Abstract
Poly(ADP-ribosyl)ation is a ubiquitous protein modification found in mammalian cells that modulates many cellular responses, including DNA repair. The poly(ADP-ribose) polymerase (PARP) family catalyze the formation and addition onto proteins of negatively charged ADP-ribose polymers synthesized from NAD+. The absence of PARP-1 and PARP-2, both of which are activated by DNA damage, results in hypersensitivity to ionizing radiation and alkylating agents. PARP inhibitors that compete with NAD+ at the enzyme’s activity site are effective chemo- and radiopotentiation agents and, in BRCA-deficient tumors, can be used as single-agent therapies acting through the principle of synthetic lethality. Through extensive drug-development programs, third-generation inhibitors have now entered clinical trials and are showing great promise. However, both PARP-1 and PARP-2 are not only involved in DNA repair but also in transcription regulation, chromatin modification, and cellular homeostasis. The impact on these processes of PARP inhibition on long-term therapeutic responses needs to be investigated.
Keywords: Poly(ADP-ribose)polymerase, PARP-1, PARP-2, Poly(ADP-ribosyl)ation, Synthetic lethality
Introduction
While deficiencies in DNA repair processes are associated with cancer susceptibility as has been seen in cancer-prone syndromes such as Xeroderma pigmentosum, Ataxia Telangiectasia (AT), Nijmegen Breakage Syndrome (NBS), and Fanconi’s anemia, the inhibition of DNA repair and other damage response proteins by small-molecule inhibitors can potentially be exploited to sensitize tumor cells when used in combination with chemo- and radiotherapy or in certain genetic backgrounds. Many therapies, including alkylating agents such as temozolomide (TMZ), camptothecins and radiation produce DNA single-strand breaks (SSBs) and inhibitors of poly(ADP-ribose)polymerase (PARP), an essential player in the repair of this type of DNA damage, have shown great potential as sensitizers in a variety of tumor types. This paper will briefly review the PARP family and its two most abundant members, PARP-1 and PARP-2, their role in DNA repair and other biological processes, and how PARP activity is being targeted for cancer therapy.
Background: the PARP family
Poly(ADP-ribosyl)ation is a ubiquitous protein modification involved in the regulation of transcription, cell proliferation, differentiation, DNA methylation and apoptosis which modulates protein function by regulating either enzymatic activities or macromolecular interactions between proteins, DNA or RNA [1–3]. This modification is carried out by the PARPs, also known as ADP-ribosyltransferases (ADPRTs) or poly(ADP-ribose) synthetases a family of abundant eukaryotic enzymes. The family members are activated in response to different cellular stresses including transient and localized DNA strand breaks caused by a variety of biological process such as DNA repair, recombination, stalled replication forks, gene rearrangements, as well as by oxidative stress, DNA-binding drugs and in certain circumstances protein–protein interactions in the absence of DNA damage. Once activated, the enzyme rapidly catalyzes the transfer of ADP-ribose moieties from NAD+ to the acceptor protein resulting in the attachment of linear or branched polymers of ADP-ribose (PAR). PARP-1 itself is the primary target for poly(ADP-ribos)ylation in vivo, with more than 90% of PAR being found on PARP-1 [4]. The modifications are heterogeneous with respect to length (from a few to 200 ADP-ribose units) and extent of branching (one branch per 20–50 ADP-ribose units). A 20 amino-acid PAR-binding motif was initially established from the analysis of several DNA-repair and checkpoint proteins [5] and more recently, using proteomic approaches, a PAR-binding motif was also identified in a group of ribonucleoproteins [6]. It was also shown that the structural macro domain is an ADP-ribose-binding module [7]. Such domains have been found in macroH2A, a histone variant involved in transcriptional repression and chromosome X inactivation [8] and PARP-9/BAL1, which is over-expressed in diffuse large B-cell lymphomas [9]. In addition to the direct covalent modifications of proteins by their PARylation, some proteins have a high affinity for the polymers themselves and this is exploited in some settings for the control of their localization and function [1].
Poly(ADP-ribosyl)ation is a dynamic process consuming substantial amounts of NAD+. The in vivo half-life of the polymer is <1 min with the steady-state levels of PAR being regulated by the catalytic reactions of poly(ADP-ribose) glycohydrolase (PARG) and possibly the ADP-ribose hydrolase ARH3. ADP-ribosyl protein lyase, which cleaves the link between the first ADP-ribose and the modified amino acids, has been described in rat tissues and might also function in human cells [10]. The degradation of PAR may begin immediately after the initiation of PAR synthesis and can be completed within minutes after the cessation of PAR synthesis has occurred [4]. This generates large amounts of AMP that in turn activates the bioenergetic sensor AMP-activated protein kinase (AMPK).
Based on a structural homology with the catalytic domain of the PARP-1 protein 17 PARP family members have been identified using bioinformatics’ approaches [3]. In addition to the catalytic domain, these proteins typically contain one or more additional motifs or domains, including zinc fingers, “BRCA1 C-terminus-like” (BRCT) motifs, ankyrin repeats, macro domains and WWE domains (involved in DNA or RNA binding, protein–protein interaction or cell signaling), conferring unique properties on each PARP protein [11]. The catalytic domain of PARP-1 contains three crucial residues: a histidine and a tyrosine that are important for NAD+ binding and a glutamic acid that is essential for polymerase activity (discussed in [10]). This latter residue has been replaced in PARPs 6–16 and raises the question as to whether these proteins have poly- or mono-(ADP-ribosyl)ating activity. For instance PARP-10 has transferase rather than polymerase activity [12]. A tentative classification of PARP-family members has been proposed according to their putative functional domains or established functions: DNA-dependent PARPs (PARP-1 and PARP-2), tankyrases, CCCH-type zinc-finger PARPs, and macroPARPS [3]. Indeed among the 17 members of the PARP family, PARP-1 and PARP-2 are the only ones reported until now to be highly stimulated by DNA damage.
PARP-1
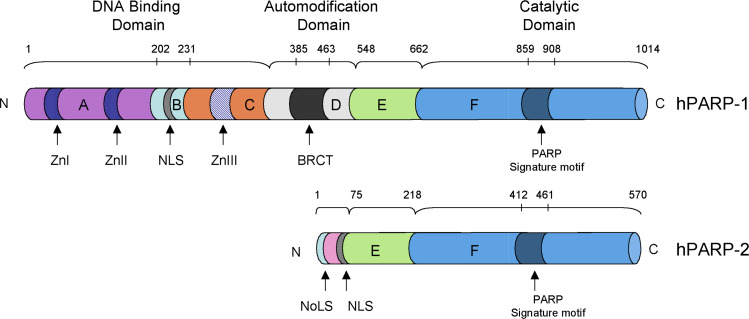
PARP-1, the founding family member, is responsible for the synthesis of the majority of PAR in eukaryotic cells and after the histones, is the most abundant nuclear protein [13]. The gene is located on chromosome 1q41-42 and the 113-kD human PARP-1 (hPARP-1) protein is organized into at least six domains, four of which have well-defined functions (Fig. 1). Domain A in the N-terminal region is the DNA-binding domain (DBD). Its affinity for damaged DNA is regulated by two zinc-finger motifs which are sufficient to target the entire protein to the damaged DNA [14]. The two PARP-1 zinc-finger motifs are unique as they recognize altered DNA structures rather than specific sequences: they are known to recognize DNA nicks, overhangs, blunt ends, and other forms of damage [14–16]. The B domain contains a bipartite nuclear localization signal (NLS) and a caspase-3 cleavage site. The auto-modification domain D contains a BRCT motif via which PARP-1 participates in various protein–protein interactions. The domain F is the catalytic C-terminal region [11]. This domain can be reduced to just a 40-kDa C-terminal polypeptide without losing the basal catalytic activity [17]. Little is known about the function of the C and E domains. However, a third zinc domain in hPARP-1 has recently been characterized in the C domain [18, 19]. This region is thought to modulate the N-terminal-to-C-terminal communication that leads to the DNA-dependent activation of PARP-1 [18]. Although this zinc ribbon domain does not bind DNA directly, it is necessary for DNA-stimulated activation of the full-length enzyme [19]. This has been recently confirmed by structural analysis of PARP-1 complexed with damaged DNA [20].
Fig. 1.
The domain structure of the two DNA damage-dependent human PARPs, PARP-1 and PARP-2, showing the DNA-binding, the automodification and catalytic domains (domains A–F). The PARP signature sequence (in dark blue within the F catalytic domain) is the most conserved between the PARPs. Zn I and Zn II: Zinc-finger motifs. Zn III: Zinc ribbon domain. NLS nuclear localization signal; BRCT BRCA1 carboxy terminus; NoLS nucleolar localization signal
Under normal conditions, PARP-1 is associated with histones, DNA, and other chromatin-associated factors [21]. Recently, it has been shown that the auto-modification of PARP-1 mediates its tight binding to the nuclear matrix in rat liver cells, creating two PARP-1 pools, and that this binding is accompanied by increased PARP-1 activity [22].
Expression patterns and genetic variation
PARP-1 and PARP-2 expression and activity have been assessed in a variety of different cells and tissues. Grube and Bürkle [23] compared PARP activities in purified leukocytes from 13 mammalian species and found a strong correlation between activity and lifespan with human cells displaying approximately five times the activity of rat cells. Intra-species comparisons revealed a decline in activity with age for PARP-1 and PARP-2 but interestingly no correlation between protein levels and enzymatic activity (reviewed in [24]). The presence of sequence variants, such as single nucleotide polymorphism (SNPs), in DNA damage detection and repair genes has been associated with altered cancer risk and responses to therapy treatment through the modulation of enzymatic activity in some cases. Five SNPs with a variant allele frequency >5% have been reported in the SNP500 study populations within the coding region of the PARP-1 gene (http://snp500cancer.nci.nih.gov/snplist.cfm). These include the Val762Ala variant (c.2285T > C rs1136410) located in exon 17 of the PARP-1 gene that has been associated with a reduced enzymatic activity [25–27] and an increased risk of developing cancers of the prostate, esophageal, lung and thyroid cancer (reviewed in [24]). A micro satellite polymorphism in the promoter, consisting of a variable number of CAs, has also been identified [28]. This is located close to the binding site of the Yin Yang 1 transcription factor that has been shown to stimulate poly(ADP-ribosy)lation and DNA repair [29–31]. Zaremba and colleagues [24] undertook a comprehensive assessment of PARP-1 expression and activity in relation to PARP-1 SNPs in a panel of 19 solid and hematological adult and pediatric human cancer cell lines and found a wide range of PARP activity which was on average higher than the mean activity measured in normal human lymphocytes from healthy volunteers and cancer patients, and supports other observations of increased PARP activity in tumors. For example, PARP-1 expression is higher in hepatocellular carcinoma vs. normal liver tissue [32]. However, no correlation was found between PARP activity and PARP-1 protein expression or an association with the status of either the Val762Ala or the (CA)n polymorphisms. Dysregulation of PARP-1 gene expression has been found in a set of 35 breast carcinomas with the majority of breast tumors strongly expressing PARP-1 mRNA showing an amplification of the 1q41-44 region where PARP-1 is localized [33]. Clearly, the regulation of PARP-1 activity is complex and needs further investigations.
