Abstract
Mass spectrometry of membrane protein complexes is still a methodological challenge due to hydrophobic and hydrophilic parts of the species and the fact that all subunits are bound non-covalently together. The present study with the novel laser induced liquid bead ion desorption mass spectrometry (LILBID-MS) reports on the determination of the subunit composition of the F1Fo-ATP synthase from Bacillus pseudofirmus OF4, that of both bovine heart and, for the first time, of human heart mitochondrial F1Fo-ATP synthases. Under selected buffer conditions the mass of the intact F1Fo-ATP synthase of B. pseudofirmus OF4 could be measured, allowing the analysis of complex subunit stoichiometry. The agreement with theoretical masses derived from sequence databases is very good. A comparison of the ATP synthase subunit composition of 5 different ATPases reveals differences in the complexity of eukaryotic and bacterial ATP synthases. However, whereas the overall construction of eukaryotic enzymes is more complex than the bacterial ones, functionally important subunits are conserved among all ATPases.
1. Introduction
F-type ATP synthases are essential enzymes ubiquitously found in bacteria, mitochondria and chloroplasts. They are ion-driven nanomotors which have two working directions. They synthesize adenosine triphosphate (ATP) from adenosine diphosphate (ADP) and a free phosphate (Pi) and therewith provide the organism with the “energy currency” of cells. The energy for this process is provided by an electrochemical gradient of protons (or less commonly Na+), which are pumped across the membrane by primary pumps such as found in the respiratory chain. In the other direction, ATP is hydrolyzed by the ATPase, which is used then as a pump for cell pH homeostasis or to acidify intracellular compartments.
The simplest form of F1Fo ATP synthases can be found in bacteria,1,2 schematically shown in Figure 1. The F1 complex (subunits α3β3γδε) is the water soluble domain of the enzyme and protrudes into the cytoplasmic reservoir of the cell. The F1 complex is connected with the Fo complex by periperal (subunits b2δ) and central stalks (subunits γε).3 The Fo complex, the “turbine-motor”, is highly hydrophobic and consists of the subunits ab2c10-15. It is responsible for ion translocation across the membrane.4 A more complex composition can be found in mitochondria. Beside the same core subunits as in the bacterial ATP synthase (a, b and c), the additional subunits d, e, f, g, 8, F6 and OSCP are associated with the mitochondrial enzyme.5 From a functional point of view, the subunits of an ATP synthase can be divided into two parts, a rotor (subunits γεc10-15) and a stator (subunits α3β3δab2).6 The rotor is driven by the ions, which are translocated through the Fo motor, and acts as a rotating camshaft within the F1 headpiece. By rotation it causes conformational changes within the three catalytic centers each formed by an α- and β-subunits protomer,7 which finally converts ADP and Pi into ATP. The overall molecular mass of the holo-F1Fo ATP synthase complex is approx. 540 kDa8 in bacteria and 570-585 kDa in mitochondria.9 Information about the mass of the functional holo complex (F1Fo) is of interest since it allows the confirmation of the correct assembly8 and, in some cases,10 the determination of subunit stoichiometries.
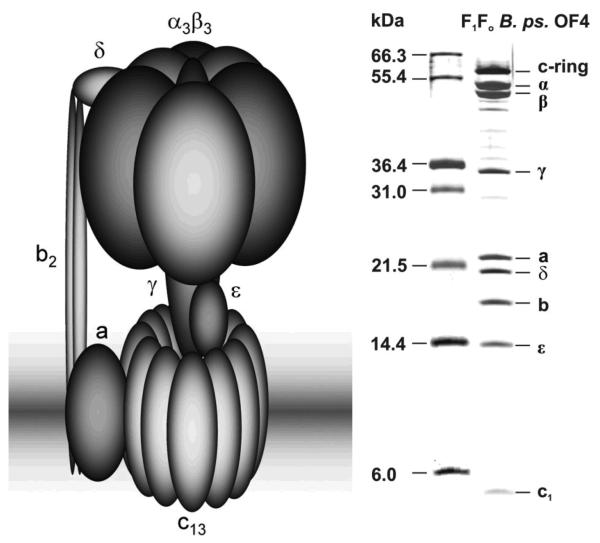
Fig. 1.
Cartoon representation of the subunit composition of a bacterial F1Fo-ATP synthase (left) and for comparison results from a 15% SDS polyacryamide gel electrophoresis35 of a sample (10μg) of purified B.pseudofirmus OF4 ATP synthase (right). A marker lane and the indicated masses in kDa are given on the left. The gel was stained with silver.36
The different biochemical conditions, needed for complex stability of the soluble and the membrane-embedded part of ATP synthases make the investigation of the intact enzyme very difficult for traditional mass spectrometric biology approaches. Non-covalent mass spectrometries such as electrospray ionization mass spectrometry (ESI-MS) and laser induced liquid bead ion desorption mass spectrometry (LILBID-MS) are well established for the investigation of soluble complexes, while membrane protein complexes remained an invincible challenge for a long time. The first intact membrane protein complexes in the gas phase were observed with LILBID11 and nano ESI (nESI)12 recently.
