Abstract
Peroxiredoxin 1 (Prx1) is an antioxidant and molecular chaperone that can be secreted from tumor cells. Prx1 is over-expressed in many cancers and elevation of Prx1 is associated with poor clinical outcome. In the current study we demonstrate that incubation of Prx1 with thioglycollate (TG)-elicited murine macrophages or immature bone marrow derived dendritic cells resulted in Toll-like receptor 4 (TLR4) dependent secretion of TNF-α and IL-6 and dendritic cell maturation. Optimal secretion of cytokines in response to Prx1 was dependent upon serum and required CD14 and MD2. Binding of Prx1 to TG-macrophages occurred within minutes and resulted in TLR4 endocytosis. Prx1 interaction with TLR4 was independent of its peroxidase activity and appeared to be dependent upon its chaperone activity and ability to form decamers. Cytokine expression occurred via the TLR-MyD88 signaling pathway, which resulted in nuclear translocation and activation of NFκB. These findings suggest that Prx1 may act as danger signal similar to other TLR4 binding chaperone molecules such as HSP72.
This is an author-produced version of a manuscript accepted for publication in The Journal of Immunology (The JI). The American Association of Immunologists, Inc. (AAI), publisher of The JI, holds the copyright to this manuscript. This version of the manuscript has not yet been copyedited or subjected to editorial proofreading by The JI; hence, it may differ from the final version published in The JI (online and in print). AAI (The JI) is not liable for errors or omissions in this author-produced version of the manuscript or in any version derived from it by the U.S. National Institutes of Health or any other third party. The final, citable version of record can be found at www.jimmunol.org.
Keywords: Peroxiredoxin 1, TLR4, NFκB
Introduction
Peroxiredoxin1 (Prx1) is a member of the typical 2-cysteine peroxiredoxin family, whose major intracellular functions are as a regulator of hydrogen peroxide signaling through its peroxidase activity and as a protein chaperone (1). Prx1 expression is elevated in various cancers, including esophageal, pancreatic, lung, follicular thyroid, and oral cancer (2–9). Elevated Prx1 levels have been linked with poor clinical outcomes and diminished overall patient survival (4,10,11). Recent studies have demonstrated that Prx1 can be secreted by non-small cell lung cancer cells, possibly via a non-classical secretory pathway (12,13).
The function of extracellular/secreted Prx1 is unknown; however a number of oxidative stress proteins, including thioredoxin and heat shock proteins, are released from stressed, transformed, and dying cells and act as “endogenous” danger signals by binding danger signal sensors/receptors in the extracellular microenvironment (14–18). Many of these “endogenous” danger signals are recognized by the danger signal receptor Toll-like receptor 4 (TLR4) (17,19). A recent study by Furuta et al. indicates that the malaria (Plasmodium berghei ANKA) homolog of Prx1/2, PbA, is a TLR4/MD2 ligand that promotes IgE-mediated protection and innate immunity (20). We hypothesize that mammalian Prx1 acts as an endogenous danger signal by binding to TLR4.
TLR4 induced gene activation is mediated through both myeloid differentiation protein 88 (MyD88) dependent and independent pathways (21). MyD88 dependent signaling causes activation of NFκB and protein kinase cascade dependent activation of AP-1, which results in the secretion of pro-inflammatory cytokines such as TNF-α and IL-6 (22,23). MyD88 independent gene activation occurs via the adaptor protein TRAM and leads to activation of Interferon Regulatory Factor 3 (IRF-3) and secretion of Type I interferons (IFN α/β) (22–24).
Our studies demonstrate that Prx1 stimulates TLR4 dependent cytokine secretion from macrophages and dendritic cells, that the interaction and subsequent cytokine secretion is peroxidase independent but chaperone/ structure dependent and that TLR4 stimulated cytokine secretion by Prx1 is optimal in the presence of CD14 and MD2 and is MyD88 dependent.
Materials and Methods
Materials
Lipopolysaccharide (LPS, Escherichia coli serotype 026:B6) polymyxin B sulfate salt, bovine serum albumin (BSA), and ovalbumin (OVA) were obtained from Sigma-Aldrich (St. Louis, MO). 7-Amino-Actinomycin D (7-AAD) and thioglycollate brewer modified media was purchased from (Becton Dickinson, La Jolla, CA). Capture and detection antibodies for IL-6 and TNF-α used in Luminex assays, as well as protein standards, were purchased from Invitrogen (Carlsbad, CA). Antibodies specific for CD11b, Gr-1, F4/80, and all isotypes were purchased from PharMingen (Mountain View, CA). Antibodies against TLR2, TLR4, and NFκB subunits were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Blocking antibodies against MD2 and CD14 were purchased from Santa Cruz Biotechnology. The phycoerythrin (PE) conjugated anti-TLR4 antibody was purchased from eBioscience (San Diego, CA). Antibodies specific for Prx1 were obtained from Lab Frontier (Seoul, South Korea); this antibody is specific for Prx1 and detects only a single band in Western analysis of cells that express Prx1 (Supplementary Figure 1A).
Animals and Cell Lines
C57BL/6NCr (TLR4+/+ and TLR2+/+), C57BL/10ScNJ (TLR4−/−), B6.129-Tlr2tm1Kir/J (TLR2−/−), C3H/HeNCr (TLR4+/+), and C3H/HeNJ (TLR4−/−) pathogen-free mice were purchased from The Jackson Laboratory (Bar Harbor, ME). Animals were housed in microisolator cages in laminar flow units under ambient light. The mice were maintained in a pathogen-free facility at Roswell Park Cancer Institute (Buffalo, NY). The Institutional Animal Care and Use Committee approved both animal care and experiments.
The role of Prx1 in vivo was determined by injecting either C57BL/6NCr or C57BL/10ScNJ mice intravenously with 90 ug Prx1 (~1000 nM). Cardiac punctures were performed 2hours later. Serum was obtained by incubation of blood at 4°C overnight then samples were centrifuged and supernatants collected.
The cultured mouse macrophage cell line (RAW264.7) was maintained in Dulbeco's Modified Eagle Media (DMEM) containing 10% defined fetal bovine serum and 100U/ml penicillin and 100 ug/ml streptomycin at 37°C and 5.0% CO2. RAW264.7 cells were transfected with the pcDNA3.1 plasmid containing either control or MyD88 dominant negative (DN) encoding oligonucleotides using FuGENE 6 (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. The transfected cells were then selected using G418 for cells expressing the control or MyD88 DN. Cells were then stimulated with buffer, Prx1, or LPS for 24h and culture media was harvested for IL-6 cytokine analysis by ELISA.
The retroviral short hairpin RNA expression constructs and retroviral infection procedure used to create a knock down of Prx1 in the lung cancer cell line (A549) were previously defined (25–27).
Macrophage and Dendritic Cell Isolation
Peritoneal elicited macrophage cells from mice were obtained by an intraperitoneal injection of 1.0 ml of 3.0 % (w/v) thioglycollate media (TG). Four days after injection, mice were sacrificed and macrophages were obtained by peritoneal lavage (28). Macrophages were enriched by adherence selection for 1 h in complete media (DMEM supplemented with 10% defined FBS, 100 U/ml penicillin and 100 μg/ml streptomycin) (28) and were characterized through FACS analysis for expression of CD11b, Gr1 and F4/80 as previously described (29); cells that were CD11b+Gr1−F4/80+ were identified as macrophages.
Immature bone marrow derived dendritic cells were generated by culture of bone marrow derived cells in GM-CSF as previously described (30,31). Dendritic cells were identified by the expression of CD11c.
Protein Purification
Recombinant human Prx1, Prx1C52S, and Prx1C83S proteins were purified as described previously (32,33). Briefly, bacterial cell extracts containing recombinant proteins were loaded onto DEAE-sepharose (GE Healthcare, USA) and equilibrated with 20 mM Tris-Cl (pH 7.5). The proteins were dialyzed with 50 mM sodium phosphate buffer (pH 6.5) containing 0.1 M NaCl. The unbound proteins from the DEAE column containing Prx1, Prx1C52S, or Prx1C83S were pooled and loaded onto a Superdex 200 (16/60, GE Healthcare, USA), and equilibrated with 50 mM sodium phosphate buffer (pH 7.0) containing 0.1 M NaCl. The fractions containing Prx1, Prx1 C52S, or Prx1C83S were pooled and stored at –80°C. Endotoxin levels of purified proteins were quantified with a Limulus Amebocyte Lysate Assay (Lonza, Walkersville, MD) according to manufacturer's directions. Prx1, Prx1C52S, and Prx1C83S were found to contain 14.14 ± 0.050 EU/ml, 14.07 ± 0.67 EU/ml, and 14.17 ± 0.025 EU/ml respectively.
