Abstract
The basal-like subtype of breast cancer is characterized by a “triple negative” (TN) phenotype (ER-, PR-, HER2/neu-). TN breast cancers share similar gene expression profiles and DNA repair deficiencies with BRCA1-associated breast cancers. BRCA1 mutant cells exhibit sensitivity to gemcitabine, cisplatin and PARP inhibition, therefore, we hypothesized that TN cancer cells may also exhibit sensitivity to these drugs. In this study, we report that TN breast cancer cells are more sensitive to these drugs compared to non-TN breast cancer cells. Moreover, combination treatments indicated that PARP inhibition by the small molecule inhibitor PJ34 or siRNA knockdown synergized with gemcitabine and cisplatin in TN cells but not in luminal cancer cells. TN cells exhibited reduced repair of UV-induced cyclobutane pyrimidine dimers after PARP inhibition, suggesting that the synergistic effect of PJ34 and gemcitabine or cisplatin reflected inefficient nucleotide excision repair. Mechanistic investigations revealed that in TN cells PJ34 reduced levels of ΔNp63α with a concurrent increase in p73 and its downstream target p21. Thus, the sensitivity to combination treatment appeared to be mediated by sustained DNA damage and inefficient DNA repair triggering p63/p73-mediated apoptosis. Our results suggest a novel therapeutic strategy to treat women with TN breast cancer, an aggressive disease which presently lacks effective treatment options.
Keywords: Basal-like Breast Cancer, Poly (ADP-ribose) Polymerase, Cisplatin, Gemcitabine, triple negative
Introduction
Breast cancer is the most common cause of malignancy and second most common cause of cancer death in women (1). This heterogeneous disease is composed of 5 major biological subtypes which are based on microarray gene classifications and include luminal A, luminal B, normal breast-like, human epidermal growth factor receptor 2 (HER2), and basal-like breast cancers (1, 2). While many of these subtypes can be treated with much success, the basal-like carcinomas are associated with high rates of relapse following chemotherapy (3, 4). Basal-like breast tumors are largely estrogen receptor (ER), progesterone receptor (PR) and HER2/neu-negative (triple-negative) and express genes characteristic of basal epithelial and normal breast myoepithelial cells (5-7). However, the genes responsible for the etiology and aggressive phenotype of basal-like breast cancers remain unknown.
As many cancer chemotherapeutic drugs and radiation therapy cause DNA damage, tumor cells defective in DNA repair pathways are predicted to be sensitive to their effects. Indeed, BRCA1 (and BRCA2) mutant cell lines have been shown to be sensitive to the DNA cross linking agents cisplatin and mitomycin C (8, 9), to the topoisomerase inhibitor etoposide (10) and to oxidative DNA damage (11). Recently, we have shown that BRCA1 deficient cells are sensitive to gemcitabine (2′, 2′-difluoro 2′-deoxycytidine, dFdC) (12) an analogue of cytosine arabinoside that exhibits anti-cancer properties due to potent inhibition of DNA synthesis (13). Gemcitabine is often used either alone or in combination with other drugs like taxanes, vinorelbine, carboplatin or trastuzumab in metastatic breast cancer. However, there are no reports of the efficacy of gemcitabine in the basal-like subtype of breast cancers.
The BRCA1 breast cancer susceptibility gene is known to be involved in a number of DNA repair pathways, including DNA double-strand break repair via homologous recombination (HR) (14), nucleotide excision repair (NER) (15) and base excision repair (BER) of oxidative DNA damage (11) . Recently, BRCA1- and BRCA2-deficient cells have also been shown to be sensitive to inhibitors of poly (ADP-ribose) polymerase (PARP) (16, 17), an enzyme involved in BER, that when inhibited leads to DNA strand breaks and cell death. In this scenario, BRCA mutant tumor cells with primary defects in DNA repair are particularly sensitive to small-molecule inhibitors of BER, such as PARP inhibitors.
Recent studies indicate that sporadic basal-like or TN tumors bear a striking resemblance to breast tumors that arise in hereditary BRCA1 mutation carriers. These similarities strongly suggest that sporadic basal-like tumors might bear defects in BRCA1-mediated DNA repair pathways (11), and exhibit similar sensitivities to DNA damaging agents and PARP inhibitors.
Therefore, using a panel of breast cancer cell lines, we examined the cytotoxic effects of gemcitabine, cisplatin and a PARP inhibitor alone and in combination. We demonstrate that like BRCA1 mutant cells, basal-like triple negative breast cancer (TNBC) cells are sensitive to the PARP inhibitor PJ34, gemcitabine and cisplatin. We further show that PJ34 acts synergistically with both gemcitabine and cisplatin in TNBC cells, but not luminal breast cancer cell lines. Moreover, we demonstrate that PJ34 and gemcitabine disrupt NER, suggesting a novel mechanism of sensitivity to these drugs in TNBC cells.
Material and Methods
Cell Lines and Reagents
All cell lines were used within 6 months of purchase. MDAMB468, hs578t, MCF7, BT549 and BT474 cell lines were obtained from ATCC and maintained in DMEM with 10% FBS. T47D and HCC1806 cells (ATCC) were maintained in RPMI1640 with 10% FBS. ATCC provides molecular authentication in support of their collection through their genomics, immunology and proteomic cores, as described, using DNA barcoding and species identification, quantitative gene expression and transcriptome analyses (18). SUM149PT cells were obtained from Asterand Plc. (Detroit, MI) where they are authenticated through gene expression data through the use of Affymetrix GeneChips, and biomolecular markers, such as estrogen receptor and Her2 status. They were maintained in Ham's F-12 media with 10% FBS. HCC1806 cells were transfected with the shRNAmir retroviral vector in pSM2 against nonspecific silencing control, PARP1 or PARP2 (Thermo Fisher Scientific, Waltham, MA). Stable clones were maintained with 1ug/ml puromycin and confirmed by Western blotting. PARP inhibitor PJ34 was purchased from EMD biosciences (Gibbstown, NJ). Methylene blue, MTT (3-(4, 5-Dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide, a tetrazole) and Cisplatin was purchased from Sigma Chemicals (St. Louis, MO). Gemcitabine was gift from Eli Lilly Pharmaceuticals (Indianapolis, IN).
Cell Viability and Colony Formation Assay
Cell viability was measured by MTT assay. For MTT assay, cells were seeded in 96-well plates, and treated with PJ34, gemcitabine, and cisplatin as indicated. MTT reagent was added to the cells, which was reduced to purple formazan crystals by the mitochondria of living cells. The reduced crystals were solubilized with dimethyl sulfoxide, and the absorbance was measured at 570 nm by spectrophotometry. All experiments were done in triplicate and also repeated three independent times and data was plotted as mean ±SD. Data from representative experiment is shown in the figures. For colony formation assay HCC1806 and MCF7 were plated at equal density and treated with either 10 μM PJ34, 0.6 nM gemcitabine or 4 μM cisplatin for 72 h. After treatment, cells were counted and either 500 or 1000 cells were plated. After 15 days, the cells were stained with methylene blue and individual colonies counted.