The particular challenge in the investigation of ATP synthases compared to smaller membrane protein complexes with smaller soluble parts, is the spatial separation of the membrane part and the soluble head that are connected by non covalently bound subunits. In the case of ESI, detergent micelles are used to protect the complex while it is transferred from solution to gas phase. These protein-containing detergent aggregates are then activated by collisions in the gas phase to release the complex from the vast detergent aggregates. The two part structure of the ATP synthase is therefore particularly challenging, since the soluble part is prone to dissociate under conditions that are needed to release the membrane-embedded region from the detergent aggregates. ATP synthases have been studied intact by nESI12,39 and LILBID.8,13
LILBID-MS has turned out to be a particularly suitable alternative to ESI for the analysis of non-covalently bound membrane protein complexes solubilized by detergents. Here, solvated biomolecules are transferred from aqueous micro droplets into vacuum by means of infrared laser pulses, the wavelength of which is tuned to the strongest absorption band of water (3 μm), to achieve an effective energy transfer into the liquid matrix rather than into the biological sample. The method has proven to be soft and sensitive allowing the investigation of detergent-solubilized samples at typically micromolar concentrations and at a desired buffer and pH.
The goal of this work was to determine and compare the masses and subunit composition of three exemplary ATP synthases; from the bacterium Bacillus pseudofirmus OF4 and from bovine and human heart mitochondria by LILBID mass spectrometry. Furthermore, we compare the masses of these subunits with those determined for the ATP synthases from the yeast Yarrowia lipolytica14 and from Bacillus sp. strain TA2.A1,8 which both have been previously measured by LILBID-MS.
2. Materials and methods
2.1 Sample preparation
Bacillus pseudofirmus OF4 F1Fo-ATP synthase
The ATP synthase was purified from Bacillus pseudofirmus OF4 cells15 in which a hexa-histidine tag was inserted after the N-terminal methionine in the chromosomal gene encoding the β-subunit of the ATP synthase. The complex was extracted from everted vesicles with 1% dodecyl maltoside (DDM) in the presence of 3 mg/ml soybean asolectin and purified by affinity chromatography on Ni-NTA agarose. The sample with a final concentration of 1 mg/ml was dialized for 24 h against 10 mM ammonium acetate buffer (NH4OAc) at pH 7.5 and 0.05% dodecyl maltoside.
Bovine heart mitochondrial F1Fo-ATP synthase
Isolation of mitochondria from Bos taurus, solubilization of mitochondrial membranes by DDM, blue native electrophoresis (BN-PAGE) and electroelution were performed as described by Wittig et al.16 Briefly: Sedimented mitochondria (4 mg protein) were solubilized by DDM setting a detergent/protein ratio of 2.5 g/g. Following 15 min of centrifugation at 100,000×g, the supernatant was recovered, supplemented with Coomassie blue G-250, and applied to a 3-13% acrylamide gradient gel for BN-PAGE. Following BN-PAGE the visible blue band of ATP synthase was cut out and the protein was electroeluted. Glycerol was added to 10% and the sample was shock frozen and stored at −80°C.
Human heart mitochondrial F1Fo-ATP synthase
Human (Homo sapiens) ATP synthase17 was prepared from post mortem heart tissue, essentially as described by Wittig et al.16 Briefly: Frozen heart tissue (105 mg; wet weight) was thawed, frayed out using tweezers, and homogenized in 1 ml of solution (250mM sucrose, 20 mM Na-phosphate, pH 7.0) using a tightly fitting glass/Teflon homogenizer. The total volume was apportioned into aliquots corresponding to 5 mg and 2×50 mg of cells, sedimented by 10 min centrifugation at 20,000×g, and frozen after removal of the supernatants or used immediately. One sediment from 50 mg heart tissue containing nuclei, larger cell fragments, and mitochondria was suspended with 450 μl of buffer (50 mM NaCl, 2 mM 6-aminohexanoic acid, 1 mM EDTA, 50 mM imidazole/HCl, pH 7.0) and solubilized by adding 40 μl DDM (10%). Following 15 min of centrifugation at 100,000×g, the supernatant was recovered, supplemented with 10 μl of 5% Coomassie blue G-250, and the total volume of 500 μl was applied to a 3-13% acrylamide gradient gel for blue native electrophoresis. The band of ATP synthase was cut out and the protein was electroeluted. 10% of glycerol was added and aliquots corresponding to 16 mg heart were shock frozen and stored at −80°C. An aliquot corresponding to 16 mg heart was used for preparation of a single sample for LILBID measurement.
In all cases proteins were kept either on ice or at 4°C during storage and preparation. Where necessary buffer exchange/desalting was performed using Zeba(™) Micro Desalt Spin Columns (Pierce) following standard procedures.
2.2 Mass spectrometry
LILBID-MS was described in detail previously.18 Briefly, the ion source contains a commercial droplet dispenser (Microdrop) which injects on demand tiny micro droplets (Ø ~ 50 μm; V ~65 pL) from 300 Torr through pressure reduction apertures into high vacuum (10−6 Torr). There they are irradiated one by one by high intensity mid-IR laser pulses (λ ~3 μm) generated by a home-built Nd:Yag pumped LiNbO3 optical parametric oscillator (OPO). Due to the absorption of the laser light by the solvent, beyond a certain intensity threshhold the droplets are disrupted, ejecting preformed analyte ions into vacuum. At low laser intensity this method functions in an “ultra soft” mode, which allows studying noncovalent solubilized integral membrane complexes. At increased laser intensity i.e. at “harsh desorption conditions” less stable complexes are thermolyzed into their subunits in a top-down fashion. The mass analysis is currently realised using a home-built time of flight (TOF) mass spectrometer of the Wiley–McLaren-type including an ion reflector (reflectron). For detecting ions of large m/z we utilize a home-built Daly-type high mass detector, with the conversion electrode being a multichannel plate, extending the detectable m/z range to about 106. The main reason for a rather moderate mass resolution is the non-compensated isotropic initial energy of the ions together with an incomplete ion desolvation. Unless otherwise stated only 200 droplets were sampled for each mass spectrum. Typically less than 10 μl of solution with an analyte concentration in the micromolar range are required for an analysis, resulting in an analyte consumption in the pmol range.
