Abstract
Background
Independent of temporal circumstances, some individuals have greater susceptibility to depressive affects, such as feelings of guilt, sadness, hopelessness, and loneliness. Identifying the genetic variants that contribute to these individual differences can point to biological pathways etiologically involved in psychiatric disorders.
Methods
Genome-wide association scans (GWA or GWAS) for the Depression scale of the Revised NEO Personality Inventory (NEO-PI-R) in community-based samples from a genetically homogeneous area of Sardinia, Italy (N = 3,972) and from the Baltimore Longitudinal Study of Aging in the US (N = 839).
Results
Meta-analytic results for genotyped or imputed single nucleotide polymorphisms (SNPs) indicate that the strongest association signals for trait depression were found in RORA (rs12912233; p= 6 × 10−7), a gene involved in circadian rhythm. A plausible biological association was also found with SNPs within GRM8 (rs17864092; p = 5 × 10−6), a metabotropic receptor for glutamate, a major excitatory neurotransmitter in the central nervous system.
Conclusions
These findings suggest shared genetic basis underlying the continuum from personality traits to psychopathology.
Keywords: GWA or GWAS, depression, neuroticism, RORA, GRM8 or mGlu8
Introduction
Depressed mood is the predominant feature in the diagnosis of mood disorders (e.g., major depressive disorder, dysthymic disorder, bipolar disorder) and is an important clinical component of many psychiatric, neurological, and physical syndromes (1). Mood disorders are one of the leading causes of disability worldwide (2, 3), with an estimated lifetime risk of 20% in the US population (4). Most mental disorders are thought to arise from the combination of multiple genes and environmental factors (5). Large, genetically informative, population-based longitudinal studies (6–8) indicate that the personality trait Neuroticism strongly reflects the genetic vulnerability to major depression, sharing an estimated 50% of the genetic liability, and consistently predicts which individuals are at greater risk for depressive illness (6, 7, 9). These links, along with evidence that personality traits are heritable (6, 10, 11) and highly stable in adulthood (12–14), indicate that Neuroticism-related personality traits are a promising endophenotype (15) for mood and other psychiatric disorders (16–18).
Neuroticism is a broad dimension that reflects the tendency to experience a wide spectrum of negative emotions. This heterogeneous trait is composed of several lower-order facets, including depression, anxiety, hostility, self-consciousness, impulsivity, and vulnerability to stress (19). These more circumscribed facets are also heritable (10, 11) and have greater predictive power for specific behavioral and health outcomes than the broader domain-level Neuroticism (20, 21).
The depression facet captures the core aspects of depressed mood, with items that assess susceptibility to feelings of sadness, hopelessness, worthlessness, discouragement, guilt, and loneliness. As such, this facet measures the characteristic psychic component of depression, not its physical symptoms (22). This enduring disposition increases vulnerability to depressive states, but also may be one common denominator underlying comorbidities among psychiatric disorders. Indeed, both clinically depressed and other psychiatric patients tend to score higher on depression compared to the other facets of Neuroticism (23, 24). Identifying genetic variants associated with trait depression may be informative not only for this personality trait, but for a wide range of psychiatric disorders that share a mood component. Moreover, as a more circumscribed facet, trait depression measures a narrower phenotype, which can increase statistical power by reducing phenotypic variability, and might thus prove advantageous in genetic association studies.
In this study, we performed two genome-wide association scans (GWA or GWAS) to search across the genome for common variants that contribute to depression vulnerability. We tested for association between trait depression and over 2 million genotyped or imputed single nucleotide polymorphisms (SNPs) in a large and homogeneous sample from Sardinia (Italy) and in a sample of participants with European ancestry from a US longitudinal study. To increase power, we meta-analytically combined the results from the two samples. Rather than focus on strong signals in one sample that might not replicate in the other, we looked for consistent effects across samples. This approach has been successful in identifying genetic variants that are reliably associated with quantitative traits, such as height, and diseases, such as diabetes (25, 26). In the field of psychiatry, there is a growing number of GWA studies for bipolar disorder (26, 27), major depression (9, 28, 29), and trait Neuroticism (16–18). This is the first GWA study for the depression facet of personality. The accumulating evidence across multiple GWA studies is providing replicable associations (30, 31), even when the primary studies did not reach statistical significance (e.g., CACNA1C)(26, 27, 32).
Methods
Sample description: SardiNIA
For the SardiNIA study, we recruited 6,148 individuals from a cluster of four towns in the Lanusei Valley, SardiNIA, Italy (11). The only exclusion criteria were being younger than 14 years old and being from regions other than Sardinia. The sample includes over 62% of the population aged 14 to 102 years; further recruitment and longitudinal testing is ongoing. Inhabitants of this area are a known founder population, descending from few ancestors with minimal admixture with other populations. Even today, most subjects are native born, and at least 95% of the SardiNIA sample have all grandparents born in the same province (11). This population was chosen because of its high genetic homogeneity, which should increase power in genetic association studies. During the first wave of assessment (2001–2004), valid personality data were obtained from 5,669 subjects, of whom 3,972 were part of the genome-wide association (GWA) scan. The sample includes 57% women and ranged in age from 14 to 90 (M = 42.8, SD = 17). Additional information on the sample has been reported elsewhere (11, 33). GWA analyses for Neuroticism and the other four domains of the Five-Factor Model in this SardiNIA sample have been previously reported for roughly 360K SNPs (17). The project was approved by institutional review boards in Italy and the USA.
Sample description: BLSA
The Baltimore Longitudinal Study of Aging (BLSA) is an ongoing multidisciplinary study of community-dwelling volunteers. The GWA analysis was restricted to subjects with European ancestry to reduce population stratification biases. A total of 839 subjects (46% women) of European descent were successfully genotyped and completed the personality questionnaire at least once. In this sample, age at first assessment ranged from 20 to 93 (M = 58.5; SD = 17). Personality traits were assessed between 1989 and 2008, and multiple assessments were available for most participants. Although personality traits are generally stable over time (13, 34), we combined all available assessments for each individual, for a total of 3,507 assessment points. By averaging across multiple time points, we reduce variability due to temporary effects and random error, thereby obtaining more reliable and robust personality score estimates. The BLSA study was approved by the local institutional review board.