Drug Combination Studies
For combination studies, BT474, MCF7, HCC1806 and MDAMB468 cells were seeded in triplicate in 96-well plates and treated with PJ34, gemcitabine or cisplatin alone or in combinations of PJ34 and gemcitabine or PJ34 and cisplatin at desired doses. MTT assays were performed after 72 h of treatment. The data was plotted using Calcusyn – Biosoft software (Cambridge, UK) (19, 20). Combination index (CI) and isobolograms were plotted using the CI equation of Chou-Talalay (21). A non-constant ratio drug combination design was used and software constructed normalized isobolograms. The experimental protocol and the isobologram plotted along with the equation for calculating CI is shown in Supplemental Figure 1. CI < 1 was synergistic, CI = 1 was additive and CI>1 was antagonistic. Study was repeated 3 independent times and representative data is shown here.
γH2AX and Rad51 staining
BT474, MDAMB468, HCC1806, HCC1806shP1 and HCC1806shP2 cells were plated overnight in chambered slides (1500 cells/ chamber). Cells were treated with 10 μM of PJ34 or 5 nM gemcitabine for either 1-4 h for Rad51 staining or 24 h for γH2AX. Controls included primary alone, isotype control and secondary alone. After treatment cells were fixed in 4% paraformaldehyde and stained overnight in primary antibody for Rad51 (H-92) (1:200 dilution) (Santa Cruz Biotechnology, Santa Cruz, CA) or γH2AX -Ser139 (1:500 dilution) (Cell Signaling, Danvers, MA). Cells were washed with TBS/ BSA and incubated for 1 h at room temperature with either, Alexa 488 or Alexa 594 (Invitrogen, Carlsbad, CA) secondary antibody for Rad51 or γH2AX respectively. Cells were fixed in Prolong gold antifade with DAPI (Invitrogen) and cured at room temperature for 24 h before visualizing. For quantification of Rad51 foci and γH2AX foci, at least 100 cells from each group were visually scored. Cells showing more than 3 foci were counted as positive for γH2AX or Rad51. The ratio of Rad51 foci in controls to treated groups was represented as fold change. For γH2AX the fold change was the ratio of γH2AX positive cells in HCC1806 control cells versus shPARP1 and shPARP2. Images from random fields were taken using a Nikon Eclipse E 800 microscope with attached camera and using Spot Software from Diagnostic Instruments (Diagnostic Instruments, Sterling Heights, MI). Images were taken with a 40X lens (40X/1.0 DIC H Plan APO oil immersion lens).
Apoptosis Detection
Apoptosis was detected by staining HCC1806 cells for Annexin V and cleaved caspase 3. Cells were plated overnight in chambered slides followed by treatment with either 10 μM PJ34, 0.6 nM gemcitabine or 4 μM cisplatin for 24 h. For Annexin V-FITC, standard manufacturers protocol was followed (Sigma). For caspase 3, cells were stained with 1:250 dilution of cleaved caspase 3 antibody (Asp175, Cell Signaling) and visualized as mentioned above.
Nucleotide excision repair assay
MDAMB468, HCC1806, HCC1806 control, HCC1806shP1 and HCC1806shP2 cells were grown overnight in 6 well plates (in triplicate). Cells were rinsed with PBS followed by exposure to 20 J/m2 of UVC irradiation. MDAMB468 and HCC1806 cells were treated with 10 μM PJ34 or 5 nM gemcitabine, while media without drugs was added to PARP1 and PARP2 knockdown cells. Genomic DNA was extracted (QIAamp DNA mini kit, Qiagen, Valencia, CA) at 0 – 24 h. Repair of cyclobutane pyrimidine dimers (CPDs) and 6-4 photoproducts (6-4PPs) was measured using an enzyme-linked immunosorbent assay (ELISA). Briefly, genomic DNA was distributed in triplicate onto microtiter plates precoated with 0.003% protamine sulfate. DNA lesions were detected with either 1:5000 TDM-2 (for CPDs) or 1:5000 64M-2 (for 6-4PPs) a gift from Dr. Toshio Mori (22). The signals were amplified and subsequently developed with 3,5,3',5'-tetramethylbenzidine (TMB) (Sigma Chemicals). Absorbance was measured at 450 nm. Each experiment was repeated 3 independent times, and representative data is shown.
Western Blot Analyses
Total cellular protein was isolated by lysing cells in modified RIPA buffer. Proteins (25 - 50 μg/lane) were separated by electrophoresis (SDS-10% PAGE) and electro-blotted onto polyvinylidene difluoride membranes (GE, Amersham place, UK). The membranes were probed with antibodies against PARP, p63, p73, p21 or actin (Santa Cruz Biotechnology), MCM (BD Biosciences, Franklin Lakes, NJ). Protein levels from the blots were evaluated using gel analysis software, ImageJ (National Institute of Health, Maryland) and ratio of protein level in controls to treated groups was represented as fold change.
Results
Sensitivity of basal-like breast cancer cells to PJ34, gemcitabine and cisplatin
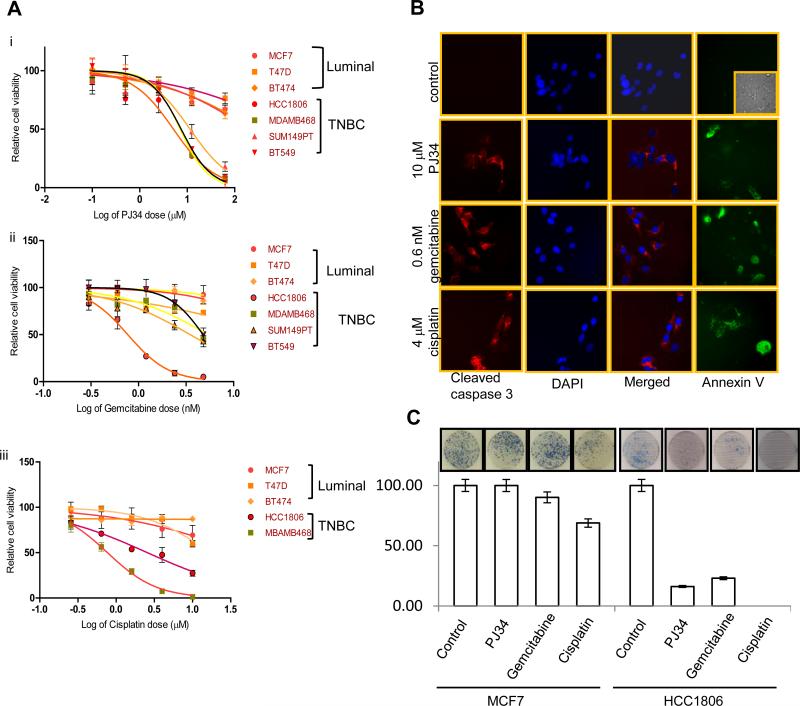
To test our hypothesis that PARP inhibitors that target BRCA1-pathway dysfunction might also be efficacious in treatment of TNBCs, a panel of sporadic TNBC cells (BT549, HCC1806 and MDAMB468) along with a BRCA1 mutant TN cell line SUM149PT, and luminal breast cancer cell lines (BT474, MCF7 and T47D) were tested for their sensitivity towards PJ34 ( 0 – 62.5 μM), gemcitabine (0 – 4.8 nM) and cisplatin (0 – 10 μM). Following 72 h treatment, cell viability was measured by MTT assay. All TNBC cell lines were significantly more sensitive to PJ34, gemcitabine and cisplatin treatment than the luminal breast cancer cell lines (Figure 1 A, Supplemental Table 1). We confirmed the sensitivity of TNBC cells to PJ34, gemcitabine and cisplatin by colony formation assay and as shown in Figure 1C, HCC1806 cells were highly sensitive to all the three drugs compared to the luminal MCF7 cells. Furthermore, all the drugs induced apoptosis in TNBC cells as evident by caspase 3 cleavage and Annexin V staining (Figure 1B). Therefore, we found that similar to BRCA1 deficient cells, TNBC cells were selectively sensitive to PARP inhibition, gemcitabine and cisplatin compared to other breast subtypes. Moreover, the resistance of luminal cell lines does not depend on HER2 status as BT474 is HER2/neu positive while MCF7 is HER2/neu negative.
