Abstract
The beating heart requires a constant flux of ATP to maintain contractile function, and there is increasing evidence that energetic defects contribute to the development of heart failure. The last ten years have seen a resurgent interest in cardiac intermediary metabolism, and a dramatic increase in our understanding of transcriptional networks that regulate cardiac energetics. The PPAR-gamma coactivator (PGC)-1 family of proteins plays a central role in these pathways. The mechanisms by which PGC-1 proteins regulate transcriptional networks and are regulated by physiological cues, and the roles they play in cardiac development and disease, are reviewed here.
Keywords: PGC-1, metabolism, heart failure, mitochondria
Introduction
The heart consumes tremendous amounts of energy. ATP consumption, per weight of tissue, is the highest in the body. Energy reserves in the heart are relatively limited, and a heart starved of its fuel and oxygen supply can only beat 20-40 times (a few seconds) before succumbing to energy deficiency. Despite this narrow window, the healthy heart will contract billions of times in the average human life. The dynamic range of cardiac activity is large, both acutely (exercise), and chronically (development, especially postnatal). Bioenergetic programs in the heart must therefore be tightly regulated.
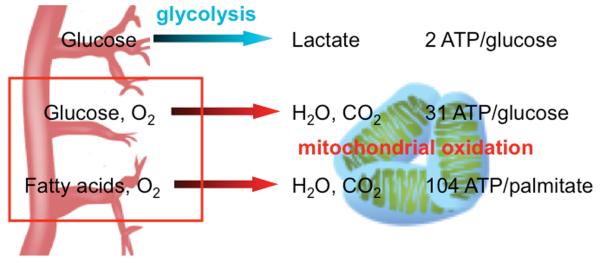
ATP is the currency of energy in the cell. Oxidative consumption of fuels in mitochondria is by far the most efficient means of generating ATP, yielding >30 ATP per molecule of glucose, compared to a net 2 ATP via anaerobic glycolysis and lactate production. It is not surprising then that the heart is highly aerobic and sustains >95% of its ATP output via oxidative breakdown of fuels. Oxidative phosphorylation of ATP occurs strictly in mitochondria, and the heart therefore maintains a high mitochondrial content. The energetic requirements of the heart increase dramatically at birth, and mitochondrial density accordingly increases sharply during the perinatal period.1-3 Mitochondrial mass makes up fully one third of the adult heart.
The PPAR-gamma coactivator (PGC)-1 transcriptional coactivators have recently emerged as powerful regulators of mitochondrial biology in the heart, by broadly regulating gene expression from both nuclear and mitochondrial genomes. The expression of PGC-1α is repressed in numerous models of heart failure, and this has been implicated as an important contributor to the maladaptive energetic profile of failing hearts. This review will focus on the PGC-1s and their role in cardiac biology.
PGC-1 coactivators
Coactivators are proteins that bind to nuclear receptors or other transcription factors and increase their ability to stimulate transcriptional activity. Most transcription factors likely require coactivators. A subset of coactivators are highly regulated, and transduce extra- and intra-cellular cues to changes in gene expression. PGC-1α is the best-studied example of such a regulated coactivator.
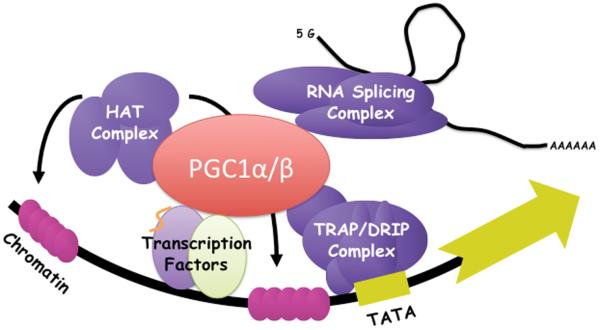
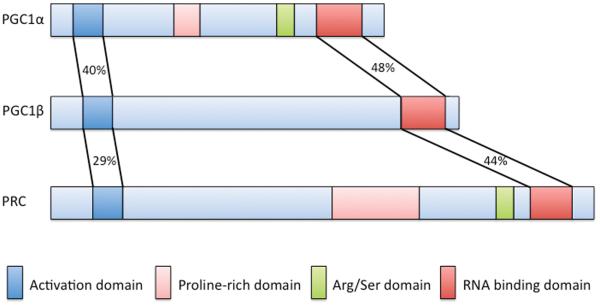
PGC-1α was originally identified in a two-hybrid screen, using a cDNA library from brown fat cells, looking for brown fat-specific interactors of the nuclear receptor PPARγ (the target of the anti-diabetic agents thiazolidinediones).4 Ectopic expression of PGC-1α in white fat cells conferred upon them some of the properties of brown fat, including induction of UCP1, a mitochondrial uncoupler and dissipator of heat. Since then, it has become clear that PGC-1α can coactivate a large repertoire of transcription factors, including most members of the nuclear receptor family.5, 6 Upon binding to transcription factors, PGC-1α interacts directly with the core transcriptional Trap/Mediator complex,7 as well as with chromatin-modifying enzymes like the p300 and SRC-1 histone acetyl transferases,8 and with the splicing machinery (Figure 1A).9 The net effect is robust activation of gene expression. The two other members of the PGC-1 family, PGC-1β and the more distant relative PRC, were identified by sequence homology to PGC-1α (Figure 1B).1
Figure 1. A.) Schematic of regulation of gene expression by PGC-1 coactivators.
PGC-1 binds directly to transcription factors and serves as a docking scaffold for histone modifying enzymes, TRAP/DRIP/Mediator complex and RNA splicing machinery. B.) The PGC-1 family of coactivators. PGC1 family members share significant primary sequence homology in their activation and RNA binding domains, as well as domain organization.
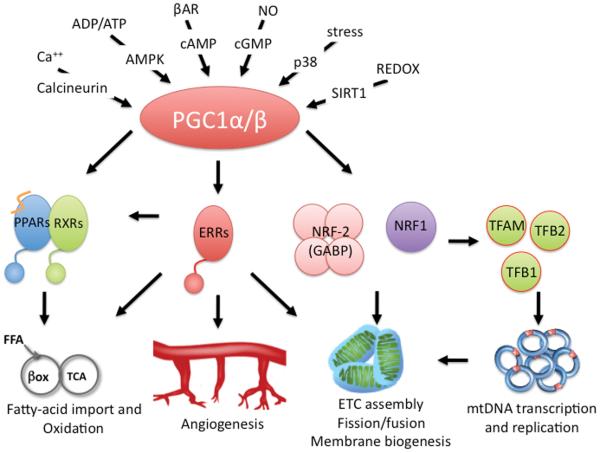
PGC-1α expression and activity are exquisitely sensitive to extracellular and physiologic cues. At the gene expression level, PGC-1α is induced in the liver and heart by fasting,1 in brown fat by cold-induced sympathetic stimulation,4 and in skeletal muscle and heart by exercise, to name a few. PGC-1β and PRC expression are generally less inducible, but changes in PGC-1β expression can for example be seen in differentiating osteoclasts,18 and PRC levels change with cell cycle progression.10 Induction of PGC-1α is often mediated by signaling by p38 MAPK and cyclic AMP (cAMP) that can be initiated by glucagon signaling (in hepatocytes) or adrenergic signaling via G-protein coupled receptors, ultimately acting on a conserved cAMP responsive element (CRE) in the PGC-1α promoter.14, 15, 19-21 An alternative promoter, generating an alternatively spliced exon 1 that changes the first 16 amino acids in the N-terminus the PGC-1α protein, is active in the heart, skeletal muscle, and brown fat, and is induced >100-fold by cAMP signaling.22, 23 PGC-1α gene expression is also sensitive to muscle-specific factors like MEF2,24, 25 calcineurin signaling,11, 26, 27 metabolic sensors like the adenosine monophosphate-activated protein kinase (AMPK),28 nitric oxide,29-31 p53, calcium/calmodulin-dependent protein kinase,27, 32 and autoregulatory positive feedback by PGC-1α itself.27 PGC-1α expression is thus affected by numerous signals, integrating important metabolic and neurohormonal states (Figure 2).
