Abstract
Botulinum neurotoxins (BoNTs) are the etiological agents responsible for botulism, a disease characterized by peripheral neuromuscular blockade and a characteristic flaccid paralysis of humans. BoNTs are the most lethal known poisons affecting humans and have been recognized as a potential bioterrorist threat. Current treatments for botulinum poisoning are predominately prophylactic in nature relying on passive immunization with antitoxins. Inhibition of the BoNT light chain metalloprotease (LC) has emerged as a new therapeutic strategy for the treatment of botulism that may provide an effective postexposure remedy. A high-throughput screening effort against the light chain of BoNT serotype A (LC/A) was conducted with the Johns Hopkins Clinical Compound Library composed of over 1,500 existing drugs. Lomofungin, a natural product first isolated in the late 1960s, was identified as an inhibitor of LC/A, displaying classical noncompetitive inhibition kinetics with a Ki of 6.7 ± 0.7 μM. The inhibition profile of lomofungin has been delineated by the use of both an active site inhibitor, 2,4-dichlorocinnamic hydroxamate, and a noncompetitive inhibitor d-chicoric acid. The inhibitor combination studies reveal that lomofungin binding is nonmutually exclusive (synergistic) with both inhibitors; the mechanistic implications of these observations are discussed. Lastly, cellular efficacy was investigated using a rat primary cell model which demonstrated that lomofungin can protect against SNAP-25 cleavage, the intracellular protein target of LC/A.
Keywords: Botulinum neurotoxin, zinc-dependent metalloprotease, high-throughput screening, small molecule inhibitor, natural product
The naturally occurring botulinum neurotoxins (BoNTs) are the causative agents of botulism, a potentially fatal food poisoning disease characterized by peripheral neuromuscular blockade and progressive flaccid paralysis. Seven antigenically distinct serotypes of the neurotoxin (A−G) are produced and secreted by the Gram-positive, anaerobic, spore-forming bacillus Clostridium botulinum, C. butyricum, C. baratii, and C. argentinense.1 BoNTs are the most poisonous substances known, with serotype A having a lethal dose for a 70 kg human of approximately 0.09−0.15 μg intravenously or intramuscularly and 0.7−0.9 μg inhalationally.2
Despite their potentially lethal toxicity, BoNTs have emerged as an extremely valuable therapeutic tool for the treatment of a variety of maladies, including strabismus, migraines, and even facial wrinkles.3,4 However, the potential use of BoNT in a bioterrorist attack remains imminent and the Centers for Disease Control and Prevention (CDC) now classifies this agent as “Category A”, placing it among the six highest-priority agents. One recent case study has predicted that malicious release of botulinum into the public’s food supply in an attack of bioterrorism could cause mass casualties if an efficient treatment is not readily available.5
BoNTs are synthesized as ∼150 kDa single-chain protoxins that are post-translationally activated by proteolytic cleavage to form mature dichain proteins consisting of a 100 kDa heavy chain (HC) and a 50 kDa light chain (LC) linked by a disulfide bond.6 The HC is responsible for the neurospecific binding, uptake, and translocation of the LC into the cytosol of neuronal cells. The LC is a Zn2+-dependent metalloprotease that cleaves one of three intracellular soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) proteins: syntaxin, vesicle-associated membrane protein (VAMP)/synaptobrevin, or synaptosomal-associated protein of 25 kDa (SNAP-25) depending on the serotype. As a consequence of protein cleavage, release of acetylcholine at the neuromuscular junction is inhibited resulting in the loss of neurotransmission.
Current treatment for botulinum poisoning relies on passive immunization along with supportive care. Equine trivalent antitoxins or human-derived immunoglobulins (BIG-IV) can be administered as a postexposure prophylaxis.2,7 However, while antibody therapy may be broadly effective, supplies are limited and use of equine antibodies can cause adverse side effects.8 Most importantly, antibodies are only capable of sequestering free circulating BoNT and as a result become ineffective once toxin has begun entry into the cell. Consequently, diagnosis and treatment must begin promptly after toxin exposure, approximately 24 h or less. Therefore, treatments that can successfully target BoNTs intracellularly and thereby efficiently alleviate botulinum symptoms postexposure are of great importance.
