Abstract
The direct detection of native proteins in heterogeneous solutions remains a challenging problem. Standard methodologies rely on a separation step to circumvent non-specific signal generation. We hypothesized that a simple and general method for the detection of native proteins in solution could be achieved through ternary complexation, where the conditional signal generation afforded by split-protein reporters could be married to the specificity afforded by either native receptors or specific antibodies. Toward this goal, we describe a solution phase split-luciferase assay for native protein detection, where we fused fragmented halves of firefly luciferase to separate receptor fragments or single-chain antibodies, allowing for conditional luciferase complementation in the presence of several biologically significant protein targets. To demonstrate the utility of this strategy, we have developed and validated assay platforms for the vascular endothelial growth factor (VEGF), the gp120 coat protein from HIV-1 and the human epidermal growth factor receptor 2 (HER2), a marker for breast cancer. The specificities of the recognition elements, CD4 and the 17b single-chain antibody, employed in the gp120 sensor allowed us to parse gp120s from different clades. Our rationally designed HER2 sensing platform was capable of discriminating between HER2 expression levels in several tumor cell lines. In addition, luminescence from reassembled luciferase was linear across a panel of cell lines with increasing HER2 expression. We envision that the proof of principle studies presented herein may allow for the potential detection of a broad range of biological analytes utilizing ternary split-protein systems.
Knowledge of aberrant protein expression patterns can be employed toward predicting outcomes to conventional therapy and aid in the development of individualized treatment (1). This is particularly evident in the treatment of breast cancer in which the expression of estrogen, progesterone and human epidermal growth factor receptor 2 (HER2) is used to stratify patients and guide the administration of both chemical and biological therapeutic agents (2). Taking a broader view, the interactions of cell surface receptors with their ligands play an important role in almost all viral infection as well as the initiation of signaling cascades. For example the interaction of the gp120 coat protein of HIV-1 with the CD4 receptor of T-lymphocytes leads to infection (3), while the interaction of the vascular endothelial growth factor (VEGF) with its receptor Flt-1 leads to angiogenesis thereby potentiating tumor growth (4). Consequently, simple and rapid methods for the direct detection of cellular receptors are desirable.
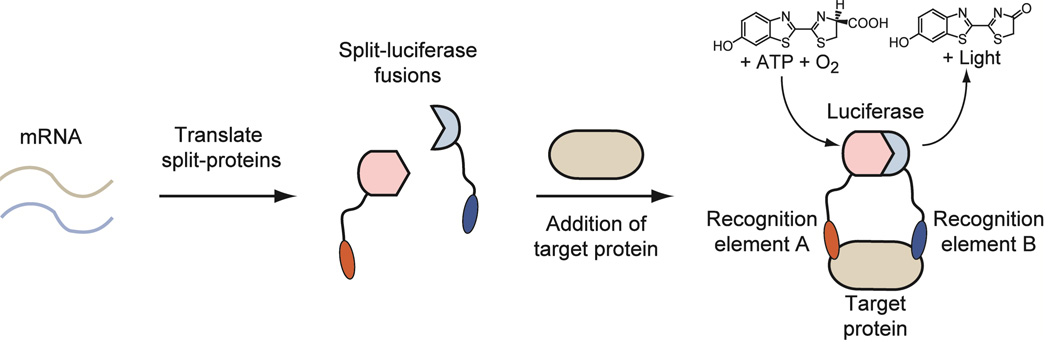
Elegant strategies for labeling and detecting native proteins in biological settings have been described, however many of these methods are encumbered by the inherent requirement for separation or chemical labeling (5). Current approaches often rely on chemical or biological derivatization and subsequent analysis by microscopy or Fluorescence-Activated Cell Sorting (FACS) (6). Alternatively, the classic Enzyme-Linked ImmunoSorbent Assay (ELISA) (7, 8) can be used to detect almost any analyte but requires that either the antigen or antibody is captured on a solid support prior to detection, followed by vigorous washing and subsequent recognition by an enzyme-secondary-antibody conjugate. This limits the utility of the ELISA for the direct detection of native proteins in complex heterogeneous fluids, such as blood or lysates. We note that techniques such as time-resolved fluorescence that exploit the long fluorescence lifetime of lanthanides such as Eu3+, also provide sensitive methods for protein detection that avoid complications associated with biological autofluorescence (9, 10). Ideally one could envision a one-step solution phase sandwich approach in which the activity of an attached split-protein reporter would depend solely upon formation of a ternary complex (Figure 1). Such a general methodology would potentially allow for the direct detection of any protein in complex environments without the need for immobilization, direct chemical derivatization or separation.
Figure 1.
A general schematic for ternary complexation mediated protein complementation is shown. mRNA encoding for split-luciferase fusions is used to initiate translation in a cell-free protein expression system. Specific recognition elements fused to the luciferase halves are used to reassemble a functional enzyme in the presence of a protein of interest leading to the generation of light. Translation, reassembly, and detection take place in the same solution.
Central to the strategy proposed above is the use of split-protein reassembly or protein complementation, which relies on a specific bimolecular interaction to drive reassembly of a fragmented reporter protein (11, 12). Johnsson and Varshavsky were the first to demonstrate this approach using split-ubiquitin (13), which has subsequently been applied to a variety of monomeric reporter proteins including dihydrofolate reductase (DHFR) (14), β-lactamase (15), GFP (16–18), Renilla luciferase (19), Gaussia luciferase (20), firefly luciferase (21), Trp1p (22), TEV protease (23) and, most recently, chorismate mutase (24). This enabling principle for the detection of bimolecular interactions has been used to delineate the yeast protein interactome (25) and could potentially lead to novel treatments for cancer (26). We have utilized this approach for the direct detection of DNA through ternary complexation (27–29), which was also utilized for the site-specific methylation of DNA (30). We sought to make use of the ternary complexation principle to develop a conditional detection platform for native extracellular proteins in heterogeneous solutions utilizing receptor fragments and antibodies as specific recognition elements.
Toward this long-term goal, we recently described a general cell-free split-protein assay for directly measuring heterodimeric protein-protein interactions (31). We identified a fragmented luciferase (32), discovered by Luker et al. through the incremental truncation approach (33), as a very sensitive protein complementation platform adaptable to a cell-free system (34, 35). Herein we adapt the cell-free split-luciferase methodology to provide a simple and potentially general solution for the rapid and direct detection of clinically relevant proteins, including growth factors as well as viral- and cell-surface receptors.
