Abstract
The short-chain fatty acid sodium butyrate (NaB), produced in the colonic lumen, induces cell cycle arrest, differentiation and/or apoptosis in colorectal carcinoma cells in vitro, establishing a potential role for NaB in colon cancer prevention. We have previously shown that butyrate decreases cyclin D1 and c-myc expression, essential for intestinal tumor development, by transcriptional attenuation. Here we determined that butyrate induced transcriptional attenuation at the cyclin D1 and c-myc genes in SW837 human colorectal adenocarcinoma cells occurs at ~100 nucleotides downstream of the transcription start site, with a similar positioning in Caco-2 cells. A concomitant decrease of RNA polymerase II occupancy at the 5′ end of each gene was observed. Since transcriptional regulation is associated with chromatin remodeling, we investigated by chromatin immunoprecipitation whether butyrate HDAC inhibitor activity altered chromatin structure at the attenuated loci. Although distributions of histone H3 trimethylated on K4 and K36 along the cyclin D1 and c-myc genes were consistent with current models, butyrate induced only modest decreases in these modifications, with a similar effect on acetylated H3, and a modest increase in H3me3K27. Finally, transcriptome analysis using novel microarrays demonstrated that butyrate-induced attenuation is widespread throughout the genome, likely independent of transcriptional initiation. We identified 42 loci potentially paused by butyrate, and showed that transcription patterns are gene-specific. The biological functions of these loci encompass a number of effects of butyrate on intestinal epithelial cell physiology.
Keywords: colon cancer, sodium butyrate, gene expression, chromatin remodeling
INTRODUCTION
The short-chain fatty acid (SCFA) butyrate is produced in the colon by fermentation of dietary fiber, which generates levels of ~20 mM in the colonic lumen, well above the 2–5 mM that induces cell cycle arrest and differentiation in culture (1–3). Colonic epithelial cells in vivo utilize these high levels as their principal energy source (4), and efficient metabolism of SCFAs is necessary for their induction of colon cell maturation (5, 6). Gene expression profiling demonstrated that in culture, butyrate altered steady state level of ~7% of the genes interrogated, either increasing or decreasing in expression monotonically with time (7). This implied an orderly cascade of events, with successive stages in cell maturation driven by sequential recruitment of genes and pathways, a hallmark of an integrated response necessary for intestinal tissue homeostasis.
c-myc and cyclin D1 are genes regulated by butyrate during colonic cell maturation. Cyclin D1 is a driving component of the cell division cycle, and c-myc stimulates cell cycling and is important in differentiation as cells migrate from the crypt towards the intestinal lumen (8). Moreover, both genes are direct targets of Wnt signaling (9, 10), a fundamental pathway deregulated in colon tumors, most commonly by mutation of Apc, which alters transcription mediated by β-catenin-Tcf targeting the cyclin D1, c-myc, and other genes (11). Targeted inactivation of either c-myc or cyclin D1 reduces Apc-initiated intestinal tumor formation, demonstrating that both are essential for tumorigenesis (12, 13). Thus, butyrate-mediated decreased expression of cyclin D1 and c-myc may be fundamental to its chemopreventive activity (14, 15).
Transcriptional attenuation or pausing, first recognized for c-fos, c-myb, adenosine deaminase, tubulin, and cyclooxygenase-2 (16–20), is now understood to be fundamental for the regulation of many inducible as well as constitutively expressed genes (21, 22). In colorectal carcinoma cells, a transcriptional block in response to butyrate was demonstrated by Heruth and co-workers (14) for c-myc, and further explored by us (23). Cyclin D1 transcriptional downregulation, however, was reported associated with transcription factors acting at the promoter in response to HDAC inhibitors and other agents (15, 24). We used imaging of growing transcripts at their site of synthesis to demonstrate that cyclin D1 was also transcriptionally paused by butyrate downstream of the transcription start site (TSS; (25)). Thus, transcriptional pausing of c-myc and cyclin D1 by butyrate can play a major role in determining steady state mRNA levels (23, 25).
Transcriptional regulation is associated with chromatin landmarks, such as specific histone modifications (reviewed in (26)). In this regard, butyrate is an inhibitor of histone deacetylase activity (HDACi), which leads to histone hyperacetylation and chromatin remodeling of induced target genes, while histone hypoacetylation has been associated with transcriptionally silent chromatin (27, 28). Here, we first determined the position near the 5′ end at which transcriptional pausing of c-myc and cyclin D1 is initiated by butyrate, and chromatin modification along the length of the gene coding regions. We then extended the investigation to a transcriptome level using novel microarrays, identifying a subset of sequences for which butyrate induced transcriptional pausing. This regulation by butyrate was highly complex and gene-specific, and the data suggest that the attenuation mechanism is independent of mechanisms that determine frequency of transcriptional initiation.
MATERIALS AND METHODS
Cells and butyrate treatment
Human colorectal adenocarcinoma SW837 cells (CCL-235) and Caco-2 cells (HTB-37) were grown as described (23, 29), and monitored for mycoplasma contamination (Mycoplasma Plus PCR Primer Set, Agilent Technologies). Exponentially growing cells at ~70% confluency were treated or not with 5 mM (SW837 cells) or 2 mM (Caco-2 cells) sodium butyrate (NaB, Sigma) for different time periods. A lower concentration of NaB was used in Caco-2 cells, as these cells are more sensitive to butyrate induction of cell death.
RNA and probe preparation
RNA was isolated from cells treated or not with NaB for different time periods.. Double-stranded cDNA for microarrays was synthesized from RNA as described (NimbleGen), using SuperScript cDNA synthesis kit (Invitrogen). Details are in Supplementary Methods.
Custom-designed oligonucleotide microarrays
cDNA gene expression arrays were designed and manufactured in collaboration with NimbleGen. Two oligonucleotide arrays were used: “5′ 3′ array” and “Tiling” array. Details are in Supplementary Methods.
Histone purification and Western blot analysis
Western blot analysis and isolation of acid soluble histones were performed as described (7, 30). Details are in Supplementary Methods.
Chromatin immunoprecipitation (ChIP)
To study Polymerase II occupancy and histone modification at a specific sequence, ChIP assay was performed as described in detail in Supplementary Methods.
Quantitative real-time PCR
cDNA from butyrate-treated and untreated cells, as well as DNA isolated by chromatin immunoprecipitation (ChIP) were analyzed by qRT-PCR using SybrGreen (Applied Biosystems), following the manufacturer’s protocol, as described in Supplementary Methods.
Analysis of gene ontology and chromosome distribution
Each sequence potentially paused by butyrate based on the 5′ 3′ array data was classified according to its gene ontology, by “DAVID Bioinformatics Resources 2008, NIAID/NIH”, as described in Supplementary Methods.
Statistical analysis
The mean value of replicates was analyzed using Student’s t-test. One-way ANOVA, and Bonferroni multiple comparison test, were employed to analyze data from the “Tiling” array. Results were considered significant when p<0.05.
