Abstract
Exposure to solar radiation can cause mortality in natural communities of pico-phytoplankton, both at the surface and to a depth of at least 30 m. DNA damage is a significant cause of death, mainly due to cyclobutane pyrimidine dimer formation, which can be lethal if not repaired. While developing a UV mutagenesis protocol for the marine cyanobacterium Prochlorococcus, we isolated a UV-hyper-resistant variant of high light-adapted strain MED4. The hyper-resistant strain was constitutively upregulated for expression of the mutT-phrB operon, encoding nudix hydrolase and photolyase, both of which are involved in repair of DNA damage that can be caused by UV light. Photolyase (PhrB) breaks pyrimidine dimers typically caused by UV exposure, using energy from visible light in the process known as photoreactivation. Nudix hydrolase (MutT) hydrolyses 8-oxo-dGTP, an aberrant form of GTP that results from oxidizing conditions, including UV radiation, thus impeding mispairing and mutagenesis by preventing incorporation of the aberrant form into DNA. These processes are error-free, in contrast to error-prone SOS dark repair systems that are widespread in bacteria. The UV-hyper-resistant strain contained only a single mutation: a 1 bp deletion in the intergenic region directly upstream of the mutT-phrB operon. Two subsequent enrichments for MED4 UV-hyper-resistant strains from MED4 wild-type cultures gave rise to strains containing this same 1 bp deletion, affirming its connection to the hyper-resistant phenotype. These results have implications for Prochlorococcus DNA repair mechanisms, genome stability and possibly lysogeny.
Introduction
Prochlorococcus, a marine cyanobacterium that numerically dominates the mid-latitude oceans, is the smallest known oxygenic phototroph and plays a central role in the oceanic carbon cycle. Its numbers can reach as high as 105 cells ml−1. The group consists of genetically distinct ecotypes that can be broadly classified as high or low light-adapted, depending on their optimal light intensity for growth (reviewed in Coleman and Chisholm, 2007). The high light-adapted ecotypes are found throughout the euphotic zone of the oceans (down to a depth of 150–200 m), reaching highest abundance in surface waters, whereas their low light-adapted counterparts dominate deeper waters.
Factors that affect mortality of Prochlorococcus include grazing, mainly from phagotrophic flagellates and microzooplankton (Guillou et al., 2001; Christaki et al., 2002; Frias-Lopez et al., 2008), and phage infection (Sullivan et al., 2003). In addition, for cells close to the surface, exposure to high solar radiation, especially in the UVB range (280–320 nm), also plays a role in mortality both at the surface and to a depth of 10–30 m (Llabrés and Agustí, 2006; Agustí and Llabrés, 2007). DNA damage, mainly due to the formation of cyclobutane pyrimidine dimers, can be lethal if not repaired and is a primary cause of UV-induced mortality (Sancar, 2000). The high light-adapted Prochlorococcus ecotypes that dominate surface waters have very small (1.66–1.74 Mb), streamlined, non-redundant genomes (Strehl et al., 1999; Kettler et al., 2007), making them potentially less able to withstand the effects of multiple mutations resulting from high doses of UV light. It is of interest therefore to determine whether the high and low light-adapted ecotypes have different susceptibilities to UV damage, and the mechanisms that might mediate these susceptibilities.
In the process of developing a mutagenesis protocol for Prochlorococcus using UV light (M.S. Osburne and B.M. Holmbeck, in prep.) we isolated a UV-hyper-resistant strain (MED4 UVR1) of high light-adapted strain Prochlorococcus MED4. We examined the differences between the hyper-resistant and wild-type (WT) cells with respect to levels of UV resistance, gene expression and genome sequence. The results allowed us to propose a mechanism for UV hyper-resistance in MED4 UVR1, and to hypothesize mechanisms by which MED4 copes with UV light in its natural environment.
Results
Isolation and initial phenotypic characterization of MED4 UV-hyper-resistant strains
A culture of high light-adapted Prochlorococcus strain MED4 was subjected to 10 rounds of UV treatment in which cells were grown to mid-log phase, treated with UV light (Experimental procedures), and allowed to recover under standard growth conditions in visible light. Recovery of culture fluorescence after UV treatment occurred more readily with increasing rounds of treatment. After 7–8 rounds of treatment and transfer, the cultures were significantly more resistant to UV light than the parent strain, as reflected in the selection for the UV-hyper-resistant strain MED4 UVR1 (Fig. 1). This general pattern was also seen during similar enrichment selections (not shown) for two additional UV-hyper-resist strains (MED4 UVR2 and MED4 UVR3), carried out at a later date. To confirm that the decrease in fluorescence caused by UV treatment was due to cell lysis, we showed that culture fluorescence of WT and hyper-resistant cells remained proportional to cell number for both UV-treated and untreated cells (Fig. S1),
Fig. 1.
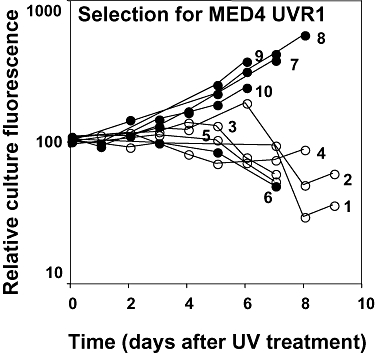
Enrichment selection for UV-hyper-resistant Prochlorococcus MED4 (MED4 UVR1). Cells from a mid-log phase culture of MED4 WT were subjected to 30 s of UV exposure (254 nm) on day 0, then returned to standard growth conditions. This was done for 10 consecutive rounds of UV exposure and regrowth (labelled 1–10). ○, rounds 1–5; •, rounds 6–10.
UV resistance
Cultures of MED4 WT and MED4 UVR1 were grown and tested for relative sensitivity to UV light at three different wavelengths: 254, 302 and 365 nm (UVC, UVB and UVA respectively). At equivalent doses of UV light, MED4 UVR1 (Fig. 2D–F) was considerably more resistant to both UVC and UVB than MED4 WT (Fig. 2A–C). However, both the hyper-resistant and WT strains were equally insensitive to UVA at the doses used in this experiment. While UVC and UVB are known to be absorbed by and cause direct damage to DNA, UVA is poorly absorbed by DNA, and its effect on cells is thought to be more complex and indirect (Kidambi et al., 1996).
Fig. 2.