Figure 2. Transcriptional networks and regulation of PGC-1α.
Multiple stimuli activate PGC-1, leading to the coactivation of key transcription factors involved in fatty-acid oxidation and import, angiogenesis, electron transport chain assembly, membrane biogenesis, and mitochondrial DNA replication and transcription.
PGC-1α is also significantly regulated post-translationally. PGC-1α protein has a relatively short half-life (20 mins), and is ubiquitylated and degraded by the proteasome.33-35 p38 MAPK phosphorylates PGC-1α on three conserved sites and inhibits its degradation.33 AMPK also directly phosphorylates PGC-1α protein, stimulating its transactivation activity.28 Conversely, phosphorylation by the Akt kinase inhibits PGC-1α activity.36 In addition, 13 conserved arginines in PGC-1α are sites of inhibitory acetylation by the GCN5 acetyl transferase (and likely other acetyl transferases as well). Deacetylation and re-activation of PGC-1α is mediated by the longevity-associated, NAD-dependent Sirt1 type III histone deacetylase.37-39 PGC-1α is also methylated,40 and can be modulated by various protein-protein interactions, including with p160myb, bcl3, prox1, and lipin.41-44 PGC-1α protein therefore directly senses key metabolic states, including ADP/ATP levels (AMPK), redox state (Sirt1), and stress pathways (p38). Most of the studies demonstrating these biochemical modifications were not performed in cardiac cells, but it is likely that the same modifications also occur in cardiomyocytes. Post-translational modifications of PGC-1β and PRC have been much less extensively studied.45
The PGC-1 proteins, and PGC-1α in particular, are thus sensitive to a vast range of signals, and are well-suited to transmit extracellular and physiologic cues to the regulation of broad genetic programs. A variety of metabolic programs in different tissues are regulated by PGC-1 coactivators, including responses to fasting and high fat feeding in liver,14, 15, 46 exercise-induced changes in skeletal muscle,21, 22, 47, 48 oxidative insults in the brain,49 and the thermogenic response to cold in brown fat.4 This review will focus on the PGC-1s in the heart, where PGC-1α and β play critical roles in maintaining energy balance.
PGC-1 transcriptional networks in the heart
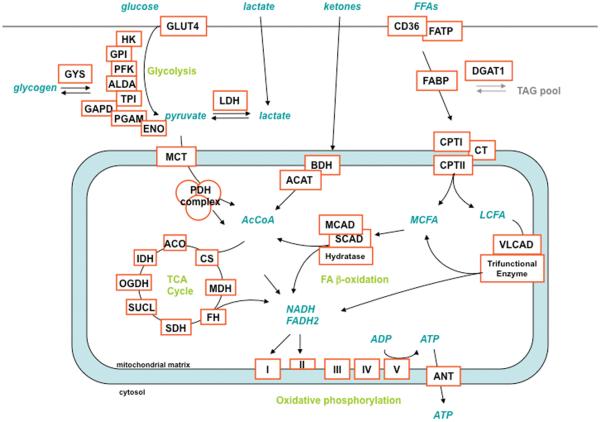
Overexpression of PGC-1α in cardiac cells induces hundreds of genes, encoding for key enzymes in all major metabolic programs needed for high-efficiency ATP production (Figure 3). >70% of the subunits of the four respiratory chain and the ATPase complexes are induced by PGC-1α, as are all 8 enzymes of the Krebs cycle. All key enzymes in fatty acid β-oxidation are induced, including fatty acid transport proteins and enzymes responsible for turnover of the intracellular triglyceride pool.50 Enzymes that mediate metabolism of lactate and ketone bodies are also induced. Induced proteins are located both in the mitochondria and in the cytosol. Overexpression of PGC-1α in cardiomyocytes in cell culture and in vivo markedly increases oxygen consumption capacity and fatty acid oxidation.16 Glucose oxidation is repressed, likely secondary to both product inhibition by fatty acids and ketones on the PDH complex, as well as induction of PDK4, a potent inhibitor of PDH activity.51 PGC-1α thus coordinates a complete program that allows cardiac cells to markedly increase fuel consumption and output of ATP. PGC-1α carries out this remarkable transcriptional induction by coactivating key families of transcription factors, each of which predominantly regulates specific subsets of genes involved in cardiac metabolism, as discussed below (Figure 2 and Table 1).
Figure 3. Substrate utilization in the heart, and metabolic enzymes induced by PGC-1α.
Neonatal rat ventricular myocytes (NRVM) were infected with adenovirus expressing PGC-1α versus GFP control, and 48hrs later gene expression was measured by qPCR. Metabolic pathways are noted in green. Induced genes are noted in red boxes. Glucose transporter 4 (GLUT4), hexokinase (HK), glucosephosphate isomerase (GPI), phosphofructose kinase (PFK), aldoase A (ALDA), triosephosphate isomerase (TPI), glyceraldehyde-3-phosphate dehydrogenase (GAPD), phosphoglycerate mutase (PGAM),enolase (ENO), glycogen synthase (GYS), lactate dehydrogenase (LDH), monocarboxylic acid transporter (MCT), pyruvate dehydrogenase (PDH), aconitase (ACO), citrate synthase (CS), isocitrate dehydrogenase (IDH), alpha-ketoglutarate dehydrogenase (OGDH), succinate-CoA-ligase (SUCL), succinate dehydrogenase (SDH), fumarase (FH), malate dehydrogenase (MDH), 3-alpha-hydroxybutyrate dehydrogenase (BDH), acetyl-CoA acetyltransferase (ACAT), medium-chain acyl-CoA dehydrogenase (MCAD), short-chain acyl-CoA dehydrogenase (SCAD), very long-chain acyl-CoA dehydrogenase (VLCAD), fatty-acid translocase (CD36), fatty-acid transport protein (FATP), fatty-acid binding protein (FABP), diacylglycerol O-acyltransferase (DGAT), carnitine palmitoyltransferase (CPT), carnitine translocase (CT), adenine nucleotide translocator (ANT), complex I (I), complex II (II), complex III (III), complex IV (IV), complex V (V), medium-chain fatty acid (MCFA), long-chain fatty acid (LCFA).
Table 1.