RESULTS
Transcription of the cyclin D1 and c-myc genes
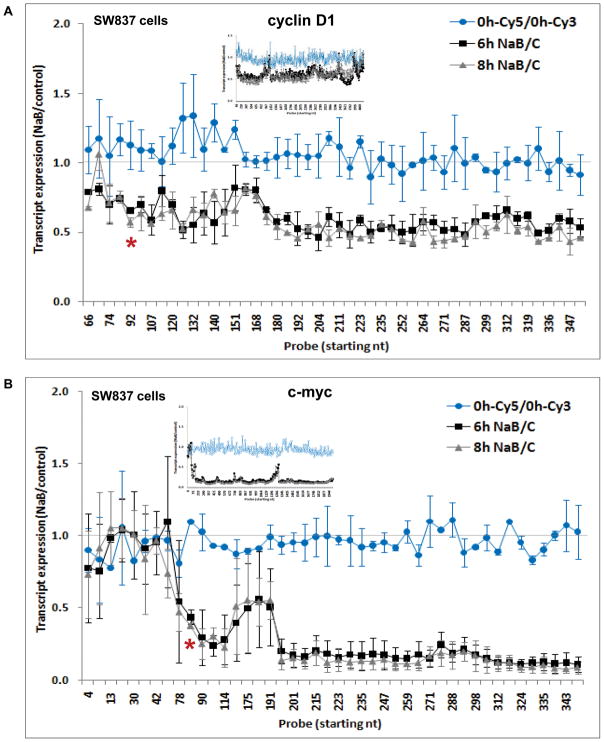
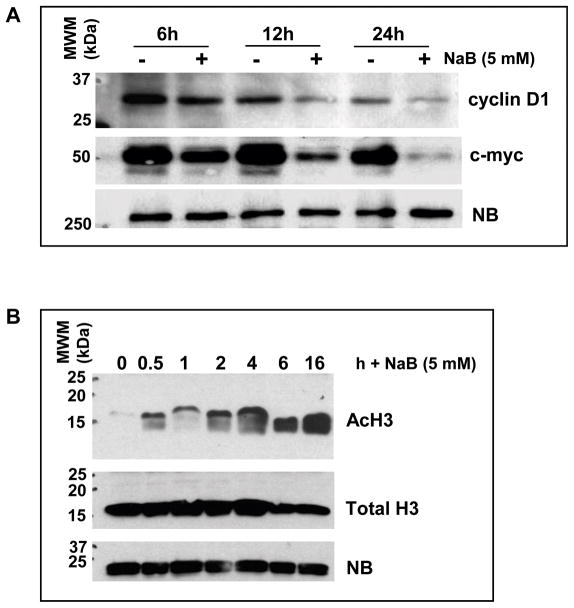
We first analyzed effects of sodium butyrate (NaB) on transcription of cyclin D1 and c-myc in human colorectal adenocarcinoma SW837 cells using a custom-designed oligonucleotide array that tiled-through the length of these sequences (Methods). These arrays were hybridized to Cy3 (untreated, cycling cells) or Cy5 (butyrate-treated cells) labeled cDNA transcribed from total, nuclear or cytoplasmic RNA. Cells were treated with 5 mM NaB for 0, 6 or 8 hrs, time points which preceded the G1 arrest and apoptosis induced by NaB at ~24 hrs (not shown). Figure 1 focuses on expression level across the first 355 nt of each sequence, shown as the average ratio of three independent experiments, and the insets show the expression throughout the entire coding regions. As expected, the ratio of hybridization of probes from untreated cells labeled with either fluorochrome was approximately 1 along each sequence (Figure 1). For cyclin D1, 6 or 8 hrs of butyrate treatment decreased its expression ~50%, beginning at nucleotide 92 (Figure 1A). There was a similar pattern for c-myc, with expression decreased 80–90% downstream of nt 84 (Figure 1B). The repressed levels of both genes generally persisted throughout the length of each sequence (Figure 1, insets). A normalization step was performed for each of the 628 and 347 probes for cyclin D1 and c-myc, respectively (see Methods). ANOVA for the NaB-treated data sets (after normalization) vs untreated cells (0 h), followed by Bonferroni multiple comparison test, showed a highly significant difference at both 6 and 8 hrs (p<0.0001), but no significant difference between 0 h-Cy3 and 0 h-Cy5. We further investigated NaB effects on cyclin D1 and c-myc transcription on another human colorectal adenocarcinoma cell line, Caco-2, and similar results were obtained. For cyclin D1, the decrease in transcript expression was again localized at nucleotide 107 (Supplementary Figure 1A). For c-myc, the decrease was somewhat further downstream, at nucleotide 186 (Supplementary Figure 1B), but still well within the 5′ UTR of the message, thus abrogating the generation of a full-length coding mRNA in Caco-2 cells, as it does in SW837 cells. In both cell lines, there were highly reproducible spikes in the generally consistent pattern of repression along both sequences, which could be related to differential hybridization of some probes on the array, despite precautions taken in designing the arrays (e.g.: insets in Figure 1, and Figure 5B). By Western blot, we confirmed that cyclin D1 and c-myc expression was ultimately reduced by NaB at the protein level, between 6 and 24 hrs of treatment (Figure 2A).
Figure 1. Cyclin D1 and c-myc transcript expression in response to butyrate.
SW837 cells were treated for 0, 6 and 8 hrs with 5 mM sodium butyrate (NaB), and cDNA was synthesized and hybridized to a “Tiling” microarray (Methods). The ratio of NaB-treated (Cy5-labeled) to untreated (Cy3-labeled) cells at each time point of treatment is shown for the first 355 nt of cyclin D1 (A) and c-myc (B) sequences, as the mean ± SD from three independent experiments. On the abscissa, the probe numbers indicate the first nucleotide of the probe. Inset: ratio of NaB-treated to untreated cells for every probe on the array for each sequence (color-coded as in the main figure). *: position where transcriptional attenuation was induced by NaB.
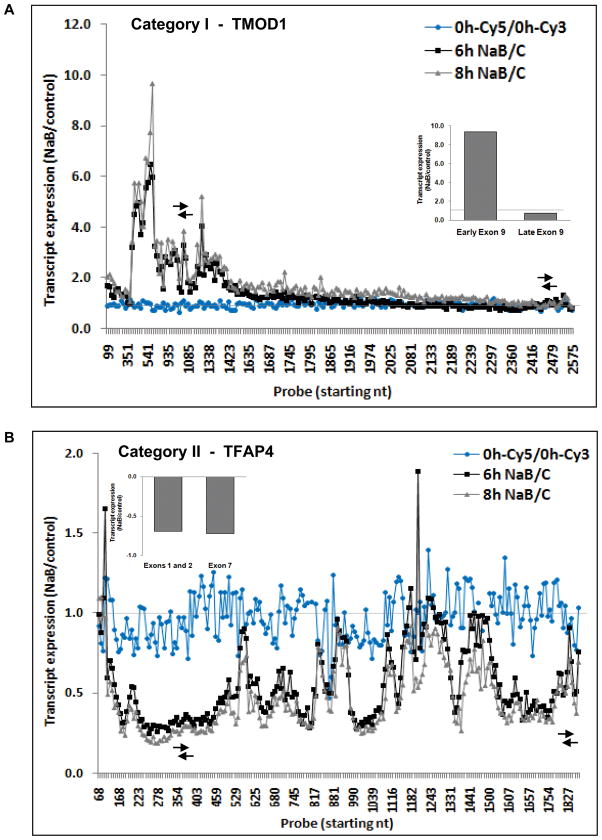
Figure 5. Transcript expression patterns of genes downregulated by butyrate, assessed by the “Tiling” microarray.
