Abstract
If not properly processed and repaired, DNA double-strand breaks (DSBs) can give rise to deleterious chromosome rearrangements, which could ultimately lead to the tumor phenotype1,2. DSB ends are resected in a 5′ to 3′ fashion in cells, to yield single-stranded DNA for the recruitment of factors critical for DNA damage checkpoint activation and repair by homologous recombination2. The resection process involves redundant pathways consisting of nucleases, DNA helicases, and associated proteins3. Being guided by recent genetic studies4–6, we have reconstituted the first eukaryotic ATP-dependent DNA end resection machinery comprising the Saccharomyces cerevisiae Mre11-Rad50-Xrs2 (MRX) complex, the Sgs1-Top3-Rmi1 (STR) complex, Dna2 protein and the heterotrimeric single-strand DNA binding protein RPA. We show that DNA strand separation during end resection is mediated by the Sgs1 helicase function, in a manner that is enhanced by Top3-Rmi1 and MRX. In congruence with genetic observations6, while the Dna2 nuclease activity is critical for resection, the Mre11 nuclease activity is dispensable. By examining the top3 Y356F allele and its encoded protein, we provide evidence that the topoisomerase activity of Top3, although critical for the suppression of crossover recombination2,7, is not needed for resection either in cells or in the reconstituted system. Our results also unveil a multi-faceted role of RPA, in the sequestration of ssDNA generated by DNA unwinding, enhancement of 5′ strand incision, and protection of the 3′ strand. Our reconstituted system should serve as a useful model for delineating the mechanistic intricacy of the DNA break resection process in eukaryotes.
The 3′ ssDNA strands derived from DSB resection attract RPA, which promotes the recruitment of checkpoint proteins to effect cell cycle arrest8. With the aid of a recombination mediator protein, such as yeast Rad52 or human BRCA2, the Rad51 recombinase displaces RPA from the ssDNA to assemble into a right-handed helical polymer capable of initiating DSB repair by homologous recombination1,2. Genetic studies in yeast have shown that DSB resection proceeds in two steps. The MRX complex plays a role in initiation, while the Sgs1 helicase, its associated proteins Top3 and Rmi1, and the helicase/nuclease Dna2, whose nuclease activity is needed for Okazaki fragment processing9,10, constitute the DNA motor-driven path of long-range resection. Exo1, a 5′-3′ exonuclease, defines a redundant resection means4–6. Here we reconstitute the Sgs1/Dna2-dependent DNA resection machinery and present results germane for understanding its mechanistic underpinnings.
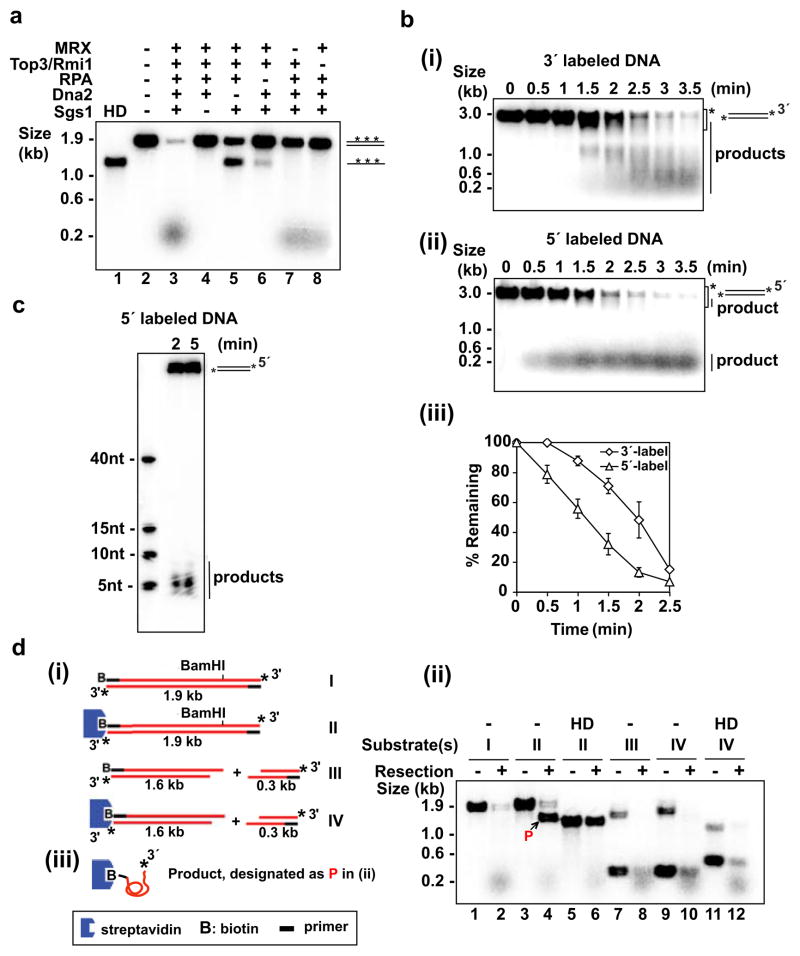
The requisite factors, namely, Sgs1, Top3-Rmi1 (TR) complex, MRX complex, Dna2, and RPA were purified and analyzed (see Supplementary Fig. 2 and the Supplementary Information). As shown in Figure 1a, the combination of these factors degraded a 1.9-kb linear, uniformly 32P-labeled duplex rapidly. Little or no digestion occurred when Dna2, Sgs1, or RPA was omitted. In the absence of Dna2, the DNA substrate was unwound completely to yield ssDNA, which was not seen upon Sgs1’s omission, indicating that the Sgs1 helicase activity but not that of Dna2 can support complete unwinding. The results also revealed a dependence of DNA unwinding on RPA. In MRX’s absence, DNA degradation was attenuated, a result similar to the delayed resection observed in mre11Δ cells3,6,11. Also in concordance with genetic results6, TR stimulated resection. The ability to degrade DNA required ATP, which could not be replaced by ATP-γ-S or AMP-PNP (Supplementary Fig. 3a). Thus, ATP hydrolysis is needed for DNA degradation. The stimulatory effects of the MRX and TR complexes will be addressed in greater details below.