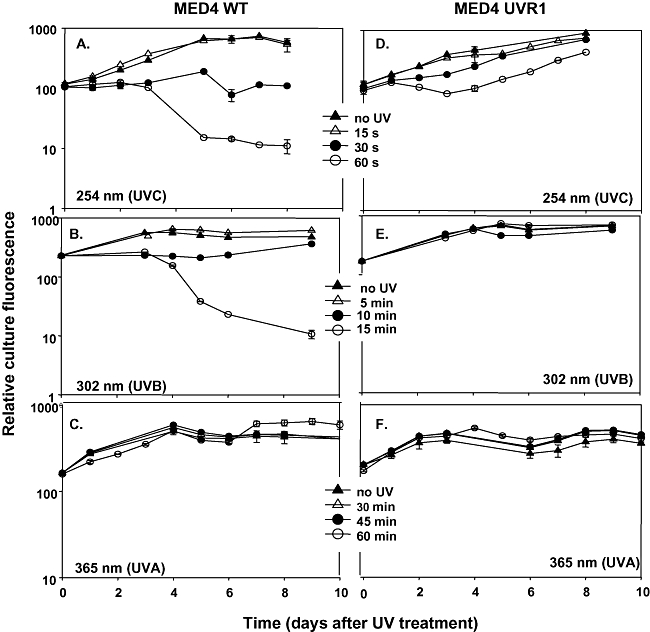
Sensitivity to UV light of MED4 WT and MED4 UVR1. MED4 WT (A, B, C) and MED4 UVR1 (D, E, F) were treated with UV light at three wavelengths: 254, 302 and 365 nm (UVC, UVB and UVA respectively), for various time intervals on day 0. Following UV treatment, cells were incubated under standard growth conditions and monitored over time, using relative culture fluorescence as a measure of cell density in the cultures. Values are the mean of measurements of duplicate cultures, with error bars showing the standard deviation.
Growth rates of MED4 WT and MED4 UVR1
To assess whether acquisition of the UV-hyper-resistant phenotype affected the growth rate under standard conditions in the absence of UV light, we measured the growth rates of WT and resistant strains in exponential phase. For MED4 WT, µ was 0.64 ± 0.01 day−1, compared with 0.67 ± 0.01 day−1 for MED4 UVR1, a significant difference according to the two tailed t-test using independent samples (P < 0.05). When the experiment was repeated, the growth rates were not significantly different, leading us to conclude that any such difference between the two strains under these conditions is small, and can only be revealed by very precise and repeated growth rate measurements. We cannot rule out that the growth rates of these strains may well vary significantly under conditions prevalent in their natural environment, where UV light is frequently present and other selection pressures may apply.
Gene expression
To begin to explore the underlying mechanisms mediating UV hyper-resistance in MED4 UVR1 we compared the complete transcriptomes of MED4 WT and MED4 UVR1 cells in mid log-phase under standard growth conditions in the absence of UV treatment, using whole genome microarrays. The only significant difference seen was in the expression of two genes, phrB and mutT, encoding photolyase and nudix hydrolase respectively, which were more highly expressed in MED4 UVR1 relative to MED4 WT ( Fig. 3). Interestingly, these two genes form a small predicted operon in MED4 WT, consistent with their apparent co-regulation in MED4 UVR1. These genes, including their order and context between the folK and degT genes, are conserved in all the sequenced high light-adapted strains of Prochlorococcus, and also in strains NATL1A and NATL2A, which show increased fitness in variable light (http://www.microbesonline.org, Kettler et al., 2007).
Fig. 3.
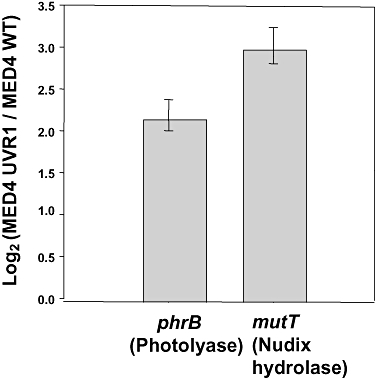
Expression of phrB and mutT in MED4 UVR1 relative to MED4 WT in log phase cells without UV treatment. Expression of phrB (encoding photolyase) and mutT (encoding nudix hydrolase) was measured using whole genome microarrays. These were the only genes that were significantly differentially expressed between MED4 WT and MED4 UVR1. Ratios are the mean values ± standard error for two microarray experiments, each carried out in duplicate.
We next used qRT-PCR to analyse a time-course of expression of several selected genes following UV treatment (primers shown in Table 1), guided in our choice of genes by the expression results from untreated MED4 WT and MED4 UVR1 cells described above. In addition to mutT and phrB, already shown to be constitutively upregulated in MED4 UVR1 cells, we examined the expression of DNA repair-related genes recA and radA, that might also be involved in the cellular response to UV light: recA is known to play a pivotal role in response to bacterial DNA damage (Michel, 2005), and while the recA analogue radA is of uncertain function in bacteria, it is thought to be involved in recombinational repair of DNA at replication forks blocked due to DNA damage [possibly resulting from pyrimidine dimer formation (Beam et al., 2002)]. radA is also known to be upregulated in response to UVB and UVC radiation in the archaeon Halobacterium sp. NRC-1 (Boubriak et al., 2008).
Table 1.
Primers used for qRT-PCR reactions.
| Gene | Primers |
|---|---|
| phrB | Forward: 5′-TGCAAGTTCAAGAGCTTGGTT-3′ |
| Reverse: 5′-CGGATCTCCTTCTTCCAAAA-3′ | |
| mutT | Forward: 5′-AACTCTTGTCCCTGCTCCTG-3′ |
| Reverse: 5′-TCTCCTTTAACCTCGGAATTGA-3′ | |
| recA | Forward: 5′-CTTGGAGATGCCTCCAGAAT-3′ |
| Reverse: 5′-CAACCACTCTTCCCTTTGGA-3′ | |
| radA | Forward: 5′-GAAGGATCGAGACCATTTGC-3′ |
| Reverse: 5′-GCTCATTCCAGTTGTTGTTCG-3′ | |
| rnpB | Forward: 5′GAGGATAGTGCCACAGAAACATACC-3′ |
| Reverse: 5′-GCTGGTGCGCTCTTACCACACCCTTG-3′ |
In the absence of UV treatment, in steady-state growth, MED4 UVR1 again had considerably higher levels of phrB and mutT transcripts relative to MED4 WT (Fig. 4A), in addition to slight constitutive upregulation of radA that was below the level of detection in the microarray experiment. Under the same conditions, recA expression was slightly lower in MED4 UVR1 as compared with MED4 WT, possibly a consequence of the constitutively high level of phrB and mutT expression (for example, excessive amounts of PhrB and MutT may reduce background levels of DNA damage, thus decreasing the need for RecA function).
