Abstract
Antimicrobial peptides (AMPs) have been studied for three decades, and yet a molecular understanding of their mechanism of action is still lacking. Here we summarize current knowledge for both synthetic vesicle experiments and microbe experiments, with a focus on comparisons between the two. Microbial experiments are done at peptide:lipid ratios that are at least 4 orders of magnitude higher than vesicle-based experiments. To close the gap between the two concentration regimes, we propose an “interfacial activity model”, which is based on an experimentally testable molecular image of AMP-membrane interactions. The interfacial activity model may be useful in driving engineering and design of novel AMPs.
Introduction
The field of antimicrobial peptide research originated in the early 1980’s when Hans Boman reported(1) that the humoral immune system of silk moths (Hyalophora cecropia) contained peptides (cecropins) with potent and broad-spectrum1 antimicrobial activity. Shortly thereafter, other AMPs were identified. For example, Shunji Natori described sarcotoxins from fly larvae(2), Robert Lehrer described defensins from mammalian macrophages (3) and Michael Zasloff described the magainins found in the skin of the frog Xenopus laevis (4). Although the primary and secondary structures of these first antimicrobial peptides are very different, the early reports described shared characteristics of biological activity and biophysical behavior that are still appropriate today to identify and categorize antimicrobial peptides; AMPs are cationic, amphipathic peptides that have broad spectrum microbicidal activity which is associated with membrane permeabilization. Unlike nonspecific membrane lytic peptide toxins, such as melittin, from the venom of the Honey Bee (apis melifera), or alamethicin from the fungus Trichoderma viride, AMPs have little cytolytic or cytotoxic activity against host cells.
Since those influential early publications, the field has undergone incredible growth, with nearly 1000 known examples of antimicrobial peptides (5–7). Naturally occurring examples are found in all classes of organisms: vertebrate animals, including humans, invertebrate animals, plants and microbes. Descriptions of newly discovered AMPs are common occurrences in the literature. In addition to the naturally occurring AMPs, peptides with the same biological activity have been designed de novo(8), engineered from natural sequences(9;10) or selected from combinatorial libraries(11;12).
Much of the vast literature on antimicrobial peptides is devoted to biophysical characterization and structure-function studies in model systems such as lipid vesicles or detergents. Yet, despite the large volume of data available, compelling structure-function relationships are very rare in antimicrobial peptide research. In fact, recent literature suggests that antimicrobial activity is not dependent on specific amino acid sequences or on specific peptide structures (9;13–16). Instead, activity depends more on the amino acid composition of a peptide and on its physical chemical properties. We have called this phenomenon “interfacial activity” and described it as “the ability of a molecule to bind to a membrane, partition into in the membrane-water interface and to alter the packing and organization of the lipids” (11;17). Interfacial activity depends mainly on the appropriate balance of hydrophobic and electrostatic interactions between peptides, water and lipids. In this paper we review the available information on the function of AMPs, with a special emphasis on correlating function observed in model systems (e.g. synthetic lipid bilayers) with biological activity. We describe a paradigm that may help bridge the gap between biophysical and biological activity: The interfacial activity model.
AMP action starts at the membrane
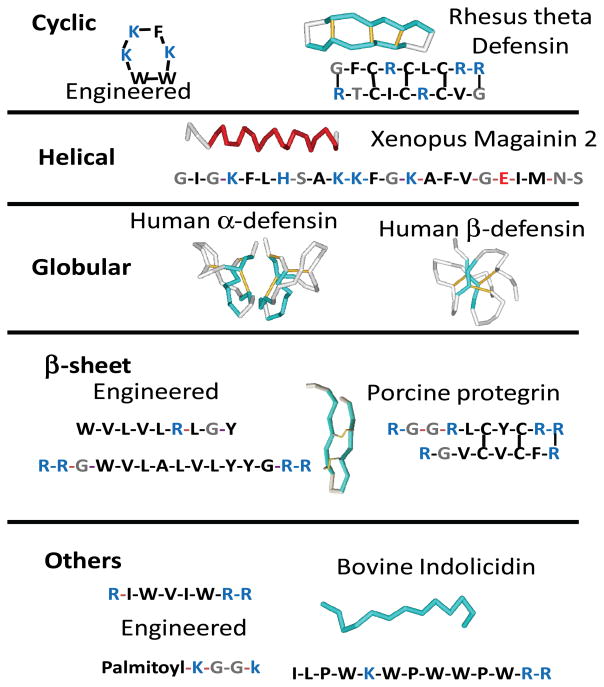
Antimicrobial peptides fall into a myriad of secondary or tertiary structure classes as shown in Figure 1. Helical and β-sheet rich AMPs are common, with many of the β-sheet examples (e.g. the defensins) having disulfide cross-linked tertiary structure. Some AMPs are cyclic(18). Some small AMPs (<~15 residues) are often poorly or irregularly structured(19). In terms of structure, AMPs are about as diverse as possible (Fig 1). However, one of the few conserved characteristics of antimicrobial peptides is their cationic and hydrophobic composition. Most AMPs are cationic overall with some examples having net charges as high as +9. From databases of AMP sequences(6;7), we observe that the basic amino acids, Arg and Lys, are roughly 50% more abundant in AMPs than in genomes overall, while the acidic amino acids, Glu and Asp, are ~75% less abundant than expected. The hydrophobic amino acids are also more abundant in AMPs, while polar residues (especially Gln) are less abundant than expected. Smaller AMPs often have higher abundances of aromatic and basic amino acids. Consistent with these observations, AMPs have been successfully engineered using reduced amino acid alphabets(17) that contain only basic and hydrophobic amino acids. Some simple cationic/hydrophobic peptides with good antimicrobial activity include RWn (n=1–5)(20), and WALRLYLVY (11). Even some cationic/hydrophobic acylated tetra-peptides(21) or modified di-peptides(20) have potent antimicrobial activity.
Figure 1.
A menagerie of antimicrobial peptide structures. AMPs range from 4 to about 40 amino acids in length, Some are linear, while others are cyclic, disulfide crosslinked or acylated. Known antimicrobial peptides have many types of secondary structure, including α-helix, β-sheet and irregular structure or random coil. Engineered AMPs have the same properties as natural examples. The one feature that unites all AMPs is their hydrophobic and cationic nature.
The mixed cationic and hydrophobic composition of AMPs makes them well suited for interacting with and perturbing microbial cytoplasmic membranes which typically present anionic surfaces, rich in lipids such as phosphatidylglycerol or cardiolipin, to the outside environment. One of the most commonly cited explanations for the selectivity of AMPs for microbes over host cells is the difference in membrane interactions due to differences in exposed anionic lipid content. Because of the contribution of electrostatic interactions, binding of AMPs to microbial membranes is significant, while binding of AMPs to the neutral phosphatidylcholine/cholesterol/sphingomyelin-rich surfaces of animal plasma membranes is weaker. There is ample direct evidence that most AMPs permeabilize microbial cytoplasmic membranes and that the membranes are often permeabilized with increasing severity with time(22). AMPs can dissipate the electrochemical gradient across microbial plasma membranes, within a few seconds of addition(11;22). Thus, AMPs must be able to rapidly pass through the thick proteoglycan layer of Gram positive bacteria and the outer membrane lipopolysaccharide (LPS) layer of Gram negative bacteria. Permeation of larger markers, including dye markers, metabolites and cytosolic proteins, through the cytoplasmic membrane occurs on the timescale of minutes to tens of minutes(11;13). After an hour or more in contact with AMPs, gross disruption of microbial membrane structure and morphology is often noted, including membrane blebbing, vesiculation, fragmentation, release of DNA, cell aggregation and destruction of cell morphology.