Figure 1. Reconstitution and 5′ polarity of DNA end resection.
a, Resection of uniformly labeled DNA. Symbol: HD, heat denaturation of DNA.
b, Time course analysis of resection with 3′-labeled (i) or 5′-labeled (ii) DNA. Mean values ±SD from three independent experiments were plotted (iii). The asterisk in (i) and (ii) marks the area that was quantified.
c, Analysis of the initial 5′ resection product in a denaturing polyacrylamide gel.
d, The biotinylated substrates (i) were incubated with the resection machinery for 5 min as in (a) and analyzed (ii). Complete digestion of the unblocked 5′ strand yielded ssDNA (designated P in (ii) and depicted in (iii)) that harbored biotin-streptavidin at the 5′ terminus and 32P at the 3′ terminus.
Importantly, supercoiled DNA was not degraded by the same combination of protein factors (Supplementary Fig. 3b), confirming that the reconstituted resection machinery is DNA end specific. The top3 Y356 mutant that lacks topoisomerase activity was used in this experiment (Supplementary Fig. 3b, c) to avoid the relaxation of the supercoiled DNA. As detailed later, the top3-Y356F mutant is differentially inactivated for the ability to attenuate mitotic crossover recombination.
We used 3-kb long 3′- and 5′-labeled linear duplex DNA substrates to test the polarity of resection. The 3′-labeled DNA was converted to intermediates that progressively decreased in size over the reaction course (Fig. 1b). In contrast, much of the product generated from 5′-labeled DNA migrated at the front of the agarose gel (Fig. 1b). The radiolabeled product derived from the 5′-labeled substrate ranged in size from 4 to 8 nucleotides, with the 5-nt size being predominant (Fig. 1c). Incubation with 3′-labeled DNA did not generate any small product until much later (Supplementary Fig. 4b). To determine whether conversion of the 3′ label into the small product stemmed from progressive digestion of the 5′ strands or from limited resection of the 3′ strands, we designed a 3′-32P-labeled substrate in which one end was tagged with biotin and could therefore be blocked from protein access with streptavidin. The results showed that a large fraction of the 3′-32P-label at the unblocked end remained intact during resection (Fig. 1d). Together, the results show that the reconstituted DNA end resection machinery has a strong specificity for the 5′ strand and incises the 3′ strand rather infrequently.
Dna2 possesses an endonuclease activity capable of digesting 3′ or 5′ single-stranded DNA12, while Mre11 harbors a 3′ to 5′ exonuclease activity and also a DNA structure-specific endonuclease activity13–15. Using the nuclease null dna2-D657A protein (Supplementary Fig. 5) and mre11-3 protein16 (Supplementary Fig. 6) with the 3′-labeled substrate, we found that extensive resection is mediated by Dna2 nuclease (Fig. 2a). Even when manganese ion was included to activate the Mre11 nuclease (Supplementary Fig. 6d, 7)13,17, 5′ cleavage was still completely dependent on Dna2 (Supplementary Fig. 7b, c). Although Sae2 protein has been implicated in the initial step of end resection5,6, its addition did not affect the efficiency of the first cleavage or product size (Supplementary Fig. 8).