Fig. 4.
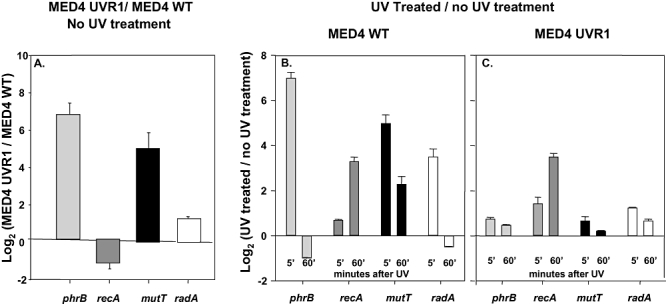
Expression of DNA repair genes determined by qRT-PCR in MED4 UVR1 and MED4 WT, with and without UV treatment. Ratios are the mean values ± standard error for two independent PCR runs, each carried out in duplicate. A. Transcript levels of four genes in MED4 UVR1 relative to MED4 WT in log phase cells in the absence of UV treatment. B and C. Transcript levels of the same four genes in log phase cultures subjected to brief UV treatment (254 nm, 30 s), relative to untreated controls. Transcript levels at 5 and 60 min after UV treatment are shown for MED4 WT (B) and MED4 UVR1 (C).
To explore the dynamics of the expression of these genes in response to UV exposure, we compared transcript levels of UV-treated and untreated MED4 WT and MED4 UVR1 cells 5 and 60 min after treatment (Fig. 4B and C). For MED4 WT, phrB, mutT and radA were rapidly upregulated, showing elevated transcript levels by 5 min post treatment. By 60 min post treatment the levels dropped – in two cases below those of the untreated cells, suggesting the need for their function had diminished. In contrast, for MED4 UVR1, phrB, mutT and radA were only slightly upregulated following UV irradiation, with transcripts decreasing only slightly between 5 and 60 min, consistent with their constitutively elevated levels. The kinetics of upregulation of the phrB-mutT operon in the WT strain (which, although rapid, takes some time as compared with constitutive expression in the mutant) provides an explanation for increased UV resistance in the mutant strain, i.e. its constitutively upregulated enzymes are already present and poised to repair damage before the onset of UV treatment, whereas in the WT these enzymes need to be expressed and synthesized following UV treatment, potentially allowing more unrepaired DNA damage to accumulate.
On the other hand, for both mutant and WT strains, recA gene expression was only slightly upregulated by 5 min post treatment, but by 60 min was present at significantly higher levels than in untreated controls. The kinetics of recA upregulation in both strains suggests that recA is likely to be normally regulated independently of the phrB-mutT operon.
Sensitivity of low light-adapted Prochlorococcus MIT9313 to UV light
With the knowledge that low light-adapted strains of Prochlorococcus encode mutT but not phrB, and thus cannot synthesize photolyase (Kettler et al., 2007), we tested the sensitivity of a low light-adapted strain (MIT9313) to UV exposure and compared it with MED4 WT (Fig. 5). As expected, MIT9313 was in fact considerably more sensitive to UVB and UVC radiation than was MED4 WT. This observation supports the contention that the photoreactivation system plays a significant role in UV resistance in the ocean, and suggests that the presence of phrB was selected for in the high light-adapted strain MED4 because it is a high light-adapted strain that occupies high light surface waters where UV radiation is present and can act as a selective agent. In contrast, photoreactivation repair would not be necessary in low light-adapted strains such as MIT9313, which occupy deeper, low light waters. The larger difference between the two strains seen with UVB radiation than with UVC radiation may reflect the fact that UVB radiation is generally less damaging than UVC, potentially allowing smaller dosage differences to be titrated more precisely.
Fig. 5.
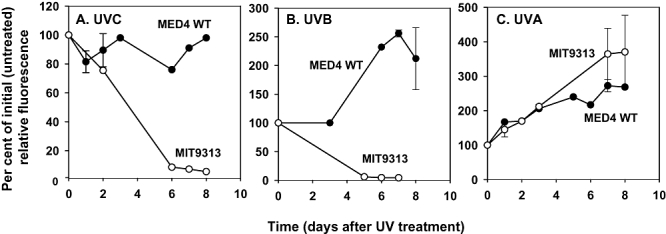
Sensitivity to UV of high light-adapted (MED4 WT) versus low light-adapted (MIT9313) strains of Prochlorococcus. Mid-log phase cells were treated with UV light for various time intervals (A: UVC for 30 s, B: UVB for 5 min, and C: UVA for 1 h) on day 0. Following UV treatment, cells were incubated under standard growth conditions and monitored for culture fluorescence over time. Fluorescence of UV-treated cultures is expressed as per cent of untreated culture fluorescence on day 0. Values are the mean of measurements of duplicate cultures, with error bars showing the standard deviation.•, MED4 WT; ○, MIT9313.
The lack of differential sensitivity to UVA treatment at the doses used is consistent with the inability of UVA radiation to cause damage that can be repaired by photolyase (Sancar, 2004)
Genome comparison of MED4 WT and MED4 UVR1
To determine the genomic underpinnings of the UVR phenotype we sequenced the genomes of both MED4 WT and MED4 UVR1. Genomes were first assembled using the sequence of a parent culture of MED4 (GenBank BX548174) as the reference, and then confirmed using a modified de novo assembly capable of detecting large insertions or deletions (Experimental procedures). MED4 WT contained 16 single nucleotide polymorphisms as compared with the published reference sequence (Rocap et al., 2003, white rows in Table 2). The stability of the MED4 WT genome over more than 5 years of growth and serial transfers in the laboratory seems remarkable.
Table 2.
DNA sequence differences between Prochlorococcus MED4 strains used in this study and the published parent MED4 genome (Rocap et al., 2003).