RESULTS AND DISCUSSION
A Cell-Free Split-Luciferase System for VEGF Detection
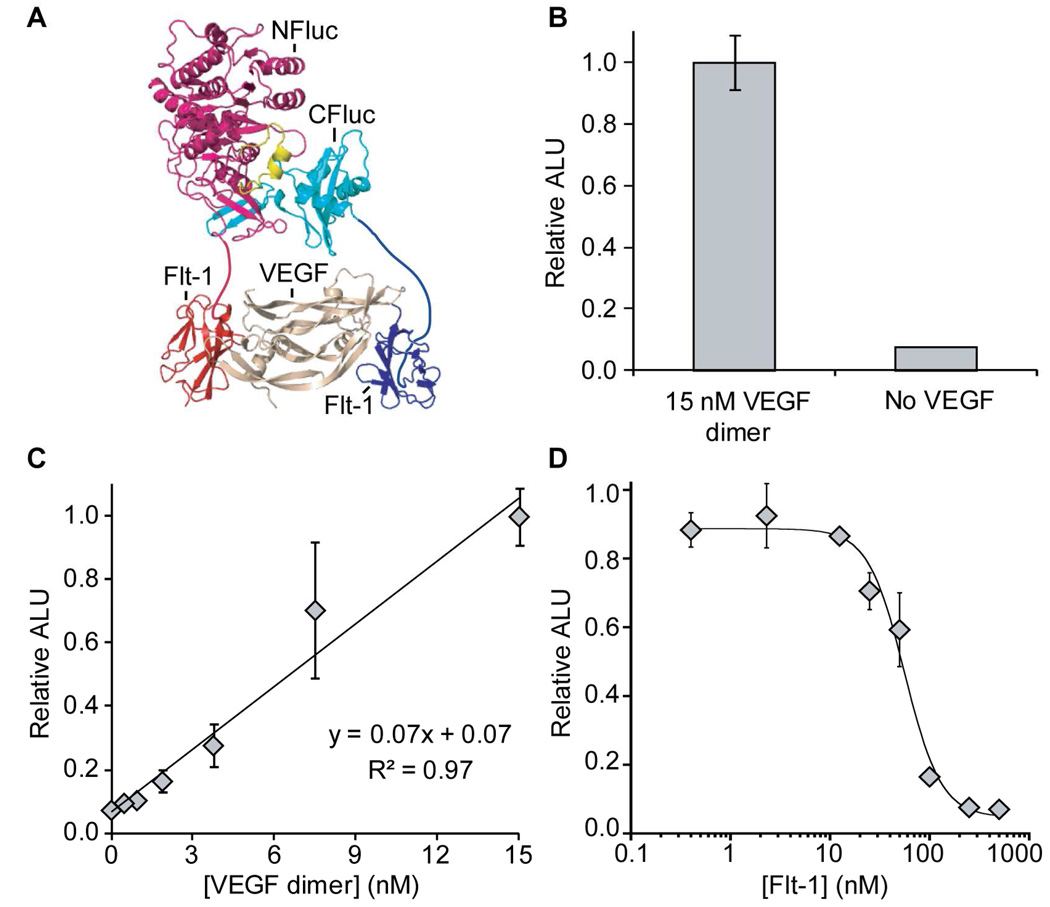
As a first test of ternary complexation, we set out to determine if a dimeric receptor fragment could be utilized to detect its extracellular ligand. We chose to target dimeric VEGF, implicated in tumor angiogenesis, which binds its extracellular receptor Flt-1 in a 1:2 stoichiometry with a reported Kd of 1.4 nM (36). We next attached the N- and C-terminal halves of luciferase (residues 2–416 and 398–550, respectively) to separate Flt-1 domain 2 fragments (Figure 2, panel a) with the expectation that a statistical distribution of Flt-1-luciferase halves bound to the VEGF dimer would still permit ~50% split-luciferase complementation. Expression of the split luciferase-Flt-1 fusion proteins in rabbit reticulocyte lysate led to an increase in luminescence of >13-fold only in the presence of 15 nM VEGF dimer, clearly demonstrating the ability to use receptor fragments to detect their ligands in this system (Figure 2, panel b). This detection system could potentially provide a means for high-throughput screening of molecules capable of disrupting the interaction of VEGF with its receptor (4). To further confirm the interaction between VEGF and Flt-1, we recombinantly expressed soluble fragments of these proteins (Figure S1) and demonstrated a 1:2 binding interaction between the VEGF dimer and Flt-1 by gel filtration (Figure S2 and S3). We additionally determined the limit of VEGF dimer detection by performing a titration, demonstrating detection of 500 pM (1.4 ng) VEGF dimer above two standard deviations from the average background signal (Figure 2, panel c). To demonstrate reversibility of VEGF/Flt-1 dependent split-luciferase reassembly we sought to compete out luciferase signal by adding free Flt-1 in trans (Figure 2, panel d). Titration with Flt-1 to a solution containing 15 nM VEGF dimer resulted in an IC50 of 56 ± 8 nM. We note that the concentration of the sensor components in the system are not known, thus this approach provides information on relative binding constants, rather than absolute equilibrium constants as in the case with fluorescence polarization or surface plasmon resonance. Finally, we tested whether the split-luciferase assay platform is compatible with the use of cell lysis reagents and protease inhibitors that may be present in certain samples and found that the luminescence signal was not significantly diminished (Figure S4). Importantly, these initial experiments with VEGF and Flt-1 established feasibility for our direct ternary complexation strategy, prompting us to further investigate the use of more general recognition scaffolds such as antibodies.
Figure 2.
Ternary complexation mediated direct detection system for the vascular endothelial growth factor (VEGF). a) A schematic of the VEGF assay is shown, where domain 2 of Flt-1 (red and blue) is attached to both the N- and C-terminal halves of luciferase to directly detect the VEGF homodimer. b) Luminescence from reassembled luciferase in the presence and absence of VEGF, total assay time of 2.5 h starting from mRNA. A >13-fold increase in luminescence is observed in the presence of 15 nM VEGF dimer. c) A titration of VEGF dimer was performed, resulting in a linear relationship to luminescence output. d) A titration of free Flt-1 was used to compete with luciferase reassembly, resulting in an IC50 of 56 ± 8 nM. ALU, arbitrary luminescence units.
Rapid Characterization of HIV-1 Clades
As our first test of antibody mediated detection, we sought to provide a rapid and sensitive method for detecting HIV-1 based on antibody specificities. Accordingly we investigated strategies for incorporating antibodies into split-protein reassembly assays. In efforts to generate antibodies from randomized libraries, the Plückthun laboratory demonstrated that split-DHFR could be reassembled in bacterial cells by attaching a protein of interest and the corresponding single-chain antibody (scFv), a covalent fusion of the variable heavy and light chains (37–39), to individual DHFR halves (40). Interaction dependent reassembly of the functional DHFR enzyme led to cell survival. Despite its appeal, the intracellular expression of functional scFvs remains challenging (41, 42). Alternatively, conditions for efficient scFv expression and refolding in cell-free translation systems supplemented with protein disulfide isomerase (PDI) have been described (43). With this in mind, we set out to develop an antibody mediated split-luciferase assay for the potential detection of HIV-1.
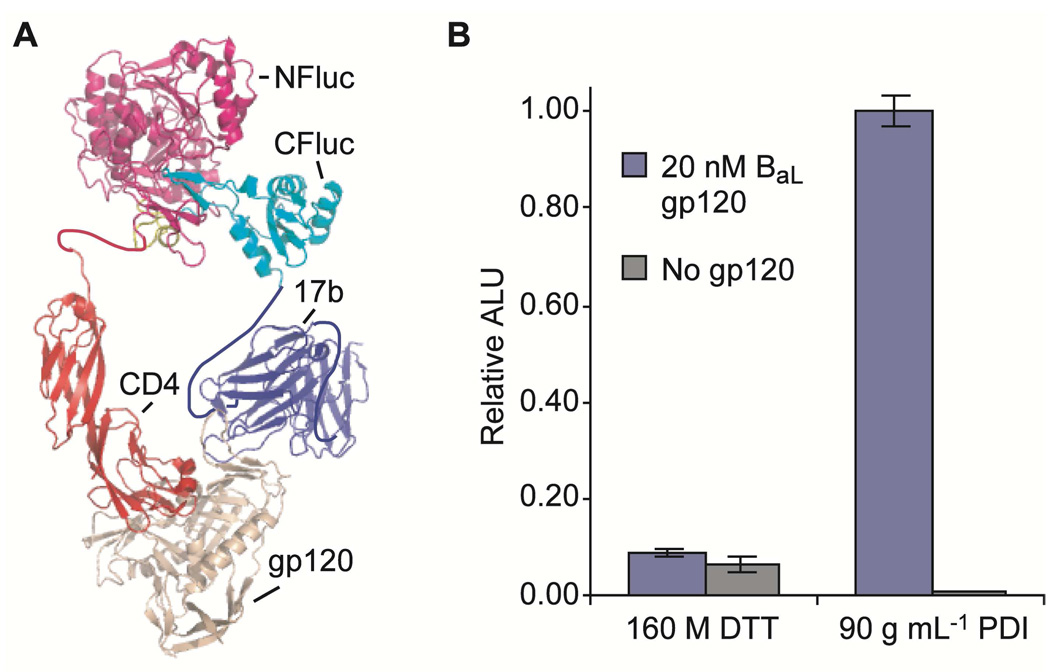
The crystal structure of the complex between CD4, gp120, and the Fab portion of a neutralizing antibody 17b (3), which has a high affinity for Clade B BaL gp120 (44), served as a model system for the development of our gp120 sandwich assay (Figure 3, panel a). We fused domain 1 and 2 (D1D2, residues 1–182) of CD4, which has been shown to bind to gp120 with a Kd of ~3 nM (45), to the N-terminal half of luciferase. As our second recognition element, we fused the C-terminal half of luciferase to the 17b scFv, which binds a CD4-induced epitope of gp120 (3, 44) (Figure 3, panel a). In accordance with previous literature results, initial experiments showed a negligible increase in luminescence for translations conducted in the presence of 20 nM BaL gp120 (Figure S5), presumably due to improper folding of the 17b scFv (43). However, the elimination of DTT and addition of PDI allowed for luminescence and the first functional demonstration of antibody mediated targeting in the split-luciferase system (Figure 3, panel b). Moreover, luminescence signal was detectable without the need for separation, washing or subsequent derivatization.