The LC cleavage of intracellular SNARE proteins inhibits fusion of synaptic vesicles, preventing acetylcholine exocytosis leading to cell intoxication. Small molecule inhibitors of the LC may provide an opportunity for development of both pre- and postexposure therapeutics. In recent years, LC of serotype A (LC/A) has been the major focus, primarily due to its potency and long duration of paralysis.9 A number of competitive inhibitors of LC/A have been reported, the most potent of which have Ki values of 0.16−12.3 μM,10−31 several of which coordinate the active site zinc cation required for catalysis.13−31 More recently, the natural product d-chicoric acid was discovered in our laboratory that putatively binds to an exosite region outside the LC/A active site, displaying noncompetitive partial inhibition.17 Nonmutually exclusive inhibition was observed with d-chicoric acid in combination with a known potent competitive active site inhibitor and demonstrated more than additive (synergistic) enzyme inhibition,17 validating that a cocktail of inhibitors with different mechanisms of inhibition may prove to be a very effective treatment for botulism.
While some success has been achieved in finding LC/A inhibitors, huge breakthroughs have yet to be realized. The enabling discoveries of inhibitors such as those cited above reflect contemporary thinking, however, emergence of more drug like molecules is needed. As a new avenue, the Johns Hopkins Clinical Compound Library (JHCCL) composed of both U.S. FDA and foreign approved drugs was analyzed against LC/A in search of novel inhibitors.18 The library was screened using a high-throughput FRET-based assay in a 96-well format.19 Initially, compounds were evaluated at a single concentration of 50 μM, followed by confirmation through dose-dependent inhibition. After chemical structure analysis, 4 potential inhibitors were moved forward.
The FRET-based assay serves as a high-throughput system, allowing for rapid and efficient screening of thousands of compounds. However, the FRET substrate SNAPtide (13 amino acids) is a very short truncated version of LC/A’s physiological substrate SNAP-25 (206 amino acids). SNAP-25 wraps around the majority of the protease’s circumference making an extensive network of protein−protein interactions important for substrate specificity and catalytic efficiency.20 Maximal catalytic efficiency of LC/A requires an optimal portion of SNAP-25 consisting of 57 amino acids (146−202).21 Consequently, validation of LC/A inhibitors and elucidation of their detailed kinetic mechanisms often require a more reliable and accurate assay. Therefore, the four candidates displaying LC/A inhibition in the FRET-based assay were further investigated using an LC MS assay with an optimized 66-mer substrate (141−206) encompassing the key recognition elements of SNAP-25.14 Upon follow-up investigations only one compound lomofungin (Figure 1) displayed significant inhibition.
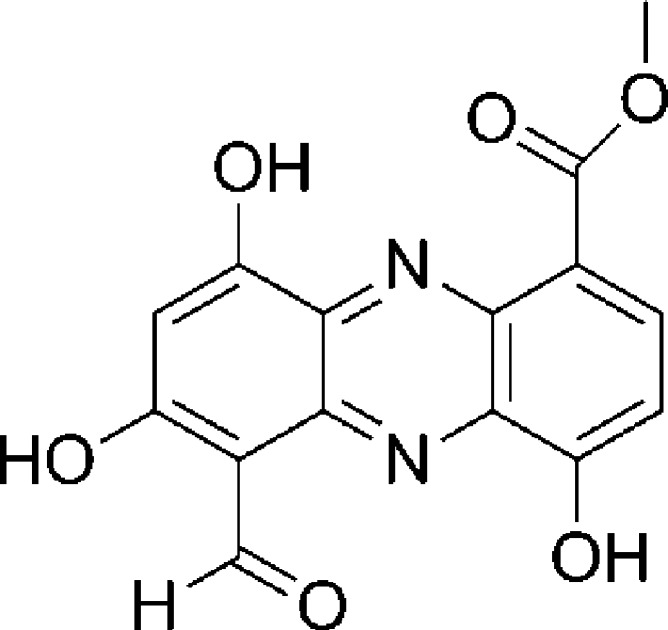
Figure 1.
