Down syndrome (trisomy 21, or DS) is the most common live-born aneuploidy in humans, occurring in approximately 1 in 700 live births (1). The inheritance of three copies of human chromosome 21 (Hsa21) results in a wide range of phenotypes including cognitive impairment, cardiac defects, and craniofacial dysmorphology (2,3). Conserved gene content and order occurs between Hsa21 and murine chromosomes Mmu16, Mmu17, and Mmu10 (4,5). To better understand the gene-phenotype relationship in DS, partial or segmental trisomies have been created in mice with triplicated portions of the murine genome that correspond to Hsa21. Numerous phenotypes associated with DS have been documented in mouse models (recently reviewed in References 6 and 7).
The most widely used and well-studied mouse model of trisomy and DS phenotypes is the Ts(1716)65Dn (written hereafter as Ts65Dn). To create this mouse, DBA/2J male mice with irradiated testes were bred to C57BL/6J female mice, and offspring carrying reciprocal translocations for Mmu16 were bred to B6C3F1 mice (8). The resultant T(16C3–4;17A2)65Dn heterozygote mice produced offspring with the small marker chromosome consisting of the telomeric 13.5 Mb of Mmu16, attached to a Mmu17 centromere with a small amount of euchromatic material (9,10). Ts65Dn mice are trisomic for orthologs of approximately half of the genes on Hsa21 and show DS-related phenotypes including reduced birthweight, perinatal lethality, craniofacial abnormalities, cognitive and behavioral impairments, cardiovascular malformations, and neurological structural deficiencies (11–14). Investigations using this mouse model have been used to predict human phenotypes as well as to examine potential therapies to correct traits associated with DS (6,15,16).
Ts65Dn mice serve as an excellent genetic and phenotypic model for DS; however, they are difficult to produce and genotype. Generating Ts65Dn mice is complicated given that males are subfertile, females are poor mothers, and offspring are limited due to perinatal loss (10,12,14). With all of these factors, trisomic pups account for ~40% of the litter on the day of birth. Physical differences have been observed between offspring containing the segmental trisomic and euploid littermates, but these phenotypes are highly variable and not reliable for positive identification of Ts65Dn mice (9,14). Ts65Dn mice were first genotyped using karyotypes from cultured lymphocytes or meiotic cells, a procedure taking 2–3 days and only done in adult mice (8). Fluorescence in situ hybridization (FISH) of nucleated blood cells at interphase has been used as the standard for identifying Ts65Dn mice, but this method is also labor-intensive and generally requires 2–3 days (17). Recently, a number of quantitative PCR methods have also been used to quantify trisomic gene copy number in Ts65Dn mice (18–21). While these processes may be relatively fast, results can be difficult to interpret, reagents are expensive, and few laboratories have the necessary equipment—thus requiring the service of a core facility (20). Erroneous inclusion of euploid mice as trisomic in assessment of a subtle phenotype can significantly reduce the accuracy of subsequent phenotyping procedures.