Cytokine Analysis
Adherent TG-elicited macrophage cells were washed 5–10 times with PBS, to remove any non-adherent cells. Once washed, complete media containing purified Prx1, Prx1C52S, Prx1C83S, or LPS at the specified concentrations were added in the presence or absence of Prx1, MD-2 and CD14 blocking or control antibodies. In the indicated experiments Prx1 proteins or LPS were incubated with polymyxin B or were boiled for 20 minutes prior to addition. After 24 h the supernatant was collected and analyzed by cytokine specific ELISA or the Luminex multiplex assay system. Serum samples were collected as indicated above and IL-6 levels were determined by ELISA. TNF-α and IL-6 ELISA kits were purchased from BD Bioscience (Franklin Lakes, NJ) and assays were completed according to manufacturer's instructions.
Luminex analyses were performed by the Institute Flow Cytometry Facility in 96-well microtiter plates (Multiscreen HV plates, Millipore, Billerica, MA) with PVDF membranes using a Tecan Genesis liquid handling robot (Research Triangle Park, NC) for all dilutions, reagent additions and manipulations of the microtiter plate. Bead sets, coated with capture antibody were diluted in assay diluents, pooled and approximately 1000 beads from each set were added per well. Recombinant protein standards were titrated from 9,000 to 1.4 pg/ml using 3-fold dilutions in diluent. Samples and standards were added to wells containing beads. The plates were incubated at ambient temperature for 120 min on a rocker, and then washed twice with diluent using a vacuum manifold to aspirate. Biotinylated detection antibodies to each cytokine were next added and the plates were incubated 60 min and washed as before. Finally, PE conjugated streptavidin was added to each well and the plates were incubated 30 min and washed. The beads were resuspended in 100 μl wash buffer and analyzed on a Luminex 100 (Luminex Corp., Austin, TX). Each sample was measured in duplicate, and blank values were subtracted from all readings. Using BeadView Software (Millipore) a log regression curve was calculated using the bead MFI values versus concentration of recombinant protein standard. Points deviating from the best-fit line, i.e. below detection limits or above saturation, were excluded from the curve. Sample cytokine concentrations were calculated from their bead's mean fluorescent intensities by interpolating the resulting best-fit line. Samples with values above detection limits were diluted and reanalyzed.
FITC Labeling of Proteins
BSA, Prx1, Prx1C52S, and Prx1C83S proteins were conjugated to FITC using a FITC conjugation kit (Sigma, St. Louis, MO). A twenty-fold excess of FITC and individual proteins were dissolved into a 0.1M sodium bicarbonate/carbonate buffer (pH adjusted to 9.0); the mix was incubated for 2 h at room temperature with gentle rocking. The excess free FITC was removed with a Sephadex G-25 column (Pharmacia, Piscataway, NJ). Proteins amounts were quantified using a standard Lowry assay. The F:P (fluorescence:protein) ratio was calculated according to the manufacturer's instructions using the optical density at 495 nm (FITC absorbance) and 280 nm (protein absorbance). FITC per nM protein for BSA, Prx1, Prx1 C52S, and Prx1 C83S were 31.00 ±1.92, 38.52 ± 2.39, 74.49 ± 2.64, and 44.44 ± 2.64 respectively.
Saturation Assay
FITC-conjugated BSA, Prx1, Prx1C52S, and Prx1C83S were diluted in 1.0 % BSA in PBS to the specified concentrations and a total reaction volume of 100 μL. These mixtures were incubated with 1.0×106 cells/mL for 20 min on ice to prevent internalization. Cells were washed twice with 1% BSA in PBS and cells were incubated to demonstrate viable from nonviable cells with 7-AAD, less than 30 min before FACsCalibur analysis. Data was acquired from a minimum of 20,000 cells, stored in collateral list mode, and analyzed using the WinList processing program (Verity Software House, Inc., Topsham, ME). Cells positive for 7-AAD (nonviable) were gated out of the events. FITC-conjugated BSA was used as a negative binding control and for mutant studies variations in FITC labeling were normalized by FITC labeling per nM proteins.
Competition Assay
Unlabeled OVA, Prx1, Prx1C52S, and Prx1C83S were briefly mixed with FITC conjugated Prx1 at the specified concentrations in 100 μL 1.0 % BSA in PBS. The mixture was incubated for 20 min on ice, before washing twice with 1.0 % BSA in PBS. Cells were then incubated with 7-AAD and analyzed within 30 min by flow cytometry. OVA was used as a negative competition control in all competition assays. Data was acquired from a minimum of 20,000 cells, stored in collateral list mode, and analyzed using the WinList processing program (Verity Software House, Inc., Topsham, ME). When using WinList to analyze results, 7-AAD positive cells were gated out of the events.
Immunoprecipitation
Immunoprecipitation was carried out with 500 μg of cell lysates and 4 μg of anti-TLR4 or anti-TLR2 overnight at 4°C. After the addition of 25 μL of Protein G-agarose (Santa Cruz Biotechnology), the lysates were incubated for an additional 4 h. To validate specific protein interactions, goat IgG (Santa Cruz Biotechnology) or mouse IgG (Santa Cruz Biotechnology) was used as negative control. The beads were washed thrice with the lysis buffer, separated by SDS-PAGE, and immunoblotted with antibodies specific for Prx1. The proteins were detected with the ECL system (Biorad).
Co-localization of Prx1/TLR4 and NFκB Translocation
Colocalization experiments were performed by the addition of 200 nM FITC-labeled Prx1 and PE-conjugated anti-TLR4 to the media of TG-elicited macrophages and kept at 37°C for the indicated times before being transferred to ice, fixed and analyzed.
Immunostaining to detect the nuclear translocation of NFκB was performed in the following manner. TG-elicited macrophages obtained from C3H/HeNCr (TLR4+/+) and C3H/HeNJ (TLR4−/−) were treated with 200nM Prx1. After the indicated times at 37°C the cells were then scraped and collected in tubes, washed twice in wash buffer (2% FBS in phosphate-buffered saline), and then fixed in fixation buffer (4% paraformaldehyde in phosphate-buffered saline) for 10 min at room temperature. After washing, the cells were re-suspended in Perm Wash buffer (0.1% Triton X-100, 3% FBS, 0.1% sodium azide in phosphate-buffered saline) containing 10 μg/ml anti-NF□B p65 antibody (Santa Cruz Biotechnology) for 20 min at room temperature. The cells were then washed with Perm Wash buffer and resuspended in Perm Wash buffer containing 7.5 μg/ml FITC conjugated F(ab′)2 donkey anti-rabbit IgG for 15 min at room temperature. Cells were washed twice in Perm Wash buffer and re-suspended in 1% paraformaldehyde containing 5 μM DRAQ5 nuclear stain (BioStatus) for 5 min at room temperature.
Image Analysis
Co-localization of Prx1 and TLR4 and nuclear translocation of NFκB were analyzed with the ImageStream® multispectral imaging flow cytometer (34) (Amnis Corp., Seattle, WA). At least 5000 events were thus acquired for each experimental condition and the corresponding images were analyzed using the IDEAS® software package. A hierarchical gating strategy was employed using image-based features of object contrast (gradient RMS) and area versus aspect ratio to select for in-focus, single cells. Co-localization and nuclear translocation was determined in each individual cell using the IDEAS® similarity feature which is a log transformed Pearson's correlation coefficient of the intensities of the spatially correlated pixels within the whole cell, of the Prx1 and TLR4 images or NFκB and DRAQ5 images, respectively The similarity score is a measure of the degree to which two images are linearly correlated.
Electrophoretic mobility shift assay (EMSA)
EMSA was performed as described previously (35). Briefly, 10 μg of nuclear protein was incubated with γ-32P-labeled double-stranded NFκB oligonucleotide (5'-AGTTGAGGGGACTTTCCCAGGC-3') in 20 μL of binding solution containing 10 mM HEPES (pH 7.9), 80 mM NaCl, 10% glycerol, 1 mM DTT, 1 mM EDTA, 100 μg/mL poly(deoxyinosinic-deoxycytidylic acid). The DNA-protein complexes were resolved on a 6% polyacrylamide gel under non-denaturing conditions at 200 V for 2 h at 4°C. Gels were dried and then subjected to autoradiography.