Cardiac and energetic phenotypes of transgenic and knockout mouse models.
| Transcription Factor or coactivator |
Gain-of-function cardiac phenotype |
Loss-of-function cardiac phenotype |
References |
|---|---|---|---|
| PPARα | ⇑FAO, lipotoxicity, HF- induced CM, ⇓-tolerance to I/R |
Mild. Resistance to I/R, ⇓work capacity |
83-87, 192 |
| PPARβ/δ | ⇑glucose utilization, resistance to I/R, ⇑glycogen content |
⇓FAO, lipotoxic CM, early lethality |
81, 89, 90 |
| PPARγ | Lipotoxic CM | Cardiac hypertrophy | 91, 92 |
| ERRα | ND | ⇓respiratory efficiency, ⇓PCr, TAC-induced CM |
154 |
| ERRγ | ND | Perinatal lethality | 68, 69 |
| TFAM | Protects from MI induced heart failure |
Spontaneous CM | 77-79 |
| PGC-1α | ⇑mitochondrial proliferation, reversible CM |
Energy deficiency (⇓ATP, ⇓PCr), TAC-induced CM, ⇓work capacity |
16, 49, 52, 153, 157 |
| PGC-1β | ND | Mild energy deficiency, Perinatal lethality and mitochondrial dysfunction when combined with PGC- 1α KO |
163-165 |
Mitochondrial biogenesis
Constitutive overexpression of PGC-1α in the heart of intact mice, under control of the cardiac-specific α-MHC promoter, leads to profound mitochondrial biogenesis, to the point of significant replacement of myofibrillar apparatus with mitochondrial matrix.16 This was the first unequivocal demonstration that PGC-1α can activate cardiac mitochondrial biogenesis in vivo. Heart failure developed early in these animals, perhaps as a consequence of myofibrillar displacement. Tetracycline-inducible PGC-1α overexpression, again under control of the α-MHC promoter, was subsequently generated in order to test the role of PGC-1α specifically in adult cardiac muscle.52 Induction of PGC-1α shortly after birth recapitulated the findings with constitutive expression: profound mitochondrial proliferation, and ensuing heart failure. Remarkably, these dramatic changes were completely reversed by subsequent re-repression of the PGC-1α transgene. Hence PGC-1α, in this context, was needed for both induction and maintenance of increased mitochondrial mass.
Interestingly, induction of PGC-1α in adult mouse hearts did not activate mitochondrial proliferation, indicating that the peri-natal period is uniquely permissive for PGC-1-induced mitochondrial proliferation. Why this is remains unclear. Despite the absence of increasing mitochondrial mass, cardiac dysfunction still occurred in the adult mice, revealing the existence of other cardiotoxic consequences of supra-physiologic doses of PGC-1α. On the other hand, mild cardiac over-expression of PGC-1α, seen in transgenic mice that express PGC-1α under control of the muscle creatine kinase (MCK) promoter, or in BAC PGC-1α transgenic mice, do not develop heart failure (53 and unpublished results), indicating that tempered PGC-1α overexpression in the heart is not toxic. It will be of interest to determine if this mild overexpression of PGC-1α in the heart can be beneficial under conditions of cardiac stress.
Transcriptional regulation of mitochondrial biogenesis and nuclear genes encoding respiratory chain subunits has been extensively studied and reviewed elsewhere.54-56 The nuclear respiratory factor (NRF) and estrogen related receptor (ERR) families of transcriptional factors are key players in the induction of these genes, and PGC-1α modulates mitochondrial biogenesis by directly coactivating these transcription factors.51, 57, 58 Binding sites for the NRF-1 monomer and NRF-2 heterotetramer (also known as GABP) are found in the promoters of most respiratory chain genes.54 The effect of overexpressing either NRF-1 or NRF-2 in cardiac tissue has not been evaluated to date, but targeted overexpression of NRF-1 in skeletal muscle did increase genes of oxidative phosphorylation (OXPHOS).59 PGC-1α physically interacts with both NRF-1 and -2, and stimulates their activity on mitochondrial genes.57
The ERRs are orphan nuclear receptors, named for their sequence similarity to the estrogen receptor (though they are not thought to bind to estrogen).60, 61 Structural studies suggest that the mechanism of transcriptional activation by ERRs may differ from that of other nuclear receptors, and that ERRs may have no natural ligands.62 All three ERRs (α, β and γ) are highly expressed in oxidative tissues, such as heart, muscle and kidney. Most of genes induced by PGC-1α also contain ERR-binding sites.58 ERRα has been studied most extensively. Overexpression of ERRα in rat neonatal cardiomyocytes results in strong induction of genes involved in glucose utilization (e.g. PDK4, HK2, GLUT4), fatty-acid oxidation (MCAD, CD36) and the OXPHOS program (ATP5b, CYCS)63. PGC-1α binds to and coactivates ERRα in cardiomyocytes, as well as other cells.58, 64, 65 The mRNA expression of ERRα is also induced by PGC1α, and ERRα also induces PPARα (see below),63 underscoring the multiple positive feedback loops that drive metabolic transcription pathways in the heart. Germline disruption of ERRα results in significant declines in the expression of genes involved in OXPHOS, and fatty-acid and glucose metabolism.
In contrast to ERRα, less is known of the role of the other ERRs in cardiac tissue. Genome wide Chip-chip studies provided evidence that ERRγ and ERRα share many targets in cardiac tissue,66 and targeted overexpression of ERRγ in skeletal muscle induces genes of fatty acid oxidation and OXPHOS (though interestingly without leading to frank increases in mitochondrial density).67 Germline disruption of ERRγ resulted in mice which die postnatally, attributed in part to a cardiac defect.68, 69 Thus ERRγ may have some redundancy with ERRα in vivo, but also plays a separate and vital role for survival. Germline disruption of ERRβ is embryonic lethal,70 attributable to placental defects. The contribution of ERRβ to cardiac physiology is thus currently unknown. To date no gain of function mouse models have been developed to address the effects of overexpressing ERRs within cardiac tissue.
PGC-1α also indirectly modulates the mitochondrial genome, in addition to its direct regulation of nuclear genes. Although >98% of the >1500 genes that encode for mitochondrial proteins are encoded by the nucleus, mitochondria do contain their own genome, encoding for 22 tRNAs, 2rRNAs, and 13 proteins that contribute to complexes I, and III-V of the electron transport chain (but not II). Mitochondrial biogenesis thus requires replication of mtDNA and the coordinated transcription of the mitochondrial genome. Cross-talk between the nuclear and mitochondrial genomes is carried out, at least in part, via the nuclear-encoded proteins TFAM, TFB1 and TFB2.71, 72 The genes for all three of these proteins are induced by PGC-1α, via the induction and co-activation of NRF-1 and NRF-2.73, 74 TFAM, a nuclear encoded high mobility group transcription factor, is responsible for both the replication and transcription of mitochondrial DNA.75, 76 Targeted disruption of TFAM specifically within cardiac tissue resulted in a significant decrease in electron transport capacity, spontaneous cardiomyopathy, and heart failure.77, 78 Conversely, increasing the expression of TFAM within cardiac tissue offered protection from heart failure induced by myocardial infarction.79 These observations underscore the importance of maintaining mitochondrial integrity and copy number as a protection against heart disease.