Category I (A) and category II (B) sequences from Table I are exemplified by TMOD1 and TFAP4 genes, respectively. Data are expressed as the ratio of NaB-treated to untreated cells. Arrows indicate the regions analyzed by qRT-PCR (inset, data as NaB-treated to untreated cells).
Figure 2. Butyrate effect on cyclin D1 and c-myc protein expression, and histone modification.
(A) Western blot for cyclin D1 and c-myc at 6–24 hrs of butyrate treatment. (B) Histone H3 acetylation (AcH3) levels in response to 30 min to 16 hrs of butyrate treatment. The total level of histone H3 is also shown. NB: nonspecific band.
RNA polymerase II occupancy and histone modification along cyclin D1 and c-myc
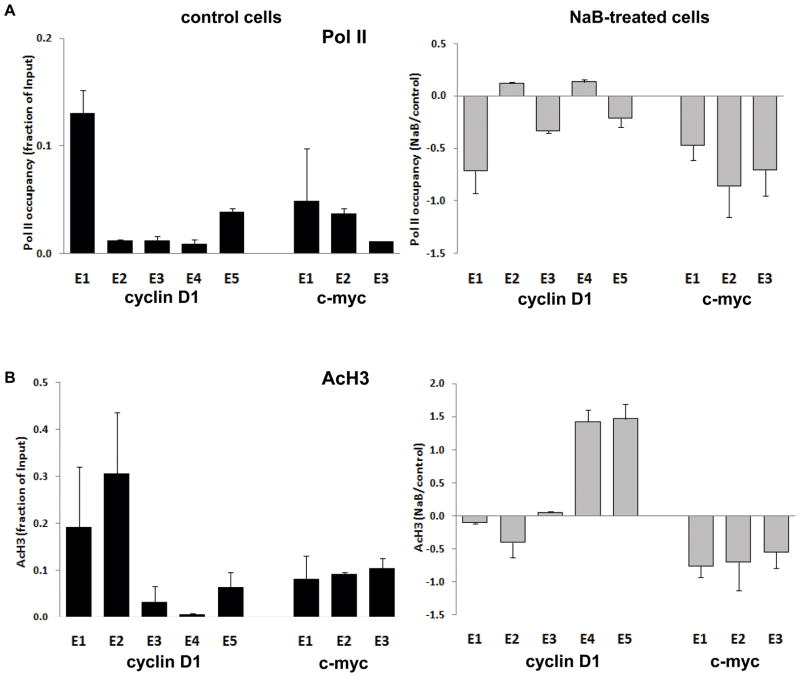
In untreated SW837 cells, analysis of Pol II by chromatin immunoprecipitation (ChIP) revealed a higher occupancy towards the 5′ end, relative to the rest of the sequence, in the cyclin D1 gene, while more similar levels were observed throughout the length of c-myc (Figure 3A, black bars). Following butyrate treatment of the cells for 6 hrs, Pol II occupancy for cyclin D1 decreased most substantially in exon 1 (nucleotides 1–407), as shown in Figure 3A (grey bars). While the butyrate-induced difference at the 5′ end of cyclin D1 was not observed downstream, for c-myc the pattern was different, with the butyrate-induced decrease being maintained at approximately the same level in exons 2 and 3 (Figure 3A).
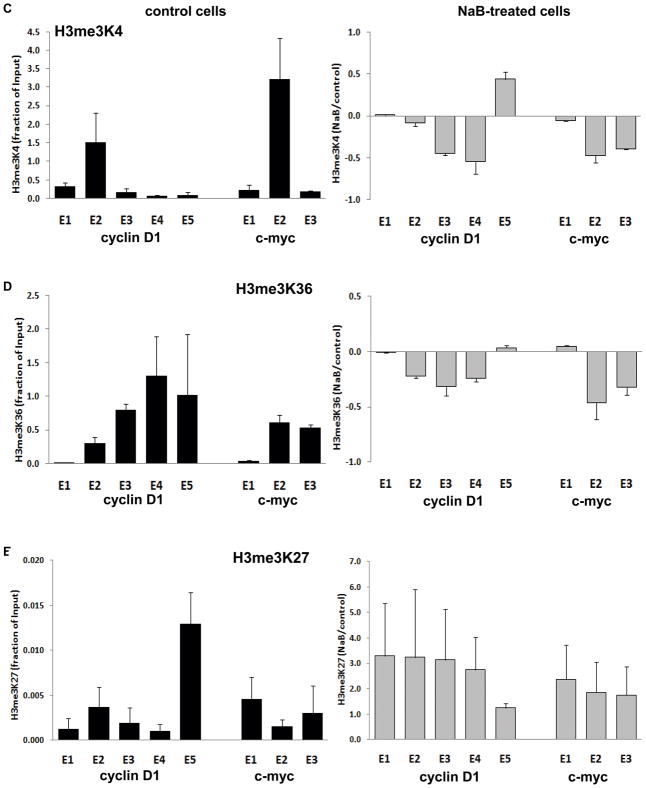
Figure 3. Chromatin immunoprecipitation (ChIP) of Pol II and modified histones.
(A) Pol II occupancy at 6 hrs of butyrate treatment (5 mM) along the five and three exons comprising the cyclin D1 and c-myc coding sequences, respectively. (B) Histone H3 acetylation (AcH3) levels throughout each sequence. (C-E) Histone H3 trimethylation on lysines 4, 36, and 27 (H3me3K4; H3me3K36; and H3me3K27). Immunoprecipitated samples were analyzed by qRT-PCR and normalized by their respective input. In each case, black bars indicate basal levels in untreated cells, with data expressed as the mean ±SD of linear (Ct) values (Methods), and grey bars indicate butyrate effects, with data expressed as the ratio of NaB-treated to untreated cells. Data are from at least three independent experiments. Similar results were found at 8 hrs of butyrate treatment (not shown).