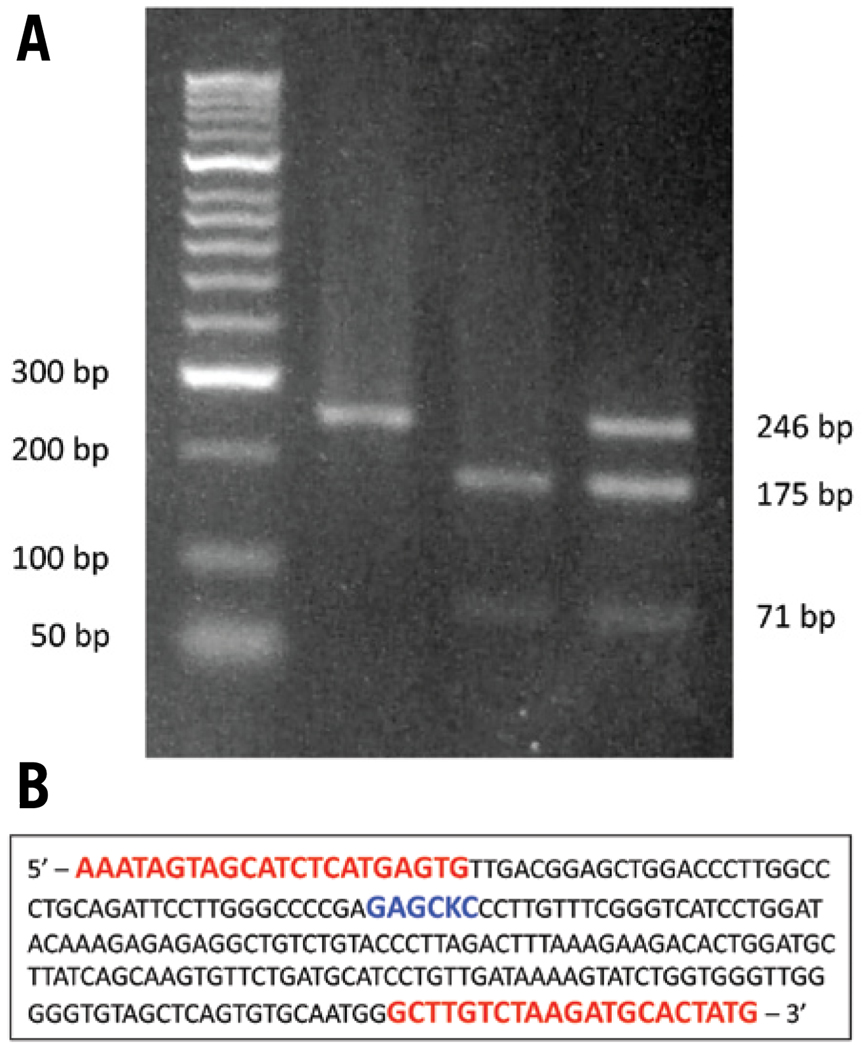
To prescreen Ts65Dn mice and reduce the time and cost of subsequent FISH identification of the trisomic segment, we have developed a simple methodology requiring the restriction digest of a PCR product amplified from the small translocation marker chromosome (1716). Our methodology utilizes a single nucleotide polymorphism (SNP) within the Zdhhc14 gene on Mmu17 near the breakpoint of the translocation chromosome. The SNP is polymorphic between the DBA/2J reciprocal translocation founder strain and the C57BL/6J and C3H/HeJ strains that contribute to the majority of the background of Ts65Dn mice. Crosses between Ts65Dn and B6C3F1 mice result in offspring with two normal Mmu17 with C57BL/6J or C3H/HeJ alleles. In most, but not all cases, trisomic offspring will have the DBA/2J Zdhhc14 allele at the Mmu17 centromeric end of the chimeric chromosome due to limited crossovers during meiosis within the region delimited by the SNP and the Mmu1716 breakpoint. PCR amplification of the area surrounding the SNP allows for the selective digest of C57BL/6J or C3H/HeJ product by SacI, which produces bands of 175 bp and 71 bp in length. Ts65Dn mice with the DBA/2J allele at the SNP will display a full-length PCR product (246 bp) in addition to the digested products from the additional copies of Mmu17 with C57BL/6J or C3H/HeJ alleles (the 71-bp band can be difficult to visualize on an agarose gel) (Figure 1).
Figure 1. PCR/restriction digest typing for the T65Dn marker chromosome using the Zdhhc14 polymorphism.
(A) PCR/restriction digest typing for the T65Dn marker chromosome in offspring from Ts65Dn × B6C3F1 matings. Euploidy or trisomy of offspring was verified by FISH. Lane 1: molecular weight markers. Lane 2: before digestion, a 246-bp PCR fragment is produced from all offspring of a Ts65Dn × B6C3F1 mating and derives from the B6, C3H, or DBA Zdhhc14 alleles. (DNA is from a Ts65Dn trisomic pup.) Lane 3: in euploid offspring, both the B6 and C3H alleles result in SacI restriction digest products of 175-bp and 71-bp. Lane 4: Ts65Dn offspring have one copy of the 246-bp band from the T65Dn marker chromosome, plus the 175- and 71-bp bands from the two normal parental chromosomes. (B) The site of Zdhhc14 polymorphism relative to primers. Location of SNP and SacI restriction endonuclease site (blue) between primer sequences (red) utilized for the PCR and restriction digest protocol.
In total, 1174 offspring from Ts65Dn × B6C3F1 matings were genotyped using this PCR and restriction enzyme methodology. Of those, 716 were sexed and were comprised of 50.3% males and 49.7% females. At weaning, 32.7% of 892 offspring carried the distinguishing DBA/2J Zdhhc14 allele on the translocation chromosome. Both positive and negative PCR results from 603 pups were confirmed using FISH with an overall 3.1% false positive rate and a 5.2% false negative rate (Table 1).
