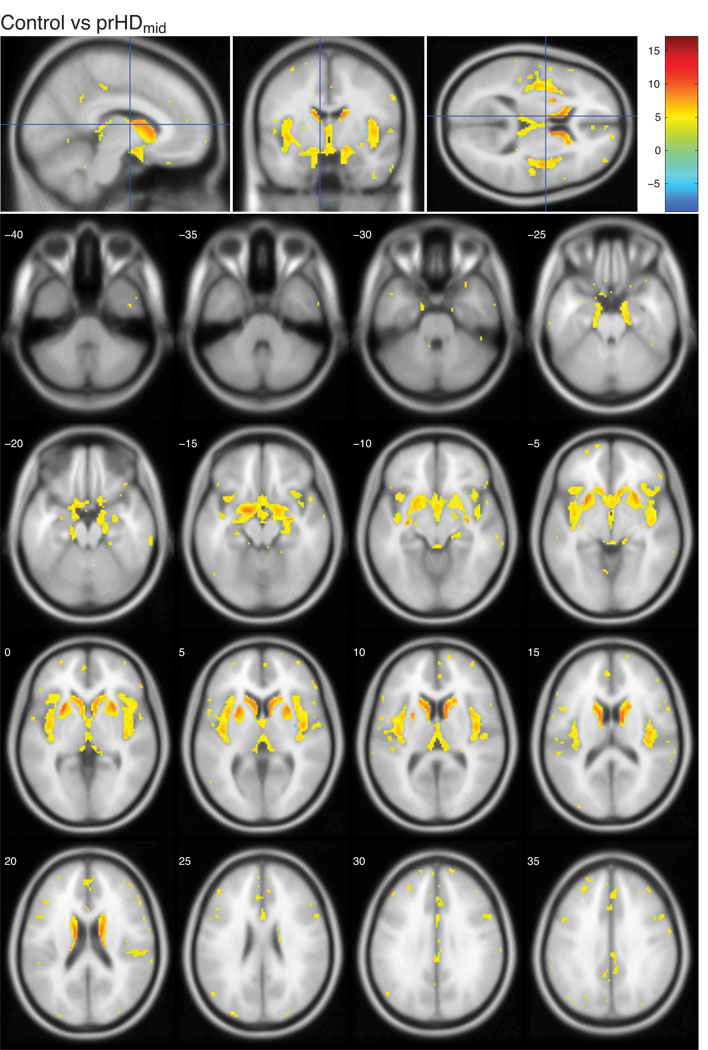
The present study was designed to investigate whether structural changes in the hypothalamic region occurred before onset of overt motor symptoms in HD using both VBM and a classification approach. It was based on T1-weighted MR images from two hundred and twenty prHD participants and seventy-five age and gender matched controls from the Predict-HD study. Prodromal HD participants were stratified into three groups based on the predicted time to clinical diagnosis (Langbehn et al., 2004); prHDfar (>15 years to predicted clinical diagnosis), prHDmid (9–15 years to predicted clinical diagnosis) and prHDnear (<9 years to predicted clinical diagnosis). We began by assessing the localization and extent of grey matter changes in the MR images using VBM. These analyses revealed regions with significant changes in local grey matter content compared to controls in all prHD groups as illustrated using statistical parametric maps (Fig. 1–3; comparisons of each prHD group to the control group are corrected for multiple comparisons using FDR and thresholded at q<0.01). In the prHDfar group, the changes were confined to a small region in the caudate (Fig. 1) while changes in the prHDmid and prHDnear groups were more widespread (Fig. 2 and 3). These changes comprised several brain regions, in particular the caudate, insula and the hypothalamic region, suggesting that these structures were all similarly affected by the disease process. In fact, 28% of the voxels in the hypothalamic region as defined by the Talairach coordinate system were significantly different from controls in the prHDnear group (Fig. 3) and 20% of the voxels were different from controls in the prHDmid group (Fig. 2). Moreover, significantly different voxel clusters were also detected in other areas such as the cerebral cortex and the cerebellum. As can be seen in Fig. 1–3, most of the significant changes found by VBM result from a decrease of grey matter in the prHD groups compared to the control group.
Figure 1. Voxel-based morphometric analysis of prHDfar and controls in the Predict-HD study.
The figures show the t-values in regions with significantly altered grey matter content in the prHDfar group compared to controls. The color bar indicates the range of t-values. Positive t-values indicate a decrease in grey matter content in prHDfar participants compared to controls, negative t-values indicate an increase in prHDfar participants compared to controls. All images are corrected for multiple comparisons using FDR and thresholded at q<0.01. Images are shown in neurological convention, overlaid on a standard MNI152 T1 template and viewed with the xjView toolbox in MATLAB (http://www.alivelearn.net/xjview8/). The top row shows the sections at MNI coordinates (−8,−4,10) mm. The following image shows the same statistical parametric map on a series of transverse slices (from MNI coordinates z = −40 to z = 35, slice thickness 5 mm).
Figure 3. Voxel-based morphometric analysis of prHDnear and controls in the Predict-HD study.
The figures show the t-values in regions with significantly altered grey matter content in the prHDnear group compared to controls. The color bar indicates the range of t-values. Positive t-values indicate a decrease in grey matter content in prHDnear participants compared to controls, negative t-values indicate an increase in prHDnear participants compared to controls. All images are corrected for multiple comparisons using FDR and thresholded at q<0.01. Images are shown in neurological convention, overlaid on a standard MNI152 T1 template and viewed with the xjView toolbox in MATLAB (http://www.alivelearn.net/xjview8/). The top row shows the sections at MNI coordinates (−8,−4, 10) mm. The following image shows the same statistical parametric map on a series of transverse slices (from MNI coordinates z = −40 to z = 35, slice thickness 5 mm).
Figure 2. Voxel-based morphometric analysis of prHDmid and controls in the Predict-HD study.
The figures show the t-values in regions with significantly altered grey matter content in the prHDmid group compared to controls. The color bar indicates the range of t-values. Positive t-values indicate a decrease in grey matter content in prHDmid participants compared to controls, negative t-values indicate an increase in prHDmid participants compared to controls. All images are corrected for multiple comparisons using FDR and thresholded at q<0.01. Images are shown in neurological convention, overlaid on a standard MNI152 T1 template and viewed with the xjView toolbox in MATLAB (http://www.alivelearn.net/xjview8/). The top row shows the sections at MNI coordinates (−8,−4,10) mm. The following image shows the same statistical parametric map on a series of transverse slices (from MNI coordinates z = −40 to z = 35, slice thickness 5 mm).
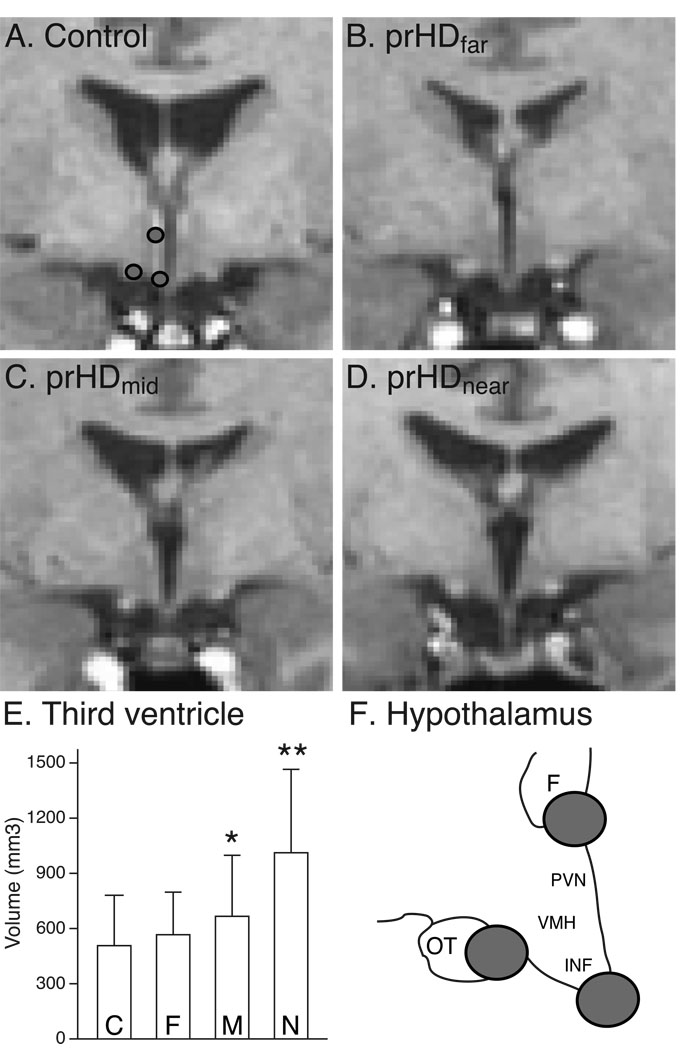
Investigations of structural changes of the hypothalamus in MR images are not trivial as no clear anatomical borders exist. Nevertheless, visual inspection of MR images demonstrated a widening of the third ventricle in at least the prHDnear and prHDmid groups (Fig. 4; compare panels C and D versus A and B). Quantitative analyses of the third ventricle volume using a one-way ANCOVA with age as a covariate showed a significant overall effect of the estimated time to onset of motor symptoms on the volume (F(3,290)=30.7, p<0.001). Including also the gender as a covariate did not significantly improve the model fit (F(1,289)=3.51, p=0.062). To localize the significant effect, we compared the adjusted means between the groups (using Bonferroni correction for multiple comparisons), and noted significant differences between the mean volume in the prHDnear group and each of the other participant groups (p<0.001), as well as between the prHDmid group and the control group (p<0.05) (Fig. 4 E).