Based on the well characterized activity of butyrate as a general inhibitor of histone deacetylases (HDACi activity, (31)), we determined butyrate effects on chromatin remodeling. An ~8-fold increase in overall levels of histone H3 acetylation (AcH3) at 30 minutes following butyrate treatment was observed, and persisted for ~6 hrs (Figure 2B). Despite this overall induction of AcH3, it has been reported that butyrate induces histone hypoacetylation on target genes downregulated in expression (32, 33). For both genes, a similar pattern of distribution for AcH3 as for Pol II was found in uninduced cells (Figure 3B). However, following butyrate treatment, AcH3 levels were differentially altered along cyclin D1, with modest decreases associated with decreased polymerase loading at the 5′ end, while for c-myc, AcH3 remained consistently decreased, similar to the persistent decrease of Pol II (Figure 3B). The increases in AcH3 in exons 4 and 5 of cyclin D1 after butyrate treatment were not associated with increases in Pol II occupancy (Figures 3A, B). This may be a technical issue, since there is a human sequence (FLJ42258) located downstream of the cyclin D1 gene on chromosome 11q13.2 and transcribed from the complementary strand, whose transcript is induced ~8-fold by butyrate (not shown). Hence, the elevation in AcH3 may be due to the overlap of this transcript that co-localizes with the 3′end of cyclin D1.
We investigated additional histone methylation marks important in transcriptional regulation. These are trimethylated histone H3 on: lysine 4 (H3me3K4), generally associated with transcription initiation; lysine 36 (H3me3K36), generally associated with elongation; and lysine 27 (H3me3K27), considered a mark or transcriptional silencing (32, 33). Figure 3C shows that in untreated cells, H3me3K4 was relatively higher at the 5′ end, consistent with its role in transcription initiation, although the highest level was in exon 2, not in exon 1. Similarly, highest level of this modification was found in exon 2 of c-myc (Figure 3C). As shown in Figure 3D, the pattern of H3me3K36 in uninduced cells was different, in each gene increasing towards the 3′ end, consistent with its association with elongation (33). While there were decreases in these histone modifications along each gene following butyrate treatment (Figure 3C, D), these were modest and generally further downstream of the marked transcriptional attenuation seen in Figure 1A, B, and thus may not account for this butyrate effect. Finally, the level of H3me3K27 was low along each gene (note scale of axis), and increased by butyrate (Figure 3E), in accordance with the role of this modification in transcriptional repression, although the data do not suggest a specific role in the attenuation of cyclin D1 and c-myc by butyrate.
Similar patterns, and modest histone modification in response to butyrate, were seen for Caco-2 cells (Supplementary Figure 2), with the exception of Pol II distribution, increased at exon 1 of c-myc, and H3me3K4, whose levels in response to butyrate where somewhat elevated in both sequences (Supplementary Figure 2).
Transcriptome analysis
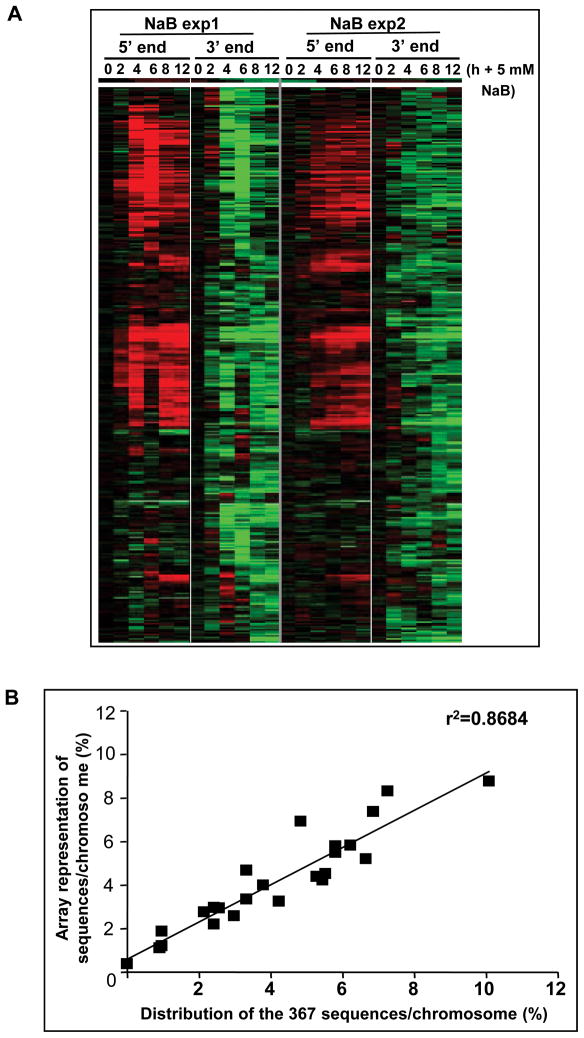
The above, and our prior work, focused on butyrate downregulation of cyclin D1 and c-myc because of the clear functional roles of these genes in colonic cell maturation and colon tumorigenesis. However, effects of butyrate on colon carcinoma cells are highly pleiotropic, altering expression of ~7% of sequences interrogated in our prior report (7). We therefore investigated the extent to which regulation of transcriptional elongation contributes to downregulation of gene expression. We first used a “5′ 3′ microarray”, with 5 probes within 100 bp of the 5′ end and 3′ end, for each of the 17,378 canonical coding sequences in GenBank (Methods). In SW837 cells untreated or treated with 5 mM NaB for 2–12 hrs, data from two independent experiments identified an overlap of 367 sequences (~2% of the sequences analyzed) for which expression was downregulated at the 3′ end following butyrate treatment (Figure 4A). Approximately 53% of these sequences were upregulated by butyrate at the 5′ end. For many of the remaining sequences, there were differences in extent of downregulation at the 5′ and 3′ ends, with a large number not altered in expression at all at the 5′ end (~43%). Each of these sequences were candidates for transcriptional attenuation downstream of the TSS.
Figure 4. Transcript expression of sequences downregulated by butyrate, assessed by the “5′ 3′ microarray”.
(A) A total of 367 sequences were downregulated by NaB at 2–12 hrs of treatment (Methods). Data are expressed as the ratio of NaB-treated to untreated cells at each time point of treatment for the 5′ end, while the 3′ end data are expressed relative to the 5′ end, as the “ratio of ratios” (Methods). A heat-map of two independent experiments is shown. Red and green indicate upregulation and downregulation in response to butyrate, respectively, while black indicates no significant change. (B) Chromosome distribution of the 367 sequences downregulated by butyrate at the 3′ end relative to the sequence representation on the array.
Since butyrate can induce chromatin modification, we investigated whether the 367 sequences downregulated at the 3′ end were clustered in regions in the human genome. Figure 4B shows that there was a linear distribution of these loci as a function of the frequency of representation of genes per chromosome on the 5′ 3′ array. Therefore, there was no overall physical clustering of these sequences. A further classification by chromosomal cytoband showed that only ~12% were clustered in groups, with a maximum of four sequences per cytoband (not shown). Therefore, as for cyclin D1 and c-myc, butyrate-induced transcriptional attenuation may be targeted, rather than involving broad regions affected by chromatin modification.