Table 1.
FISH confirmation of PCR genotyped Ts65Dn offspring
| Agea | Status | n | FISH-positive | FISH-negative | False type |
|---|---|---|---|---|---|
| P0 | PCR-positive | 59 | 59 | 0 | 0% |
| P0 | PCR-negative | 93 | 4 | 89 | 4.3% |
| P5-P10 | PCR-positive | 168 | 159 | 7 | 4.2% |
| P5-P10 | PCR-negative | 119 | 7 | 112 | 5.9% |
| P21-P30 | PCR-positive | 70 | 68 | 2 | 2.9% |
| P21-P30 | PCR-negative | 153 | 8 | 145 | 5.2% |
Age, in post-natal (P) days
Our methodolgy is inexpensive, identifies trisomic mice with more than 90% concordance to FISH genotyping, can be done in most laboratories, and takes ~6 h to complete (instrument run times and incubations take the majority of this time). We have utilized this methodology to reduce the subsequent FISH analysis by 60–70%. For example, in a subset of 338 female mice from a breeding colony that were genotyped using the described PCR protocol, 101 were confirmed by FISH results. Additional negative mice were selected and FISH-genotyped to serve as controls. This corresponds to a concomitant reduction in time and cost of the subsequent genotyping, though the exact cost may vary depending on the laboratory. The methodology can only prescreen Ts65Dn offspring due to the small frequency of recombination events between the small centromeric end of Mmu17 on the marker chromosomes (originally derived from DBA/2J) and corresponding regions on the two full-length copies of Mmu17 (largely derived from the B6C3F1 males after generations of crossing onto this background). We have utilized this methodology in our colonies as a cost effective and efficient method to prescreen Ts65Dn mice for experimental procedures and continued mating, with subsequent FISH identification of the small marker chromosome.
Acknowledgments
We thank members of the Roper Lab and Yan Xiang of the Reeves Lab who performed PCR and FISH genotyping on Ts65Dn mice. This work was partially supported by a Research Support Funds Grant (to R.J.R.) from Indiana University-Purdue University Indianapolis and by The U.S. Public Health Service (PHS; grant no. R01 HD38384 to R.H.R.). This paper is subject to the NIH Public Access Policy.
Footnotes
Competing interests
The authors declare no competing interests.
Supplementary material for this article is available at www.BioTechniques.com/article/113342.
References
- 1.Christianson A, Howson CP, Modell B. March of Dimes Birth Defects Foundation. White Plains, NY: 2006. March of Dimes Global Report on Birth Defects: The Hidden Toll of Dying and Disabled Children; pp. 1–98. [Google Scholar]
- 2.Epstein CJ. Down Syndrome (Trisomy 21) In: Scriver CR, Beaudet AL, Sly WS, Valle D, editors. The Metabolic & Molecular Bases of Inherited Disease. New York: McGraw-Hill; 2001. pp. 1223–1256. [Google Scholar]
- 3.Van Cleve SN, Cohen WI. Part I: clinical practice guidelines for children with Down syndrome from birth to 12 years. J. Pediatr. Health Care. 2006;20:47–54. doi: 10.1016/j.pedhc.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 4.Hattori M, Fujiyama A, Taylor TD, Watanabe H, Yada T, Park HS, Toyoda A, Ishii K, et al. The DNA sequence of human chromosome 21. Nature. 2000;405:311–319. doi: 10.1038/35012518. [DOI] [PubMed] [Google Scholar]
- 5.Mural RJ, Adams MD, Myers EW, Smith HO, Miklos GL, Wides R, Halpern A, Li PW, et al. A comparison of whole-genome shotgun-derived mouse chromosome 16 and the human genome. Science. 2002;296:1661–1671. doi: 10.1126/science.1069193. [DOI] [PubMed] [Google Scholar]
- 6.Salehi A, Faizi M, Belichenko PV, Mobley WC. Using mouse models to explore genotype-phenotype relationship in Down syndrome. Ment. Retard. Dev. Disabil. Res. Rev. 2007;13:207–214. doi: 10.1002/mrdd.20164. [DOI] [PubMed] [Google Scholar]