3. Results and Discussion
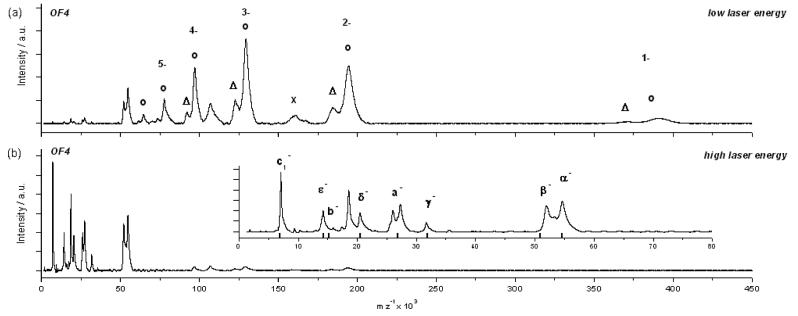
We first studied the F1Fo-ATP synthase from Bacillus pseudofirmus OF4, a facultative alkaliphilic strain, which grows at a pH up to 11.2 but still uses an exclusively proton-coupled ATP synthase.15 It was prepared as described in Materials and Methods. Theoretically it consists of 8 different proteins (Table 1) in a stoichiometry of α3β3γδεab2c13 making a total of 25 proteins for one F1Fo complex (chemical composition: C24′356H39′393N6′407O7′082S154; with a total number of atoms of 77,392 and with 5,008 amino acids as calculated with: www.expasy.org/cgi-bin/protparam). The overall theoretical calculated mass of the holo enzyme is 540,231.6 Da. The schematically drawn common structure of a bacterial F1Fo-ATP synthase is illustrated together with its subunit composition on an SDS polyacrylamide gel in Figure 1. Our aim was to determine the total mass of the complex, the masses of its subcomplexes (F1 and Fo) and of the single subunits by LILBID mass spectrometry. The mass spectrum (Fig. 2(a)) recorded under soft conditions, i.e. low laser intensity, shows two distinct charge distributions (n=1-6) of anions, which can be very likely assigned to the water soluble F1 subcomplex and the F1 subcomplex, lacking the δ-subunit (for a comparison of measured and expected masses see Table 1). Additionally, signals of the highly hydrophobic Fo subcomplex are visible. Thus in the low m/z range charge distributions of the α- and β-subunits can be observed. These findings suggest that dissociation already took place in solution phase.At high laser energy (Fig. 2(b)) almost all the signals of the subcomplexes are strongly diminished, while signals for the single subunits appear. As shown in the enlarged inset of Fig. 2(b), all 8 expected subunits (α, β, γ, δ, ε, a, b and c) were present in the enzyme preparation (Fig. 1 and 2(a)). The observed masses of the subunits are in good agreement with the calculated values from a sequence database (Table 1). Thus, the LILBID spectra do not only reflect the exact single subunit masses but also the composition of the enzyme's subcomplexes. The dissociation observed under the buffer conditions used for recording these spectra was surprising. It resulted in the loss of the intact F1Fo complex. For this reason we also measured this sample in Tris/HCl buffer containing 1 mM of MgCl2.
Table 1.
Calculated and experimental masses of Bacillus OF4 F1Fo-ATP synthase (as determined from GenBank Locus AF330160.1)
| Subunit / subcomplex |
Uniprot | Calc. Mass [Da]a |
LILBID mass [kDa] |
|---|---|---|---|
| a | AAG48358.1 | 26863.77 | 26.8 (± 0.3) |
| b | AAG48359.1 | 15212.75 | 15.8 (± 0.2) |
| c | AAC08039.1 | 6956.06 | 7.0 (± 0.1) |
| α | AAG48361.1 | 54674.48 | 54.1 (± 0.5) |
| β b | AAG48363.1 | 51752.14 | 51.3 (± 0.5) |
| γ | AAG48362.1 | 31835.49 | 31.5 (± 0.5) |
| δ | AAG48360.1 | 20534.63 | 20.3 (± 0.2) |
| ε | AAG48364.1 | 14327.68 | 14.4 (± 0.2) |
| F1 (α3β3γδε) |
385833.50 | 384.0 (± 3) | |
| F1-δ (α3β3γε) |
365298.87 | 363.0 (± 3) | |
| Fo (ab2c13)c |
154367.77 | 154.0 (± 2) | |
| F1Fo (α3β3γδεab2c13)c |
540201.27 | 542.0 (± 5) |
calculations were performed with BioEdit (www.mbio.ncsu.edu/BioEdit/bioedit.html)
all β-subunits contain a hexa-histidine tag at the N-terminus (840,86 Da), which is included in the calculation, see Materials and Methods for details
The tridecameric rotor ring stoichiometry of Bacillus pseudofirmus OF4 was determined in Preiss et al.38
Fig. 2.