The chemical structure of lomofungin.
Lomofungin, a secondary metabolite, was first isolated from the soil-dwelling Gram-positive bacteria Streptomyces lomodensis in the late 1960s.22 Lomofungin possesses a broad range of biological activities including antibacterial and antifungal properties. This natural product was shown to be effective against yeast, fungi, and both Gram-positive and Gram-negative bacteria.22 Lomofungin or 1-carbomethoxy-5-formyl-4,6,8-trihydroxyphenazine is a member of the phenazine class of natural products and contains five substituents appended upon a three-ring phenazine core (Figure 1).23
To elucidate the kinetic mechanism of LC/A inhibition by lomofungin, inhibition studies were conducted at varying concentrations of the 66-mer substrate and lomofungin in our LC MS assay.14 A Ki of 6.7 ± 0.7 μM was obtained for lomofungin, and classical noncompetitive inhibition was most consistent with the results. Mechanistically, lomofungin decreases the maximal velocity (Vmax) of the enzymatic reaction but does not affect Km.24 Noncompetive inhibition predicts that lomofungin and the substrate bind independently and rever sibly to distinct sites of LC/A. Thus, lomofungin binds with equal avidity to the free enzyme (E) as well as the enzyme substrate complex (ES); and the substrate binds with equal avidity to E as well as the enzyme inhibitor complex (EI). That said, the ESI complex is catalytically inert.24
The noncompetitive behavior of lomofungin was unexpected since we anticipated binding within the active site. As such, additional properties of inhibitor binding were revealed through inhibitor combination studies. In brief, properly constructed inhibitor combination studies will reveal whether two inhibitors may bind to the enzyme simultaneously. Two active site inhibitors may not bind simultaneously since they both occupy the same space on the enzyme. The interaction of two such inhibitors is called mutually exclusive binding and will produce an additive effect on enzyme inhibition when used in combination. On the other hand, if each inhibitor binds to distinct positions on the enzyme surface such that each inhibitor may bind simultaneously, the interaction is called nonmutually exclusive binding and the impact on enzyme inhibition will be synergistic when used in combination. Plots of 1/velocity versus inhibitor concentrations at fixed concentrations of the second inhibitor are diagnostic. If the inhibitors are mutually exclusive, a family of parallel lines is observed. On the other hand, if the inhibitors are nonmutually exclusive, a pattern of intersecting lines is observed.
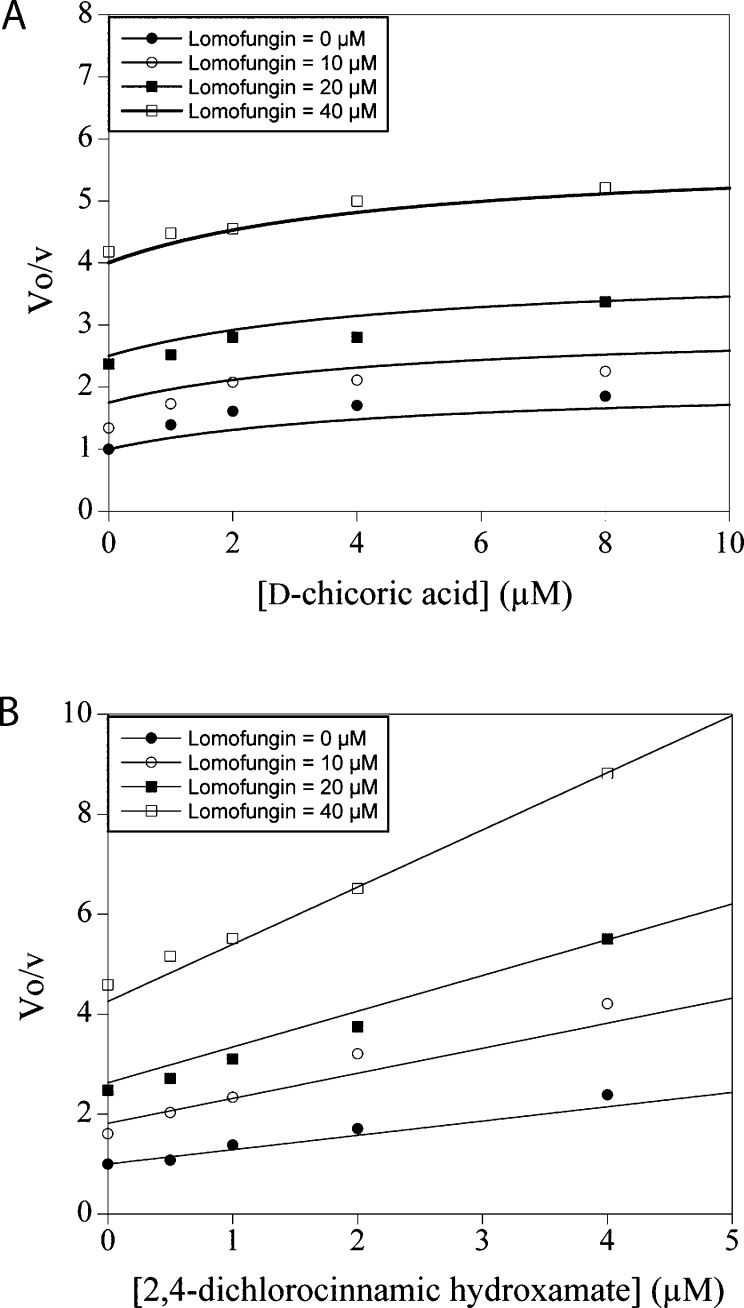