PARP-1 and DNA repair
A number of parp-1 knockout mice have been generated (for instance [34–36]). These animals are viable and fertile, suggesting that PARP-1 activity is not essential for viability in the absence of genotoxic stress. However, the mice and derived cells are hypersensitive to ionizing radiation and alkylating agents, such as MNNG, MNU, or MMS [37]. Thus although PARP-1 appears to be dispensable for normal cellular activity, it would appear to be an essential survival factor for recovery from DNA damage. A second phenotype observed in these knockout animals is increased genomic instability. A four- to fivefold increase in the frequency of sister chromatid exchange has been reported in the PARP-1 knockout derived cells [38].
In vitro, the first role that was described for PARP-1 was in the response to DNA damage. As discussed above PARP-1 is a molecular sensor of DNA strand breaks and through its ribosylating activity plays a key role in the spatial and temporal organization of their repair. The main targets for this activity are PARP-1 itself, histones H1 and H2B, high mobility group proteins (HMGP) (non-histone components of chromatin), topoisomerases I and II, DNA helicases, SSB repair (SSBR) and base-excision repair (BER) proteins and various transcription factors with proteins either directly interacting with the ADP-ribosylated PARP or via the PARs. Through its own poly(ADP-ribosyl)ation and that of histones, PARP-1 loosens the chromatin allowing access to DNA strand breaks and is also involved in the recruitment of the components of the BER/SSBR pathways [39]. The automodification of PARP-1 itself promotes its interaction with XRCC1 through its BRCT domain. This leads to the recruitment of DNA polymerase β and DNA ligase III thus completing the repair of the single-strand break. PARP-1 has also been shown to be involved in human flap endonuclease 1 (FEN-1)-dependent strand displacement during DNA synthesis by DNA polymerase β [40]. In addition to its role in BER, PARP-1 is involved in several other DNA damage-repair processes, including the repair of DNA double-strand breaks (DSBs). For instance, the recruitment of MRE11, NBS1, and ATM to DSBs has been reported to depend on PARP-1 activity [41] and PARP-1 is a component of an alternative pathway of non-homologous end joining (NHEJ) [42]. When essential components of the classical pathway of NHEJ are absent, PARP-1 is recruited for DSB repair with the ligase III and probably with histone H1 [43]. The presence of poly(ADP-ribose) chains at the site of DNA damage also acts as an anti-recombinogenic factor, preventing the inappropriate recombination of homologous DNA [44].
PARP-1 and chromatin structure
In the context of the cellular response to DNA damage, histone modifications also play an important role in the recognition and accessibility of sites to the DNA-repair machinery. Early biochemical studies suggested that PARP-1 could disrupt chromatin structure by poly(ADP-ribosyl)ation of histones (mainly histone H1) and destabilization of nucleosomes [45–47]. As mentioned above, chromatin poly(ADP-ribosyl)ation leads to its decondensation, followed by the release of PARP-1 from the DNA allowing the recruitment and access of various DNA repair enzymes [48].
PARP-1 binds to histones H1, H2A, and H2B. In addition, the core histones H3 and H4 are essential for PARP-1’s interaction with nucleosomal particles [49]. While histone H4 activates PARP-1 activity, histone H2A completely inhibits PARP-1 activity [49]. Non-histone chromosomal proteins, including HMGP and the heterochromatin proteins HP1α and HP1β are also poly(ADP-ribosyl)ated [50, 51]. In addition to its role in DNA repair, PARP-1 also plays a role in chromatin repair after oxidative stress. Nucleosomal histones are known to protect DNA from free radical-mediated damage. This protection is dependent on nucleosome assembly and does not reflect the presence of scavenging proteins [52]. Oxidatively damaged histones are able to cross-link with DNA and would impair the detachment-reassembly process. Thus, to maintain genomic integrity, the oxidized histones must be degraded by the 20S proteasome and this degradation is highly selective following oxidation. While histones H1 and H4 have the highest degradation rates, PARylated histone H2A is at the other end of the spectrum [53]. The 20S proteasome is known to interact with PARylated PARP-1 leading to the specific stimulation of its proteolytic activity [54]. It has been shown recently that poly(ADP-ribosyl)ation protects histones from degradation and that oxidized histones are less PARylated, making them more prone to proteosomal degradation by the activated proteasome and thus protecting the integrity of the chromatin [55]. The damage-induced phosphorylation of histone variant H2AX may serve as a signal for the timely recruitment and/or retention of DNA repair and checkpoints proteins in the vicinity of DNA lesions. The H2AX-associated factors promote both integration and dissociation of H2AX and exchange with conventional H2A histone. These factors include FACT (Spt16/SSRP1), DNA-PK and PARP-1. It has been shown that FACT, involved in the H2AX exchange process, is stimulated by phosphorylation and inhibited by ADP-ribosylation [56]. More recently, the chromatin remodeling enzyme ALC1 (amplified in liver cancer 1), a member of the SNF2 superfamily of ATPases, has been shown to be rapidly recruited to DNA damage sites via an interaction with poly(ADP)ribosylated PARP-1, activating its ATPase and chromatin remodeling activities and catalyzing PARP1-stimulated nucleosome sliding [57, 58].
The combined effects of (ADP-ribosyl)ation and other post-translational modifications have also been described. For instance (ADP-ribosyl)ation can occur on acetylated core histones and newly synthesized core histones can be reversibly acetylated and (ADP-ribosyl)ated, probably to facilitate their assembly into histone complexes and accessibility during replication [59].
PARP-1 and PARG have also been shown to influence chromatin domain structure through their interaction with the NAD+-dependent histone-deacetylase silent information repressor protein-2 (SIR2) resulting in the silencing of specific chromosomal domains in drosophila [60]. The molecular dissection of these interactions is facilitated in this model system that only has a single Parp gene which encodes a protein with the same domain structure as PARP-1, a single tankyrase gene and a single gene predicted to encode a PARG protein and in which Parp mutations are lethal and drastically alter many aspects of developmental physiology [61]. The activation and ADP-ribosylation of PARP (and possibly other local chromatin proteins) loosens chromatin structure early in the silencing process and this facilitates SIR2 access to acetylated histone tails. PARG contributes to this process by degrading protein bound ADP-ribose moieties, and thus controlling SIR2 activity and localization [60].
Another dimension to the regulatory functions of PARP-1 is its role in the regulation of DNA methylation patterns, gene silencing, and imprinting (reviewed by [62]). CTCF, a nuclear factor involved in imprinting and insulator processes, activates PARP-1 activity inducing PARP-1 auto-modification. These polymers non-covalently interact with the DNA methyl-transferase 1 (Dnmt1), inhibiting its enzymatic activity. In the absence of PARylated PARP-1, the Dnmt1 is free to methylate DNA, whereas if the levels of PARylated PARP-1 remain high (for instance if PARG is absent), the stable inhibition of Dnmt1 would prevent DNA methylation resulting in DNA hypomethylation. It would seem that not only can PARylated PARP-1 bind Dnmt1 but it can also bind to the Dnmt1 promoter, maintaining this region unmethylated [63]. Thus the deregulation of PAR levels, which is dependent on the balance between PARP and PARG activities, could have a major impact on cellular methylation patterns.
Other cellular functions of PARP-1
As discussed above, PARP-1 can influence transcription and gene expression via several non-exclusive mechanisms including its histone-modifying enzymatic activity promoting the dissociation of nucleosomes and the decondensation of chromatin and regulating genome methylation patterns. In addition, PARP-1 can act through its direct interaction with transcription factors and their binding sites. PARP-1 has been shown to function as a transcriptional co-activator of NF-κB, the transcription factor that regulates immune and inflammatory response genes [64].
The discovery that PARP-1 can also be activated by certain types of DNA structures, such as hairpins, cruciforms, and supercoiled DNA [15], invalidated the dogma that DNA beaks are essential for its activation. Protein–protein interactions in the absence of DNA in vivo and in vitro, as reported for phosphorylated ERK2 [65] and for the CCCTC-binding factor [66], have also been shown to result in PARP-1 activation. The elucidation of the mechanism that leads to this activation remains to be established.
PARP-2
Cells derived from parp-1 knockout mice were shown to be capable of synthesizing poly(ADP-ribose) in response to DNA damage [67]. Such studies led to the discovery of a second DNA-damage-activated PARP, PARP-2, that is also a nuclear protein and forms PAR in response to DNA damage. The activity of PARP-2 accounts for approximately 10% of the total PARP activity of human cells [68]. Mice in which the parp-2 gene is deleted are viable, however if both parp-1 and parp-2 are deleted embryonic lethality is observed, suggesting that there is some overlap in their functional activities [69]. Although Parp-2 knockout mice are generally less sensitive to DNA damage produced by IR than parp-1 knockout mice [69], parp-2 knockout cells are more sensitive to doses below 2 Gy, raising the possibility that PARP-2 plays a role in the G2/M phase cell-cycle checkpoint activated in response to low doses of ionizing radiation [70].
Among the PARP family members, PARP-2 is the closest relative of PARP-1 with their catalytic F-domains having approximately 69% similarity. The N-terminal domain of murine PARP-2 does not contain zinc-finger motifs but a highly basic DBD, a NLS and a nucleolar localization signal (NoLS; Fig. 1) which displays homology with the SAP domain found in various nuclear proteins like APE-1 and Ku-70, involved in chromosomal organization or DNA repair. The PARP-2 DBD is structurally different from that of PARP-1, probably reflecting differences in the DNA structures recognized by each enzyme [11, 68]. The PARP-2 E domain acts both as the interacting interface with various partners and as an auto-modification domain [71]. The border between the E and F domains contains a caspase-8 cleavage site [72].