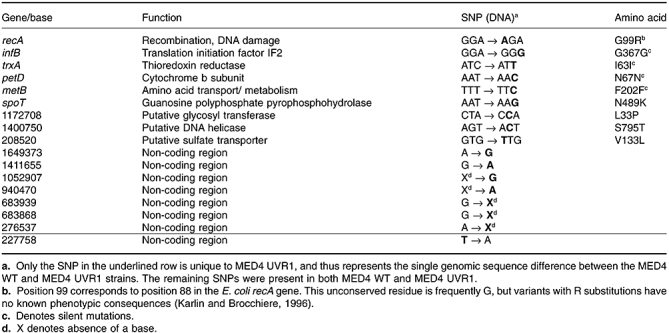 |
Relative to the reference sequence, MED4 UVR1 contained the same 16 SNPs as MED4 WT; in addition, however, it contained a single base pair deletion (underlined row, Table 2), located just upstream of the mutT gene, in a putative regulatory region of the mutT-phrB operon (Fig. 6). This result was confirmed by Sanger sequencing of a PCR fragment encompassing the 1 bp deletion (Experimental procedures), and strongly suggests that this mutation, located in a possible regulatory region for the downstream operon, is responsible for the high-level constitutive expression of the operon in the mutant, thus conferring UV hyper-resistance.
Fig. 6.
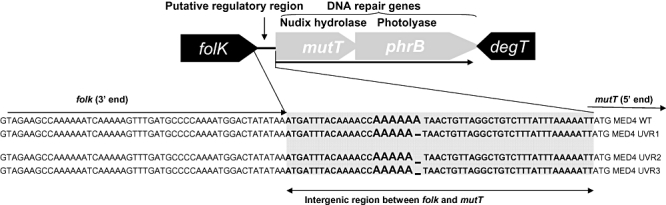
The MED4 mutT-phrB operon and location of UV hyper-resistance mutations in hyper-resistant mutants. The mutT-phrB operon of Prochlorococcus MED4 WT is flanked by two unrelated structural genes, folK and degT (http://www.microbesonline.org/cgi-bin/fetchLocus.cgi?locus=565621&disp=1). The highlighted sequence shows the intergenic region between folk and mutT, including the location of the homopolymer region (enlarged font) containing the 1 bp deletion (underlined) in the MED4 UVR strains as compared with MED4 WT. The intergenic region of MED4 WT was sequenced twice, first as the parent of MED4 UVR1, and then verified a year later as the parent of MED4 UVR2 and MED4 UVR3.
Because we do not have robust genetic tools for Prochlorococcus, we were unable to test directly whether this deletion would give rise to the UV-hyper-resist phenotype when transferred to a naïve WT strain. To further substantiate our hypothesis therefore we sequenced this region of the genome (containing the single base pair deletion in MED4 UVR1) in the other UV-hyper-resist strains, MED4 UVR2 and MED4 UVR3. Remarkably, both MED4 UVR2 and MED4 UVR3 carried the identical 1 bp deletion upstream of the mutT-phrB operon (Fig. 6). Although the mechanism by which this sequence may control expression of the operon is not clear, this result provides compelling evidence that the deletion is responsible for the UVR phenotype, as the strong UV challenge repeatedly enriched for cells containing the identical deletion mutation. Although there are no obvious regulatory motifs present in the intergenic sequence, and such sequences have not yet been defined for Prochlorococcus, a conceivable scenario is that a transcriptional activator or repressor might normally bind to the upstream region to regulate expression of the mutT-phrB operon in the WT case. The deletion in the mutant strain may disrupt the binding site, either preventing binding of the repressor, or causing the activator to bind more efficiently, resulting in constitutive expression. The role of radA in this model is unclear, as the function of its encoded enzyme remains to be elucidated. Additional studies of global transcription following UV treatment should clarify these issues further.
We note that MED4 UVR1 has not reverted to UV sensitivity despite being maintained under non-selective conditions for several years, which might be explained both by the fact that spontaneous compensatory 1 bp insertion mutations are relatively rare (Schaaper and Dunn, 1991; Mira et al., 2001), and that MED4 WT does not appear to grow faster than the mutant under laboratory conditions.
Analysis of the mutT genomic region in sequences from wild Prochlorococcus
Because increased resistance to UV radiation could conceivably confer an advantage to Prochlorococcus that dominate surface waters, it was of interest to determine whether the mutant sequence could be found in the wild. We searched the GOS (Global Ocean Survey) metagenomic database (Experimental procedures) to see whether we could find any incidence of the single base deletion in fragments of Prochlorococcus genomes that could be identified as coming from directly upstream of mutT. The search revealed 31 sequences with high similarity to MED4, one of which contained the SNP found in our mutant strains (JCVI_READ_1103359078035). Seven additional sequences contained ‘G’ in place of ‘A’ at the same position as the deletion mutation [i.e. AAAAGA instead of AAAA_A (mutant) or AAAAAA (WT) – see Table 3]. The remaining 23 sequences contained the WT sequence AAAAAA at the same location (Table 3). While there are not enough data to make statistical arguments, and we have not taken sequencing error into consideration – especially for this homopolymeric stretch of DNA – we find these data provocative albeit inconclusive with regard to the relevance of our laboratory observations to wild populations of Prochlorococcus. We also emphasize that the number of sequences analysed is vanishingly small relative to the 104–105Prochlorococcus cells ml−1 populating many of the stations sampled in GOS. Thus even if the mutation were present at a high frequency, we would not necessarily expect to see it in a sample of this size.
Table 3.
Sequences upstream of the mutT-phrB operon in wild Prochlorococcus strains.
| Strain or GOSa read number | Relevant sequence upstream of mutT-phrB |
|---|---|
| MED4 WT | AAAAAA |
| MED4 UVR1, UVR2, UVR3 | AAAA_A |
| JCVI_READ_1103359078035 | AAAA_A |
| JCVI_READ_1093017692266 | AAAAGA |
| JCVI_READ_1093018630368 | AAAAGA |
| JCVI_READ_1103769464916 | AAAAGA |
| JCVI_READ_1103769440186 | AAAAGA |
| JCVI_READ_1103769744552 | AAAAGA |
| JCVI_READ_1103242864663 | AAAAGA |
| JCVI_READ_1103359190125 | AAAAGA |
| JCVI_READ_ 630691 | AAAAAA |
| JCVI_READ_ 843287 | AAAAAA |
| JCVI_READ_1519837 | AAAAAA |
| JCVI_READ_1568009 | AAAAAA |
| JCVI_READ_1653138 | AAAAAA |
| JCVI_READ_1664920 | AAAAAA |
| JCVI_READ_1091141237359 | AAAAAA |
| JCVI_READ_1092256175212 | AAAAAA |
| JCVI_READ_1092963601548 | AAAAAA |
| JCVI_READ_1093017692266 | AAAAAA |
| JCVI_READ_1092344011058 | AAAAAA |
| JCVI_READ_1092347061561 | AAAAAA |
| JCVI_READ_1092344012813 | AAAAAA |
| JCVI_READ_1103769374878 | AAAAAA |
| JCVI_READ_1102140312344 | AAAAAA |
| JCVI_READ_1105333561959 | AAAAAA |
| JCVI_READ_1103668709694 | AAAAAA |
| JCVI_READ_1103242175527 | AAAAAA |
| JCVI_READ_1105333435622 | AAAAAA |
| JCVI_READ_1104230100283 | AAAAAA |
| JCVI_READ_1103242847341 | AAAAAA |
| JCVI_READ_1108840035938 | AAAAAA |
| JCVI_READ_1108830123070 | AAAAAA |
The GOS database can be accessed at http://camera.calit2.net/index.php.
