Abstract
Amino acid starvation by asparaginase (ASNase) enhances phosphorylation of eukaryotic initiation factor 2 (eIF2) by general control nonderepressible 2 (GCN2) kinase, leading to reduced global mRNA translation rates. This conserves energy and allows cells time to reprogram stress-related gene expression to alleviate cell injury. This study addressed the importance of GCN2 for the immune system to adapt to amino acid starvation by ASNase. GCN2+/+ and GCN2−/− mice were injected once daily with ASNase or saline for up to 7 d. In both thymus and spleen, activation of amino acid stress response genes to ASNase, such as asparagine synthetase and CAAT enhancer binding protein homologous protein, required GCN2. ASNase reduced food intake and body weight in both genotypes, but spleen and thymus wet weights and total cell numbers in thymus, spleen, bone marrow, and mesenteric lymph nodes were less in GCN2−/− mice treated with ASNase (genotype x ASNase, P < 0.05). In the thymus, GCN2−/− mice treated with ASNase demonstrated enhanced apoptosis and fewer cells in all subpopulations examined (CD3+, CD4–8-, CD4+8+, CD4+8-, CD4–8+) compared with GCN2+/+ mice treated with ASNase (genotype x ASNase, P < 0.05). In the spleen, GCN2 deletion magnified ASNase-induced reductions in CD4+ T cells, CD8+ T cells, CD19+ B cells, and CD11b+ leukocytes (genotype x ASNase, P < 0.05). These results indicate that loss of GCN2 enhances immunosuppression by ASNase and that this eIF2 kinase is broadly required for amino acid stress management in the immune system.
Introduction
Asparaginase (ASNase)7 is an effective treatment for acute lymphoblastic leukemia, the most common childhood cancer (1). ASNase catalyzes the hydrolysis of amino acid asparagine to aspartic acid and ammonia, exploiting an Achilles heel in the leukemic lymphoblast, low or absent levels of asparagine synthetase (ASNS) (2). The potency of the drug to induce cell death is a consequence of asparagine starvation and is strengthened by its intrinsic glutaminase activity, which lowers the levels of glutamine, an essential substrate in the synthesis of asparagine by ASNS (glutamine + aspartate → glutamate + asparagine) (3–5). Previous studies published by this laboratory and others demonstrated that the secondary glutaminase activity of ASNase is detrimental to the patient, activating amino acid stress responses in liver, pancreas, and spleen and resulting in hepatotoxicity and other complications (6, 7).
Amino acids are the building blocks of life. As such, their intracellular availability can directly affect the rate of protein synthesis. Deprivation of amino acids can also limit transfer RNA (tRNA) aminoacylation, i.e. the pairing of an amino acid to its cognate tRNA (a process also known as tRNA charging). Reductions in tRNA charging proximal to the ribosome are sensed by general control nonderepressible kinase 2 (GCN2), a protein kinase with a single known substrate, the α-subunit of eukaryotic initiation factor 2 (eIF2) (8, 9). The attachment of phosphate to eIF2 via GCN2 initiates a cascade of events that results in the reduction of general protein synthesis concomitant with the preferential translation of Activating Transcription Factor 4 (ATF4) mRNA (10). Subsequent binding of ATF4 protein to promoter regions of DNA functions to reconfigure gene expression to alleviate the particular amino acid stress. For example, ATF4 is a transcriptional activator of ASNS expression, which would lead to increased asparagine synthesis (11). However, if the amino acid stress is unrecoverable, the eIF2/ATF4 pathway then switches to one that promotes programmed cell death by a mechanism involving extended expression of the proapoptotic transcription factor C/EBP Homologous Protein (CHOP). This eIF2/ATF4 pathway can recognize and be activated by many different environmental and physiological stresses; hence, this pathway has been referred to as the integrated stress response (ISR) (12). Various forms of amino acid deprivation activate the ISR in vivo, including dietary leucine and threonine deprivation (13, 14), sulfur amino acid deficiency (15, 16), tryptophan catabolism by indoleamine 2, 3 dioxygenase (17), halofuginone treatment (18), aminopeptidase inhibitors (19), and ASNase (7, 20).
Previously, we and others reported that mice deleted for GCN2 are unable to effectively sense and manage dietary essential amino acid deprivation, resulting in altered food intake (14, 21) and increased mortality (13). Recent work from our laboratory demonstrates that ASNase acutely (≤8 h) activates the ISR in liver via GCN2 and not by one of the other protein kinases that phosphorylate the α-subunit of eIF2 (20). In this study, we addressed the role of GCN2 in the immunosuppression caused by ASNase. We hypothesized that loss of GCN2 function would render the immune system more sensitive to the cytotoxic effects of amino acid deprivation, leading to enhanced cell death of lymphocytes.
Experimental Procedures
Measurement of l-ASNase activity.
The activity of ASNase derived from Escherichia coli (Elspar, Merck & Co.) was determined by the Nesslerization technique, as previously described (7). During the course of these experiments, it was found that the enzyme activity of ASNase in solutions of PBS declined steadily over time, with little measureable enzyme activity after 48 h. This loss of activity was observed even when the enzyme was stored at 4°C. Freezing aliquots of ASNase in solution prevented total loss of enzyme activity, maintaining ∼50% upon thaw. To ensure appropriate ASNase doses over time, once the ASNase was dissolved in PBS, the entire volume was divided into small aliquots and immediately frozen at −80°C. The enzyme activity was then determined from a thawed aliquot before administration. Measurements from multiple thawed aliquots over time were determined to contain similar enzyme activities. As such, the enzyme activity of the first thawed aliquot was assigned to represent all frozen aliquots from the same batch. Once thawed, the entire contents of each aliquot were immediately administered and any remaining volume was discarded.
Mice.
The following study protocol was approved by the Institutional Care and Use Committee at the Indiana University School of Medicine-Evansville. Young adult (8–12 wk old) male and female C57BL/6J mice (also referred to as GCN2+/+) and GCN2 null mice [also referred to as GCN2−/−; backcrossed onto the C57BL/6J genetic background 8–10 generations (13)] bred in-house were maintained on a 12-h-light:-dark cycle and provided free access to commercial rodent unpurified diet consisting of 18% protein and 4% fat (7017 NIH-31 Open Formula Mouse/Rat Sterilizable Diet, Harlan Teklad) and tap water throughout the experiment. Mice were injected intraperitoneally with either ASNase (3 IU/g body weight) or an equivolume of PBS once daily for up to 7 d.