Previously we reported on the distinct inhibition of LC/A by d-chicoric acid.17 Similar to lomofungin, d-chicoric acid displayed noncompetitive inhibition although d-chicoric acid fails to fully inhibit the enzyme even at saturating concentrations. From inhibitor combination studies we demonstrated that d-chicoric acid binding is nonmutually exclusive with the active site competitive inhibitor 2,4-dichlorocinnamic hydroxamate.17 Since lomofungin also inhibits noncompetitively, we evaluated LC/A inhibition when lomofungin was used in combination with d-chicoric acid and when used in combination with 2,4-dichlorocinnamic hydroxamate. The results of these combination studies are presented in Figure 2. In panel A nonmutually exclusive binding is observed between lomofungin and d-chicoric acid. The slight curvature of the data and the best fit lines result from the partial inhibition displayed by d-chicoric acid (Supporting Information). More obviously, the intersecting lines presented in panel B by the combination of lomofunginin with 2,4-dichlorocinnamic hydroxamate indicate a nonmutually exclusive binding interaction.
Figure 2.
(A) Lomofungin in combination with d-chicoric acid displaying nonmutually exclusive inhibition compounded with partial inhibition. (B) Lomofungin in combination with 2,4-dichlorocinnamic hydroxamate displaying nonmutually exclusive inhibition.
We offer the following mechanistic interpretation of these results: the inhibition, and by inference binding, of lomofungin is distinct from active site targeted inhibitor, i.e. 2,4-dichlorocinnamic hydroxamate.13 We shall use the term exosite to distinguish a position on the enzyme that is nonoverlapping with the active site catalytic machinery and the volume/area containing the binding region of P1−P1′. Within the botulinum literature, two distinct exosites have been identified for LC/A.20 One is referred to as the α-exosite, so named because the recognition elements of it bind the 147−167 amino acids of SNAP-25 which are in an alpha helical conformation. The α-exosite is remote from the active site, essentially on the opposite face of LC/A. The second exosite (β-exosite) occurs just beyond the scissile bond and binds SNAP-25 residues 201−204 which form a β-strand. From our mechanistic studies, we are unable to unambiguously assign a specific location on the enzyme surface where inhibitor binding occurs and caution the reader that exosites in this discussion are not necessarily identified as the α- and β-exosites already termed. That said, the results from lomofungin combination studies with d-chicoric acid are more consistent with unique binding sites for each compound although the partial inhibition observed with d-chicoric acid makes this conclusion more tenuous.