Figure 3.
An antibody enabled split-luciferase assay for direct detection of gp120. a) A schematic of the solution phase detection system for gp120 is shown. b) The specificity of the solution phase gp120 detection system is shown, emphasizing the luminescence signal generated from the assay when either DTT or PDI are included during translation. ALU, arbitrary luminescence units. DTT, dithiothreitol, PDI, protein disulfide isomerase.
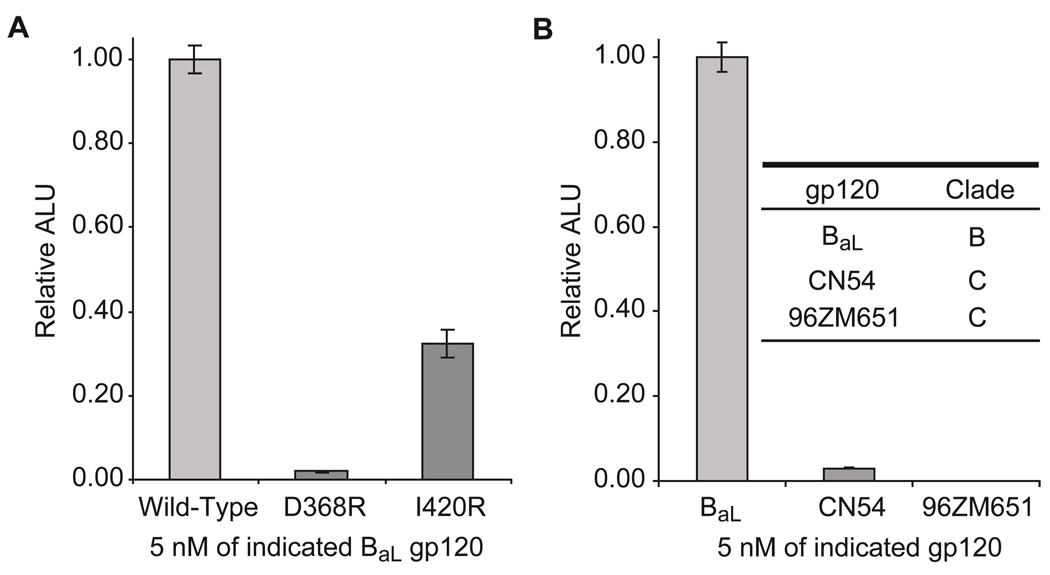
Having established conditions for favorable protein folding, we sought to verify the specificity of our gp120 assay. Accordingly, we first investigated luciferase reassembly in the presence of different BaL gp120s containing single amino acid mutations, D368R and I420R, which are known to reduce either CD4 (46) or 17b (47) binding, respectively. Indeed these mutant gp120s considerably reduce luminescence relative to the wild-type, confirming that both functional CD4 and 17b binding are required for luciferase reassembly (Figure 4, panel a). We also interrogated split-luciferase activity as a function of gp120 concentration (Figure S6), demonstrating that our assay system is capable of reporting on the presence of at least 12 ng mL−1 of BaL gp120, which is comparable to commercially available gp120 ELISAs and is likely a function of antibody/D1D2 affinities. This titration experiment also indicated that ~5 nM of active complex (folding capable split-halves) is translated utilizing our current cell-free conditions.
Figure 4.
Antibody enabled split-luciferase assay for direct detection of gp120 and verification of clades. a) The specificity of the solution phase gp120 detection system is shown. Assays were performed on the indicated wild-type or mutant gp120s; D368R and I420R mutations are known to decrease CD4 or 17b binding respectively (46, 47). b) The specificity of the antibody mediated gp120 detection system, as determined by luciferase reassembly, across a panel of gp120s from the indicated clades is shown. The observed luminescence highlights the ability to rapidly categorize HIV-1 clades using scFv-based split-luciferase complementation. ALU, arbitrary luminescence units.
To use our sandwich assay for the potential characterization of HIV-1 clades, we investigated gp120s from isolates CN54 and 96ZM651, both of which are clade C viruses. Maximal luciferase signal was observed only in the presence of BaL gp120 (clade B), while a slight increase in luminescence was observed for CN54 gp120, and no detectable signal was generated for 96ZM651 gp120 (Figure 4, panel b). This highlights the potential feasibility of this complexation approach for providing a method for distinguishing HIV-1 clades and sub-types using known antibody specificities without the need for DNA sequencing. In the long-term to accurately profile HIV-1, a panel of known gp120 antibodies with predetermined specificities is necessary that can be attached to the two halves of luciferase (44, 48). An unknown sample would be interrogated against the split-luciferase panel to potentially determine the specific HIV-1 clade.
Probing HER2 Expression in Human Breast Cancer Cells
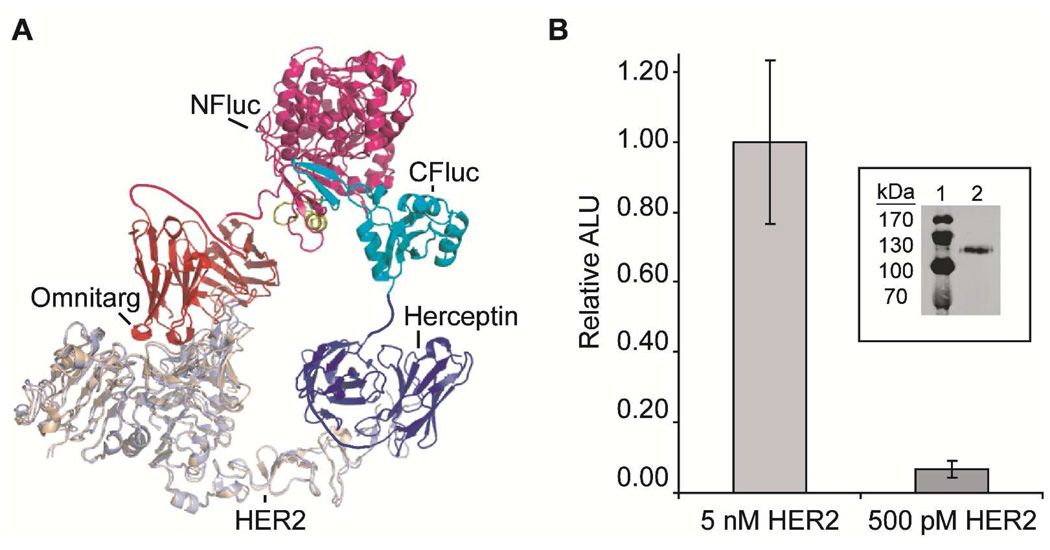
Having identified suitable expression conditions for using scFvs in our split-luciferase system, we turned to establishing whether this assay can be utilized for the determination of the relative levels of cell surface proteins present in human cells. This is particularly relevant to breast cancer treatment where the expression of estrogen receptors, progesterone receptors and HER2 in tumors is used to stratify patients and guide treatment strategies (2). Therefore we chose to develop a general method for detecting extracellular receptors using scFvs as protein recognition elements. Specifically we chose the extracellular domain (ECD, residues 1–631) of HER2, which is over-expressed in ~30% of human breast cancers and is directly correlated with poor clinical outcomes. Genentech has described two antibodies Herceptin and Omnitarg which bind distinct epitopes of the HER2 ECD. Overlaying the crystal structures of these bound antibodies indicates that they are likely capable of binding HER2 simultaneously (49, 50). Moreover the reported binding constants for an scFv version of Herceptin and the Fab portion of Omnitarg for the HER2 ECD are 150 pM (51) and 8.5 nM (52), respectively, which is well within our assay’s detection limits. Thus we constructed mRNAs in which the scFv of Omnitarg was fused to the N-terminal portion of luciferase and the C-terminal portion of luciferase was fused to the scFv of Herceptin (Figure 5, panel a). As an initial test of the HER2 sandwich assay, the HER2 ECD was expressed, purified, and added at varying concentrations to the two split-luciferase scFv fusions after translation in rabbit reticulocyte lysate. Within 30 minutes a concentration dependent increase in luminescence in the presence of the HER2 ECD was observed (Figure 5, panel b), indicating that this new antibody enabled sandwich assay was indeed capable of reporting on the presence of HER2 at sub-nanomolar levels. Preliminary experiments indicate that these cell-free translations can be stored at least 7 days at –80 °C prior to the addition of HER2 (Figure S7), potentially allowing for the long-term storage of reagents and the detection of clinically relevant analytes at the point of care.