Discussion
Using visible light, especially in the blue-violet range, as a source of energy, photolyases catalyse the repair of cyclobutane dipyrimidine dimers formed by UV damage to DNA (Sancar, 2004). Metagenomics studies have shown that photolyase and cryptochrome gene homologues have been found to be highly represented in surface seawater (DeLong et al., 2006; Frias-Lopez et al., 2008; Singh et al., 2009), in one instance, representing ∼0.1% of the genes present in a sample from a high-UV ocean environment (Singh et al., 2009). Such environments are also enriched for transcripts of these genes (Frias-Lopez et al., 2008). Results of these studies are consistent with the high levels of UV radiation to which the ocean surface is exposed daily, and which, according to recent measurements, may penetrate to depths of tens of metres (Tedetti and Sempere, 2006). Moreover, photolyase is encoded on the genomes of only a subset of the sequenced Prochlorococcus strains, specifically those that are high light-adapted or display increased relative fitness in fluctuating light (Kettler et al., 2007). Thus photolyase, specifically needed to repair radiation damage to DNA, appears to be maintained when there is a strong selection for its activity. This strong selection is reflected by the increased sensitivity to UVB and C of low light-adapted strain MIT9313, which does not encode phrB, as compared with MED4 WT. Further, this difference in sensitivity between MIT9313 and MED4 WT was not observable in response to UVA radiation, which does not cause the formation of pyrimidine dimers. Whereas UVC and UVB can cause direct damage to DNA, UVA (320–400 nm) causes only indirect damage through reactive oxygen intermediates (Kidambi et al., 1996).
Nudix hydrolase, on the other hand, is encoded by the ‘core’ gene mutT, found in all the Prochlorococcus genomes sequenced to date (Kettler et al., 2007). This is consistent with its function to repair damage to GTP caused by reactive oxygen species formed not only as a result of oxidizing radiation such as UV light, but also formed as a result of oxidative stress from activities such as photosynthesis (Latifi et al., 2009). It is noteworthy that although UVC radiation does not normally penetrate the earth's atmosphere, the levels of UVA and UVB radiation measured at the surface of the ocean and possibly tens of metres below may reach or greatly exceed the doses used in these experiments (Buma et al., 2001).
Interestingly, photolyase and nudix hydrolase repair activities are not error prone and are thereforenon-mutagenic, in contrast to SOS repair (a form of dark repair) that allows survival at the cost of mutagenesis. Despite the fact that MED4 and other strains of Prochlorococcus encode a number of SOS-type genes including recA and lexA (the canonical repressor of SOS genes), the potential advantage of using error-free repair is apparent, as it would seem that the small, streamlined genomes of the high light-adapted ecotypes of Prochlorococcus would be adversely affected by high mutation rates. In support of this idea, Prochlorococcus strains also lack low-fidelity DNA polymerases II and IV, which are known to replicate across lesions, thereby leading to mutations (Napolitano et al., 2000; Michel, 2005). They do, however, encode one of the low-fidelity DNA polymerases, PolV (encoded by umuCD). It is noteworthy that selection for resistance to repeated UV light exposure in E. coli has produced mutants that are typically affected in several different ‘dark repair’ genes involved in DNA replication and repair (Alcantara-Diaz et al., 2004), including radA uvrA, and polA, whereas enrichment for UV hyper-resistance in MED4 resulted in a lesion affecting photoreactivation repair. Although the specific E. coli UV-resistance mutations isolated may at least partly be an artefact of their selection, i.e. they were selected in the dark in order to avoid photoreactivation repair following mutagenesis, it is tempting to speculate that with a larger and less streamlined genome, E. coli might be better able to tolerate mutations caused by error-prone repair mechanisms and may therefore make more use of these mechanisms.
Photolyase has been shown to be the major UV-resistance factor for another cyanobacterium, Synechocystis PCC 6803 (Ng and Pakrasi, 2001). This was shown genetically, i.e. a knockout of the phrB gene conferred increased UV sensitivity to the organism, whereas a recA knockout mutant was still relatively resistant to UVC (Minda et al., 2005). The authors suggested that RecA may be used primarily for other functions in this organism, such as recombination repair, which is error-free (Sargentini and Smith, 1985; Cox, 1999). Although Synechocystis PCC 6803 also encodes a lexA gene, it may also be used for functions other than the regulation of SOS repair. Our data show that the Prochlorococcus recA gene was upregulated in both MED4 WT and MED4 UVR1 following UV irradiation, but only after the phrB and mutT genes were upregulated. It is not known whether MED4 or any Prochlorococcus strain makes use of an SOS repair system, and if so, under what circumstances. However, regardless of whether the 1 bp deletion was a new mutation caused by UV treatment or a pre-existing mutant in the original MED4 batch culture, it is striking that three enrichments for UV hyper-resistance, carried out at different times from the same MED4 WT strain using 10 rounds of treatment with UVC radiation of sufficient intensity to kill most of the cells, selected for this same 1 bp deletion mutation – perhaps suggesting there may be limited ways of becoming UV hyper-resistant. Further, a comparison of the genome sequences of MED4 WT and MED4 UVR1 points to high genome stability in the surviving cells. Again, with frequent and long-lasting environmental UV irradiation exposure, it would seem advantageous for Prochlorococcus to make use primarily of error-free photoreactivation repair systems, especially with the abundance of visible light available to power the system.