Two mechanistic classes of LC/A inhibitors are now known. The distinguishing properties are competitive or noncompetitive kinetics versus a large substrate able to engage the α- and β-exosites and whether they are capable of nonmutually exclusive binding with a known active site inhibitor. Our kinetic data identify lomofungin as a non-active site based inhibitor and likely nonoverlapping with d-chicoric acid. In support of distinct binding locations for d-chicoric acid and lomofungin are the interactions observed when the smaller substrate SNAPtide is employed. Lomofungin, like active site based inhibitors, is inhibitory, but d-chicoric acid is not inhibitory to SNAPtide hydrolysis. From such observations it is tempting to speculate that d-chicoric acid may be binding in a region some distance from the active site, in fact, so far remote that it has no overlap with the SNAPtide substrate, yet it still interferes with larger substrates such as the 66-mer. On the other hand, lomofungin is inhibitory against both large and small peptide substrates, by default it should bind considerably closer to the active site and overlap with SNAPtide.
Analysis of the chemical structure reveals that lomofungin contains two 8-hydroxyquinoline motifs known to chelate bivalent metals. LC/A requires Zn2+ for catalysis, and compounds capable of chelating the tightly bound active site Zn2+ have proven to be potent inhibitors.13 A number of quinolinol derivatives have been identified as LC/A inhibitors, and their mechanism of inhibition has been proposed.25−27 One study deemed compounds containing this motif as not suitable for further development as LC/A inhibitors due to chelation of the active site zinc ion.25 However, a more recent study has yielded an effective quinolinol inhibitor of LC/A, and it was determined that inhibition of the protease was not due to chelation or sequestering of Zn2+ from LC/A.26
To further define lomofungin’s mechanism of inhibition we investigated this compound's metallospecificity. The simple compound 8-hydroxyquinoline showed no significant inhibition of LC/A even at the highest concentration tested (500 μM). To further validate that 8-hydroxyquinoline and lomofungin were not simply sequestering Zn2+ from the active site, both compounds were evaluated with BoNT LC/B using a FRET-based assay developed in our laboratory.28 8-Hydroxyquinoline did not show any significant inhibition of the LC/B, and lomofungin displayed weak inhibition with an IC50 ≥ 50 μM. The lack of inhibition by 8-hydroxyquinoline is not totally unexpected; the formation constant, (log Kav), of 8-hydroxyquinoline for Zn2+ is approximately 9.4,29 which is significantly weaker than EDTA, a high-affinity metal chelator known to inhibit metalloproteases via sequestering of Zn2+ from the active site.30 Based on the compilation of the lomogungin, 8-hydroxyquinoline, and inhibition combination studies we propose that LC/A inhibition by lomofungin does not proceed through direct chelation and/or sequestering of the active site Zn2+ and that lomofungin is an exclusive inhibitor for the LC/A serotype.