Figure 4. Representative images of the hypothalamic region.
Representative native-space T1-weighted images from participants in the control (A), prHDfar (B), prHDmid (C) and prHDnear (D) groups at the plane where the six different points on the borders of the hypothalamic region were identified for validation of the normalization process. The locations of the three points in the left hemisphere are shown in (A) and schematically in (F). The images illustrate a widening of the third ventricle in the groups closer to estimated onset of motor symptoms which is confirmed by a volumetric analysis of the third ventricle (E). Data is presented as mean ± SD. C= control, F= prHDfar, M = prHDmid and N = prHDnear . **= p<0.001 (adjusting for age effect and using Bonferroni correction) compared to all other groups; * = p<0.05 (adjusting for age effect and using Bonferroni correction) compared to the control group; F= fornix; IFN= the infundibular nucleus; OT= optical tract; PVN= the paraventricular nucleus of the hypothalamus; VMH= the ventromedial nucleus of the hypothalamus.
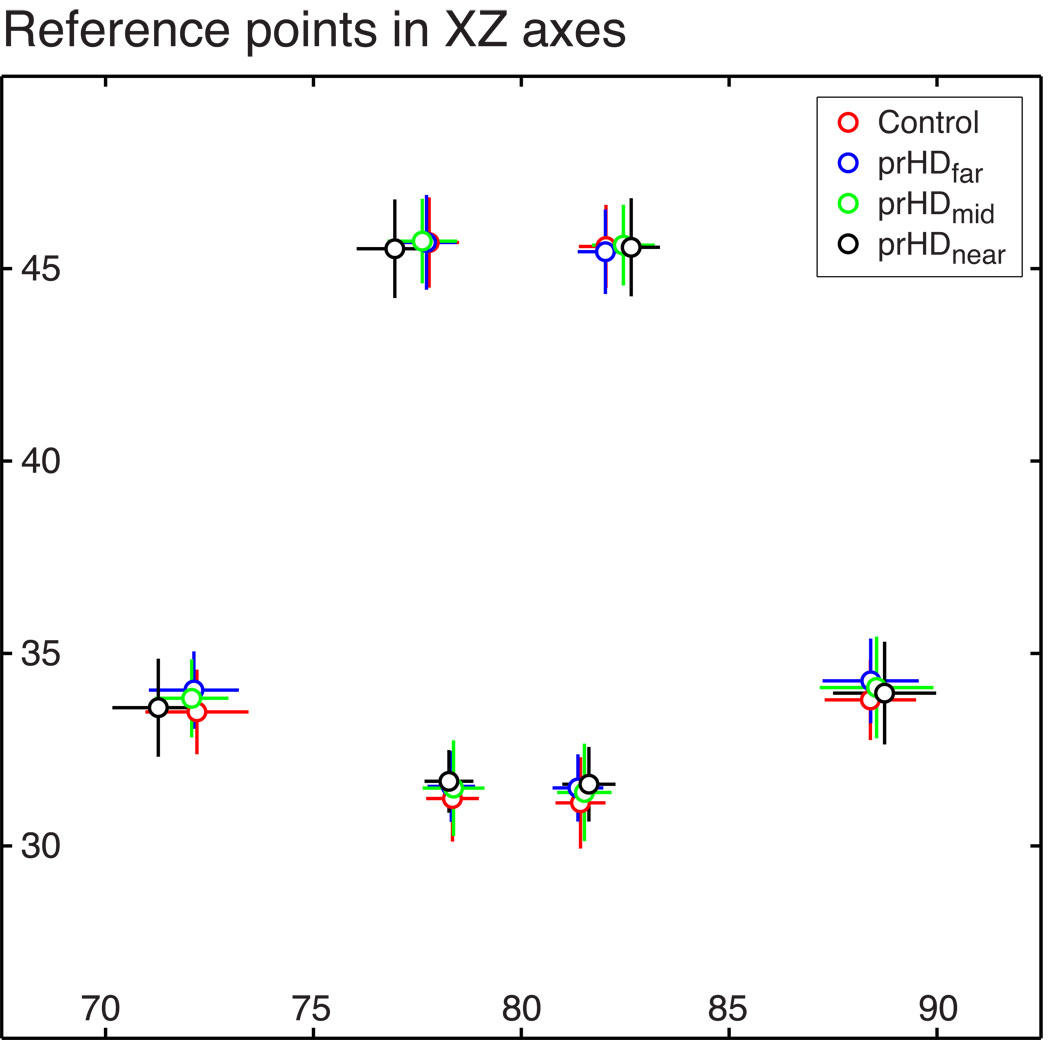
As correct interpretation of VBM results requires accurate alignment, we validated that the hypothalamic region overlapped between groups after the normalization process. For this purpose, we examined whether six points of reference on the border of the hypothalamic region in each of the native T1-weighted images indeed overlapped between the different groups after mapping to the standard template by the transformations determined by SPM5. The points were placed in the medial and ventral edge of the fornix, the medial and ventral edge of the optic tract and in the inferior and medial edge of the hypothalamus, which were landmarks that could be identified in all images in the four different groups (Fig. 4 A and F). We then compared the location of the points in the MNI space after normalization across groups and individuals and found that only minor shifts of small magnitude occurred between the groups (Fig. 5). In fact, the largest group mean difference for any point in any of the three coordinate directions was less than 1 mm, i.e. less than half the voxel size in the normalized images. The standard deviation across all participants was less than 1.25 mm, or 0.625 voxel, for each point in each of the three coordinate directions. The dorsal and lateral points in the left hemisphere appear to have been shifted systematically outward in the prHDnear group compared to the control group, which could potentially affect the VBM analysis. However, the regions of statistically significant grey matter alterations in the prHDnear group compared to the control group in the VBM analysis parallel those found when contrasting the prHDmid group and the control group. At the same time, the ventral points appeared not to have been significantly shifted in the prHDmid group compared to the control group (Fig. 5). Therefore, we expect the potential confounding effects of the shift on the VBM results in this case to be small.
Figure 5. Validation of the spatial alignment of the hypothalamic region.
The mean positions in the normalized images of the six different points of reference on the hypothalamic borders in a plane 2.7 mm posterior to the anterior commissure did not differ by more than 1 mm in any coordinate direction between any pair of participant groups, which is illustrated in a medio-lateral and dorsal-ventral plane. Data is represented as mean ± SD.