Trait depression assessment
Trait depression was assessed using the English (19) and Italian (35) versions of the Revised NEO Personality Inventory (NEO-PI-R). The Depression scale consists of 8 items, including two reversed scored items to reduce the effects of acquiescence. The items are answered on a five-point Likert scale, from strongly disagree to strongly agree. Scores followed a normal distribution and were standardized (M = 50, SD = 10) using American combined gender norms (19). The Depression scores ranged from 29 to 86 (M = 54, SD = 9) in the SardiNIA sample and from 27 to 86 (M = 48, SD = 10) in the BLSA sample, both of which are in the range observed in non-clinical populations. No structured psychiatric evaluation was available in either sample.
There is a large body of evidence that personality scores are both reliable and valid across cultures (10, 36). Indeed, the NEO-PI-R has a robust factor structure that has been replicated in Italy (35) and in more than 50 cultures (36), even at the genetic level (11, 33). In both samples, the depression facet scale has good psychometric properties: internal consistency reliability was .73 in the SardiNIA sample and .80 in the BLSA sample. In the BLSA, available longitudinal data (13) indicate that corrected stability coefficient for the depression facet is .86 over an interval of 10 years.
Genetic assays and imputation
DNA was extracted from blood. Genotyping was performed in the BLSA sample with the 550k Illumina platform, and in the SardiNIA sample with the 10K and 500K Affymetrix mapping array set (see below and previous reports for additional information)(17, 25). The genotype calling algorithm used was the BRLMM for the SardiNIA sample and the Beadstudio for the BLSA sample. Genotype data from both samples passed quality controls. For the SardiNIA cohort, sample call rate was > 95%, and SNPs exclusions criteria were Hardy-Weinberg equilibrium (HWE) <= 10−6, SNP call rate <= 90%, and minor allele frequency (MAF) < 5%. For the BLSA cohort, sample call rate was > 98.5%, SNPs exclusions criteria were HWE < 10−4, SNP call rate < 99%, and MAF < 5%. The genotyping approach used in the SardiNIA study takes advantage of the large number of multigenerational families in this relatively homogeneous sample from a founder population. Related individuals, such as siblings and parents/offspring, share long multi-megabase stretches of chromosome. If these shared stretches are genotyped with high density array in only a few individuals, the information from these individuals can be propagated to their relatives who inherit shared chromosome stretches with them (37–39). Thus, data from Sardinian individuals genotyped with the Affymetrix Mapping 500K Array Set (N = 1,412) was used to infer missing genotypes in their offspring or siblings genotyped with the 10K Array Set (N =2,893), for a total of 4,305 samples available for GWA analyses (personality data were available for 3,972 of these participants). This within-family imputation method, based on “identical-by-descent” sharing and implemented by the MERLIN program (38), has enabled full GWA scans in the SardiNIA sample. The results from the SardiNIA sample have been combined successfully with other GWA studies of physical and mental traits, such as height, weight, lipid levels, and cigarette smoking (25, 40–42).
To combine the data from the different array sets in the two cohorts, and to increase the overall coverage of the genome to up to 2.5 million SNPs, we imputed autosomal SNPs reported in the HapMap CEU sample, using the imputation program MACH v1.015 (http://www.sph.umich.edu/csg/abecasis/MACH/index.html ). Markers showing low imputation quality (r2 ≤ 0.3) were discarded from the analysis.
Statistical Analyses
To account for family structure in the SardiNIA sample, we used the program MERLIN (38) to evaluate the additive effect of all genotyped or imputed SNPs on trait depression. Imputed SNP dosages were coded using fractional counts between 0 and 2 according to the estimated number of copies of each allele. In MERLIN, regression coefficients are estimated in the context of a variance component model to adjust for relatedness among individuals (38). The same association test was carried out in the BLSA sample. An inverse normal transformation was applied to personality traits to avoid inflated type I error. Sex, age, and age squared were included as covariates in all analyses to account for sex and age differences in personality traits (34, 43).
In the SardiNIA sample, we checked the genomic control value for our genome-wide association analyses (44), and we did a principal component analysis of genome-wide SNP data in a subset of unrelated individuals (45). Neither analysis suggested evidence of population substructure or genetic outliers. Analyses in the BLSA were restricted to the European-American subsample. Self-reported ethnicity was confirmed by comparisons between the BLSA and HapMap genomic data. To account for population structure, in the BLSA we further adjusted for the first two principal components derived from an EIGENSTRAT analysis utilizing ~10,000 randomly selected SNPs from the 550K SNP panel (45).
A weighted z score based fixed effects meta-analysis method was used to combine results from the SardiNIA and BLSA samples using the program METAL (http://www.sph.umich.edu/csg/abecasis/metal/). In brief, for every SNP, a reference allele was identified and a z statistic summarizing the magnitude of the P-value for the association and direction of effect was generated in each sample. An overall z statistic was then computed as a weighted average of the individual statistics, and a corresponding P-value for that statistic was computed. The weights were proportional to the square root of the number of individuals in each study and scaled such that the squared weights summed to 1. Genomic control correction was applied to the test statistics in each study when appropriate. A Bonferroni corrected p-value of 5x10−8 was considered genome-wide significant.
Results
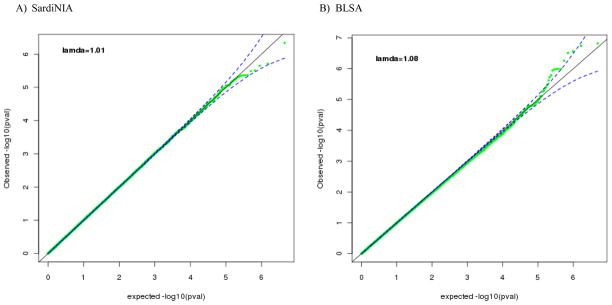
GWA analyses for the depression personality trait were carried out on 3,972 individuals from the SardiNIA sample and on 839 individuals from the BLSA sample, all of European ancestry. The Q-Q plots (Figure 1) indicate that there may be an excess of significant associations in the BLSA (λ=1.08), but not in the SardiNIA sample (λ=1.01). To identify SNPs associated with trait depression, we combined the results from the GWA analyses in the SardiNIA and BLSA samples in a meta-analysis (total N = 4,811). There were no genome-wide significant findings (threshold: P < 5x10−8). The top 25 SNPs, ranked by p-value, are presented in Table 1. In addition, the SNPs with p ≤ 10−4 are presented in Supplement: Table S1.