The membrane is not the only site of AMP action
While most AMPs interact with and influence the integrity of microbial membranes, it is not known if membrane permeabilization is always the lethal event or if the membrane is the only site of action. There are AMPs that may have alternate modes of action. For example the bovine neutrophil defensin indolicidin, a 13-mer rich in basic and aromatic residues, has typical low μM microbicidal effectiveness (23) and yet does not always permeabilize microbial membranes(11). Likewise, in a set of ten similar AMPs that we selected from a combinatorial library (11;17) we found some peptides that rapidly and completely permeabilized microbial membranes while others had only small effects on microbial membrane integrity(24;25). Antimicrobial activity and the degree of membrane permeabilization were essentially uncorrelated, yet all of the peptides had similar low μM broad-spectrum antimicrobial potency. Although potent biomembrane permeabilization is often associated with AMP activity, it is apparently not always required for activity.
It is not known whether small amounts of membrane permeabilization, alone, can account for lethality of AMPs. Those peptides that do not extensively permeabilize microbe membranes may be having effects on microbe viability that depend on interactions with intracellular components, perhaps in addition to effects on the membrane. Given the similarity of physical chemistry between AMPs and some cell penetrating peptides (26–28) it should not be surprising that AMPs can also translocate across microbial membranes, and can sometimes do so without extensive permeabilization. Although translocation of peptides into microbes has not often been studied carefully, there are reports that peptide translocation is coupled to leakage. For example the human defensin cryptdin-4 and the frog AMP magainin 2 both translocate across bilayers with a halftime of about 10 minutes(24;29). It seems likely that many AMPs will translocate across microbial membranes. Axelsen reported changes in gene transcription in multiple genes in microorganisms treated with sub-permeabilizing concentrations of cecropin A (30) and concluded that systemic metabolic effects were taking place prior to membrane permeabilization. This work suggested that translocation is occurring at concentrations that do not induce permeabilization. Other intracellular targets of AMPs have been proposed, including DNA and chaperonins.
Almeida and Pokorny have suggested that it may be appropriate to recognize a very broad overlap between membrane permeabilization and membrane translocation activity(31;32). They have described a quantitative, predictive model based on hydrophobicity scales(31). It is likely that most AMPs will also be cell penetrating peptides when interacting with living microbes, and will have access to the cytoplasm. Multiple simultaneous mechanisms of AMP action (e.g. membrane permeabilization as well as intracellular effects) may help explain their broad-spectrum activity and the rarity of inducible resistance.
Model membrane studies
There are countless studies showing that AMPs interact with and perturb lipid bilayer membranes in vitro. Within these many experiments, lipid compositions, buffer conditions, peptide and lipid concentrations vary widely. Even the phase state of the lipids and the degree of bilayer hydration vary between experiments. Combined with the inherently variable nature of AMP activity, these factors confuse the interpretation and comparison of results. While lipid compositions vary widely, mixtures of anionic and zwitterionic lipids are often used to mimic microbial membranes. For example, some laboratories use mixtures of phosphatidylglycerol and phosphatidylethanolamine to specifically mimic the E. coli inner membrane(33), while others use pure phosphatidylglycerol to specifically mimic Gram positive bacteria(34). Other commonly used systems include phosphatidylcholine mixed with phosphatidylglycerol, phosphatidylserine or cardiolipin to broadly mimic the anionic surface of a microbe(12;17). Experiments have also been performed with lipids extracted from E. coli or other organisms(35). To mimic exposed mammalian or host membranes, zwitterionic phosphatidylcholine (PC) or PC-cholesterol membranes are often used(8). Because of the variety of protocols and experimental conditions used, a single most appropriate lipid composition has never emerged from the literature.
In model membranes and living cells, lipid composition will affect peptide binding as well as the inherent susceptibility of the bilayer to permeabilization. Other than electrostatic effects, there are few, if any, examples of specific binding of an AMP to a particular lipid species. Cationic/hydrophobic peptides with good interfacial activity are expected to perturb any fluid phase lipid bilayer membrane to which they bind well enough, although bilayers with some lipid compositions may be inherently more or less stable than others. In practical terms AMPs that are less hydrophobic and more cationic will require anionic lipids for binding. For example, human defensins, which are highly cationic, must be studied in bilayers with anionic lipids, or else they are inactive due to poor binding(36). The more hydrophobic AMPs, such as the magainins, are active in zwitterionic lipid bilayers as well as anionic bilayers. It is likely that any model system that comprises a fully hydrated, fluid phase bilayer to which one can achieve reasonable bound peptide concentration is appropriate as a model system. Recently we showed that large unilamellar vesicles(11;12;25) composed of 90% phosphatidylcholine and 10% phosphatidylglycerol in 50 mM sodium phosphate buffer could be used to specifically select non-hemolytic, broad-spectrum antimicrobial peptides from combinatorial libraries, suggesting that this may be a good consensus membrane composition to study AMPs.
Upon binding of AMPs to anionic lipid vesicles through electrostatic and hydrophobic interactions, many types of perturbations can occur, including membrane permeabilization. But AMPs can also drive vesicle aggregation, vesicle fusion, formation of non-bilayer phases, lipid phase separation, transbilayer movement (flip-flop) of lipids and complete solubilization of membranes. Some of these effects may not relate to antimicrobial activity and should be considered experimental artifacts. Although not often reported in the literature, vesicle aggregation and fusion are very common occurrences when cationic peptides are added to anionic vesicles(13), as evidenced by rapid increases in turbidity upon peptide addition. It is likely that some (or many) reports of peptide-induced “leakage” from anionic vesicles observed at very high P:L ratios (such as P:L = 1:10) include leakage that is due to leaky fusion/aggregation events in addition to (or instead of) direct membrane permeabilization. Few attempts have been made to distinguish leakage that is purely a consequence of fusion from legitimate leakage that occurs without vesicle fusion. In a recent paper(13) we examined AMP-induced fusion and leakage independently and found that leakage and vesicle fusion both occurred at P:L ≥ 1:50 while only leakage occurred at lower P:L (up to 1:500). We concluded that leakage measured at P:L = 1:50 may have included a contribution from fusion as well as membrane permeabilization.
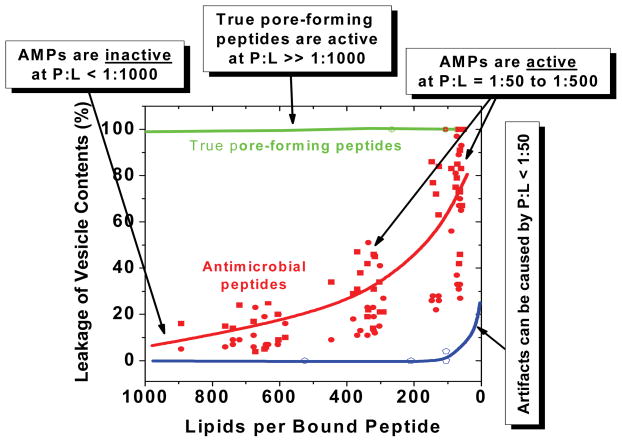
As shown by the example data in Figure 3, most antimicrobial peptides are active against vesicles when bound peptide to lipid ratios are from about 1:500 to 1:50, or about 200–2000 peptides per vesicle. In contrast, ideal pore-forming peptides can permeabilize lipid vesicle with as few as 10 peptides bound per vesicle, or 1 peptide per 10,000 lipids(37). We speculate that AMPs are not more potent because the very potent peptides lack selectivity. On the other end of the spectrum, any membrane-interacting molecule can disrupt membranes at very high concentrations. For this reason, it is likely that some published results for putative AMPs permeabilizing vesicles are essentially artifacts that arise from extremely high peptide:lipid ratios. The literature contains many experiments which show significant leakage of vesicle entrapped contents at P:L of 1:10, 1:1 or even at P:L = 10:1. Any in vitro membrane permeabilization measurements that show activity only at more than 1 peptide bound per 50 lipids (P:L = 1:50) should be viewed with caution.