Figure 5.
A dual antibody mediated split-luciferase sandwich assay for the direct detection of purified human epidermal growth factor receptor 2 (HER2). a) An overlay of HER2 (tan and light blue) with the bound luciferase fusion proteins is shown. b) A HER2 sandwich assay performed on purified HER2 expressed from Lec1 cells. The inset shows a western blot analysis of the purified HER2 protein, lane 1 molecular weight standards and lane 2 purified HER2 protein. ALU, arbitrary luminescence units.
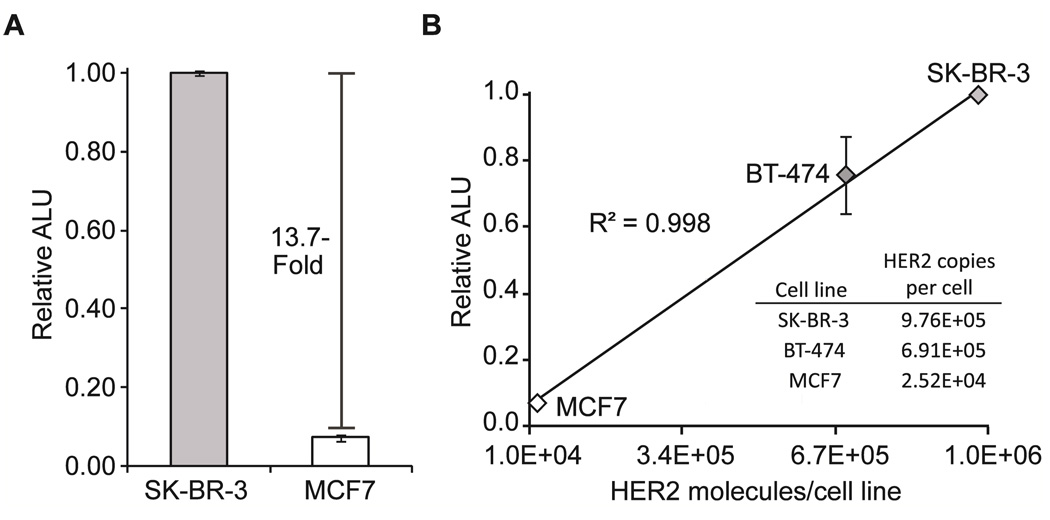
Finally we sought to determine if our assay could directly detect differential levels of expression of HER2 in human breast cancer cells. We chose the SK-BR-3, BT-474, and MCF7 cell lines which have been shown to produce approximately 1 × 106, 7 × 105 and 2.5 × 104 copies of HER2 per cell, respectively (53). Cells were added directly to completed translation reactions and incubated at room temperature for 40 minutes. In the presence of SK-BR-3 cells (2.5 × 104 cells, theoretically ~400 pM HER2) a 13.7-fold increase in luminescence was observed with respect to the MCF7 cells (2.5 × 104 cells, theoretically ~11 pM HER2), indicating that our sandwich assay is capable of directly reporting on the differential expression levels of HER2 in human breast cancer cell lines (Figure 6, panel a). Finally, in the presence of the BT-474 cell line an intermediate signal was observed, which was proportional to the theoretical amount of HER2 in the assay, 280 pM. These combined trends in luminescence signal directly correlated to the known expression levels of HER2 in these cell lines (Figure 6, panel b) (53). These results indicate that this rapid assay is capable of directly determining the relative expression levels of HER2 in human cells without the need for separation or FACS analysis. Additional experiments indicate that as few as 2,600 SK-BR-3 cells can be detected using this assay format (Figure S8). These results highlight the potential utility of this assay platform for the rapid stratification of breast cancer patients based on receptor expression.
Figure 6.
A dual antibody mediated split-luciferase sandwich assay for the direct detection of human epidermal growth factor receptor 2 (HER2) in human breast cancer cells. a) A HER2 sandwich assay performed on human breast cancer cells, SK-BR-3 or MCF7 cells were added after translation and luminescence was monitored after 40 min (2.5 × 104 cells during luminescence assay). b) The luminescence signal obtained from the HER2 sandwich assay correlates with literature values for HER2 expression in the corresponding cell lines (47). ALU, arbitrary luminescence units.
Conclusions
We have demonstrated the feasibility of ternary protein detection utilizing split-protein reassembly. Using this approach, we initially attached the receptor fragments of Flt-1 to the fragmented luciferase halves to detect VEGF, which could potentially be applied to identify molecules with anti-angiogenic properties. Our current sensitivity for this assay is 1.4 ng of VEGF dimer, which is on the same scale as physiological concentrations of VEGF in the sera or effusions from cancer patients reported to be as high as 50 ng/mL (54). In order to develop a more general recognition strategy, we have successfully employed single-chain antibodies as specific recognition elements. This approach was validated utilizing a single-chain antibody and a cellular receptor fragment as targeting domains, which were utilized to demonstrate the long-term potential for the characterization of HIV-1 clades by means of specific antibody-gp120 mediated recognition panels. Finally, we extended this ternary approach for the determination of the relative levels of HER2 in human breast cancer cells using an entirely antibody-based recognition system. This strategy requires the identification of dual recognition elements capable of simultaneously binding to the target of interest, which may be generally accessible through elegant methods for the evolution of antibody fragments (55). We envision that this simple and potentially general methodology will provide an approach for the rapid detection of a broad range of native proteins in complex heterogeneous systems, including blood and tissues. Furthermore, this assay might be useful for monitoring protein complex formation to follow aspects of stem cell differentiation or cancer cell progression, utilizing the unique cell-surface markers of stem cells that can provide a handle for following mechanisms of pluripotency maintenance and lineage commitment (56). The use of the firefly luciferase reporter with a broad luminescence profile with a maximum at 560 nm (range ~ 500 – 650) is particularly attractive in a biological context, considering that emission wavelengths greater than 600 nm are more easily transmitted through mammalian tissues (57, 58). However, a further red-shifted luciferase variant, such as railroad worm luciferase with a maximum emission at 630 nm, may prove even more appropriate for biological imaging (59). More generally, this ternary capture approach demonstrates the ability to generate conditionally activated split-proteins dependent upon native protein abundance, which may be potentially redesigned for therapeutic or drug delivery applications.