LILBID anion spectra of B. pseudofirmus OF4 F1Fo-ATP synthase after dialysis in a 10 mM ammonium acetate buffer. (a): recorded under soft conditions. Shown are the charge distributions of F1 (circles), F1 without δ subunit (triangles) and Fo (cross). (b): recorded under harsh conditions, with a magnification given in the inset. The subunits are labelled with the black bars indicating the respective calculated masses.
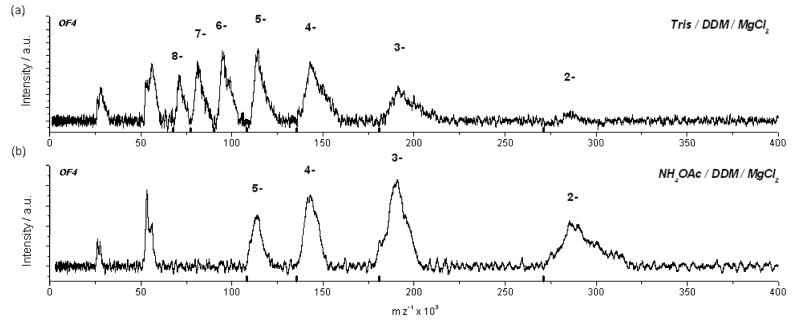
Fig. 3(a) shows the LILBID mass spectrum measured under the same soft desorption conditions as for the spectrum in Fig. 2(a). Here the main peaks correspond to a mass of approximately 542 kDa, visible in various charged states (N=2-8) matching the theoretically calculated mass of the intact F1Fo complex (with the mass of the complex corresponding to the onset of the peak). The peak broadening is due to nonspecific association of Tris buffer, lipids and detergent molecules. Furthermore, the increased stability of the intact complex due to the addition of Mg2+ reflects the relative solution phase stability in the mass spectra as has already been shown for nucleic acids.19 Moreover an additional charge distribution of the highly charged3 α- and β-subunits are observed, which indicates that dissociation still takes place to some minor extent. To investigate whether the Tris buffer or the Mg2+ ions are responsible for the increased stability of the whole complex, we transferred the sample into an ammonium acetate (NH4OAc) buffer containing 1 mM of MgCl2. Fig. 3(b) shows the corresponding LILBID mass spectrum recorded using the same conditions as for the sample in Tris/HCl buffer. The signal-to-noise ratio considerably increased as compared with the spectrum in Fig. 3(a) while the resolution under soft desorption conditions hardly changed. The charge distribution of the anions is now shifted to lower charged states compared to the spectrum in Fig. 3(a). This difference in the charge distribution of the complex is attributed to the protonating effect of the NH4+ ions. Moreover it was not possible to obtain signals of the intact complex when transferring the sample into a NH4OAc buffer without MgCl2. This further demonstrates that Mg2+ ions are crucial for complex stability and function, a fact, which was investigated very early in ATP synthase research20 and confirmed many times since. Our results demonstrate that LILBID-MS is able to analyse the mass of an intact ATP synthase.
Fig. 3.
LILBID anion spectrum of B. pseudofirmus OF4 F1Fo-ATP synthase recorded under soft conditions. (a): Sample buffer was 10 mM Tris/HCl of pH 7.5, 0.08% DDM and 1 mM of MgCl2. (b): Sample buffer was 10 mM NH4OAc of pH 7.5, 0.08% DDM and 1 mM of MgCl2. Depicted are the charge states of the bacterial F1Fo complex.
Human and bovine F1Fo
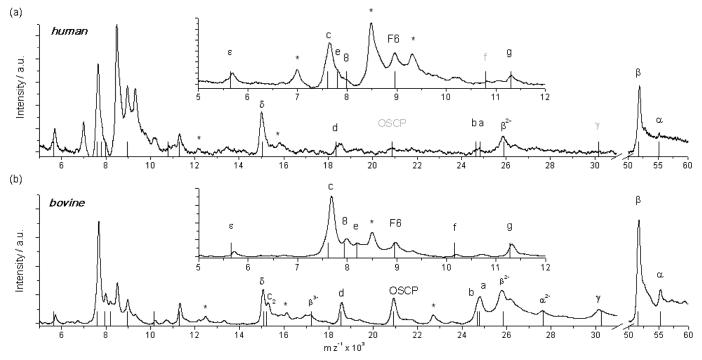
Bovine and human samples from Blue Native Electrophoresis (BNE) were transfered into 30 mM NH4HCO3, 0.05% DDM and measured in a harsh desorption mode. Final concentrations of the bovine and human samples were approximately in the sub-micromolar range in both cases. While the mass spectrum of bovine ATPase was recently reported by Walker et al.21 the mass spectrum of human ATP synthase presented in this work is the first mass spectrum of human ATP synthase reported so far. Both spectra (Fig. 4) show great similarity especially regarding the most intense peaks of the β-, δ- and c subunits, however subtle differences are expected and also observed for other subunits (see Table 2). The peaks labelled with an asterisk appeared repeatedly for both samples from different preparations of bovine ATP synthase and also for that of human ATP synthase. Most likely these peaks originate from foreign proteins, since the preparative conditions used for both samples are not sufficient to completely remove contaminations. When the spectrum of bovine ATP synthase is compared to a previously reported MS analysis,21 the masses agree within ±150 Da with the masses reported before and with the theoretical masses. All subunits except for the AGP and MLQ proteins (named after amino-terminal sequencing) are detected. These proteins are missing, since DDM, a detergent that is known to strip both proteins from the complex,22 was used for solubilisation and also for a buffer exchange in this work.