Figure 1. Cell viability of breast cancer cells after PJ34, gemcitabine or cisplatin treatment.
Cells were treated with (Ai) 0.01- 62.5 μM of PJ34 or (Aii) 0.15 – 4.8 nM gemcitabine or (Aiii) 0.256 – 10 μM cisplatin for 72 h and cell viability was determined by MTT assay. (B) Apoptosis in HCC1806 cells as shown by cleaved caspase 3 and Annexin V staining (inset is brightfield image) (C) HCC1806 and MCF7 cells were stained with methylene blue for Colony formation assay after 15 days. Number of colonies are shown and the inset shows representative figure. Experiments were done 3 independent times and in triplicate and average cell, viability in log scale (± SD) is shown.
Synergism between PJ34 and gemcitabine or cisplatin
Our results show that TNBC cells are selectively sensitive to PJ34, gemcitabine and cisplatin when used individually. However, to determine whether inhibition of PARP by PJ34 acts in a synergistic, additive, or antagonistic fashion with gemcitabine or cisplatin, we treated BT474, MCF7, HCC1806 and MDAMB468 cell lines with each agent alone or in combination for 72 h. Cells were either treated with 1μM of PJ34 and 0.0016 – 1μM gemcitabine or 1.875 - 15 μM PJ34 and 10 μM cisplatin. CalcuSyn software was used to calculate the CI, and plot normalized isobologram (supplemental Figure 2 and 3) (19, 21). CI < 1, CI = 1, and CI > 1 quantitatively indicate synergism, additivity, and antagonism, respectively. We also calculated the linear coefficient “r value” to estimate the accuracy of measurement and all our experiments had an r value > 0.90 for the median-effect plot.
As shown in Table 1 Ai and ii, TNBC cell lines HCC1806 and MDAMB468 cells exhibited synergism for all the different combinations of PJ34 and gemcitabine doses, whereas the luminal BT474 and MCF7 cell lines (Table 1A iii, iv) exhibited antagonistic effects at most dose combinations. Similarly, PJ34 and cisplatin had additive to synergistic effects in both HCC1806 and MDAMB468 cells (Table 1 B i and ii) where as antagonism was observed in the luminal BT474 and MCF7 cell lines (Table 1 B iii and iv).
Table 1.
Synergistic effect of PJ34 with either gemcitabine or cisplatin in breast cancer cell lines. HCC1806, MDAMB468, MCF7 and BT474 cells were treated with PJ34, gemcitabine or cisplatin or a combination of the drugs at desired concentrations for 72 h and MTT assay was performed. CI was calculated by Calcusyn – Biosoft software. Table (A) show the CI values of PJ34 in combination with gemcitabine and table (B) shows CI values of PJ34 in combination with cisplatin.
| A | B | ||||||
|---|---|---|---|---|---|---|---|
| i | i | ||||||
| HCC1806 | HCC1806 | ||||||
| Gemcitabine (μM) | PJ34 (μM) | CI | Effect | PJ34 (μM) | Cisplatin (μM) | CI | Effect |
| 0.0016 | 1 | 0.88 | synergism | 1.875 | 10 | 1.09 | additive |
| 0.008 | 1 | 0.95 | synergism | 3.75 | 10 | 0.85 | synergism |
| 0.04 | 1 | 0.97 | synergism | 7.5 | 10 | 0.83 | synergism |
| 0.2 | 1 | 0.98 | synergism | 15 | 10 | 0.646 | synergism |
| 1 | 1 | 0.98 | synergism | ||||
| ii | ii | ||||||
| MDAMB468 | MDAMB468 | ||||||
| Gemcitabine (μM) | PJ34 (μM) | CI | Effect | PJ34 (μM) | Cisplatin (μM) | CI | Effect |
| 0.0016 | 1 | 0.74 | synergism | 1.875 | 10 | 0.83 | synergism |
| 0.008 | 1 | 0.48 | synergism | 3.75 | 10 | 0.76 | synergism |
| 0.04 | 1 | 0.25 | synergism | 7.5 | 10 | 0.82 | synergism |
| 0.2 | 1 | 0.72 | synergism | 15 | 10 | 0.91 | synergism |
| 1 | 1 | 0.90 | synergism | ||||
| iii | iii | ||||||
| BT474 | BT474 | ||||||
| Gemcitabine (μM) | PJ34 (μM) | CI | Effect | PJ34 (μM) | Cisplatin (μM) | CI | Effect |
| 0.0016 | 1 | 1.34 | antagonism | 1.875 | 10 | 74 | antagonism |
| 0.008 | 1 | 3.51 | antagonism | 3.75 | 10 | 1.31 | antagonism |
| 0.04 | 1 | 1.33 | antagonism | 7.5 | 10 | 1.53 | antagonism |
| 0.2 | 1 | 18.30 | antagonism | 15 | 10 | 1.54 | antagonism |
| 1 | 1 | 50.22 | antagonism | ||||
| iv | iv | ||||||
| MCF7 | MCF7 | ||||||
| Gemcitabine (μM) | PJ34 (μM) | CI | Effect | PJ34 (μM) | Cisplatin (μM) | CI | Effect |
| 0.0016 | 1 | 1.29 | antagonism | 1.875 | 10 | 1.71 | antagonism |
| 0.008 | 1 | 2.93 | antagonism | 3.75 | 10 | 1.28 | antagonism |
| 0.04 | 1 | 7.79 | antagonism | 7.5 | 10 | 1.20 | antagonism |
| 0.2 | 1 | 0.74 | synergism | 15 | 10 | 0.89 | synergism |
| 1 | 1 | 1.19 | antagonism | ||||
We also treated cells with varied concentrations of PJ34 and kept the concentration of gemcitabine constant, and conversely kept the concentration of PJ34 constant while changing the concentration of cisplatin. Again, TNBC cell lines exhibited additive to synergistic effects, while antagonism was observed in BT474 and MCF7 cell lines (data not shown).