All mice were injected in the morning and were killed 6 h after injection on d 2, 3, 4, and 6, and 12 h after injection on d 7. The number of daily injections is based on the work of Durden et al. (22, 23), which demonstrates that 4–7 d of ASNase treatment results in maximal cytotoxicity and immunosuppression. Mice were decapitated and the spleen and thymus were dissected carefully, rinsed in ice-cold PBS, weighed, and then either frozen immediately in liquid nitrogen, transferred to ice-cold fluorescence-activated cell sorting (FACS) buffer (PBS with 1% fetal bovine serum and 0.05% sodium azide), or fixed in 4% paraformaldehyde. The bone marrow was isolated from the femur and tibia by flushing with a needle into FACS buffer on ice.
Cellular composition of bone marrow, spleens, and thymii.
Major cell types within spleens, thymii, and bone marrow were determined using flow cytometric analysis protocols as previously described (24, 25). Cell surface markers were detected on a FACScan flow cytometer (Becton Dickinson). Data were analyzed using CellQuest Pro software (Becton Dickinson), gating on 10,000 lymphocytes as defined by forward and side scatter. Markers of T cell development and function included CD3 [fluorescein isothiocyanate (FITC)-17A2], CD4 [phycoerythrin (PE)-GK1.5], and CD8 (FITC-53.6.72); B cell markers included CD19 (FITC-1D3), surface IgM (sIgM) (FITC-μ-chain, MP Biomedicals), and B220 (PE-RA3–6B2); and CD11b (PE-Mac-1αchain) was used as a marker of leukocytes in spleen. Isotype-matched antibodies against IgG2a (B39–4), IgG2b (A95–1), and IgM (R4–22) were used as controls. The percent positive cells for each subpopulation of the thymus and spleen were adjusted based on total cell numbers in that tissue and results are presented as 106 cells per organ. Bone marrow data are presented as percent positive cells for each subpopulation. Total cell count of each organ or tissue was determined using a Coulter Counter.
Immunoblot analysis.
Weighed, frozen tissues were transferred to microfuge tubes on ice and homogenized in 7 volumes of lysis buffer (13). The homogenates were immediately centrifuged at 10,000 × g for 10 min at 4°C for analysis of protein levels and phosphorylation state as previously described (13). Phosphorylation of eIF2 was assessed using an antibody that recognizes the α-subunit only when it is phosphorylated at serine 51 (Cell Signaling Technology) and normalized for total eIF2α (Santa Cruz Biotechnology). Phosphorylation of eIF 4E binding protein 1 and ribosomal protein S6 kinase 1 (S6K1) were measured using polyclonal antibodies (Bethyl). Caspase-3 cleavage was evaluated by expressing the amount of cleaved form as a ratio to the uncleaved protein, using polyclonal antibodies (Cell Signaling Technology). Blots were developed with enhanced chemiluminescence (Amersham Biosciences). Protein expression was analyzed using a Kodak 4000MM multimodal imager and band density was quantitated using Kodak Molecular Imaging Software (version 4.0.4).
RT and real-time PCR.
Total RNA was extracted from frozen tissue using TriReagent (Molecular Research Center) followed by DNase treatment (VersaGene DNase kit, Gentra Systems). To inactivate the reaction, the samples were heated to 70°C for 5 min. The A260:280 ratio was between 1.8 and 2.0 following RNA purification using the RNeasy mini kit (QIAGEN). mRNA levels were determined by quantitative PCR using TaqMan reagents. One microgram of RNA solutions was used for RT using a High-Capacity cDNA Reverse Transcription kit (Applied Biosystems) according to the manufacturer’s instructions. TaqMan Gene Expression Master Mix and TaqMan Gene Expression Assays (Applied Biosystems) were used for the PCR step. Amplification and detection were performed using the Step-OnePlus Real-Time PCR System (Applied Biosystems) (20). Each mRNA from a single sample was measured in triplicate. 18S ribosomal RNA was chosen as the normalization control gene. Results were obtained by the comparative Ct method and are expressed as fold of the experimental control (PBS-treated GCN2+/+).
Histology.
Tissues fixed in 4% paraformaldehyde were frozen and then sectioned (10 μm) using a cryostat. Terminal deoxynucleotidyl transferase mediated dUTP nick end labeling (TUNEL) assays were performed according to the manufacturer’s instructions (Trevigen TACS 2 TdT-Blue Label In Situ Apoptosis Detection kit, R&D Systems). TUNEL-positive cells were measured from 3 equal-sized areas per section. Digital images of selected areas captured at 200× were imported into Scion Image for Windows (Scion) and TUNEL-positive cells were manually marked and counted using the counting tool. Frozen sections were also stained with Oil Red O and counter stained with hematoxylin to visualize lipid content and tissue histology.
Statistics.
All data were analyzed by the STATISTICA statistical software package for the Macintosh, volume II (StatSoft). Data were analyzed for homogeneity of variance and data sets with unequal variance underwent log transformation before ANOVA. Data were analyzed using 2-way ANOVA to assess main and interaction effects, with genotype and ASNase treatment as independent variables. When a significant main or interaction effect was detected, Tukey’s post hoc analysis was used to identify the means that differ. The data presented are expressed as means ± SEM. The level of significance was set at P < 0.05 for all statistical tests.
Results
Food intake, body weight, tissue weights, and total cell numbers.