Fig. 4.
LILBID anion spectrum of human (a) and bovine (b) F1Fo-ATP synthases recorded at harsh conditions using 1000 and 900 droplets respectively. The insets show a magnification of the lower mass range. The black bars represent calculated masses of the subunits as stated in Table 2. Missing peaks of subunits are marked gray. An asterisk marks a contamination.
Table 2.
Calculated and experimental masses of bovine and human mitochondrial ATP synthase
| Subunit | bovine |
human |
|||||
|---|---|---|---|---|---|---|---|
| Uniprot | Calc. Mass [Da]a |
ESI mass [Da]20 |
LILBID mass [kDa] |
Uniprot | Calc. Mass [Da]a | LILBID mass [kDa] |
|
| α | P19483 | 55263.39 | 55244.6 | 55.3 (±0.2) | P25705 | 55209.32 | 55.2 (±0.21 |
| β | P00829 | 51562.97 | 51709.1 | 51.7 (±0.3) | P06576 | 51769.25 | 51.9 (±0.3) |
| γ | P05631 | 30255.71 | 30141.7 | 30.2 (±0.1) | P36542 | 30165.73 | not detected |
| δ | P05630 | 15064.93 | 15066.1 | 15.1 (±0.1) | P30049 | 15019.93 | 15 (±0.1) |
| ε | P05632 | 5651.67 | 5651.8 | 5.7 (±0.05) | P56381 | 5648.57 | 5.7 (±0.05) |
| b | P13619 | 24668.72 | 24670.7 | 24.7 (±0.3)b | P24539 | 24625.61 | 24.5 (±0.3)b |
| OSCP | P13621 | 20929.75 | 20931.6 | 20.9 (±0.1) | P48047 | 20875.49 | not detected |
| a |
P00847 Q8SFX8 |
24787.91 | 24817.6 | 24.7 (±0.2) | P00846 | 24817.21 | 24.8 (±0.3)b |
| d | P13620 | 18561.28 | 18604.6 | 18.6 (±0.1) | O75947 | 18360.02 | 18.5 (±0.2) |
| e | Q00361 | 8189.47 | 8189.8 | 8.2 (±0.05) | P56385 | 7802.02 | 7.8 (±0.3)b |
| f | Q28851 | 10165.99 | 10208.6 | 10.2 (±0.1) | P56134 | 10786.67 | not detected |
| g | Q28852 | 11286.26 | 11327.6 | 11.3 (±0.1) |
O75964 Q8N5M1 |
11428.48 11036.99 11386.44 |
11,3±(0,05) |
| 8 | P03929 | 7936.56 | 7964.4 | 8 (±0.05) | P03928 | 7991.65 | 8 (±0.3)b |
| c |
P32876 P07926 Q3ZC75 |
7608.01 | 7650.0 | 7.7 (±0.1) |
P05496 Q06055 P48201 |
7608.01 | 7.6 (±0.1) |
| F6 | P02721 | 8958.09 | 8959.0 | 9 (±0.05) | P18859 | 8960.15 | 8.9 (±0.1) |
As published in Meyer et al. 22
These signals occur in a shoulder only and hence the error range is untypical large.
The spectrum of human ATP synthase measured at an even lower protein concentration than that of the bovine ATP synthase is characterized by a reduced overall signal intensity compared to the bovine system. However, 8 subunits from the 15 expected ones can be clearly assigned. Only the subunits f and γ presumably disappeared in the noise of the spectrum. They are also of low intensity in the spectrum of the bovine ATPase, recorded at a slightly higher concentration. Another 5 peaks appear as shoulders of intense peaks.
The absence of the OSCP peak, which is intense in the bovine ATPase spectrum, is surprising. In addition it was not possible to record spectra of the intact complexes which most likely is a result of the rather low concentration of these samples. The unusual high amount of droplets needed to obtain the signals of the subunits (1000 for human and 900 for bovine ATP synthase) is a further indication of an insufficient sample concentration. In general respiratory complexes can be recovered in native state from BN gels as shown for bovine complex I23 and for the ATP synthase from yeast.24 Furthermore the detection of intact complexes with LILBID-MS after electro-elution from BN gels was already demonstrated for complex I from Yarrowia lipolytica.33 Nevertheless we think that the lack of detectable native ATP synthase complexes is mainly a result of our sample preparation protocol applied for LILBID analysis using detergent to remove Coomassie-dye from the protein which is delipidating and denaturing especially when handling very low protein amounts.