PARP knockdown sensitizes cells to gemcitabine and cisplatin
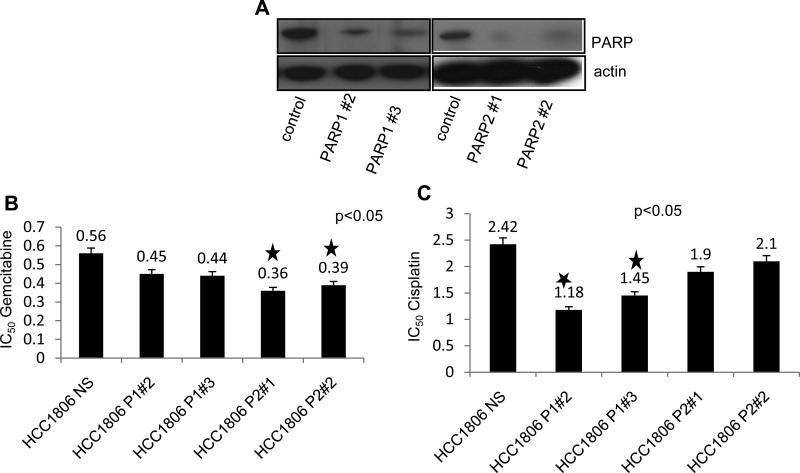
Many PARP inhibitors are known to inhibit PARP1 and PARP2, both of which are involved in DNA repair pathways. Therefore, to investigate the role of PARP inhibition in sensitizing TNBC cells to gemcitabine and cisplatin, we generated stable clones of PARP1 and PARP2 knockdown cells in TN HCC1806 cell line. Decreased protein expression of PARP1 and PARP2 protein was confirmed by immunoblotting (Figure 2 A). We then treated HCC1806 non silencing control , HCC1806 shPARP1 and shPARP2 clones (2 each) with either 0.15 – 4.8 nM of gemcitabine or 0.25 – 8 μM of cisplatin for 72 h, measured cell viability by MTT assay and calculated IC50 values by Prism software. As shown in Figure 2 B and 2 C, both PARP1 and PARP2 knockdown sensitized HCC1806 cells to gemcitabine and cisplatin. PARP2 knockdown significantly (p<0.05) sensitized cells to gemcitabine compared to HCC1806 (Figure 2 B). Conversely PARP1 knockdown significantly (p<0.05) sensitized cells to cisplatin (Figure 2 C).
Figure 2. Cell viability of PARP1 and PARP2 knockdown cells after gemcitabine and cisplatin treatment.
(A) Western blot analysis of PARP in HCC1806 control, shPARP1, and shPARP2 knockdown cells. Cells were treated with increasing concentrations of either gemcitabine or cisplatin and MTT assay was performed after 72 h. IC50 values were determined by Prism and are represented as graphs for cells treated with either (B) gemcitabine or (C) cisplatin. Average cell viability in triplicate (± SD) is shown.
DNA damage in basal-like breast cancer cell lines after PJ34 and gemcitabine treatment
Since PARP and gemcitabine play a major role in DNA repair and inhibition of DNA synthesis respectively, we investigate the effect of PJ34 and gemcitabine on DNA damage by staining for Rad51 foci and γH2AX, which accumulate at sites of broken DNA.
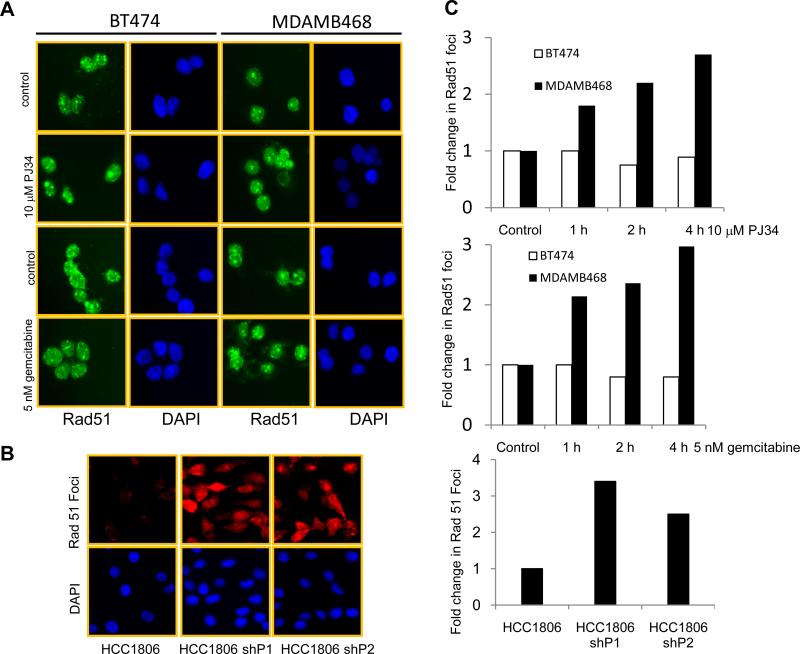
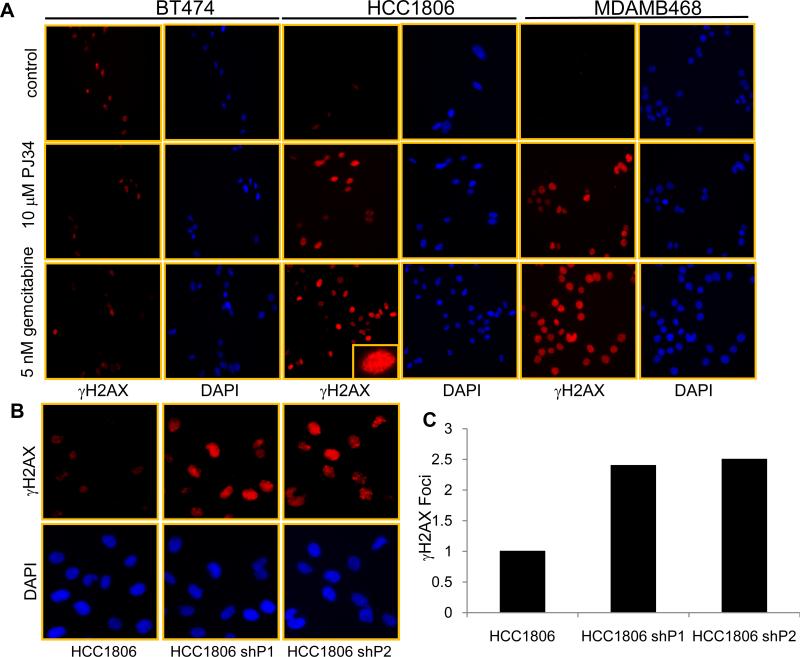
To study Rad51 foci formation, luminal BT474 and TN MDAMB468 cell lines were treated with either 10 μM of PJ34 or 5 nM of gemcitabine for 1- 4 h. As shown in Figure 3 A, 4 hr of PJ34 or gemcitabine treatment increased the Rad51 foci in MDAMB468 cells. BT474 cells had a higher basal level of Rad51 foci, but PJ34 and gemcitabine treatment did not further increase the number of foci. We next counted the number of Rad51 foci and observed a 2-fold increase after PJ34 and gemcitabine treatment (Figure 3 C upper and middle panels). Similar to PJ34 treatment, knockdown of PARP1 and PARP2 also increased the number of Rad51 foci in compared to HCC1806 control cells as seen and quantified in Figure 3B and C (lower panel). We next treated BT474, MDAMB468 and HCC1806 cells with 10 μM PJ34 or 5 nM gemcitabine for 24 h and stained cells for γH2AX. As shown in Figure 4 A, following treatment there was no increase in the number of γH2AX -positive cells in BT474 cell line. On the other hand, HCC1806 and MDAMB468 cell lines showed a significant increase in the number of γH2AX -positive cells. PARP1 and 2 knockdown cells also exhibited 2 fold increases in γH2AX positive cells compared to HCC1806 control cells as shown and quantified in Figure 4 B and C. These observations suggest that TNBC cells accumulate DNA damage after inhibition of PARP activity or due to inhibition of DNA synthesis by gemcitabine. Moreover, cell cycle analysis (data not shown) illustrates that PJ34 treatment arrested cells in the G2/M and gemcitabine induced an S-phase block in TNBC cells, consistent with cell cycle arrest after DNA damage.