The role of an SOS repair system in Prochlorococcus has yet to be established. Future experiments that measure expression of relevant candidate SOS and other DNA repair genes under conditions designed to simulate exposures in the natural environment will address this question. In this context it is interesting to note that true temperate phage that can be excised from the bacterial genome require SOS-type processes, initiated by DNA damage, for their excision (Rokney et al., 2008). To date, despite the isolation of many Prochlorococcus cyanophage and despite strong evidence of the presence of phage genes in both low and high light-adapted Prochlorococcus genomes, temperate phage have not yet been isolated for this organism. An increased understanding of the role of SOS and photoreactivation in Prochlorococcus DNA damage repair in low and high light-adapted strains may help to elucidate possibilities for lysogeny in Prochlorococcus.
Experimental procedures
Growth conditions
All Prochlorococcus cultures were grown at 22°C under constant light provided by cool-white fluorescent lamps at irradiances of ∼25 mol Q m−2 s−1 for MED4 strains, and ∼16 mol Q m−2 s for MIT9313. Cultures were grown in Pro99 medium, consisting of sterile (0.2 µm filtered, autoclaved) Sargasso Sea water supplemented with 800 µM NH4Cl, 50 µM NaH2PO4, and trace metals (Moore et al., 2007). Growth was monitored using Turner Design fluorometer 10-AU to measure bulk chlorophyll fluorescence, used as a proxy for culture cell density (Moore et al., 2007). Bulk chlorophyll fluorescence has been shown to be directly proportional to cell concentration for cells growing in exponential phase (Moore et al., 2007), as determined by measuring cell number throughout log phase by means of flow cytometry.
UV treatment and enrichment for hyper-resistant mutants
In all cases, UV treatment was applied using a UVLMS-38 8-W UV lamp (UVP, Upland, CA), generating wavelengths of 365/302/254 nm, with cells at a distance of 22 cm below the light source. Cells were slowly rotated using a mechanical rotator platform during all UV treatments. For resistance enrichment, cells were irradiated at room temperature for 30 s at 254 nm (UV fluence 9.2 J m−2) in an uncovered sterile glass Petri dish (150 mm diameter) containing 25 ml of culture grown to a fluorescence of ∼100 (early- to mid-log phase). Following treatment, cells were immediately returned to standard growth conditions. In cases where fluorescence decreased following UV treatment, cells were maintained under standard growth conditions until they recovered (i.e. reached a fluorescence of 100). After recovery cells were diluted 1:10 into Pro99 medium and again grown to fluorescence of ∼100 for the next round of UV irradiation. To confirm that culture fluorescence was proportional to cell number following UV treatment, cell counts on selected samples were carried out by means of flow cytometry, using 2 µm fluorescent beads as a size reference (Moore et al., 2007). Results are shown in Fig. S1.
Once isolated, UV-hyper-resist strains were purified at least 20 times by dilution to very low titre (10 cells ml−1), followed by regrowth. This process was necessary because it is difficult, and in this case not possible, to isolate and regrow pure single colonies of Prochlorococcus. In addition, at least 10 cells ml−1 were required for these cultures to grow in liquid. However, DNA sequencing of mutant versus WT batch cultures confirmed the appropriate genotypes, i.e. all three mutant cultures contained the same deletion mutation upstream of the mutT-phrB operon. Furthermore, in spite of continuous culturing, involving cycles of dilution and regrowth, for periods longer than 2 years, the mutants have retained their UV-hyper-resistant genotype and phenotype.
RNA preparation
Cells (UV-treated or untreated) were collected by centrifugation (7 min, 12 000 g, 22°C), resuspended in 1 ml of RNA later (Ambion, Austin, TX, USA), quick frozen and stored at −80°C. RNA was isolated using the Mirvana miRNA isolation kit (Ambion) according to the manufacturer's instructions. DNA was removed using Turbo DNase treatment (Ambion) according to the manufacturer's instructions and DNA removal was confirmed by PCR using the primers shown in Table 1 for qRT-PCR. RNA was then ethanol precipitated and resuspended to a concentration of 100 ng ml−1.
qRT-PCR reactions
RNA was isolated as described above, reverse-transcribed, and subjected to triplicate real-time PCR reactions as described previously (Frias-Lopez et al., 2008), using the primer sets shown in Table 1. We calculated gene expression levels using the Comparative CT (2−ΔΔCT) method (Livak and Schmittgem, 2001; Frias-Lopez et al., 2008), where CT is the threshold cycle at which fluorescence rises above background levels, and ΔΔCT = ΔCT, sample − ΔCT, reference. Expression of gene rnpB was used as the endogenous reference.
Microarray hybridization and analysis
For microarray hybridizataion, 100 ng of RNA was first amplified using the MessageAmp II-Bacteria Kit (Ambion) according to the manufacturer's instructions, as previously described (Frias-Lopez et al., 2008).
cDNA was synthesized, labelled, and hybridized to customized MD4-9313 microarrays (Affymetrix, Santa Clara, CA), and scanned, all as previously described (Lindell et al., 2005). Hybridizations were visualized using GeneSpring software (version 7.3.1; Silicon Genetics, Palo Alto, CA), initially normalized using the Robust Multichip Average algorithm (Bolstad et al., 2003) implemented in GeneSpring, and were later normalized using the Loess correction performed by using the open-source Statistical Data Analysis software R version 2.5.0 (http://www.r-project.org/). We used the program QVALUE, which measures significance in terms of the false discovery rate (Storey and Tibshirani, 2003). A gene was considered as differentially expressed if the q-value was < 0.05.
Genome sequencing
Library preparation
Genomic DNA libraries suitable for sequencing on the Illumina Genome Analyzer were prepared as described (manufacturer's document ‘11251892_Genomic_DNA_Sample_Prep.pdf’) except that genomic DNA (30 µg in 300 µl TE, in an ice bath solution) was fragmented using a model UCD-200 Bioruptor sonicator (Diagenode): low power, 15 s on/15 s off cycle, for a total of 10 min. Adaptor–DNA constructs were purified on a 2% agarose-TAE gel, run at 120 V for 60 min, with a low-molecular-weight DNA ladder (New England BioLabs) run in parallel. Blank wells between each sample prevented DNA cross contamination. DNA of size 150–200 bp was excised from the gel and purified using a QIAGEN Gel Extraction Kit. Samples were eluted into 50 µl elution buffer, then extracted using the Qiagen MinElute Gel Extraction Kit and eluted into 30 µl of elution buffer.