PARP-1 and PARP-2 can homo-and hetero-dimerize and poly(ADP-ribose)ate each other [71]. PARP-2 was found to interact with the BER proteins XRCC1, DNA polymerase β and DNA ligase III, all of which are PARP-1 partners [71]. PARP-2 is a component of a functional BER complex in vivo, however, PARP-1 and PARP-2 were shown to accumulate with different kinetics at laser-induced DNA damaged sites, leading to an implication of PARP-2 at later steps in the BER/SSB repair process [73]. PARP-2 was recently shown to be critical for PARP-1 mediated activation of homologous recombination at stalled replication forks [74]. Some other specific functions have been shown for PARP-2, for instance a role in genome surveillance, the maintenance of telomere integrity, translocations during immunoglobulin class switch recombination, spermatogenesis, adipogenesis and T cell development, and the maintenance of centromeric heterochromatin integrity (reviewed in [72]). PARP-2 has been proposed to play a role in the G2/M phase cell-cycle checkpoint in response to low-dose ionizing radiation, pointing to possible clinical applications of specific PARP-2 inhibitors in combination with continuous low-dose radiotherapy [72].
The cellular consequences of PARP inhibition
PARP inhibitors and chemopotentiation
One of the first reports that a PARP inhibitor enhanced cytoxicity came 30 years ago from the group of Sydney Shall [75] who demonstrated that 3-aminobenzamide (3-AB) prevented the rejoining of DNA strand breaks caused by the alkylating agent dimethyl sulphate and increased its toxicity in L1210 mouse leukemia lymphoblast cells. 3-AB is a simple analogue of nicotinamide, the other product of PARP-1 mediated catalysis of NAD+, itself a weak PARP inhibitor. Most inhibitors act through binding to the catalytic domain of the enzyme and thus while are specific for PARP activity, are not selective for the different members of the PARP superfamily. The benzamides are approximately 10–20 fold more potent than nicotinamide but lack the specificity and potency to be of use in preclinical and clinical studies. These observations lead several groups to embark on PARP inhibitor development programs. For instance, Banasik and colleagues [76] identified a number of more potent inhibitors including 1,5-dihydroisoquinoline, 2-methyquinazolin-4-(3H)-one, 4-amino-1-8-napthalimide and 2-nitro-6-(5H)-phenanthridinone, which have been used as lead compounds for subsequent drug development and the identification of the essential features for efficient PARP inhibition through structure–activity studies (see [77]). Inhibitor design has also been aided by crystallographic studies of inhibitors bound to PARP catalytic sites, such as those recently published for the catalytic domain of human PARP-2 in complex with the PARP inhibitor ABT-888 [78]. The results contribute to the understanding of the structural similarities and differences that can be exploited to design more selective inhibitors but for most cross-reactivity between different members of the human PARP family is observed. These proteins share only between 18 and 45% homology in their catalytic domains, but crystal structures indicate their structure is conserved and the mode of NAD+ cofactor binding is very similar.
Using this rational drug design approach, more potent inhibitors have been identified and that had low toxicity and were active in combination studies with anticancer chemotherapies in xenograft models (see [79]). For instance, Miknyocksli et al. [80] showed that CEP-6800, a 3-amonomethyl carbazole imide inhibitor of PARP-1 and PARP-2, potentiated the antitumor activity of TMZ, irinotecan, and cisplatin against tumor xenografts in mouse, Tentori et al. [81] that GP1 15427 could enhance the antitumor efficacy of TMZ against melanoma, glioblastoma multiforme, and lymphoma growing in the mouse brain, and Calabrese and colleagues found an improved therapeutic index with AG14361, an inhibitor 1,000 times more potent than 3-AB, in combination with TMZ, irinotecan, and radiation in a human colon tumor xenograft model [82]. The monofunctional alkylating agent TMZ generates three major modified bases in DNA, N7-methylguanine, 3-methyladenine and O6-methylguanine, the latter being repaired by the O6-methylguanine-DNA methyltransferase (AGT). If not repaired by AGT, the mismatch repair (MMR) pathway intervenes, but as the modified base is located in the template strand, this triggers repeated cycles of MMR-mediated excision/resynthesis, which eventually leads to the generation of single-stand breaks, growth arrest, and apoptosis. The enhancement of the antitumor activity of TMZ by the presence of a PARP inhibitor comes from the blocking of BER that removes the N-methylpurines (reviewed in [79]). However, recent data from Horton et al. [83] has shown that potentiation can be found in a variety of repair backgrounds in leukemia cells. They found that while the inhibitor ABT-888 enhanced TMZ growth inhibition most effectively in MMR-deficient cells with low MGMT activity, it also potentiated TMZ activity in MMR-deficient cells with elevated MGMT activity and unexpectedly in MMR-proficient leukemia cells. Clearly the mechanistic basis of these observations needs further investigation but they do suggest that the potentiation might be independent of MGMT status opening up the possibility of using this drug combination in other clinical settings.
The first phase 1 clinical trial of a PARP inhibitor was carried out between 2003 and 2005 with AGO14699, a potent tricyclic indole inhibitor, in combination with TMZ in patients with advanced solid tumors [84] and paved the way for other trials with other PARP inhibitors in combination with chemotherapeutic agents (www.clinicaltrials.gov; recently reviewed in [10] and [85]). A total of 48 clinical trials involving PARP inhibitors in a cancer setting are registered: 24 phase I, 2 phase I/II, 2I phase II and one phase III study involving over 4,500 patients. The majority of these trials will combine a PARP inhibitor with standard chemotherapeutic protocols, for instance with TMZ in patients with skin and solid tumors, with the largest being a phase III trial of gemcitabine/carboplatin with or without the PARP inhibitor BS1-201 in patients with previously untreated advanced squamous cell lung cancer, the ECLIPSE trial, which has a start date of March 2010. The remaining trials are either combining PARP inhibitors with radiation or are targeting tumors, which as discussed below, are genetically predisposed to die when PARP activity is inactivated or share characteristics of such tumors.
PARP inhibitors and radiosensitization
Early experimental evidence had also shown that PARP inhibitors acted as radiosensitizers in rodent cells, but their effect against human cells was often only marginal ([86] and refs therein). To elucidate the origin of this apparent differential radiosensitization, our group assessed the radiation survival of a panel of human and rodent cell lines including PARP-1 repair-deficient and repair-proficient mouse 3T3 fibroblasts cell lines, in the absence and presence of the PARP inhibitor 4-amino-1,8-naphthalimide (ANI). The results suggested that radiosensitization by ANI depends primarily on the percentage of cells engaged in DNA replication. Indeed, in synchronized HeLa cells, radiosensitization by ANI was found to occur specifically in the S phase of the cell cycle [86].
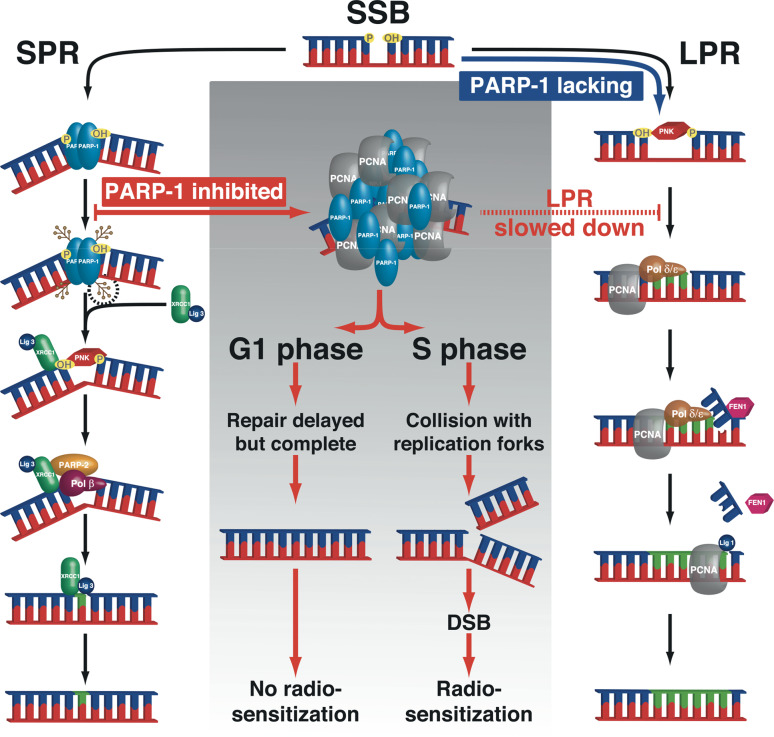
The consequences of PARP-1 disruption or inhibition on DNA SSBR and radio-induced lethality were further investigated in synchronized, isogenic HeLa cells stably silenced or not for PARP-1 (PARP-1-KD) or XRCC1 (XRCC1-KD) [87]. PARP inhibition prevented the recruitment of XRCC1 at sites of 405 nm laser micro irradiation, slowed SSBR tenfold and triggered the accumulation of large persistent foci of PARP-1 and PCNA at photo-damaged sites which we believe hinder the recruitment of other effectors of the BER pathway. Under these experimental conditions where PARP was inhibited, radiosensitization occurs only in those cells treated in S phase (Fig. 2) due to the collision of the persisting SSBs with replication forks and the formation of a lethal DNA double strand breaks. The inhibitor used in this study, as in many, targets both PARP-1 and PARP-2 activity and thus formally a role of PARP-2 in this radiosensitization can not be ruled out. The development of inhibitors with a greater specificity for PARP-2 compared to PARP-1 and cell line models lacking just PARP-2 will allow us to address its relative contribution to this response.
Fig. 2.