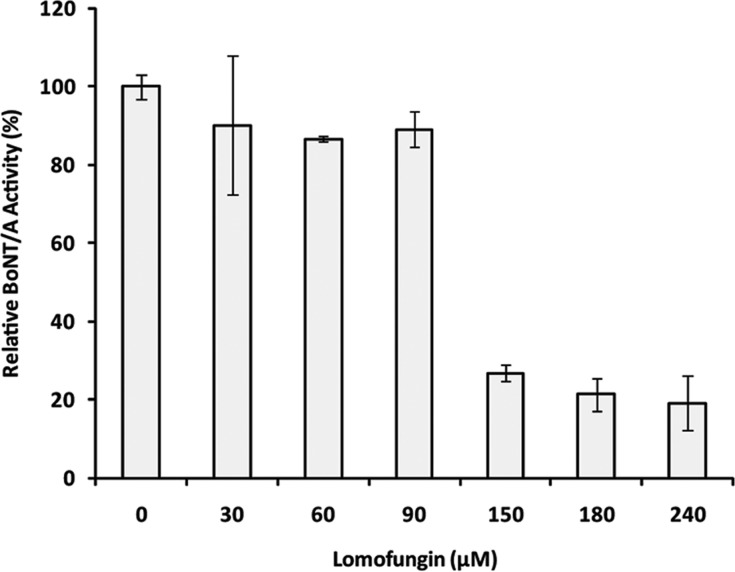
A variety of LC/A inhibitors have been identified through cell free in vitro experiments using recombinant forms of the enzyme and the activated holotoxin. However, cellular intoxication is the result of cytosolic cleavage of SNAP-25 by LC/A that has been translocated into the neuronal cell. Therefore, the therapeutic potential of LC/A inhibitors may be better evaluated with cell-based assays which monitor intracellular cleavage of SNAP-25. The cellular efficacy of lomofungin was investigated using primary rat cerebellar neurons. Lomofungin was added to the cells at varying concentrations (30 μM to 240 μM) followed by the addition of the BoNT/A holotoxin (0.2 nM) without preequilibration of compound and toxin.27 The relative activity of BoNT/A, correlating to the amount of cellular protection by lomofungin, is represented in Figure 3. At 90 μM lomofungin provided only modest protection (∼10%); however, greater than 70% protection was observed at the higher concentration of 150 μM (Figure 3). The calculated EC50 for lomofungin in this cellular assay is 131 ± 16 μM. This value is approximately 20-fold greater than the inhibition constant determined against the recombinant LC/A; however it is important to note that lomofungin was administered as only a single dose for a 4 h cellular assay. In addition, cells are complex biological systems and, therefore, other factors such as cell permeability influence the efficacy of drugs within cellular models.
Figure 3.
Lomofungin inhibition of SNAP-25 cleavage in primary rat cerebellar neurons displayed as relative BoNT/A activity, correlating to the amount of cellular protection.
In summary, a research approach incorporating high-throughput screening, compound validation, kinetic characterization, and in vitro cell-based testing was used to successfully identify a previously undisclosed pharmacophore for inhibition of BoNT intoxication. Lomofungin effectively inhibits LC/A protease activity and offers some protection of rat cerebellar neurons against BoNT/A intoxication. We consider lomofungin an interesting lead with a rich chemical framework that will serve as a precursor for the development of more potent analogues. Finally, we highlight the distinct binding of lomofungin to LC/A. It is tantalizing to speculate based on our kinetic data that lomofungin would map closer to the β-exosite on the protease while d-chicoric acid would map closer to the α-exosite. However, equally possible is that these molecules are binding at yet to be defined locations on the enzyme. Crystallographic studies are ongoing and should provide more detailed binding information for these two unusual LC/A inhibitors.
Supporting Information Available
Detailed information on the experimental procedures for enzymatic and cell-based assays, as well as kinetic data analysis. This material is available free of charge via the Internet at http://pubs.acs.org.
This project was supported with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under Contract No. N01-AI-30050 (C.B.S.) and Award No. AI082190 (K.D.J.).