Regions of interest in the caudate, insula and hypothalamus can classify prHD
To support the interpretation that the hypothalamus, as well as the caudate and the insula are centrally involved in the disease process from early on in the course of the illness, we investigated their ability to correctly distinguish prHD participants from control participants, as well as from prHD participants with a different predicted time to clinical onset, compared to that of the whole brain. In the comparison, we also added ROIs from the cerebral cortex and the cerebellum, as these regions both showed significant changes in the VBM analysis and contained tissue-CSF borders. Hence, five ROIs of equal size in the hypothalamic region, the left caudate and the left insula, as well as the cortex and cerebellum were chosen (Fig. 6). Using an L2-regularized logistic regression model, we found that all ROIs, as well as the whole brain, discriminated well the control group from the prHDnear group (Table 2; the sensitivity and specificity for all classification analyses are shown in Supplementary Table 1). The highest classification accuracy in the comparison between the prHDnear group and controls was achieved with data from the insula (89.0% correctly classified), followed by ROIs from the caudate (88.3%), the whole brain (87.6%) and hypothalamic region (86.2%) (Table 2). In the comparison between controls and the prHDfar group, the ROIs in the insula, cerebral cortex and cerebellum failed to classify participants better than chance. Interestingly, two of the main areas identified in the VBM analysis, namely the caudate and the hypothalamic region, retained their ability to distinguish better than chance in all comparisons against the control group. They remained similar to analysis performed using the whole brain images. Although the insula performed the best when contrasting the control and the prHDnear group, its ability to distinguish the control group from the prHDfar group was not significantly different from chance. The fact that better than chance classification accuracies can be achieved when the prHDfar group is compared to controls using ROIs in the hypothalamic region and the caudate argue that changes in these regions are part of the early pathological events in HD.
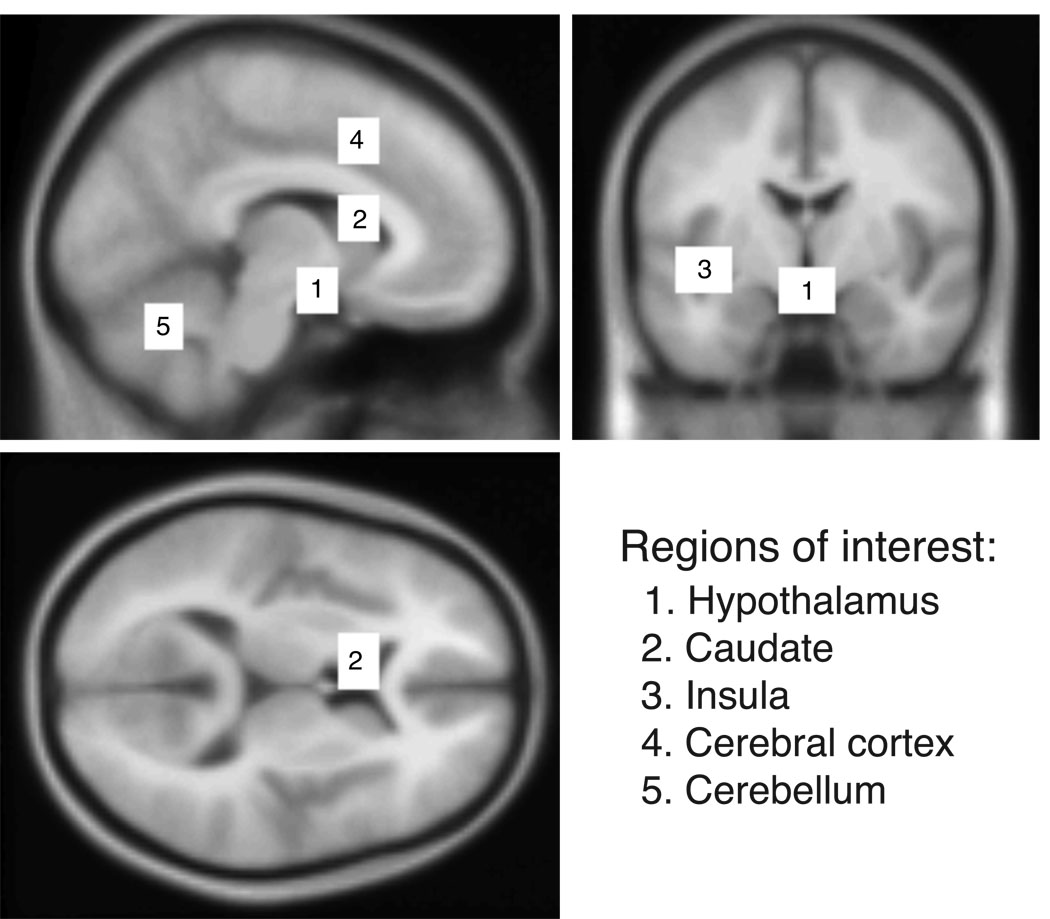
Figure 6. Region of interests used for classification analyses in prHD.
The anatomical localizations of the five equally sized regions of interests (22 mm × 16 mm × 18 mm, 792 voxels) in the hypothalamic region (1), the left caudate (2), the left insula (3), the cerebral cortex (4), and the cerebellum (5) that were used for the classification analyses.
Table 2. Classification accuracy.
Estimated classification accuracy (95% CI) of an L2-regularized logistic regression model, trained with whole brain images and regions of interests in five different brains region. Accuracies significantly different from chance (lower 95% CI limit >0.50) are indicated in bold.
| Whole brain | Hypothalamic region |
Caudate | Insula | Cortex | Cerebellum | |
|---|---|---|---|---|---|---|
|
Control vs prHDnear |
87.6 (80.8,92.3) |
86.2 (79.3,91.2) |
88.3 (81.6,92.8) |
89.0 (82.4,93.4) |
69.7 (61.4,76.9) |
64.8 (56.4,72.4) |
|
Control vs prHDmid |
72.0 (64.0,78.9) |
74.7 (66.8,81.2) |
76.7 (68.9,83.0) |
75.3 (67.5,81.8) |
64.0 (55.7,71.6) |
59.3 (51.0,67.2) |
|
Control vs prHDfar |
60.7 (52.3,68.4) |
62.7 (54.4,70.3) |
60.7 (52.3,68.4) |
52.0 (43.7,60.2) |
57.3 (49.0,65.3) |
50.7 (42.4,58.9) |
|
prHDfar vs prHDnear |
80.0 (72.4,86.0) |
78.6 (70.9,84.8) |
85.5 (78.5,90.6) |
76.6 (68.7,83.0) |
58.6 (50.1,66.6) |
62.1 (53.6,69.9) |
|
prHDmid vs prHDnear |
69.0 (60.7,76.2) |
64.8 (56.4,72.4) |
65.5 (57.1,73.1) |
59.3 (50.8,67.3) |
55.2 (46.7,63.4) |
64.8 (56.4,72.4) |
|
prHDfar vs prHDmid |
56.7 (48.3,64.7) |
62.7 (54.4,70.3) |
63.3 (55.0,70.9) |
59.3 (51.0,67.2) |
60.0 (51.7,67.8) |
46.0 (37.9,54.3) |
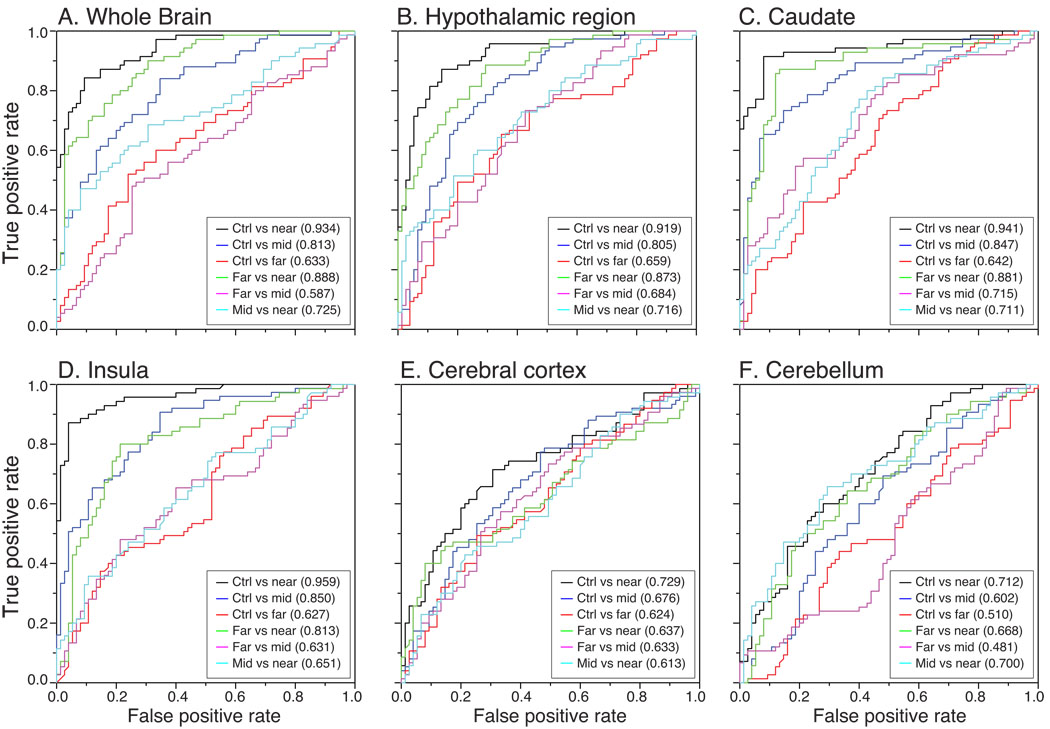
Next we investigated whether the prHD groups could be distinguished from each other using L2-regularized logistic regression, with the intention to explore if disease progression could be tracked using structural MRI. The caudate, the hypothalamic region and the insula showed a better than chance accuracy in all comparisons between prHD groups, suggesting that disease progression could be reflected in changes in MR signals in these areas (Table 2; Supplementary Table 1). Combining all five ROIs provided a slightly higher classification accuracy than the individual ROIs in all comparisons except prHDfar vs prHDnear, where the caudate region alone was superior (data not shown). The receiver operating characteristic (ROC) curves for all ROIs and whole brain images in all classifications as well as areas under the ROC curves are shown in Fig. 7. The ROC curves illustrate the relationship between true positive rates (sensitivity) and false positive rates (1-specificity) for different values of the cut-off parameter, which was set to 0.5 in our classification analyses.