Figure 1.
Quantile-Qualtile plot of observed vs expected log P-value for the SardiNIA (A) and the Baltimore Longitudinal Study of Aging (B).
Table 1.
SardiNIA and BLSA meta-analysis of association results for trait depression.
| Imputation quality (r2) | Effect-allele frequency | Effect size | p-value | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Gene | chr | position | Effect-allele | SardiNIA | BLSA | SardiNIA | BLSA | SardiNIA | BLSA | SardiNIA | BLSA | Meta-analysis |
| rs12912233 | RORA | 15 | 59.05 | T/C | 0.54 | 0.96 | .46 | .46 | 0.15 | 0.09 | 1.8 ×10−6 | 6.1 ×10−2 | 6.3 ×10−7 |
| rs8070473 | SLFN12L | 17 | 30.87 | T/G | 0.81 | 0.99 | .32 | .32 | −0.16 | −0.07 | 1.1 ×10−6 | 2.3 ×10−1 | 1.5 ×10−6 |
| rs349475 | CDH18* | 5 | 19.48 | T/C | 0.66 | 0.86 | .29 | .29 | 0.18 | 0.02 | 2.4 ×10−7 | 7.3 ×10−1 | 2.4 ×10−6 |
| rs12420464 | EIF3F | 11 | 7.97 | T/G | 0.37 | GN | .04 | .04 | −0.38 | −0.08 | 1.3 ×10−6 | 3.8 ×10−1 | 3.3 ×10−6 |
| rs8068353 | SLFN12L | 17 | 30.86 | C/G | 0.78 | 0.93 | .32 | .32 | −0.15 | −0.08 | 4.4 ×10−6 | 1.6 ×10−1 | 3.4 ×10−6 |
| rs1927745 | FAM155A* | 13 | 106.58 | A/G | 0.66 | GN | .24 | .24 | −0.16 | −0.06 | 3.5 ×10−6 | 2.4 ×10−1 | 4.7 ×10−6 |
| rs10514585 | CDH13 | 16 | 81.84 | A/G | GN | 0.99 | .31 | .31 | 0.15 | 0.06 | 3.1 ×10−6 | 2.8 ×10−1 | 4.9 ×10−6 |
| rs11009175 | ITGB1* | 10 | 33.33 | A/G | 0.86 | 0.96 | .17 | .17 | 0.16 | 0.16 | 4.2 ×10−5 | 2.3 ×10−2 | 5.4 ×10−6 |
| rs17864092 | GRM8 | 7 | 126.23 | T/C | 0.65 | GN | .90 | .90 | −0.17 | −0.25 | 1.9 ×10−4 | 2.2 ×10−3 | 5.5 ×10−6 |
| rs9634463 | FAM155A* | 13 | 106.57 | T/C | 0.89 | 0.90 | .24 | .24 | −0.16 | −0.05 | 2.5 ×10−6 | 3.7 ×10−1 | 5.7 ×10−6 |
| rs7329003 | FAM155A* | 13 | 106.57 | A/G | GN | 0.90 | .75 | .75 | −0.16 | −0.05 | 2.5 ×10−6 | 3.7 ×10−1 | 5.8 ×10−6 |
| rs713548 | FAM155A* | 13 | 106.57 | T/G | 0.91 | 0.90 | .25 | .25 | −0.16 | −0.05 | 2.6 ×10−6 | 3.7 ×10−1 | 5.9 ×10−6 |
| rs4775340 | RORA | 15 | 59.06 | A/G | 0.55 | GN | .43 | .43 | 0.14 | 0.09 | 2.1 ×10−5 | 6.5 ×10−2 | 6.3 ×10−6 |
| rs1449984 | - | 2 | 23.33 | A/G | 0.96 | GN | .72 | .72 | −0.15 | −0.06 | 6.7 ×10−6 | 2.0 ×10−1 | 6.6 ×10−6 |
| rs9301191 | FAM155A* | 13 | 106.57 | T/C | 0.85 | 0.90 | .79 | .79 | −0.17 | −0.05 | 2.9 ×10−6 | 3.9 ×10−1 | 7.1 ×10−6 |
| rs8028646 | RORA | 15 | 59 | A/T | GN | 0.99 | .25 | .25 | −0.14 | −0.15 | 1.6 ×10−4 | 4.6 ×10−3 | 7.2 ×10−6 |
| rs1924397 | FAM155A* | 13 | 105.41 | A/C | 0.54 | 0.80 | .96 | .96 | 0.32 | 0.15 | 2.0 ×10−5 | 8.3 ×10−2 | 7.6 ×10−6 |
| rs10744304 | - | 12 | 126.34 | A/G | 0.68 | GN | .63 | .63 | −0.15 | −0.05 | 5.0 ×10−6 | 3.2 ×10−1 | 8.7 ×10−6 |
| rs2017305 | DDX21* | 10 | 70.38 | A/G | 1.00 | 0.97 | .92 | .92 | −0.22 | −0.21 | 6.9 ×10−5 | 2.3 ×10−2 | 9.0 ×10−6 |
| rs3740596 | DDX50 | 10 | 70.34 | A/G | 1.00 | 0.97 | .92 | .92 | −0.22 | −0.20 | 6.8 ×10−5 | 2.6 ×10−2 | 9.5 ×10−6 |
| rs12638201 | CCR9 | 3 | 45.91 | A/G | 0.82 | GN | .14 | .14 | 0.17 | 0.19 | 4.9 ×10−5 | 4.9 ×10−2 | 1.1 ×10−5 |
| rs4796093 | SLFN12L | 17 | 30.87 | A/G | 0.89 | GN | .64 | .64 | −0.14 | −0.06 | 9.3 ×10−6 | 2.6 ×10−1 | 1.2 ×10−5 |
| rs4885589 | - | 13 | 78.45 | T/C | 0.86 | 0.96 | .60 | .60 | 0.10 | 0.18 | 1.1 ×10−3 | 2.4 ×10−4 | 1.3 ×10−5 |
| rs1539163 | DDX21 | 10 | 70.41 | T/C | 1.00 | 0.96 | .08 | .08 | −0.21 | −0.21 | 1.1 ×10−4 | 2.4 ×10−2 | 1.4 ×10−5 |
| rs8023563 | RORA | 15 | 59 | A/T | GN | 0.98 | .76 | .76 | −0.13 | −0.15 | 2.9 ×10−4 | 5.4 ×10−3 | 1.5 ×10−5 |
Nearest gene. GN = Genotyped. The first allele in the Effect-allele column is the reference allele for allele frequency and effect size direction.