Figure 3.
Peptide activity against lipid vesicles. True transmembrane pore-forming peptides, such as alamethicin, permeabilize vesicles at very low peptide:lipid ratios. The green line is based on experimental measurements. Antimicrobial peptides, on the other hand, are active against lipid vesicles only at high peptide:lipid ratios. Data points in red are actual data from several recent publications(13;17). Almost any peptide that binds to membranes can cause leakage at very high concentration, shown schematically in blue.
The prevailing transmembrane pore models
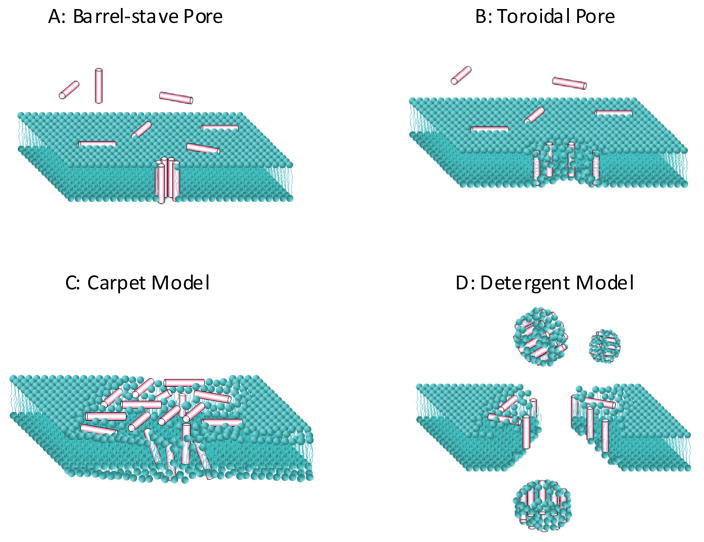
The simplest models of membrane permeation by peptides involve the formation of transbilayer pores or channels through the membrane as shown by the models in Figure 2. In a barrel stave pore, peptides interact laterally with one another to form a specific structure that is reminiscent of a membrane protein ion channel. In the toroidal pore model, specific peptide-peptide interactions are not present. Instead, peptides affect the local curvature of the bilayer in a cooperative manner such that a toroid of high curvature forms. These two structures are fundamentally different. Barrel stave pores work with the bilayer hydrocarbon core, using it as a template for peptide-self assembly, while toroidal pores work against the hydrocarbon core, disrupting the normal segregation of polar and non-polar parts of the membrane by providing alternate surfaces for lipid hydrocarbon and headgroups to interact favorably with.
Figure 2.
Commonly cited models for antimicrobial peptide activity. Barrel-stave and toroidal pores are membrane-spanning aqueous channels. Antimicrobial peptides are described with the carpet model. Such peptides permeabilize membranes by “carpeting” the bilayer with peptides. At high concentrations carpet model peptides can behave more like detergents.
A classical example of a lytic peptide toxin that almost certainly forms transmembrane pores is alamethicin, a 20 residue fungal peptide that folds into an amphipathic α-helix. Alamethicin can exist, depending on hydration and concentration, either parallel or perpendicular to the lipid bilayer normal(38). The perpendicular structure (39;40) is consistent with transmembrane barrel stave or toroidal pore. Huang has given evidence that the pore is a barrel-stave pore(39), however we have shown that alamethicin catalyzes very rapid transbilayer equilibration of lipids, which is consistent with a toroidal pore (41). In any case, the evidence clearly suggests a transmembrane pore for alamethicin. The lytic peptide toxin melittin, from honey bee venom, is another classical example of a pore-forming peptide which forms a membrane-spanning pore. But in the realm of membrane permeabilizing peptides, these classical “examples” are actually exceptions. In any case, alamethicin and melittin are not really antimicrobial peptides, which are defined by their host defense function, but instead are nonspecific, membrane-permeabilizing peptide toxins. For the vast majority of so called “pore forming” antimicrobial peptides, evidence for water-filled, transmembrane pore is scarce or lacking entirely, as we discuss below.
The prevailing non-pore models
In addition to the pore models described above, AMP activity has also been described using some common non-pore “models” that have been proposed to explain or categorize the mechanism of action of AMPs. The so called “carpet model” is the most commonly cited phenomenological model, and was proposed in 1996 by Shai (42) to explain the mechanism of action of mammalian cecropin P1 on model membranes. Cecropin P1 is always oriented parallel to the membrane surface and is active only at high P:L ratios. Shai and coworkers concluded that the peptide is active only when it forms a “carpet” on the bilayer surface. The “detergent model” is also often cited to explain the catastrophic collapse of membrane integrity and probe-size independent leakage observed with some AMPs at high peptide concentration(35).
While these catchphrases, and others, have been useful towards providing a common lexicon to a diverse field, they lack a specific molecular or physical-chemical basis for membrane-permeabilizing activity. While these models may have broadly inspired successful design efforts(16), they have rarely been used to make specific molecular predictions, or drive engineering and design. In contrast some recent discussions have a laid a physical chemical foundation that should be much more useful. For example, Bechinger and Lohner (43;44) recently presented a model based on molecular shape to describe antimicrobial peptide activity. They suggested that the physical chemistry of AMPs and membranes could be described with phase diagrams, as one would describe mixtures of lipids. Recently, Epand and colleagues(45;46) have proposed that lateral lipid phase separation, or lipid clustering, induced by cationic antimicrobial peptides explains their biophysical properties as well as their selectivity and biological activity. They have provided experimental evidence to support this idea suggesting that leakage could occur at phase boundary defects (45). Almeida and colleagues have described AMP activity in terms of binding, insertion and perturbation (32;47–49) using kinetic and thermodynamic models. Such quantitative descriptions of AMP activity, once they are parameterized to correlate peptide sequence or composition to activity, could allow for predictions of activity to be made and thus could be useful for engineering. We suggest here that the physical chemical concept of “interfacial activity”, perhaps in conjunction with these other concepts, will be useful to explain, predict and engineer the activity of AMPs.
AMP may not form transmembrane pores
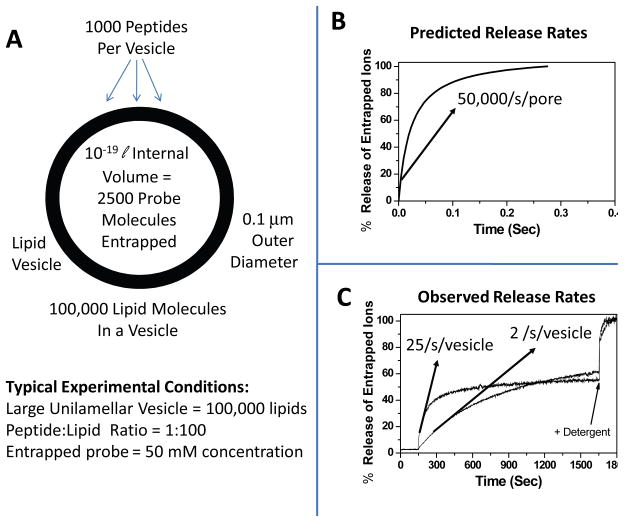
Despite innumerable studies of AMP activity in cells, vesicles and other synthetic bilayer systems, compelling evidence for specific, transmembrane, pores or channels in vesicle bilayers, even transient pores, is rare. While some structural studies have indirectly supported the idea of discreet pores(50–52), vesicle-based leakage experiments are mostly inconsistent with this idea. AMPs bind to vesicles rapidly (<30 sec) (17) and remain bound at equilibrium. Because large unilamellar vesicles (LUV) contain roughly 100,000 lipids each, in a typical experiment (peptide:lipid ratio, P:L, of 1:100) there are about 1000 peptides bound to each vesicle, capable of forming a hundred or more small pores. To examine what would happen to a vesicle’s contents if a pore were present, we have performed numerical simulations of uncharged small molecule release from single vesicles based on random walk diffusion through single pores. These simulations predict the release kinetics of entrapped small probes from a large unilamellar vesicle(53) through a single water-filled channel of 10 Å diameter (Figure 4). The calculated release rate of 50,000 ions per second per pore is consistent with the known ability of protein ion channels to pass as many as 107 ions per second (54). Because the interior of a “large” unilamellar vesicle is only about 10−19 liters there are only a few hundred to a few thousand probe molecules inside each vesicle. The simulation results, in figure 4, show that a single aqueous channel through a bilayer will release all the contents of a vesicle in a few tenths of a second, at most. A vesicle with 1000 bound peptides (P:L = 1:100) could conceivably contain more than 100 pores. In reality, vesicles with 1000 bound antimicrobial peptides typically release only a portion of their contents over tens of minutes, with maximum initial rates rates from 0.1 to 100 ions per second per vesicle. Commonly, initial rates are 1-10 ions per second per vesicle (i.e. per 1000 peptides). The observed release rates are thus at least three orders of magnitude slower than predicted for even a single aqueous channel. Furthermore, leakage from vesicles caused by AMPs is often incomplete, suggesting that true pores do not form, even transiently. This analysis suggests that most AMPs do not ever form true pores in large unilamellar vesicles and leakage occurs by a general disruption of membrane integrity.