Purified adaptor–DNA constructs were enriched for fragments having the proper adaptor molecules on both ends using PCR (1 µl DNA, 25 µl Phusion DNA polymerase (Finnzymes Oy), 1 µl each of PCR primers 1.1 and 2.1 (© 2006 and 2008 Illumina), and 22 µl water). The PCR protocol was: 30 s at 98°C, 18 cycles of: 10 s at 98°C, 30 s at 65°C, 30 s at 72°C, followed by a final extension of 5 min at 72°C. Amplified DNA was purified using the Qiagen QIAquick PCR Purification Kit, and eluted into 50 µl of elution buffer. The DNA library concentration was determined using a NanoDrop ND8000 spectrophotometer, followed by analysis on an Agilent 2100 Bioanalyzer using the Agilent RNA 6000 Pico Chip.
Sequencing methods
DNA library samples were diluted to a concentration of 10 nM, and were clustered at a final concentration of 2.0 pM each on an Illumina GA flow cell, according to the manufacturer's standard protocols. Sequencing of the samples on the clustered flow cell was performed on the Genome Analyzer using a single read 36 cycle protocol, following the manufacturer's standard procedures and protocols.
Data analysis
Each sample (MED4 WT and MED4 UVR1) was run on one flow cell lane of an Illumina GA I, yielding 5.8 and 7.8 million 36-base sequence reads respectively. Raw images were processed using the Genome Analyzer Pipeline Software v0.3 to produce DNA sequence files in FASTQ format. Output files were imported and assembled using the SeqMan NGEN v1.2 program from DNAStar with the original published MED4 genome (BX548174) as the reference sequence. To check for possible large insertions, deletions, inversions, or genomic rearrangements, further assembly analysis was carried out using NGen's Split Reference Sequence Strategy (http://www.dnastar.com/products/ReferenceGuidedAssembly.php). Resulting genome assemblies were imported into DNAStar LaserGene8, where mutations were identified.
Sequencing of PCR fragments
DNA fragments spanning the putative regulatory region upstream of mutT were PCR-amplified using forward and reverse primers: (5′-GGTGGGAAGATTTTTATACGGTTGACGAG-3′, and 5′-AATGCTGCGCCAGGATGC-3′) respectively. Amplified DNA fragments were treated with ExoSAP-IT (USB, Cleveland, OH) and submitted for pyrosequencing (Amplicon Express, Pullman, WA).
Analysis of GOS database
The GOS database was searched to recruit reads and their paired ends homologous to a 30 bp stretch of the 51 bp intergenic region (Fig. 6) between folK and mutT. This search yielded 175 sequences that matched at least 30 bp of the intergenic region. To select those with high similarity (at least 95%) to high-light Prochlorococcus strains over the entire length of the read (normally ∼700–1000 bp), blastn was used to compare the reads against the microbial genomes database in Microbes Online (http://www.microbesonline.org). Thirty-one reads had high similarity to Prochlorococcus, and these were analysed for the presence of SNPs in the intergenic region, as described above. Reads of interest can be accessed in the CAMERA (Community Cyberinfrastructure for Advanced Marine Microbial Ecology Research and Analysis) database (http://camera.calit2.net/index.php).
Acknowledgments
We thank D. Sher and A. Thompson for assistance with data analysis, Q. Zeng and D. Sher for reviewing the manuscript, and all members of the Chisholm laboratory for interesting and helpful discussions. Thanks also to J. Engelking of DNAStar for his work on the genome assemblies. This research was supported by grants to SWC from the Gordon and Betty Moore Foundation, the Department of Energy Genomics GTL program, the National Science Foundation, and grants to BMH from the Howard Hughes Medical Institute and the Lord Foundation.
Supporting information
Additional Supporting Information may be found in the online version of this article.
Fig. S1. Cell fluorescence following UV treatment is proportional to cell number. Cells from mid-log phase cultures of MED4 WT and MED4 UVR1 were subjected to 30 s of UV exposure (254 nm) on Day 0, then returned to standard growth conditions. Untreated (○) and UV-treated (•) cultures were sampled for culture fluorescence (A and B), and for cell number using flow cytometry (C and D). Values are the mean of measurements of duplicate cultures, with error bars showing the standard deviation.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Agustí S, Llabrés M. Solar radiation-induced mortality of marine pico-phytoplankton in the oligotrophic ocean. Photochem Photobiol. 2007;83:793–801. doi: 10.1111/j.1751-1097.2007.00144.x. [DOI] [PubMed] [Google Scholar]
- Alcantara-Diaz D, Brena-Valle M, Serment-Guerrero J. Divergent adaptation of Escherichia coli to cyclic ultraviolet light exposures. Mutagenesis. 2004;19:349–354. doi: 10.1093/mutage/geh039. [DOI] [PubMed] [Google Scholar]
- Beam C, Saveson C, Lovett S. Role for radA/sms in recombination intermediate processing in Escherichia coli. J Bacteriol. 2002;184:6836–6844. doi: 10.1128/JB.184.24.6836-6844.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolstad B, Irizarry R, Astrand M, Speed T. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Boubriak I, Ng W, Dassarma PS, Dassar A, Crowley D, McCready S. Transcriptional responses to biologically relevant doses of UV-B radiation in the model archaeon, Halobacterium sp. NRC-1. Saline Systems. 2008;4:13–26. doi: 10.1186/1746-1448-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buma A, Helbling E, Boer MD, Villafañe V. Patterns of DNA damage and photoinhibition in temperate South-Atlantic picophytoplankton exposed to solar ultraviolet radiation. J Photochem Photobiol B. 2001;62:9–18. doi: 10.1016/s1011-1344(01)00156-7. [DOI] [PubMed] [Google Scholar]
- Christaki U, Courties C, Karayanni H, Giannakourou A, Maravelias C, Kormas K, Lebaron P. Dynamic characteristics of Prochlorococcus and Synechococcus consumption by bacterivorous nanoflagellates. Microb Ecol. 2002;43:341–352. doi: 10.1007/s00248-002-2002-3. [DOI] [PubMed] [Google Scholar]