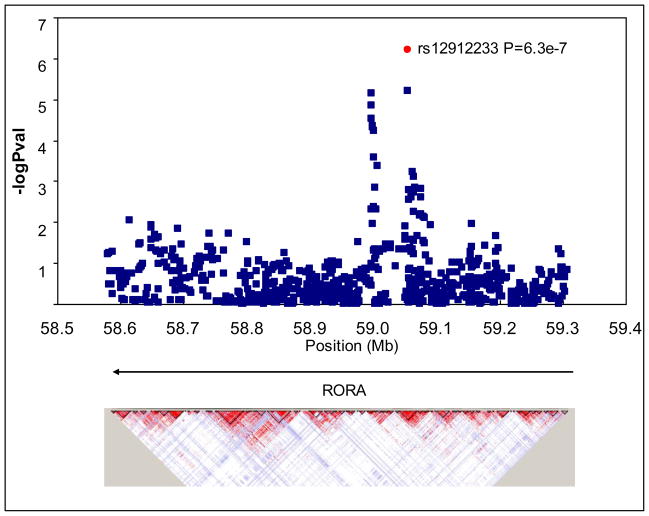
The meta-analysis indicates that the SNP with the lowest p-value maps within an intron of the RORA gene (rs12912233; p=6 × 10−7). As shown in Figure 2, a number of other SNPs within RORA show strong associations with trait depression. Some of these SNPs had a stronger association in the SardiNIA sample (e.g., rs12912233, rs4775340), but for others the effects were quite similar across the two samples (e.g., rs8028646, rs8023563). For rs12912233, and most of the other significant SNPs in RORA, the allele with the lower frequency was associated with roughly 0.15 SD higher depression scores (see Table 1).
Figure 2.
Plot of -log10 P values for depression trait meta-analysis for SNPs mapping within the RORA gene. Bottom panel presents patterns of linkage disequilibrium (r2) for the region in the HapMap CEPH population.
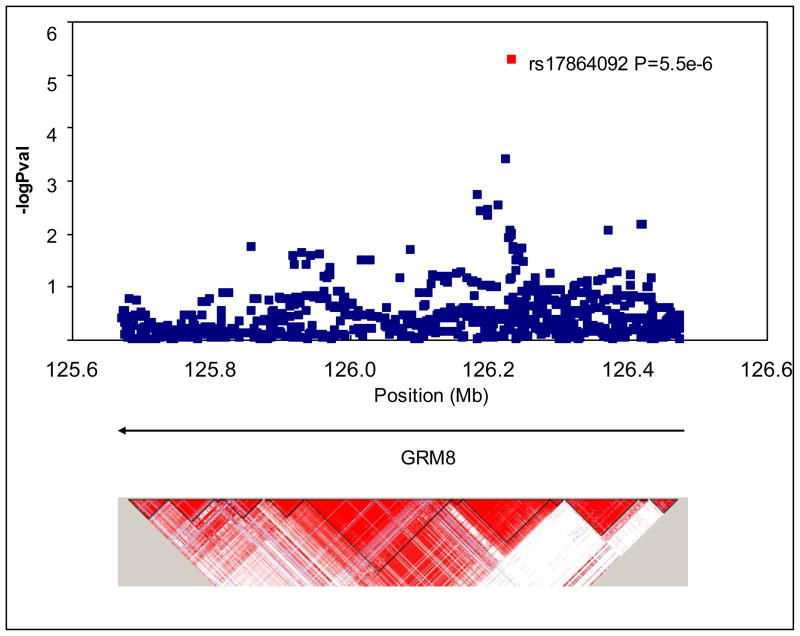
Among the top hits, the most biologically plausible finding was the association between trait depression and SNPs within the metabotropic glutamate receptor type 8 gene (GRM8 or mGlu8)(see Figure 3). The strongest effect was observed for the intronic SNP rs17864092 (p = 5.5x10−6); the allele T (frequency 90% in both SardiNIA and BLSA samples) was associated with lower depression scores. In terms of effect size, individuals with the risk allele (C) scored roughly two T-score points (0.2 SD) higher on depression, compared to the homozygous TT. Interestingly, a pharmacogenetic study of antipsychotic response implicated rs17864092 in verbal fluency scores (46). Other SNPs in GRM8 that were associated with trait depression in our two samples (e.g., rs17867725, p = 4.2x10−4; rs11563409, p = .019) are associated with cognitive phenotypes of psychotic patients (46). In addition, other GRM8 SNPs (rs2299495, rs1361995, rs10487457, rs10487459) that show some evidence of association in either the SardiNIA or the BLSA sample have been associated with alcohol dependence and other psychiatric disorders in previous research (47).
Figure 3.
Plot of -log10 P values for depression trait meta-analysis for SNPs mapping within the GRM8 gene. Bottom panel presents patterns of linkage disequilibrium (r2) for the region in the HapMap CEPH population.
Other SNPs in Table 1 were strongly associated in the SardiNIA sample but showed weak or non-significant p-values in the BLSA. Although many of these SNPs mapped within or near genes of unknown function (e.g., SLFN12L and FAM155A), in the SardiNIA sample we found strong associations between the depression personality trait and SNPs within the CDH13 gene (rs10514585) and near the CDH18 gene (rs349475). These Cadherin genes encode for cell adhesion proteins, which may play a role in regulating synapse formation, function and plasticity (48–50). CDH13 is expressed in the heart and several brain tissues, whereas CDH18 is expressed specifically in the brain (51, 52). GWA studies have implicated SNPs within the large CDH13 gene in several traits and diseases, such as introversion (17), substance abuse (53, 54), and attention deficit hyperactivity disorder (55).
Discussion
We report results from the first GWA study of the depression personality trait, the facet of Neuroticism most closely related to the core component of mood disorders. We genotyped or imputed over 2 million SNPs in a large homogeneous sample from Sardinia (Italy) and in a longitudinal sample from the US, for a total of 4,811 individuals. The imputation of genetic information is now a common practice in GWA studies (25–27, 30, 56), but it was not used in our previous GWA study of the five major dimensions of personality (17) or by the previous GWA studies of Neuroticism (16, 18). Although we found no genome wide statistical significant associations, our GWA results point to genes involved in brain function, behavior, and psychopathology, and can provide useful insight in the biology of depression. Specifically, we found the SNPs most strongly associated with trait depression were within the RORA and GRM8 genes.