Fatty acid import and utilization
Mitochondrial function also requires various ancillary, non-mitochondrial programs, such as the transport and initial breakdown of nutrients. Normal adult cardiac muscle relies primarily on fatty acids as an energy source. Interestingly, most fatty acids are shuttled through the intracellular triglyceride pool before being transported to mitochondria for oxidation.50 Fatty acid import, shuttling, and oxidation must thus be coordinated, under both basal and stimulated conditions. Overexpression of PGC-1α in cardiomyocytes in cell culture and in vivo markedly increases fatty acid oxidation.16 The PPAR family of transcription factors plays a prominent role in this process. There are three isoforms of PPARs (α, β/δ and γ), all of which form heterodimers with retinoic acid-activated receptors (RXRs) to bind DNA. Fatty-acids and their catabolic byproducts act as low/medium-affinity ligands for PPARs, thus likely transmitting information about the intracellular lipid milieu to gene regulation. Natural, high-affinity, physiological ligands for the PPARs have yet to be identified, and might not exist. On the other hand, high-affinity pharmacological agents that can modulate the activity of the PPARs exist, including fibrates for PPARα and thiazolidinediones for PPARγ.80
PPARα has been studied more extensively in cardiac tissue than the other PPARs. PPARα expression is high in tissues that consume fatty acids (heart, BAT, kidney, liver). PGC1α binds to, and coactivates PPARα in cardiac cells, thereby inducing numerous genes critical for fatty acid handling, including CD36 (import into cell), CPT1b (import into mitochondria), PDK4 (reciprocal inhibition of pyruvate entry into mitochondria), and MCAD (rate-limiting step in medium chain fatty acid β oxidation).81, 82 Hearts of PPARα null mice exhibit a decrease in fatty-acid oxidation and concomitant increase in glucose oxidation, coincident with decreased expression of fatty-acid oxidation genes.83, 84 The animals showed improved tolerance to ischemia/reperfusion injury ex vivo, likely in part due to decreased fatty acid oxidation/import.85 Conversely, overexpression of PPARα within cardiac tissue, using the myosin heavy chain (MHC) promoter, resulted in induced fatty-acid oxidation and repressed glycolysis.86, 87 High expression of PPARα in the heart led to spontaneous left-ventricular dysfunction and lipotoxicity,86 suggesting that increases in fatty oxidation were insufficient to match the increase in fatty acid uptake. Consistent with this notion, feeding a chow enriched in long-chain fatty acids to lower expressing transgenic animals also precipitated cardiac dysfunction.86, 87 Crossing PPARα overexpressing mice with mice lacking cardiac CD36 or LPL, both of which are critical for import of fatty acids into cardiomyocytes, rescued the cardiac toxicity of PPARα overexpression.82, 88 Together, these data support the notion that PPARα overexpression leads to intracellular lipid accumulation that is toxic to cardiac function.
PPARβ/δ has also been relatively well studied in the heart. Overexpression of PPARβ/δ in neonatal cardiomyocytes, or transgenic overexpression in the heart, induced genes involved in fatty-acid oxidation, and this was further enhanced with the addition of PPARβ/δ ligands.81, 89 In contrast to PPARα overexpressors, however, the MHC-PPARβ/δ hearts exhibited increased glucose utilization, did not develop cardiomyopathy even on high fat diet, and were resistant to myocardial infarct. The difference between these two transgenics may reflect the lack of induction of fatty acid import genes (e.g. FATP, CD36, ACS, FAS) in the PPARβ/δ overexpressors,81 thus averting toxic accumulation of intracellular lipids. Conversely, cardiac-specific disruption of PPARβ/δ led to a decrease in genes involved in fatty-acid oxidation,90 but also increased cardiac lipid accumulation and lipotoxic cardiomyopathy. Again, this contrast with the PPARα KO mice is probably the result of unchanged fatty-acid import in the PPARβ/δ mice, in combination with decreased fatty acid oxidation.
The 3rd PPAR, PPARγ, is expressed at substantially lower levels in the heart than in other tissues like fat and liver, and its role in the heart remains somewhat controversial. Clinical modulation of PPARγ activity with thiazolidinediones can reduce fatty acid oxidation in the heart, but this likely occurs in large part indirectly by sequestering fatty acids in fat depots and reducing the exposure of the heart to circulating fatty acid levels. Nevertheless, cardiomyocyte-specific deletion of PPARγ does result in cardiac hypertrophy,91 indicating an important role for PPARγ in these cells. Overexpression of PPARγ under the control of the αMHC promoter resulted in increased lipid uptake and oxidation in the heart, increased expression of glucose metabolism genes, and systolic dysfunction.92 PGC-1α is a potent activator of PPARγ activity in brown fat biology,4 and this is likely true in cardiomyocytes as well.
Angiogenesis
The vasculature transports oxygen and nutrients and thus plays a critical role in mitochondrial metabolism. The heart is highly vascular, consistent with its high oxidative and metabolic needs. Recent findings in skeletal muscle have implicated PGC-1α in the regulation of blood vessel formation. PGC-1α expression is induced by ischemia-like conditions in cell culture, and in turn PGC-1α activates a broad program of angiogenic factors, including VEGF.93 The induction of VEGF occurs independently of the canonical Hypoxia-Inducible Factor (HIF) pathway, and instead is mediated by coactivation of ERRα on a novel enhancer in the first intron of the VEGF gene. Transgenic overexpression of PGC-1α in skeletal muscle induces robust angiogenesis, and accelerates recovery of blood flow after surgically-induced hindlimb ischemia, demonstrating the functional capacity of newly formed vessels.93 Endurance exercise induces both mitochondrial proliferation and new blood vessel formation in skeletal muscle, and is one of the few examples of physiological angiogenesis in adult tissues.94 PGC-1α is strongly induced in skeletal muscle by exercise in rodents and humans,22, 95-99 likely in part by adrenergic activation and induction of the alternative promoter of PGC-1α discussed above.22, 23 Mice lacking PGC-1α specifically in skeletal muscle fail to expand their vascular network in response to exercise, demonstrating that PGC-1α mediates exercise-induced angiogenesis.22 PGC-1α thus coordinates the delivery of oxygen and nutrients to myocytes (angiogenesis) with their transport across the cell and their consumption in mitochondria. It will now be of great interest to determine if PGC-1α plays a similar role in the heart. Microvascular rarefaction has recently been implicated in the development of heart failure.100-102 Decreased activity of PGC-1α may thus be contributing to this decrease in vessel density and subsequent heart failure.
NRFs, ERRs, and PPARs thus form together a coordinated transcriptional network orchestrated by PGC1α to regulate energy homeostasis within cardiac tissue. Each transcription factor controls a subset of genes, encompassing specific metabolic pathways. PGC-1α orchestrates these various metabolic pathways by directly regulating each of the transcription factors. A large number of other transcription factors can also be bound by PGC-1 coactivators, including MEF2 and FOXO1 family members, and most nuclear receptors.5, 6 These factors have mostly not been studied in the context of cardiac metabolism. As outlined in the previous section, PGC-1α is also exquisitely sensitive to various physiologic cues, both at the level of mRNA expression and post-translational modification. PGC-1α thus likely acts as an integrator of metabolic and neurohormonal information, and uses that information to appropriately modulate metabolic pathways in cardiomyocytes (Figure 2). It will be of great interest to investigate if, and how, upstream signals can confer specificity on PGC-1α and differently influence its pleitropic downstream pathways.