Figure 7. ROC curves for the classification accuracy in prHD and control participants.
ROC curves obtained using a L2-regularized logistic regression model when contrasting controls participants to the different prHD groups, as well as when contrasting prHD groups to each other for the whole brain (A), and regions of interests in the hypothalamic region (B), the caudate nucleus (C), the insula (D), the cerebral cortex (E) and the cerebellum (F). Areas under the ROC curves are indicated in each graph. In each binary comparison, the “positive” group is considered the one closest to expected clinical onset (with the control group considered further from onset than any of the prHD groups).
Discussion
We designed the present study to investigate whether changes in the hypothalamic region were detectable in structural MR images from HD gene carriers already before the onset of overt motor symptoms. For this purpose we took advantage of a large data set which was generated as part of a unique multi-center study (Paulsen et al., 2006; 2008), which consisted of MR images from 220 HD gene carriers and 75 controls. We found that changes in the hypothalamic region were present in prHD at least a decade before predicted time of clinical diagnosis using VBM analyses. The changes in this region were in fact among the earliest features that could be detected. These findings are consistent with and extend those reported previously in early stages of clinical HD using similar techniques (Kassubek et al., 2004; Douaud et al., 2006). The alterations in the hypothalamic region paralleled changes in the caudate nucleus and the insula, and to some extent the cerebral cortex, all of which are regions known to be affected in prHD (e.g. Thieben et al., 2002; Aylward et al., 2004; Kipps et al., 2005; 2007; Rosas et al., 2005; 2006; 2008; Paulsen et al., 2006b; 2010; Henley et al., 2008; Jurgens et al., 2008; Tabrizi et al., 2009).
In the present study, we extended our analysis to investigate whether MR images could be used to discriminate prHD of different expected time to clinical diagnosis from controls as well as from each other, as a high classification accuracy of a region would indicate that the region is indeed involved in the disease process. The highest estimated classification accuracies were obtained between controls and prHDnear using data from the whole brain or ROIs in the caudate, insula or the hypothalamus (80–90%), whereas ROIs located in the cerebral cortex or the cerebellum performed less well (<70%). Overall similar results were obtained in the comparisons between the prHDfar and prHDnear groups for most regions. On the other hand, for comparisons between prHDmid and control or prHDfar groups, the estimated classification accuracy obtained using the whole brain data fell 15–24%. Although the accuracy for the caudate and hypothalamic ROIs showed a similar drop, they remained powerful regions for discrimination between any two groups across all analyses. Hence, the classification accuracy using the hypothalamic ROI followed closely the results obtained with the caudate especially when contrasting against the prHDmid group, suggesting that important changes occurred in this area during the preclinical progression of the disease. A recent study has used a similar automated method based on a support vector machine to evaluate the classification potential of prHD gene carriers against controls using MR data from a smaller group of individuals (Klöppel et al., 2009a). In that study, classification accuracies of 69% and 83% were obtained with data from the whole brain or caudate, respectively. The same group has also shown that a pattern of structural white matter changes in the putaminal region and corpus callosum measured by diffusion tensor imaging could classify 82% of prHD cases correctly (Klöppel et al., 2008). Taken together, these studies suggest that classification strategies based on structural imaging can distinguish prHD from controls.
Although VBM provides a tool to investigate whether a region is affected in different groups in an un-biased fashion, there are limitations with this method (Ridgway et al., 2008; Klöppel et al., 2009b). These limitations include the normalization process of the images, the issue of performing multiple statistical tests and difficulties in detecting multivariate relationships. Control experiments were therefore undertaken in the present study that confirmed that the coordinates of voxels on the ventral-dorsal borders of the hypothalamic region agreed well between the different groups of participants after the normalization. In fact, the largest group difference between any two points in any of the three directions was less than half the size of the voxels, which is likely to be less than the accuracy of the identification method for the reference points. Hence, the confounding effects of these shifts on the VBM results are expected to be small. Taken together, this strengthens the interpretation that the detected changes in the hypothalamic region constitute true alterations in tissue composition. However, the great variations between different studies using VBM as well as in the reporting of different parameters render the published data sometimes difficult to interpret and reproduce in all cases, and has been a source of criticism (Henley et al., 2009a). Several user-specified VBM parameters including the level of statistical correction, modulation, smoothing kernel size and software version have a significant impact on the data (recently reviewed in Klöppel et al., 2009b; Henley et al., 2009a). Guidelines on how to report VBM analyses are now available and these have been considered in our study in order to facilitate the interpretation of our data (Ridgway et al., 2008).
The search for pathological changes detected by imaging techniques before onset of motor symptoms and that track with progression is an important focus for the development of novel therapies in HD (recently reviewed in Bohanna et al., 2008; Klöppel et al., 2009b; Paulsen, 2009). Specifically, recent studies based on MRI and DTI analyses have suggested that whole brain atrophy, reduction in striatal volume, or white matter atrophy occur in prodromal HD (Aylward, 2007; Rosas et al., 2006; Henley et al., 2009b; Tabrizi et al., 2009). Although these findings are not specific for HD, they may be used to define the state of disease progression. Such markers of state including classification techniques such as the one presented in this study can potentially be used to enrich the inclusion criteria in clinical trials testing efficacy of treatments in HD. The imaging studies in prHD cases will be important also in identifying early structural and functional changes that may be involved in causing the non-motor symptoms and signs that may occur many years before motor onset. The early changes in the hypothalamic region detected in the present study might have implications towards this end. It is indeed plausible that early symptoms and signs such as depression, alterations in the circadian rhythm and metabolic dysregulation might be related to changes in the hypothalamic region. This interpretation is supported by recent observations that microglia are activated in the hypothalamus in prHD and that dopamine D2 receptor levels are reduced (Politis et al., 2008). Future studies should, therefore, investigate whether causative links exist between hypothalamic dysfunction and the development of non-motor symptoms, and may take advantage of mammalian model systems where consequences of region and cell specific expression and deletion of mutant huntingtin can be elucidated.
Supplementary Material
Acknowledgements
This work was supported by grants to the Swedish Research Council (M2006-6238 and K2009- 61X-21520-01-1 to Å.P; K2009-61P-20945-03-1 to D.K.); the Torsten och Ragnar Soderberg foundation (Å.P.); the Bagadilico network (YZ, ÅP, DK); the Crafoord Foundation (Å.P.); Jeanssons Foundations (Å.P.); The Swedish National Board of Health and Welfare (Å.P.); Åke Wiberg Foundation (Å.P.); the province of Skane state grants (Å.P.); and NeuroFortis (C.S.). The Predict-HD study is supported by grants to J.S.P. from the National Institutes of Health through the National Institute of Neurological Disorders and Stroke (grant number 40068); the National Institutes of Mental Health (grant number 01579); the Roy J. and Lucille Carver Trust; the Howard Hughes Medical Institute; the Huntington Disease Society of America; and the CHDI Foundation, Inc.