Comparison
A comparison of the ATP synthase subunit compositions in Table 3 reveals differences in the complexity of eukaryotic and bacterial ATP synthases.22 However, whereas the overall construction of eukaryotic enzymes is more complex than the bacterial ones, selected but functionally important subunits remain conserved among all ATP synthases. This includes, for example, the catalytically active β-subunits; these are known to show subtle but nevertheless important differences in structure3: These subunits maintain different conformational states and nucleotide affinities for ADP+Pi to ATP conversion, as proposed for the enzyme's binding change mechanism7. A high conservation can also be found in the subunits α and γ of the F1 subcomplex, a fact which is also reflected by the masses obtained in LILBID-MS. In the Fo motor, the most highly conserved subunit is the subunit c, which is known to form the rotor ring in all ATP synthases. High-resolution structures of several rotor rings are available.26,27,28,29,38 From these structures it has become clear that the overall construction of rotor rings is highly conserved, whereas subtle variations within the rotor ring shape and construction of their ion binding sites are responsible for the specificity of the coupling ions (H+ or Na+),28,30,38 which drive the Fo motor. In the context of this study of the ATP synthase rotor rings, LILBID-MS proved to be a novel tool for the determination of rotor ring stoichiometries (cn), of which n represents the number subunits necessary to form one c-ring.10 Interestigly, the number of c-subunits in principle equals the number of ions (n) transported across the membrane26,29 for every 360° rotation of the rotor1,3,31 in which three ATP molecules are synthesized in the three β-subunits of the F1 complex. Hence the ion-to-ATP ratio of the ATP synthase can be expressed by n/3. LILBID-MS has been proven to be applicable for the assignment of rotor ring stoichiometries10 and hence also represents a relatively easy-and-fast but nevertheless accurate method for the determination of ion-to-ATP ratios in ATP synthases,32 which represents an important parameter in cell bioenergetics. In addition it should be pointed out, that the relative signal intensities of the single charged α and β subunits may be different for different ATP synthases. In bacterial F1Fo-ATP synthases the intensities of these two subunits usually are approximately equal as can be seen for OF4 (Fig. 2(b)) and as observed for TA2.8 This is not surprising when comparing the theoretical isoelectric points (pI): in OF4 the pI for α is 5.11 and that for β is 5.01 (calculated with “compute pI/Mw tool” at http://www.expasy.ch/tools/pi_tool.html for all values). In the mammalian F1Fo-ATP synthases presented in this work the β subunits occur with a relative high signal intensity whereas the α subunits are hardly visible. For the human complex the pIs for α and β subunits are 8.28 and 5.0 while for the bovine complex the pIs are 8.27 and 5.0, respectively. Thus at the given buffer conditions the α subunits carry less negative charges than β and hence are only weakly detected in anion mode. In F1Fo-ATP synthases from Yarrowia lipolytica14 the α subunit (pI 6.6) occurs at about half the intensity of the β subunit (pI 4.81).14 Though calculated pIs may not reflect the accurate charge state in the functioning proteins, the LILBID spectra correctly reflect the relative charge states in solution for the presented complexes.
Table 3.
Comparison of subunit masses of F1Fo ATP-synthases measured by LILBID
| Subunit |
Bacillus pseudofirmus OF4 |
Bacillus sp. strain TA2.A18,36 |
Yarrowia lipolytica14 |
Bos taurus F1Fo (excerpt of Tab. 2) |
Homo sapiens F1Fo (excerpt of Tab. 2) |
|---|---|---|---|---|---|
| a | 26.8 | 26.6 | 24.7 | 27.8 | |
| b | 15.8 | 19.6 | 22.4 | 24.7 | 24.6 |
| c | 7.0 | 7.1 | 7.7 | 7.7 | 7.6 |
| α | 54.1 | 55.0 | 55.6 | 55.3 | 55.3 |
| β | 51.3 | 51.2 | 51.1 | 51.7 | 51.7 |
| γ | 31.5 | 31.9 | 30.1 | 30.2 | 30.2 |
| δ | 20.3 | 20.3 | 6.7 | 15.1 | 15.1 |
| ε | 14.4 | 14.9 | 14.7 | 5.7 | 5.7 |
Conclusions
We determined the molecular masses of the subunits of three different F-type ATP synthases, i.e. a bacterial complex from Bacillus pseudofirmus OF4 and an eukarotic ATP synthase from Bos taurus (bovine) and, for the first time, of human (Homo sapiens) heart mitochondria, by laser induced liquid bead ion desorption-mass spectrometry (LILBID-MS). This novel method allows the mass determination of non-covalently assembled membrane protein complexes up to the MDa-range with high accuracy. Low amounts of protein complexes (micrograms or less) can be dissolved in detergent solution at easily manageable concentrations. The method can be applied in several modes, from soft laser desorption, giving the intact macromolecular complexes to medium to harsh laser intensity, which disassembles the protein complexes partially into subcomplexes. At even higher laser intensities, the subcomplexes become disassembled and the single subunit composition can be determined. By combining these top-down mass data for those cases where all subunits and the mass of the integral complex are identified, the determination of the stoichiometry of the fully assembled complexes is possible.