Recent data demonstrate that transcriptional units, previously thought to have a straight-forward organization that generates a single canonical pre-mRNA transcript, are in fact much more complex. In particular, multiple short transcripts are generated at the 5′ end as well as the 3′ end of most genes (34). Thus, there was likely to be underlying complexity to the structure and regulation of individual transcripts identified in Figure 4A.
We therefore analyzed the 367 sequences downregulated at the 3′ end in response to NaB using another custom-designed microarray with probes that tile-through each of these sequences, as was done for cyclin D1 and c-myc (Figure 1). Focusing on the 6–8 hrs time period that showed the greatest changes in expression, there were 85 sequences for which the 3′ end of the canonical transcript was expressed at a lower level than the 5′ end, based on the 5′3′ array analysis. Studies in four independent experiments using the “Tiling” array confirmed that 42 of these sequences were expressed to a significantly lower extent (>1.5-fold) towards the 3′ end than at the 5′ end, and thus potentially paused by butyrate. Closer examination revealed that these 42 sequences fell into two general classes, represented equally, as shown in Table I and Figure 5. Category I sequences were upregulated at the 5′ end by butyrate but either downregulated or not significantly altered in expression further downstream, illustrated by the TMOD1 gene (Figure 5A). Twenty eight of the twenty nine sequences comprising category I showed similar patterns of graded decrease in expression along the coding sequence, although they varied in the relative position at which expression was downregulated by butyrate. TMEM49 sequence (NM_030938) was the exception, showing increased expression by butyrate for the first half of the sequence but an abrupt decrease at the junction of exons 10 and 11 (Supplementary Figure 3), which could suggest induction of alternative splicing by butyrate. However, by RT-PCR and qRT-PCR, we have not detected any alternatively spliced form of this gene in SW837 cells in response to butyrate (data not shown). Category II genes (Table I) were not altered by butyrate in expression at the very 5′ end, as is the case for cyclin D1 and c-myc (Figure 1), but then showed heterogeneous patterns among the sequences, highly reproducible across independent experiments. These patterns encompassed decreased expression as for TFAP4 (Figure 5B), or increased expression before being downregulated by butyrate, as delineated in Table I. These patterns may result from complex transcription, characterized by multiple sites of initiation and termination as well as pausing.
Table I. Classification of transcript patterns in SW837 cells in response to butyrate.
(A) Forty two sequences found to be potentially paused by the “5′ 3′ array” and the “Tiling” array were classified according to NaB effect throughout the canonical coding region. Up and Down indicate upregulation and downregulation, respectively (>1.5-fold change), while NS indicates no significant change in response to butyrate. Examples of each category are shown in Figure 5.
| Coding region | ||||
|---|---|---|---|---|
| Category | 5′ end | middle | 3′ end | Sequences (%) |
| I | UP | UP | NS | 47.6 |
| UP | DOWN | NS | 2.4 | |
| UP | UP/DOWN | DOWN | 2.4 | |
| II | NS | DOWN | DOWN | 21.4 |
| NS | UP | NS | 16.7 | |
| NS | DOWN | NS | 9.5 | |
The distribution of these two categories with respect to their gene ontology is shown in Table II, with their gene ID in Supplementary Table IA. The repressed targets in category I were enriched in “Cytoskeleton organization” (~18%, Table IIA), and in “Apoptosis” (~15%), possibly contributing to the induction by butyrate of cell cycle arrest and differentiation in the cells analyzed (2, 3). In contrast, category II sequences were associated with the terms “Zinc ion binding”, a characteristic of many transcriptional factors involved also in transcriptional elongation (35, 36); and indeed, the term “Regulation of transcription” was also linked to this category (Table IIB). However, for category II, the input number of genes analyzed (Methods) was below the threshold required for assigning a P value. Nevertheless, the data suggest a differential pattern of transcriptional downregulation by butyrate, which may be related to different biological functions.
Table II. Functional classification of transcript patterns in SW837 cells in response to butyrate.
The biological function and gene ontology of Category I (A) and Category II (B) sequences were determined by DAVID Bioinformatic database (see Methods).
| A | ||
|---|---|---|
| Gene Ontology, Category I sequences | Percentage | P Value |
| Cytoskeleton organization and biogenesis | 18.5 | 0.001 |
| Apoptosis | 14.8 | 0.038 |
| Enzyme binding | 11.1 | 0.030 |
| Actin binding | 11.1 | 0.036 |
| Caspase activation | 7.4 | 0.036 |
| B | |
|---|---|
| Gene Ontology, Category II sequences | Percentage |
| Zinc ion binding, zinc finger | 33.3 |
| Intracellular organelle | 33.3 |
| Regulation of transcription | 22.2 |
| Protein transport and binding | 22.2 |
| Peptidase activity | 22.2 |
| tRNA metabolic process | 11.1 |
DISCUSSION
Butyrate, a natural dietary product and an HDAC inhibitor fundamental to normal colonic cell maturation and with potential activity in chemoprevention and chemotherapy, has extensive effects on gene expression. Butyrate alters gene expression as well as underlying epigenetic mechanisms of transcriptional regulation (reviewed in (37)). We first focused on effects on two specific genes, cyclin D1 and c-myc, fundamental in intestinal cell maturation and tumorigenesis, and then expanded analysis to a transcriptome level that revealed a complexity of response to butyrate not previously appreciated.
Transcriptional pausing was previously reported at exon 1/intron 1 boundary of c-myc in hematopoietic tumor cells carrying translocations involving this gene (38, 39). Here, by employing high-resolution custom-designed tiling arrays, we determined that in colon carcinoma cells, after 6–8 hours of butyrate treatment, there is almost a complete decrease in c-myc transcript expression, and marked downregulation of cyclin D1 expression (~50%), due to a decreased steady state level of the transcripts that begin at ~100 nucleotides downstream of the TSS (i.e. <1% and ~2% of cyclin D1 and c-myc coding sequences, respectively; Figure 1), in agreement with previous reports that transcriptional attenuation of genes occurs close to the transcription start site (40). The more complete decrease in steady state level of c-myc than of cyclin D1 transcripts may be related to the fundamental role that alterations in c-myc play in colon tumor development (12, 41), as well as the critical role of c-myc in reprogramming normal intestinal cell maturation (8). It should also be noted that deletion of only a single allele of cyclin D1 was sufficient to decrease intestinal tumor formation in ApcMin/+ mice, suggesting that effects of this locus may be particularly sensitive to more modest changes in gene expression (13).
Transcriptional attenuation in the c-myc gene has been attributed to paused RNA polymerase II (Pol II) and/or premature termination in the promoter-proximal region (21, 39). While in Caco-2 cells, Pol II distribution increased by butyrate at the very 5′ end of c-myc, in accord with the induction of transcriptional attenuation by butyrate in this locus, in SW837 cells butyrate inhibited Pol II occupancy towards the 5′ end of cyclin D1, and throughout c-myc. In SW837 cells, butyrate-induced attenuation could be associated with a decrease in elongating polymerase complexes, as described before for other cell types (21, 42, 43). Butyrate, a well-characterized inhibitor of HDAC activity, induced a large increase in histone H3 acetylation, a mark of transcriptional activity, as we determined occurs, for example, on CDKN1A (not shown), confirming published data (44).