- Coleman M, Chisholm S. Code and context: Prochlorococcus as a model for cross-scale biology. Trends Microbiol. 2007;15:398–407. doi: 10.1016/j.tim.2007.07.001. [DOI] [PubMed] [Google Scholar]
- Cox M. Recombinational DNA repair in bacteria and the RecA protein. Prog Nucleic Acid Res Mol Biol. 1999;63:311–366. doi: 10.1016/s0079-6603(08)60726-6. [DOI] [PubMed] [Google Scholar]
- DeLong E, Preston C, Mincer T, Rich V, Hallam S, Frigaard N, et al. Community genomics among stratified microbial assemblages in the ocean's interior. Science. 2006;311:496–503. doi: 10.1126/science.1120250. [DOI] [PubMed] [Google Scholar]
- Frias-Lopez J, Shi Y, Tyson G, Coleman M, Schuster S, Chisholm S, Delong E. Microbial community gene expression in ocean surface waters. Proc Natl Acad Sci USA. 2008;105:3805–3810. doi: 10.1073/pnas.0708897105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillou L, Jacquet S, Chretiennot-Dinet M-J, Vaulot D. Grazing impact of two small heterotrophic flagellates on Prochlorococcus and Synechococcus. Aquat Microb Ecol. 2001;26:201–207. [Google Scholar]
- Karlin S, Brocchiere L. Evolutionary conservation of RecA genes in relation to protein structure and function. J Bacteriol. 1996;178:1881–1894. doi: 10.1128/jb.178.7.1881-1894.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettler G, Martiny A, Huang K, Zucker J, Coleman M, Rodrigue S, et al. Patterns and implications of gene gain and loss in the evolution of Prochlorococcus. PLoS Genet. 2007;3:e231. doi: 10.1371/journal.pgen.0030231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidambi S, Booth M, Kokjohn T, Miller R. recA-dependence of the response of Pseudomonas aeruginosa to UVA and UVB irradiation. Microbiology. 1996;142:1033–1040. doi: 10.1099/00221287-142-4-1033. [DOI] [PubMed] [Google Scholar]
- Latifi A, Ruiz M, Zhang C. Oxidative stress in cyanobacteria. FEMS Microbiol Rev. 2009;33:258–278. doi: 10.1111/j.1574-6976.2008.00134.x. [DOI] [PubMed] [Google Scholar]
- Lindell D, Jaffe J, Johnson Z, Church G, Chisholm S. Photosynthesis genes in marine viruses yield proteins during host infection. Nature. 2005;438:86–89. doi: 10.1038/nature04111. [DOI] [PubMed] [Google Scholar]
- Livak K, Schmittgem T. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Llabrés M, Agustí S. Picophytoplankton cell death induced by UV radiation: evidence for oceanic Atlantic communities. Limnol Oceanogr. 2006;51:21–29. [Google Scholar]
- Michel B. After 30 years of study, the bacterial SOS response still surprises us. PLoS Biol. 2005;3:e255. doi: 10.1371/journal.pbio.0030255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minda R, Ramchandani J, Joshi V, Bhattacharjee S. A homozygous recA mutant of Synechocystis PCC6803: construction strategy and characteristics eliciting a novel RecA independent UVC resistance in dark. Mol Genet Genomics. 2005;274:616–624. doi: 10.1007/s00438-005-0054-z. [DOI] [PubMed] [Google Scholar]
- Mira A, Ochman H, Moran N. Deletional bias and the evolution of bacterial genomes. Trends Genet. 2001;17:589–596. doi: 10.1016/s0168-9525(01)02447-7. [DOI] [PubMed] [Google Scholar]
- Moore L, Coe A, Zinser E, Saito M, Sullivan M, Lindell D, et al. Culturing the marine cyanobacterium Prochlorococcus. Limnol Oceanogr: Methods. 2007;5:353–362. [Google Scholar]
- Napolitano R, Janel-Bintz R, Wagner J, Fuchs R. All three SOS-inducible DNA polymerases (Pol II, Pol IV and Pol V) are involved in induced mutagenesis. EMBO J. 2000;19:6259–6265. doi: 10.1093/emboj/19.22.6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng W, Pakrasi H. DNA photolyase homologs are the major UV resistance factors in the cyanobacterium Synechocystis sp. PCC 6803. Mol Gen Genet. 2001;264:924–930. doi: 10.1007/s004380000383. [DOI] [PubMed] [Google Scholar]
- Rocap G, Larimer F, Lamerdin J, Malfatti S, Chain P, Ahlgren N, et al. Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature. 2003;424:1042–1047. doi: 10.1038/nature01947. [DOI] [PubMed] [Google Scholar]
- Rokney A, Kobiler O, Amir A, Court D, Stavans J, Adhya S, Oppenheim A. Host responses influence on the induction of lambda prophage. Mol Microbiol. 2008;68:29–36. doi: 10.1111/j.1365-2958.2008.06119.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A. Photolyase and cryptochrome blue-light photoreceptors. Adv Protein Chem. 2004;69:73–100. doi: 10.1016/S0065-3233(04)69003-6. [DOI] [PubMed] [Google Scholar]
- Sancar G. Enzymatic photoreactivation: 50 years and counting. Mutat Res. 2000;451:25–37. doi: 10.1016/s0027-5107(00)00038-5. [DOI] [PubMed] [Google Scholar]
- Sargentini N, Smith K. Spontaneous mutagenesis: the roles of DNA repair, replication, and recombination. Mutat Res. 1985;154:1–27. doi: 10.1016/0165-1110(85)90007-7. [DOI] [PubMed] [Google Scholar]
- Schaaper R, Dunn R. Spontaneous mutation in the Escherichia coli lacI gene. Genet. 1991;129:317–326. doi: 10.1093/genetics/129.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Doerks T, Letunic I, Raes J, Bork P. Discovering functional novelty in metagenomes: examples from light-mediated processes. J Bacteriol. 2009;191:32–41. doi: 10.1128/JB.01084-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey J, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strehl B, Holtzendorff J, Partensky F, Hess W. A small and compact genome in the marine cyanobacterium Prochlorococcus marinus CCMP 1375: lack of an intron in the gene for tRNA(Leu)(UAA) and a single copy of the rRNA operon. FEMS Microbiol Lett. 1999;181:261–266. doi: 10.1111/j.1574-6968.1999.tb08853.x. [DOI] [PubMed] [Google Scholar]
- Sullivan M, Waterbury J, Chisholm S. Cyanophages infecting the oceanic cyanobacterium Prochlorococcus. Nature. 2003;424:1047–1051. doi: 10.1038/nature01929. [DOI] [PubMed] [Google Scholar]
- Tedetti M, Sempere R. Penetration of ultraviolet radiation in the marine environment. A review. Photochem Photobiol. 2006;82:389–397. doi: 10.1562/2005-11-09-IR-733. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