Statistical Analysis
Statistical analyses were performed using a standardized t-test with Welch's correction, where equal variances were not assumed, to compare experimental groups. Differences were considered significant when P values were ≤ 0.05.
RESULTS
Prx1 stimulation of cytokine secretion from DCs and TG-macrophages and maturation of DCs is dependent upon TLR4
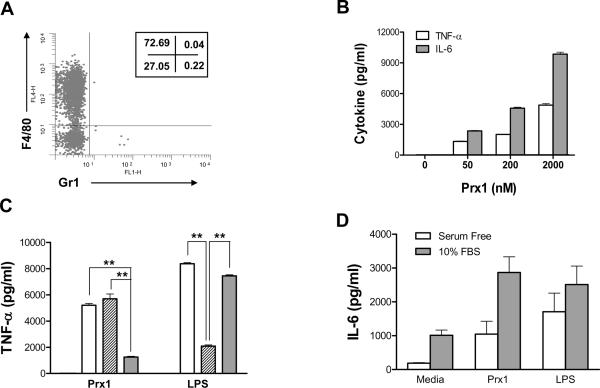
Thioglycolate (TG)-elicited murine macrophages were used to assess the ability of Prx1 to stimulate cytokine secretion. Macrophage phenotype was assessed by analysis of peritoneal exudate cell populations for CD11b, Gr1, and F4/80 expression. The isolated populations were greater than 99% CD11b+ and of the CD11b+ cell population a majority were Gr1−, F4/80+ (Figure 1A). Stimulation of TG-elicited macrophages with Prx1 resulted in the dose dependent secretion of TNF-α and IL-6 that was significantly greater than that observed in unstimulated cells at all doses (P≤0.01; Figure 1B). Pre-incubation of Prx1 with the endotoxin inactivator polymixin B had no significant effect on Prx1 stimulation of cytokine secretion (Figure 1C); in contrast, denaturing of Prx1 significantly reduced its ability to stimulate cytokine secretion (P<0.01).
Figure 1. Prx1 stimulates cytokine secretion from macrophages.
(A) TG-elicited macrophages were analyzed by flow cytometry for expression of CD11b, Gr1, and F4/80. A representative histogram of 3 independent isolations is shown and depicts Gr1 and F4/80 expression by CD11b+ cells. Numbers in the insets indicate the percentages of CD11b+ cells in each quadrant. (B) TG-elicited macrophages were incubated with stimulants for 24 h; supernatants were harvested and analyzed for TNF-α (open bars) and IL-6 levels (gray bars). Results are shown as pg/ml and are representative of three independent experiments; error bars represent standard deviation. (C) TG-elicited macrophages were incubated for 24h with media only (black bars), 100 nM LPS or 2000 nM Prx1 (open bars), 100 nM LPS or 2000 nM Prx1 pre-incubated for 20 minutes with 10 ug/mL polymyxin B (hatched bars), or 100 nM LPS or denatured 2000 nM Prx1 (gray bars). Asterisks indicate P≤0.01 as compared to cells treated with Prx1 or LPS alone. (D) TG-elicited macrophages were incubated with media alone, Prx1 (50 nM) or LPS (100 nM) for 24 h in the presence (gray bars) or absence (open bars) of 10% FBS. Supernatants were harvested and analyzed for IL-6 levels. Results are shown as pg/ml; error bars represent standard deviation.
Stimulation of cytokine secretion by TG-elicited macrophages following incubation with Prx1 was significantly diminished in the absence of serum (P≤0.01; Figure 1D); however even in serum free conditions, incubation of TG-elicited macrophages with Prx1 significantly increased IL-6 secretion (P≤0.005 when compared to secretion by cells incubated in serum free media). Prx1 was also able to stimulate cytokine secretion from the cultured dendritic cell line, DC1.2, and the murine macrophage cell line, RAW264.7 (data not shown).
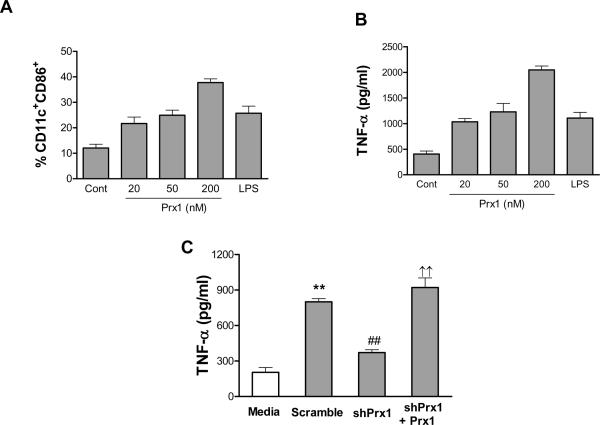
Exogenous Prx1 was able to induce maturation and activation of immature bone marrow derived DCs (iBMDCs). iBMDCs were incubated with increasing concentrations of Prx1 for 24h and examined for cell surface expression of co-stimulatory molecules and secretion of TNF-α. Addition of Prx1 led to significant dose dependent increase in cell surface expression of the co-stimulatory molecule, CD86 (Figure 2A) and TNF-α secretion (Figure 2B) at all doses tested (P≤0.01 when compared to control).
Figure 2. Prx1 stimulates dendritic cell maturation and activation.
(A and B) Immature bone marrow derived dendritic cells (iBMDCs) were incubated with media alone, 20–200 nM Prx1 or 100 nM LPS for 24h. (A) Following incubation cells were analyzed by flow cytometry for expression of CD11c and CD86. Results are shown as percent total cells; error bars represent standard deviation. (B) Supernatants were harvested and analyzed for TNF-α. Results are shown as pg/ml and are representative of three independent experiments; error bars represent standard deviation. (C) TG-elicited macrophages were incubated with media harvested from prostate tumor cell lines that were transfected with cDNA encoding for either control shRNA (Scramble) or shRNA specific for Prx1 (shPrx1) or in media harvested from cells expressing Prx1 specific shRNA to which 50 nM exogenous Prx1 had been added (shPrx1 + Prx1). Following 24h incubation, supernatants were harvested and analyzed for TNF-α. Results are shown as pg/ml and are representative of three independent experiments; error bars represent standard deviation. **: P≤0.01 when compared to TNF-α levels secreted by cells incubated with media alone; ##: P≤0.01 when compared to TNF-α levels secreted by cells incubated with media from cells expressing control shRNA; ††: P≤0.01 when compared to TNF-α levels secreted by cells incubated with media from cells expressing shRNA specific for Prx1.
It is possible that enhanced secretion of cytokines from iBMDCs and TG-elicited macrophages upon addition of exogenous recombinant Prx1 is a phenomena of the recombinant protein and not physiologically relevant. To begin to determine whether Prx1 could promote cytokine secretion in a physiologic context, TG-elicited macrophages were incubated for 24h in the presence of supernatant collected from Prx1-secreting tumor cells or supernatant collected from tumor cells engineered to express shRNA specific for Prx1. Expression of shRNA resulted in reduced expression of Prx1, but not Prx2 (Supplementary Figure 1B). Incubation of TG-elicited macrophages with supernatants of tumor cells engineered to express a non-specific shRNA, resulted in enhanced expression of TNF-α (Sc, Figure 2C; P≤0.0001 when compared to media). In contrast, TG-elicited macrophages incubated with supernatants collected from tumor cells expressing reduced levels of Prx1 secreted significantly lower levels of TNF-α (P≤0.0001 when compared to incubation with supernatant harvested from cells expressing control shRNA; Figure 2C); addition of exogenous Prx1 to these supernatants restored TNF-α secretion from TG-elicited macrophages (shPrx1 + Prx1; P≤0.003 when compared to incubation with supernatant harvested from cells expressing shRNA specific for Prx1).