Scheme summarizing the differential effect of PARP-1 inhibition versus silencing on SSB repair via the BER pathway. The experimental evidence would suggest that SSB rejoining proceeds via the LPR sub-pathway when the SPR is deficient because of a lack of PARP-1. In contrast, inhibition of poly(ADP-ribosyl)ation in PARP-1-proficient cells was found to result in the accumulation of PARP-1 and PCNA in the vicinity of DNA damaged sites, with a tenfold reduction of the bulk rate of SSBR. This does not impact on radiosensitivity in the G1 phase of the cell cycle, probably because cells have enough time to perform SSB repair. However, under such conditions in S phase collision of unrepaired SSBs with replication forks results in a increase of radiosensitivity due to the formation of a large number of DSBs (reproduced with permission from [87])
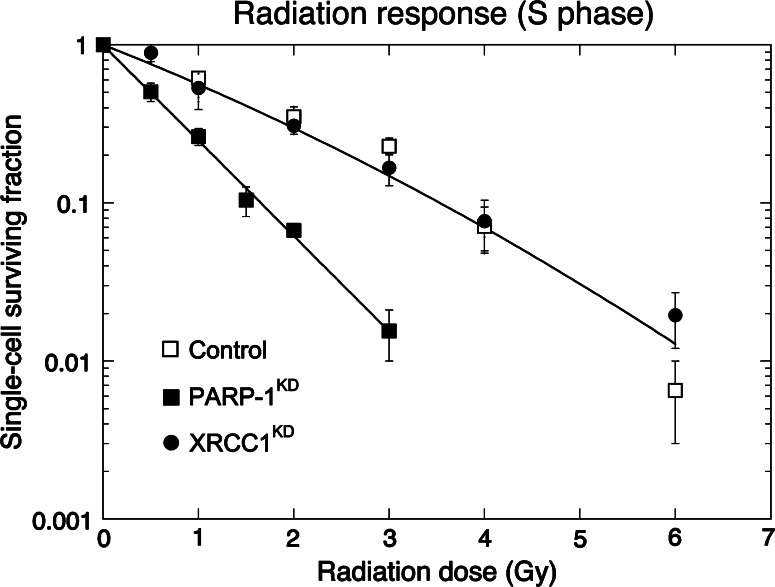
PARP-1 silencing also prevented XRCC1 recruitment to damage sites but did not lengthen the lifetime of PCNA foci and we found that PARP-1-KD and XRCC1-KD cells in S phase completed SSBR as measured by alkaline elution as rapidly as in controls, while SSBR was delayed in G1. Taken together, the data demonstrate that a PARP-1 and XRCC1-independent SSBR pathway operates when the short patch repair (SPR) branch of BER is absent. The likely mechanism is a switch from SPR to long patch repair branch as PCNA recruitment at photo-damaged sites was normal in PARP-1-KD cells (Fig. 2). PARP-1 silencing, however, under these conditions elicited hyper-radiosensitivity, which is not seen in cells in which XRCC1 is depleted (Fig. 3). As SSBR is completed in both cell types, it would suggest that some other activity of PARP-1, other than its role in SSBR, is the underlying cause of this radiation sensitivity. As discussed above, PARP-1 plays a role in the condensation of chromatin and transcriptional repression and our working model is that altered regulation of chromatin conformation following suppression of PARP-1 results in enhanced susceptibility to radiation-induced damage. However, it can not be excluded that PARP-1 plays a role in the processing of DSBs. Studies from our group had shown that the incidence and the rejoining kinetics of neocarzinostatin-induced DNA double-strand breaks was identical in PARP-1+/+ and PARP-1−/− 3T3 cells. This would suggest that PARP-1 is not a major determinant of DNA double-strand break recovery with either strand break rejoining or cell survival as an endpoint [88]. However, recent studies that investigated the activation of PARP-1 and DNA-dependent protein kinase (DNA-PK), a key player in DSB repair, by different types of DNA ends and the interaction of these two proteins in the cellular response to ionizing radiation do suggest that PARP-1 is required for a pathway that facilitates the fast repair of DNA DSBs [89]. Making use of the PARP inhibitor (KU-0058684) and the DNA-PK inhibitor (NU7441) Mitchell and colleagues, found that the inactivation of the two proteins was not additive, suggesting that they co-operate within the same pathway and that they showed similar and overlapping affinities for oligonucleotides with blunt, 3′GGG or 5′GGG overhanging termini. The exact molecular mechanisms of this interaction clearly remain to be established. One possible link between PARP-1 and DNA-PK is the cyclin-dependent kinase 5 (CDK5). Identified in a synthetic lethal siRNA screen for genes that mediate sensitivity to PARP inhibitors [90], CDK5 is a proline-directed serine/threonine kinase activated by the non-cyclin proteins p35 and p39. It is a very abundant protein in the nervous system and has been implicated in neurodegeneration and neurodegenerative diseases where it is believed to play a role in the changes in cell-cycle regulation which precede neuron death (for review see [91]). The underlying mechanisms have been suggested to involve the inhibition of chromatin condensation as well as modulation of the activity of the DNA DSB repair protein ataxia-telangiectasia mutated (ATM) that was found to be phosphorylated by CDK5 in the nucleus of neuronal cells following exposure to camptothecin [92]. O’Hare and colleagues [93] showed a reduction in nuclear p35, a CDK5 cofactor, and an increase in nuclear CDK5 with increasing time of exposure to camptothecin in primary neuronal cultures suggesting that its relocation is important in the damage responses and the inhibition of CDK5 activity has also been shown to increase DNA damage-induced neuronal death [94].
Fig. 3.
Radiation survival in control, PARP-1-KD, and XRCC1-KD HeLa cells synchronized in S phase. Cells were synchronized by a double thymidine block, allowed to progress in S phase for 2 h and exposed to increasing doses of gamma rays. Following treatment, the cells were allowed to grow as colonies for 10–15 days, fixed, stained with Coomassie Blue, and then counted. The experimental data for control and XRCC1-KD lay within the same envelope of statistical deviation whereas the PARP-1-KD cells showed an enhanced radiation sensitivity (reproduced with permission from [87])
Turner and colleagues showed that CDK5 functions in the DNA damage response in human breast cancer cells. In these cells, CDK5 silencing induced spontaneous formation of DNA DSBs and an increased sensitivity to camptothecin [90]. Additionally, an increase in endogenous CDK5 activity after irradiation was observed in this cell type. They hypothesized that the mechanism of sensitivity to PARP inhibitor is a failure of the G2/M cell-cycle checkpoint. It is possible that in the presence of greatly increased level of SSBs, failure of this checkpoint leads to replication fork collapse and subsequent cell death. Interestingly, both ATM and PARP-1 have been shown to be phosphorylated by CDK5 together with other factors implicated in chromatin modifications [95]. Therefore, based on these observations an alternative hypothesis for the increased radiation sensitivity seen in the absence of PARP-1 is via a link with CDK5 and through it a modulation of DSB repair and activation of cell-cycle checkpoints.
The implication of these results is that both the absence of PARP-1 and the inhibition of PARP activity will enhance the cytoxicity of radiotherapy particularly in rapidly dividing tumors and this working hypothesis has been supported by results from a number of groups using different PARP inhibitors and experimental models. Chalmers and colleagues showed that the PARP-1 and PARP-2 inhibitor KU-0059436/AZD2281/olaprib increases the radiosensitivity of human glioma cell lines by a replication-dependent mechanism that generates persistent DSBs [96]. They have since shown that the simultaneous inhibition of homologous recombination further enhances the radiosensitizing effect of PARP inhibitors in replicating human glioma cells. The inhibition of the chaperone protein heat shock protein 90 results in the downregulation of BRCA2 and Rad51 protein levels and additive radiosensitization was observed [97]. Whether the radiation sensitivity observed when a PARP inhibitor is combined with radiation is exclusively due to compromised SSBR that results in an increase in DSBs or whether DSB repair is also modulated remains to be fully established. The PARP inhibitor E7016 (formerly known as GPI21016) has also been shown to enhance tumor cell radiosensitivity in vitro and in vivo and enhanced growth delay seen with the addition of E7016 to temozolomide and radiotherapy in a glioma mouse model suggests a potential role for this drug combination in the treatment of glioblastoma multiforme [98]. Preclinical data showing an enhancement of the response of tumor cells to radiation has now been documented for several PARP inhibitors including AG14361 [82], ABT-888 [99, 100], INO-1001 [101], and radiosensitization of human and rodent cell lines by INO-1001 and GPI15427 (MGI Pharma) [102] has been reported. These preclinical studies have lead to date to the start of three phase I clinical trials which will evaluate the safety of PARP inhibitors in addition to radiotherapy. One will test ABT-888 in combination with whole-brain radiation therapy in subjects with brain metastases. The two others will test either ABT-888 or BSI-201 in combination with TMZ and radiation therapy in subjects with newly diagnosed glioblastoma.
PARP inhibitors as single therapeutic agents
In 2005, the groups of Ashworth and Helleday [103, 104] showed preclinically that BRCA1 and BRCA2-deficient cells, in which the homologous recombination DSB repair pathway was compromised, were hypersensitive to the blockade of SSB repair by inhibition of PARP activity and demonstrated for the first time the potential use of PARP inhibitors as single therapeutic agents. The mechanistic basis of this cytoxicity is that blocking SSB repair leads to the formation of DSBs at the replication forks. In normal or heterozygote cells, the intact DSB repair can process these lethal lesions, but DSB repair is compromised in tumor cells that have lost the remaining BRCA1 or BRCA2 wild-type allele. The loss of both SSB and DSB repair pathways causes cell death. This is an example of the concept of synthetic lethality, defined as the situation when mutation in either of two genes individually has no effect but combining the mutations leads to death, that is proving to be a promising approach in the development of cancer treatments.
A number of clinical trials have been initiated to test the efficacy of this approach. The orally active PARP inhibitor olaparib (previously known as KU0059436) was tested in a phase 1 trial in a study population of 60 patients that was enriched for carriers of a BRCA1 or BRCA2 mutation [105]. The trial showed that olaparib had an acceptable side-effect profile and did not have the toxic effects commonly associated with conventional chemotherapy. Pharmacokinetic data indicated rapid absorption and elimination and pharmacodynamic studies confirmed PARP inhibition in surrogate samples and tumor tissues and a clinical benefit was observed but only in mutation carriers. Over 650 individuals will be included in the registered phase I and II trials of PARP inhibitors as a single agent for the treatment of breast, ovarian, prostate and pancreatic cancers in BRCA1 and BRCA2 mutation carriers, which are underway or in the planning stages (www.clinicaltrials.gov; recently reviewed by [106]).
Effective HR requires many proteins and this opens the possibility that synthetic lethality can be achieved in pathological settings where other components of the HR pathway are lacking through germline and/or somatic mutations or via other mechanisms that control protein expression in combination with PARP inhibitors. For example, recent results from Mendes-Pereira et al. [107] have shown that cells harboring PTEN mutations are sensitive to PARP inhibitors. Similarly, ATM deficiency, which is often found in mantle cell lymphoma, confers sensitivity to PARP inhibitors [108].
PARP inhibitors in other clinical settings
As discussed [109], PARP-1 functions as a double-edged sword: on the one hand, activation of PARP is essential for DNA repair, but on the other hand, over-activation represents an important mechanism of tissue damage in various pathological conditions associated with oxidative and nitrosative stress, including myocardial reperfusion injury, heart transplantation, and autoimmune B-cell destruction associated with diabetes mellitus and hepatic injury. Continuous or excessive activation of PARP produces a depletion of NAD+ and ATP, slowing glycolysis and mitochondrial respiration, which could eventually lead to cellular dysfunction and necrotic cell death. PARP activation also has consequences on the transcriptional regulation of several proteins including other transcriptional factors, such as NF-κB, implicated in inflammation. The beneficial effects of the inhibition of PARP have been demonstrated in a number of cardiovascular situations and there is considerable expectation that the use of PARP inhibitors will be an efficient therapy (see recent reviews by [109–112]).