The strongest meta-analytic signal was found for a number of closely linked SNPs within the RORA gene, particularly in the SardiNIA sample. RORA, or retinoic acid receptor-related orphan receptor alpha, is a member of the nuclear hormone-receptor superfamily. In the mouse nervous system, RORA is localized in the cerebellum, thalamus, cerebral cortex, superchiasmatic nucleus and other structures (57). The function of this gene appears to be complex. Deletion within the RORA gene causes the staggerer mouse phenotype, which is characterized by severe cerebellar ataxia due to a defect in the development of Purkinje cells (58). RORA also seems to play a role in immunity (59) and has emerged as an important component of mammalian circadian rhythms (60, 61). As recently reviewed (62), multiple lines of evidence from animal models, GWA, and linkage studies converge on variants in the RORA gene that may be linked to bipolar disorder. A recent study that examined circadian candidate genes in a Swedish population-based sample also found RORA to be associated with clinical depression (63). This evidence, together with our GWA results, supports a role of RORA in trait depression, and given its function as a circadian gene, may be implicated in the cyclic nature of mood disorders, especially seasonal and bipolar disorder (64, 65).
Glutamate is a widespread excitatory neurotransmitter involved in multiple brain functions (e.g., synaptic plasticity) and has been implicated in neuropathology (e.g., Alzheimer’s disease, addictions)(47, 66, 67). Glutamate activates a number of ionotropic (NMDA, AMPA, kainate, and delta) and metabotropic (mGlu) receptors. In this study, the strongest effect was found for GRM8 (or mGlu8), a group III metabotropic glutamate receptor, a subgroup known to modulate glutamatergic neurotransmission via presynaptic inhibition of glutamate release (68). The group III metabotropic receptors are part of the control system that maintain glutamate levels within normal boundaries, as excessive levels of glutamate in the synaptic space have excitotoxic effects, triggering cellular damage, neuronal atrophy and loss. Growing evidence suggests that the glutamatergic system plays a major role in the pathophysiology of neuropsychiatric disorders; glutamate receptors are seen as promising therapeutic targets (67–70). In animal models, agonists for type III mGlu receptors produce anxiolytic-like effects, but the evidence is mixed for the antidepressant-like effects across behavioral tests (70–72). In addition, a GRM8 agonist has been shown to suppress alcohol self-administration and cue-induced reinstatement of alcohol-seeking behaviors (73). At the genetic level, GRM8 has been tested as a candidate gene for Schizophrenia (74) and alcohol-dependence phenotypes (47) and was part of a network of glutamate receptor genes that emerged from a GWA study of cigarette smoking (56). Thus, the results of our GWA study that implicate GRM8 suggest that genetic variants in a key component of the glutamate neurotransmission system may contribute to risk of depression and other psychiatric disorders.
The GRM8 and the RORA are promising candidate genes, but larger samples are required to obtain definitive evidence. Consistent with most other studies of quantitative traits and disorders (16–18, 25, 53–55), the variants we identified explained a small portion of variance (1% or less), which suggests that common SNPs with large effects on trait depression are unlikely to exist. Although GWA have been successful in identifying common variants associated with various complex traits, the full genetic component of complex traits will require the examination of other types of variants, such as rare variants and copy number variants. Large-scale sequencing projects are one approach to address some of the limitations of the current GWA studies and move the field forward. Sequence data would provide virtually complete genetic coverage and allow the assessment of the effect of rarer variants. Other approaches are also needed to investigate epigenetic effect and the role of environmental factors that contribute to psychiatric disorders. Epigenetic regulations of gene expression such as DNA methylation, histone modifications, DNA rearrangement, and RNA inhibition, have been implicated in complex behaviors and psychiatric disorders (75, 76). Still, the role of epigenetic phenomena is complex and will require new methods to fully evaluate the biological mechanisms that contribute to the etiology of complex disorders. To date, the GWA can provide an unbiased examination across the genome for common variants that contribute to quantitative traits and diseases. Even small effects can point to genes that may harbor rarer variants with larger effects, and may elucidate the role of biological pathways.
Our top results are likely to be enriched with SNPs that are truly associated with the depression personality trait, and can contribute to the accumulation of evidence in support or against any particular gene in association with depression and related phenotypes. If these findings are confirmed, they would support the hypothesis behind GWA studies that common variants contribute to disease liability. There is also growing evidence that different psychiatric disorders share common genetic loci (30–32). This study extends these findings suggesting that common variants associated with psychiatric disorders in clinical studies contribute to individual differences on trait depression in the general population.
Supplementary Material
Acknowledgments
We would like to thank SardiNIA study participants for their volunteerism and the local civil and religious authorities in Sardinia for their support. We thank Prof. Antonio Cao for his leadership of the SardiNIA project. Antonio Terracciano had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. Funding: This research was supported in part by the Intramural Research Program of the NIH, National Institute on Aging.