APPENDIX
PREDICT-HD Investigators, Coordinators, Motor Raters, Cognitive Raters (October/November 2008 data cut)
Henry Paulson, MD, Mackenzie Elbert, BS, Peg Nopoulos, MD, Robert Rodnitzky, MD, Ergun Uc, MD, BA, Leigh Beglinger, PhD, Kevin Duff, PhD, Vincent A. Magnotta, PhD, Stephen Cross, BA, Nicholas Doucette, BA, Andrew Juhl, BS, Jessica Schumacher, BA, Mycah Kimble, BA, Pat Ryan, MS, MA, Jessica Wood, MD, PhD, Eric Epping, MD, PhD, and Teri Thomsen, MD (University of Iowa Hospitals and Clinics, Iowa City, Iowa, USA);
David Ames, MD, Edmond Chiu, MD, Phyllis Chua, MD, Olga Yastrubetskaya, PhD, Phillip Dingjan, M.Psych., Kristy Draper D.Psych, Nellie Georgiou-Karistianis PhD, Anita Goh, D.Psych, and Angela Komiti (The University of Melbourne, Kew, Victoria, Australia);
Lynn Raymond, MD, PhD, Rachelle Dar Santos, BSc, and Joji Decolongon, MSC (University of British Columbia, Vancouver, British Columbia, Canada);
Adam Rosenblatt, MD, Christopher A. Ross, MD, PhD, Abhijit Agarwal, MBBS, MPH, Barnett Shpritz, BS, MA, OD, and Claire Welsh (Johns Hopkins University, Baltimore, Maryland, USA);
William M. Mallonee, MD, and Greg Suter, BA (Hereditary Neurological Disease Centre, Wichita, Kansas, USA);
Ali Samii, MD, Hillary Lipe, ARNP, and Kurt Weaver, PhD (University of Washington and VA Puget Sound Health Care System, Seattle, Washington, USA);
Randi Jones, PhD, Joan Harrison, RN, Carol Ingram, RN, Cathy Wood-Siverio, MS, Stewart A. Factor, DO, J. and Claudia Testa MD, PhD (Emory University School of Medicine, Atlanta, Georgia, USA);
Roger A Barker, BA, MBBS, MRCP, Sarah Mason, BSC, (Cambridge Centre for Brain Repair, Cambridge, UK);
Elizabeth McCusker, MD, Jane Griffith, RN, Bernadette Bibb, PhD, and Kylie Richardson (Westmead Hospital, Sydney, Australia);
Bernhard G. Landwehrmeyer, MD, Daniel Ecker, MD, Patrick Weydt, MD, Michael Orth MD PhD, Sigurd Süβmuth, MD RN, Katrin Barth, RN, and Sonja Trautmann, RN, (University of Ulm, Ulm, Germany);
Kimberly Quaid, PhD, Melissa Wesson, MS and Joanne Wojcieszek, MD (Indiana University School of Medicine, Indianapolis, IN);
Mark Guttman, MD, Alanna Sheinberg, BA, Adam Singer, and Janice Stober, BA, BSW (Centre for Addiction and Mental Health, University of Toronto, Markham, Ontario, Canada);
Susan Perlman, MD and Arik Johnson, PsyD (University of California, Los Angeles Medical Center, Los Angeles, California, USA);
Michael D. Geschwind, MD, PhD, Jon Gooblar, and Mira Guzijan, MA, (University of California San Francisco, California, USA);
Tom Warner, MD, PhD, Stefan Kloppel, MD, Maggie Burrows, RN, BA, Thomasin Andrews, MD, BSC, MRCP, Elisabeth Rosser, MBBS, FRCP, Sarah Tabrizi, BSC, PhD (National Hospital for Neurology and Neurosurgery, London, UK);
Anne Rosser, MD, PhD, MRCP, Jenny Naji, PhD, BSC, Kathy Price, RN and Olivia Jane Handley, PhD, BS (Cardiff University, Cardiff, Wales, UK);
Peter Como, PhD, Amy Chesire, Frederick Marshall, MD, and Mary Wodarski, BA (University of Rochester, Rochester, New York, USA);
Oksana Suchowersky, MD, FRCPC, Sarah Furtado, MD, PhD, FRCPC, Mary Lou Klimek, RN, BN, MA, (University of Calgary, Calgary, Alberta, Canada);
Diana Rosas, MD, MS, Anne Young, MD, PhD, Alex Bender and Alexandra Zaleta (Massachusetts General Hospital, Boston, Massachusetts, USA);
Peter Panegyres, MB, BS, PhD, Carmela Connor, BP, MP, DP, Elizabeth Vuletich, BSC, Mark Woodman BSC and Rachel Zombor (Neurosciences Unit, Graylands, Selby-Lemnos & Special Care Health Services, Perth, Australia);
Joel Perlmutter, MD and Stacey Barton, MSW, LCSW (Washington University, St. Louis, Missouri, USA);
Sheila A Simpson, MD, Kirsty Matheson, Alexandra Ure, BSC (Clinical Genetics Centre, Aberdeen, Scotland, UK);
David Craufurd, MD, Ruth Fullam, BSC, Rhona Macleod, RN, PhD, Andrea Sollom, MA and Elizabeth Howard, MD (University of Manchester, Manchester, UK)
Pietro Mazzoni, MD, PhD, Karen Marder, MD, MPH, Carol Moskowitz, MS, and Paula Wasserman, MA (Columbia University Medical Center, New York, New York, USA);
Lauren Seeberger, MD, Alan Diamond, DO, Diane Erickson, RN, Dawn Miracle, BS, MS, Sherrie Montellano, MA, Rajeev Kumar, MD (Colorado Neurological Institute, Englewood, Colorado, USA);
Vicki Wheelock, MD, Terry Tempkin, RNC, MSN, Joseph Marsano, Margaret Sanders, and Kathleen Baynes, PhD (University of California Davis, Sacramento, California, USA);
Joseph Jankovic, MD, Christine Hunter, RN, CCRC, William Ondo, MD, (Baylor College of Medicine, Houston, Texas, USA);
Justo Garcia de Yebenes, MD, Monica Bascunana Garde, Marta Fatas, Asuncion Martinez. (Hospital Ramón y Cajal, Madrid, Spain);
Martha Nance, MD, Dawn Radtke, RN, and David Tupper, PhD (Hennepin County Medical Center, Minneapolis, Minnesota, USA);
Wayne Martin, MD, Pamela King, BScN, RN, Marguerite Wieler, MSc, PT, and Satwinder Sran, BSC (University of Alberta, Edmonton, Alberta, Canada);
Steering Committee
Jane Paulsen, PhD, Principal Investigator, Kevin Duff, PhD, Douglas Langbehn, MD, PhD, Hans Johnson, PhD, Vince Magnotta, MD, Peg Nopoulos, MD, Ron Pierson, Janet Williams (University of Iowa Hospitals and Clinics, Iowa City, IA); Elizabeth Aylward, PhD (University of Washington and VA Puget Sound Health Care System, Seattle, WA); Kevin Biglan, MD (University of Rochester, Rochester, NY); Cheryl Erwin, JD, PhD (McGovern Center for Health, Humanities and the Human Spirit); Mark Guttman, MD (The Centre for Addiction and Mental Health, University of Toronto, Markham, ON, Canada); Michael Hayden, MD, PhD, Blair Leavitt, MD (University of British Columbia, Vancouver, BC, Canada); Marcy MacDonald, PhD (Massachusetts General Hospital); Martha Nance, MD (Hennepin County Medical Center, Minneapolis, MN); Steve Rao, PhD (Cleveland Clinic), Christopher A. Ross, MD, PhD (John Hopkins University, Baltimore, MD); Julie Stout, PhD (Indiana University, Bloomington, IN, USA and Monash University, Victoria, Australia).
Study Coordination Center
Steve Blanchard, MSHA, Ann Dudler, Machelle Henneberry, Kelsey Montross, BA, Philip O’Brien, MA, James A. Mills, MEd, MS, Chiachi Wang, MS, Christine Werling-Witkoske, William Adams, BA, Karla Anderson, BS, Jamy Schumacher and Sean Thompson, BA (University of Iowa).