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Johnson E. A.; Bradshaw M. Clostridium botulinum and its neurotoxins: a metabolic and cellular perspective. Toxicon 2001, 39, 1703–22. [DOI] [PubMed] [Google Scholar]
- Arnon S. S.; Schechter R.; Inglesby T. V.; Henderson D. A.; Bartlett J. G.; Ascher M. S.; Eitzen E.; Fine A. D.; Hauer J.; Layton M.; Lillibridge S.; Osterholm M. T.; O’Toole T.; Parker G.; Perl T. M.; Russell P. K.; Swerdlow D. L.; Tonat K. Botulinum toxin as a biological weapon: medical and public health management. JAMA, J. Am. Med. Assoc. 2001, 285, 1059–70. [DOI] [PubMed] [Google Scholar]
- Hackett R.; Kam P. C. Botulinum toxin: pharmacology and clinical developments: a literature review. Med. Chem. 2007, 3, 333–45. [DOI] [PubMed] [Google Scholar]
- Truong D. D.; Jost W. H. Botulinum toxin: clinical use. Parkinsonism Relat. Disord. 2006, 12, 331–55. [DOI] [PubMed] [Google Scholar]
- Wein L. M.; Liu Y. Analyzing a bioterror attack on the food supply: the case of botulinum toxin in milk. Proc. Natl. Acad. Sci. U.S.A. 2005, 102, 9984–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oguma K.; Fujinaga Y.; Inoue K. Structure and function of Clostridium botulinum toxins. Microbiol. Immunol. 1995, 39, 161–8. [DOI] [PubMed] [Google Scholar]
- Arnon S. S.; Schechter R.; Maslanka S. E.; Jewell N. P.; Hatheway C. L. Human botulism immune globulin for the treatment of infant botulism. N. Engl. J. Med. 2006, 354, 462–71. [DOI] [PubMed] [Google Scholar]
- Black R. E.; Gunn R. A. Hypersensitivity reactions associated with botulinal antitoxin. Am. J. Med. 1980, 69, 567–70. [DOI] [PubMed] [Google Scholar]
- Willis B.; Eubanks L. M.; Dickerson T. J.; Janda K. D. The strange case of the botulinum neurotoxin: using chemistry and biology to modulate the most deadly poison. Angew. Chem., Int. Ed. 2008, 47, 8360–79. [DOI] [PubMed] [Google Scholar]
- Burnett J. C.; Ruthel G.; Stegmann C. M.; Panchal R. G.; Nguyen T. L.; Hermone A. R.; Stafford R. G.; Lane D. J.; Kenny T. A.; McGrath C. F.; Wipf P.; Stahl A. M.; Schmidt J. J.; Gussio R.; Brunger A. T.; Bavari S. Inhibition of metalloprotease botulinum serotype A from a pseudo-peptide binding mode to a small molecule that is active in primary neurons. J. Biol. Chem. 2007, 282, 5004–14. [DOI] [PubMed] [Google Scholar]
- Burnett J. C.; Wang C.; Nuss J. E.; Nguyen T. L.; Hermone A. R.; Schmidt J. J.; Gussio R.; Wipf P.; Bavari S. Pharmacophore-guided lead optimization: the rational design of a non-zinc coordinating, sub-micromolar inhibitor of the botulinum neurotoxin serotype a metalloprotease. Bioorg. Med. Chem. Lett. 2009, 19, 5811–3. [DOI] [PubMed] [Google Scholar]
- Pang Y. P.; Vummenthala A.; Mishra R. K.; Park J. G.; Wang S.; Davis J.; Millard C. B.; Schmidt J. J. Potent new small-molecule inhibitor of botulinum neurotoxin serotype A endopeptidase developed by synthesis-based computer-aided molecular design. PLoS One 2009, 4, e7730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldt G. E.; Kennedy J. P.; Janda K. D. Identification of a potent botulinum neurotoxin a protease inhibitor using in situ lead identification chemistry. Org. Lett. 2006, 8, 1729–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capkova K.; Hixon M. S.; McAllister L. A.; Janda K. D. Toward the discovery of potent inhibitors of botulinum neurotoxin A: development of a robust LC MS based assay operational from low to subnanomolar enzyme concentrations. Chem. Commun. (Cambridge) 2008, 3525–7. [DOI] [PubMed] [Google Scholar]
- Park J. G.; Sill P. C.; Makiyi E. F.; Garcia-Sosa A. T.; Millard C. B.; Schmidt J. J.; Pang Y. P. Serotype-selective, small-molecule inhibitors of the zinc endopeptidase of botulinum neurotoxin serotype A. Bioorg. Med. Chem. 2006, 14, 395–408. [DOI] [PubMed] [Google Scholar]