Methods
General Materials
Flexi-Rabbit Reticulocyte Lysate, RNasin, Steady-Glo Luciferase Assay System, and the T7 Ribomax Transcription Kit were purchased from Promega. G50 ProbeQuant columns were obtained from GE Healthcare. XL-1 Blue E. coli cells were purchased from Stratagene. Ni-NTA agarose resin was purchased from Qiagen. BaL, CN54, and 96ZM651 gp120s were obtained from the NIH AIDS Reference and Reagent Program, catalog numbers 4961, 7749, and 10080 respectively. Wild-type BaL gp120 and the BaL gp120 D368R and I420R mutants used in Figure 4, panel a were a generous gift of R. Wyatt. PDI, the Ala-Gln dipeptide, and Trypan Blue were purchased from Sigma. SK-BR-3 and BT-474 cells were obtained from ATCC (HTB-30 and HTB-20). MCF7 cells were a generous gift of the B. Olenyuk laboratory. Cell culture media and reagents were purchased from Hyclone. Plasmids encoding the VH and VL regions of both Herceptin (60) and Omnitarg (52) separated by a (GGGGS)3 linker were purchased from Bio Basic.
VEGF Expression and Refolding
A pQE30-VEGF expression plasmid was transformed into XL-1 Blue E. coli by electroporation according to the manufacturer’s instructions. An overnight culture of these cells was used to inoculate a 1 L culture of 2xYT media supplemented with 100 µg mL−1 ampicillin at an OD600 of 0.05. Protein expression was induced at an OD600 of 0.8 with 1 mM IPTG. Protein expression was allowed to proceed overnight at 37 °C. Cells were pelleted by centrifugation and resuspended in lysis buffer (Tris-HCl at pH = 8 containing 8 M Urea). Resuspended cells were lysed by sonication. The lysate was cleared by centrifugation at 18,000 rcf for 30 min. His-tagged VEGF was purified under denaturing conditions using Ni-NTA resin using the manufacturer’s instructions. Imidazole wash fractions were collected, pooled, and stored at −20 °C until required. Collected fractions were thawed on ice, concentrated and FPLC purified using a preparative Hi-Load 16/60 Superdex™ 75 column (Pharmacia Biotech) equilibrated with denaturing buffer (Tris-HCl at pH = 8 containing 6 M Urea). Full length monomeric VEGF was isolated, pooled, and stored at −20 °C until required for refolding.
The pooled fractions containing full-length monomeric VEGF were diluted to 50 µg mL−1 with buffer containing 6M Urea, 0.1 M Na2HPO4, 10 mM Tris-HCl at pH = 8.5, 1 mM EDTA, and 20 mM DTT. This solution was incubated for 3 hrs at room temperature to facilitate reduction. Reduced monomeric VEGF was then dialyzed against 100 mM Tris-HCl at pH = 8.5, 5 mM cysteine, 1 mM cystine, 0.5 M Urea, and 2 mM EDTA overnight at room temperature.
To separate dimeric VEGF from monomeric and multimeric species the refolded VEGF was concentrated and purified by FPLC using a Superdex 75 column equilibrated with PBS. Fractions containing refolded dimeric VEGF were collected, pooled, concentrated, and reapplied to the Superdex 75 column. Refolded VEGF was characterized by SDS-PAGE under reducing and non-reducing conditions to visualize the monomeric versus dimeric form. Concentrations were obtained by UV absorbance.
Flt-1 Expression and Refolding
A pRSFDuet-Flt-1 expression plasmid was transformed into XL-1 Blue E. coli by electroporation according to the manufacturer’s instructions. An overnight culture of these cells was used to inoculate a 1 L culture of 2xYT media supplemented with 35 µg mL−1 kanamycin at an OD600 of 0.05. Protein expression was induced at an OD600 of 0.8 with 1 mM IPTG. Cells were pelleted by centrifugation and resuspended followed by lysis by sonication. The lysate was cleared by centrifugation at 18,000 rcf for 30 min. His-tagged Flt-1 was purified under denaturing conditions using Ni-NTA resin according to the manufacturer’s instructions. Imidazole wash fractions were collected, pooled, and stored at −20 °C until required. Collected fractions were thawed on ice, concentrated and FPLC purified using a preparative Hi-Load 16/60 Superdex™ 75 column (Pharmacia Biotech) equilibrated with denaturing buffer (6 M urea, 10 mM Tris-HCl, 100 mM Na2HPO4, pH 8.0). Flt-1 was refolded overnight at 4 °C in 2 L of refolding buffer (0.3 M urea, 10 mM Tris-HCl, 100 mM Na2HPO4, 5 mM cysteine and 1 mM cystine) system in a 3 kDa MWCO dialysis snake-skin tubing (Pierce) at a final concentration of 50 – 100 µg mL−1. After refolding, Flt-1 was subsequently purified in phosphate buffered saline (pH 7.4) using an analytical Superdex™ 75 column (Pharmacia Biotech).
Flt-1 Luciferase Fusion mRNA Production
Open reading frames encoding for domain 2 of the Flt-1 receptor were cloned into bacterial vectors containing either the N- or C-terminal portions of firefly luciferase, residues 2–416 and 398–550 respectively (32), separated by a flexible amino acid linker. These plasmid sequences were confirmed by the University of Arizona DNA Sequencing Facility. These constructs were PCR amplified using a 5’ primer encoding a T7 promoter and Kozak sequence and 3’ primer containing a stem loop. mRNA was generated using the T7 Ribomax Transcription Kit and purified using a G50 ProbeQuant column. Concentrations of each mRNA were determined by UV absorbance.
VEGF-Flt-1 Sandwich Assay
Translations using Flexi-Rabbit Reticulocyte Lysate were carried out according to the manufacture’s procedure using 2 pmols of each mRNA encoding for the Flt-1 fusions, 400 µM DTT, 70 mM KCl, 200 µM of each amino acid, 66% Lysate in 25 µL. Reactions were incubated at 30 °C for 90 min after which 75 nM VEGF dimer or an equivalent volume of PBS was added and incubated for 1 hour at room temperature. Luminescence was monitored on a Turner TD20e luminometer by mixing 10 µL of translation with 40 µL of Steady-Glo Luciferase Assay System giving a final concentration of 15 nM VEGF dimer. Luminescence was monitored 1 min after mixing with a 10 sec integration. Reactions were performed in duplicate and averaged.
VEGF Titration
To determine the assay sensitivity, translations using Flexi-Rabbit Reticulocyte Lysate were carried out according to the manufacture’s procedure using 2 pmols of each mRNA encoding for the Flt-1 fusions, 400 µM DTT, 70 mM KCl, 200 µM of each amino acid, and 66% Lysate in a 25 µL reaction. Translations proceeded at 30 °C for 90 minutes, followed by addition of varying concentrations of VEGF dimer or an equivalent volume of PBS. After binding at room temperature for 1 hour, 10 µL of the reaction was added to 40 µL of Steady-Glo Luciferase Assay System, giving final VEGF dimer concentrations of 15, 7.5, 3.8, 1.9, 0.9, or 0.5 nM or no VEGF. Luminescence was monitored 1 min after mixing with a 10 sec integration. Reactions were performed in duplicate and averaged.
Flt-1 Titration
Translations using Flexi-Rabbit Reticulocyte Lysate were carried out according to the manufacture’s procedure using 2 pmols of each mRNA encoding for the Flt-1 fusions, 400 µM DTT, 70 mM KCl, 200 µM of each amino acid, and 66% Lysate in a 25 µL reaction. Reactions were incubated at 30 °C for 90 min after which 22.5 µL of the reaction was added to 75 nM VEGF dimer in the presence of varying concentrations of free Flt-1. Binding was allowed to achieve equilibrium at room temperature for 1 hour, followed by addition of 10 µL of the reaction to 40 µL of Steady-Glo Luciferase Assay System, resulting in final concentrations of 15 nM VEGF dimer and 500, 250, 100, 50, 25, 12.5, 2.3, or 0.4 nM or no Flt-1. Luminescence was monitored 1 min after mixing with a 10 sec integration. Reactions were performed in duplicate and averaged, followed by normalization to the sample containing 15 nM VEGF and no Flt-1.