Scientific Sections
Bio Markers
Blair Leavitt (Chair), MDCM, FRCPC (University of British Columbia); Stefano DiDonato, MD (Neurological Insitute “C. Besta”, Italy); Andrew Juhl, BS (University of Iowa); Wayne Mattson, PhD (VA Medical Center, Bedford, MA); Asa Petersen, MD, PhD (Lund University, Sweden), Sarah Tabrizi, PhD (National Hospital for Neurology and Neurology and Neurosurgery, London).
Cognitive
Deborah Harrington, PhD (Chair, Cognitive Science Battery Development, University of California, San Diego); Holly Westervelt, PhD (Quality Control and Training, Brown University); David Moser, PhD, Megan Smith, PhD, Stephen Cross, BA, and James Mills, MEd, MS (University of Iowa); Julie Stout PhD, Colin Campbell, BS and John Davison, PhD (Monash University); and Sarah Queller, PhD (Indiana University);
Functional Assessment
Janet Williams, PhD (Co-Chair), Lee Anna Clark, PhD, Anne Leserman, MSW LISW, Justin O’Rourke, Bradley Brossman, MA, Eunyoe Ro, MA (University of Iowa); Rebecca Ready, PhD (University of Massachusetts); Anthony Vaccarino, PhD (Ontario Cancer Biomarker Network); Sarah Farias, PhD (University of California, Davis); Noelle Carlozzi, PhD (Kessler Medical Rehabilitation Research & Education Center); and Carissa Nehl, PhD (VA Medical Center, Iowa City, IA).
Genetics
Marcy MacDonald, PhD (Co-Chair), Jim Gusella, PhD, and Rick Myers, PhD (Massachusetts General Hospital), Michael Hayden, PhD (University of British Columbia); Tom Wassink, MD (Co-Chair) and Eric Epping, MD, PhD (University of Iowa).
Imaging
Administrative
Ron Pierson PhD (Chair, University of Iowa); Steve Potkin, MD (University of California, Irvine); and Arthur Toga, PhD (University of California, Los Angeles).
Striatal
Elizabeth Aylward PhD and Kurt Weaver, PhD (Chair, Seattle Children's Research Institute); Mirza Faisal Beg, PhD (Simon Fraser University); and Michael Miller, PhD, Sarah Reading, MD and Christopher A. Ross, MD, PhD (Johns Hopkins University).
Surface Analysis
Peg Nopoulos (Chair), MD, Eric Axelson, Jeremy Bockholt and Kelsey Vitense (University of Iowa).
Shape Analysis
Christopher A. Ross (Chair), MD, PhD. (Johns Hopkins University).
DTI
Vincent Magnotta (Chair), PhD, Karl Helmer, PhD (Massachusetts General Hospital); Kelvin Lim, MD (University of Ulm, Germany); Sasumu Mori, PhD (Johns Hopkins University); Allen Song, PhD (Duke University); and Jessica Turner, PhD (University of California, Irvine).
fMRI
Steve Rao (Chair), Erik Beall, PhD, Katherine Koenig, PhD, Mark Lowe, PhD, Michael Phillips, MD, Christine Reece, BS and Jan Zimbelman, PhD, PT (Cleveland Clinic).
Motor
Kevin Biglan, MD (Chair, University of Rochester).
Psychiatric
Kevin Duff (Chair) PhD, Karen Anderson, PhD, Jess Fedorowicz, MD and Robert Robinson, MD (University of Iowa); David Craufurd, MD (Manchester University); and Eric van Duijn, MD (Leiden University Medical Center, Netherlands).
Core Sections
Statistics
Douglas Langbehn, MD, PhD (Chair), James Mills, MEd, MS and Chiachi Wang, MS (University of Iowa); David Oakes, PhD and Keith Bourgeois, BS (University of Rochester); and Kathryn Whitlock, MS (Indiana University).
Recruitment/Retention
Martha Nance, MD (Co-Chair, University of Minnesota); Stacie Vik, BA (Co-Chair), Anne Leserman, MSW, LISW, (Co-Chair), William Adams, BA, Christine Anderson, BA, Jessica Schumaker, BA, Nick Doucette, BA (University of Iowa); and Norm Reynolds, MD (University of Wisconsin, Milwaukee).
Ethics
Cheryl Erwin, JD, PhD, (Chair, McGovern Center for Health, Humanities and the Human Spirit); William Coryell, MD and Janet Williams, PhD (University of Iowa); and Martha Nance, MD (University of Minnesota).
IT/Management
Hans Johnson, PhD (Chair) Jim Smith, AS, R.J. Connell, BS, and Jeremy Bockholt, B.S. (University of Iowa) and Joseph Weber, BS (University of Rochester).
Program Management
Administrative
Chris Werling (Co-Chair), Karla Anderson, BS, Ann Dudler, Jamy Schumacher, Sean Thompson, BA (University of Iowa); and Elise Kayson (Co-Chair) and Elaine Julian-Baros, BS, CCRC (University of Rochester).
Financial
Steve Blanchard, MSHA (Co-Chair), Philip O’Brien, MA (Co-Chair), Machelle Henneberry and Kelsey Montross, BA (University of Iowa).
Clinical Trials Coordination Center
Keith Bourgeois, BS, Catherine Covert, MA, Susan Daigneault, Elaine Julian-Baros, BS, CCRC, Kay Meyers, BS, Karen Rothenburgh, Beverly Olsen, BA, Constance Orme, BA, Tori Ross, MA, Megan Simone, MOL, Joseph Weber, BS, and Hongwei Zhao, PhD (University of Rochester, Rochester, NY.)
Cognitive Coordination Center
Julie C. Stout, PhD, Sarah Queller, PhD, Shannon A. Johnson, PhD, J. Colin Campbell, BS, Eric Peters, BS, Noelle E. Carlozzi, PhD, and Terren Green, BA, Shelley N. Swain, MA, David Caughlin, BS, Bethany Ward-Bluhm, BS and Kathryn Whitlock, MS (Indiana University, Bloomington, Indiana, USA; Monash University.
Recruitment and Retention Committee
Jane Paulsen, PhD, Stacie Vik, BA, Christine Anderson, BA (University of Iowa, USA); Amy Chesire (University of Rochester, USA); Abhijit Agarwal, MBBS, MPH; Jane Griffith, RN (Westmead Hospital, AU); Katrin Barth and Sonja Trautman (University of Ulm, GE); Mira Guzijan, MA (University of California- San Francisco, USA); Jenny Naji, PhD (Cardiff University, UK); Elaine Julian-Baros, BS, CCRC, Elise Kayson, MS, RNC (University of Rochester, USA); Norm Reynolds, and Julie Stout, PhD (Indiana University, Bloomington, IN, USA and Monash University, Victoria, Australia).
Event Monitoring Committee
Jane Paulsen, PhD, William Coryell, MD (University of Iowa, USA); Julie Stout, PhD (Indiana University and Monash University, Australia); Cheryl Erwin, JD, PhD (McGovern Center for Health, Humanities and the Human Spirit, USA); and Steven Hersch, MD, PhD (Massachusetts University).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Members of “the Huntington study group PREDICT-HD investigators” are listed as an Appendix.