Figure 4.
Release of vesicle-entrapped probes. A: A large unilamellar vesicle of 0.1 μm LUV, the type used in most experiments, is made of roughly 105 lipids enclosing a volume of about 10−19 liter. B: Predicted kinetics of probe release from large unilamellar vesicles containing a single pore of 10 Å diameter. Complete release occurs in a few tenths of a second. C: Actual release kinetics observed in experiments with AMPs. Note the difference in X-axis scale indicating that actual release data is 3 orders of magnitude slower than predicted. Furthermore, the simulated release goes to completion, while the actual release is incomplete.
Giant Unilamellar Vesicle Studies
Studies of AMP activity in giant unilamellar vesicles (GUV) are revealing because they allow individual vesicles and individual events to be monitored. Not surprisingly, different AMPs studied in GUVs in different laboratories behave quite variably. In some cases, individual vesicles accumulate AMPs on their surface until a catastrophic burst event occurs (55). Bursting occurs only above a threshold peptide concentration, and is a stochastic event in a collection of vesicles. This behavior is consistent with the all or none release behavior observed in LUVs (described above). In other GUV experiments, vesicles remain intact after AMP addition but steadily lose entrapped contents over the course of a few hundred seconds(56;57). Toroidal or barrel stave pores have been invoked to explain this type of observation. Given the 100 fold larger size of a GUV relative to an LUV, leakage through a single pore is expected to occur in the observed time range (Figure 4). But a single GUV actually has enough bound peptide to have millions of peptide pores. In a typical GUV experiment, a small concentration of GUVs (very low lipid concentration, see Fig. 5) are exposed to 1-10 μM peptide. The bound peptide:lipid ratio will be high, minimally P:L = 1:50 for a peptide that binds moderately well (see Figure 5). A 10 μm GUV will contain about 109 lipids (compared to 105 for an LUV) meaning that a GUV will have roughly 2×107 peptides bound to it in a typical experiment. That is enough peptide bound to a single giant vesicle to form about two million pores, assuming decameric pores. The fact that release of entrapped contents is very slow even when there are twenty million peptides bound to each vesicle indicates that transmembrane pore formation, if it occurs at all, is a very rare event. It is much more likely that there is a low flux of markers through the membrane caused by the global disruption of the lipid packing caused by the peptides. Tamba and Yamazaki called this a “two-state transition” model of external binding followed by critical destabilization of the bilayer. This is the same idea used to explain “carpet model” leakage from LUVs.
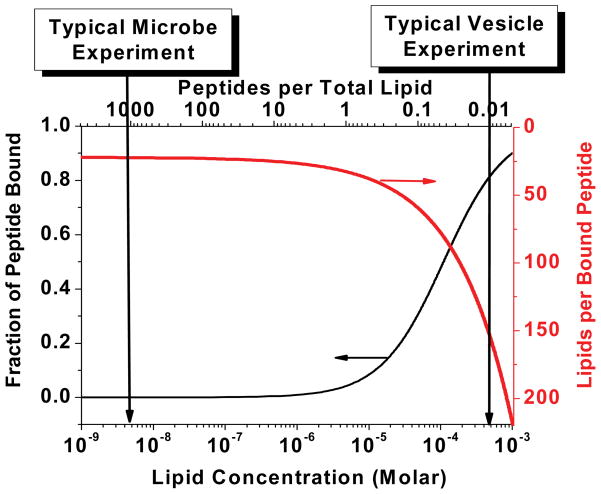
Figure 5.
Typical peptide:lipid ratios in vesicle leakage experiments and microbe sterilization assays. Microbe sterilization assays are done under conditions of peptide excess over lipid such that the membrane is saturated by peptide and there is a large reservoir of unbound peptide. Vesicle leakage experiments are done under conditions where lipid is in excess and most peptide is bound.
The Enigma of Partial Transient Release
Studies of vesicle permeabilization by AMPs (e.g. Fig. 4C) frequently show an enigmatic and still unexplained behavior: Upon addition of peptide to a homogeneous population of vesicles (or vesicles to peptide) a rapid burst of leakage occurs which then stops with a halftime of just a few minutes. Leakage often ceases before complete loss of entrapped contents has taken place. A second addition of peptide will cause a second burst of leakage which also stops (WCW unpublished observations). Increasing the peptide concentration will increase the fractional loss of contents. Perhaps even more enigmatic is the observation that partial transient release is sometimes graded(13), in which all of the vesicles release a fraction of their contents, and is sometimes all-or-none(17), in which a fraction of the vesicles release all of their contents and the remainder release none of their contents. Furthermore, AMP-induced leakage of probes is sometimes probe-size dependent and sometimes probe-size independent. For many AMPs the rate of leakage after the rapid initial burst is very slow, sometimes immeasurably slow. For example, we have shown that some antimicrobial peptides (13;17) release almost no measurable amount of entrapped markers for as long as 24 hours after the initial burst, despite the fact that hundreds of peptides remain bound to each vesicle and that the peptides have the same secondary structure as during the initial leakage phase. See Figure 4C for example of actual release data. Published leakage curves for many AMPs are very similar to these curves. In sharp contrast, true pore-forming peptides, such as alamethicin, cause rapid and continual leakage of vesicle contents which does not stop until complete release has occurred. As shown diagrammatically in Figure 3, leakage caused by true pore forming peptides occurs at P:L ratios as low as 1:10000 (10 peptide/vesicle) whereas most AMPs have good activity only in the range of P:L = 1:100 (1000 peptides per vesicle) or higher.
Transient failure
Partial transient release shows that vesicle permeabilization by most AMPs must be driven by non-equilibrium events. It has been suggested that it is driven by the disequilibrium state that immediately follows the addition of peptide to the vesicles in which only the outer monolayer of the vesicle has peptide bound to it. The resulting imbalance of charge, area, and surface tension may drive the transient, sometimes catastrophic, failure of the bilayer structure and concomitant leakage of entrapped contents. Depending on the magnitude and lifetime of the failure, it is possible to imagine either graded or all-or-none release, and it is possible to imagine size-dependent or size independent release.