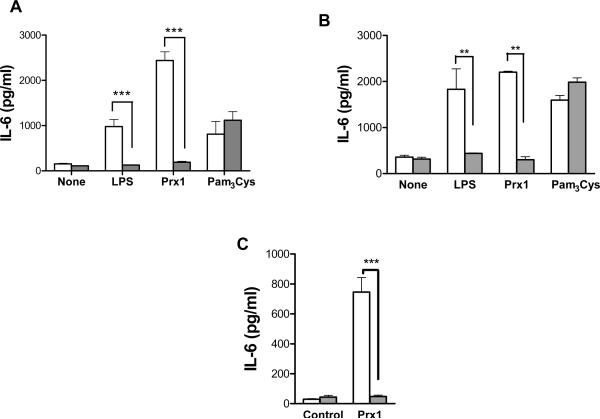
An evolutionary homolog of Prx1 interacts with TLR4 to induce inflammation (20). To test whether Prx1 activation of iBMDCs and TG-elicited macrophages was dependent upon TLR4, iBMDCs and TG-elicited macrophages were isolated from C57BL/6NCr (TLR4+/+) and C57BL/10ScNJ (TLR4−/−) mice and stimulated with Prx1, LPS or Pam3Cys, a TLR2 agonist. The results indicate that Prx1, LPS, and Pam3Cys stimulate cytokine secretion from iBMDCs (Figure 3A) and macrophages isolated from C57BL/6NCr mice (Figure 3B); only Pam3Cys stimulated cytokine secretion from iBMDCs and macrophages isolated from C57BL/10ScNJ mice (P≤0.01 when compared to cytokine secretion by cells isolated form C57BL/NCr mice).
Figure 3. Prx1 induced cytokine secretion is TLR4 dependent.
(A) iBMDCs were isolated from C57BL/6 (TLR4+/+; open bars) and C57BL/10ScNJ (TLR4−/−; closed bars) mice and stimulated with 200 nM Prx1, 100 nM LPS, or 100 mM Pam3Cys. Supernatants were collected and analyzed by IL-6 ELISA kits. (B) TG-elicited macrophages were isolated from C57BL/6 (TLR4+/+; open bars) and C57BL/10ScNJ (TLR4−/−; closed bars) mice and stimulated with 200 nM Prx1, 100 nM LPS, or 100 mM Pam3Cys. Supernatants were collected and analyzed by IL-6 ELISA kits. Results are presented as pg/ml; error bars represent standard deviation; asterisks indicate P values less that 0.01. (C) Naïve C57BL/6 (TLR4+/+; open bars) and C57BL/10ScNJ (TLR4−/−; closed bars) mice were injected i.p. with 200 nm Prx1. Six hours later, blood was collected and analyzed by ELISA for the presence of IL-6. Results are presented as pg/ml; error bars represent standard deviation; asterisks indicate P≤0.0002.
The ability of Prx1 to induce TLR4 dependent inflammation in vivo was tested by i.p. injection of recombinant Prx1 into either C57BL/6NCr (TLR4+/+) or C57BL/10ScNJ (TLR4−/−) mice. Blood was collected 2h post injection and the extent of systemic inflammation was determined by assessing the level of systemic IL-6 (Figure 3C). Injection of Prx1 resulted in a significant increase in systemic IL-6 levels (P≤0.0002) in C57BL/6NCr (TLR4+/+) mice, but had no significant effect on systemic IL-6 levels in C57BL/10ScNJ (TLR4−/−) mice.
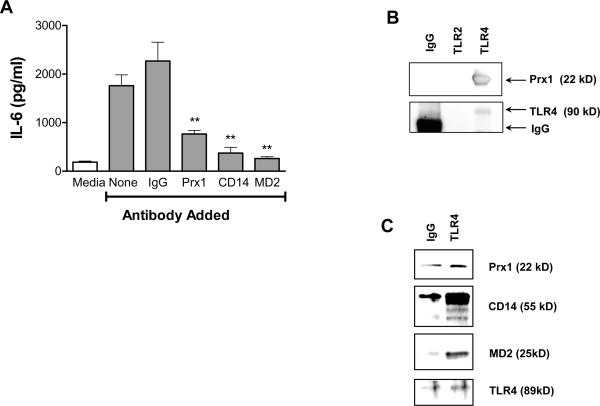
The reduced expression of cytokines by TG-elicited macrophages following incubation with Prx1 in the absence of serum (Figure 1D) suggests that serum proteins may contribute to optimal Prx1/TLR4 interaction. Many TLR4 ligands interact with TLR4 as part of a larger complex that can include CD14 and/or MD2; the evolutionary homolog of Prx1, PbA, interacts with TLR4 in an MD2-dependent manner (20). To determine whether Prx1 enhancement of cytokine secretion from TG-elicited macrophages involves CD14 or MD2, cells were incubated with Prx1 or LPS in the presence of blocking antibodies to MD2, CD14 or control IgG (Figure 4A). Addition of blocking antibodies to Prx1, CD14 or MD2 significantly inhibited the ability of Prx1 to stimulate IL-6 secretion from TG-elicited macrophages when compared to that induced by Prx1 in the presence of control IgG (P≤0.01). Blocking antibodies to CD14 and MD2 also blocked cytokine secretion in LPS stimulated cells (Supplementary Figure 1C).
Figure 4. Interaction of Prx1 with TLR4 is dependent upon CD14 and MD2.
(A) TG-elicited macrophages were isolated from C57BL/6 mice and stimulated with 50 nM Prx1 in the presence or absence of control or blocking antibodies to Prx1, CD14 or MD2 for 24h. Supernatants were collected and analyzed by IL-6 ELISA kits. Results are presented as pg/ml; error bars represent SEM; asterisks indicate P values less that 0.01. (B) TG-elicited macrophages were harvested and cell lysates were precipitated with antibodies to TLR4, TLR2, and mouse/goat IgG as described in Materials and Methods; resulting precipitates were separated by SDS-PAGE and probed by Western blot analysis for the presence of Prx1. Blots were also probed with antibodies to TLR4 or TLR2 as a loading control. (C) TG-elicited macrophages were harvested and cell lysates were incubated with antibodies to TLR4 or mouse/goat IgG as described in Materials and Methods; resulting precipitates were separated by SDS-PAGE and probed by Western blot analysis for the presence of Prx1, CD14 and MD2. Blots were also probed with antibodies to TLR4 as a loading control.
To further demonstrate the interaction Prx1 and TLR4/MD2/CD14, TG-elicited macrophage cell lysates were incubated with isotype control antibodies or antibodies specific for TLR4 or TLR2 (Figure 4B). The antibody complexes were isolated and immunoblotting was performed using antibodies to Prx1; Prx1 was only found in the lysates immunoprecipitated with TLR4 (Figure 4B). The TLR4/Prx1 complexes isolated from Prx1 treated cells also contained CD14 and MD2 (Figure 4C), confirming the finding that Prx1 interacts with TLR4 in a complex that contains both CD14 and MD2.
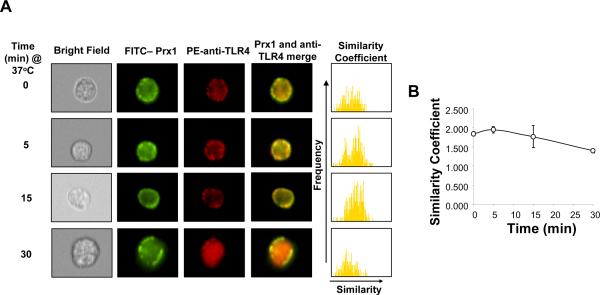
The kinetics of the Prx1 and TLR4 interaction was determined using image stream analysis (Amnis) to examine co-localization of the two molecules. TG-elicited macrophages were incubated with FITC-labeled Prx1 and PE-conjugated anti-TLR4 antibodies. The merged images of representative cells indicate that Prx1 and TLR4 localize together on the membrane of the macrophage within 5 minutes and that by 30 min, TLR4 and a portion of the Prx1 molecules have been internalized (Figure 5A). The histograms to the right of the merged images are a statistical analysis of the similarity of FITC-Prx1 and PE-anti-TLR4 in 5,000 cells on a pixel-by-pixel basis. A shift of this distribution to the right indicates a greater degree of similarity. The average similarity coefficient at each time point was demonstrated in Figure 5B. At all time points there was a high similarity of Prx1 and TLR4 staining (similarity coefficients > 1), indicating a co-localization Prx1 and TLR4. These results confirm that Prx1 and TLR4 interact on the cell surface and that at least of portion of the Prx1 is internalized with TLR4.
Figure 5. Kinetics of TLR4/Prx1 Interaction.
(A) TG-elicited macrophages were stimulated with 200 nM FITC-Prx1 or PE-conjugated anti-TLR4 (PE-TLR4). Samples were harvested at the indicated times samples and cell populations were analyzed by Amnis technology. Representative examples of immunostained cells and a merged image of the two stains for each time point are shown. The far right column shows a histogram of the pixel by pixel statistical analysis of each cell (n=5,000) analyzed in which the y-axis is number of cells and the x-axis is the similarity coefficient between Prx1 and TLR4. (B) The average similarity coefficient of all cells for each time point is shown; error bars represent standard deviation.