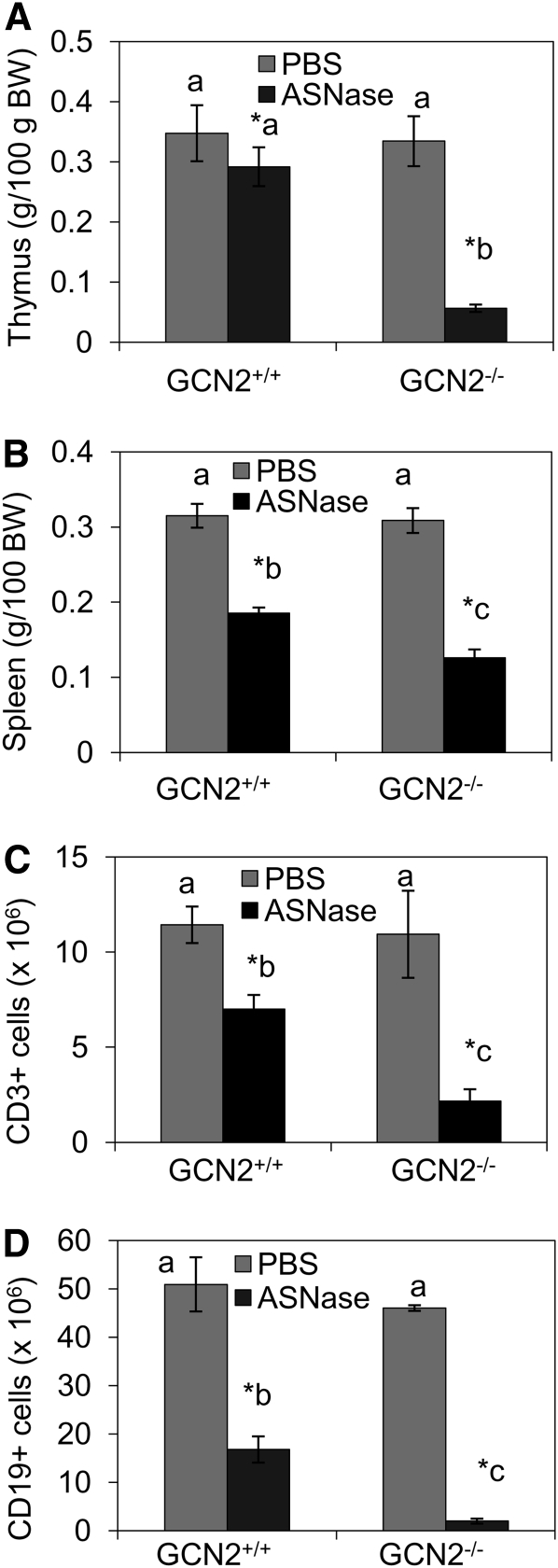
No mice died or were moribund during the course of this study. ASNase reduced food intake in both strains of mice (Table 1), resulting in loss of body weight on d 3, 4, and 6 (Table 1; Supplemental Table 1). A different response pattern was noted in the immune tissues. Following 3 daily injections of ASNase, GCN2 null mice possessed substantially smaller thymii compared with GCN2−/− PBS controls and GCN2+/+ mice injected with ASNase (genotype × ASNase, P < 0.02; Supplemental Table 1). By d 6, the wet weights of both spleen and thymus from GCN2−/− mice treated with ASNase were smaller than those from ASNase-treated GCN2+/+ mice (genotype × ASNase, P < 0.05; Fig. 1A,B; Supplemental Fig. 1). No other body organs in the GCN2−/− mice had this pattern of reduction in wet weight following administration of ASNase (data not shown).
TABLE 1.
Body weights of GCN2+/+ and GCN2#x2212/#x2212 mice administered ASNase at 3 IU/g body weight or equivolume PBS for 6 d1
| GCN2+/+ |
GCN2−/− |
P-value |
|||||
| PBS | ASNase | PBS | ASNase | Genotype | ASNase | G × A | |
| Body weight | |||||||
| Initial, g | 17.43 ± 1.26 | 18.18 ± 0.89 | 18.63 ± 0.90 | 18.66 ± 0.68 | ns2 | ns | ns |
| Final, g | 18.02 ± 1.37 | 17.37 ± 0.70 | 18.49 ± 1.02 | 16.08 ± 0.80 | ns | ns | ns |
| % change | 3.2 ± 1.1 | −4.2 ± 2.1* | −0.8 ± 1.4# | −14.2 ± 1.9#* | <0.001 | <0.001 | ns |
| Food intake, g/d | 4.4 ± 0.2 | 4.2 ± 0.2* | 4.6 ± 0.2 | 4.0 ± 0.3* | ns | 0.001 | ns |
Data are means ± SEM, = 6. #Different from corresponding GCN2+/+, P < 0.05; *different from corresponding PBS, P < 0.05.
ns, Not significant > 0.05.
FIGURE 1.
Changes in thymus (A) and spleen (B) wet weights and numbers of CD3+ thymocytes (C) and splenic CD19+ B cells (D) in GCN2+/+ and GCN2−/− mice administered ASNase at 3 IU/g body weight or PBS for 6 d. In C, numbers of CD3+ thymocytes are shown. In D, numbers of splenic CD19+ B cells are shown. Values are means ± SEM, n = 6. *Different from corresponding PBS, P < 0.05. When the genotype × ASNase interaction was significant, labeled means without a common letter differ, P < 0.05.
Measurements of cell loss were consistent with tissue weights. Specifically, ASNase reduced the total numbers of cells in the thymus (Table 2) and spleen (Table 3) in both strains of mice (P < 0.001). GCN2 deletion augmented cell losses by ASNase in thymus and spleen, reducing cell numbers to just 3.4 and 8.5% of PBS-injected GCN2−/− mice, respectively (genotype × ASNase, P < 0.05). Total numbers of CD3+ thymocytes and CD19+ splenic B cells were also reduced to a greater extent by the combination of ASNase and GCN2 deletion (genotype × ASNase, P < 0.05; Fig. 1C,D). Furthermore, ASNase more substantially reduced the total number of cells in the bone marrow (genotype × ASNase, P < 0.05; Table 4) and mesenteric lymph nodes (genotype × ASNase, P < 0.05; Supplemental ) in GCN2−/− mice. Thus, the functional absence of GCN2 enhanced cell losses by ASNase in all immune tissues studied.
TABLE 2.