Figure 2. Requirement for Sgs1 helicase and Dna2 nuclease activities.
a, The dna2-D657A (DA) mutant and mutant MRX harboring mre11-3 (11-3) were tested with 3′-labeled substrate. Symbol: HD, heat-denaturation of DNA.
b, The dna2-K1080E (KE) and sgs1-K706A (KA) mutants were tested.
c, Srs2 helicase, Fen1 nuclease, or their combination failed to substitute for Sgs1 and Dna2. Symbol: DA, dna2-D657A.
d, RAD27 over-expression (YEp24) failed to suppress the end resection defect of dna2Δ cells. Initial resection next to the break (MAT probe) and long-range resection (FEN2 probe) were analyzed6, as depicted (i). A set of primary data and the mean values ±SD from three independent experiments are presented (ii).
The dependence of DNA resection on ATP hydrolysis (Supplementary Fig. 3a, b) implies that either or both of the DNA motor functions residing in Sgs1 and Dna2 might be indispensable. We therefore purified the ATPase defective variants, dna2-K1080E9 and sgs1-K706A18 (Supplementary Fig. 5, 9), and tested them with 3′-labeled DNA. Substitution of Sgs1 with sgs1-K706A led to ablation of resection, but resection occurred normally with dna2-K1080E replacing Dna2 (Fig. 2b). We note that these biochemical results are consistent with genetic observations that the Sgs1 helicase activity, but not the Dna2 helicase activity, as being indispensable for DSB resection5,6.
We investigated the specificity of some of the protein factors by replacing Sgs1 with the Srs2 helicase that functions in DNA damage repair and response1 and Dna2 with Fen1 (product of the RAD27 gene), a 5′ FLAP endonuclease involved in Okazaki fragment processing19. We found that Srs2 and Fen1 are non-functional in resection (Fig. 2c). These biochemical results are in congruence with genetic data showing that deletion of SRS2 has no impact on the rate and extent of DSB resection6, and that RAD27 fails to suppress the resection defect of pif1-m2 dna2Δ cells (Fig. 2d). Our results also suggest that suppression of the dna2 mutant phenotype by RAD27 over-expression20 stems from the restoration of Okazaki fragment processing.
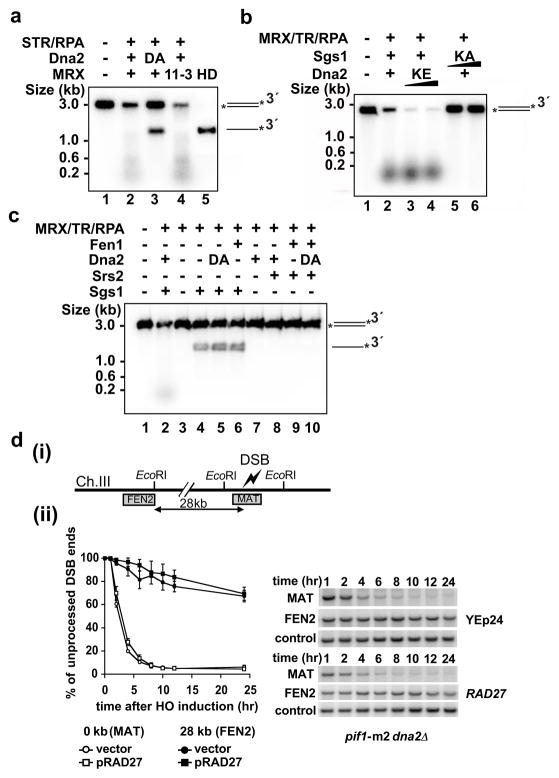
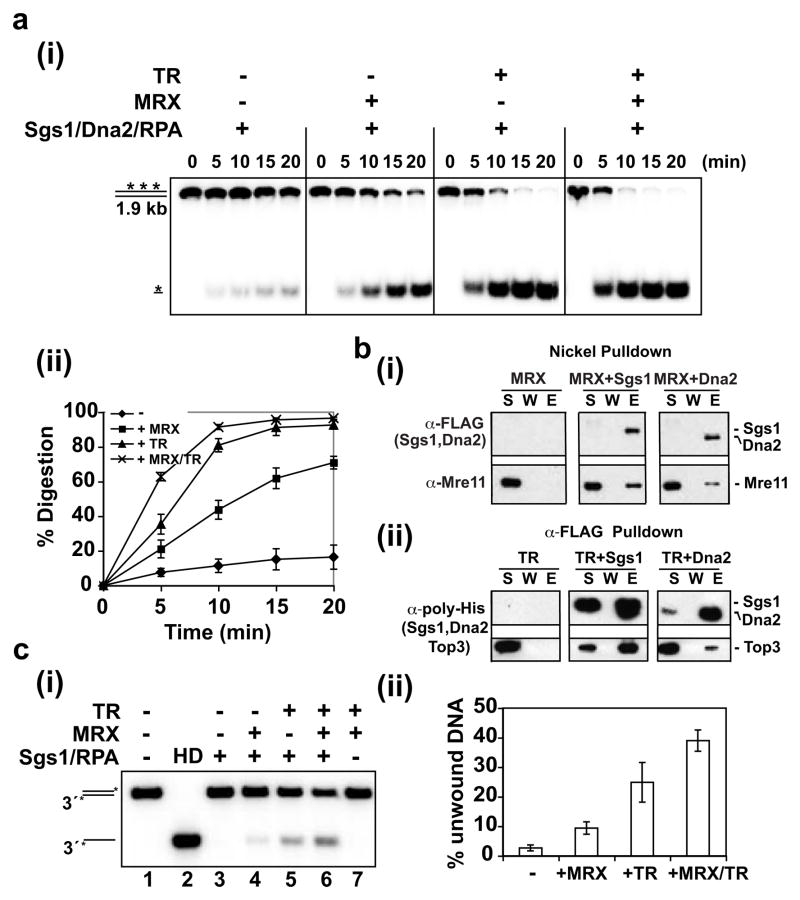
The effects of MRX and TR on resection were further defined with a limiting amount of Dna2 and Sgs1 and uniformly labeled DNA. While the combination of Sgs1/Dna2/RPA was able to mediate resection, the addition of MRX or TR stimulated the reaction by about 4 fold or 8 fold, respectively (Fig. 3a). Consistent with results shown earlier (Fig. 1a), the most efficient resection was seen when both MRX and TR were included (Fig. 3a). Interestingly, MRX or TR alone could stimulate the Sgs1-mediated unwinding of a 3-kb substrate, by about 4 or 8 