Stimulation of cytokine secretion and binding to TLR4 depends upon Prx1 structure
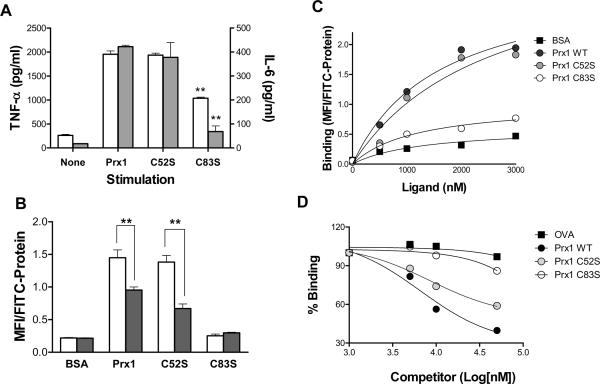
Prx1 acts as both a peroxidase and a protein chaperone (1). To determine whether the ability of Prx1 to stimulate cytokine secretion from TG-elicited macrophages was related to its peroxidase activity and/or chaperone activity, two Prx1 mutants were examined. The Prx1C52S mutant lacks peroxidase activity but retains the decamer structure needed for chaperone activity; Prx1C83S exists mainly as a dimer, has reduced chaperone activity and intact peroxidase activity (32,33,36). Cytokine secretion following Prx1C52S stimulation of TG-elicited macrophages was not significantly distinct from that observed following stimulation with Prx1 (Figure 6A); however, TG-elicited macrophages stimulated with Prx1C83S displayed a significant reduction in cytokine secretion (P≤0.01).
Figure 6. Prx1 Binding to TLR4 is Structure Dependent.
(A) TG-elicited macrophages isolated from TLR4+/+ (white bars) or TLR4−/− macrophages (filled bars) and incubated with media (None), Prx1, Prx1C52S, or Prx1C83S at 200nM for 24h and supernatants were harvested and analyzed for the presence of TNF-α and IL-6. (B) TG-elicited macrophages isolated from TLR4+/+ (white bars) or TLR4−/− macrophages (filled bars) and incubated with 2000 nM of FITC-labeled proteins for 20 minutes, followed by analysis by flow cytometry. Viable cells were selected for analysis by elimination of 7-AAD high populations. Results were normalized for any differences in FITC-labeling and reported in MFI/FITC per nM protein; error bars represent standard deviation. Asterisks indicate a P value ≤ 0.01. (C) TG-elicited macrophages were incubated with FITC-BSA (squares), Prx1 (dark circles), Prx1C52S (gray circles), and Prx1 C83S (open circles) at various concentrations for 20 min and analyzed by flow cytometry. Results are normalized for differences in FITC-labeling and reported in MFI/FITC per nM protein. Each curve is representative of three individual trials. (D) TG-elicited macrophages were incubated with 1000 nM Prx1, washed and incubated with increasing concentrations of competitors: OVA (squares), Prx1 (dark circles), Prx1C52S (gray circles), Prx1C83S (open circles). Results are shown as a percentage MFI of FITC-Prx1 with no competitor; error bars represent standard deviation. All experiments were performed in triplicate and the combined results are presented.
Prx1 binding to TG-elicited macrophages was dependent upon the presence of TLR4 as binding of Prx1 and the enzymatic null mutant (Prx1C52S) was significantly decreased in the absence of TLR4 (Figure 6B). Prx1C83S binding was minimal to either TLR4 expressing or non-expressing macrophages, confirming that Prx1 interaction with TLR4 is peroxidase independent and structure dependent.
Saturation binding (Figure 6C) and competition analyses (Figure 6D) were used to determine the Kd, and Ki values for Prx1 binding to the surface of TG-elicited macrophages. The Kd for Prx1 binding to TG-elicited macrophages was 1.6 mM and the Ki was 4.1 mM (Table 1).
Table 1.
Binding Constants for Prx1
| BSA | Prx1 | Prx1C52S | Prx1C83S | |
|---|---|---|---|---|
| Bmax (MFI/FITC Protein) | 0.6143 | 3.148 | 3.607 | 1.033 |
| Kd (mM) | 1.3 | 1.6 | 2.5 | 1.2 |
| Ki (mM) | 1.1 × 107 | 4.1 | 5.2 | 4.5 × 105 |
|
| ||||
| Log (Ki) | 10.0 | 3.6 | 3.7 | 8.6 |
Prx1 stimulation of cytokine secretion is MyD88-dependent and leads to TLR4-dependent translocation of NFκB to the nucleus
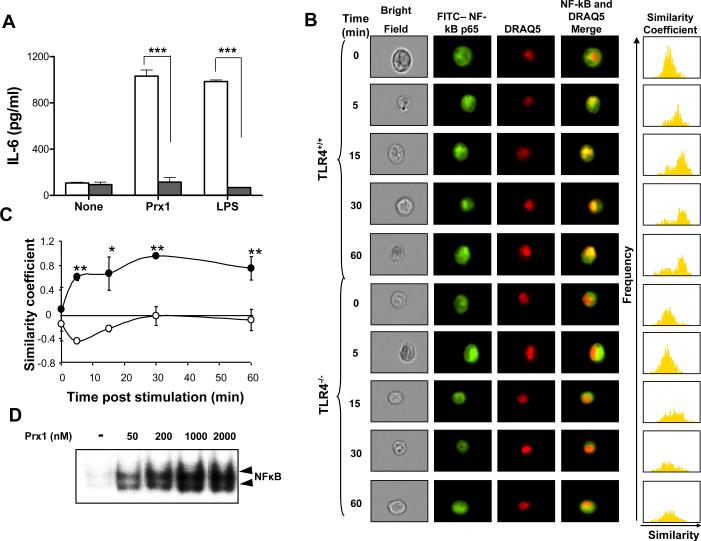
The consequential downstream signaling events of ligand-mediated activation of TLR4 can be MyD88 dependent or independent. Prx1 was used to stimulate cytokine expression from RAW264.7 cells expressing dominant negative (DN) MyD88 protein. IL-6 secretion following Prx1 stimulation is dependent on MyD88 function (Figure 7A), indicating that Prx1 activates the MyD88 signaling cascade, which can lead to activation of NFκB (20, 32).
Figure 7. Prx1 stimulation of macrophages is MyD88 dependent and leads nuclear translocation of NFκB.
(A) Stable transfectants of the RAW264.7 macrophage cell line containing control (open bar) or MyD88 DN (filled bars) expressing plasmids were stimulated with 100 nM LPS or 1000 nM Prx1 for 24 h and the resulting supernatants were assayed for IL-6 expression by ELISA. ELISA analysis was performed in three independent experiments; error bars represent standard deviation. Asterisks indicate a P value ≤ 0.001. (B) TG-elicited macrophages isolated from C3H/HeNCr (TLR4+/+) and C3H/HeNJ (TLR4−/−) mice were stimulated with 200 nM Prx1 in complete media. At the indicated time points cells were stained with FITC conjugated antibodies to NFκB p65 and DRAQ5 (nuclear stain) for 10 min and analyzed using Amnis technology. The furthest right column shows a pixel by pixel statistical analysis of the similarity of NFκB and nuclear staining. (C) The average numerical value of the overall similarity coefficients for each time point in both C3H/HeNCr (filled circles) and C3H/HeNJ (open circles) macrophages is; error bars represent standard deviation. (D) TG-elicited macrophages were incubated with the indicated concentrations of Prx1 for 1 hour. EMSA analysis was performed as described in Materials and Methods.
To determine if Prx1/TLR4 interaction leads to NFκB activation, NFκB translocation following Prx1 stimulation was analyzed in macrophages isolated from C3H/HeNCr and C3H/HeNJ mice. C3H/HeNJ mice have a mutation in the TLR4 ligand binding domain that prevents ligand binding (37). TG-elicited macrophages from C3H/HeNCr and C3H/HeNJ mice were incubated with 200 nM Prx1 at 37°C for the indicated times, transferred to ice and incubated with antibodies against NFκB p65; the nuclear stain DRAQ5 was added 15 minutes prior to image stream analysis. Prx1 incubation with macrophages isolated from C3H/HeNCr mice triggered NFκB translocation within 5 min and nuclear localization was apparent for up to 60 min (Figure 7B). In contrast Prx1 incubation with macrophages isolated from C3H/HeNJ mice did not trigger NFκB translocation (Figure 7B). The histogram to the right of the merged image column depicts the similarity of NFκB and the nuclear stain on a pixel-by-pixel basis. Prx1 stimulation led to NFκB translocation to the nucleus in a TLR4 dependent manner as demonstrated by the positive similarity coefficient observed following Prx1 stimulation of C3H/H3NCr TG-elicited macrophages, which was decreased following Prx1 stimulation of C3H/HeNJ TG-elicited macrophages (Figure 7C). The ability of Prx1 to activate NF-κB was confirmed by EMSA, which indicated that incubation of macrophages with Prx1 resulted in a dose dependent increase in NFκB DNA binding activity (Figure 7D).