Figure 3. Increase in Rad51 foci after PARP inhibition or gemcitabine treatment in TNBC cells.
Representative image of Rad51 foci in (A) BT474 and MDAMB468 cells treated with either 10 μM PJ34 or 5 nM gemcitabine for 4 h. (B) HCC1806 control and PARP1 and 2 knockdown cells. (C) Quantification of Rad51 foci after 1, 2 or 4 h of either PJ34 (top) or gemcitabine treatment (middle), or in PARP1 and PARP2 knockdown cells (bottom). Cells with 3 foci or more were counted as positive.
Figure 4. DNA damage in cells after PARP inhibition or gemcitabine treatment.
(A) A representative image of γH2AX in BT474, MDAMB468 and HCC1806 cells treated with 10 μM PJ34 or 5 nM gemcitabine for 24 h. (Inset) magnified image shows distinct punctate staining. (B) γH2AX positive cells in HCC1806 control and HCC1806 shPARP1 and shPARP2 knockdown cells. (C) Quantification of γH2AX foci in HCC1806 control, PARP1 and PARP2 knockdown cells.
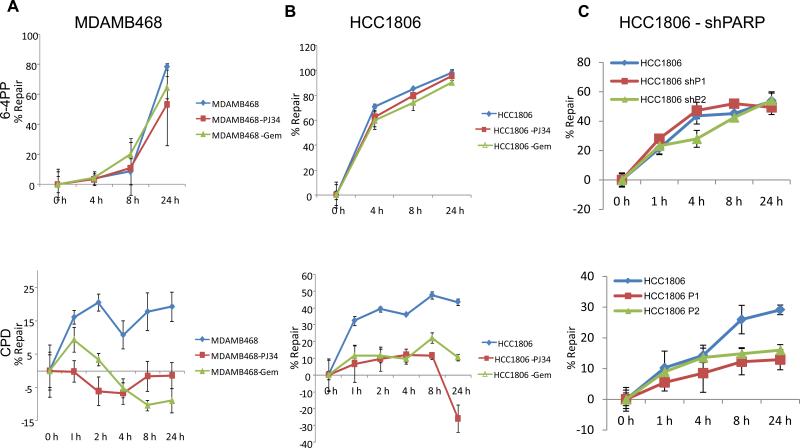
Inefficient repair of UV-induced DNA damage after PJ34 and gemcitabine treatment
Recent studies have shown that PARP is activated by UV-irradiation (23, 24); however, the role of PARP after UV-induced DNA damage is not clear. Previously, our laboratory demonstrated that human and mouse BRCA1 mutant cells are defective in global genomic repair (GGR) compared to BRCA1 wild-type and luminal breast cancer cells (15). Since TNBC cells bear resemblance to BRCA mutant cells, we investigated UV-induced DNA damage repair via the GGR pathway in TNBC cells treated with either PJ34 or gemcitabine and in PARP1 and PARP2 knockdown cells.
As shown in Figure 5 A and B both MDAMB468 and HCC1806 cells treated with either PJ34 or gemcitabine were efficient in repairing 6-4PPs (top panel). However, treatment of cells with PJ34 or gemcitabine completely inhibited repair of CPDs (bottom panel). Similarly, knockdown of both PARP1 and PARP2 rendered the cells inefficient in repairing CPDs with no change in the ability to repair 6-4PPs (Figure 5 C). Therefore, the synergistic effect observed in TNBC cells between PJ34 and cisplatin or gemcitabine may be in part due to inhibition of the GGR pathway along with defects in other DNA repair pathways.
Figure 5. Decreased NER in TNBC cells after PARP inhibition or gemcitabine treatment.
Cells were irradiated with 20 J/m2 UVC (A-C) and then treated with either 10 μM PJ34 or 5 nM gemcitabine for 1 - 24 h (A and B). Upper panel shows repair of 6-4PPs in MDAMB468, HCC1806 and HCC1806 shPARP1 and shPARP2 respectively. While the lower panel demonstrates the repair of CPDs.