Total cell counts and thymocyte subpopulations in GCN2+/+ and GCN2#x2212/#x2212 mice administered ASNase at 3 IU/g body weight or equivolume PBS for 6 d1
| GCN2+/+ |
GCN2−/− |
P-value |
|||||
| PBS | ASNase | PBS | ASNase | Genotype | ASNase | G × A | |
| cells x 106 | |||||||
| Total | 78.28 ± 9.49a | 41.23 ± 8.29*b | 69.68 ± 11.60 #a | 2.36 ± 0.66#*c | <0.001 | <0.001 | <0.001 |
| CD4-CD8- | 1.53 ± 0.20a | 1.50 ± 0.17*a | 1.32 ± 0.20#a | 0.25 ± 0.04#*b | <0.001 | <0.001 | <0.001 |
| CD4+CD8+ | 66.31 ± 8.72a | 33.18 ± 7.46*b | 57.66 ± 7.74#a | 0.10 ± 0.05#*c | <0.001 | <0.001 | <0.001 |
| CD4+CD8- | 8.08 ± 0.75a | 5.05 ± 0.65*b | 8.21 ± 1.86#a | 1.49 ± 0.45#*c | <0.001 | 0.003 | 0.007 |
| CD4-CD8+ | 2.47 ± 0.51a | 1.50 ± 0.14*b | 2.49 ± 0.65#a | 0.48 ± 0.14#*c | <0.001 | 0.005 | 0.006 |
Data are means ± SEM, = 6. #Different from corresponding GCN2+/+, P < 0.05; *different from corresponding PBS, P < 0.05. When the genotype × ASNase interaction was significant, labeled means without a common letter differ, P < 0.05.
TABLE 3.
Total cell counts and lymphocyte subpopulations in the spleen of GCN2+/+ and GCN2#x2212/#x2212 mice administered ASNase at 3 IU/g body weight or equivolume PBS for 6 d1
| GCN2+/+ |
GCN2−/− |
P-value |
|||||
| PBS | ASNase | PBS | ASNase | Genotype | ASNase | G × A | |
| cells x 106 | |||||||
| Total | 91.89 ± 8.78a | 43.48 ± 4.76*b | 84.69 ± 2.08#a | 7.23 ± 2.26#*c | <0.001 | <0.001 | 0.001 |
| CD4+ | 17.36 ± 1.82a | 10.10 ± 0.70*b | 17.07 ± 0.84#a | 2.97 ± 0.80#*c | <0.001 | <0.001 | 0.005 |
| CD8+ | 9.23 ± 1.33a | 5.48 ± 0.77*b | 8.28 ± 0.32#a | 0.79 ± 0.26#*c | <0.001 | <0.001 | 0.015 |
| CD4/CD8 | 1.88 ± 0.12b | 1.85 ± 0.12*b | 2.06 ± 0.08#b | 4.06 ± 0.32#*a | <0.001 | <0.001 | <0.001 |
| CD11b+ | 7.12 ± 0.98a | 3.72 ± 0.29*b | 4.13 ± 0.60#b | 0.24 ± 0.04#*c | <0.001 | <0.001 | <0.001 |
Data are means ± SEM, = 6. #Different from corresponding GCN2+/+, P < 0.05; *different from corresponding PBS, P < 0.05. When the genotype × ASNase interaction was significant, labeled means without a common letter differ, P < 0.05.
TABLE 4.
Total cell counts and B lymphocyte subpopulations in the bone marrow of GCN2+/+ and GCN2#x2212/#x2212 mice administered ASNase at 3 IU/g body weight or equivolume PBS for 6 d1
| GCN2+/+ |
GCN2−/− |
P-value |
|||||
| PBS | ASNase | PBS | ASNase | Genotype | ASNase | G × A | |
| cells x 106 | |||||||
| Total | 18.19 ± 1.74a | 16.35 ± 1.33*a | 15.85 ± 0.73#a | 5.08 ± 0.38*#b | <0.001 | <0.001 | 0.001 |
| B220- sIgM- | 8.19 ± 0.90a | 10.01 ± 1.00a | 7.79 ± 0.74a | 3.98 ± 0.29b | ns2 | ns | 0.002 |
| B220+ sIgM- | 5.26 ± 0.48a | 3.14 ± 0.45*b | 4.16 ± 0.60#a | 0.14 ± 0.02*#c | <0.001 | <0.001 | < 0.001 |
| B220+ sIgM+ | 4.62 ± 0.74a | 3.06 ± 0.60*b | 3.82 ± 0.15#a | 0.92 ± 0.10*#c | <0.001 | <0.001 | 0.003 |
Data are means ± SEM, = 6. #Different from corresponding GCN2+/+, P < 0.05; *different from corresponding PBS, P < 0.05. When the genotype × ASNase interaction was significant, labeled means without a common letter differ, P < 0.05; 2ns, Not significant, P > 0.05.
Initial experiments testing once daily injections of ASNase for 7 d included inactivated ASNase as the excipient control, which, importantly, did not negatively affect body weights, tissue weights, or cell counts following 7 daily injections (Supplemental Tables 2–4). Administration of low-dose ASNase (<2 IU/g body weight) modestly reduced body weight gain and wet weights of the spleen and thymus aftr 7 d (P < 0.03; Supplemental Table 2). Administration of low-dose ASNase to GCN2−/− mice treated with ASNase resulted in additional reductions in spleen and thymus weights (genotype × ASNase, P < 0.05) and cell numbers in the thymus (Supplemental Table 3) and spleen (Supplemental Table 4) (genotype × ASNase, P < 0.05). Thus, loss of GCN2 function rendered lower doses of ASNase more cytotoxic to lymphocytes in the thymus and spleen.
Mechanism of cell death in thymus.
To address the underlying basis for the severe reduction in thymic cell number in GCN2−/− mice by ASNase, the proportions of maturing thymocytes expressing the T cell surface markers CD4 (T helper cell) and/or CD8 (cytotoxic T lymphocyte) were examined (Table 2). In all subpopulations of maturing thymocytes, GCN2−/− mice treated with ASNase had substantially fewer numbers compared with GCN2+/+ mice treated with ASNase (genotype × ASNase, P < 0.05). However, the immature CD4+CD8+ subpopulation in GCN2−/− mice was especially affected, with over 99% reduction in cell number.