Footnotes
Financial disclosure: Paul T. Costa, Jr., receives royalties from the Revised NEO Personality Inventory. All other authors report no biomedical financial interests or potential conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.American Psychiatric Association. Diagnostic and statistical manual of mental disorders. 4. American Psychiatric Association; Washington, DC: 1994. [Google Scholar]
- 2.Murray CJ, Lopez AD. Evidence-based health policy–lessons from the Global Burden of Disease Study. Science. 1996;274:740–743. doi: 10.1126/science.274.5288.740. [DOI] [PubMed] [Google Scholar]
- 3.Lopez AD, Mathers CD, Ezzati M, Jamison DT, Murray CJ. Global and regional burden of disease and risk factors, 2001: systematic analysis of population health data. Lancet. 2006;367:1747–1757. doi: 10.1016/S0140-6736(06)68770-9. [DOI] [PubMed] [Google Scholar]
- 4.Kessler RC, Berglund P, Demler O, Jin R, Merikangas KR, Walters EE. Lifetime prevalence and age-of-onset distributions of DSM-IV disorders in the National Comorbidity Survey Replication. Arch Gen Psychiatry. 2005;62:593–602. doi: 10.1001/archpsyc.62.6.593. [DOI] [PubMed] [Google Scholar]
- 5.Risch N, Herrell R, Lehner T, Liang KY, Eaves L, Hoh J, et al. Interaction between the serotonin transporter gene (5-HTTLPR), stressful life events, and risk of depression: a meta-analysis. JAMA. 2009;301:2462–2471. doi: 10.1001/jama.2009.878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kendler KS, Gatz M, Gardner CO, Pedersen NL. Personality and major depression: a Swedish longitudinal, population-based twin study. Arch Gen Psychiatry. 2006;63:1113–1120. doi: 10.1001/archpsyc.63.10.1113. [DOI] [PubMed] [Google Scholar]
- 7.Kendler KS, Neale MC, Kessler RC, Heath AC, Eaves LJ. A longitudinal twin study of personality and major depression in women. Archives of General Psychiatry. 1993;50:853–862. doi: 10.1001/archpsyc.1993.01820230023002. [DOI] [PubMed] [Google Scholar]
- 8.Fanous A, Gardner CO, Prescott CA, Cancro R, Kendler KS. Neuroticism, major depression and gender: a population-based twin study. Psychol Med. 2002;32:719–728. doi: 10.1017/s003329170200541x. [DOI] [PubMed] [Google Scholar]
- 9.Sullivan PF, de Geus EJ, Willemsen G, James MR, Smit JH, Zandbelt T, et al. Genome-wide association for major depressive disorder: a possible role for the presynaptic protein piccolo. Mol Psychiatry. 2009;14:359–375. doi: 10.1038/mp.2008.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jang KL, McCrae RR, Angleitner A, Riemann R, Livesley WJ. Heritability of facet-level traits in a cross-cultural twin sample: Support for a hierarchical model of personality. Journal of Personality and Social Psychology. 1998;74:1556–1565. doi: 10.1037//0022-3514.74.6.1556. [DOI] [PubMed] [Google Scholar]
- 11.Pilia G, Chen WM, Scuteri A, Orrú M, Albai G, Dei M, et al. Heritability of Cardiovascular and Personality Traits in 6,148 Sardinians. PloS Genetics. 2006;2:e132. doi: 10.1371/journal.pgen.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Viken RJ, Rose RJ, Kaprio J, Koskenvuo M. A developmental genetic-analysis of adult personality: Extroversion and neuroticism from 18 to 59 years of age. Journal of Personality and Social Psychology. 1994;66:722–730. doi: 10.1037//0022-3514.66.4.722. [DOI] [PubMed] [Google Scholar]
- 13.Terracciano A, Costa PT, Jr, McCrae RR. Personality plasticity after age 30. Personality and Social Psychology Bulletin. 2006;32:999–1009. doi: 10.1177/0146167206288599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Terracciano A, McCrae RR, Costa PT., Jr Intra-individual change in personality stability and age. Journal of Research in Personality. doi: 10.1016/j.jrp.2009.1009.1006. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gottesman, Gould TD. The endophenotype concept in psychiatry: Etymology and strategic intentions. American Journal of Psychiatry. 2003;160:636–645. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- 16.van den Oord EJCG, Kuo P-H, Hartmann AM, Webb BT, Moller H-J, Hettema JM, et al. Genomewide Association Analysis Followed by a Replication Study Implicates a Novel Candidate Gene for Neuroticism. Arch Gen Psychiatry. 2008;65:1062–1071. doi: 10.1001/archpsyc.65.9.1062. [DOI] [PubMed] [Google Scholar]
- 17.Terracciano A, Sanna S, Uda M, Deiana B, Usala G, Busonero F, et al. Genome-wide association scan for five major dimensions of personality. Mol Psychiatry. 2008 doi: 10.1038/mp.2008.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shifman S, Bhomra A, Smiley S, Wray NR, James MR, Martin NG, et al. A whole genome association study of neuroticism using DNA pooling. Mol Psychiatry. 2008 doi: 10.1038/sj.mp.4002048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Costa PT, Jr, McCrae RR. Revised NEO Personality Inventory (NEO-PI-R) and NEO Five-Factor Inventory (NEO-FFI) professional manual. Psychological Assessment Resources; Odessa, FL: 1992. [Google Scholar]
- 20.Paunonen SV, Haddock G, Forsterling F, Keinonen M. Broad versus narrow personality measures and the prediction of behaviour across cultures. European Journal of Personality. 2003;17:413–433. [Google Scholar]
- 21.Terracciano A, Lockenhoff CE, Crum RM, Bienvenu OJ, Costa PT., Jr Five-Factor Model personality profiles of drug users. BMC Psychiatry. 2008;8:22. doi: 10.1186/1471-244X-8-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Costa PT, Jr, McCrae RR. Depression as an enduring disposition. In: Schneider LS, Reynolds CF III, Lebowitz BD, Friedhoff AJ, editors. Diagnosis and treatment of depression in late life: Results of the NIH consensus development conference. American Psychiatric Press; Washington, DC: 1993. pp. 173–187. [Google Scholar]
- 23.Costa PT, Jr, Bagby RM, Herbst JH, McCrae RR. Personality self-reports are concurrently reliable and valid during acute depressive episodes. J Affect Disord. 2005;89:45–55. doi: 10.1016/j.jad.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 24.Bagby RM, Bindseil K, Schuller DR, Rector NA, Young LT, Cooke RG, et al. Relationship between the Five-Factor Model of personality and unipolar, bipolar and schizophrenic patients. Psychiatry Research. 1997;70:83–94. doi: 10.1016/s0165-1781(97)03096-5. [DOI] [PubMed] [Google Scholar]