fold, respectively, and an additive effect was seen (Fig. 3c). The results from in vitro pulldown revealed that MRX interacts with Sgs1 at physiological ionic strength (Fig. 3b). This agrees with a previous study showing co-fractionation and co-immunoprecipitation of Sgs1 with Mre1121. MRX also associates with Dna2 (Fig. 3b), although it does not affect Dna2’s nuclease activity (Supplementary Fig. 10c). Since MRX binds DNA ends22, it could be involved in the DNA end recruitment of Sgs1 and Dna2. This premise is supported by the finding that DNA which had been pre-processed to generate 3′ ssDNA tails could be resected efficiently by Sgs1-Dna2-RPA with or without MRX (Supplementary Fig. 11). We also verified that the TR complex binds Sgs123 and noted a weak interaction between TR and Dna2 as well (Fig. 3b). We did not detect any significant physical interaction of TR with MRX and, using protection of 5′-labeled DNA against lambda exonuclease22 as assay, found no specific affinity of TR for DNA ends (data not shown).
Figure 3. Role of MRX and TR in DNA resection and unwinding.
a, The resection role of MRX and TR was analyzed with uniformly labeled DNA. Reactions were resolved in 10% polyacrylamide gels (i), and the mean values ±SD from three independent experiments were plotted (ii).
b, Pulldown through the (His)6 tag (Nickel) or FLAG tag (α-FLAG) on FLAG-(His)6-tagged Dna2 or Sgs1 to test for their interaction with MRX (i) or (His)6-Top3-Rmi1 complex (ii). The supernatant (S), wash (W) and SDS eluate (E) fractions were immunoblotted with the indicated antibodies.
c. Sgs1 and RPA were examined for the unwinding of a 3 kb DNA in the absence or presence of MRX and/or TR (i), and the mean values ±SD from three independent experiments were plotted (ii).
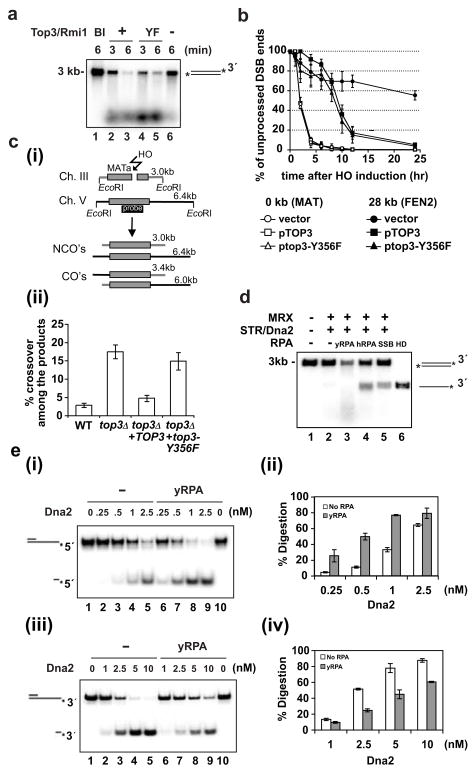
Top3 co-operates with Rmi1 and Sgs1 in the suppression of mitotic crossover formation7, presumably via dissolution of the double Holliday Junction (dHJ)24. Results shown earlier (Supplementary Fig. 3b) suggested that the catalytically null top3-Y356F mutant could function in resection. Indeed, a direct comparison with wild type Top3 revealed no deficiency in the top3-Y356F mutant in resection (Fig. 4a). Likewise, top3-Y356F mutant cells are proficient in resecting the HO-induced DSB (Fig. 4b). However, while dHJ dissolution occurs efficiently with Sgs1, Rmi1, and Top3 in vitro, top3-Y356F is completely defective in this regard (Supplementary Fig. 3d). Consistent with the biochemical data, the top3-Y356F allele is unable to suppress the high level of mitotic crossovers in top3Δ cells (Fig. 4c). These results indicate that the catalytic activity of Top3 is critical for crossover control but dispensable for DSB resection.