Butyrate also induces histone hypoacetylation close to the TSS in downregulated genes (45). The butyrate-induced differences in levels of AcH3 that we detected across the exons of both cyclin D1 and c-myc were generally related to Pol II occupancy. However, although decreased towards the TSS, the AcH3 changes were modest. Similarly, basal levels of H3me3K4 were higher nearer the 5′ end of each gene, while H3me3K36 increased towards the 3′ end, consistent with reports that these modifications associate with transcriptional initiation and elongation, respectively, in actively transcribed genes (46, 47). However, butyrate again only modestly decreased the levels of these two modifications in both genes analyzed. On the other hand, there was a moderate increase in the levels of H3me3K27 by butyrate throughout cyclin D1 and c-myc, consistent with its role in transcriptional silencing (33). Thus, although the alterations we detected in these histone marks were consistent with gene silencing by butyrate, direct correspondence with the region of attenuation was not definitive. This is consistent with the proposal that these histone modifications are not rigidly associated with transcriptional state (26), and that the functional epigenetic landscape is more complex than previously thought (48).
Our previous data have shown that butyrate is highly pleiotropic in its effects on gene expression, with a cascade of changes that increases as a function of time following exposure, a hallmark of sequential recruitment of genes and pathways with cell maturation (7). Therefore, we pursued these widespread effects of butyrate using a whole transcriptome analysis, identifying a subset of 42 genes potentially attenuated by butyrate. Transcription of these genes, as well as of cyclin D1 and c-myc, was decreased by NaB downstream of the TSS, suggesting that the initiation step of the transcriptional process was not the principal target, consistent with other work demonstrating transcriptional elongation as a rate-limiting step of transcription (49). Moreover, a recent report showed that in developmental systems, many genes that do not produce full-length mRNAs nonetheless are transcriptionally initiated (47).
Our transcriptome analysis revealed that while genes responding to butyrate fell into general categories based on the location and nature of altered expression along the coding sequence (Table I and Figure 5), the pattern of transcription was distinct and highly reproducible for each gene, reflecting the high complexity of genome transcription recently elucidated (50). High-throughput parallel sequencing of transcripts should be informative in understanding this heterogeneity along these specific loci.
There are two important points regarding the complexity of transcriptional regulation by butyrate. First, butyrate is a short-chain fatty acid, consumed as a nutrient and generated by fermentation of dietary fiber, and the primary energy source for colonic epithelial cells (4). Therefore, the intestine evolved in the presence of this compound, and the complex patterns of gene expression triggered by butyrate may be key to generating the highly coordinated response that establishes and maintains homeostasis of the colon. Second, butyrate may also exhibit anti-tumor properties, and the control of expression of proto-oncogenes such as c-myc, as well as of genes which directly promote selective advantage of tumor cells (e.g.: cell growth, survival, vascularization) by transcriptional attenuation is broader than originally thought.
In conclusion, transcriptional attenuation of the cyclin D1 and c-myc genes in response to butyrate begins at about nucleotide 100 downstream of the transcription start site. The role of chromatin remodeling and histone modification in response to butyrate is complex, may be independent of the HDAC inhibitory activity of butyrate, and may follow a gene specific pattern. It has been proposed that control of transcriptional elongation could be a widespread mechanism of gene regulation in eukaryotes (38). Our results demonstrate that this is the case for the effects of the short-chain fatty acid butyrate, and identify gene targets regulated by this mechanism.
Supplementary Material
Supplementary Figure 1. Cyclin D1 and c-myc transcript expression in response to butyrate in Caco-2 cells.
“Tiling” microarray data for Caco-2 cells at 0, 6 and 8 hrs of butyrate treatment (2 mM). The ratio of NaB-treated to untreated cells at each time point of treatment is shown for the first 355 nt of cyclin D1 (A) and c-myc (B) sequences. On the abscissa, the probe numbers indicate the first nucleotide of the probe. Inset: ratio of NaB-treated to untreated cells for every probe on the array (color-coded as in the main figure). *: position where transcriptional attenuation was induced by NaB.
Supplementary Figure 2. Chromatin immunoprecipitation (ChIP) of modified histones and Pol II in Caco-2 cells.
(A) Pol II occupancy along the five and three exons of cyclin D1 and c-myc, respectively, at 6 hrs of butyrate treatment (2 mM). (B) Histone H3 acetylation (AcH3) levels throughout each sequence. (C-E) Histone H3 trimethylation on lysines 4, 36, and 27 (H3me3K4; H3me3K36; and H3me3K27). Immunoprecipitated samples were analyzed by qRT-PCR and normalized by their respective input. In each case, black bars indicate basal levels in untreated cells, with data expressed as the mean ±SD of linear (Ct) values (Methods), and grey bars indicate butyrate effects, with data expressed as the ratio of NaB-treated to untreated cells. Data are from at least three independent experiments. Similar results were found at 8 hrs of butyrate treatment (not shown).
Supplementary Figure 3. Transcriptional regulation of TMEM49 by butyrate, assessed by the “Tiling” microarray.
Transcript expression of TMEM49 in SW837 cells at 0, 6 or 8 hrs of butyrate treatment (5 mM). The ratio of NaB-treated to untreated cells at each time point of treatment is shown for the exonic regions, and data are representative of four independent experiments (Methods). On the abscissa, the probe numbers indicate the first nucleotide of the probe. The bar on the abscissa indicates the junction between exons 10 and 11, where butyrate-induced pause was observed. Arrows indicate the regions analyzed by qRT-PCR (inset, data as NaB-treated to untreated cells).
Supplementary Table I. Identification of sequences transcriptionally paused by butyrate
The sequence ID (GenBank) of forty two sequences paused by butyrate, and a brief description of the sequence name and nature, are shown for Category I (A) and Category II (B) sequences (see Table I).
Supplementary Table II. Primer sequences designed for quantitative RT-PCR.
The human cyclin D1, c-myc, TMOD1, TFAP4, and TMEM49 genes were analyzed by qRT-PCR. Primers were obtained from Sigma.
Acknowledgments
supported in part by grants from the American Institute for Cancer Research to MCD, and from the National Cancer Institute U54CA100926, CA123473, and CA135561 to LHA, and Cancer Center support grant, P30CA13330. The authors wish to thank Paolo Norio, John Mariadason, Anna Velcich and Georgia Corner for help in conducting these experiments.
Footnotes
The authors declare no conflict of interest.