PGC-1β has been investigated less extensively, but likely shares many of the properties of PGC-1α. NRF's, PPAR's, and ERR's can all be targeted by PGC-1β. Certain post-translational modifications seen on PGC-1α, including acetylation and deacetylation, are also present on PGC-1β.45 Pathways downstream of PGC-1β appear to be quite similar to those of PGC-1α,103 though important subtleties can be discerned. For example, overexpression of PGC-1β in skeletal myotubes leads to increased respiration, as does PGC-1α, but less proton leak is seen than with PGC-1α.104 How this occurs remains unclear. As noted above, PGC-1β gene expression is generally less easily modulated than that of PGC-1α. PRC has been much less investigated (in fact not at all in cardiomyocytes), but PRC can coactivate NRF's,10 and knockdown of PRC by siRNA in human osteosarcoma cells significantly inhibits mitochondrial genes and activity,105 suggesting that the same might be true in cardiac cells or tissue.
PGC-1s and cardiac energetic failure
Human genetic diseases poignantly demonstrate the impact of energetic deficiencies on cardiac function. More than 50% of disease-causing mutations in mitochondrial DNA lead to cardiomyopathy in humans,68, 106-114 many in the context of complex syndromes like Kearns Sayre and Leigh Syndromes. Mutations in nuclear-encoded genes of the respiratory chain also lead to cardiac dysfunction 115-119. In addition, genetic defects in mitochondrial fuel handling, most notably in fatty acid transport and β-oxidation, can also lead to profound cardiomyopathy (CM).116, 120-124 The use of genetic manipulations in mice has extensively supported these clinical observations. Targeted mutations affecting processes including fatty acid transport and oxidation, high energy phosphate transport and shuttling, protection from mitochondrial reactive oxygen species (ROS), and mitochondrial DNA (mtDNA) proofreading activity, all lead to often profound cardiac dysfunction.76, 78, 123, 125-130
These observations leave little doubt, therefore, that defects in mitochondrial function can lead to heart disease. Most cases of heart failure, however, are not caused by rare genetic mutations. Mounting evidence points to the “generically” failing heart, of diverse etiologies, as also energy-starved.109-111, 131 ATP concentrations are decreased by up to 25% in failing hearts.110, 132, 133 Normally, cardiomyocytes tightly maintain their ATP concentration, and a 25% decrease in ATP marks advanced disease, much like elevated glucose levels do in diabetes. This “snapshot” value also likely belies much more profound decreases in fluxes of ATP generation, as well as increases in ADP (and thus loss of the phosphorylation potential, the driving force for ATPases). Phosphocreatine (PCr)-to-ATP ratios, a measure of high energy phosphate buffering capacity and an indirect measure of ADP, is decreased as much 60% in heart failure,110 and ATP flux through the creatine kinase reaction has also been shown to be reduced.134 In fact, the PCr/ATP ratio is a better predictor of cardiovascular mortality than is ejection fraction.135 The efficiency of ATP production, as defined by ATP flux/oxygen consumption, also declines in heart failure.136
How does the failing heart become energy starved? Acquired defects in mitochondrial respiration likely play a large part. Mitochondrial dysfunction is seen in human cardiomyopathy 134, 137-141 and in most animal models of heart failure.77, 142-145 Mutations in mtDNA have been demonstrated in humans after treatment with cardiotoxic therapies like doxorubicin or nucleoside reverse transcriptase inhibitors (NRTIs),112, 146 and in rodents after myocardial infarction.142 Replication of the mitochondrial genome is blunted in human heart failure.147 In addition, declines in the expression of numerous nuclear genes encoding for mitochondrial proteins are seen in both human heart failure and in rodent models.148-151 There appears, therefore, to be a coordinated down-regulation of mitochondrial pathways during heart failure. How this occurs, however, is not clear. The notion that dysregulation of PGC-1 coactivators may play a role was first suggested by the observation in rodents that the expression of PGC-1α is reduced in pressure overload-induced cardiac hypertrophy.149 Since that initial observation, repression of PGC-1α has been seen in a number of rodent models of cardiac hypertrophy or failure152-156, suggesting that decreased PGC-1α is a common signature of acquired cardiac disease.
The consequences of decreased PGC-1 activity in the heart have been investigated extensively, using genetically modified mouse models. Two groups generated PGC-1α knockout mice, with slightly different results. Spiegelman's group reported pronounced brain defects in PGC-1α −/− mice, with a strong sensitivity to neurotoxins, possibly due to decreased protection against reactive oxygen species.49, 157 At baseline, these animals did not exhibit an overt cardiac phenotype, and the hearts had normal mitochondrial content.158 However, gene expression analyses did reveal reactivation of “embryonic” markers, including ANP, BNP, and β-MHC, suggesting the presence of cardiac dysfunction. Nuclear magnetic resonance studies revealed pronounced decreases in ATP concentrations, suggesting significant decreases in energy reserves. Consistent with this, PGC-1α −/− hearts were unable to increase work output in response to inotropic stimulation.158 With aging, the animals spontaneously developed mild cardiac dysfunction. More strikingly, hemodynamic challenge in the form of transverse aortic banding (TAB) led to pronounced cardiac failure in PGC-1α −/− mice.153 Cardiac failure only occurred after 2 months, indicating that PGC-1α is dispensable for the acute response to aortic banding, but it is critical for maintaining the subsequent compensated state. Interestingly, ERRα −/− mice revealed a very similar phenotype after TAB: impaired energetics, including decreased levels of phosphocreatine and ATP, and the progressive development of heart failure154, underscoring the central role that ERRα plays in PGC-1α biology.
The PGC-1α −/− mice generated by Kelly's group did not display the same extensive neurological defects described above.159 This difference may stem from the retention in these mice of alternatively splice forms of PGC-1α. For example, a short form of PGC-1α that is highly expressed in the brain was recently described.160, 161 Nevertheless, cardiac inotropic and chronotropic responses to exercise were both blunted in these PGC-1α −/− mice, and cardiac hemodynamics measured in isolated working hearts demonstrated mild but significantly decreased cardiac work and output. A subsequent meticulous examination of cardiac fuel preference and ATP-producing capacity in these PGC-1α −/− hearts revealed significant defects.162
Permeabilized myocardial fibers isolated from PGC-1α −/− hearts had reduced fatty acid oxidation, decreased ATP synthesis rates, and blunted efficiency (i.e. ATP produced per oxygen consumed). Isolated working hearts from PGC-1α −/− mice had decreased fatty acid oxidation and diminished cardiac power. Structural studies showed abnormal mitochondrial cristae density and cytoplasmic accumulation of neutral lipids, suggesting that the reduction in fatty acid consumption was not matched by a reduction in fatty acid import.