Comparing LILBID with nESI
LILBID and nESI, which may be considered as native mass spectrometries, can be used on biological samples with similar sensitivity. Nevertheless they are complementary in respects, so it is quite likely that LILBID will continue to contribute value to the investigation of noncovalent complexes and especially membrane complexes. For example, while detergents are needed for solubilisation of hydrophobic complexes and remain a challenge for ESI, they seem to have a less critical impact for LILBID. LILBID is also more tolerant to addition of salt or various buffers other than ammonium acetate. Therefore non-covalent membrane complexes, or complexes that contain soluble and hydrophobic parts can reliably be detected without dissociation with LILBID, while the ESI results mirror the stability of a membrane complex, giving the method a strong bias towards soluble complexes. The softness of LILBID results in ions, which are still protected by a shell of buffer/detergent molecules, which may even be an entire micelle surrounding the protein. This leads to a noticeable broadening of the peaks towards higher masses and hence hampers the determination of the exact mass of the complex, more than for a well-resolved ESI spectrum. This drawback of LILBID-MS is evident when comparing the results presented in the present work with the analysis of a V-type ATP synthase by ESI-MS.39 On the other hand the assignment challenges for the highly charged ESI ions rather lie in determination of the right charge states as well as overlapping peak series, which my cause congestion of the spectra. Ions in LILBID generally carry much less charges than in ESI and show an enhanced propensity for the detection of negative ions. This may be an advantage in fragmentation studies due to a reduced Coulombic repulsion.
Both methods offer soft and harsh mode for measurements in a top-down fashion. While CID (collision induced dissociation) for nESI leads to the dissociation of usually one or two proteins in peripheral position, harsh laser conditions of LILBID will result in a complete disassembly of all subunits by thermolysis. Their charge states will mirror their net charge in solution, while in ESI the charging is mainly caused by the ammonium acetate buffer. Medium laser conditions in LILBID can allow the survival of strongly interacting subcomplexes. So the different fragmentation pathways of both methods can deliver truly complementary results. The mass resolution of LILBID is presently not yet satisfactory, but the next generation of a LILBID mass spectrometer with improved mass resolution is currently under development.
Outlook
LILBID-MS offers a novel way to determine masses of hydrophobic membrane protein complexes at low concentrations and in detergent solution. The applicability of the method encompasses not only selected mid-sized membrane complexes such as ATP synthases (complex V) reported here, but also other even larger membrane complexes such as Complex I from the mitochondrial respiratory chain with 41 subunits and a total mass of 960 kDa.33 Furthermore, the stoichiometry in the quaternary structure of rhodopsins and a potassium channel have been recently analyzed by this method.34
Acknowledgements/Financial disclosures
This work was supported in parts by the research grant GM28454 from the National Institute of General Medical Sciences (to TAK), the Cluster of Excellence “Macromolecular Complexes” at the Goethe University Frankfurt (DFG Project EXC 115) (to BB and TM) and the DFG Collaborative Research Center (SFB) 807 (to TM). I.W was supported by the Deutsche Forschungsgemeinschaft, Sonderforschungsbereich 815, Project Z1 (Redox-Proteomics).
Notes and references
- 1.Boyer PD. Ann. Rev. Biochem. 1997;66:717–749. doi: 10.1146/annurev.biochem.66.1.717. [DOI] [PubMed] [Google Scholar]
- 2.Junge W, Sielaff H, Engelbrecht S. Nature. 2009;459:364–370. doi: 10.1038/nature08145. [DOI] [PubMed] [Google Scholar]
- 3.Abrahams JP, Leslie AGW, Lutter R, Walker JE. Nature. 1994;370:621–628. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- 4.Dimroth P, Ballmoos C.v., Meier T. EMBO Rep. 2006;7:276–282. doi: 10.1038/sj.embor.7400646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collinson IR, Skehel JM, Fearnley IM, Runswick MJ, Walker JE. Biochem. 1996;35:12640–12646. doi: 10.1021/bi960969t. [DOI] [PubMed] [Google Scholar]
- 6.Capaldi RA, Aggerle R. Trends Biochem. Sc. 2002;27:154–160. doi: 10.1016/s0968-0004(01)02051-5. [DOI] [PubMed] [Google Scholar]
- 7.Boyer PD. Biochim. Biophys. Acta. 1993;1140:215–250. doi: 10.1016/0005-2728(93)90063-l. [DOI] [PubMed] [Google Scholar]
- 8.Morgner N, Hoffmann J, Barth H-D, Meier T, Brutschy B. Int. J. Mass Spectr. 2008;277:309–313. [Google Scholar]