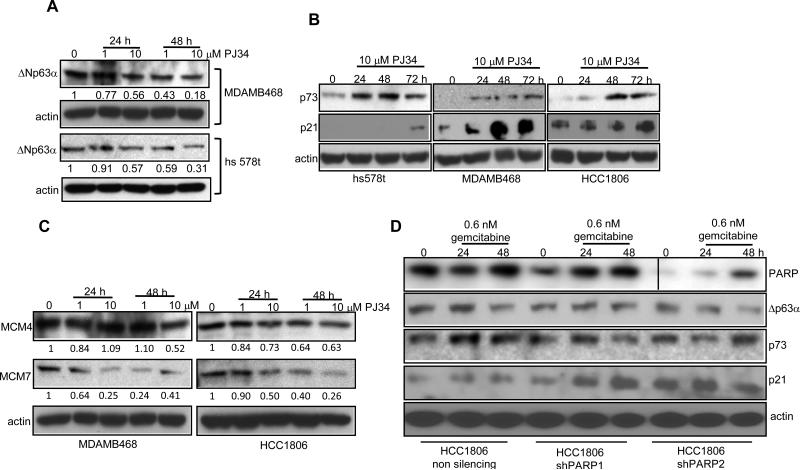
Role of p63 and p73 in sensitizing basal-like breast cancer cells to PJ34 and gemcitabine
Studies have suggested that 0 – 30% of invasive ductal breast carcinomas express ΔNp63α protein (25-27). p73 can induce apoptosis by p53-independent mechanisms, making it particularly important for therapeutics in basal-like breast carcinomas, which are most often null or mutant for p53. We found that p63 is significantly overexpressed in TNBC cell lines by analyzing gene expression data from a study published by Neve et al (28). Since p63 can act as an oncogene (25), the correlation between p63 expression and PJ34 and gemcitabine mediated cell death becomes even more important.
As shown in Figure 6 A, PJ34 treated hs568t, MDAMB486 and HCC1806 TN cells exhibit more than 3-fold decrease in expression of ΔNp63α. ΔNp63α binds to p73 and prevents its proapoptotic activity. We therefore analyzed levels of p73 protein and observed an increase in expression of p73 in PJ34 treated cells with a concurrent increase in the expression of p21 (Figure 6 B). A recent study showed that p53 and p73 via p21 can repress minichromosome maintenance proteins (MCM) (29). Since analysis of the published microarray data (28) revealed that MCM proteins are overexpressed in TNBC cells, we examined the expression of MCM proteins after PJ34 treatment. As shown in Figure 6 C, PJ34 treatment decreased the levels of MCM4 and 7 in TNBC cells. We next treated PARP1 and PARP2 knockdown cells with gemcitabine for 24 or 48 h and as shown in Figure 5 D gemcitabine treatment down regulated the protein level of ΔNp63α with a concurrent increase in p73 and p21. Thus, the results suggest that DNA damage may trigger the p63-p73 pathway to induce cell cycle arrest and apoptosis.
Figure 6. Modulation of p63, p73, p21 and MCMs in TNBC cells.
Western analysis of (A) ΔNp63α in MDAMB468 and hs578t cells treated with PJ34 (B) p73 and p21 in hs578t, HCC1806 and MDAMB468 cells treated with PJ34 (C) MCM4 and MCM7 in MDAMB468 and HCC1806 cells treated with PJ34. (D) Expression of PARP, ΔNp63α, p73 and p21 in HCC1806, HCC1806 shPARP1 and shPARP2 cells treated with 0.6 nM gemcitabine.
Discussion
The discovery of molecular subclasses of breast cancer suggests that treatments may be targeted more selectively with improved outcomes. Currently, a major challenge is to identify such targets and more effective therapeutic regimens for TNBCs that are not responsive to endocrine therapy or trastuzumab. BRCA1 mutation carriers commonly develop basal-like breast tumors with defects in DNA repair and have been shown to have altered sensitivity to certain cytotoxic DNA damaging agents. In the current study, we explored whether agents known to selectively target DNA repair deficient BRCA1 mutant cells would also be effective in TNBC cells.
Previously we have established that BRCA1 mutant cell lines are more sensitive to cisplatin and gemcitabine than matched BRCA1 wild-type cells (12). Mechanisms to explain this observation include defects in DNA repair pathways involved in HR, NER and resolution of the intra- and interstrand DNA crosslinks induced by cisplatin. In our current study, we found that TNBC cell lines are sensitive to cisplatin compared to luminal cells. We also report for the first time that TNBC cell lines exhibit profound sensitivity to gemcitabine, compared to luminal types which were not sensitive to gemcitabine. These novel findings suggest a targeted chemotherapeutic approach to TN and BRCA1 deficient breast cancer.
PARP-1 and -2 are involved in various DNA repair mechanisms. They bind to the DNA damage sites and activate themselves by automodification. Recent studies have suggested that decreasing PARP expression by RNA interference or by chemical inhibitors sensitizes BRCA1 and BRCA2 deficient cells to cell death through synthetic lethality with their DNA repair defects (16, 17). Our finding that TNBC cells share DNA repair defects with BRCA1 mutant cells (11) suggest PARP inhibitors may be effective in treating these tumors, as well. In fact, our results show that TNBC cells are more susceptible to PARP inhibition than the luminal type of breast cancer cells. Moreover, the sensitivity to PARP inhibitors is similar to BRCA1 mutant cells.
Since PARP plays a major role in the response to DNA damage, we also wished to examine whether inhibitor of PARP acts synergistically with DNA damaging cytotoxic agents. Therefore, to explore the effects of drug combinations in breast cancer subtypes, we performed isobologram analyses and found that the combination of PJ34 along with gemcitabine or cisplatin had a synergistic effect in TNBC cells. Remarkably, however, the combination proved antagonistic in repair proficient luminal breast cancer cell lines.
A recent study (30) showed that PARP-1 and -2 are required to reactivate replication at stalled DNA forks. Thus, synergism observed between gemcitabine and PARP inhibition in TNBC cells may be attributed to stalled replication forks caused by incorporation of gemcitabine into replicating DNA and failure to reactivate replication at the stalled fork due to inhibition of PARP activity. These results have clinical significance, and suggest a regimen combining a platinum agent with gemcitabine and a PARP inhibitor may have unique efficacy in TNBC but may not prove effective in other subtypes of breast cancers. Interestingly, knocking down PARP2 further sensitized TN cells to gemcitabine while PARP1 knockdown cells were sensitive to cisplatin. It is therefore possible that PARP1 and PARP2 responded preferentially to DNA damage caused by different DNA damaging agents and this observation is being further investigated.
Our work shows that H2AX is phosphorylated and forms distinct nuclear foci in response to gemcitabine in TNBC cell lines similar to other deoxycytidine nucleoside analogues, like 1-h-D-arabinofuranosylcytosine and troxacitabine (31). Additionally, consistent with previous studies (32, 33) we found that gemcitabine treatment caused the accumulation of Rad51 nuclear foci. It is not clear if these foci result from gemcitabine-induced stalled replication forks and/or gemcitabine-induced accumulation of cells in S phase. However, it is possible that stalled replication forks caused by gemcitabine trigger HR repair of chemotherapy-induced DNA damage.
Similarly, inhibition of PARP either by drugs or by RNAi also led to increase γH2AX foci and Rad51 foci, consistent with the idea that loss of PARP might increase the formation of DNA strand breaks that are repaired by HR (34, 35). Recently, we (11) have shown that TNBC cells are defective in BER , moreover these cells may also be defective in HR, thus inhibition of PARP along with defective DNA repair mechanisms may lead to synthetic lethality.