Recently, Quiles-Perez et al. [32] have shown that PARP inhibition is capable of controlling hepatocellular carcinoma xenograft growth, protected against diethylnitrosamine-induced hepato-carcinogenesis and prevented tumor vasculogenesis by transcriptional regulation of both transcription factors and the expression of genes involved in tumor progression. These changes in the tumor microenvironment brought about by PARP inhibition can also have an impact on responses to radiotherapy and drug delivery. Vasoactivity has been reported for the PARP inhibitors AG14361 [82] and AG0014699 (Pfizer Oncology) [113] and differences were noted between the chemopotentiating effect of AG0014699 in vitro and in vivo models: the sensitivity of SW620 cells to TMZ was not enhanced by AG0014699 in vitro but in vivo the combination of AG0014699 and TMZ gave rise to pronounced tumor growth delays at doses of TMZ that alone gave only transient delays. Another tumor microenvironment that influences the chemo- and radioresistance of tumors is hypoxia. Hypoxic cells are 2.5 to threefold more radioresistant than oxic cells and intra-tumoral hypoxia can be a significant cause of treatment failure after radical radiotherapy. The group of Bristow [114] hypothesized that PARP inhibition may be useful in radiosensitizing hypoxic tumor cells, which they demonstrated using in vitro models and the inhibitor ABT-888, making these agents attractive adjuncts for radiotherapy via mechanisms other than the inhibition of DNA repair.
Conclusions
There is a growing body of literature and editorials documenting and discussing the impact of PARP inhibitors as effective chemo- and radiopotentiation agents and, in BRCA-deficient tumors, as single-agent therapies. Clearly, their potential has caught the attention of researchers and industrialists, but there are many aspects of their use that still need to be established. For instance, PARP inhibitor-chemotherapy drug combinations for different tumor types remain to be fully optimized, as does the dose and schedule to maximize efficacy while minimizing side-effects. The same considerations apply to using PARP inhibitors to potentiate the effects of radiotherapy, where the complex interaction of dose and schedule may also influence the risk of developing adverse secondary reactions such as radiation-induced fibrosis and secondary cancers. Emerging evidence suggests that many sporadic tumors display a certain degree of “BRCAness” [115], that is they share expression profiles that are found in tumors from BRCA1 and BRCA2 mutation carriers. The phenotypic similarities between some “triple-negative” breast cancers and the most prevalent type of breast cancer seen in BRCA1 mutation carriers has lead to suggestions that these difficult-to-treat breast tumors would be sensitive to PARP inhibition (recently reviewed in [116]). Indeed there are five registered clinical trails in which this will be assessed and preliminary results from a phase II trial presented at ASCO and reported in JNCI [117] are very encouraging. The challenge remains to identify those subgroups of patients who could benefit from single-agent PARP inhibition.
The development of resistance to targeted therapy must also be considered. PARP-inhibitor-resistant clones have been derived from the human BRCA2-deficient cell line CAPAN1 by a deletion of a mutation in BRCA2 [118]. A similar mechanism seems to be associated with carboplatin resistance in some BRCA2 mutation carriers with ovarian cancer (reviewed in [119]). Whether all mutations will revert at the same frequency remains to be established, but this eventuality has to be taken into consideration when determining treatment options. Over the last decade, certain biological functions and the mode of action of some of the PARP family members have been deciphered, however, it has become increasingly clear that family members participate in a wide range of cellular processes. For instance, PARP-1 not only plays a role in DNA repair but is an important transcriptional activator, and in particular of the NF-κB stress-inducible transcription factors. What the long-term effects are on cellular homeostasis, including the epigenetic control of gene expression, of the inhibition of PARP activity and deciphering how this might impact long-term therapeutic responses will be a challenge for the next decade.
Acknowledgments
Research in INSERM U612 is supported by the Institut National de la Santé et de la Recherche Médicale and Institut Curie. Grants from EDF for research on PARP activity and CDK5 are also gratefully acknowledged. The authors thank Dr. Vincent Pennaneach and Dr. Vincent Favaudon for their helpful comments on this manuscript.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- 1.Hakme A, Wong HK, Dantzer F, Schreiber V. The expanding field of poly(ADP-ribosyl)ation reactions. ‘protein modifications: beyond the usual suspects’ review series. EMBO Rep. 2008;9:1094–1100. doi: 10.1038/embor.2008.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim MY, Zhang T, Kraus WL. Poly(ADP-ribosyl)ation by PARP-1: ‘PAR-laying’ NAD+ into a nuclear signal. Genes Dev. 2005;19:1951–1967. doi: 10.1101/gad.1331805. [DOI] [PubMed] [Google Scholar]
- 3.Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 4.D’Amours D, Desnoyers S, D’Silva I, Poirier GG. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J. 1999;342(Pt 2):249–268. [PMC free article] [PubMed] [Google Scholar]
- 5.Pleschke JM, Kleczkowska HE, Strohm M, Althaus FR. Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. J Biol Chem. 2000;275:40974–40980. doi: 10.1074/jbc.M006520200. [DOI] [PubMed] [Google Scholar]
- 6.Gagne JP, Hunter JM, Labrecque B, Chabot B, Poirier GG. A proteomic approach to the identification of heterogeneous nuclear ribonucleoproteins as a new family of poly(ADP-ribose)-binding proteins. Biochem J. 2003;371:331–340. doi: 10.1042/BJ20021675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karras GI, Kustatscher G, Buhecha HR, Allen MD, Pugieux C, Sait F, Bycroft M, Ladurner AG. The macro domain is an ADP-ribose binding module. EMBO J. 2005;24:1911–1920. doi: 10.1038/sj.emboj.7600664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ladurner AG. Inactivating chromosomes: a macro domain that minimizes transcription. Mol Cell. 2003;12:1–3. doi: 10.1016/s1097-2765(03)00284-3. [DOI] [PubMed] [Google Scholar]
- 9.Aguiar RC, Takeyama K, He C, Kreinbrink K, Shipp MA. B-aggressive lymphoma family proteins have unique domains that modulate transcription and exhibit poly(ADP-ribose) polymerase activity. J Biol Chem. 2005;280:33756–33765. doi: 10.1074/jbc.M505408200. [DOI] [PubMed] [Google Scholar]
- 10.Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, Poirier GG (2010) PARP inhibition: PARP1 and beyond. Nat Rev Cancer [DOI] [PMC free article] [PubMed]
- 11.Ame JC, Spenlehauer C, de Murcia G. The PARP superfamily. Bioessays. 2004;26:882–893. doi: 10.1002/bies.20085. [DOI] [PubMed] [Google Scholar]
- 12.Kleine H, Poreba E, Lesniewicz K, Hassa PO, Hottiger MO, Litchfield DW, Shilton BH, Luscher B. Substrate-assisted catalysis by PARP10 limits its activity to mono-ADP-ribosylation. Mol Cell. 2008;32:57–69. doi: 10.1016/j.molcel.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 13.Virag L, Szabo C. The therapeutic potential of poly(ADP-ribose) polymerase inhibitors. Pharmacol Rev. 2002;54:375–429. doi: 10.1124/pr.54.3.375. [DOI] [PubMed] [Google Scholar]
- 14.Gradwohl G, Menissier de Murcia JM, Molinete M, Simonin F, Koken M, Hoeijmakers JH, de Murcia G. The second zinc-finger domain of poly(ADP-ribose) polymerase determines specificity for single-stranded breaks in DNA. Proc Natl Acad Sci USA. 1990;87:2990–2994. doi: 10.1073/pnas.87.8.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lonskaya I, Potaman VN, Shlyakhtenko LS, Oussatcheva EA, Lyubchenko YL, Soldatenkov VA. Regulation of poly(ADP-ribose) polymerase-1 by DNA structure-specific binding. J Biol Chem. 2005;280:17076–17083. doi: 10.1074/jbc.M413483200. [DOI] [PubMed] [Google Scholar]
- 16.Petrucco S, Percudani R. Structural recognition of DNA by poly(ADP-ribose)polymerase-like zinc finger families. FEBS J. 2008;275:883–893. doi: 10.1111/j.1742-4658.2008.06259.x. [DOI] [PubMed] [Google Scholar]
- 17.Simonin F, Poch O, Delarue M, de Murcia G. Identification of potential active-site residues in the human poly(ADP-ribose) polymerase. J Biol Chem. 1993;268:8529–8535. [PubMed] [Google Scholar]
- 18.Langelier MF, Servent KM, Rogers EE, Pascal JM. A third zinc-binding domain of human poly(ADP-ribose) polymerase-1 coordinates DNA-dependent enzyme activation. J Biol Chem. 2008;283:4105–4114. doi: 10.1074/jbc.M708558200. [DOI] [PubMed] [Google Scholar]
- 19.Tao Z, Gao P, Hoffman DW, Liu HW. Domain C of human poly(ADP-ribose) polymerase-1 is important for enzyme activity and contains a novel zinc-ribbon motif. Biochemistry. 2008;47:5804–5813. doi: 10.1021/bi800018a. [DOI] [PubMed] [Google Scholar]