DISCUSSION
We present compelling evidence that Prx1 stimulates TLR4-dependent secretion of TNF-α and IL-6 from TG-elicited macrophages and DCs. Cytokine secretion was the result of TLR4 stimulation of the MyD88-dependent signaling cascade and resulted in activation and translocation of NFκB. Prx1 is an intercellular protein that is secreted from tumor cells and activated T cells (12,13,38). The ability of Prx1 to interact with TLR4 and stimulate the release of pro-inflammatory cytokines suggests that it may also act as an endogenous damage-associated molecular pattern molecule (DAMP).
HSP72 and HMGB1, which have also been classified as endogenous DAMPs, have been shown to interact with TLR4 (17,19,39,40). Saturation and competition studies indicate that Prx1 has a Kd of ~1.3 mM and a Ki of ~4.1 mM; extrapolation of data presented by Binder et al. (41) implies that HSP72 has a Kd of 2.1–4.4 mM and a Ki of 10–21.8 mM, suggesting that Prx1 interaction with TLR4 is stronger than that of HSP72. Binding affinities are not available for HMGB1.
Identification of TLR4 as a receptor for a recombinant protein is complicated by the potential of the presence of LPS within the recombinant protein preparation. To account for this possibility in the results presented here, two controls were included in all of the performed studies. In the first control, recombinant proteins were combined with polymixin B prior to their addition to immune cells. Polymixin B is a powerful inactivator of LPS; pre-incubation of recombinant Prx1 with polymixin B had no effect on the ability of Prx1 to stimulate cytokine expression (Figure 1). However pre-incubation of LPS with the same concentration of polymixin B significantly inhibited its ability to stimulate cytokine release. As a second control, Prx1 and LPS were boiled prior to addition to immune cells; denaturing Prx1 significantly inhibited its ability to stimulate cytokine release, but boiling had no effect on the ability of LPS to stimulate cytokine release. Finally, all of the recombinant proteins used in this study were prepared in the same fashion and following purification all were found to have equivalent levels of endotoxin (~14 EU/ml), yet Prx1C83S stimulated significantly lower cytokine secretion and did not appear to bind to TLR4 expressing cells. Thus it appears as though the results demonstrating that Prx1 interacts with TLR4 are not due to the presence of LPS contamination.
Prx1, HSP72 and HMGB1 not appear to have significant structural similarity nor do these molecules appear to share homology with LPS (22,42). Prx1, HSP72 and HMGB1 are molecular chaperones and the lack of structural homology between HSP72/HMGB1 and other TLR4 ligands has led some to speculate that the chaperone cargo rather than the chaperone is being recognized by TLR4 (43,44). In support of this hypothesis, recent studies have shown that HMGB1 binding to TLR9 is a result of TLR9 recognition of HMGB1/DNA complexes (45). Extracellular Prx1 is present as a decamer, which is associated with Prx1 chaperone activity (46) and our studies indicate that Prx1 binding to TLR4 was dependent upon the ability to form decamers (Figures 3 and 4B). Thus it is possible that Prx1 binding of TLR4 is due to recognition of its cargo rather than of Prx1 itself.
The Prx1C83S mutant, which lacks chaperone activity and exists primarily as a dimer (46), did not appear to bind to TLR4 (Figure 4B); however the purified mutant protein was able to stimulate cytokine secretion from macrophages (Figure 4A). Assays for biological function are traditionally more sensitive than binding assays and it is possible that the interaction of the dimeric form of Prx1 with TLR4 was below the level of detection in the binding assay employed in these studies. A small portion of Prx1C83S is present as a tetramer (46), which may also be able to interact with TLR4 at a level that is below detection, but that is sufficient to stimulate cytokine secretion.
Prx1 stimulation of cytokine secretion was dependent on TLR4 and MyD88 (Figure 3, 4 and 5); however, FITC-labeled Prx1 did bind to macrophages isolated from TLR4−/− (B10ScNJ) mice (Figure 4B), albeit at a lower level than bound to macrophages isolated from TLR4+/+ (B6) mice. Examination of the interaction of Prx1 with TLR4 at a cellular level indicated that while a majority of the TLR4 was internalized upon Prx1 binding, at least a portion of the Prx1 remained on the cell surface (Figure 3B/C). These findings could be the result of excess Prx1 or alternatively that Prx1 is binding to additional receptors. Other TLR4 binding DAMPs have been shown to bind to multiple danger receptors (14,17,19,28,47–49) and in some cases DAMP binding to TLR4 requires co-receptors. PbA, the malaria homolog of Prx1 requires MD2 to bind to TLR4 (20); our studies indicate that Prx1 stimulation of cytokine secretion is optimal in the presence of serum and that antibodies to CD14 and MD2 block cytokine secretion from Prx1 stimulated cells. Furthermore, immunoprecipated complexes of TLR4 and Prx1 contain MD2 and CD14, suggesting that these proteins contribute to the binding of Prx1 to TLR4.
Numerous studies have shown that activation of TLRs expressed on tumor cells can act to promote tumor survival, chemoresistance, progression and metastasis (50–52). Furthermore, inflammation, such as that which occurs during chronic infection, has been shown to promote carcinogenesis primarily through the generation of a tumor permissive microenvironment and recruitment of tumor promoting macrophages (52,53). In contrast there is evidence suggesting that TLR4 induction of IL-10-producing T cells acts to regulate the destructive tendencies of inflammation and that the incidence of gastric cancer is increased in the absence of TLR4 (54,55). However, the presence of LPS, the prototypical TLR4 ligand, has been shown to accelerate tumor growth in both clinical and preclinical studies (50). Prx1 expression is elevated in various cancers and cancer cell lines (2–9,26) and elevated Prx1 levels have been linked with poor clinical outcomes and diminished overall patient survival (4,10,11). Thus it is possible that release of Prx1 from tumor cells, as has been shown to occur in lung cancer cells (12,13), and subsequent interaction with both TLR4 expressing tumor cells and innate immune cells may promote tumor growth.
In conclusion we have made the novel observation that extracellular recombinant and tumor cell released Prx1 stimulate TNF-α and IL-6 secretion from macrophages and dendritic in a TLR4, MyD88 dependent fashion. The physiological consequence of the presence of extracellular Prx1 in the tumor microenvironment is unknown; however our studies suggest that Prx1 may contribute to the generation of chronic inflammation and establishment of a microenvironment that supports tumor growth and immune evasion.
Supplementary Material
ACKNOWLEDGMENTS
We greatly appreciate the critical comments provided by Drs. Barbara W. Henderson, David Bellnier (Department of Cellular Stress Biology) and Sally Schneider (Department of Immunology) prior to publication. We also acknowledge the intellectual input provided by Dr. Young-Mee Park at the initial stages of this work.
The work was supported by NIH Grants CA109480, CA111846, CA98156, CA126667 (HM), and CA129111 (XYW) and in part by the Roswell Park Cancer Center Support Grant CA16056.