Figure 4. Role of Top3 and RPA in resection.
a, Top3-Rmi1 and top3-Y356F-Rmi1 were tested in the resection system with 3′-labeled substrate. Symbol: Bl, no protein added.
b, Analysis of 5′ resection in top3Δ cells supplemented with the low copy pRS315 vector carrying TOP3 or top3-Y356F gene. Analysis was done as in Figure 2d.
c, Crossover (CO’s) and noncrossover (NCO’s) recombinants are distinguished based on their size (i). The frequencies of CO’s 8 hr after break induction were determined7 and the mean values ±SD from three independent experiments were plotted (ii).
d, SSB or hRPA supports DNA unwinding but not end resection. Symbol: HD, heat denaturation of DNA.
e, Yeast RPA stimulates 5′ end digestion (i) but attenuates 3′ end digestion (iii) by Dna2. The mean values ±SD from three independent experiments were plotted in (ii) and (iv).
Earlier we saw a strict dependence of DNA resection on RPA (Fig. 1a), which is expected to facilitate the DNA unwinding step by sequestering ssDNA generated by Sgs1. Our finding of DNA unwinding in the absence of Dna2 as being strongly reliant upon RPA (data not shown) and the fact that human RPA (hRPA) or E. coli SSB is able to support DNA unwinding (Fig. 4d) are consistent with this premise. However, little or no resection occurred with either hRPA or SSB (Fig. 4d), indicating that yeast RPA (yRPA) must fulfill another role in resection. That yRPA, but neither hRPA nor SSB, physically interacts with Dna2 reinforces this idea10 (Supplementary Fig. 10a). Alternatively, or in addition, hRPA and SSB may inhibit the nuclease activity of yeast Dna2. To understand the specificity of yRPA in resection, we used partially duplex DNA substrates that bore either a labeled 3′ or 5′ ssDNA tail to examine how yRPA, hRPA, and SSB affect the nuclease attribute of Dna2. Interestingly, yRPA enhanced 5′ DNA incision by Dna2 but, in reproducible fashion, attenuated the 3′ DNA cleavage reaction (Fig. 4e). We note that yRPA was previously found to reduce the cleavage of the 3′ tail of a substrate bearing a G4 DNA structure25. Importantly, strong inhibition of the Dna2 nuclease function by hRPA and SSB was seen (Supplementary Fig. 10b). The dna2-Δ405N mutant protein, which is partially defective in RPA interaction and Dna2 nuclease enhancement26 (Supplementary Fig. 12a, b), is stimulated by yRPA to a lesser degree (Supplementary Fig. 12c) and, accordingly, is significantly less effective than Dna2 in DNA resection (Supplementary Fig. 12d).
Our studies have clarified the role of the Sgs1 helicase and Dna2 nuclease activities and of the MRX and TR complexes in DSB resection in a reconstituted system. RPA is found to be an essential component of the Sgs1/Dna2 resection pathway, by supporting the DNA unwinding step and enhancing the 5′ endonuclease activity of Dna2. Importantly, RPA attenuates 3′ DNA cleavage by Dna2, an attribute that likely serves to enforce the 5′ strand specificity of the resection machinery. The mechanistic details of the DNA resection machinery are depicted in our working model (Supplementary Fig. 1).
The reconstituted DNA motor-driven resection machinery described herein and in the companion article by Cejka et al27 comprises ten polypeptides, which is much more complex than the resection systems from prokaryotes and archaea, such as E. coli RecBCD (three polypeptides), AdnAB (two polypeptides) from mycobacteria, or Mre11-Rad50-HerA-NurA (four polypeptides) from Pyrococcus furiosu28. Moreover, the dependence on a single-strand DNA binding protein (i.e. RPA) is not evident in these other systems. The DNA damage repair process is linked to checkpoint-mediated cell-cycle control, wherein RPA-coated ssDNA activates the ATR/Mec1 kinase through ATRIP/Ddc28. Based on our results, we propose that RPA plays an even earlier role in checkpoint activation by promoting resection. As such, RPA could provide a means for coupling the decision of DSB end processing with checkpoint activation. The reconstituted system should be beneficial for understanding the mechanism of DSB resection in humans, as there is evidence that the Sgs1 orthologue, BLM, which is mutated in the cancer-prone Bloom syndrome, is similarly involved in DSB processing4. It will be of great interest to test whether human DNA2, found in both mitochondria and the nucleus29,30, also helps mediate chromosomal end resection in a manner analogous to yeast Dna2.