Although UV-induced activation of PARP has been reported, the possible role of PARP in NER has not received much attention. Studies have shown that repair of 8-oxoG is stimulated by XPC (36) and CSB (37, 38) and since our laboratory has demonstrated that BRCA1 mutant cells are defective in GGR pathway of NER, but not transcription coupled repair (TCR) (15), and in BER (11) we investigated the role of PARP in GGR. We showed that PARP inhibition chemically or by RNAi decreased the capacity of TNBC cells to remove UVC-induced CPD lesions, a result consistent with previous reports of PARP playing a role in GGR (39, 40). NER is known to be involved in platinum-DNA adduct repair. Therefore, the synergism observed with PARP inhibition and cisplatin may be partly due to inhibition of NER. However, if the repair is facilitated by the TCR pathway of NER is not established by this study and further investigation into the mechanism is underway.
Understanding the molecular mechanism behind drug treatment is critical to predict the clinical efficacy of treatment. Meta-analyses of published microarray data (28) for gene expression changes common to the basal-like and BRCA1-mutated cell lines identified p63 and MCM family members to be significantly overexpressed (p<0.05) in both groups. Our study showed that the treatment of TNBC cells with PJ34 or gemcitabine resulted in a decreased expression of ΔNp63α with concurrent increase in expression of p73 and cyclin dependent kinase inhibitor p21 that in turn represses the MCM proteins (41). MCMs are required for licensing of origins, providing a signal for initiating replication in S phase and are frequently overexpressed in cancer cells (42-44). Our data demonstrated that MCM 4/7 are downregulated in TNBC cells treated with PJ34. Based on these lines of evidence, we hypothesize that PJ34 treatment leads to downregulation of ΔNp63α with concurrent increase in expression of p73 and p21, which in turn decreases expression of MCM 4/7 leading to cell cycle arrest and cell death. Overall, we found that human TNBC cells, similar to BRCA1-deficient cell lines, were more sensitive to PJ34, gemcitabine and cisplatin, and exhibited synergistic responses to combinations of these agents. This sensitivity seems to be dependent on inefficient DNA repair mechanisms causing sustained DNA damage that may trigger the p63/p73 mediated apoptotic signaling cascade. These data suggest novel options for targeted treatment of TNBCs.
Supplementary Material
Acknowledgments
Financial Support: U.S. National Institute of Health grant R01 CA108794, Breast Cancer Research Foundation (to JMF), and Susan G. Komen for the Cure Postdoctoral Fellowships (to KH and EA).
Abbreviations
- TN
triple negative
- TNBC
triple negative breast cancer
Reference
- 1.Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 2.Sorlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:10869–74. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carey LA, Dees EC, Sawyer L, et al. The triple negative paradox: primary tumor chemosensitivity of breast cancer subtypes. Clin Cancer Res. 2007;13:2329–34. doi: 10.1158/1078-0432.CCR-06-1109. [DOI] [PubMed] [Google Scholar]
- 4.Liedtke C, Mazouni C, Hess KR, et al. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J Clin Oncol. 2008;26:1275–81. doi: 10.1200/JCO.2007.14.4147. [DOI] [PubMed] [Google Scholar]
- 5.Yehiely F, Moyano JV, Evans JR, Nielsen TO, Cryns VL. Deconstructing the molecular portrait of basal-like breast cancer. Trends Mol Med. 2006;12:537–44. doi: 10.1016/j.molmed.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 6.Rakha EA, Tan DS, Foulkes WD, et al. Are triple-negative tumours and basal-like breast cancer synonymous? Breast Cancer Res. 2007;9:404. doi: 10.1186/bcr1827. author reply 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nielsen TO, Hsu FD, Jensen K, et al. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res. 2004;10:5367–74. doi: 10.1158/1078-0432.CCR-04-0220. [DOI] [PubMed] [Google Scholar]
- 8.Moynahan ME, Cui TY, Jasin M. Homology-directed dna repair, mitomycin-c resistance, and chromosome stability is restored with correction of a Brca1 mutation. Cancer Res. 2001;61:4842–50. [PubMed] [Google Scholar]
- 9.Bhattacharyya A, Ear US, Koller BH, Weichselbaum RR, Bishop DK. The breast cancer susceptibility gene BRCA1 is required for subnuclear assembly of Rad51 and survival following treatment with the DNA cross-linking agent cisplatin. The Journal of biological chemistry. 2000;275:23899–903. doi: 10.1074/jbc.C000276200. [DOI] [PubMed] [Google Scholar]
- 10.Treszezamsky AD, Kachnic LA, Feng Z, Zhang J, Tokadjian C, Powell SN. BRCA1- and BRCA2-deficient cells are sensitive to etoposide-induced DNA double-strand breaks via topoisomerase II. Cancer Res. 2007;67:7078–81. doi: 10.1158/0008-5472.CAN-07-0601. [DOI] [PubMed] [Google Scholar]
- 11.Alli E, Sharma VB, Sunderesakumar P, Ford JM. Defective repair of oxidative dna damage in triple-negative breast cancer confers sensitivity to inhibition of poly(ADP-ribose) polymerase. Cancer Res. 2009;69:3589–96. doi: 10.1158/0008-5472.CAN-08-4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharma VBHA, Cowan K, Ford JM. Enhanced sensitivity to cisplatin and gemcitabine in DNA repair deficient Brca1 null mouse embryonic fibroblasts. Proc Am Assoc Cancer Res. 2005:4390. [Google Scholar]
- 13.Huang P, Chubb S, Hertel LW, Grindey GB, Plunkett W. Action of 2',2'-difluorodeoxycytidine on DNA synthesis. Cancer Res. 1991;51:6110–7. [PubMed] [Google Scholar]
- 14.Zhang J, Powell SN. The role of the BRCA1 tumor suppressor in DNA double-strand break repair. Mol Cancer Res. 2005;3:531–9. doi: 10.1158/1541-7786.MCR-05-0192. [DOI] [PubMed] [Google Scholar]
- 15.Hartman AR, Ford JM. BRCA1 induces DNA damage recognition factors and enhances nucleotide excision repair. Nat Genet. 2002;32:180–4. doi: 10.1038/ng953. [DOI] [PubMed] [Google Scholar]
- 16.Farmer H, McCabe N, Lord CJ, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–21. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 17.Bryant HE, Schultz N, Thomas HD, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–7. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 18.ATCC Bulletin Maintaining high standards in cell culture. 2010 [Google Scholar]
- 19.Chou TC, Stepkowski SM, Kahan BD. Computerized quantitation of immunosuppressive synergy for clinical protocol design. Transplantation proceedings. 1994;26:3043–5. [PubMed] [Google Scholar]