REFERENCES
- 1.Wood ZA, Schroder E, Robin HJ, Poole LB. Structure, mechanism and regulation of peroxiredoxins. Trends Biochem. Sci. 2003;28:32–40. doi: 10.1016/s0968-0004(02)00003-8. [DOI] [PubMed] [Google Scholar]
- 2.Kim HJ, Chae HZ, Kim YJ, Kim YH, Hwangs TS, Park EM, Park YM. Preferential elevation of Prx I and Trx expression in lung cancer cells following hypoxia and in human lung cancer tissues. Cell Biol. Toxicol. 2003;19:285–298. doi: 10.1023/b:cbto.0000004952.07979.3d. [DOI] [PubMed] [Google Scholar]
- 3.Noh DY, Ahn SJ, Lee RA, Kim SW, Park IA, Chae HZ. Overexpression of peroxiredoxin in human breast cancer. Anticancer Res. 2001;21:2085–2090. [PubMed] [Google Scholar]
- 4.Quan C, Cha EJ, Lee HL, Han KH, Lee KM, Kim WJ. Enhanced expression of peroxiredoxin I and VI correlates with development, recurrence and progression of human bladder cancer. J. Urol. 2006;175:1512–1516. doi: 10.1016/S0022-5347(05)00659-2. [DOI] [PubMed] [Google Scholar]
- 5.Yanagawa T, Ishikawa T, Ishii T, Tabuchi K, Iwasa S, Bannai S, Omura K, Suzuki H, Yoshida H. Peroxiredoxin I expression in human thyroid tumors. Cancer Lett. 1999;145:127–132. doi: 10.1016/s0304-3835(99)00243-8. [DOI] [PubMed] [Google Scholar]
- 6.Yanagawa T, Iwasa S, Ishii T, Tabuchi K, Yusa H, Onizawa K, Omura K, Harada H, Suzuki H, Yoshida H. Peroxiredoxin I expression in oral cancer: a potential new tumor marker. Cancer Lett. 2000;156:27–35. doi: 10.1016/s0304-3835(00)00434-1. [DOI] [PubMed] [Google Scholar]
- 7.Chang JW, Jeon HB, Lee JH, Yoo JS, Chun JS, Kim JH, Yoo YJ. Augmented expression of peroxiredoxin I in lung cancer. Biochem. Biophys. Res. Commun. 2001;289:507–512. doi: 10.1006/bbrc.2001.5989. [DOI] [PubMed] [Google Scholar]
- 8.Qi Y, Chiu JF, Wang L, Kwong DL, He QY. Comparative proteomic analysis of esophageal squamous cell carcinoma. Proteomics. 2005;5:2960–2971. doi: 10.1002/pmic.200401175. [DOI] [PubMed] [Google Scholar]
- 9.Shen J, Person MD, Zhu J, Abbruzzese JL, Li D. Protein expression profiles in pancreatic adenocarcinoma compared with normal pancreatic tissue and tissue affected by pancreatitis as detected by two-dimensional gel electrophoresis and mass spectrometry. Cancer Res. 2004;64:9018–9026. doi: 10.1158/0008-5472.CAN-04-3262. [DOI] [PubMed] [Google Scholar]
- 10.Kim JH, Bogner PN, Ramnath N, Park Y, Yu J, Park YM. Elevated peroxiredoxin 1, but not NF-E2-related factor 2, is an independent prognostic factor for disease recurrence and reduced survival in stage I non-small cell lung cancer. Clin. Cancer Res. 2007;13:3875–3882. doi: 10.1158/1078-0432.CCR-06-2893. [DOI] [PubMed] [Google Scholar]
- 11.Chen MF, Keng PC, Shau H, Wu CT, Hu YC, Liao SK, Chen WC. Inhibition of lung tumor growth and augmentation of radiosensitivity by decreasing peroxiredoxin I expression. Int. J. Radiat. Oncol. Biol. Phys. 2006;64:581–591. doi: 10.1016/j.ijrobp.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 12.Chang JW, Lee SH, Lu Y, Yoo YJ. Transforming growth factor-beta1 induces the non-classical secretion of peroxiredoxin-I in A549 cells. Biochem. Biophys. Res. Commun. 2006;345:118–123. doi: 10.1016/j.bbrc.2006.04.073. [DOI] [PubMed] [Google Scholar]
- 13.Chang JW, Lee SH, Jeong JY, Chae HZ, Kim YC, Park ZY, Yoo YJ. Peroxiredoxin-I is an autoimmunogenic tumor antigen in non-small cell lung cancer. FEBS Lett. 2005;579:2873–2877. doi: 10.1016/j.febslet.2005.04.028. [DOI] [PubMed] [Google Scholar]
- 14.Bianchi ME, Manfredi AA. High-mobility group box 1 (HMGB1) protein at the crossroads between innate and adaptive immunity. Immunol. Rev. 2007;220:35–46. doi: 10.1111/j.1600-065X.2007.00574.x. [DOI] [PubMed] [Google Scholar]
- 15.Yoshida S, Katoh T, Tetsuka T, Uno K, Matsui N, Okamoto T. Involvement of thioredoxin in rheumatoid arthritis: its costimulatory roles in the TNF-alpha-induced production of IL-6 and IL-8 from cultured synovial fibroblasts. J. Immunol. 1999;163:351–358. [PubMed] [Google Scholar]
- 16.Hunter-Lavin C, Davies EL, Bacelar MM, Marshall MJ, Andrew SM, Williams JH. Hsp70 release from peripheral blood mononuclear cells. Biochem. Biophys. Res. Commun. 2004;324:511–517. doi: 10.1016/j.bbrc.2004.09.075. [DOI] [PubMed] [Google Scholar]
- 17.Asea A, Rehli M, Kabingu E, Boch JA, Bare O, Auron PE, Stevenson MA, Calderwood SK. Novel signal transduction pathway utilized by extracellular HSP70: role of toll-like receptor (TLR) 2 and TLR4. J. Biol. Chem. 2002;277:15028–15034. doi: 10.1074/jbc.M200497200. [DOI] [PubMed] [Google Scholar]
- 18.Lotze MT, Zeh HJ, Rubartelli A, Sparvero LJ, Amoscato AA, Washburn NR, Devera ME, Liang X, Tor M, Billiar T. The grateful dead: damage-associated molecular pattern molecules and reduction/oxidation regulate immunity. Immunol. Rev. 2007;220:60–81. doi: 10.1111/j.1600-065X.2007.00579.x. [DOI] [PubMed] [Google Scholar]
- 19.Apetoh L, Ghiringhelli F, Tesniere A, Criollo A, Ortiz C, Lidereau R, Mariette C, Chaput N, Mira JP, Delaloge S, Andre F, Tursz T, Kroemer G, Zitvogel L. The interaction between HMGB1 and TLR4 dictates the outcome of anticancer chemotherapy and radiotherapy. Immunol. Rev. 2007;220:47–59. doi: 10.1111/j.1600-065X.2007.00573.x. [DOI] [PubMed] [Google Scholar]
- 20.Furuta T, Imajo-Ohmi S, Fukuda H, Kano S, Miyake K, Watanabe N. Mast cell-mediated immune responses through IgE antibody and Toll-like receptor 4 by malarial peroxiredoxin. Eur. J. Immunol. 2008;38:1341–1350. doi: 10.1002/eji.200738059. [DOI] [PubMed] [Google Scholar]
- 21.Lin WW, Karin M. A cytokine-mediated link between innate immunity, inflammation, and cancer. J. Clin. Invest. 2007;117:1175–1183. doi: 10.1172/JCI31537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fitzgerald KA, Chen ZJ. Sorting out Toll signals. Cell. 2006;125:834–836. doi: 10.1016/j.cell.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 23.O'Neill LA. When signaling pathways collide: positive and negative regulation of toll-like receptor signal transduction. Immunity. 2008;29:12–20. doi: 10.1016/j.immuni.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 24.Bowie AG. Insights from vaccinia virus into Toll-like receptor signalling proteins and their regulation by ubiquitin: role of IRAK-2. Biochem. Soc. Trans. 2008;36:449–452. doi: 10.1042/BST0360449. [DOI] [PubMed] [Google Scholar]
- 25.Kim YJ, Ahn JY, Liang P, Ip C, Zhang Y, Park YM. Human prx1 gene is a target of Nrf2 and is up-regulated by hypoxia/reoxygenation: implication to tumor biology. Cancer Res. 2007;67:546–554. doi: 10.1158/0008-5472.CAN-06-2401. [DOI] [PubMed] [Google Scholar]
- 26.Park SY, Yu X, Ip C, Mohler JL, Bogner PN, Park YM. Peroxiredoxin 1 interacts with androgen receptor and enhances its transactivation. Cancer Res. 2007;67:9294–9303. doi: 10.1158/0008-5472.CAN-07-0651. [DOI] [PubMed] [Google Scholar]
- 27.Park SY, Kim YJ, Gao AC, Mohler JL, Onate SA, Hidalgo AA, Ip C, Park EM, Yoon SY, Park YM. Hypoxia increases androgen receptor activity in prostate cancer cells. Cancer Res. 2006;66:5121–5129. doi: 10.1158/0008-5472.CAN-05-1341. [DOI] [PubMed] [Google Scholar]