References
- 1.Barnard JA, Warwick G. Butyrate rapidly induces growth inhibition and differentiation in HT-29 cells. Cell Growth Differ. 1993;4:495–501. [PubMed] [Google Scholar]
- 2.Hague A, Manning AM, Hanlon KA, Huschtscha LI, Hart D, Paraskeva C. Sodium butyrate induces apoptosis in human colonic tumour cell lines in a p53-independent pathway: implications for the possible role of dietary fibre in the prevention of large-bowel cancer. Int J Cancer. 1993;55:498–505. doi: 10.1002/ijc.2910550329. [DOI] [PubMed] [Google Scholar]
- 3.Heerdt BG, Houston MA, Augenlicht LH. Potentiation by specific short-chain fatty acids of differentiation and apoptosis in human colonic carcinoma cell lines. Cancer Res. 1994;54:3288–93. [PubMed] [Google Scholar]
- 4.Roediger WE. Role of anaerobic bacteria in the metabolic welfare of the colonic mucosa in man. Gut. 1980;21:793–8. doi: 10.1136/gut.21.9.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Augenlicht LH, Anthony GM, Church TL, et al. Short-chain fatty acid metabolism, apoptosis, and Apc-initiated tumorigenesis in the mouse gastrointestinal mucosa. Cancer Res. 1999;59:6005–9. [PubMed] [Google Scholar]
- 6.Heerdt BG, Augenlicht LH. Effects of fatty acids on expression of genes encoding subunits of cytochrome c oxidase and cytochrome c oxidase activity in HT29 human colonic adenocarcinoma cells. J Biol Chem. 1991;266:19120–6. [PubMed] [Google Scholar]
- 7.Mariadason JM, Corner GA, Augenlicht LH. Genetic reprogramming in pathways of colonic cell maturation induced by short chain fatty acids: comparison with trichostatin A, sulindac, and curcumin and implications for chemoprevention of colon cancer. Cancer Res. 2000;60:4561–72. [PubMed] [Google Scholar]
- 8.Mariadason JM, Nicholas C, L’Italien KE, et al. Gene expression profiling of intestinal epithelial cell maturation along the crypt-villus axis. Gastroenterology. 2005;128:1081–8. doi: 10.1053/j.gastro.2005.01.054. [DOI] [PubMed] [Google Scholar]
- 9.He TC, Sparks AB, Rago C, et al. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–12. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 10.Tetsu O, McCormick F. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature. 1999;398:422–6. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- 11.Sancho E, Batlle E, Clevers H. Live and let die in the intestinal epithelium. Curr Opin Cell Biol. 2003;15:763–70. doi: 10.1016/j.ceb.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 12.Sansom OJ, Meniel VS, Muncan V, et al. Myc deletion rescues Apc deficiency in the small intestine. Nature. 2007;446:676–9. doi: 10.1038/nature05674. [DOI] [PubMed] [Google Scholar]
- 13.Hulit J, Wang C, Li Z, et al. Cyclin D1 genetic heterozygosity regulates colonic epithelial cell differentiation and tumor number in ApcMin mice. Mol Cell Biol. 2004;24:7598–611. doi: 10.1128/MCB.24.17.7598-7611.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heruth DP, Zirnstein GW, Bradley JF, Rothberg PG. Sodium butyrate causes an increase in the block to transcriptional elongation in the c-myc gene in SW837 rectal carcinoma cells. J Biol Chem. 1993;268:20466–72. [PubMed] [Google Scholar]
- 15.Lallemand F, Courilleau D, Sabbah M, Redeuilh G, Mester J. Direct inhibition of the expression of cyclin D1 gene by sodium butyrate. Biochem Biophys Res Commun. 1996;229:163–9. doi: 10.1006/bbrc.1996.1774. [DOI] [PubMed] [Google Scholar]
- 16.Bender TP, Thompson CB, Kuehl WM. Differential expression of c-myb mRNA in murine B lymphomas by a block to transcription elongation. Science. 1987;237:1473–6. doi: 10.1126/science.3498214. [DOI] [PubMed] [Google Scholar]
- 17.Chen Z, Harless ML, Wright DA, Kellems RE. Identification and characterization of transcriptional arrest sites in exon 1 of the human adenosine deaminase gene. Mol Cell Biol. 1990;10:4555–64. doi: 10.1128/mcb.10.9.4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mechti N, Piechaczyk M, Blanchard JM, Jeanteur P, Lebleu B. Sequence requirements for premature transcription arrest within the first intron of the mouse c-fos gene. Mol Cell Biol. 1991;11:2832–41. doi: 10.1128/mcb.11.5.2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Middleton KM, Morgan GT. Premature termination of transcription can be induced on an injected alpha-tubulin gene in Xenopus oocytes. Mol Cell Biol. 1990;10:727–35. doi: 10.1128/mcb.10.2.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tong X, Yin L, Joshi S, Rosenberg DW, Giardina C. Cyclooxygenase-2 regulation in colon cancer cells: modulation of RNA polymerase II elongation by histone deacetylase inhibitors. J Biol Chem. 2005;280:15503–9. doi: 10.1074/jbc.M411978200. [DOI] [PubMed] [Google Scholar]
- 21.Krumm A, Meulia T, Brunvand M, Groudine M. The block to transcriptional elongation within the human c-myc gene is determined in the promoter-proximal region. Genes Dev. 1992;6:2201–13. doi: 10.1101/gad.6.11.2201. [DOI] [PubMed] [Google Scholar]
- 22.Raschke EE, Albert T, Eick D. Transcriptional regulation of the Ig kappa gene by promoter-proximal pausing of RNA polymerase II. J Immunol. 1999;163:4375–82. [PubMed] [Google Scholar]
- 23.Wilson AJ, Velcich A, Arango D, et al. Novel detection and differential utilization of a c-myc transcriptional block in colon cancer chemoprevention. Cancer Res. 2002;62:6006–10. [PubMed] [Google Scholar]
- 24.Zhang ZK, Davies KP, Allen J, et al. Cell cycle arrest and repression of cyclin D1 transcription by INI1/hSNF5. Mol Cell Biol. 2002;22:5975–88. doi: 10.1128/MCB.22.16.5975-5988.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maier S, Daroqui MC, Scherer S, et al. Butyrate and vitamin D3 induce transcriptional attenuation at the cyclin D1 locus in colonic carcinoma cells. J Cell Physiol. 2009;218:638–42. doi: 10.1002/jcp.21642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–19. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 27.Boffa LC, Vidali G, Mann RS, Allfrey VG. Suppression of histone deacetylation in vivo and in vitro by sodium butyrate. J Biol Chem. 1978;253:3364–6. [PubMed] [Google Scholar]
- 28.Turner BM. Histone acetylation and an epigenetic code. Bioessays. 2000;22:836–45. doi: 10.1002/1521-1878(200009)22:9<836::AID-BIES9>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 29.Mariadason JM, Rickard KL, Barkla DH, Augenlicht LH, Gibson PR. Divergent phenotypic patterns and commitment to apoptosis of Caco-2 cells during spontaneous and butyrate-induced differentiation. J Cell Physiol. 2000;183:347–54. doi: 10.1002/(SICI)1097-4652(200006)183:3<347::AID-JCP7>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 30.Daroqui MC, Puricelli LI, Urtreger AJ, Elizalde PV, Lanuza GM, Bal de Kier Joffe E. Involvement of TGF-beta(s)/T(beta)Rs system in tumor progression of murine mammary adenocarcinomas. Breast Cancer Res Treat. 2003;80:287–301. doi: 10.1023/a:1024910332621. [DOI] [PubMed] [Google Scholar]