- Tang J.; Park J. G.; Millard C. B.; Schmidt J. J.; Pang Y. P. Computer-aided lead optimization: improved small-molecule inhibitor of the zinc endopeptidase of botulinum neurotoxin serotype A. PLoS One 2007, 2, e761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowe G. N.; Silhar P.; Hixon M. S.; Silvaggi N. R.; Allen K. N.; Moe S. T.; Jacobson A. R.; Barbieri J. T.; Janda K. D. Chirality holds the key for potent inhibition of the botulinum neurotoxin serotype A protease. Org. Lett. 2010, 12, 756–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silhar P.; Capkova K.; Salzameda N. T.; Barbieri J. T.; Hixon M. S.; Janda K. D. Botulinum neurotoxin A protease: discovery of natural product exosite inhibitors. J. Am. Chem. Soc. 2010, 132, 2868–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong C. R.; Chen X.; Shi L.; Liu J. O.; Sullivan D. J. Jr. A clinical drug library screen identifies astemizole as an antimalarial agent. Nat. Chem. Biol. 2006, 2, 415–6. [DOI] [PubMed] [Google Scholar]
- Boldt G. E.; Kennedy J. P.; Hixon M. S.; McAllister L. A.; Barbieri J. T.; Tzipori S.; Janda K. D. Synthesis, characterization and development of a high-throughput methodology for the discovery of botulinum neurotoxin a inhibitors. J. Comb. Chem. 2006, 8, 513–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger A. T.; Rummel A. Receptor and substrate interactions of clostridial neurotoxins. Toxicon 2009, 54, 550–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S.; Barbieri J. T. Unique substrate recognition by botulinum neurotoxins serotypes A and E. J. Biol. Chem. 2006, 281, 10906–11. [DOI] [PubMed] [Google Scholar]
- Johnson L. E.; Dietz A. Lomofungin, a new antibiotic produced by Streptomyces lomondensis sp. n. Appl. Microbiol. 1969, 17, 755–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tipton C. D.; Rinehart K. L. Jr. Lomofungin. I. Degradative studies of a new phenazine antibiotic. J. Am. Chem. Soc. 1970, 92, 1425–6. [DOI] [PubMed] [Google Scholar]
- Segel I. H.Enzyme Kinetics: Behavior and Analysis of Rapid Equilibrium and Steady-State Enzyme Systems; Wiley & Sons: Hoboken, NJ, 1975. [Google Scholar]
- Burnett J. C.; Schmidt J. J.; Stafford R. G.; Panchal R. G.; Nguyen T. L.; Hermone A. R.; Vennerstrom J. L.; McGrath C. F.; Lane D. J.; Sausville E. A.; Zaharevitz D. W.; Gussio R.; Bavari S. Novel small molecule inhibitors of botulinum neurotoxin A metalloprotease activity. Biochem. Biophys. Res. Commun. 2003, 310, 84–93. [DOI] [PubMed] [Google Scholar]
- Lai H.; Feng M.; Roxas-Duncan V.; Dakshanamurthy S.; Smith L. A.; Yang D. C. Quinolinol and peptide inhibitors of zinc protease in botulinum neurotoxin A: effects of zinc ion and peptides on inhibition. Arch. Biochem. Biophys. 2009, 491, 75–84. [DOI] [PubMed] [Google Scholar]
- Roxas-Duncan V.; Enyedy I.; Montgomery V. A.; Eccard V. S.; Carrington M. A.; Lai H.; Gul N.; Yang D. C.; Smith L. A. Identification and biochemical characterization of small-molecule inhibitors of Clostridium botulinum neurotoxin serotype A. Antimicrob. Agents Chemother. 2009, 53, 3478–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzameda N. T.; Barbieri J. T.; Janda K. D. Synthetic substrate for application in both high and low throughput assays for botulinum neurotoxin B protease inhibitors. Bioorg. Med. Chem. Lett. 2009, 19, 5848–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fresier H. Structure and behavior of organic analytical reagents. III. Stability of chelates of 8-hydroxyquinoline and analogous reagents. J. Am. Chem. Soc. 1952, 74, 5239–42. [Google Scholar]
- Schiavo G.; Rossetto O.; Santucci A.; DasGupta B. R.; Montecucco C. Botulinum neurotoxins are zinc proteins. J. Biol. Chem. 1992, 267, 23479–83. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.