- 20.Lilyestrom W, van der Woerd MJ, Clark N, Luger K. Structural and biophysical studies of human PARP-1 in complex with damaged DNA. J Mol Biol. 2010;395:983–994. doi: 10.1016/j.jmb.2009.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tulin A, Chinenov Y, Spradling A. Regulation of chromatin structure and gene activity by poly(ADP-ribose) polymerases. Curr Top Dev Biol. 2003;56:55–83. doi: 10.1016/s0070-2153(03)01007-x. [DOI] [PubMed] [Google Scholar]
- 22.Zaalishvili G, Margiani D, Kutalia K, Suladze S, Zaalishvili T. Automodification of PARP-1 mediates its tight binding to the nuclear matrix. Biochem Biophys Res Commun. 2010;393:123–125. doi: 10.1016/j.bbrc.2010.01.096. [DOI] [PubMed] [Google Scholar]
- 23.Grube K, Bürkle A. Poly(ADP-ribose) polymerase activity in mononuclear leukocytes of 13 mammalian species correlates with species-specific life span. Proc Natl Acad Sci USA. 1992;89:11759–11763. doi: 10.1073/pnas.89.24.11759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zaremba T, Ketzer P, Cole M, Coulthard S, Plummer ER, Curtin NJ. Poly(ADP-ribose) polymerase-1 polymorphisms, expression and activity in selected human tumour cell lines. Br J Cancer. 2009;101:256–262. doi: 10.1038/sj.bjc.6605166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cottet F, Blanche H, Verasdonck P, Le Gall I, Schachter F, Burkle A, Muiras ML. New polymorphisms in the human poly(ADP-ribose) polymerase-1 coding sequence: lack of association with longevity or with increased cellular poly(ADP-ribosyl)ation capacity. J Mol Med. 2000;78:431–440. doi: 10.1007/s001090000132. [DOI] [PubMed] [Google Scholar]
- 26.Lockett KL, Hall MC, Xu J, Zheng SL, Berwick M, Chuang SC, Clark PE, Cramer SD, Lohman K, Hu JJ. The ADPRT V762A genetic variant contributes to prostate cancer susceptibility and deficient enzyme function. Cancer Res. 2004;64:6344–6348. doi: 10.1158/0008-5472.CAN-04-0338. [DOI] [PubMed] [Google Scholar]
- 27.Wang XG, Wang ZQ, Tong WM, Shen Y. PARP1 Val762Ala polymorphism reduces enzymatic activity. Biochem Biophys Res Commun. 2007;354:122–126. doi: 10.1016/j.bbrc.2006.12.162. [DOI] [PubMed] [Google Scholar]
- 28.Fougerousse F, Meloni R, Roudaut C, Beckmann JS. Dinucleotide repeat polymorphism at the human poly(ADP-ribose) polymerase gene (PPOL) Nucleic Acids Res. 1992;20:1166. doi: 10.1093/nar/20.5.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gordon S, Akopyan G, Garban H, Bonavida B. Transcription factor YY1: structure, function, and therapeutic implications in cancer biology. Oncogene. 2006;25:1125–1142. doi: 10.1038/sj.onc.1209080. [DOI] [PubMed] [Google Scholar]
- 30.Oei SL, Shi Y. Poly(ADP-ribosyl)ation of transcription factor Yin Yang 1 under conditions of DNA damage. Biochem Biophys Res Commun. 2001;285:27–31. doi: 10.1006/bbrc.2001.5115. [DOI] [PubMed] [Google Scholar]
- 31.Oei SL, Shi Y. Transcription factor Yin Yang 1 stimulates poly(ADP-ribosyl)ation and DNA repair. Biochem Biophys Res Commun. 2001;284:450–454. doi: 10.1006/bbrc.2001.4985. [DOI] [PubMed] [Google Scholar]
- 32.Quiles-Perez R, Munoz-Gamez JA, Ruiz-Extremera A, O’Valle F, Sanjuan-Nunez L, Martin-Alvarez AB, Martin-Oliva D, Caballero T, Munoz de Rueda P, Leon J, Gonzalez R, Muntane J, Oliver FJ, Salmeron J. Inhibition of poly adenosine diphosphate-ribose polymerase decreases hepatocellular carcinoma growth by modulation of tumor-related gene expression. Hepatology. 2010;51:255–266. doi: 10.1002/hep.23249. [DOI] [PubMed] [Google Scholar]
- 33.Bieche I, de Murcia G, Lidereau R. Poly(ADP-ribose) polymerase gene expression status and genomic instability in human breast cancer. Clin Cancer Res. 1996;2:1163–1167. [PubMed] [Google Scholar]
- 34.de Murcia JM, Niedergang C, Trucco C, Ricoul M, Dutrillaux B, Mark M, Oliver FJ, Masson M, Dierich A, LeMeur M, Walztinger C, Chambon P, de Murcia G. Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc Natl Acad Sci USA. 1997;94:7303–7307. doi: 10.1073/pnas.94.14.7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Masutani M, Nozaki T, Nishiyama E, Shimokawa T, Tachi Y, Suzuki H, Nakagama H, Wakabayashi K, Sugimura T. Function of poly(ADP-ribose) polymerase in response to DNA damage: gene-disruption study in mice. Mol Cell Biochem. 1999;193:149–152. [PubMed] [Google Scholar]
- 36.Wang ZQ, Stingl L, Morrison C, Jantsch M, Los M, Schulze-Osthoff K, Wagner EF. PARP is important for genomic stability but dispensable in apoptosis. Genes Dev. 1997;11:2347–2358. doi: 10.1101/gad.11.18.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shall S, de Murcia G. Poly(ADP-ribose) polymerase-1: what have we learned from the deficient mouse model? Mutat Res. 2000;460:1–15. doi: 10.1016/s0921-8777(00)00016-1. [DOI] [PubMed] [Google Scholar]
- 38.Trucco C, Oliver FJ, de Murcia G, Menissier-de Murcia J. DNA repair defect in poly(ADP-ribose) polymerase-deficient cell lines. Nucleic Acids Res. 1998;26:2644–2649. doi: 10.1093/nar/26.11.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.El-Khamisy SF, Masutani M, Suzuki H, Caldecott KW. A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res. 2003;31:5526–5533. doi: 10.1093/nar/gkg761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prasad R, Lavrik OI, Kim SJ, Kedar P, Yang XP, Vande Berg BJ, Wilson SH. DNA polymerase beta-mediated long patch base excision repair. Poly(ADP-ribose)polymerase-1 stimulates strand displacement DNA synthesis. J Biol Chem. 2001;276:32411–32414. doi: 10.1074/jbc.C100292200. [DOI] [PubMed] [Google Scholar]
- 41.Haince JF, McDonald D, Rodrigue A, Dery U, Masson JY, Hendzel MJ, Poirier GG. PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. J Biol Chem. 2008;283:1197–1208. doi: 10.1074/jbc.M706734200. [DOI] [PubMed] [Google Scholar]
- 42.Wang M, Wu W, Wu W, Rosidi B, Zhang L, Wang H, Iliakis G. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res. 2006;34:6170–6182. doi: 10.1093/nar/gkl840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iliakis G. Backup pathways of NHEJ in cells of higher eukaryotes: cell cycle dependence. Radiother Oncol. 2009;92:310–315. doi: 10.1016/j.radonc.2009.06.024. [DOI] [PubMed] [Google Scholar]
- 44.Yang YG, Cortes U, Patnaik S, Jasin M, Wang ZQ. Ablation of PARP-1 does not interfere with the repair of DNA double-strand breaks, but compromises the reactivation of stalled replication forks. Oncogene. 2004;23:3872–3882. doi: 10.1038/sj.onc.1207491. [DOI] [PubMed] [Google Scholar]
- 45.Huletsky A, de Murcia G, Muller S, Hengartner M, Menard L, Lamarre D, Poirier GG. The effect of poly(ADP-ribosyl)ation on native and H1-depleted chromatin. A role of poly(ADP-ribosyl)ation on core nucleosome structure. J Biol Chem. 1989;264:8878–8886. [PubMed] [Google Scholar]
- 46.Mathis G, Althaus FR. Release of core DNA from nucleosomal core particles following (ADP-ribose)n-modification in vitro. Biochem Biophys Res Commun. 1987;143:1049–1054. doi: 10.1016/0006-291x(87)90358-5. [DOI] [PubMed] [Google Scholar]
- 47.Poirier GG, de Murcia G, Jongstra-Bilen J, Niedergang C, Mandel P. Poly(ADP-ribosyl)ation of polynucleosomes causes relaxation of chromatin structure. Proc Natl Acad Sci USA. 1982;79:3423–3427. doi: 10.1073/pnas.79.11.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de Murcia G, Huletsky A, Lamarre D, Gaudreau A, Pouyet J, Daune M, Poirier GG. Modulation of chromatin superstructure induced by poly(ADP-ribose) synthesis and degradation. J Biol Chem. 1986;261:7011–7017. [PubMed] [Google Scholar]
- 49.Pinnola A, Naumova N, Shah M, Tulin AV. Nucleosomal core histones mediate dynamic regulation of poly(ADP-ribose) polymerase 1 protein binding to chromatin and induction of its enzymatic activity. J Biol Chem. 2007;282:32511–32519. doi: 10.1074/jbc.M705989200. [DOI] [PubMed] [Google Scholar]
- 50.Quenet D, Gasser V, Fouillen L, Cammas F, Sanglier-Cianferani S, Losson R, Dantzer F. The histone subcode: poly(ADP-ribose) polymerase-1 (Parp-1) and Parp-2 control cell differentiation by regulating the transcriptional intermediary factor TIF1beta and the heterochromatin protein HP1alpha. Faseb J. 2008;22:3853–3865. doi: 10.1096/fj.08-113464. [DOI] [PubMed] [Google Scholar]
- 51.Tanuma S, Johnson GS. ADP-ribosylation of nonhistone high mobility group proteins in intact cells. J Biol Chem. 1983;258:4067–4070. [PubMed] [Google Scholar]
- 52.Enright HU, Miller WJ, Hebbel RP. Nucleosomal histone protein protects DNA from iron-mediated damage. Nucleic Acids Res. 1992;20:3341–3346. doi: 10.1093/nar/20.13.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ullrich O, Sitte N, Sommerburg O, Sandig V, Davies KJ, Grune T. Influence of DNA binding on the degradation of oxidized histones by the 20S proteasome. Arch Biochem Biophys. 1999;362:211–216. doi: 10.1006/abbi.1998.1031. [DOI] [PubMed] [Google Scholar]
- 54.Mayer-Kuckuk P, Ullrich O, Ziegler M, Grune T, Schweiger M. Functional interaction of poly(ADP-ribose) with the 20S proteasome in vitro. Biochem Biophys Res Commun. 1999;259:576–581. doi: 10.1006/bbrc.1999.0824. [DOI] [PubMed] [Google Scholar]