References
- Arnulf I, Nielsen J, Lohmann E, Schiefer J, Wild E, Jennum P, Konofal E, Walker M, Oudiette D, Tabrizi S, Durr A. Rapid eye movement sleep disturbances in Huntington disease. Arch. Neurol. 2008;65:482–488. doi: 10.1001/archneur.65.4.482. [DOI] [PubMed] [Google Scholar]
- Ashburner J, Friston KJ. Voxel-based morphometry--the methods. Neuroimage. 2000;11:805–821. doi: 10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]
- Ashburner J, Friston KJ. Unified segmentation. Neuroimage. 2005;26:839–851. doi: 10.1016/j.neuroimage.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Aylward EH, Sparks BF, Field KM, Yallapragada V, Shpritz BD, Rosenblatt A, Brandt J, Gourley LM, Liang K, Zhou H, Margolis RL, Ross CA. Onset and rate of striatal atrophy in preclinical Huntington disease. Neurology. 2004;63:66–72. doi: 10.1212/01.wnl.0000132965.14653.d1. [DOI] [PubMed] [Google Scholar]
- Aylward EH. Change in MRI striatal volumes as a biomarker in preclinical Huntington's disease. Brain Res. Bull. 2007;72:152–158. doi: 10.1016/j.brainresbull.2006.10.028. [DOI] [PubMed] [Google Scholar]
- Aziz A, Fronczek R, Maat-Schieman M, Unmehopa U, Roelandse F, Overeem S, van Duinen S, Lammers GJ, Swaab D, Roos R. Hypocretin and melanin-concentrating hormone in patients with Huntington disease. Brain Pathol. 2008;18:474–483. doi: 10.1111/j.1750-3639.2008.00135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz NA, Pijl H, Frölich M, van der Graaf AW, Roelfsema F, Roos RA. Increased hypothalamic-pituitary-adrenal axis activity in Huntington's disease. J. Clin. Endocrinol. Metab. 2009;94:1223–1228. doi: 10.1210/jc.2008-2543. [DOI] [PubMed] [Google Scholar]
- Björkqvist M, Petersen A, Bacos K, Isaacs J, Norlen P, Gil J, Popovic N, Sundler F, Bates GP, Tabrizi SJ, Brundin P, Mulder H. Progressive alterations in the hypothalamic-pituitary-adrenal axis in the R6/2 transgenic mouse model of Huntington's disease. Hum. Mol. Genet. 2006;15:1713–1721. doi: 10.1093/hmg/ddl094. [DOI] [PubMed] [Google Scholar]
- Bohanna I, Georgiou-Karistianis N, Hannan AJ, Egan GF. Magnetic resonance imaging as an approach towards identifying neuropathological biomarkers for Huntington's disease. Brain Res. Rev. 2008;58:209–225. doi: 10.1016/j.brainresrev.2008.04.001. [DOI] [PubMed] [Google Scholar]
- Douaud G, Gaura V, Ribeiro MJ, Lethimonnier F, Maroy R, Verny C, Krystkowiak P, Damier P, Bachoud-Levi AC, Hantraye P, Remy P. Distribution of grey matter atrophy in Huntington's disease patients: a combined ROI-based and voxel-based morphometric study. Neuroimage. 2006;32:1562–1575. doi: 10.1016/j.neuroimage.2006.05.057. [DOI] [PubMed] [Google Scholar]
- Duff K, Paulsen JS, Beglinger LJ, Langbehn DR, Stout JC Predict-HD Investigators of the Huntington Study Group. Psychiatric symptoms in Huntington's disease before diagnosis: the predict-HD study. Biol. Psychiatry. 2007;62:1341–1346. doi: 10.1016/j.biopsych.2006.11.034. [DOI] [PubMed] [Google Scholar]
- Fan RE, Chang KW, Hsieh CJ, Wang XR, Lin CJ. LIBLINEAR: A library for large linear classification. Journal of Machine Learning Research. 2008;9:1871–1874. Software available at http://www.csie.ntu.edu.tw/~cjlin/liblinear. [Google Scholar]
- Henley SM, Wild EJ, Hobbs NZ, Warren JD, Frost C, Scahill RI, Ridgway GR, MacManus DG, Barker RA, Fox NC, Tabrizi SJ. Defective emotion recognition in early HD is neuropsychologically and anatomically generic. Neuropsychologia. 2008;46:2152–2160. doi: 10.1016/j.neuropsychologia.2008.02.025. [DOI] [PubMed] [Google Scholar]
- Henley SM, Ridgway GR, Scahill RI, Klöppel S, Tabrizi SJ, Fox NC, Kassubek J for the EHDN Imaging Working Group. Pitfalls in the Use of Voxel-Based Morphometry as a Biomarker: Examples from Huntington Disease. Am. J. Neuroradiol. 2009a doi: 10.3174/ajnr.A1939. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henley SM, Wild EJ, Hobbs NZ, Frost C, MacManus DG, Barker RA, Fox NC, Tabrizi SJ. Whole-brain atrophy as a measure of progression in premanifest and early Huntington's disease. Mov. Disord. 2009b;24:932–936. doi: 10.1002/mds.22485. [DOI] [PubMed] [Google Scholar]
- Huntington's Disease Collaborative Research Group. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- Julien CL, Thompson JC, Wild S, Yardumian P, Snowden JS, Turner G, Craufurd D. Psychiatric disorders in preclinical Huntington's disease. J. Neurol. Neurosurg. Psychiatry. 2007;78:939–943. doi: 10.1136/jnnp.2006.103309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurgens CK, van de Wiel L, van Es AC, Grimbergen YM, Witjes-Ané MN, van der Grond J, Middelkoop HA, Roos RA. Basal ganglia volume and clinical correlates in 'preclinical' Huntington's disease. J. Neurol. 2008;255:1785–1791. doi: 10.1007/s00415-008-0050-4. [DOI] [PubMed] [Google Scholar]
- Kassubek J, Juengling FD, Kioschies T, Henkel K, Karitzky J, Kramer B, Ecker D, Andrich J, Saft C, Kraus P, Aschoff AJ, Ludolph AC, Landwehrmeyer GB. Topography of cerebral atrophy in early Huntington's disease: a voxel based morphometric MRI study. J. Neurol. Neurosurg. Psychiatry. 2004;75:213–220. [PMC free article] [PubMed] [Google Scholar]
- Kipps CM, Duggins AJ, Mahant N, Gomes L, Ashburner J, McCusker EA. Progression of structural neuropathology in preclinical Huntington's disease: a tensor based morphometry study. J. Neurol. Neurosurg. Psychiatry. 2005;76:650–655. doi: 10.1136/jnnp.2004.047993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipps CM, Duggins AJ, McCusker EA, Calder AJ. Disgust and happiness recognition correlate with anteroventral insula and amygdala volume respectively in preclinical Huntington's disease. J. Cogn. Neurosci. 2007;19:1206–1217. doi: 10.1162/jocn.2007.19.7.1206. [DOI] [PubMed] [Google Scholar]
- Klöppel S, Draganski B, Golding CV, Chu C, Nagy Z, Cook PA, Hicks SL, Kennard C, Alexander DC, Parker GJ, Tabrizi SJ, Frackowiak RS. White matter connections reflect changes in voluntary-guided saccades in pre-symptomatic Huntington's disease. Brain. 2008;131:196–204. doi: 10.1093/brain/awm275. [DOI] [PubMed] [Google Scholar]
- Klöppel S, Chu C, Tan GC, Draganski B, Johnson H, Paulsen JS, Kienzle W, Tabrizi SJ, Ashburner J, Frackowiak RS PREDICT-HD Investigators of the Huntington Study Group. Automatic detection of preclinical neurodegeneration: presymptomatic Huntington disease. Neurology. 2009a;72:426–431. doi: 10.1212/01.wnl.0000341768.28646.b6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klöppel S, Henley S, Hobbs N, Wolf RC, Kassubek J, Tabrizi SJ, Frackowiak RS. Magnetic resonance imaging of Huntington's Disease: Preparing for clinical trials. Neuroscience. 2009b;164:205–219. doi: 10.1016/j.neuroscience.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langbehn DR, Brinkman RR, Falush D, Paulsen JS, Hayden MR International Huntington's Disease Collaborative Group. A new model for prediction of the age of onset and penetrance for Huntington's disease based on CAG length. Clin. Genet. 2004;65:267–277. doi: 10.1111/j.1399-0004.2004.00241.x. [DOI] [PubMed] [Google Scholar]
- Lin CJ, Weng RC, Keerthi S. Trust region newton method for large-scale logistic regression. Journal of Machine Learning Research. 2008;9:627–650. [Google Scholar]
- Mai JK, Paxinos G, Voss T. Atlas of the human brain. third edition. Academic Press; 2007. [Google Scholar]