In any case, release driven by AMPs over a wide range of peptide concentration usually stops or slows significantly before all vesicle contents have been released. This probably means that once transbilayer equilibrium of peptides has been reached, the driving force for large-scale bilayer destabilization is decreased. Recently, we reported a family of AMPs that causes partial, all-or-none release of vesicle contents(17). Assuming that all peptides ultimately reach transbilayer equilibrium, this observation suggests that in addition to the stochastic, transient event that relieves transbilayer asymmetry of peptides and concomitantly causes complete release of entrapped contents, there can also be a “silent” non permeabilizing pathway to equilibrium. At least for this family of peptides, a fraction of the vesicles reach equilibrium by a catastrophic event, losing all their contents, while the remaining vesicles reach equilibrium by a process that involves no leakage whatsoever. The silent pathway to equilibrium is presumably peptide translocation across the bilayer. At higher peptide concentration, the fraction of vesicles releasing all their contents increases, consistent with an increase in probability of catastrophic bilayer permeabilization.
A comparison of biophysical and biological experiments
While the biological activity of AMPs and their vesicle permeabilizing activity are correlated, it is not known how closely the mechanisms of action overlap in the two systems. For example, there are no data to suggest that transient release phenomena occur in microbial permeabilization. The overlap in mechanism is a critical question in engineering and design studies, because these are almost always based on vesicle permeabilization assays. To begin comparing mechanisms of action we have contrasted a typical in vitro experiment using synthetic lipid bilayers with a typical biological experiment measuring on microbe sterilization. Experimentally verified stoichiometry information from the two classes of experiments is shown in Table 1. In both experiments it is common to use peptide concentrations in the low micromolar range, ~ 5 μM. Vesicle experiments are typically carried out at 0.1 – 1 millimolar lipid, which gives system P:L ratios roughly P:L = 1:100. Biological assays are carried out at 104–106 bacterial cells per ml (58). Assuming that an E. coli cell is tubular and is 1 μm wide and 2 μm long, and taking into account the area of a lipid molecule (~0.7 nm2)(59) and the fact that there are two membranes, we calculate roughly 2.5×107 lipids in bacterial cell. Thus, at 105 cells per ml the total lipid concentration is about 5 nanomolar. This is 4–5 orders of magnitude lower lipid concentration than a typical vesicle-based experiment. The total (system) peptide to lipid ratio in a microbe sterilization experiment is as high as 1000:1, compared to P:L = 1:100 in a typical vesicle leakage experiment.
Table 1.
Statistics of peptide and lipid stoichiometry in vesicle permeabilization and microbe sterilization assays calculated for typical experimental conditions. Bound peptide:lipid ratios are conservative estimates based on direct, experimentally measured binding (see text).
| Property | Large Unilamellar Vesicle | E. coli sterilization assay |
|---|---|---|
| Dimension | 0.1 μm diameter | 1 μm × 2 μm diameter |
| Aqueous Volume | 10−19 liter | 10−15 liter |
| Number of Lipids | 100,000/vesicle | 25,000,000/cell |
| Typical Concentrations | 500 μM lipid 1×1012 vesicles/ml |
105 cells/ml 0.004 μM lipid |
| 5 μM peptide | 5 μM peptide | |
| Relative Peptide | 1×103 peptides/vesicle | 1 ×109 peptides/cell |
| Total Peptide:Lipid | 1 peptide:100 lipids | 1000 peptide : 1 lipid |
| Bound Peptide:Lipid | 1 peptide : 200 lipids | 100 peptides : 1 lipid |
In order to compare these experiments directly, we must know how much of the peptide is actually bound. In vesicle permeabilization experiments using anionic vesicles, peptide binding, when measured, is usually significant; 10 to 100% of the total peptide is bound to the vesicles(11). For this comparison, we assume 50% binding under typical conditions. The few available direct measurements of AMP binding to microbes show that they bind much better than one would predict from lipid vesicle binding. Despite the fact that total lipid concentration of a microbe experiment is nM, 10–95% of a μM peptide is bound to microbes within 10–15 minutes of addition, (60)(R. Rathinakumar and W.C. Wimley, unpublished observations). Thus, based on direct measurements of binding, in a typical microbe sterilization assay there are roughly 10–100 peptides bound for each bacterial lipid molecule (Table 1) amounting to as many as 109 peptides bound to every cell. Importantly, such high concentrations of bound peptide are required for activity; a ten-fold decrease in peptide concentration (e.g. from 5 μM to 0.5 μM peptide) eliminates antimicrobial activity for most AMPs despite the fact that there are still huge numbers of peptides bound to each cell. Of course, it is not physically possible to have 100 peptides bound per lipid in a membrane; peptide must be binding in large amounts to other cellular components. AMPs are known to bind lipopolysaccharide, cell wall components, and DNA (61;62). Whatever the actual membrane binding, it is reasonable to assume the microbial membranes are saturated with peptide and that there is a large reservoir of non-membrane peptide bound to each cell. With as many as 109 peptides bound to each cell, AMPs will actually outnumber all proteins, ATP, and most electrolytes and metabolites. (For example, there are probably several million ATP molecules per bacterial cell). The practical consequence of these numbers is that microbe killing can result from leakage that is very inefficient. For example, one critical molecule like ATP released for every 100 bound peptides is enough to deplete ATP completely. Given the conditions described in Table 1, it should not be surprising that bacteria do not frequently develop inducible resistance to AMPs, and when they do it involves changes in gross membrane architecture(63). Most importantly, these numbers show that the formation of explicit transmembrane pores is not necessary to explain the biological activity of AMPs.
For comparison, a similar calculation can be done for a typical erythrocyte hemolysis assay (e.g. 50 μM peptide added to 5×108 red blood cells (8)). This calculation gives a system P:L ratio of about 1:1. Because binding of AMPs to erythrocytes is weak, the bound P:L ratio may be 1:20 or less. Thus, in terms of overall peptide to lipid ratio, a typical hemolysis assay is performed under conditions that are comparable to lipid vesicle leakage assays. While the overall peptide to lipid ratio in a hemolysis assay is quite dissimilar to the conditions of a microbe sterilization assay, it is likely that the membranes in both assays are close to saturation, and thus may be comparable to one another.
The Interfacial Activity Model
The hydrocarbon core of an unperturbed lipid bilayer membrane is one of the most hydrophobic microenvironments found in nature, with physical-chemical properties that are very similar to a liquid alkane phase(64). Accordingly, the hydrocarbon core (25 to 30 Å thick) normally imparts a strict barrier to the permeation of polar or charged solutes through the bilayer. Yet, it is positioned between the two bilayer interfacial zones, called “zones of tumultuous chemical heterogeneity” by Wiener and White (64)). Each interfacial zone is 10–15 Å thick and is comprised of the lipid polar groups, water and solution counter ions, as well as small amounts of the hydrocarbon groups.
The property that we define as “interfacial activity” is the ability of a peptide to perturb the permeability barrier imposed by the hydrocarbon core by partitioning into the interfacial region of the bilayer and driving local rearrangements in vertical lipid packing (i.e. normal to the bilayer plane) that alter the segregation between the hydrocarbon core and the interfacial groups. This activity is driven by peptides that bind to membranes, and are amphipathic, but with imperfect segregation of polar and nonpolar groups. Unlike a very hydrophobic peptide, or one with ideal amphipathicity, when an interfacially active peptide is bound to a bilayer, the bilayer must be deformed, and the hydrocarbon disrupted (i.e. intermingled with polar lipid headgroup moieties), to simultaneously accommodate the nonpolar and polar/charged groups of the peptide (11). Examination of the structures and sequences of AMPs (Figure 1) reveals that most of them have a hydrophobic segment or patch, but one that is not nearly large enough to span a bilayer, and is interrupted by, or bounded by at least one polar residue, frequently arginine or lysine. Such “imperfect amphipathicity” is necessary for interfacial activity. Much of the available data suggests that the exact structure or spatial arrangement of the hydrophobic and polar groups is not as important as the physical chemical balance between the two types of interactions. When such a peptide binds to a bilayer, the hydrophobic portion, often dominated by aromatic residues drives a deep partitioning into the interfacial zones. But the nearby polar residues promote the incursion of lipid polar groups deeper in the membrane, along with the polar residues of the peptide. We suggest that the signature of an interfacially active peptide in a lipid bilayer experiment will be bilayer translocation of peptide and lipid, even at low peptide concentration. At higher peptide concentrations, the non-equilibrium distribution causes a transient, cooperative transbilayer movement of peptide, lipids and polar solutes, which defines the leakage activity of the peptide.