- 20.Chou TC, Talalay P. Generalized equations for the analysis of inhibitions of Michaelis-Menten and higher-order kinetic systems with two or more mutually exclusive and nonexclusive inhibitors. European journal of biochemistry / FEBS. 1981;115:207–16. doi: 10.1111/j.1432-1033.1981.tb06218.x. [DOI] [PubMed] [Google Scholar]
- 21.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Advances in enzyme regulation. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 22.Mori T, Nakane M, Hattori T, Matsunaga T, Ihara M, Nikaido O. Simultaneous establishment of monoclonal antibodies specific for either cyclobutane pyrimidine dimer or (6-4)photoproduct from the same mouse immunized with ultraviolet-irradiated DNA. Photochem Photobiol. 1991;54:225–32. doi: 10.1111/j.1751-1097.1991.tb02010.x. [DOI] [PubMed] [Google Scholar]
- 23.Chang H, Sander CS, Muller CS, Elsner P, Thiele JJ. Detection of poly(ADP-ribose) by immunocytochemistry: a sensitive new method for the early identification of UVB- and H2O2-induced apoptosis in keratinocytes. Biological chemistry. 2002;383:703–8. doi: 10.1515/BC.2002.072. [DOI] [PubMed] [Google Scholar]
- 24.Vodenicharov MD, Ghodgaonkar MM, Halappanavar SS, Shah RG, Shah GM. Mechanism of early biphasic activation of poly(ADP-ribose) polymerase-1 in response to ultraviolet B radiation. Journal of cell science. 2005;118:589–99. doi: 10.1242/jcs.01636. [DOI] [PubMed] [Google Scholar]
- 25.Ribeiro-Silva A, Ramalho LN, Garcia SB, Brandao DF, Chahud F, Zucoloto S. p63 correlates with both BRCA1 and cytokeratin 5 in invasive breast carcinomas: further evidence for the pathogenesis of the basal phenotype of breast cancer. Histopathology. 2005;47:458–66. doi: 10.1111/j.1365-2559.2005.02249.x. [DOI] [PubMed] [Google Scholar]
- 26.Wang X, Mori I, Tang W, et al. p63 expression in normal, hyperplastic and malignant breast tissues. Breast Cancer. 2002;9:216–9. doi: 10.1007/BF02967592. [DOI] [PubMed] [Google Scholar]
- 27.Leong CO, Vidnovic N, DeYoung MP, Sgroi D, Ellisen LW. The p63/p73 network mediates chemosensitivity to cisplatin in a biologically defined subset of primary breast cancers. J Clin Invest. 2007;117:1370–80. doi: 10.1172/JCI30866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neve RM, Chin K, Fridlyand J, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–27. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scian MJ, Carchman EH, Mohanraj L, et al. Wild-type p53 and p73 negatively regulate expression of proliferation related genes. Oncogene. 2008;27:2583–93. doi: 10.1038/sj.onc.1210898. [DOI] [PubMed] [Google Scholar]
- 30.Bryant HE, Petermann E, Schultz N, et al. PARP is activated at stalled forks to mediate Mre11-dependent replication restart and recombination. The EMBO journal. 2009;28:2601–15. doi: 10.1038/emboj.2009.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ewald B, Sampath D, Plunkett W. H2AX phosphorylation marks gemcitabine-induced stalled replication forks and their collapse upon S-phase checkpoint abrogation. Mol Cancer Ther. 2007;6:1239–48. doi: 10.1158/1535-7163.MCT-06-0633. [DOI] [PubMed] [Google Scholar]
- 32.Parsels LA, Morgan MA, Tanska DM, et al. Gemcitabine sensitization by checkpoint kinase 1 inhibition correlates with inhibition of a Rad51 DNA damage response in pancreatic cancer cells. Mol Cancer Ther. 2009;8:45–54. doi: 10.1158/1535-7163.MCT-08-0662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wachters FM, van Putten JW, Maring JG, Zdzienicka MZ, Groen HJ, Kampinga HH. Selective targeting of homologous DNA recombination repair by gemcitabine. Int J Radiat Oncol Biol Phys. 2003;57:553–62. doi: 10.1016/s0360-3016(03)00503-0. [DOI] [PubMed] [Google Scholar]
- 34.Schultz N, Lopez E, Saleh-Gohari N, Helleday T. Poly(ADP-ribose) polymerase (PARP-1) has a controlling role in homologous recombination. Nucleic acids research. 2003;31:4959–64. doi: 10.1093/nar/gkg703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang YG, Cortes U, Patnaik S, Jasin M, Wang ZQ. Ablation of PARP-1 does not interfere with the repair of DNA double-strand breaks, but compromises the reactivation of stalled replication forks. Oncogene. 2004;23:3872–82. doi: 10.1038/sj.onc.1207491. [DOI] [PubMed] [Google Scholar]
- 36.D'Errico M, Parlanti E, Teson M, et al. New functions of XPC in the protection of human skin cells from oxidative damage. The EMBO journal. 2006;25:4305–15. doi: 10.1038/sj.emboj.7601277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sunesen M, Stevnsner T, Brosh RM, Jr, Dianov GL, Bohr VA. Global genome repair of 8-oxoG in hamster cells requires a functional CSB gene product. Oncogene. 2002;21:3571–8. doi: 10.1038/sj.onc.1205443. [DOI] [PubMed] [Google Scholar]
- 38.Tuo J, Chen C, Zeng X, Christiansen M, Bohr VA. Functional crosstalk between hOgg1 and the helicase domain of Cockayne syndrome group B protein. DNA repair. 2002;1:913–27. doi: 10.1016/s1568-7864(02)00116-7. [DOI] [PubMed] [Google Scholar]
- 39.Flohr C, Burkle A, Radicella JP, Epe B. Poly(ADP-ribosyl)ation accelerates DNA repair in a pathway dependent on Cockayne syndrome B protein. Nucleic acids research. 2003;31:5332–7. doi: 10.1093/nar/gkg715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ghodgaonkar MM, Zacal N, Kassam S, Rainbow AJ, Shah GM. Depletion of poly(ADP-ribose) polymerase-1 reduces host cell reactivation of a UV-damaged adenovirus-encoded reporter gene in human dermal fibroblasts. DNA repair. 2008;7:617–32. doi: 10.1016/j.dnarep.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 41.De Laurenzi V, Melino G. Evolution of functions within the p53/p63/p73 family. Annals of the New York Academy of Sciences. 2000;926:90–100. doi: 10.1111/j.1749-6632.2000.tb05602.x. [DOI] [PubMed] [Google Scholar]
- 42.Blow JJ, Laskey RA. A role for the nuclear envelope in controlling DNA replication within the cell cycle. Nature. 1988;332:546–8. doi: 10.1038/332546a0. [DOI] [PubMed] [Google Scholar]
- 43.Blow JJ, Dutta A. Preventing re-replication of chromosomal DNA. Nat Rev Mol Cell Biol. 2005;6:476–86. doi: 10.1038/nrm1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsuyama T, Tada S, Watanabe S, Seki M, Enomoto T. Licensing for DNA replication requires a strict sequential assembly of Cdc6 and Cdt1 onto chromatin in Xenopus egg extracts. Nucleic acids research. 2005;33:765–75. doi: 10.1093/nar/gki226. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.