- Mochel F, Charles P, Seguin F, Barritault J, Coussieu C, Perin L, Le Bouc Y, Gervais C, Carcelain G, Vassault A, Feingold J, Rabier D, Durr A. Early energy deficit in Huntington disease: identification of a plasma biomarker traceable during disease progression. PLoS ONE. 2007;2:e647. doi: 10.1371/journal.pone.0000647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton AJ, Wood NI, Hastings MH, Hurelbrink C, Barker RA, Maywood ES. Disintegration of the sleep-wake cycle and circadian timing in Huntington's disease. J. Neurosci. 2005;25:157–163. doi: 10.1523/JNEUROSCI.3842-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen JS, Hayden M, Stout JC, Langbehn DR, Aylward E, Ross CA, Guttman M, Nance M, Kieburtz K, Oakes D, Shoulson I, Kayson E, Johnson S, Penziner E Predict-HD Investigators of the Huntington Study Group. Preparing for preventive clinical trials: the Predict-HD study. Arch. Neurol. 2006a;63:883–890. doi: 10.1001/archneur.63.6.883. [DOI] [PubMed] [Google Scholar]
- Paulsen JS, Magnotta VA, Mikos AE, Paulson HL, Penziner E, Andreasen NC, Nopoulos PC. Brain structure in preclinical Huntington's disease. Biol Psychiatry. 2006b;59:57–63. doi: 10.1016/j.biopsych.2005.06.003. [DOI] [PubMed] [Google Scholar]
- Paulsen JS, Langbehn DR, Stout JC, Aylward E, Ross CA, Nance M, Guttman M, Johnson S, MacDonald M, Beglinger LJ, Duff K, Kayson E, Biglan K, Shoulson I, Oakes D, Hayden MR Predict-HD Investigators and Coordinators of the Huntington Study Group. Detection of Huntington's disease decades before diagnosis: the Predict-HD study. J. Neurol. Neurosurg. Psychiatry. 2008;79:874–880. doi: 10.1136/jnnp.2007.128728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen JS. Functional imaging in Huntington's disease. Exp. Neurol. 2009;216:272–277. doi: 10.1016/j.expneurol.2008.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen JS, Nopoulos PC, Aylward E, Ross CA, Johnson H, Magnotta VA, Juhl A, Pierson RK, Mills J, Langbehn D, Nance M PREDICT-HD Investigators and Coordinators of the Huntington's Study Group (HSG) Striatal and white matter predictors of estimated diagnosis for Huntington disease. Brain Res. Bull. 2010;82:201–207. doi: 10.1016/j.brainresbull.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersén A, Gil J, Maat-Schieman ML, Björkqvist M, Tanila H, Araújo IM, Smith R, Popovic N, Wierup N, Norlén P, Li JY, Roos RA, Sundler F, Mulder H, Brundin P. Orexin loss in Huntington's disease. Hum. Mol. Genet. 2005;14:39–47. doi: 10.1093/hmg/ddi004. [DOI] [PubMed] [Google Scholar]
- Petersén A, Hult S, Kirik D. Huntington disease-new perspectives based on neuroendocrine changes in rodent models. Neurodegen. Dis. 2009;6:54–64. doi: 10.1159/000225377. [DOI] [PubMed] [Google Scholar]
- Phillips W, Shannon KM, Barker RA. The current clinical management of Huntington's disease. Mov. Disord. 2008;23:1491–1504. doi: 10.1002/mds.21971. [DOI] [PubMed] [Google Scholar]
- Politis M, Pavese N, Tai YF, Tabrizi SJ, Barker RA, Piccini P. Hypothalamic involvement in Huntington's disease: an in vivo PET study. Brain. 2008;131:2860–2869. doi: 10.1093/brain/awn244. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2009. ISBN 3-900051-07-0. http://www.R-project.org. [Google Scholar]
- Ridgway GR, Henley SM, Rohrer JD, Scahill RI, Warren JD, Fox NC. Ten simple rules for reporting voxel-based morphometry studies. Neuroimage. 2008;40:1429–1435. doi: 10.1016/j.neuroimage.2008.01.003. [DOI] [PubMed] [Google Scholar]
- Ridgway GR, Omar R, Ourselin S, Hill DL, Warren JD, Fox NC. Issues with threshold masking in voxel-based morphometry of atrophied brains. Neuroimage. 2009;44:99–111. doi: 10.1016/j.neuroimage.2008.08.045. [DOI] [PubMed] [Google Scholar]
- Rosas HD, Hevelone ND, Zaleta AK, Greve DN, Salat DH, Fischl B. Regional cortical thinning in preclinical Huntington disease and its relationship to cognition. Neurology. 2005;65:745–747. doi: 10.1212/01.wnl.0000174432.87383.87. [DOI] [PubMed] [Google Scholar]
- Rosas HD, Tuch DS, Hevelone ND, Zaleta AK, Vangel M, Hersch SM, Salat DH. Diffusion tensor imaging in presymptomatic and early Huntington's disease: Selective white matter pathology and its relationship to clinical measures. Mov. Disord. 2006;21:1317–1325. doi: 10.1002/mds.20979. [DOI] [PubMed] [Google Scholar]
- Rosas HD, Salat DH, Lee SY, Zaleta AK, Pappu V, Fischl B, Greve D, Hevelone N, Hersch SM. Cerebral cortex and the clinical expression of Huntington's disease: complexity and heterogeneity. Brain. 2008;131:1057–1068. doi: 10.1093/brain/awn025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh N, Moutereau S, Durr A, Krystkowiak P, Azulay JP, Tranchant C, Broussolle E, Morin F, Bachoud-Lévi AC, Maison P. Neuroendocrine disturbances in Huntington's disease. PLoS ONE. 2009;4:e4962. doi: 10.1371/journal.pone.0004962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawiak SJ, Wood NI, Williams GB, Morton AJ, Carpenter TA. Voxel-based morphometry in the R6/2 transgenic mouse reveals differences between genotypes not seen with manual 2D morphometry. Neurobiol Dis. 2009;33:20–27. doi: 10.1016/j.nbd.2008.09.016. [DOI] [PubMed] [Google Scholar]
- Tabrizi SJ, Langbehn DR, Leavitt BR, Roos RA, Durr A, Craufurd D, Kennard C, Hicks SL, Fox NC, Scahill RI, Borowsky B, Tobin AJ, Rosas HD, Johnson H, Reilmann R, Landwehrmeyer B, Stout JC TRACK-HD investigators. Biological and clinical manifestations of Huntington's disease in the longitudinal TRACK-HD study: cross-sectional analysis of baseline data. Lancet Neurol. 2009;8:791–801. doi: 10.1016/S1474-4422(09)70170-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thieben MJ, Duggins AJ, Good CD, Gomes L, Mahant N, Richards F, McCusker E, Frackowiak RS. The distribution of structural neuropathology in pre-clinical Huntington's disease. Brain. 2002;125:1815–1828. doi: 10.1093/brain/awf179. [DOI] [PubMed] [Google Scholar]
- Tisserand DJ, Visser PJ, van Boxtel MPJ, Jolles J. The relation between global and limbic brain volumes on MRI and cognitive performance in healthy individuals across the age range. Neurobiol. Aging. 2000;21:569–576. doi: 10.1016/s0197-4580(00)00133-0. [DOI] [PubMed] [Google Scholar]
- Underwood BR, Broadhurst D, Dunn WB, Ellis DI, Michell AW, Vacher C, Mosedale DE, Kell DB, Barker RA, Grainger DJ, Rubinsztein DC. Huntington disease patients and transgenic mice have similar pro-catabolic serum metabolite profiles. Brain. 2006;129:877–886. doi: 10.1093/brain/awl027. [DOI] [PubMed] [Google Scholar]
- van Duijn E, Kingma EM, van der Mast RC. Psychopathology in verified Huntington's disease gene carriers. J. Neuropsychiatry. Clin. Neurosci. 2007;19:441–448. doi: 10.1176/jnp.2007.19.4.441. [DOI] [PubMed] [Google Scholar]
- Videnovic A, Leurgans S, Fan W, Jaglin J, Shannon KM. Daytime somnolence and nocturnal sleep disturbances in Huntington disease. Parkinsonism Relat. Disord. 2009;15:471–474. doi: 10.1016/j.parkreldis.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Wilson EB. Probable inference, the law of succession, and statistical inference. Journal of the American Statistical Association. 1927;158:209–212. [Google Scholar]
- Wolf RC, Vasic N, Schönfeldt-Lecuona C, Landwehrmeyer GB, Ecker D. Dorsolateral prefrontal cortex dysfunction in presymptomatic Huntington's disease: evidence from event-related fMRI. Brain. 2007;130:2845–2857. doi: 10.1093/brain/awm210. [DOI] [PubMed] [Google Scholar]
- Wright IC, McGuire PK, Poline JB, Travere JM, Murray RM, Frith CD, Frackowiak RS, Friston KJ. A voxel-based method for the statistical analysis of gray and white matter density applied to schizophrenia. NeuroImage. 1995;2:244–252. doi: 10.1006/nimg.1995.1032. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.