Production of mRNA Encoding for the CD4 and 17b Split-Luciferase Fusions
Open reading frames encoding for residues 1–182 of CD4 and the VH and VL regions of 17b separated by a (GGGGS)3 linker were cloned into vectors containing the N- and C-terminal portions of luciferase respectively. These plasmid sequences were confirmed by the University of Arizona DNA Sequencing Facility. These constructs were PCR amplified using a 5’ primer encoding a T7 promoter and Kozak sequence and 3’ primer containing a stem loop. mRNA was generated using the T7 Ribomax Transcription Kit and purified using a G50 ProbeQuant column. Concentrations of each mRNA were determined by UV absorbance.
The Effect of DTT and PDI on the gp120 Sandwich Assay
Translations using Flexi-Rabbit Reticulocyte Lysate were carried out according to the manufacture’s procedure using 2 pmols of each of the mRNAs encoding the CD4-NFluc and CFluc-17b fusions, 70 mM KCl, 20 µM of each amino acid, 66% Lysate, 0.5 µL RNasin (resulting in 160 µM DTT during translation, from the RNasin storage buffer, during translation) where indicated, 90 µg mL−1 PDI where indicated, and either 100 nM BaL gp120 or an equivalent volume of PBS in a 25 µL reaction. Reactions were incubated at 30 °C for 90 min after which luminescence was monitored on a Turner TD20e luminometer by mixing 20 µL of translation with 80 µL of Steady-Glo Luciferase Assay System giving a final concentration of 20 nM BaL gp120. Luminescence was monitored 1 min after mixing with a 10 sec integration. Reactions were performed in duplicate and averaged.
Specificity of the gp120 Sandwich Assay
Translations and luciferase detection were carried out as described above, using 90 µg mL−1 PDI and no RNasin, except that 25 nM of the indicated gp120 was added during translation, giving a final concentration of 5 nM gp120.
Production of mRNA Encoding for the Herceptin and Omnitarg Split-Luciferase Fusions
Open reading frames encoding for the VH and VL regions of Omnitarg and Herceptin separated by a (GGGGS)3 linker were cloned into vectors containing the N- and C-terminal portions of luciferase respectively. These plasmid sequences were confirmed by the University of Arizona DNA Sequencing Facility. These constructs were PCR amplified using a 5’ primer encoding a T7 promoter and Kozak sequence and 3’ primer containing a stem loop. mRNA was generated using the T7 Ribomax Transcription Kit and purified using a G50 ProbeQuant column. Concentrations of each mRNA were determined by UV absorbance.
Expression, Purification, and Western Blot Analysis of the HER2 ECD
Lec1 cells stably expressing a human growth hormone-histidine tagged-HER2 ECD protein (49) were grown in 95% αMEM (without nucleotides or L-Gln) and 5% FBS supplemented with 100 nM methotrexate, 0.5 mg mL−1 G418, 584 mg L−1 Ala-Gln, 100 units mL−1 penicillin, and 100 µg mL−1 streptomycin. Cell cultures were allowed to grow for three days after which protein was purified from 50 mLs of culture media using Ni-NTA affinity chromatography. Protein was eluted with 10 mM Tris-HCl at pH = 7.5 containing 50 mM NaCl and 500 mM Imidazole. This solution was used directly for the experiments described below.
Western blot analysis was performed using a rabbit anti-his-tag polyclonal primary antibody (QED Biosciences, 18814) and an IR dye conjugated anti-rabbit secondary goat antibody (Licor Biosciences, IgG IRDye 800CW, 926-32211). A Licor Biosciences Odyssey scanner was used for imaging. HER2 ECD concentration was estimated from SDS-PAGE analysis.
HER2 Sandwich Assay using Purified HER2 ECD
Translations using Flexi-Rabbit Reticulocyte Lysate were carried out according to the manufacture’s procedure using 2 pmols of each of the mRNAs encoding the Omnitarg-NFluc and CFluc-Herceptin fusions, 70 mM KCl, 20 µM of each amino acid, 66% Lysate, and 90 µg mL−1 PDI in a 25 µL reaction. Reactions were incubated at 30 °C for 90 min after which purified HER2 ECD or an equivalent volume of storage buffer (10 mM Tris-HCl at pH = 7.5, 50 mM NaCl, and 500 mM Imidazole) was added to the translation. These solutions were allowed to equilibrate at room temperature for 30 min. Luminescence was monitored on a Turner TD20e luminometer by mixing 20 µL of translation with 80 µL of Steady-Glo Luciferase Assay System. Readings were taken 1 min after mixing with a 10 sec integration. Reactions were performed in duplicate, background subtracted (using samples containing no HER2 ECD), and averaged. HER2 ECD concentrations after rapid dilution are shown.
HER2 Sandwich Assay using Human Breast Cancer Cells
SK-BR-3, BT-474, and MCF7 cells were grown in 90% RPMI 1640 and 10% FBS supplemented with 100 units mL−1 penicillin, 100 µg mL−1 streptomycin, and 0.1% fungizone. Cells were detached using PBS containing 25 mM EDTA, washed, and resuspended in PBS; after which they were counted by Trypan Blue exclusion. Cells were diluted to 1.4 × 104 cells µL−1 prior to use in the assay.
Translations using Flexi-Rabbit Reticulocyte Lysate were carried out according to the manufacture’s procedure using 2 pmols of each of the mRNAs encoding the Omnitarg-NFluc and CFluc-Herceptin fusions, 70 mM KCl, 20 µM of each amino acid, and 66% Lysate per 25 µL reaction. Reactions were incubated at 30 °C for 90 min after which 5 µL of cells or an equivalent volume of PBS was added. These solutions were allowed to equilibrate at room temperature for 40 min with gentle shaking. Luminescence was monitored on a Turner 20/20n or TD20e luminometer by mixing 20 µL of translation with 80 µL of Steady-Glo Luciferase Assay System. Luminescence was monitored 1 min after mixing with a 10 sec integration. The luminescence readings are from assays which were performed in duplicate on cells grown in separate flasks, background subtracted (using samples containing no cells), and averaged. The number of cells in the luminescence assay is reported.
Supplementary Material
ACKNOWLEDGMENTS
J.L. Furman and J.R. Porter were supported by an NIH Institutional Training Grant (5T32GM008804-05). We thank the NIH for support (R01AI068414). We thank P. Longo, K. Block, and B. Fritz for helpful discussions, S. Rana for initial Flt-1 constructs, B. Olenyuk for providing the MCF7 cell line, D. Leahy for providing the HER2 ECD expressing Lec1 cell line, and the NIH AIDS repository for HIV-1 reagents.
Footnotes
Supporting Information Available. This material is available free of charge via the Internet.