Methods Summary
The proteins and protein complexes (i.e. Mre11-Rad50-Xrs2, Dna2, yeast and human RPA proteins, Sgs1, Top3-Rmi1, Srs2, Fen1) used in this study were expressed in insect, yeast, or E. coli cells and purified as detailed in the Supplementary Information. E. coli SSB protein was purchased (New England BioLabs). For the end resection reactions, linear DNA was 3′ or 5′ labeled with 32P using standard methods, and the internally 32P-labeled DNA was generated by PCR. Other 32P-labeled DNA substrates for testing Sgs1 and Dna2 DNA helicase and Dna2 nuclease activities were prepared as described in the Supplementary Information. The supercoiled φX174 DNA was purchased (New England BioLabs) and its linearization was by digestion with the restriction enzyme StuI. The DNA resection reactions were analyzed in agarose or polyacrylamide gels under denaturing or non-denaturing conditions, followed by phosphorimaging analysis of the dried gel or ethidium bromide treatment of the gel to visualize DNA species. The pulldown experiments to test for protein-protein interactions made use of affinity tags on the indicated proteins. Visualization of proteins was by the staining of SDS polyacrylamide gels with Coomassie Blue or by immunoblotting. Experimental details for the Dna2 nuclease, double Holliday Junction dissolution, DNA helicase, and ATPase assays, and pre-resection of uniformly 32P-labeled DNA can be found in the Methods section or Supplementary Information. Genetic assays to measure the distribution of recombinant products of the gene conversion and crossover types or the kinetics of the resection of a site-specific DNA double strand break were conducted as described6,7.
Methods
DNA end resection substrates
The 3-kb pBluescript SK(−) was linearized with EcoRV and then 5′-labeled with [γ-32P]-ATP (PerkinElmer) and T4 polynucleotide kinase (New England Biolabs) or 3′-labeled with [α-32P] Cordycepin 5′-triphosphate (PerkinElmer) and terminal deoxytransferase (Roche). The 1.9-kb internally labeled substrate was generated by PCR in the presence of [α-32P]-dCTP (PerkinElmer) and 5′-GCCGAGCCATATGTCAAGTGAGTCAACAACT TTCATCGTGG-3′ and 5′-GCACGACTCGAGTTAATTATTGCTATTGTTTGGACTT CCCC-3′ as primers and the S. cerevisiae HDF2 gene as template. The substrates were purified from 1% agarose gels with the Gel Extraction kit (Qiagen). The substrate in Figure 1d was generated using the same template and primers, except with the first primer being biotinylated at the 5′ end. To block the DNA end that harbored the biotin label, a 10 μl reaction (50 mM Tris-HCl, pH 7.9, 100 mM NaCl, 5 mM MgCl2, 1 mM DTT) containing substrate (5 nM ends) and 0.5 mg/ml streptavidin (Invitrogen) was incubated at 25°C for 20 min. Where indicated, the substrate was treated with BamHI to yield two fragments (1.6 kb and 0.3 kb), the larger of which bore the biotin. Biotinylation of the 5′ terminus reduced the efficiency at which the companion 3′ terminus was labeled with 32P (see Fig. 1d, lane 7).
DNA end resection assays
Reactions were conducted in 10 μl buffer (40 mM Tris-HCl, pH 7.5, 2 mM ATP, 4 mM MgCl2, 50 mM KCl, 1 mM DTT, 100 μg/ml BSA, an ATP-regenerating system of 20 mM creatine phosphate and 20 μg/ml creatine kinase) and the specified DNA substrate (0.5 nM ends) at 30ºC. Unless specified otherwise, each reaction contained 10 nM MRX complex, 10 nM Sgs1, 10 nM TR complex, 20 nM Dna2 and 100 nM RPA. The reaction was deproteinized by SDS (0.2%) and proteinase K (0.5 mg/ml) at 37ºC for 10 min before analysis in an agarose gel (1 or 1.2%) in TAE (35 mM Tris-acetate, pH 7.4, 0.5 mM EDTA) buffer. Alternatively, reaction mixtures were resolved in a 10% polyacrylamide gel in TBE (89 mM Tris-borate, pH 8.0, 2 mM EDTA) buffer or an 18% polyacrylamide gel under denaturing (7 M Urea in TBE buffer) conditions. Gels were dried onto DE3 paper (Whatman) before phosphorimaging analysis. For analyzing the resection product size (Fig. 1c), the amount of DNA substrate was increased five fold (2.5 nM ends). For time course analysis testing MRX and TR effects (Fig. 4a), we set up 30 μl reactions containing Sgs1 (1 nM), Dna2 (1 nM), RPA (100 nM) without or with MRX (2 nM), TR (1 nM) or both MRX and TR, and internally labeled DNA (0.13 nM DNA ends) as substrate.