Both PGC-1α −/− models thus show significantly impaired bioenergetics and moderate defects in cardiac function, which are markedly worsened by hemodynamic and metabolic challenges. PGC-1α is thus felt to be critical for myocardial metabolic flexibility. Interpretations from both of these mouse models must be tempered by the fact that PGC-1α was absent in the entire body, and that PGC-1α clearly has pleitropic roles in numerous extra-cardiac tissues. Cardiac-specific deletion of PGC-1α has not yet been reported.
The cardiac phenotype of PGC-1β −/− mice appears to be significantly less pronounced, though it has been studied less extensively. Three groups have generated mice either lacking or bearing deletions in PGC-1β.163-165 The Kelly group reported mild reductions in cardiac mitochondrial density in PGC-1β −/− mice, and a mild chronotropic defect in response to dobutamine stimulation in vivo. Cardiac contractility appeared normal.163 The other two groups reported no cardiac abnormalities,164, 165 and the response of PGC-1β −/− mice to hemodynamic stresses like TAC have not been reported.
Despite the mild phenotype of PGC-1β −/− mice, the importance of PGC-1β in cardiac energetics was made clear by the simultaneous deletion of both PGC-1α and β. PGC-1α and PGC-1β both powerfully induce mitochondrial biogenesis,104 and regulate sets of genes that largely coincide. The coactivators thus provide extensive redundancy. To test this notion directly, Kelly's group generated PGC-1α/β −/− double knockout mice.103 Almost all double KO mice died in the immediate post-natal period, with evidence of significant cardiac dysfunction. Mice bearing total-body deletion of PGC-1α but only cardiac-specific deletion of PGC-1β also died perinatally with the same cardiac phenotype, elegantly demonstrating that the cause of death in these animals was cardiac dysfunction. Heart from double KO animals had markedly diminished mitochondrial mass, and density.103
Interestingly, hearts from PGC-1α/β double KO animals had apparently normal mitochondrial mass until late in gestation. PGC-1α and β are thus both largely dispensable for mitochondrial biogenesis during prenatal development. Marked proliferation of cardiac mitochondria normally occurs immediately after birth, both in rodents and larger animals, coinciding with increasing workload and a switch to fatty acid consumption at birth. At the same time, there is a marked induction of PGC-1α expression.2, 16 PGC-1α and β likely mediate this event together, since no increase in cardiac mitochondrial proliferation is seen in the double KO mice. Post-natal mitochondrial proliferation is thought to be mediated in large part by thyroid hormone signaling,166, 167 and PGC-1α and β can coactivate the thyroid hormone receptor,168 suggesting a likely, but untested, mechanism.
In addition to changes in mitochondrial respiratory function, cardiomyopathy and heart failure are also typically marked by important shifts in fuel use. The heart is a fuel omnivore, capable of consuming glucose, fatty acids of various lengths, lactate, pyruvate, ketones, and amino acids (Figure 3). During fetal development, when oxygen tension and fatty acid levels are lower, the heart primarily consumes glucose and lactate. Shortly after birth, coincident with sharp increases in cardiac work and with lactation, numerous genes of fatty acid transport and oxidation are induced, and cardiac substrate use largely switches to fatty acids.1, 3 The adult heart generates 60% of its ATP from oxidation of fatty acids, primarily long-chain. It is likely that the PGC-1s mediate in large part this perinatal switch in substrate use, given the strong coactivation of PPARs by PGC-1s on genes of fatty oxidation and the pronounced induction of PGC-1alpha expression at birth, although this remains to be formally proven. Conversely, cardiac hypertrophy and heart failure, of almost any cause, are marked by an incomplete “switch” back to glucose consumption and away from fatty acid oxidation.141, 169 An increasing level of evidence points to this switch as an important adaptive response.141 Again, declines in PGC-1 activity during cardiac remodeling may contribute to this substrate switch. Interestingly, some cardiac pathologies such as that associated with diabetes have an early increase in fatty acid use, prior to the development of frank systolic dysfunction. Whether a transient increase in PGC-1 activity contributes to this observation is not known.
Taken together, these studies demonstrate the key role that PGC-1α and β play in regulating mitochondrial mass, oxygen consumption, respiratory efficiency, and fatty acid oxidation in the normal heart. Overall, the coactivators appear dispensable for developmental processes, at least until shortly before birth; instead, they appear critical for environmental and physiological modulation of cardiac energetic balance. PGC-1α likely, for example, mediates exercise-induced mitochondrial proliferation in the heart (though this has not yet been formally tested). Conversely, a substantial body of evidence suggests that PGC-1α down-regulation during cardiac failure may play an important maladaptive role.
PGC-1α in the vasculature
In addition to its role in myocytes and their communication with the vasculature, PGC-1α appears to also have important functions in the vascular wall itself. Endothelial cells mediate local tissue homeostasis by regulating blood flow, coagulation, metabolic exchange between the circulation and tissue, and trafficking of inflammatory cells. Endothelium dysfunction is an early feature of chronic cardiovascular diseases,170 and is usually associated with excess levels of ROS.171 Anti-oxidant pathways are thus critical for protection from endothelial dysfunction. A number of proteins can decrease ROS, either by limiting ROS production, such as the mitochondrial uncoupling proteins (UCP2 and 3) and the adenine nucleotide translocator (ANT), or by directly scavenging ROS, such as manganese superoxide dismutase (MnSOD), catalase, peroxiredoxin 3 (Prx3), Prx5, and thioredoxin 2 (Trx2). PGC-1α directly regulates this anti-ROS program, thus offseting the increase in ROS production that would otherwise occur with mitochondrial biogenesis.49 Overexpression of PGC-1α in endothelial cells induces the expression of MnSOD, catalase, Prx3, Prx5, Trx2, and ANT,172, 173 likely in part via coactivation of the O subfamily of forkhead transcription factor 3a (Foxo3a) transcription factor.174 Overexpression of PGC-1α in endothelial cells reduces levels of ROS, and rescues ROS-mediated mitochondrial toxicity and cellular apoptosis.172, 173, 175 Activation of AMPK in endothelial cells also prevents oxidative cell injury through a PGC-1α-mediated mechanism.175 In intact animals, chronic angiotensin II administration induces endothelial dysfunction, as measured by reduced endothelium-dependent relaxation to acetylcholine, and this was reversible by activation of AMPK in wild-type but not in PGC-1α−/− mice.175 The reduction of ROS by PGC-1α in endothelial cells is also associated with reduced expression of chemokines and adhesion molecules including vascular cell adhesion molecule-1 (VCAM-1) and monocyte chemoattractant protein-1 (MCP-1), as well as reduced activity of the redox-sensitive transcription factor NF-kappa B activity, suggesting a role of PGC-1α in endothelium-related inflammation.176 Finally, shear stress, a potent beneficial stimulus in endothelial cells, appears to induce PGC-1α, possibly through SIRT1.177 Together, these data strongly suggest that PGC-1α plays an important role in endothelial redox homeostasis.