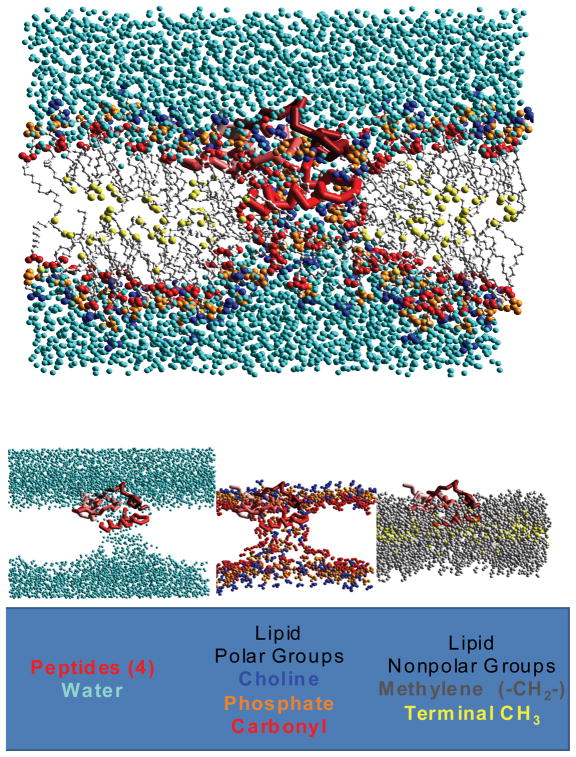
In Figure 6, we show the results of molecular dynamics simulations of a peptide “pore” conducted by Sengupta and Marrink (65). These simulations capture the essence of the interfacial activity model in several aspects. The presence of peptides, which are mostly partitioned into the interfacial zone, dramatically perturbs the normally strict segregation of polar and nonpolar moieties across the bilayer. Compare, for example, the distribution of water and lipid polar groups in the vicinity of the peptides (bilayer middle) and away from the peptides (bilayer left/right). The leakage of polar solutes across the bilayer occurs because they are carried across the bilayer along with peptide and lipid molecules when the perturbed bilayer structure is formed. The most significant difference between the simulation results and the interfacial activity model is that some interfacially active antimicrobial peptides can act without self-assembly, as shown experimentally by Almeida (49).
Figure 6.
Molecular dynamics simulation of peptide pore formation from Sengupta and Marrink (65). Top: The peptide-lipid bilayer with CH2 groups removed. Blue spheres are water molecules and yellow spheres are the terminal methyl groups. Other color spheres are the lipid polar groups. Bottom: The same peptide-lipid bilayer separated into groups. Notice in the vicinity of the peptide “pore” the strict segregation between polar and non-polar is broken down.
When one considers interfacial activity in the context of a peptide-saturated bacterial membrane, the details will be different from a lipid vesicle experiment, but the interactions are driven by the same basic physical chemistry. This is the reason there is a strong correlation between the two experiments, but not an exact overlap in mechanism. We have proposed here that the interfacial activity of imperfectly amphipathic peptides, as measured in lipid vesicle experiments, is a reasonable surrogate measurement for the likelihood that a peptide will have broad-spectrum antimicrobial activity.
Experimentally Testable Predictions
The interfacial activity model can be expressed in terms of experimentally testable predictions.
Interfacially active peptides partition into the bilayer interface and locally perturb polar-nonpolar segregation of lipid moieties.
Interfacial activity requires imperfect amphipathicity and the proper balance of hydrophobic and polar amino acids.
Interfacial activity will depend more on amino acid composition than peptide structure or sequence.
Interfacial activity does not require peptide self-assembly.
The permeation pathway is mostly made of lipid and peptide, not an aqueous channel.
Only a few molecules are transported across the membrane per peptide.
At similar bound peptide to lipid ratios, an interfacially active peptide will behave similarly in all fluid phase membranes.
At low bound peptide to lipid ratios AMPs will translocate across membranes.
Lipid translocation, peptide translocation and membrane leakage will always be coupled.
Leakage from vesicles will depend on the rate of peptide addition.
Non-peptide molecules (peptide mimetics or other polymers) which are imperfectly amphipathic can have AMP-like activity.
Conclusions
In terms of detailed molecular mechanisms, multiple overlapping mechanisms of AMP activity probably exist and it may not be possible to define a single unifying description. What we have done here is to define the physical chemical commonalities that seem to be important in antimicrobial peptide activity. While there is overlap between the interfacial activity model, and others in the literature, we think it is beneficial to begin to think about the problem of antimicrobial peptide mechanism, engineering and design using an experimentally testable, mechanistic and semi-molecular model of AMP action.
Supplementary Material
Acknowledgments
For many interesting antimicrobial peptide discussions over the course of many years, I thank Kalina Hristova, Paulo Almeida, Antje Pokorny, Mikhail Merzliakov, Yechiel Shai, Jack Blazyk, Bill Walkenhorst, Josh Rausch, Chris Bishop, Ramesh Rathinakumar, Aram Krauson, Jessica Marks, Thomas Freeman, Drew Hoffmann, Jing He, Andrew Wimley and the members of the New Orleans Protein Folding Intergroup. This work is supported by the National Institutes of Health GM060000 and the Louisiana Board of Regents Support Fund RC/EEP-05 (2007–2010).
Footnotes
Active against multiple distinct classes of microbes; e.g. Gram positive bacteria, Gram negative bacteria and fungi.
References
- 1.Steiner H, Hultmark D, Engstrom A, Bennich H, Boman HG. Sequence and specificity of two antibacterial proteins involved in insect immunity. Nature (London) 1981;292:246–248. doi: 10.1038/292246a0. [DOI] [PubMed] [Google Scholar]
- 2.Okada M, Natori S. Purification and characterization of an antibacterial protein from haemolymph of Sarcophaga peregrina (flesh-fly) larvae. Biochem J. 1983;211:727–734. doi: 10.1042/bj2110727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Patterson-Delafield J, Szklarek D, Martinez RJ, Lehrer RI. Microbicidal cationic proteins of rabbit alveolar macrophages: Amino acid composition and functional attributes. Infect Immun. 1981;31:723–731. doi: 10.1128/iai.31.2.723-731.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zasloff M. Magainins, a class of antimicrobial peptides from Xenopus skin: isolation, characterization of two active forms, and partial cDNA sequence of a precursor. Proc Natl Acad Sci U S A. 1987;84:5449–5453. doi: 10.1073/pnas.84.15.5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tossi A. Antimicrobial peptides datatbase. 2005 http://www.bbcm.units.it/~tossi/. Web Site.