REFERENCES
- 1.Santarius T, Shipley J, Brewer D, Stratton MR, Cooper CS. A census of amplified and overexpressed human cancer genes. Nat. Rev. Cancer. 2010;10:59–64. doi: 10.1038/nrc2771. [DOI] [PubMed] [Google Scholar]
- 2.Musgrove EA, Sutherland RL. Biological determinants of endocrine resistance in breast cancer. Nat. Rev. Cancer. 2009;9:631–643. doi: 10.1038/nrc2713. [DOI] [PubMed] [Google Scholar]
- 3.Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fischer C, Mazzone M, Jonckx B, Carmeliet P. FLT1 and its ligands VEGFB and PlGF: drug targets for anti-angiogenic therapy? Nat. Rev. Cancer. 2008;8:942–956. doi: 10.1038/nrc2524. [DOI] [PubMed] [Google Scholar]
- 5.Chen I, Ting AY. Site-specific labeling of proteins with small molecules in live cells. Curr. Opin. Biotechnol. 2005;16:35–40. doi: 10.1016/j.copbio.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 6.Parks DR, Herzenberg LA. Fluorescence-activated cell sorting - Theory, experimental optimization, and applications in lymphoid-cell biology. Methods Enzymol. 1984;108:197–241. doi: 10.1016/s0076-6879(84)08086-1. [DOI] [PubMed] [Google Scholar]
- 7.Engvall E, Perlmann P. Enzyme-Linked Immunosorbent Assay (ELISA) quantitative assay of immunoglobulin-G. Immunochemistry. 1971;8:871–874. doi: 10.1016/0019-2791(71)90454-x. [DOI] [PubMed] [Google Scholar]
- 8.Van Weeme BK, Schuurs AHW. Immunoassay using antigen-enzyme conjugates. FEBS Lett. 1971;15:232–236. doi: 10.1016/0014-5793(71)80319-8. [DOI] [PubMed] [Google Scholar]
- 9.Selvin PR. Principles and biophysical applications of lanthanide-based probes. Annu. Rev. Biophys. Biomol. Struct. 2002;31:275–302. doi: 10.1146/annurev.biophys.31.101101.140927. [DOI] [PubMed] [Google Scholar]
- 10.Inglese J, Johnson RL, Simeonov A, Xia MH, Zheng W, Austin CP, Auld DS. High-throughput screening assays for the identification of chemical probes. Nat. Chem. Biol. 2007;3:466–479. doi: 10.1038/nchembio.2007.17. [DOI] [PubMed] [Google Scholar]
- 11.Michnick SW, Ear PH, Manderson EN, Remy I, Stefan E. Universal strategies in research and drug discovery based on protein-fragment complementation assays. Nat. Rev. Drug Discovery. 2007;6:569–582. doi: 10.1038/nrd2311. [DOI] [PubMed] [Google Scholar]
- 12.Muller J, Johnsson N. Split-ubiquitin and the split-protein sensors: Chessman for the endgame. Chembiochem. 2008;9:2029–2038. doi: 10.1002/cbic.200800190. [DOI] [PubMed] [Google Scholar]
- 13.Johnsson N, Varshavsky A. Split ubiquitin as a sensor of protein interactions in-vivo. Proc. Natl. Acad. Sci. USA. 1994;91:10340–10344. doi: 10.1073/pnas.91.22.10340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pelletier JN, Campbell-Valois FX, Michnick SW. Oligomerization domain-directed reassembly of active dihydrofolate reductase from rationally designed fragments. Proc. Natl. Acad. Sci. USA. 1998;95:12141–12146. doi: 10.1073/pnas.95.21.12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galarneau A, Primeau M, Trudeau LE, Michnick SW. beta-Lactamase protein fragment complementation assays as in vivo and in vitro sensors of protein-protein interactions. Nat. Biotechnol. 2002;20:619–622. doi: 10.1038/nbt0602-619. [DOI] [PubMed] [Google Scholar]
- 16.Ghosh I, Hamilton AD, Regan L. Antiparallel leucine zipper-directed protein reassembly: Application to the green fluorescent protein. J. Am. Chem. Soc. 2000;122:5658–5659. [Google Scholar]
- 17.Hu CD, Kerppola TK. Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis. Nat. Biotechnol. 2003;21:539–545. doi: 10.1038/nbt816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MacDonald ML, Lamerdin J, Owens S, Keon BH, Bilter GK, Shang Z, Huang Z, Yu H, Dias J, Minami T, Michnick SW, Westwick JK. Identifying off-target effects and hidden phenotypes of drugs in human cells. Nat. Chem. Biol. 2006;2:329–337. doi: 10.1038/nchembio790. [DOI] [PubMed] [Google Scholar]
- 19.Paulmurugan R, Gambhir SS. Monitoring protein-protein interactions using split synthetic renilla luciferase protein-fragment-assisted complementation. Anal. Chem. 2003;75:1584–1589. doi: 10.1021/ac020731c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Remy I, Michnick SW. A highly sensitive protein-protein interaction assay based on Gaussia luciferase. Nat. Methods. 2006;3:977–979. doi: 10.1038/nmeth979. [DOI] [PubMed] [Google Scholar]
- 21.Paulmurugan R, Umezawa Y, Gambhir SS. Noninvasive imaging of protein-protein interactions in living subjects by using reporter protein complementation and reconstitution strategies. Proc. Natl. Acad. Sci. USA. 2002;99:15608–15613. doi: 10.1073/pnas.242594299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tafelmeyer P, Johnsson N, Johnsson K. Transforming a (beta/alpha)(8)-barrel enzyme into a split-protein sensor through directed evolution. Chem. Biol. 2004;11:681–689. doi: 10.1016/j.chembiol.2004.02.026. [DOI] [PubMed] [Google Scholar]
- 23.Wehr MC, Laage R, Bolz U, Fischer TM, Grunewald S, Scheek S, Bach A, Nave K-A, Rossner MJ. Monitoring regulated protein-protein interactions using split TEV. Nat. Methods. 2006;3:985–993. doi: 10.1038/nmeth967. [DOI] [PubMed] [Google Scholar]
- 24.Muller MM, Kries H, Csuhai E, Kast P, Hilvert D. Design, selection, and characterization of a split chorismate mutase. Protein Sci. 2010;19:1000–1010. doi: 10.1002/pro.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tarassov K, Messier V, Landry CR, Radinovic S, Molina MMS, Shames I, Malitskaya Y, Vogel J, Bussey H, Michnick SW. An in vivo map of the yeast protein interactome. Science. 2008;320:1465–1470. doi: 10.1126/science.1153878. [DOI] [PubMed] [Google Scholar]
- 26.Varshavsky A. Targeting the absence: Homozygous DNA deletions as immutable signposts for cancer therapy. Proc. Natl. Acad. Sci. USA. 2007;104:14935–14940. doi: 10.1073/pnas.0706546104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Furman JL, Badran AH, Shen SY, Stains CI, Hannallah J, Segal DJ, Ghosh I. Systematic evaluation of split-fluorescent proteins for the direct detection of native and methylated DNA. Bioorg. Med. Chem. Lett. 2009;19:3748–3751. doi: 10.1016/j.bmcl.2009.04.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stains CI, Furman JL, Segal DJ, Ghosh I. Site-specific detection of DNA methylation utilizing mCpG-SEER. J. Am. Chem. Soc. 2006;128:9761–9765. doi: 10.1021/ja060681j. [DOI] [PubMed] [Google Scholar]
- 29.Stains CI, Porter JR, Ooi AT, Segal DJ, Ghosh I. DNA sequence-enabled reassembly of the green fluorescent protein. J. Am. Chem. Soc. 2005;127:10782–10783. doi: 10.1021/ja051969w. [DOI] [PubMed] [Google Scholar]
- 30.Nomura W, Barbas CF. In vivo site-specific DNA methylation with a designed sequence-enabled DNA methylase. J. Am. Chem. Soc. 2007;129:8676–8677. doi: 10.1021/ja0705588. [DOI] [PubMed] [Google Scholar]
- 31.Porter JR, Stains CI, Jester BW, Ghosh I. A General and Rapid Cell-Free Approach for the Interrogation of Protein–Protein, Protein–DNA, and Protein–RNA Interactions and their Antagonists Utilizing Split-Protein Reporters. J. Am. Chem. Soc. 2008;130:6488–6497. doi: 10.1021/ja7114579. [DOI] [PubMed] [Google Scholar]