- 25.Sanna S, Jackson AU, Nagaraja R, Willer CJ, Chen WM, Bonnycastle LL, et al. Common variants in the GDF5-UQCC region are associated with variation in human height. Nat Genet. 2008;40:198–203. doi: 10.1038/ng.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferreira MA, O'Donovan MC, Meng YA, Jones IR, Ruderfer DM, Jones L, et al. Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat Genet. 2008;40:1056–1058. doi: 10.1038/ng.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muglia P, Tozzi F, Galwey NW, Francks C, Upmanyu R, Kong XQ, et al. Genome-wide association study of recurrent major depressive disorder in two European case-control cohorts. Mol Psychiatry. 2008 doi: 10.1038/mp.2008.131. [DOI] [PubMed] [Google Scholar]
- 29.Shyn SI, Shi J, Kraft JB, Potash JB, Knowles JA, Weissman MM, et al. Novel loci for major depression identified by genome-wide association study of Sequenced Treatment Alternatives to Relieve Depression and meta-analysis of three studies. Mol Psychiatry. 2009 doi: 10.1038/mp.2009.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Purcell SM, Wray NR, Stone JL, Visscher PM, O'Donovan MC, Sullivan PF, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stefansson H, Ophoff RA, Steinberg S, Andreassen OA, Cichon S, Rujescu D, et al. Common variants conferring risk of schizophrenia. Nature. 2009;460:744–747. doi: 10.1038/nature08186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Green EK, Grozeva D, Jones I, Jones L, Kirov G, Caesar S, et al. The bipolar disorder risk allele at CACNA1C also confers risk of recurrent major depression and of schizophrenia. Mol Psychiatry. 2009 doi: 10.1038/mp.2009.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Costa PT, Jr, Terracciano A, Uda M, Vacca L, Mameli C, Pilia G, et al. Personality traits in Sardinia: testing founder population effects on trait means and variances. Behav Genet. 2007;37:376–387. doi: 10.1007/s10519-006-9103-6. [DOI] [PubMed] [Google Scholar]
- 34.Terracciano A, McCrae RR, Brant LJ, Costa PT., Jr Hierarchical linear modeling analyses of NEO-PI-R scales in the Baltimore Longitudinal Study of Aging. Psychology and Aging. 2005;20:493–506. doi: 10.1037/0882-7974.20.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Terracciano A. The Italian version of the NEO PI-R: conceptual and empirical support for the use of targeted rotation. Personality and Individual Differences. 2003;35:1859–1872. doi: 10.1016/S0191-8869(03)00035-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCrae RR, Terracciano A 78 Members of the Personality Profiles of Cultures Project. Universal features of personality traits from the observer's perspective: Data from 50 cultures. Journal of Personality and Social Psychology. 2005;88:547–561. doi: 10.1037/0022-3514.88.3.547. [DOI] [PubMed] [Google Scholar]
- 37.Burdick JT, Chen WM, Abecasis GR, Cheung VG. In silico method for inferring genotypes in pedigrees. Nat Genet. 2006;38:1002–1004. doi: 10.1038/ng1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen WM, Abecasis GR. Family-based association tests for genomewide association scans. Am J Hum Genet. 2007;81:913–926. doi: 10.1086/521580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Y, Willer C, Sanna S, Abecasis G. Genotype imputation. Annu Rev Genomics Hum Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scuteri A, Sanna S, Chen WM, Uda M, Albai G, Strait J, et al. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 2007;3:e115. doi: 10.1371/journal.pgen.0030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, Clarke R, et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008;40:161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu JZ, Tozzi F, Waterworth DM, Pillai SG, Muglia P, Middleton L, et al. Meta-analysis and imputation refines the association of 15q25 with smoking quantity. Nat Genet. 2010;42:436–440. doi: 10.1038/ng.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Costa PT, Jr, Terracciano A, McCrae RR. Gender differences in personality traits across cultures: Robust and surprising findings. Journal of Personality and Social Psychology. 2001;81:322–331. doi: 10.1037/0022-3514.81.2.322. [DOI] [PubMed] [Google Scholar]
- 44.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 45.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 46.Need AC, Keefe RS, Ge D, Grossman I, Dickson S, McEvoy JP, et al. Pharmacogenetics of antipsychotic response in the CATIE trial: a candidate gene analysis. Eur J Hum Genet. 2009;17:946–957. doi: 10.1038/ejhg.2008.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen AC, Tang Y, Rangaswamy M, Wang JC, Almasy L, Foroud T, et al. Association of single nucleotide polymorphisms in a glutamate receptor gene (GRM8) with theta power of event-related oscillations and alcohol dependence. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:359–368. doi: 10.1002/ajmg.b.30818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Togashi H, Abe K, Mizoguchi A, Takaoka K, Chisaka O, Takeichi M. Cadherin regulates dendritic spine morphogenesis. Neuron. 2002;35:77–89. doi: 10.1016/s0896-6273(02)00748-1. [DOI] [PubMed] [Google Scholar]
- 49.Bozdagi O, Valcin M, Poskanzer K, Tanaka H, Benson DL. Temporally distinct demands for classic cadherins in synapse formation and maturation. Mol Cell Neurosci. 2004;27:509–521. doi: 10.1016/j.mcn.2004.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hirano S, Suzuki ST, Redies C. The cadherin superfamily in neural development: diversity, function and interaction with other molecules. Front Biosci. 2003;8:d306–355. doi: 10.2741/972. [DOI] [PubMed] [Google Scholar]
- 51.Takeuchi T, Misaki A, Liang SB, Tachibana A, Hayashi N, Sonobe H, et al. Expression of T-cadherin (CDH13, H-Cadherin) in human brain and its characteristics as a negative growth regulator of epidermal growth factor in neuroblastoma cells. J Neurochem. 2000;74:1489–1497. doi: 10.1046/j.1471-4159.2000.0741489.x. [DOI] [PubMed] [Google Scholar]
- 52.Shibata T, Shimoyama Y, Gotoh M, Hirohashi S. Identification of human cadherin-14, a novel neurally specific type II cadherin, by protein interaction cloning. J Biol Chem. 1997;272:5236–5240. doi: 10.1074/jbc.272.8.5236. [DOI] [PubMed] [Google Scholar]