- 7.Wiseman FK, Alford KA, Tybulewicz VL, Fisher EM. Down syndrome—recent progress and future prospects. Hum. Mol. Genet. 2009;18:R75–R83. doi: 10.1093/hmg/ddp010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davisson MT, Schmidt C, Reeves RH, Irving NG, Akeson EC, Harris BS, Bronson RT. Segmental trisomy as a mouse model for Down syndrome. Prog. Clin. Biol. Res. 1993;384:117–133. [PubMed] [Google Scholar]
- 9.Akeson EC, Lambert JP, Narayanswami S, Gardiner K, Bechtel LJ, Davisson MT. Ts65Dn—localization of the translocation breakpoint and trisomic gene content in a mouse model for Down syndrome. Cytogenet. Cell Genet. 2001;93:270–276. doi: 10.1159/000056997. [DOI] [PubMed] [Google Scholar]
- 10.Reeves RH, Irving NG, Moran TH, Wohn A, Kitt C, Sisodia SS, Schmidt C, Bronson RT, et al. A mouse model for Down syndrome exhibits learning and behaviour deficits. Nat. Genet. 1995;11:177–184. doi: 10.1038/ng1095-177. [DOI] [PubMed] [Google Scholar]
- 11.Delabar JM, Aflalo-Rattenbac R, Creau N. Developmental defects in trisomy 21 and mouse models. ScientificWorldJournal. 2006;6:1945–1964. doi: 10.1100/tsw.2006.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moore CS. Postnatal lethality and cardiac anomalies in the Ts65Dn Down syndrome mouse model. Mamm. Genome. 2006;17:1005–1012. doi: 10.1007/s00335-006-0032-8. [DOI] [PubMed] [Google Scholar]
- 13.Richtsmeier JT, Baxter LL, Reeves RH. Parallels of craniofacial maldevelopment in Down syndrome and Ts65Dn mice. Dev. Dyn. 2000;217:137–145. doi: 10.1002/(SICI)1097-0177(200002)217:2<137::AID-DVDY1>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 14.Roper RJ, St John HK, Philip J, Lawler A, Reeves RH. Perinatal loss of Ts65Dn Down syndrome mice. Genetics. 2006;172:437–443. doi: 10.1534/genetics.105.050898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baxter LL, Moran TH, Richtsmeier JT, Troncoso J, Reeves RH. Discovery and genetic localization of Down syndrome cerebellar phenotypes using the Ts65Dn mouse. Hum. Mol. Genet. 2000;9:195–202. doi: 10.1093/hmg/9.2.195. [DOI] [PubMed] [Google Scholar]
- 16.Reeves RH, Garner CC. A year of unprecedented progress in Down syndrome basic research. Ment. Retard. Dev. Disabil. Res. Rev. 2007;13:215–220. doi: 10.1002/mrdd.20165. [DOI] [PubMed] [Google Scholar]
- 17.Moore CS, Lee JS, Birren B, Stetten G, Baxter LL, Reeves RH. Integration of cytogenetic with recombinational and physical maps of mouse chromosome 16. Genomics. 1999;59:1–5. doi: 10.1006/geno.1999.5812. [DOI] [PubMed] [Google Scholar]
- 18.Hewitt CA, Carmichael CL, Wilkins EJ, Cannon PZ, Pritchard MA, Scott HS. Multiplex ligation-dependent probe amplification (MLPA) genotyping assay for mouse models of down syndrome. Front. Biosci. 2007;12:3010–3016. doi: 10.2741/2291. [DOI] [PubMed] [Google Scholar]
- 19.Isaksson M, Stenberg J, Dahl F, Thuresson AC, Bondeson ML, Nilsson M. MLGA—a rapid and cost-efficient assay for gene copy-number analysis. Nucleic Acids Res. 2007;35:e115. doi: 10.1093/nar/gkm651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu DP, Schmidt C, Billings T, Davisson MT. Quantitative PCR genotyping assay for the Ts65Dn mouse model of Down syndrome. BioTechniques. 2003;35:1170–1178. doi: 10.2144/03356st02. [DOI] [PubMed] [Google Scholar]
- 21.Ramakrishna N, Meeker C, Li S, Jenkins EC, Currie JR, Flory M, Lee B, Liu MS, et al. Polymerase chain reaction method to identify Down syndrome model segmentally trisomic mice. Anal. Biochem. 2005;340:213–219. doi: 10.1016/j.ab.2005.02.002. [DOI] [PubMed] [Google Scholar]