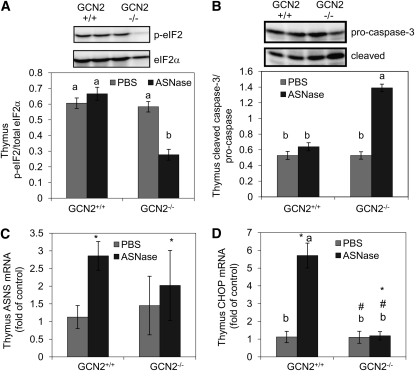
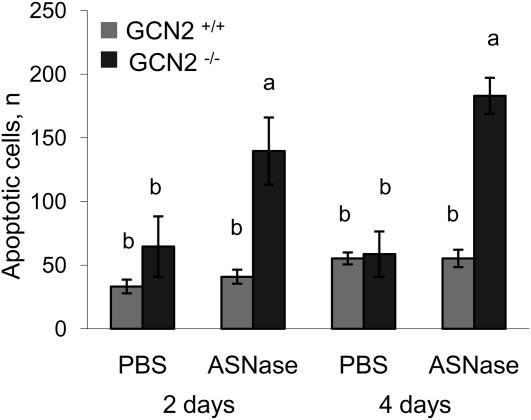
In the thymus of GCN2+/+ mice, the phosphorylation of eIF2 was not altered by ASNase at the time points measured (Fig. 2A). Nevertheless, both ASNS and CHOP mRNA were robustly induced by ASNase in GCN2+/+ mice (P < 0.05; Fig. 2B,C), whereas cleavage of caspase-3, a marker of apoptosis, was not (Fig. 2D). In contrast, the phosphorylation of eIF2 in the thymus of GCN2−/− mice was reduced (genotype × ASNase, P < 0.05), whereas ASNS and CHOP mRNA expression remained unchanged. Additionally, caspase-3 cleavage was increased (genotype × ASNase, P < 0.05), suggesting increased apoptosis. To confirm this, a TUNEL assay performed on frozen sections of thymus, which showed an increased number of apoptotic cells in GCN2−/− mice administered ASNase at 2 and 4 d following treatment (genotype × ASNase, P < 0.05; Fig. 3; Supplemental Fig. 2A). Frozen sections of thymus from GCN2−/− mice treated with ASNase for 4 d also demonstrated increased abundance of neutral lipids not detected in GCN2+/+ mice treated with ASNase for the same number of days (Supplemental Fig. 2B). Fatty infiltration of the atrophic thymus is reported under conditions of severe malnutrition and stress, but the mechanism is unknown (26). These results support the idea that GCN2 enhances the expression of key ISR genes that are required to alleviate amino acid stress in the thymus. The inability of GCN2−/− mice to invoke the ISR upon ASNase treatment reduces cell survival of these immune cells.
FIGURE 2.
Western blots (A,B, upper panel) and mean values (A,B, lower panel) for protein expression levels of eIF2 phosphorylation (A) and caspase-3 cleavage (B) and mRNA expression levels of ASNS (C) and CHOP (D) in the thymus of GCN2+/+ and GCN2−/− mice administered ASNase at 3 IU/g body weight or PBS for 6 d. Values are means ± SEM, n = 6. #Different from corresponding GCN2+/+, P < 0.05; *different from corresponding PBS, P < 0.05. When the genotype × ASNase interaction was significant, labeled means without a common letter differ, P < 0.05.
FIGURE 3.
Frozen sections of thymus evaluated for DNA fragmentation by TUNEL assay in GCN2+/+ and GCN2−/− mice administered ASNase at 3 IU/g body weight or PBS for 2 and 4 d. Values are means ± SEM, n = 6. When the effects of the genotype × ASNase interaction was significant, labeled means without a common letter differ, P < 0.05.
The mammalian target of rapamycin complex 1 (mTORC1) signaling pathway was also assessed by examining the phosphorylation states of S6K1 and eIF 4E binding protein 1. (Supplemental Fig. 4A,B). ASNase by itself did not dampen mTORC1 signaling in thymus. However, GCN2−/− mice treated with ASNase demonstrated reduced phosphorylation of both proteins (genotype × ASNase, P < 0.05).
Mechanism of toxicity in the spleen and bone marrow.
Injection of ASNase into GCN2−/− mice resulted in a considerable decline in both CD4+ and CD8+ cells, exceeding that in GCN2+/+ mice treated with ASNase (genotype × ASNase, P < 0.05) (Table 3). ASNase also affected the CD4:CD8 ratio in the spleen of GCN2−/− mice, suggesting that fewer CD8+ T cells survived relative to CD4+ T cells (genotype × ASNase, P = 0.05) (Table 3). Furthermore, the proportion of cells expressing CD11b, a marker found on granulocytes, monocytes, natural killer cells, and subsets of T and B cells, was also substantially depleted in the spleens of GCN2−/− mice treated with ASNase (genotype × ASNase, P < 0.05), suggesting broad immunosuppression as opposed to 1 or 2 specifically targeted immune cell subpopulations.
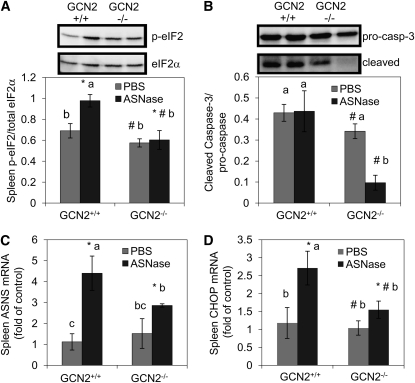
To understand the biochemical mechanism behind the changes in spleen, key markers of the ISR and mTORC1 pathways were again examined. ASNase-induced activation of the ISR in spleen was reflected by increased eIF2 phosphorylation and mRNA expression of ASNS and CHOP (P < 0.05; ). These events were absent in the spleens of GCN2−/− mice administered ASNase (genotype × ASNase, P < 0.05). Cleavage of caspase-3 was unchanged in the spleens from GCN2+/+ mice treated with ASNase, similar to the thymus. On the other hand, caspase-3 cleavage and both expression and phosphorylation of S6K1 were reduced in the spleens of ASNase-treated GCN2−/− mice (genotype × ASNase, P < 0.05) (Fig. 4B; Supplemental Fig. 4C,D). The reduction in caspase-3 cleavage may suggest differential activation of apoptotic pathways between tissues or could simply be a reflection of the very small numbers of remaining splenic lymphocyte populations (Fig. 1D; Table 3).
FIGURE 4.
Western blots and mean values of eIF2 phosphorylation (A), caspase-3 cleavage (B), ASNS mRNA expression (C), and CHOP mRNA expression (D) in the spleen of GCN2+/+ and GCN2−/− mice administered ASNase at 3 IU/g body weight or PBS for 6 d. Values are means ± SEM, n = 6. #Different from corresponding GCN2+/+, P < 0.05; * different from corresponding PBS, P < 0.05. When the genotype × ASNase interaction was significant, labeled means without a common letter differ, P < 0.05.