Effect of MRX and TR on Sgs1-mediated DNA unwinding
The 3-kb 3′-labeled DNA substrate (0.5 nM ends) was incubated with Sgs1 (5 nM) and RPA (100 nM) in 10 μl buffer (30 mM Tris-HCl, pH 7.5, 1 mM DTT, 100 μg/ml BSA, 2 mM MgCl2, 2 mM ATP, and the ATP regenerating system) at 30°C for 5 min. The reaction mixtures were deproteinized, resolved in a 1.2% agarose gel in TAE buffer, which were dried onto DE3 paper and subject to phosphorimaging analysis.
Dna2 nuclease assay
Dna2 was incubated with or without yRPA, hRPA or SSB (20 nM each) at 4°C for 5 min in 9.5 μl buffer (25 mM Tris-HCl, pH 7.5, 1 mM DTT, 100 μg/ml BSA, 100 mM KCl, 2 mM MgCl2). Following the addition of the indicated DNA substrate (5 nM), the completed reaction (10 μl) was incubated at 37°C for 20 min. The reaction mixtures were deproteinized and resolved in a 10% polyacrylamide gel at 4°C in TBE buffer, and the gel was dried onto DE3 paper before phosphorimaging analysis.
Affinity pull-down assays
To test for MRX-Sgs1 or MRX-Dna2 interaction, MRX (750 ng) was incubated with FLAG-(His)6-tagged Sgs1 (250 ng) or Dna2 (200 ng) at 4°C for 30 min in 30 μl buffer (10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.4, 137 mM NaCl, 3 mM KCl, 0.05% Igepal CA-630 (Sigma), 1 mM DTT, 20 mM imidazole). After mixing with 8 μl of Nickel-NTA resin at 4°C for 30 min, the resin was washed three times with 300 μl of buffer, and 20 μl of 2% SDS was used to elute proteins. The supernatant (S), last wash (W) and SDS eluate (E) fractions, 5 μl each, were resolved by 8% SDS-PAGE and immunoblotted with either anti-FLAG or anti-Mre11 antibodies.
To test for TR-Sgs1 or TR-Dna2 interaction, TR (100 ng) and Sgs1 (250 ng) or Dna2 (200 ng) were incubated as above, except that anti-Flag-M2 resin (Sigma) was used to capture protein complexes. Analysis was as above, with anti-poly-histidine antibodies being used to reveal Sgs1, Dna2 and Top3.
Supplementary Material
Acknowledgments
We thank Judith Campbell, Steven Brill, and Lorraine Symington for providing materials, Xiaoyu Xue for the dHJ substrate, and Stephen Kowalczykowski for communicating results. This work was supported by grants from the US National Institutes of Health and by a postdoctoral fellowship from the Susan G. Komen for the Cure Foundation.
Footnotes
Author Contributions:
H.Y.N, G.I. and P.S. designed the experiments and wrote the paper. H.Y.N., W.H.C., Z.Z., Y.H.K., P.C., W.X.Z. and R.P. conducted the experiments. L.L. and D.L. provided key materials and technical expertise.
The Supplementary Information is linked to the online version of this paper at www.nature.com/nature. A figure summarizing the main result of this paper is available in supplementary information.