- 55.Catalgol B, Wendt B, Grimm S, Breusing N, Ozer NK, Grune T. Chromatin repair after oxidative stress: role of PARP-mediated proteasome activation. Free Radic Biol Med. 2010;48:673–680. doi: 10.1016/j.freeradbiomed.2009.12.010. [DOI] [PubMed] [Google Scholar]
- 56.Heo K, Kim H, Choi SH, Choi J, Kim K, Gu J, Lieber MR, Yang AS, An W. FACT-mediated exchange of histone variant H2AX regulated by phosphorylation of H2AX and ADP-ribosylation of Spt16. Mol Cell. 2008;30:86–97. doi: 10.1016/j.molcel.2008.02.029. [DOI] [PubMed] [Google Scholar]
- 57.Ahel D, Horejsi Z, Wiechens N, Polo SE, Garcia-Wilson E, Ahel I, Flynn H, Skehel M, West SC, Jackson SP, Owen-Hughes T, Boulton SJ. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science. 2009;325:1240–1243. doi: 10.1126/science.1177321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gottschalk AJ, Timinszky G, Kong SE, Jin J, Cai Y, Swanson SK, Washburn MP, Florens L, Ladurner AG, Conaway JW, Conaway RC. Poly(ADP-ribosyl)ation directs recruitment and activation of an ATP-dependent chromatin remodeler. Proc Natl Acad Sci USA. 2009;106:13770–13774. doi: 10.1073/pnas.0906920106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Faraone-Mennella MR. Chromatin architecture and functions: the role(s) of poly(ADP-RIBOSE) polymerase and poly(ADPribosyl)ation of nuclear proteins. Biochem Cell Biol. 2005;83:396–404. doi: 10.1139/o05-042. [DOI] [PubMed] [Google Scholar]
- 60.Tulin A, Naumova NM, Menon AK, Spradling AC. Drosophila poly(ADP-ribose) glycohydrolase mediates chromatin structure and SIR2-dependent silencing. Genetics. 2006;172:363–371. doi: 10.1534/genetics.105.049239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tulin A, Spradling A. Chromatin loosening by poly(ADP)-ribose polymerase (PARP) at Drosophila puff loci. Science. 2003;299:560–562. doi: 10.1126/science.1078764. [DOI] [PubMed] [Google Scholar]
- 62.Caiafa P, Guastafierro T, Zampieri M. Epigenetics: poly(ADP-ribosyl)ation of PARP-1 regulates genomic methylation patterns. Faseb J. 2009;23:672–678. doi: 10.1096/fj.08-123265. [DOI] [PubMed] [Google Scholar]
- 63.Zampieri M, Passananti C, Calabrese R, Perilli M, Corbi N, De Cave F, Guastafierro T, Bacalini MG, Reale A, Amicosante G, Calabrese L, Zlatanova J, Caiafa P. Parp1 localizes within the Dnmt1 promoter and protects its unmethylated state by its enzymatic activity. PLoS One. 2009;4:e4717. doi: 10.1371/journal.pone.0004717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hassa PO, Hottiger MO. The functional role of poly(ADP-ribose)polymerase 1 as novel coactivator of NF-kappaB in inflammatory disorders. Cell Mol Life Sci. 2002;59:1534–1553. doi: 10.1007/s00018-002-8527-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cohen-Armon M, Visochek L, Rozensal D, Kalal A, Geistrikh I, Klein R, Bendetz-Nezer S, Yao Z, Seger R. DNA-independent PARP-1 activation by phosphorylated ERK2 increases Elk1 activity: a link to histone acetylation. Mol Cell. 2007;25:297–308. doi: 10.1016/j.molcel.2006.12.012. [DOI] [PubMed] [Google Scholar]
- 66.Guastafierro T, Cecchinelli B, Zampieri M, Reale A, Riggio G, Sthandier O, Zupi G, Calabrese L, Caiafa P. CCCTC-binding factor activates PARP-1 affecting DNA methylation machinery. J Biol Chem. 2008;283:21873–21880. doi: 10.1074/jbc.M801170200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shieh WM, Ame JC, Wilson MV, Wang ZQ, Koh DW, Jacobson MK, Jacobson EL. Poly(ADP-ribose) polymerase null mouse cells synthesize ADP-ribose polymers. J Biol Chem. 1998;273:30069–30072. doi: 10.1074/jbc.273.46.30069. [DOI] [PubMed] [Google Scholar]
- 68.Ame JC, Rolli V, Schreiber V, Niedergang C, Apiou F, Decker P, Muller S, Hoger T, Menissier-de Murcia J, de Murcia G. PARP-2, A novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase. J Biol Chem. 1999;274:17860–17868. doi: 10.1074/jbc.274.25.17860. [DOI] [PubMed] [Google Scholar]
- 69.Menissier de Murcia J, Ricoul M, Tartier L, Niedergang C, Huber A, Dantzer F, Schreiber V, Ame JC, Dierich A, LeMeur M, Sabatier L, Chambon P, de Murcia G. Functional interaction between PARP-1 and PARP-2 in chromosome stability and embryonic development in mouse. EMBO J. 2003;22:2255–2263. doi: 10.1093/emboj/cdg206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schreiber V, Ricoul M, Amé J-C, Dantzer F, Meder V, Spenlehauer C, Steigler P, Niedergang C, Sabatier L, Favaudon V, Ménissier-de-Murcia J, de Murcia G (2004) Poly(ADP-ribosyl)ation, Landes Bioscience
- 71.Schreiber V, Ame JC, Dolle P, Schultz I, Rinaldi B, Fraulob V, Menissier-de Murcia J, de Murcia G. Poly(ADP-ribose) polymerase-2 (PARP-2) is required for efficient base excision DNA repair in association with PARP-1 and XRCC1. J Biol Chem. 2002;277:23028–23036. doi: 10.1074/jbc.M202390200. [DOI] [PubMed] [Google Scholar]
- 72.Yelamos J, Schreiber V, Dantzer F. Toward specific functions of poly(ADP-ribose) polymerase-2. Trends Mol Med. 2008;14:169–178. doi: 10.1016/j.molmed.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 73.Mortusewicz O, Ame JC, Schreiber V, Leonhardt H. Feedback-regulated poly(ADP-ribosyl)ation by PARP-1 is required for rapid response to DNA damage in living cells. Nucleic Acids Res. 2007;35:7665–7675. doi: 10.1093/nar/gkm933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bryant HE, Petermann E, Schultz N, Jemth AS, Loseva O, Issaeva N, Johansson F, Fernandez S, McGlynn P, Helleday T. PARP is activated at stalled forks to mediate Mre11-dependent replication restart and recombination. EMBO J. 2009;28:2601–2615. doi: 10.1038/emboj.2009.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Durkacz BW, Omidiji O, Gray DA, Shall S. (ADP-ribose)n participates in DNA excision repair. Nature. 1980;283:593–596. doi: 10.1038/283593a0. [DOI] [PubMed] [Google Scholar]
- 76.Banasik M, Komura H, Shimoyama M, Ueda K. Specific inhibitors of poly(ADP-ribose) synthetase and mono(ADP-ribosyl)transferase. J Biol Chem. 1992;267:1569–1575. [PubMed] [Google Scholar]
- 77.Curtin NJ. PARP inhibitors for cancer therapy. Expert Rev Mol Med. 2005;7:1–20. doi: 10.1017/S146239940500904X. [DOI] [PubMed] [Google Scholar]
- 78.Karlberg T, Hammarstrom M, Schutz P, Svensson L, Schuler H. Crystal structure of the catalytic domain of human PARP2 in complex with PARP inhibitor ABT-888. Biochemistry. 2010;49:1056–1058. doi: 10.1021/bi902079y. [DOI] [PubMed] [Google Scholar]
- 79.Tentori L, Graziani G. Chemopotentiation by PARP inhibitors in cancer therapy. Pharmacol Res. 2005;52:25–33. doi: 10.1016/j.phrs.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 80.Miknyoczki SJ, Jones-Bolin S, Pritchard S, Hunter K, Zhao H, Wan W, Ator M, Bihovsky R, Hudkins R, Chatterjee S, Klein-Szanto A, Dionne C, Ruggeri B. Chemopotentiation of temozolomide, irinotecan, and cisplatin activity by CEP-6800, a poly(ADP-ribose) polymerase inhibitor. Mol Cancer Ther. 2003;2:371–382. [PubMed] [Google Scholar]
- 81.Tentori L, Leonetti C, Scarsella M, D’Amati G, Vergati M, Portarena I, Xu W, Kalish V, Zupi G, Zhang J, Graziani G. Systemic administration of GPI 15427, a novel poly(ADP-ribose) polymerase-1 inhibitor, increases the antitumor activity of temozolomide against intracranial melanoma, glioma, lymphoma. Clin Cancer Res. 2003;9:5370–5379. [PubMed] [Google Scholar]
- 82.Calabrese CR, Almassy R, Barton S, Batey MA, Calvert AH, Canan-Koch S, Durkacz BW, Hostomsky Z, Kumpf RA, Kyle S, Li J, Maegley K, Newell DR, Notarianni E, Stratford IJ, Skalitzky D, Thomas HD, Wang LZ, Webber SE, Williams KJ, Curtin NJ. Anticancer chemosensitization and radiosensitization by the novel poly(ADP-ribose) polymerase-1 inhibitor AG14361. J Natl Cancer Inst. 2004;96:56–67. doi: 10.1093/jnci/djh005. [DOI] [PubMed] [Google Scholar]
- 83.Horton TM, Jenkins G, Pati D, Zhang L, Dolan ME, Ribes-Zamora A, Bertuch AA, Blaney SM, Delaney SL, Hegde M, Berg SL. Poly(ADP-ribose) polymerase inhibitor ABT-888 potentiates the cytotoxic activity of temozolomide in leukemia cells: influence of mismatch repair status and O6-methylguanine-DNA methyltransferase activity. Mol Cancer Ther. 2009;8:2232–2242. doi: 10.1158/1535-7163.MCT-09-0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Plummer R, Jones C, Middleton M, Wilson R, Evans J, Olsen A, Curtin N, Boddy A, McHugh P, Newell D, Harris A, Johnson P, Steinfeldt H, Dewji R, Wang D, Robson L, Calvert H. Phase I study of the poly(ADP-ribose) polymerase inhibitor, AG014699, in combination with temozolomide in patients with advanced solid tumors. Clin Cancer Res. 2008;14:7917–7923. doi: 10.1158/1078-0432.CCR-08-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Drew Y, Plummer R. PARP inhibitors in cancer therapy: two modes of attack on the cancer cell widening the clinical applications. Drug Resist Updat. 2009;12:153–156. doi: 10.1016/j.drup.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 86.Noel G, Godon C, Fernet M, Giocanti N, Megnin-Chanet F, Favaudon V. Radiosensitization by the poly(ADP-ribose) polymerase inhibitor 4-amino-1, 8-naphthalimide is specific of the S phase of the cell cycle and involves arrest of DNA synthesis. Mol Cancer Ther. 2006;5:564–574. doi: 10.1158/1535-7163.MCT-05-0418. [DOI] [PubMed] [Google Scholar]