- 53.Uhl GR, Drgon T, Liu QR, Johnson C, Walther D, Komiyama T, et al. Genome-wide association for methamphetamine dependence: convergent results from 2 samples. Arch Gen Psychiatry. 2008;65:345–355. doi: 10.1001/archpsyc.65.3.345. [DOI] [PubMed] [Google Scholar]
- 54.Treutlein J, Cichon S, Ridinger M, Wodarz N, Soyka M, Zill P, et al. Genome-wide association study of alcohol dependence. Arch Gen Psychiatry. 2009;66:773–784. doi: 10.1001/archgenpsychiatry.2009.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lasky-Su J, Neale BM, Franke B, Anney RJ, Zhou K, Maller JB, et al. Genome-wide association scan of quantitative traits for attention deficit hyperactivity disorder identifies novel associations and confirms candidate gene associations. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:1345–1354. doi: 10.1002/ajmg.b.30867. [DOI] [PubMed] [Google Scholar]
- 56.Vink JM, Smit AB, de Geus EJ, Sullivan P, Willemsen G, Hottenga JJ, et al. Genome-wide association study of smoking initiation and current smoking. Am J Hum Genet. 2009;84:367–379. doi: 10.1016/j.ajhg.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ino H. Immunohistochemical characterization of the orphan nuclear receptor ROR alpha in the mouse nervous system. J Histochem Cytochem. 2004;52:311–323. doi: 10.1177/002215540405200302. [DOI] [PubMed] [Google Scholar]
- 58.Hamilton BA, Frankel WN, Kerrebrock AW, Hawkins TL, FitzHugh W, Kusumi K, et al. Disruption of the nuclear hormone receptor RORalpha in staggerer mice. Nature. 1996;379:736–739. doi: 10.1038/379736a0. [DOI] [PubMed] [Google Scholar]
- 59.Yang XO, Pappu BP, Nurieva R, Akimzhanov A, Kang HS, Chung Y, et al. T helper 17 lineage differentiation is programmed by orphan nuclear receptors ROR alpha and ROR gamma. Immunity. 2008;28:29–39. doi: 10.1016/j.immuni.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ko CH, Takahashi JS. Molecular components of the mammalian circadian clock. Hum Mol Genet 15 Spec No. 2006;2:R271–277. doi: 10.1093/hmg/ddl207. [DOI] [PubMed] [Google Scholar]
- 61.Sato TK, Panda S, Miraglia LJ, Reyes TM, Rudic RD, McNamara P, et al. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron. 2004;43:527–537. doi: 10.1016/j.neuron.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 62.Le-Niculescu H, Patel SD, Bhat M, Kuczenski R, Faraone SV, Tsuang MT, et al. Convergent functional genomics of genome-wide association data for bipolar disorder: comprehensive identification of candidate genes, pathways and mechanisms. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:155–181. doi: 10.1002/ajmg.b.30887. [DOI] [PubMed] [Google Scholar]
- 63.Lavebratt C, Sjoholm LK, Partonen T, Schalling M, Forsell Y. PER2 variantion is associated with depression vulnerability. Am J Med Genet B Neuropsychiatr Genet. 2009 doi: 10.1002/ajmg.b.31021. [DOI] [PubMed] [Google Scholar]
- 64.Soria V, Martinez-Amoros E, Escaramis G, Valero J, Perez-Egea R, Garcia C, et al. Differential Association of Circadian Genes with Mood Disorders: CRY1 and NPAS2 are Associated with Unipolar Major Depression and CLOCK and VIP with Bipolar Disorder. Neuropsychopharmacology. doi: 10.1038/npp.2009.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McClung CA. Circadian genes, rhythms and the biology of mood disorders. Pharmacol Ther. 2007;114:222–232. doi: 10.1016/j.pharmthera.2007.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hynd MR, Scott HL, Dodd PR. Glutamate-mediated excitotoxicity and neurodegeneration in Alzheimer's disease. Neurochem Int. 2004;45:583–595. doi: 10.1016/j.neuint.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 67.Sanacora G, Zarate CA, Krystal JH, Manji HK. Targeting the glutamatergic system to develop novel, improved therapeutics for mood disorders. Nat Rev Drug Discov. 2008;7:426–437. doi: 10.1038/nrd2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schoepp DD. Unveiling the functions of presynaptic metabotropic glutamate receptors in the central nervous system. J Pharmacol Exp Ther. 2001;299:12–20. [PubMed] [Google Scholar]
- 69.Lavreysen H, Dautzenberg FM. Therapeutic potential of group III metabotropic glutamate receptors. Curr Med Chem. 2008;15:671–684. doi: 10.2174/092986708783885246. [DOI] [PubMed] [Google Scholar]
- 70.Cryan JF, Kelly PH, Neijt HC, Sansig G, Flor PJ, van Der Putten H. Antidepressant and anxiolytic-like effects in mice lacking the group III metabotropic glutamate receptor mGluR7. Eur J Neurosci. 2003;17:2409–2417. doi: 10.1046/j.1460-9568.2003.02667.x. [DOI] [PubMed] [Google Scholar]
- 71.Palucha A, Tatarczynska E, Branski P, Szewczyk B, Wieronska JM, Klak K, et al. Group III mGlu receptor agonists produce anxiolytic- and antidepressant-like effects after central administration in rats. Neuropharmacology. 2004;46:151–159. doi: 10.1016/j.neuropharm.2003.09.006. [DOI] [PubMed] [Google Scholar]
- 72.Stachowicz K, Klodzinska A, Palucha-Poniewiera A, Schann S, Neuville P, Pilc A. The group III mGlu receptor agonist ACPT-I exerts anxiolytic-like but not antidepressant-like effects, mediated by the serotonergic and GABA-ergic systems. Neuropharmacology. 2009;57:227–234. doi: 10.1016/j.neuropharm.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 73.Backstrom P, Hyytia P. Suppression of alcohol self-administration and cue-induced reinstatement of alcohol seeking by the mGlu2/3 receptor agonist LY379268 and the mGlu8 receptor agonist (S)-3,4-DCPG. Eur J Pharmacol. 2005;528:110–118. doi: 10.1016/j.ejphar.2005.10.051. [DOI] [PubMed] [Google Scholar]
- 74.Takaki H, Kikuta R, Shibata H, Ninomiya H, Tashiro N, Fukumaki Y. Positive associations of polymorphisms in the metabotropic glutamate receptor type 8 gene (GRM8) with schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2004;128B:6–14. doi: 10.1002/ajmg.b.20108. [DOI] [PubMed] [Google Scholar]
- 75.Mill J, Petronis A. Molecular studies of major depressive disorder: the epigenetic perspective. Mol Psychiatry. 2007;12:799–814. doi: 10.1038/sj.mp.4001992. [DOI] [PubMed] [Google Scholar]
- 76.Tsankova N, Renthal W, Kumar A, Nestler EJ. Epigenetic regulation in psychiatric disorders. Nat Rev Neurosci. 2007;8:355–367. doi: 10.1038/nrn2132. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.