- 6.Wang Z, Wang G. APD: the Antimicrobial Peptide Database. Nucleic Acids Res. 2004;32:590–592. doi: 10.1093/nar/gkh025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fjell CD, Hancock RE, Cherkasov A. AMPer: a database and an automated discovery tool for antimicrobial peptides. Bioinformatics. 2007;23:1148–1155. doi: 10.1093/bioinformatics/btm068. [DOI] [PubMed] [Google Scholar]
- 8.Blazyk J, Wiegand R, Klein J, Hammer J, Epand RM, Epand RF, Maloy WL, Kari UP. A novel linear amphipathic beta-sheet cationic antimicrobial peptide with enhanced selectivity for bacterial lipids. J Biol Chem. 2001;276:27899–27906. doi: 10.1074/jbc.M102865200. [DOI] [PubMed] [Google Scholar]
- 9.Hilpert K, Elliott MR, Volkmer-Engert R, Henklein P, Donini O, Zhou Q, Winkler DF, Hancock RE. Sequence requirements and an optimization strategy for short antimicrobial peptides. Chem Biol. 2006;13:1101–1107. doi: 10.1016/j.chembiol.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 10.Avrahami D, Shai Y. Conjugation of a magainin analogue with lipophilic acids controls hydrophobicity, solution assembly, and cell selectivity. Biochemistry. 2002;41:2254–2263. doi: 10.1021/bi011549t. [DOI] [PubMed] [Google Scholar]
- 11.Rathinakumar R, Walkenhorst WF, Wimley WC. Broad-spectrum Antimicrobial Peptides by Rational Combinatorial Design and High-throughput Screening: The Importance of Interfacial Activity. J Am Chem Soc. 2009;131:7609–7617. doi: 10.1021/ja8093247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rausch JM, Marks JR, Wimley WC. Rational combinatorial design of pore-forming beta-sheet peptides. Proc Natl Acad Sci U S A. 2005;102:10511–10515. doi: 10.1073/pnas.0502013102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rausch JM, Marks JR, Rathinakumar R, Wimley WC. Beta-sheet pore-forming peptides selected from a rational combinatorial library: mechanism of pore formation in lipid vesicles and activity in biological membranes. Biochemistry. 2007;46:12124–12139. doi: 10.1021/bi700978h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hilpert K, Volkmer-Engert R, Walter T, Hancock RE. High-throughput generation of small antibacterial peptides with improved activity. Nat Biotechnol. 2005;23:1008–1012. doi: 10.1038/nbt1113. [DOI] [PubMed] [Google Scholar]
- 15.Jin Y, Hammer J, Pate M, Zhang Y, Zhu F, Zmuda E, Blazyk J. Antimicrobial activities and structures of two linear cationic peptide families with various amphipathic beta-sheet and alpha-helical potentials. Antimicrob Agents Chemother. 2005;49:4957–4964. doi: 10.1128/AAC.49.12.4957-4964.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mowery BP, Lee SE, Kissounko DA, Epand RF, Epand RM, Weisblum B, Stahl SS, Gellman SH. Mimicry of antimicrobial host-defense peptides by random copolymers. J Am Chem Soc. 2007;129:15474–15476. doi: 10.1021/ja077288d. [DOI] [PubMed] [Google Scholar]
- 17.Rathinakumar R, Wimley WC. Biomolecular engineering by combinatorial design and high-throughput screening: small, soluble peptides that permeabilize membranes. J Am Chem Soc. 2008;130:9849–9858. doi: 10.1021/ja8017863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang YQ, Yuan J, Osapay G, Osapay K, Tran D, Miller CJ, Ouellette AJ, Selsted ME. A cyclic antimicrobial peptide produced in primate leukocytes by the ligation of two truncated alpha-defensins. Science. 1999;286:498–502. doi: 10.1126/science.286.5439.498. [DOI] [PubMed] [Google Scholar]
- 19.Ladokhin AS, Selsted ME, White SH. CD spectra of indolicidin antimicrobial peptides suggest turns, not polyproline helix. Biochemistry. 1999;38:12313–12319. doi: 10.1021/bi9907936. [DOI] [PubMed] [Google Scholar]
- 20.Liu Z, Brady A, Young A, Rasimick B, Chen K, Zhou C, Kallenbach NR. Length effects in antimicrobial peptides of the (RW)n series 12. Antimicrob Agents Chemother. 2007;51:597–603. doi: 10.1128/AAC.00828-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Makovitzki A, Avrahami D, Shai Y. Ultrashort antibacterial and antifungal lipopeptides. Proc Natl Acad Sci U S A. 2006;103:15997–16002. doi: 10.1073/pnas.0606129103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsuzaki K, Sugishita K-I, Harada M, Fujii N, Miyajima K. Interactions of an antimicrobial peptide, magainin 2, with outer and inner membranes of Gram-negative bacteria. Biochim Biophys Acta. 1997;1327:119–130. doi: 10.1016/s0005-2736(97)00051-5. [DOI] [PubMed] [Google Scholar]
- 23.Van Abel RJ, Tang YQ, Rao VSV, Dobbs CH, Tran D, Barany G, Selsted ME. Synthesis and characterization of indolicidin, a tryptophan-rich antimicrobial peptide from bovine neutrophils. Int J Pept Protein Res. 1995;45:401–409. doi: 10.1111/j.1399-3011.1995.tb01055.x. [DOI] [PubMed] [Google Scholar]
- 24.Zhang G, Lin X, Long Y, Wang Y, Zhang Y, Mi H, Yan H. A peptide fragment derived from the T-cell antigen receptor protein alpha-chain adopts beta-sheet structure and shows potent antimicrobial activity. Peptides. 2009;30:647–653. doi: 10.1016/j.peptides.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 25.Rathinakumar R, Wimley WC. High-throughput discovery of broad-spectrum peptide antibiotics. FASEB J. 2010 doi: 10.1096/fj.10-157040. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heitz F, Morris MC, Divita G. Twenty years of cell-penetrating peptides: from molecular mechanisms to therapeutics. Br J Pharmacol. 2009;157:195–206. doi: 10.1111/j.1476-5381.2009.00057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park N, Yamanaka K, Tran D, Chandrangsu P, Akers JC, de Leon JC, Morrissette NS, Selsted ME, Tan M. The cell-penetrating peptide, Pep-1, has activity against intracellular chlamydial growth but not extracellular forms of Chlamydia trachomatis. J Antimicrob Chemother. 2009;63:115–123. doi: 10.1093/jac/dkn436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yesylevskyy S, Marrink SJ, Mark AE. Alternative mechanisms for the interaction of the cell-penetrating peptides penetratin and the TAT peptide with lipid bilayers. Biophys J. 2009;97:40–49. doi: 10.1016/j.bpj.2009.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsuzaki K. Control of cell selectivity of antimicrobial peptides. Biochim Biophys Acta. 2008;1788:687–1692. doi: 10.1016/j.bbamem.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 30.Hong RW, Shchepetov M, Weiser JN, Axelsen PH. Transcriptional profile of the Escherichia coli response to the antimicrobial insect peptide cecropin A. Antimicrob Agents Chemother. 2003;47:1–6. doi: 10.1128/AAC.47.1.1-6.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Almeida PF, Pokorny A. Mechanisms of antimicrobial, cytolytic, and cell-penetrating peptides: from kinetics to thermodynamics. Biochemistry. 2009;48:8083–8093. doi: 10.1021/bi900914g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pokorny A, Kilelee EM, Wu D, Almeida PF. The activity of the amphipathic peptide delta-lysin correlates with phospholipid acyl chain structure and bilayer elastic properties. Biophys J. 2008;95:4748–4755. doi: 10.1529/biophysj.108.138701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hunter HN, Jing W, Schibli DJ, Trinh T, Park IY, Kim SC, Vogel HJ. The interactions of antimicrobial peptides derived from lysozyme with model membrane systems. Biochim Biophys Acta. 2005;1668:175–189. doi: 10.1016/j.bbamem.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 34.Cronan JE. Bacterial membrane lipids: where do we stand? Annu Rev Microbiol. 2003;57:203–224. doi: 10.1146/annurev.micro.57.030502.090851. [DOI] [PubMed] [Google Scholar]