In the bone marrow, the cell surface markers B220 and sIgM were used to delineate preferential differentiation of bone marrow precursors into B lymphocytes, with B220 expression signifying lineage commitment to B cells and additional sIgM expression representing mature B cells. Gated cells not expressing either marker would also preferentially, but not exclusively, represent early pro-B cells. ASNase reduced B220+sIgM- and B220+sIgM+ cell populations in GCN2+/+ mice to 60 and 66% of PBS-injected GCN2+/+ mice, respectively (P < 0.05) while not altering the double negative B220-sIgM- population (Table 4). ASNase in GCN2−/− mice caused further reductions in total cell numbers and all bone marrow subpopulations, reducing the B220+sIgM- and B220+sIgM+ cell populations to 3.4 and 24% of PBS-injected GCN2−/− mice, respectively (genotype × ASNase, P < 0.05).
Discussion
In this study, we found that the ability of ASNase to reduce immune cell populations in the spleen, thymus, and bone marrow of mice is broadly augmented in the absence of GCN2. As a consequence, loss of GCN2 enhances immunosuppression by ASNase, emphasizing the essential role for GCN2 in the ability of the immune system to adapt to amino acid stress. This is the first report to our knowledge to document an essential role for GCN2 in the intact immune system, revealing a broader importance than that previously described in T cells (17). This work also emphasizes that the amino acid deprivation response is important to study with respect to ASNase and similar drugs used to treat cancer. Previously, we reported that GCN2, but not another eIF2 kinase called PERK, mediated the effects of ASNase on the ISR (20). These data, along with the current results, collectively establish ASNase as a valid model of amino acid deprivation. These findings also have important implications for development of anticancer therapies that rely on nutritional stress for targeted cell death.
In response to amino acid deprivation, GCN2 phosphorylates eIF2 to lower general protein synthesis while increasing the translation of a select group of mRNA. Reduced protein synthesis conserves energy, while preferential translation of the transcriptional activator ATF4 activates stress-related genes in the ISR, which function to reestablish amino acid homeostasis. Failure of ISR to alleviate cell injury, either because the nutrient stress is too intense and/or prolonged or because the ISR machinery is defective, can drive cells toward apoptosis.
GCN2 is required for adaptation to dietary amino acid starvation (13, 15) and for ISR activation by ASNase in liver (7, 20). This in combination with our findings that ASNase increases the ISR in spleen led to the hypothesis that GCN2 plays a vital role in the survival of immune cells when challenged with ASNase. Indeed, the current data shows that GCN2 is critically important for leukocytes and especially for lymphocytes to survive amino acid deprivation by ASNase. Loss of GCN2 blocks induction of the ISR to ASNase, eliciting cell death of immune cells but not causing overall morbidity as previously reported in leucine deprivation (13). This is because whereas a leucine-devoid diet starves for an amino acid that cannot be made de novo in any tissue, ASNase depletes amino acids that can be synthesized in the body. Thus, it is those tissues that are most sensitive to loss of an exogenous supply of asparagine and/or glutamine that rely on GCN2-activated amino acid stress responses. These results establish that GCN2 is central for cell survival in the immune system in response to amino acid stress by ASNase, influencing the survival of both B and T lymphocytes as well as other leukocytes and perhaps even early hematopoietic progenitor cells.
Several reports have suggested an important role for GCN2 in T helper cell proliferation and differentiation (17, 18, 27). Activation of the enzyme indoleamine 2,3-dioxygenase (IDO), produced by immunoregulatory dendritic cells, inhibits activation of responding T cells, favoring the induction of T regulatory cells. The generation of tolerogenic properties by IDO is associated with GCN2 activation of the ISR, which is activated in response to IDO-mediated tryptophan catabolism (17). Recently, it was also discovered that treatment of CD4+ T helper cells with halifugnone (a drug used to treat fibrotic diseases such as scleroderma) inhibits TH17 cell differentiation in association with activation of the GCN2-ATF4 ISR (18). The current study bolsters the idea that GCN2 is vital to the T cell and extends this concept to all immune cell populations. Indeed, the current data show that when faced with amino acid deprivation by ASNase, all immune cell populations from GCN2−/− mice demonstrate enhanced cell death. Thus, the role of GCN2 in immunity is much broader than regulating T cell anergy.
Lymphocytes, when differentiating or proliferating, are exquisitely sensitive to amino acid supply (28, 29). Early literature reports concluded that glutamine is essential for both lymphoblastic transformation and plasma cell differentiation (30, 31). In addition, many immune cells require an exogenous supply of glutamine as an energy source and precursor for DNA synthesis (32). This creates a conundrum with respect to ASNase chemotherapy, because although glutamine hydrolysis augments asparagine depletion to enhance the killing of leukemic lymphoblasts, it also starves normal, healthy leukocytes of an essential nutrient, leading to a generalized immunosuppression that can increase patient morbidity and mortality (33). Previous work testing a glutaminase-free form of ASNase that retains antilymphoma activity demonstrated reduced phosphorylation of eIF2 in the spleen of healthy mice (7). We speculate that GCN2 inhibition in combination with this drug could further improve tumor killing. Considering that chemical and small molecule inhibitors of GCN2 are currently being developed (34), the testing of this combination in anticancer regimens is promising.
It is also noted that the T and B lymphocyte subpopulations most negatively affected were those at an early stage of differentiation. Lymphocyte development is a process in which proliferation is coupled to differentiation. To undergo efficient proliferation, lymphocytes must coordinate entry into the cell cycle with increased metabolism (35). This is consistent with the idea that amino acid deprivation would be particularly stressful during lymphocyte development. Phosphorylation of eIF2 allows the cell to coordinate rates of protein synthesis with nutrient and energy stores to survive nutrient-limiting conditions during cell differentiation (9). Thus, loss of this checkpoint would be disadvantageous for the cell at a most critical point in its life cycle. Additional loss of ATF4 expression and the ISR would further compromise the ability of cells to reprogram the transcriptome that is required to reestablish metabolic homeostasis. Considering the reported defects in hematopoiesis identified in the ATF4 knockout mouse (36), it is possible that the GCN2-directed ISR is vital for managing amino acid stress across all leukocyte lineages.