The vascular wall also contains vascular smooth muscle cells (VSMCs) and pericytes, which surround endothelial cells and contribute significantly to the function of the vasculature. PGC-1α is expressed in VSMCs, and can be upregulated by AMPK activators.176 Conversely, treatment of VSMCs with angiotensin II activates Akt, which phosphorylates PGC-1alpha on serine 570 and inhibits it36, leading to repression of PGC-1alpha-dependent genes like catalase. Overexpression of PGC-1alpha in these cells inhibits angiotensin II-induced increases in ROS and [3H]leucine incorporation, markers of vascular hypertrophy.178 Overexpression of PGC-1α in VSMCs also reduces TNF-induced ROS generation, VCAM-1 and MCP-1 level, and NF-kappa B activity.179 Migration and proliferation of VSMCs are also key components of intimal hyperplasia following endovascular injury, such as after coronary stenting. Overexpression of PGC-1α significantly inhibits migration of VSMCs, while knockdown of PGC-1α enhances migration. In a carotid balloon injury rat model, PGC-1α overexpression inhibited neointimal formation, likely through upregulation of SOD2.179
There is thus a burgeoning understanding of the role of PGC-1α in endothelial and perivascular cells, with a strong indication that PGC-1α regulates redox homeostasis in these cells. No data are available on PGC-1β or PRC, and no vascular-specific mouse models of PGC-1 coactivators have yet been reported. It will be of great interest to know the effects of endothelial PGC-1 coactivators on angiogenesis and atherosclerosis, two critical endothelial processes of high relevance to cardiac disease. In this context, it is also important to note that PGC-1β and PPARs appear to play important roles in macrophage activation, a key step in the development of atherosclerosis. This is discussed in detail by Dr. Chawla in his accompanying contribution to this series.180
Conclusions, and human relevance
It has become abundantly clear that the PGC-1 coactivators are key regulators of metabolism in the heart, both in cardiomyocytes and likely in other cardiac cell types. In the context of highly metabolically active cells like cardiomyocytes, PGC-1α coordinates broad genetic programs spanning the entire metabolism of preferred substrates: blood vessel recruitment for transport of oxygen and nutrients, transport of fatty acids into the cell and the mitochondrion, oxidation of fatty acids via β-oxidation and the TCA cycle, generation of ATP via the respiratory chain complex, transport of ATP back to the cytoplasm, and protection against ROS generated by the respiratory chain. PGC-1α appears to be more responsive to extracellular and metabolic cues than are PGC-1β and PRC, and it has been studied more extensively. Nevertheless, important redundancies exist between PGC-1α and β. Little is known of the role PRC in the heart.
Does abnormal activity of PGC-1α and/or β contribute to cardiac remodeling and heart failure? This was first suggested by the observation in rodents that the expression of PGC-1α and known targets of PGC-1α are reduced in a number of rodent models of cardiac dysfunction, as discussed above. The situation in humans is less clear. Some have reported decreased expression of PGC-1α in failing human hearts,181, 182 while others have not.147, 151 Interpretations of these findings are invariably difficult, because patients differ widely in treatment and other critical clinical variables. It may also be that PGC-1α expression is not altered in human heart failure, but rather that its intrinsic activity is decreased. As noted above, PGC-1α is heavily modified post-translationally. Evaluating this possibility has been difficult, however, in part due to poor antibodies to PGC-1s, and to the short half-life of PGC-1α protein.33 What is clear is that numerous mitochondrial genes and other known targets of PGC-1 like glycolytic and FAO genes are repressed in human heart failure,147, 151 strongly suggesting that PGC-1α may be playing a role.
It is also possible that, in certain circumstances, PGC-1α is at first repressed but then induced in terminal heart failure as a compensatory response to accelerated energetic collapse. Indeed, the expression of PGC-1α can be induced by ischemia93, 180 and ATP wasting.28, 183 Mitochondrial proliferation has been observed in certain models of cardiac dysfunction, most notably diabetic models,184-187 and in human mitochondrial myopathy.181 Mitochondrial proliferation may be mediated by PGC-1α in some cases,184 but perhaps not all.188, 189 Conclusions drawn thus differ depending on the etiology of heart disease studied, and what time point in the process of cardiac adaptation and maladaptation is investigated. In short, it remains unclear to what extent PGC-1α activity is altered in human heart failure, and, although the hypothesis is highly plausible, the jury is still out on whether alterations in PGC-1 activity play a causal role in heart failure.
Are the PGC-1 proteins possible therapeutic targets? Whether the PGC-1s contribute to heart failure or not, they represent appealing nodal points for modulation of the cardiac energy state. Transcription factors other than nuclear receptors are historically regarded are “nondruggable”, although this dogma is increasingly being contested. Small molecules that specifically modulate PGC-1α/ERRα interaction have been developed, for example.190 Pharmaceuticals that target pathways upstream of the PGC-1s may also provide interesting opportunities, without having to target PGC-1 proteins directly.191 Gene therapy may afford another approach, though targeting vectors for the heart remain a challenge. It is also important to appreciate that a blanket activation of the pleitropic effects of PGC-1α on metabolism may not be ideal. Excessive activation of PGC-1α leads to cardiac dysfunction,52 suggesting that remaining within a therapeutic window will be important. Alternatively, activation of specific PGC-1 pathways might avert toxic side effects. This could be done either by pharmaceutical targeting of specific PGC-1/transcription factor interactions, or by introduction of genetically modified PGC-1 molecules. Clearly much remains to be learned. The PGC-1 coactivators are undisputedly key regulators of cardiac bioenergetic, and understanding the mechanisms by which they do so will undoubtedly continue to yield new and important insights.
Figure 4. Coregulation of mitogenesis and vasculogenesis.
Vessels provide a constant supply of glucose, oxygen and fatty acids which are metabolized to produce ATP as an energy source. Net 2 ATP via anaerobic glycolysis and lactate production per glucose molecule or 30 ATP per molecule of glucose via oxidative consumption within the mitochondria. Catabolism of long chain fatty-acids such as palmitate within the mitochondria can yield 104 ATP per molecule.
Acknowledgments
Sources of Funding: GCR has received support from NIH (5T32HL007374-31) and UNCF-Merck Science Initiative (post-doctoral fellowship), and ZA is supported by the NHLBI, the Smith Family Foundation, the Ellison Foundation, and the Harvard Stem Cell Institute.