References
- 1.Sung P, Klein H. Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nat Rev Mol Cell Biol. 2006;7:739–750. doi: 10.1038/nrm2008. [DOI] [PubMed] [Google Scholar]
- 2.Krogh BO, Symington LS. Recombination proteins in yeast. Annu Rev Genet. 2004;38:233–271. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- 3.Mimitou EP, Symington LS. Nucleases and helicases take center stage in homologous recombination. Trends Biochem Sci. 2009;34:264–272. doi: 10.1016/j.tibs.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 4.Gravel S, Chapman JR, Magill C, Jackson SP. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 2008;22:2767–2772. doi: 10.1101/gad.503108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ira G, Malkova A, Liberi G, Foiani M, Haber JE. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell. 2003;115:401–411. doi: 10.1016/s0092-8674(03)00886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 9.Budd ME, Choe WC, Campbell JL. DNA2 encodes a DNA helicase essential for replication of eukaryotic chromosomes. J Biol Chem. 1995;270:26766–26769. doi: 10.1074/jbc.270.45.26766. [DOI] [PubMed] [Google Scholar]
- 10.Bae SH, Bae KH, Kim JA, Seo YS. RPA governs endonuclease switching during processing of Okazaki fragments in eukaryotes. Nature. 2001;412:456–461. doi: 10.1038/35086609. [DOI] [PubMed] [Google Scholar]
- 11.Ivanov EL, Sugawara N, White CI, Fabre F, Haber JE. Mutations in XRS2 and RAD50 delay but do not prevent mating-type switching in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:3414–3425. doi: 10.1128/mcb.14.5.3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bae SH, Seo YS. Characterization of the enzymatic properties of the yeast Dna2 Helicase/endonuclease suggests a new model for Okazaki fragment processing. J Biol Chem. 2000;275:38022–38031. doi: 10.1074/jbc.M006513200. [DOI] [PubMed] [Google Scholar]
- 13.Trujillo KM, et al. Yeast Xrs2 binds DNA and helps target Rad50 and Mre11 to DNA ends. J Biol Chem. 2003;278:48957–48964. doi: 10.1074/jbc.M309877200. [DOI] [PubMed] [Google Scholar]
- 14.Furuse M, et al. Distinct roles of two separable in vitro activities of yeast Mre11 in mitotic and meiotic recombination. EMBO J. 1998;17:6412–6425. doi: 10.1093/emboj/17.21.6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moreau S, Ferguson JR, Symington LS. The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol Cell Biol. 1999;19:556–566. doi: 10.1128/mcb.19.1.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bressan DA, Olivares HA, Nelms BE, Petrini JH. Alteration of N-terminal phosphoesterase signature motifs inactivates Saccharomyces cerevisiae Mre11. Genetics. 1998;150:591–600. doi: 10.1093/genetics/150.2.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paull TT, Gellert M. The 3′ to 5′ exonuclease activity of Mre11 facilitates repair of DNA double-strand breaks. Mol Cell. 1998;1:969–979. doi: 10.1016/s1097-2765(00)80097-0. [DOI] [PubMed] [Google Scholar]
- 18.Lu J, et al. Human homologues of yeast helicase. Nature. 1996;383:678–679. doi: 10.1038/383678a0. [DOI] [PubMed] [Google Scholar]
- 19.Liu Y, Kao HI, Bambara RA. FLAP endonuclease 1: a central component of DNA metabolism. Annu Rev Biochem. 2004;73:589–615. doi: 10.1146/annurev.biochem.73.012803.092453. [DOI] [PubMed] [Google Scholar]
- 20.Budd ME, Campbell JL. A yeast replicative helicase, Dna2 helicase, interacts with yeast FEN-1 nuclease in carrying out its essential function. Mol Cell Biol. 1997;17:2136–2142. doi: 10.1128/mcb.17.4.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chiolo I, et al. Srs2 and Sgs1 DNA helicases associate with Mre11 in different subcomplexes following checkpoint activation and CDK1-mediated Srs2 phosphorylation. Mol Cell Biol. 2005;25:5738–5751. doi: 10.1128/MCB.25.13.5738-5751.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen L, Trujillo K, Ramos W, Sung P, Tomkinson AE. Promotion of Dnl4-catalyzed DNA end-joining by the Rad50/Mre11/Xrs2 and Hdf1/Hdf2 complexes. Mol Cell. 2001;8:1105–1115. doi: 10.1016/s1097-2765(01)00388-4. [DOI] [PubMed] [Google Scholar]
- 23.Chen CF, Brill SJ. Binding and activation of DNA topoisomerase III by the Rmi1 subunit. J Biol Chem. 2007;282:28971–28979. doi: 10.1074/jbc.M705427200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu L, Hickson ID. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- 25.Masuda-Sasa T, Polaczek P, Peng XP, Chen L, Campbell JL. Processing of G4 DNA by Dna2 helicase/nuclease and replication protein A (RPA) provides insights into the mechanism of Dna2/RPA substrate recognition. J Biol Chem. 2008;283:24359–24373. doi: 10.1074/jbc.M802244200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bae KH, et al. Bimodal interaction between replication-protein A and Dna2 is critical for Dna2 function both in vivo and in vitro. Nucleic Acids Res. 2003;31:3006–3015. doi: 10.1093/nar/gkg422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cejka P, et al. DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2. Nature. 2010 doi: 10.1038/nature09355. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Niu H, Raynard S, Sung P. Multiplicity of DNA end resection machineries in chromosome break repair. Genes Dev. 2009;23:1481–1486. doi: 10.1101/gad.1824209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng L, et al. Human DNA2 is a mitochondrial nuclease/helicase for efficient processing of DNA replication and repair intermediates. Mol Cell. 2008;32:325–336. doi: 10.1016/j.molcel.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duxin JP, et al. Human Dna2 is a nuclear and mitochondrial DNA maintenance protein. Mol Cell Biol. 2009;29:4274–4282. doi: 10.1128/MCB.01834-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.