- 28.Berwin B, Hart JP, Rice S, Gass C, Pizzo SV, Post SR, Nicchitta CV. Scavenger receptor-A mediates gp96/GRP94 and calreticulin internalization by antigen-presenting cells. EMBO J. 2003;22:6127–6136. doi: 10.1093/emboj/cdg572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gollnick SO, Liu X, Owczarczak B, Musser DA, Henderson BW. Altered expression of interleukin 6 and interleukin 10 as a result of photodynamic therapy in vivo. Cancer Res. 1997;57:3904–3909. [PubMed] [Google Scholar]
- 30.Inaba J, Inaba M, Romani N, Aya H, Deguchi M, Ikehara S, Muramatsu S, Steinman RM. Generation of large numbers of dendritic cells from mouse bone marrow cultures supplemented with granulocyte/macrophage colony-stimulating factor. J. Exp. Med. 1992;176:1693–1702. doi: 10.1084/jem.176.6.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang XY, Facciponte J, Chen X, Subjeck JR, Repasky EA. Scavenger receptor-A negatively regulates antitumor immunity. Cancer Res. 2007;67:4996–5002. doi: 10.1158/0008-5472.CAN-06-3138. [DOI] [PubMed] [Google Scholar]
- 32.Kim YJ, Lee WS, Ip C, Chae HZ, Park EM, Park YM. Prx1 suppresses radiation-induced c-Jun NH2-terminal kinase signaling in lung cancer cells through interaction with the glutathione S-transferase Pi/c-Jun NH2-terminal kinase complex. Cancer Res. 2006;66:7136–7142. doi: 10.1158/0008-5472.CAN-05-4446. [DOI] [PubMed] [Google Scholar]
- 33.Lee W, Choi KS, Riddell J, Ip C, Ghosh D, Park JH, Park YM. Human peroxiredoxin 1 and 2 are not duplicate proteins: the unique presence of CYS83 in Prx1 underscores the structural and functional differences between Prx1 and Prx2. J. Biol. Chem. 2007;282:22011–22022. doi: 10.1074/jbc.M610330200. [DOI] [PubMed] [Google Scholar]
- 34.George TC, Fanning SL, Fitzgeral-Bocarsly P, Medeiros RB, Highfill S, Shimizu Y, Hall BE, Frost K, Basiji D, Ortyn WE, Morrissey PJ, Lynch DH. Quantitative measurement of nuclear translocation events using similarity analysis of multispectral cellular images obtained in flow. J. Immunol. Methods. 2006;311:117–129. doi: 10.1016/j.jim.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 35.Suh J, Payvandi F, Edelstein LC, Amenta PS, Zong WX, Gelinas C, Rabson AB. Mechanisms of constitutive NF-kappaB activation in human prostate cancer cells. Prostate. 2002;52:183–200. doi: 10.1002/pros.10082. [DOI] [PubMed] [Google Scholar]
- 36.Matsumura T, Okamoto K, Iwahara S, Hori H, Takahashi Y, Nishino T, Abe Y. Dimer-oligomer interconversion of wild-type and mutant rat 2-Cys peroxiredoxin: disulfide formation at dimer-dimer interfaces is not essential for decamerization. J. Biol. Chem. 2008;283:284–293. doi: 10.1074/jbc.M705753200. [DOI] [PubMed] [Google Scholar]
- 37.Triantafilou M, Triantafilou K. Lipopolysaccharide recognition: CD14, TLRs and the LPS-activation cluster. Trends Immunol. 2002;23:301–304. doi: 10.1016/s1471-4906(02)02233-0. [DOI] [PubMed] [Google Scholar]
- 38.Geiben-Lynn R, Kursar M, Brown NV, Addo MM, Shau H, Lieberman J, Luster AD, Walker BD. HIV-1 antiviral activity of recombinant natural killer cell enhancing factors, NKEF-A and NKEF-B, members of the peroxiredoxin family. J. Biol. Chem. 2003;278:1569–1574. doi: 10.1074/jbc.M209964200. [DOI] [PubMed] [Google Scholar]
- 39.Park JS, Gamboni-Robertson F, He Q, Svetkauskaite D, Strassheim D, Sohn JW, Yamada S, Maruyama I, Banerjee A, Ishizaka A, Abraham E. High mobility group box 1 protein interacts with multiple Toll-like receptors. Am. J. Physiol Cell Physiol. 2006;290:C917–C924. doi: 10.1152/ajpcell.00401.2005. [DOI] [PubMed] [Google Scholar]
- 40.Park JS, Svetkauskaite D, He Q, Kim JY, Strassheim D, Ishizaka A, Abraham E. Involvement of toll-like receptors 2 and 4 in cellular activation by high mobility group box 1 protein. J. Biol. Chem. 2004;279:7370–7377. doi: 10.1074/jbc.M306793200. [DOI] [PubMed] [Google Scholar]
- 41.Binder RJ, Harris ML, Menoret A, Srivastava PK. Saturation, competition, and specificity in interaction of heat shock proteins (hsp) gp96, hsp90, and hsp70 with CD11b+ cells. J. Immunol. 2000;165:2582–2587. doi: 10.4049/jimmunol.165.5.2582. [DOI] [PubMed] [Google Scholar]
- 42.Kim HM, Park BS, Kim JI, Kim SE, Lee J, Oh SC, Enkhbayar P, Matsushima N, Lee H, Yoo OJ, Lee JO. Crystal structure of the TLR4-MD-2 complex with bound endotoxin antagonist Eritoran. Cell. 2007;130:906–917. doi: 10.1016/j.cell.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 43.Asea A, Kabingu E, Stevenson MA, Calderwood SK. HSP70 peptidembearing and peptide-negative preparations act as chaperokines. Cell Stress. Chaperones. 2000;5:425–431. doi: 10.1379/1466-1268(2000)005<0425:hpbapn>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Williams JH, Ireland HE. Sensing danger--Hsp72 and HMGB1 as candidate signals. J. Leukoc. Biol. 2008;83:489–492. doi: 10.1189/jlb.0607356. [DOI] [PubMed] [Google Scholar]
- 45.Tian J, Avalos AM, Mao SY, Chen B, Senthil K, Wu H, Parroche P, Drabic S, Golenbock D, Sirois C, Hua J, An LL, Audoly L, La RG, Bierhaus A, Naworth P, Marshak-Rothstein A, Crow MK, Fitzgerald KA, Latz E, Kiener PA, Coyle AJ. Toll-like receptor 9-dependent activation by DNA-containing immune complexes is mediated by HMGB1 and RAGE. Nat. Immunol. 2007;8:487–496. doi: 10.1038/ni1457. [DOI] [PubMed] [Google Scholar]
- 46.Wood ZA, Poole LB, Hantgan RR, Karplus PA. Dimers to doughnuts: redox-sensitive oligomerization of 2-cysteine peroxiredoxins. Biochemistry. 2002;41:5493–5504. doi: 10.1021/bi012173m. [DOI] [PubMed] [Google Scholar]
- 47.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 48.Harris HE, Raucci A. Alarmin(g) news about danger: workshop on innate danger signals and HMGB1. EMBO Rep. 2006;7:774–778. doi: 10.1038/sj.embor.7400759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kono H, Rock KL. How dying cells alert the immune system to danger. Nat. Rev. Immunol. 2008;8:279–289. doi: 10.1038/nri2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Killeen SD, Wang JH, Andrews EJ, Redmond HP. Exploitation of the Toll-like receptor system in cancer: a doubled-edged sword? Br. J. Cancer. 2006;95:247–252. doi: 10.1038/sj.bjc.6603275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen K, Huang J, Gong W, Iribarren P, Dunlop NM, Wang JM. Toll-like receptors in inflammation, infection and cancer. Int. Immunopharmacol. 2007;7:1271–1285. doi: 10.1016/j.intimp.2007.05.016. [DOI] [PubMed] [Google Scholar]
- 52.Chen R, Alvero AB, Silasi DA, Steffensen KD, Mor G. Cancers take their Toll--the function and regulation of Toll-like receptors in cancer cells. Oncogene. 2008;27:225–233. doi: 10.1038/sj.onc.1210907. [DOI] [PubMed] [Google Scholar]
- 53.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.El-Omar EM, Ng MT, Hold GL. Polymorphisms in Toll-like receptor genes and risk of cancer. Oncogene. 2008;27:244–252. doi: 10.1038/sj.onc.1210912. [DOI] [PubMed] [Google Scholar]
- 55.Higgins SC, Lavelle EC, McCann C, Keogh B, McNeela E, Byrne P, O'Gorman B, Jarnicki A, McGuirk P, Mills KH. Toll-like receptor 4-mediated innate IL-10 activates antigen-specific regulatory T cells and confers resistance to Bordetella pertussis by inhibiting inflammatory pathology. J. Immunol. 2003;171:3119–3127. doi: 10.4049/jimmunol.171.6.3119. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.