Based on the results of this study, we suggest that targeted inhibition of GCN2 may serve to enhance the efficacy of current formulations of ASNase and other anticancer drugs that involve deprivation of amino acids [e.g. aminopeptidase inhibitors (19)]. Considering that the absence of GCN2 increased cell losses following lower doses of ASNase (Supplemental Tables 2–4), we predict that treatment with a GCN2 inhibitor targeted to lymphocytes would render a hyperproliferating lymphocyte much more sensitive to lower doses of ASNase. This approach could reduce overall cytotoxicity and complications associated with ASNase, currently a major problem in older patients. Support for this idea is found in the recent report demonstrating that inhibition of ATF4 expression blocks proliferation and survival of transformed cells, and abrogation of GCN2 or ATF4 expression significantly inhibits tumor growth in mice (37).
Supplementary Material
Acknowledgments
We thank Diana Fuqua and Debbie Wagner for superior animal care and maintenance of the mouse colonies and Gary White for his advice concerning the sectioning and staining of the thymus. T.G.A. and C.J.A. designed the research; P.B. and J.K.C. conducted the research with assistance from R.R., C.J.A. and T.G.A.; P.B. and T.G.A. wrote the paper with scientific counsel and support from C.J.A. and R.C.W; and T.G.A. had primary responsibility for final content. All authors read and approved the final manuscript.
Footnotes
Supported by the American Institute for Cancer Research (T.G.A.), Indiana University School of Medicine (T.G.A.), and the NIH (R01 GM49164; R.C.W.).
Supplemental Tables 1–4 and are available with the online posting of this paper at jn.nutrition.org.
ASNase, asparaginase; ASNS, asparagine synthetase; ATF4, activating transcription factor 4; CHOP, CAAT enhancer binding protein homologous protein; eIF2, eukaryotic initiation factor 2; FACS, fluorescence-activated cell sorting; FITC, fluorescein isothiocyanate; GCN2, general control nonderepressible 2; IDO, indoleamine 2,3-dioxygenase; ISR, integrated stress response; mTORC1, mammalian target of rapamycin complex 1; PE, phycoerythrin; S6K1, ribosomal protein S6 kinase 1; sIgM, surface immunoglobin M; tRNA, transfer RNA; TUNEL, terminal deoxynucleotidyl transferase mediated dUTP nick end labeling.
Literature Cited
- 1.Pui CH, Evans WE. Treatment of acute lymphoblastic leukemia. N Engl J Med. 2006;354:166–78 [DOI] [PubMed] [Google Scholar]
- 2.Leslie M, Case MC, Hall AG, Coulthard SA. Expression levels of asparagine synthetase in blasts from children and adults with acute lymphoblastic leukaemia. Br J Haematol. 2006;132:740–2 [DOI] [PubMed] [Google Scholar]
- 3.Panosyan EH, Grigoryan RS, Avramis IA, Seibel NL, Gaynon PS, Siegel SE, Fingert HJ, Avramis VI. Deamination of glutamine is a prerequisite for optimal asparagine deamination by asparaginases in vivo (CCG-1961). Anticancer Res. 2004;24:1121–5 [PubMed] [Google Scholar]
- 4.Rotoli BM, Uggeri J, Dall'Asta V, Visigalli R, Barilli A, Gatti R, Orlandini G, Gazzola GC, Bussolati O. Inhibition of glutamine synthetase triggers apoptosis in asparaginase-resistant cells. Cell Physiol Biochem. 2005;15:281–92 [DOI] [PubMed] [Google Scholar]
- 5.Cory JG, Cory AH. Critical roles of glutamine as nitrogen donors in purine and pyrimidine nucleotide synthesis: asparaginase treatment in childhood acute lymphoblastic leukemia. In Vivo. 2006;20:587–9 [PubMed] [Google Scholar]
- 6.Ollenschlager G, Roth E, Linkesch W, Jansen S, Simmel A, Modder B. Asparaginase-induced derangements of glutamine metabolism: the pathogenetic basis for some drug-related side-effects. Eur J Clin Invest. 1988;18:512–6 [DOI] [PubMed] [Google Scholar]
- 7.Reinert RB, Oberle LM, Wek SA, Bunpo P, Wang XP, Mileva I, Goodwin LO, Aldrich CJ, Durden DL, et al. Role of glutamine depletion in directing tissue-specific nutrient stress responses to L-asparaginase. J Biol Chem. 2006;281:31222–33 [DOI] [PubMed] [Google Scholar]
- 8.Kimball S, Anthony T, Cavener D, Jefferson L. Nutrient signaling through mammalian GCN2. : Winderickx P, Taylor J, Topics in current genetics: nutrient-induced responses in eukaryotic cells. Berlin and Heidelberg: Springer-Verlag; 2004. p. 113–30 [Google Scholar]
- 9.Wek RC, Jiang HY, Anthony TG. Coping with stress: eIF2 kinases and translational control. Biochem Soc Trans. 2006;34:7–11 [DOI] [PubMed] [Google Scholar]
- 10.Kilberg MS, Shan J, Su N. ATF4-dependent transcription mediates signaling of amino acid limitation. Trends Endocrinol Metab. 2009;20:436–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siu F, Bain PJ, LeBlanc-Chaffin R, Chen H, Kilberg MS. ATF4 is a mediator of the nutrient-sensing response pathway that activates the human asparagine synthetase gene. J Biol Chem. 2002;277:24120–7 [DOI] [PubMed] [Google Scholar]
- 12.Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell. 2003;11:619–33 [DOI] [PubMed] [Google Scholar]
- 13.Anthony TG, McDaniel BJ, Byerley RL, McGrath BC, Cavener DR, McNurlan MA, Wek RC. Preservation of liver protein synthesis during dietary leucine deprivation occurs at the expense of skeletal muscle mass in mice deleted for eIF2 kinase GCN2. J Biol Chem. 2004;279:36553–61 [DOI] [PubMed] [Google Scholar]