- 31.Sealy L, Chalkley R. The effect of sodium butyrate on histone modification. Cell. 1978;14:115–21. doi: 10.1016/0092-8674(78)90306-9. [DOI] [PubMed] [Google Scholar]
- 32.Barski A, Cuddapah S, Cui K, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–37. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 33.Vakoc CR, Sachdeva MM, Wang H, Blobel GA. Profile of histone lysine methylation across transcribed mammalian chromatin. Mol Cell Biol. 2006;26:9185–95. doi: 10.1128/MCB.01529-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carninci P, Sandelin A, Lenhard B, et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat Genet. 2006;38:626–35. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- 35.Price DH. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol Cell Biol. 2000;20:2629–34. doi: 10.1128/mcb.20.8.2629-2634.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tan S, Aso T, Conaway RC, Conaway JW. Roles for both the RAP30 and RAP74 subunits of transcription factor IIF in transcription initiation and elongation by RNA polymerase II. J Biol Chem. 1994;269:25684–91. [PubMed] [Google Scholar]
- 37.Kim YS, Milner JA. Dietary modulation of colon cancer risk. J Nutr. 2007;137:2576S–9S. doi: 10.1093/jn/137.11.2576S. [DOI] [PubMed] [Google Scholar]
- 38.Bentley DL, Groudine M. Sequence requirements for premature termination of transcription in the human c-myc gene. Cell. 1988;53:245–56. doi: 10.1016/0092-8674(88)90386-8. [DOI] [PubMed] [Google Scholar]
- 39.Keene RG, Mueller A, Landick R, London L. Transcriptional pause, arrest and termination sites for RNA polymerase II in mammalian N- and c-myc genes. Nucleic Acids Res. 1999;27:3173–82. doi: 10.1093/nar/27.15.3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wright S, Mirels LF, Calayag MC, Bishop JM. Premature termination of transcription from the P1 promoter of the mouse c-myc gene. Proc Natl Acad Sci U S A. 1991;88:11383–7. doi: 10.1073/pnas.88.24.11383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ignatenko NA, Holubec H, Besselsen DG, et al. Role of c-Myc in intestinal tumorigenesis of the ApcMin/+ mouse. Cancer Biol Ther. 2006;5:1658–64. doi: 10.4161/cbt.5.12.3376. [DOI] [PubMed] [Google Scholar]
- 42.Bentley DL, Groudine M. A block to elongation is largely responsible for decreased transcription of c-myc in differentiated HL60 cells. Nature. 1986;321:702–6. doi: 10.1038/321702a0. [DOI] [PubMed] [Google Scholar]
- 43.Nepveu A, Marcu KB. Intragenic pausing and anti-sense transcription within the murine c-myc locus. Embo J. 1986;5:2859–65. doi: 10.1002/j.1460-2075.1986.tb04580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hinnebusch BF, Meng S, Wu JT, Archer SY, Hodin RA. The effects of short-chain fatty acids on human colon cancer cell phenotype are associated with histone hyperacetylation. J Nutr. 2002;132:1012–7. doi: 10.1093/jn/132.5.1012. [DOI] [PubMed] [Google Scholar]
- 45.Rada-Iglesias A, Enroth S, Ameur A, et al. Butyrate mediates decrease of histone acetylation centered on transcription start sites and down-regulation of associated genes. Genome Res. 2007;17:708–19. doi: 10.1101/gr.5540007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bannister AJ, Schneider R, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T. Spatial distribution of di- and tri-methyl lysine 36 of histone H3 at active genes. J Biol Chem. 2005;280:17732–6. doi: 10.1074/jbc.M500796200. [DOI] [PubMed] [Google Scholar]
- 47.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–12. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 49.Saunders A, Core LJ, Lis JT. Breaking barriers to transcription elongation. Nat Rev Mol Cell Biol. 2006;7:557–67. doi: 10.1038/nrm1981. [DOI] [PubMed] [Google Scholar]
- 50.Birney E, Stamatoyannopoulos JA, Dutta A, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Cyclin D1 and c-myc transcript expression in response to butyrate in Caco-2 cells.
“Tiling” microarray data for Caco-2 cells at 0, 6 and 8 hrs of butyrate treatment (2 mM). The ratio of NaB-treated to untreated cells at each time point of treatment is shown for the first 355 nt of cyclin D1 (A) and c-myc (B) sequences. On the abscissa, the probe numbers indicate the first nucleotide of the probe. Inset: ratio of NaB-treated to untreated cells for every probe on the array (color-coded as in the main figure). *: position where transcriptional attenuation was induced by NaB.
Supplementary Figure 2. Chromatin immunoprecipitation (ChIP) of modified histones and Pol II in Caco-2 cells.
(A) Pol II occupancy along the five and three exons of cyclin D1 and c-myc, respectively, at 6 hrs of butyrate treatment (2 mM). (B) Histone H3 acetylation (AcH3) levels throughout each sequence. (C-E) Histone H3 trimethylation on lysines 4, 36, and 27 (H3me3K4; H3me3K36; and H3me3K27). Immunoprecipitated samples were analyzed by qRT-PCR and normalized by their respective input. In each case, black bars indicate basal levels in untreated cells, with data expressed as the mean ±SD of linear (Ct) values (Methods), and grey bars indicate butyrate effects, with data expressed as the ratio of NaB-treated to untreated cells. Data are from at least three independent experiments. Similar results were found at 8 hrs of butyrate treatment (not shown).
Supplementary Figure 3. Transcriptional regulation of TMEM49 by butyrate, assessed by the “Tiling” microarray.
Transcript expression of TMEM49 in SW837 cells at 0, 6 or 8 hrs of butyrate treatment (5 mM). The ratio of NaB-treated to untreated cells at each time point of treatment is shown for the exonic regions, and data are representative of four independent experiments (Methods). On the abscissa, the probe numbers indicate the first nucleotide of the probe. The bar on the abscissa indicates the junction between exons 10 and 11, where butyrate-induced pause was observed. Arrows indicate the regions analyzed by qRT-PCR (inset, data as NaB-treated to untreated cells).
Supplementary Table I. Identification of sequences transcriptionally paused by butyrate
The sequence ID (GenBank) of forty two sequences paused by butyrate, and a brief description of the sequence name and nature, are shown for Category I (A) and Category II (B) sequences (see Table I).
Supplementary Table II. Primer sequences designed for quantitative RT-PCR.
The human cyclin D1, c-myc, TMOD1, TFAP4, and TMEM49 genes were analyzed by qRT-PCR. Primers were obtained from Sigma.