- 87.Godon C, Cordelieres FP, Biard D, Giocanti N, Megnin-Chanet F, Hall J, Favaudon V. PARP inhibition versus PARP-1 silencing: different outcomes in terms of single-strand break repair and radiation susceptibility. Nucleic Acids Res. 2008;36:4454–4464. doi: 10.1093/nar/gkn403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Noel G, Giocanti N, Fernet M, Megnin-Chanet F, Favaudon V. Poly(ADP-ribose) polymerase (PARP-1) is not involved in DNA double-strand break recovery. BMC Cell Biol. 2003;4:7. doi: 10.1186/1471-2121-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mitchell J, Smith GC, Curtin NJ. Poly(ADP-Ribose) polymerase-1 and DNA-dependent protein kinase have equivalent roles in double strand break repair following ionizing radiation. Int J Radiat Oncol Biol Phys. 2009;75:1520–1527. doi: 10.1016/j.ijrobp.2009.07.1722. [DOI] [PubMed] [Google Scholar]
- 90.Turner NC, Lord CJ, Iorns E, Brough R, Swift S, Elliott R, Rayter S, Tutt AN, Ashworth A. A synthetic lethal siRNA screen identifying genes mediating sensitivity to a PARP inhibitor. EMBO J. 2008;27:1368–1377. doi: 10.1038/emboj.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dhariwala FA, Rajadhyaksha MS. An unusual member of the Cdk family: Cdk5. Cell Mol Neurobiol. 2008;28:351–369. doi: 10.1007/s10571-007-9242-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tian B, Yang Q, Mao Z. Phosphorylation of ATM by Cdk5 mediates DNA damage signalling and regulates neuronal death. Nat Cell Biol. 2009;11:211–218. doi: 10.1038/ncb1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.O’Hare MJ, Kushwaha N, Zhang Y, Aleyasin H, Callaghan SM, Slack RS, Albert PR, Vincent I, Park DS. Differential roles of nuclear and cytoplasmic cyclin-dependent kinase 5 in apoptotic and excitotoxic neuronal death. J Neurosci. 2005;25:8954–8966. doi: 10.1523/JNEUROSCI.2899-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Anne SL, Saudou F, Humbert S. Phosphorylation of huntingtin by cyclin-dependent kinase 5 is induced by DNA damage and regulates wild-type and mutant huntingtin toxicity in neurons. J Neurosci. 2007;27:7318–7328. doi: 10.1523/JNEUROSCI.1831-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gagne JP, Isabelle M, Lo KS, Bourassa S, Hendzel MJ, Dawson VL, Dawson TM, Poirier GG. Proteome-wide identification of poly(ADP-ribose) binding proteins and poly(ADP-ribose)-associated protein complexes. Nucleic Acids Res. 2008;36:6959–6976. doi: 10.1093/nar/gkn771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dungey FA, Loser DA, Chalmers AJ. Replication-dependent radiosensitization of human glioma cells by inhibition of poly(ADP-Ribose) polymerase: mechanisms and therapeutic potential. Int J Radiat Oncol Biol Phys. 2008;72:1188–1197. doi: 10.1016/j.ijrobp.2008.07.031. [DOI] [PubMed] [Google Scholar]
- 97.Dungey FA, Caldecott KW, Chalmers AJ. Enhanced radiosensitization of human glioma cells by combining inhibition of poly(ADP-ribose) polymerase with inhibition of heat shock protein 90. Mol Cancer Ther. 2009;8:2243–2254. doi: 10.1158/1535-7163.MCT-09-0201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Russo AL, Kwon HC, Burgan WE, Carter D, Beam K, Weizheng X, Zhang J, Slusher BS, Chakravarti A, Tofilon PJ, Camphausen K. In vitro and in vivo radiosensitization of glioblastoma cells by the poly(ADP-ribose) polymerase inhibitor E7016. Clin Cancer Res. 2009;15:607–612. doi: 10.1158/1078-0432.CCR-08-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Albert JM, Cao C, Kim KW, Willey CD, Geng L, Xiao D, Wang H, Sandler A, Johnson DH, Colevas AD, Low J, Rothenberg ML, Lu B. Inhibition of poly(ADP-ribose) polymerase enhances cell death and improves tumor growth delay in irradiated lung cancer models. Clin Cancer Res. 2007;13:3033–3042. doi: 10.1158/1078-0432.CCR-06-2872. [DOI] [PubMed] [Google Scholar]
- 100.Donawho CK, Luo Y, Penning TD, Bauch JL, Bouska JJ, Bontcheva-Diaz VD, Cox BF, DeWeese TL, Dillehay LE, Ferguson DC, Ghoreishi-Haack NS, Grimm DR, Guan R, Han EK, Holley-Shanks RR, Hristov B, Idler KB, Jarvis K, Johnson EF, Kleinberg LR, Klinghofer V, Lasko LM, Liu X, Marsh KC, McGonigal TP, Meulbroek JA, Olson AM, Palma JP, Rodriguez LE, Shi Y, Stavropoulos JA, Tsurutani AC, Zhu GD, Rosenberg SH, Giranda VL, Frost DJ. ABT-888, an orally active poly(ADP-ribose) polymerase inhibitor that potentiates DNA-damaging agents in preclinical tumor models. Clin Cancer Res. 2007;13:2728–2737. doi: 10.1158/1078-0432.CCR-06-3039. [DOI] [PubMed] [Google Scholar]
- 101.Brock WA, Milas L, Bergh S, Lo R, Szabo C, Mason KA. Radiosensitization of human and rodent cell lines by INO-1001, a novel inhibitor of poly(ADP-ribose) polymerase. Cancer Lett. 2004;205:155–160. doi: 10.1016/j.canlet.2003.10.029. [DOI] [PubMed] [Google Scholar]
- 102.Khan K, Araki K, Wang D, Li G, Li X, Zhang J, Xu W, Hoover RK, Lauter S, O’Malley B, Jr, Lapidus RG, Li D. Head and neck cancer radiosensitization by the novel poly(ADP-ribose) polymerase inhibitor GPI-15427. Head Neck. 2010;32:381–391. doi: 10.1002/hed.21195. [DOI] [PubMed] [Google Scholar]
- 103.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 104.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, Martin NM, Jackson SP, Smith GC, Ashworth A. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 105.Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, Mortimer P, Swaisland H, Lau A, O’Connor MJ, Ashworth A, Carmichael J, Kaye SB, Schellens JH, de Bono JS. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 106.Sandhu SK, Yap TA, de Bono JS. Poly(ADP-ribose) polymerase inhibitors in cancer treatment: a clinical perspective. Eur J Cancer. 2010;46:9–20. doi: 10.1016/j.ejca.2009.10.021. [DOI] [PubMed] [Google Scholar]
- 107.Mendes-Pereira AM, Martin SA, Brough R, McCarthy A, Taylor JR, Kim JS, Waldman T, Lord CJ, Ashworth A. Synthetic lethal targeting of PTEN mutant cells with PARP inhibitors. EMBO Mol Med. 2009;1:315–322. doi: 10.1002/emmm.200900041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Williamson CT, Muzik H, Turhan AG, Zamo A, O’Connor MJ, Bebb DG, Lees-Miller SP. ATM deficiency sensitizes mantle cell lymphoma cells to poly(ADP-ribose) polymerase-1 inhibitors. Mol Cancer Ther. 2010;9:347–357. doi: 10.1158/1535-7163.MCT-09-0872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Peralta-Leal A, Rodriguez-Vargas JM, Aguilar-Quesada R, Rodriguez MI, Linares JL, de Almodovar MR, Oliver FJ. PARP inhibitors: new partners in the therapy of cancer and inflammatory diseases. Free Radic Biol Med. 2009;47:13–26. doi: 10.1016/j.freeradbiomed.2009.04.008. [DOI] [PubMed] [Google Scholar]
- 110.Jagtap P, Szabo C. Poly(ADP-ribose) polymerase and the therapeutic effects of its inhibitors. Nat Rev Drug Discov. 2005;4:421–440. doi: 10.1038/nrd1718. [DOI] [PubMed] [Google Scholar]
- 111.Moroni F. Poly(ADP-ribose)polymerase 1 (PARP-1) and postischemic brain damage. Curr Opin Pharmacol. 2008;8:96–103. doi: 10.1016/j.coph.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 112.Pacher P, Szabo C. Role of poly(ADP-ribose) polymerase 1 (PARP-1) in cardiovascular diseases: the therapeutic potential of PARP inhibitors. Cardiovasc Drug Rev. 2007;25:235–260. doi: 10.1111/j.1527-3466.2007.00018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ali M, Telfer BA, McCrudden C, O’Rourke M, Thomas HD, Kamjoo M, Kyle S, Robson T, Shaw C, Hirst DG, Curtin NJ, Williams KJ. Vasoactivity of AG014699, a clinically active small molecule inhibitor of poly(ADP-ribose) polymerase: a contributory factor to chemopotentiation in vivo? Clin Cancer Res. 2009;15:6106–6112. doi: 10.1158/1078-0432.CCR-09-0398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Liu SK, Coackley C, Krause M, Jalali F, Chan N, Bristow RG. A novel poly(ADP-ribose) polymerase inhibitor, ABT-888, radiosensitizes malignant human cell lines under hypoxia. Radiother Oncol. 2008;88:258–268. doi: 10.1016/j.radonc.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 115.Turner N, Tutt A, Ashworth A. Hallmarks of ‘BRCAness’ in sporadic cancers. Nat Rev Cancer. 2004;4:814–819. doi: 10.1038/nrc1457. [DOI] [PubMed] [Google Scholar]
- 116.Comen EA, Robson M. Poly(ADP-ribose) polymerase inhibitors in triple-negative breast cancer. Cancer J. 2010;16:48–52. doi: 10.1097/PPO.0b013e3181cf01eb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tuma RS. PARP inhibitors: will the new class of drugs match the hype? J Natl Cancer Inst. 2009;101:1230–1232. doi: 10.1093/jnci/djp315. [DOI] [PubMed] [Google Scholar]
- 118.Edwards SL, Brough R, Lord CJ, Natrajan R, Vatcheva R, Levine DA, Boyd J, Reis-Filho JS, Ashworth A. Resistance to therapy caused by intragenic deletion in BRCA2. Nature. 2008;451:1111–1115. doi: 10.1038/nature06548. [DOI] [PubMed] [Google Scholar]
- 119.Ashworth A. Drug resistance caused by reversion mutation. Cancer Res. 2008;68:10021–10023. doi: 10.1158/0008-5472.CAN-08-2287. [DOI] [PubMed] [Google Scholar]