- 9.Wittig I, Schägger H. Biochim. Biophys. Acta. 2008;1777:592–598. doi: 10.1016/j.bbabio.2008.04.027. [DOI] [PubMed] [Google Scholar]
- 10.Meier T, Morgner N, Matthies D, Pogoryelov D, Keis S, Cook GM, Dimroth P, Brutschy B. Mol. Microbiol. 2007;65:1181–1192. doi: 10.1111/j.1365-2958.2007.05857.x. [DOI] [PubMed] [Google Scholar]
- 11.Morgner N, Kleinschroth T, Barth H-D, Ludwig B, Brutschy B. J. Am. Soc. Mass Spectrom. 2007;18:1429–1438. doi: 10.1016/j.jasms.2007.04.013. [DOI] [PubMed] [Google Scholar]
- 12.Barrera NP, Di Bartolo N, Robinson CV. Science. 2008;321:243–246. doi: 10.1126/science.1159292. [DOI] [PubMed] [Google Scholar]
- 13.Vonck J, Pisa KY, Morgner N, Brutschy B, Müller V. J. Biol. Chem. 2009;284:10110–10119. doi: 10.1074/jbc.M808498200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sokolova L, Wittig I, Barth H-D, Schägger H, Brutschy B, Brandt U. Proteomics. 2010;10:1401–1407. doi: 10.1002/pmic.200900756. [DOI] [PubMed] [Google Scholar]
- 15.Hicks DB, Krulwich TA. J. Biol. Chem. 1990;265:20547–20554. [PubMed] [Google Scholar]
- 16.Wittig I, Braun H-P, Schägger H. Nature Prot. 2006;1:418–428. doi: 10.1038/nprot.2006.62. [DOI] [PubMed] [Google Scholar]
- 17.Aggeler R, Coons J, Taylor SW, Gosh SS, Garcia JC, Capaldi RA, Marusich MF. J. Biol. Chem. 2002;277:33906–33912. doi: 10.1074/jbc.M204538200. [DOI] [PubMed] [Google Scholar]
- 18.Morgner N, Barth H-D, Brutschy B. Austr. J. Chem. 2006;59:109–114. [Google Scholar]
- 19.Hoffmann J, Schmidt TL, Heckel A, Brutschy B. Rapid Com. Mass Spectr. 2009;23:2176–2180. doi: 10.1002/rcm.4129. [DOI] [PubMed] [Google Scholar]
- 20.Pullman ME, Penefsky HS, Datta A, Racker E. J. Biol. Chem. 1960;235:3322–3329. [PubMed] [Google Scholar]
- 21.Carroll J, Fearnley IM, Wang1 Q, Walker JE. Anal.Biochem. 2009;395:249–255. doi: 10.1016/j.ab.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 22.Meyer B, Wittig I, Trifilieff E, Karas M, Schägger H. Mol. Cell. Proteomics. 2007;6:1215–1225. doi: 10.1074/mcp.M700076-MCP200. [DOI] [PubMed] [Google Scholar]
- 23.Clason T, Ruiz T, Schägger H, Peng G, Zickermann V, Brandt U, Michel H, Radermacher M. J. Struct. Biol. 2010;169:81–88. doi: 10.1016/j.jsb.2009.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wittig I, Stuart RA, Velours J, Schägger H. Mol. Cell. Proteomics. 2008;7:995–1004. doi: 10.1074/mcp.M700465-MCP200. [DOI] [PubMed] [Google Scholar]
- 25.Nakamoto RK, Baylis Scanlon JA, Al-Shawi MK. Arch. Biochem. Biophys. 2008;476:43–50. doi: 10.1016/j.abb.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meier T, Polzer P, Diederichs K, Welte W, Dimroth P. Science. 2005;308:659–662. doi: 10.1126/science.1111199. [DOI] [PubMed] [Google Scholar]
- 27.Murata T, Yamato I, Kakinuma Y, Leslie AG, Walker JE. Science. 2005;308:654–659. doi: 10.1126/science.1110064. [DOI] [PubMed] [Google Scholar]
- 28.Meier T, Krah A, Bond PJ, Pogoryelov D, Diederichs K, Faraldo-Gómez JD. J. Mol. Biol. 2009;391:498–507. doi: 10.1016/j.jmb.2009.05.082. [DOI] [PubMed] [Google Scholar]
- 29.Pogoryelov D, Yildiz Ö, Faraldo-Gômez JD, Meier T. Nat. Struct. Mol. Biol. 2009;16:1068–1073. doi: 10.1038/nsmb.1678. [DOI] [PubMed] [Google Scholar]
- 30.Krah A, Pogoryelov D, Langer JD, Bond PJ, Meier T, Faraldo-Gómez JD. Biochim. Biophys. Acta. 1797:763–772. doi: 10.1016/j.bbabio.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 31.Noji H, Yasuda R, Yoshida M, Kinosita K., Jr. Nature. 1997;386:299–302. doi: 10.1038/386299a0. [DOI] [PubMed] [Google Scholar]
- 32.Ferguson SJ. Curr. Biol. 2000;10:R804–R808. doi: 10.1016/s0960-9822(00)00765-x. [DOI] [PubMed] [Google Scholar]
- 33.Morgner N, Zickermann V, Kerscher S, Wittig I, Abdrakhmanova A, Barth H-D, Brutschy B, Brandt U. BBA-Bioenergetics. 2008;1777:1384–1391. doi: 10.1016/j.bbabio.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 34.Hoffmann J, Aslimovska L, Bamann C, Glaubitz C, Bamberg E, Brutschy B. Phys. Chem. Chem. Phys. 2010;12:3480–3485. doi: 10.1039/b924630d. [DOI] [PubMed] [Google Scholar]
- 35.Schägger H, von Jagow G. Anal. Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 36.Nesterenko MV, Tilley M, Upton SJ. J. Biochem. Biophys. Methods. 1994;28:239–242. doi: 10.1016/0165-022x(94)90020-5. [DOI] [PubMed] [Google Scholar]
- 37.Matthies D, Preiss L, Klyszejko AL, Muller D, Cook GM, Vonck J, Meier T. J. Mol. Biol. 2009;388:611–618. doi: 10.1016/j.jmb.2009.03.052. [DOI] [PubMed] [Google Scholar]
- 38.Preiss L, Yildiz Ö, Hicks DB, Krulwich TA, Meier T. PLoS Biology. doi: 10.1371/journal.pbio.1000443. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang ZY, Zheng Y, Mazon H, Milgrom E, Kitagawa N, Kish-Trier E, Heck AJR, Kane PM, Wilkens S. J. Biol. Chem. 2008;283:35983–35995. doi: 10.1074/jbc.M805345200. [DOI] [PMC free article] [PubMed] [Google Scholar]