- 32.Luker KE, Smith MCP, Luker GD, Gammon ST, Piwnica-Worms H, Piwnica-Worms DP. Kinetics of regulated protein-protein interactions revealed with firefly luciferase complementation imaging in cells and living animals. Proc. Natl. Acad. Sci. USA. 2004;101:12288–12293. doi: 10.1073/pnas.0404041101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ostermeier M, Nixon AE, Shim JH, Benkovic SJ. Combinatorial protein engineering by incremental truncation. Proc. Natl. Acad. Sci. USA. 1999;96:3562–3567. doi: 10.1073/pnas.96.7.3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goto Y, Ohta A, Sako Y, Yamagishi Y, Murakami H, Suga H. Reprogramming the translation Initiation for the synthesis of physiologically stable cyclic peptides. ACS Chem. Biol. 2008;3:120–129. doi: 10.1021/cb700233t. [DOI] [PubMed] [Google Scholar]
- 35.Shimizu Y, Inoue A, Tomari Y, Suzuki T, Yokogawa T, Nishikawa K, Ueda T. Cell-free translation reconstituted with purified components. Nat. Biotechnol. 2001;19:751–755. doi: 10.1038/90802. [DOI] [PubMed] [Google Scholar]
- 36.Wiesmann C, Fuh G, Christinger HW, Eigenbrot C, Wells JA, deVos AM. Crystal structure at 1.7 angstrom resolution of VEGF in complex with domain 2 of the Flt-1 receptor. Cell. 1997;91:695–704. doi: 10.1016/s0092-8674(00)80456-0. [DOI] [PubMed] [Google Scholar]
- 37.Bird RE, Hardman KD, Jacobson JW, Johnson S, Kaufman BM, Lee SM, Lee T, Pope SH, Riordan GS, Whitlow M. Single-chain antigen-binding proteins. Science. 1988;242:423–426. doi: 10.1126/science.3140379. [DOI] [PubMed] [Google Scholar]
- 38.Holliger P, Hudson PJ. Engineered antibody fragments and the rise of single domains. Nat. Biotechnol. 2005;23:1126–1136. doi: 10.1038/nbt1142. [DOI] [PubMed] [Google Scholar]
- 39.Huston JS, Levinson D, Mudgetthunter M, Tai MS, Novotny J, Margolies MN, Ridge RJ, Bruccoleri RE, Haber E, Crea R, Oppermann H. Protein engineering of antibody-binding sites - Recovery of specific activity in an anti-digoxin single-chain Fv analog produced in Escherichia-Coli. Proc. Natl. Acad. Sci. USA. 1988;85:5879–5883. doi: 10.1073/pnas.85.16.5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mössner E, Koch H, Plückthun A. Fast selection of antibodies without antigen purification: adaptation of the protein fragment complementation assay to select antigen-antibody pairs. J. Mol. Biol. 2001;308:115–122. doi: 10.1006/jmbi.2001.4575. [DOI] [PubMed] [Google Scholar]
- 41.Biocca S, Ruberti F, Tafani M, Pierandrel-Amaldi P, Cattaneo A. Redox state of single chain Fv fragments targeted to the endoplasmic reticulum, cytosol and mitochondria. Nat. Biotechnol. 1995;13:1110–1115. doi: 10.1038/nbt1095-1110. [DOI] [PubMed] [Google Scholar]
- 42.Koch H, Gräfe N, Schiess R, Plückthun A. Direct selection of antibodies from complex libraries with the protein fragment complementation assay. J. Mol. Biol. 2006;357:427–441. doi: 10.1016/j.jmb.2005.12.043. [DOI] [PubMed] [Google Scholar]
- 43.Ryabova LA, Desplancq D, Spirin AS, Pluckthun A. Functional antibody production using cell-free translation: Effects of protein disulfide isomerase and chaperones. Nat. Biotechnol. 1997;15:79–84. doi: 10.1038/nbt0197-79. [DOI] [PubMed] [Google Scholar]
- 44.Lagenaur LA, Villarroel VA, Bundoc V, Dey B, Berger EA. sCD4-17b bifunctional protein: Extremely broad and potent neutralization of HIV-1 Env pseudotyped viruses from genetically diverse primary isolates. Retrovirology. 2010;7 doi: 10.1186/1742-4690-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ryu SE, Kwong PD, Truneh A, Porter TG, Arthos J, Rosenberg M, Dai XP, Xuong NH, Axel R, Sweet RW, Hendrickson WA. Crystal-structure of an HIV-binding recombinant fragment of human CD4. Nature. 1990;348:419–426. doi: 10.1038/348419a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Olshevsky U, Helseth E, Furman C, Li J, Haseltine W, Sodroski J. Identification of individual human immunodeficiency virus type 1 gp120 amino acids important for CD4 receptor binding. J. Virol. 1990;64:5701–5707. doi: 10.1128/jvi.64.12.5701-5707.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thali M, Moore JP, Furman C, Charles M, Ho DD, Robinson J, Sodroski J. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J. Virol. 1993;67:3978–3988. doi: 10.1128/jvi.67.7.3978-3988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Binley JA, Wrin T, Korber B, Zwick MB, Wang M, Chappey C, Stiegler G, Kunert R, Zolla-Pazner S, Katinger H, Petropoulos CJ, Burton DR. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J. Virol. 2004;78:13232–13252. doi: 10.1128/JVI.78.23.13232-13252.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cho HS, Mason K, Ramyar KX, Stanley AM, Gabelli SB, Denney DW, Leahy DJ. Structure of the extracellular region of HER2 alone and in complex with the Herceptin Fab. Nature. 2003;421:756–760. doi: 10.1038/nature01392. [DOI] [PubMed] [Google Scholar]
- 50.Franklin MC, Carey KD, Vajdos FF, Leahy DJ, de Vos AM, Sliwkowski MX. Insights into ErbB signaling from the structure of the ErbB2-pertuzumab complex. Cancer Cell. 2004;5:317–328. doi: 10.1016/s1535-6108(04)00083-2. [DOI] [PubMed] [Google Scholar]
- 51.Kubetzko S, Balic E, Waibel R, Zangemeister-Wittke U, Pluckthun A. PEGylation and multimerization of the anti-p185(HER-2) single chain Fv fragment 4D5 - Effects on tumor targeting. J. Biol. Chem. 2006;281:35186–35201. doi: 10.1074/jbc.M604127200. [DOI] [PubMed] [Google Scholar]
- 52.Adams CW, Allison DE, Flagella K, Presta L, Clarke J, Dybdal N, McKeever K, Sliwkowski MX. Humanization of a recombinant monoclonal antibody to produce a therapeutic HER dimerization inhibitor, pertuzumab. Cancer Immunol. Immunother. 2006;55:717–727. doi: 10.1007/s00262-005-0058-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Prang N, Preithner S, Brischwein K, Goster P, Woppel A, Muller J, Steiger C, Peters M, Baeuerle PA, da Silva AJ. Cellular and complement-dependent cytotoxicity of Ep-CAM-specific monoclonal antibody MT201 against breast cancer cell lines. Br. J. Cancer. 2005;92:342–349. doi: 10.1038/sj.bjc.6602310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kraft A, Weindel K, Ochs A, Marth C, Zmija J, Schumacher P, Unger C, Marme D, Gastl G. Vascular endothelial growth factor in the sera and effusions of patients with malignant and nonmalignant disease. Cancer. 1999;85:178–187. [PubMed] [Google Scholar]
- 55.Winter G, Griffiths AD, Hawkins RE, Hoogenboom HR. Making Antibodies by Phage Display Technology. Annu. Rev. Immunol. 1994;12:433–455. doi: 10.1146/annurev.iy.12.040194.002245. [DOI] [PubMed] [Google Scholar]
- 56.Pera MF, Tam PPL. Extrinsic regulation of pluripotent stem cells. Nature. 2010;465:713–720. doi: 10.1038/nature09228. [DOI] [PubMed] [Google Scholar]
- 57.Rice BW, Cable MD, Nelson MB. In vivo imaging of light-emitting probes. J. Biomed. Opt. 2001;6:432–440. doi: 10.1117/1.1413210. [DOI] [PubMed] [Google Scholar]
- 58.Hida N, Awais M, Takeuchi M, Ueno N, Tashiro M, Takagi C, Singh T, Hayashi M, Ohmiya Y, Ozawa T. High-Sensitivity Real-Time Imaging of Dual Protein-Protein Interactions in Living Subjects Using Multicolor Luciferases. Plos One. 2009;4 doi: 10.1371/journal.pone.0005868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li XY, Nakajima Y, Niwa K, Viviani VR, Ohmiya Y. Enhanced red-emitting railroad worm luciferase for bioassays and bioimaging. Protein Sci. 2010;19:26–33. doi: 10.1002/pro.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Carter P, Presta L, Gorman CM, Ridgway JBB, Henner D, Wong WLT, Rowland AM, Kotts C, Carver ME, Shepard HM. Humanization of an anti-P185Her2 antibody for human cancer therapy. Proc. Natl. Acad. Sci. USA. 1992;89:4285–4289. doi: 10.1073/pnas.89.10.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.