- 35.Hristova K, Selsted ME, White SH. Critical role of lipid composition in membrane permeabilization by rabbit neutrophil defensins. J Biol Chem. 1997;272:24224–24233. doi: 10.1074/jbc.272.39.24224. [DOI] [PubMed] [Google Scholar]
- 36.Wimley WC, Selsted ME, White SH. Interactions between human defensins and lipid bilayers: Evidence for the formation of multimeric pores. Protein Sci. 1994;3:1362–1373. doi: 10.1002/pro.5560030902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parente RA, Nir S, Szoka F. Mechanism of leakage of phospholipid vesicle contents induced by the peptide GALA. Biochemistry. 1990;29:8720–8728. doi: 10.1021/bi00489a031. [DOI] [PubMed] [Google Scholar]
- 38.He K, Ludtke SJ, Heller WT, Huang HW. Mechanism of alamethicin insertion into lipid bilayers. Biophys J. 1996;71:2669–2679. doi: 10.1016/S0006-3495(96)79458-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qian S, Wang W, Yang L, Huang HW. Structure of the Alamethicin Pore Reconstructed by X-ray Diffraction Analysis. Biophys J. 2008;94:3512–3522. doi: 10.1529/biophysj.107.126474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.North CL, Barranger-Mathys M, Cafiso DS. Membrane orientation of the N-terminal segment of alamethicin determined by solid-state 15N NMR. Biophys J. 1995;69:2392–2397. doi: 10.1016/S0006-3495(95)80108-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wimley WC, White SH. Determining the membrane topology of peptides by fluorescence quenching. Biochemistry. 2000;39:161–170. doi: 10.1021/bi991836l. [DOI] [PubMed] [Google Scholar]
- 42.Gazit E, Miller IR, Biggin PC, Sansom MSP, Shai Y. Structure and orientation of the mammalian antibacterial peptide cecropin P1 within phospholipid membranes. J Mol Biol. 1996;258:860–870. doi: 10.1006/jmbi.1996.0293. [DOI] [PubMed] [Google Scholar]
- 43.Bechinger B, Lohner K. Detergent-like actions of linear amphipathic cationic antimicrobial peptides. Biochim Biophys Acta. 2006;1758:1529–1539. doi: 10.1016/j.bbamem.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 44.Bechinger B. Rationalizing the membrane interactions of cationic amphipathic antimicrobial peptides by their molecular shape. Current Opinion in Colloid & Interface Science. 2009;14:349–355. [Google Scholar]
- 45.Epand RF, Maloy WL, Ramamoorthy A, Epand RM. Probing the “charge cluster mechanism” in amphipathic helical cationic antimicrobial peptides. Biochemistry. 2010;49:4076–4084. doi: 10.1021/bi100378m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Epand RM, Epand RF. Lipid domains in bacterial membranes and the action of antimicrobial agents. Biochim Biophys Acta. 2009;1788:289–294. doi: 10.1016/j.bbamem.2008.08.023. [DOI] [PubMed] [Google Scholar]
- 47.Pokorny A, Almeida PF. Kinetics of dye efflux and lipid flip-flop induced by delta-lysin in phosphatidylcholine vesicles and the mechanism of graded release by amphipathic, alpha-helical peptides. Biochemistry. 2004;43:8846–8857. doi: 10.1021/bi0497087. [DOI] [PubMed] [Google Scholar]
- 48.Gregory SM, Pokorny A, Almeida PF. Magainin 2 Revisited: A Test of the Quantitative Model for the All-or-None Permeabilization of Phospholipid Vesicles. Biophys J. 2009;96:116–131. doi: 10.1016/j.bpj.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gregory SM, Cavenaugh A, Journigan V, Pokorny A, Almeida PF. A quantitative model for the all-or-none permeabilization of phospholipid vesicles by the antimicrobial peptide cecropin A. Biophys J. 2008;94:1667–1680. doi: 10.1529/biophysj.107.118760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mani R, Cady SD, Tang M, Waring AJ, Lehrer RI, Hong M. Membrane-dependent oligomeric structure and pore formation of a beta-hairpin antimicrobial peptide in lipid bilayers from solid-state NMR. Proc Natl Acad Sci U S A. 2006;103:16242–16247. doi: 10.1073/pnas.0605079103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mani R, Buffy JJ, Waring AJ, Lehrer RI, Hong M. Solid-state NMR investigation of the selective disruption of lipid membranes by protegrin-1. Biochemistry. 2004;43:13839–13848. doi: 10.1021/bi048650t. [DOI] [PubMed] [Google Scholar]
- 52.Yang L, Weiss TM, Lehrer RI, Huang HW. Crystallization of antimicrobial pores in membranes: magainin and protegrin. Biophys J. 2000;79:2002–2009. doi: 10.1016/S0006-3495(00)76448-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mayer LD, Hope MJ, Cullis PR. Vesicles of variable sizes produced by a rapid extrusion procedure. Biochim Biophys Acta. 1986;858:161–168. doi: 10.1016/0005-2736(86)90302-0. [DOI] [PubMed] [Google Scholar]
- 54.Hille B. Ionic channels in nerve membranes. Prog Biophys Mol Biol. 1970;21:3–32. doi: 10.1016/0079-6107(70)90022-2. [DOI] [PubMed] [Google Scholar]
- 55.Domingues TM, Riske KA, Miranda A. Revealing the Lytic Mechanism of the Antimicrobial Peptide Gomesin by Observing Giant Unilamellar Vesicles. Langmuir. 2010;26:11077–11084. doi: 10.1021/la100662a. [DOI] [PubMed] [Google Scholar]
- 56.Tamba Y, Yamazaki M. Magainin 2-induced pore formation in the lipid membranes depends on its concentration in the membrane interface. J Phys Chem B. 2009;113:4846–4852. doi: 10.1021/jp8109622. [DOI] [PubMed] [Google Scholar]
- 57.Tamba Y, Yamazaki M. Single giant unilamellar vesicle method reveals effect of antimicrobial peptide magainin 2 on membrane permeability. Biochemistry. 2005;44:15823–15833. doi: 10.1021/bi051684w. [DOI] [PubMed] [Google Scholar]
- 58.Wiegand I, Hilpert K, Hancock RE. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc. 2008;3:163–175. doi: 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
- 59.Marsh D. CRC Handbook of Lipid Bilayers. CRC Press; Boca Raton: 1990. [Google Scholar]
- 60.Tran D, Tran PA, Tang YQ, Yuan J, Cole T, Selsted ME. Homodimeric theta-defensins from rhesus macaque leukocytes: isolation, synthesis, antimicrobial activities, and bacterial binding properties of the cyclic peptides. J Biol Chem. 2002;277:3079–3084. doi: 10.1074/jbc.M109117200. [DOI] [PubMed] [Google Scholar]
- 61.Papo N, Shai Y. Can we predict biological activity of antimicrobial peptides from their interactions with model phospholipid membranes? Peptides. 2003;24:1693–1703. doi: 10.1016/j.peptides.2003.09.013. [DOI] [PubMed] [Google Scholar]
- 62.Matsuzaki K, Sugishita K, Miyajima K. Interactions of an antimicrobial peptide, magainin 2, with lipopolysaccharide-containing liposomes as a model for outer membranes of gram-negative bacteria. FEBS Lett. 1999;449:221–224. doi: 10.1016/s0014-5793(99)00443-3. [DOI] [PubMed] [Google Scholar]
- 63.Peschel A, Sahl HG. The co-evolution of host cationic antimicrobial peptides and microbial resistance. Nat Rev Microbiol. 2006;4:529–536. doi: 10.1038/nrmicro1441. [DOI] [PubMed] [Google Scholar]
- 64.Wiener MC, White SH. Structure of a fluid dioleoylphosphatidylcholine bilayer determined by joint refinement of x-ray and neutron diffraction data. III. Complete structure. Biophys J. 1992;61:434–447. doi: 10.1016/S0006-3495(92)81849-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sengupta D, Leontiadou H, Mark AE, Marrink SJ. Toroidal pores formed by antimicrobial peptides show significant disorder. Biochim Biophys Acta. 2008;1778:2308–2317. doi: 10.1016/j.bbamem.2008.06.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.