- 14.Hao S, Sharp JW, Ross-Inta CM, McDaniel BJ, Anthony TG, Wek RC, Cavener DR, McGrath BC, Rudell JB, et al. Uncharged tRNA and sensing of amino acid deficiency in mammalian piriform cortex. Science. 2005;307:1776–8 [DOI] [PubMed] [Google Scholar]
- 15.Sikalidis AK, Stipanuk MH. Growing rats respond to a sulfur amino acid-deficient diet by phosphorylation of the alpha subunit of eukaryotic initiation factor 2 heterotrimeric complex and induction of adaptive components of the integrated stress response. J Nutr. 2010;140:1080–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee JI, Dominy JE, Jr, Sikalidis AK, Hirschberger LL, Wang W, Stipanuk MH. HepG2/C3A cells respond to cysteine deprivation by induction of the amino acid deprivation/integrated stress response pathway. Physiol Genomics. 2008;33:218–29 [DOI] [PubMed] [Google Scholar]
- 17.Munn DH, Sharma MD, Baban B, Harding HP, Zhang Y, Ron D, Mellor AL. GCN2 kinase in T cells mediates proliferative arrest and anergy induction in response to indoleamine 2,3-dioxygenase. Immunity. 2005;22:633–42 [DOI] [PubMed] [Google Scholar]
- 18.Sundrud MS, Koralov SB, Feuerer M, Calado DP, Kozhaya AE, Rhule-Smith A, Lefebvre RE, Unutmaz D, Mazitschek R, et al. Halofuginone inhibits TH17 cell differentiation by activating the amino acid starvation response. Science. 2009;324:1334–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krige D, Needham LA, Bawden LJ, Flores N, Farmer H, Miles LE, Stone E, Callaghan J, Chandler S, et al. CHR-2797: an antiproliferative aminopeptidase inhibitor that leads to amino acid deprivation in human leukemic cells. Cancer Res. 2008;68:6669–79 [DOI] [PubMed] [Google Scholar]
- 20.Bunpo P, Dudley A, Cundiff JK, Cavener DR, Wek RC, Anthony TG. GCN2 protein kinase is required to activate amino acid deprivation responses in mice treated with the anti-cancer agent L-asparaginase. J Biol Chem. 2009;284:32742–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maurin AC, Jousse C, Averous J, Parry L, Bruhat A, Cherasse Y, Zeng H, Zhang Y, Harding HP, et al. The GCN2 kinase biases feeding behavior to maintain amino acid homeostasis in omnivores. Cell Metab. 2005;1:273–7 [DOI] [PubMed] [Google Scholar]
- 22.Distasio JA, Durden DL, Paul RD, Nadji M. Alteration in spleen lymphoid populations associated with specific amino acid depletion during L-asparaginase treatment. Cancer Res. 1982;42:252–8 [PubMed] [Google Scholar]
- 23.Durden DL, Salazar AM, Distasio JA. Kinetic analysis of hepatotoxicity associated with antineoplastic asparaginases. Cancer Res. 1983;43:1602–5 [PubMed] [Google Scholar]
- 24.Coligan J, Kruisbeek A, Margulies D, Shevach E, Strober W, Current protocols in immunology. Hoboken (NJ): John Wiley and Sons, Inc.; 2006 [Google Scholar]
- 25.Bunpo P, Murray B, Cundiff J, Brizius E, Aldrich CJ, Anthony TG. Alanyl-glutamine consumption modifies the suppressive effect of L-asparaginase on lymphocyte populations in mice. J Nutr. 2008;138:338–43 [DOI] [PubMed] [Google Scholar]
- 26.Prentice AM. The thymus: a barometer of malnutrition. Br J Nutr. 1999;81:345–7 [PubMed] [Google Scholar]
- 27.Brenk M, Scheler M, Koch S, Neumann J, Takikawa O, Hacker G, Bieber T, von Bubnoff D. Tryptophan deprivation induces inhibitory receptors ILT3 and ILT4 on dendritic cells favoring the induction of human CD4+CD25+ Foxp3+ T regulatory cells. J Immunol. 2009;183:145–54 [DOI] [PubMed] [Google Scholar]
- 28.Yaqoob P, Calder PC. Glutamine requirement of proliferating T lymphocytes. Nutrition. 1997;13:646–51 [DOI] [PubMed] [Google Scholar]
- 29.Newsholme EA, Crabtree B, Ardawi MS. Glutamine metabolism in lymphocytes: its biochemical, physiological and clinical importance. Q J Exp Physiol. 1985;70:473–89 [DOI] [PubMed] [Google Scholar]
- 30.Crawford J, Cohen HJ. The essential role of L-glutamine in lymphocyte differentiation in vitro. J Cell Physiol. 1985;124:275–82 [DOI] [PubMed] [Google Scholar]
- 31.Kitoh T, Asai S, Akiyama Y, Kubota M, Mikawa H. The inhibition of lymphocyte blastogenesis by asparaginase: critical role of glutamine in both T and B lymphocyte transformation. Acta Paediatr Jpn. 1992;34:579–83 [DOI] [PubMed] [Google Scholar]
- 32.Wu GY, Field CJ, Marliss EB. Glutamine and glucose metabolism in rat splenocytes and mesenteric lymph node lymphocytes. Am J Physiol. 1991;260:E141–7 [DOI] [PubMed] [Google Scholar]
- 33.Kafkewitz D, Bendich A. Enzyme-induced asparagine and glutamine depletion and immune system function. Am J Clin Nutr. 1983;37:1025–30 [DOI] [PubMed] [Google Scholar]
- 34.Robert F, Williams C, Yan Y, Donohue E, Cencic R, Burley SK, Pelletier J. Blocking UV-induced eIF2alpha phosphorylation with small molecule inhibitors of GCN2. Chem Biol Drug Des. 2009;74:57–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aifantis I, Sawai C. Energy addiction and lymphocyte differentiation: a new role for the liver kinase B1 kinase. Eur J Immunol. 2010;40:19–21 [DOI] [PubMed] [Google Scholar]
- 36.Masuoka HC, Townes TM. Targeted disruption of the activating transcription factor 4 gene results in severe fetal anemia in mice. Blood. 2002;99:736–45 [DOI] [PubMed] [Google Scholar]
- 37.Ye J, Kumanova M, Hart LS, Sloane K, Zhang H, De Panis DN, Bobrovnikova-Marjon E, Diehl JA, Ron D, et al. The GCN2–ATF4 pathway is critical for tumour cell survival and proliferation in response to nutrient deprivation. EMBO J. 2010;29:2082–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.