Non-standard Abbreviations and Acronyms
- PPAR
Peroxisome proliferator-activated receptor
- PGC-1
PPAR-gamma coactivator
- PRC
PGC-1 related coactivator
- MAPK
Mitogen-activated protein kinase
- AMPK
adenosine monophosphate-activated protein kinase
- SIRT
silent mating type information regulation 2 homolog
- NAD
Nicotinamide adenine dinucleotide
- PDK
Pyruvate dehydrogenase kinase
- MHC
Myosin heavy chain
- MCK
Muscle creatine kinase
- NRF
Nuclear respiratory factor
- ERR
Estrogen related receptor
- GABP
GA-binding protein
- OXPHOS
oxidative phosphorylation
- mtDNA
mitochondrial DNA
- TFAM
Transcription factor A, mitochondrial
- TFBM
transcription factor B, mitochondrial
- KO
Knockout
- HIF
Hypoxia-inducible factor
- ROS
Reactive oxygen species
- PCr
Phosphocreatine
- VSMC
Vascular smooth muscle cell
Footnotes
Disclosures: none
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lopaschuk GD, Spafford MA. Energy substrate utilization by isolated working hearts from newborn rabbits. Am J Physiol. 1990;258:H1274–1280. doi: 10.1152/ajpheart.1990.258.5.H1274. [DOI] [PubMed] [Google Scholar]
- 2.Buroker NE, Ning XH, Portman M. Cardiac pparalpha protein expression is constant as alternate nuclear receptors and pgc-1 coordinately increase during the postnatal metabolic transition. PPAR Res. 2008;2008:279531. doi: 10.1155/2008/279531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lopaschuk GD, Collins-Nakai RL, Itoi T. Developmental changes in energy substrate use by the heart. Cardiovasc Res. 1992;26:1172–1180. doi: 10.1093/cvr/26.12.1172. [DOI] [PubMed] [Google Scholar]
- 4.Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 5.Lin J, Handschin C, Spiegelman BM. Metabolic control through the pgc-1 family of transcription coactivators. Cell Metab. 2005;1:361–370. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 6.Finck BN, Kelly DP. Pgc-1 coactivators: Inducible regulators of energy metabolism in health and disease. J Clin Invest. 2006;116:615–622. doi: 10.1172/JCI27794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wallberg AE, Yamamura S, Malik S, Spiegelman BM, Roeder RG. Coordination of p300-mediated chromatin remodeling and trap/mediator function through coactivator pgc-1alpha. Mol Cell. 2003;12:1137–1149. doi: 10.1016/s1097-2765(03)00391-5. [DOI] [PubMed] [Google Scholar]
- 8.Puigserver P, Adelmant G, Wu Z, Fan M, Xu J, O'Malley B, Spiegelman BM. Activation of ppargamma coactivator-1 through transcription factor docking. Science. 1999;286:1368–1371. doi: 10.1126/science.286.5443.1368. [DOI] [PubMed] [Google Scholar]
- 9.Monsalve M, Wu Z, Adelmant G, Puigserver P, Fan M, Spiegelman BM. Direct coupling of transcription and mrna processing through the thermogenic coactivator pgc-1. Mol Cell. 2000;6:307–316. doi: 10.1016/s1097-2765(00)00031-9. [DOI] [PubMed] [Google Scholar]
- 10.Andersson U, Scarpulla RC. Pgc-1-related coactivator, a novel, serum-inducible coactivator of nuclear respiratory factor 1-dependent transcription in mammalian cells. Mol Cell Biol. 2001;21:3738–3749. doi: 10.1128/MCB.21.11.3738-3749.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin J, Puigserver P, Donovan J, Tarr P, Spiegelman BM. Peroxisome proliferator-activated receptor gamma coactivator 1beta (pgc-1beta), a novel pgc-1-related transcription coactivator associated with host cell factor. J Biol Chem. 2002;277:1645–1648. doi: 10.1074/jbc.C100631200. [DOI] [PubMed] [Google Scholar]
- 12.Lin J, Tarr PT, Yang R, Rhee J, Puigserver P, Newgard CB, Spiegelman BM. Pgc-1beta in the regulation of hepatic glucose and energy metabolism. J Biol Chem. 2003;278:30843–30848. doi: 10.1074/jbc.M303643200. [DOI] [PubMed] [Google Scholar]
- 13.Kressler D, Schreiber SN, Knutti D, Kralli A. The pgc-1-related protein perc is a selective coactivator of estrogen receptor alpha. J Biol Chem. 2002;277:13918–13925. doi: 10.1074/jbc.M201134200. [DOI] [PubMed] [Google Scholar]
- 14.Yoon JC, Puigserver P, Chen G, Donovan J, Wu Z, Rhee J, Adelmant G, Stafford J, Kahn CR, Granner DK, Newgard CB, Spiegelman BM. Control of hepatic gluconeogenesis through the transcriptional coactivator pgc-1. Nature. 2001;413:131–138. doi: 10.1038/35093050. [DOI] [PubMed] [Google Scholar]
- 15.Herzig S, Long F, Jhala US, Hedrick S, Quinn R, Bauer A, Rudolph D, Schutz G, Yoon C, Puigserver P, Spiegelman B, Montminy M. Creb regulates hepatic gluconeogenesis through the coactivator pgc-1. Nature. 2001;413:179–183. doi: 10.1038/35093131. [DOI] [PubMed] [Google Scholar]
- 16.Lehman JJ, Barger PM, Kovacs A, Saffitz JE, Medeiros DM, Kelly DP. Peroxisome proliferator-activated receptor gamma coactivator-1 promotes cardiac mitochondrial biogenesis. J Clin Invest. 2000;106:847–856. doi: 10.1172/JCI10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ishii KA, Fumoto T, Iwai K, Takeshita S, Ito M, Shimohata N, Aburatani H, Taketani S, Lelliott CJ, Vidal-Puig A, Ikeda K. Coordination of pgc-1beta and iron uptake in mitochondrial biogenesis and osteoclast activation. Nat Med. 2009;15:259–266. doi: 10.1038/nm.1910. [DOI] [PubMed] [Google Scholar]
- 18.Wei W, Wang X, Yang M, Smith LC, Dechow PC, Wan Y. Pgc1beta mediates ppargamma activation of osteoclastogenesis and rosiglitazone-induced bone loss. Cell Metab. 2010;11:503–516. doi: 10.1016/j.cmet.2010.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cao W, Daniel KW, Robidoux J, Puigserver P, Medvedev AV, Bai X, Floering LM, Spiegelman BM, Collins S. P38 mitogen-activated protein kinase is the central regulator of cyclic amp-dependent transcription of the brown fat uncoupling protein 1 gene. Mol Cell Biol. 2004;24:3057–3067. doi: 10.1128/MCB.24.7.3057-3067.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robidoux J, Cao W, Quan H, Daniel KW, Moukdar F, Bai X, Floering LM, Collins S. Selective activation of mitogen-activated protein (map) kinase kinase 3 and p38alpha map kinase is essential for cyclic amp-dependent ucp1 expression in adipocytes. Mol Cell Biol. 2005;25:5466–5479. doi: 10.1128/MCB.25.13.5466-5479.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pogozelski AR, Geng T, Li P, Yin X, Lira VA, Zhang M, Chi JT, Yan Z. P38gamma mitogen-activated protein kinase is a key regulator in skeletal muscle metabolic adaptation in mice. PLoS One. 2009;4:e7934. doi: 10.1371/journal.pone.0007934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chinsomboon J, Ruas J, Gupta RK, Thom R, Shoag J, Rowe GC, Sawada N, Raghuram S, Arany Z. The transcriptional coactivator pgc-1{alpha} mediates exercise-induced angiogenesis in skeletal muscle. Proc Natl Acad Sci U S A. 2009;106:21401–21406. doi: 10.1073/pnas.0909131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miura S, Kai Y, Kamei Y, Ezaki O. Isoform-specific increases in murine skeletal muscle peroxisome proliferator-activated receptor-{gamma} coactivator-1{alpha} (pgc-1{alpha}) mrna in response to {beta}2-adrenergic receptor activation and exercise. Endocrinology. 2008 doi: 10.1210/en.2008-0466. [DOI] [PubMed] [Google Scholar]
- 24.Czubryt MP, McAnally J, Fishman GI, Olson EN. Regulation of peroxisome proliferator-activated receptor gamma coactivator 1 alpha (pgc-1 alpha) and mitochondrial function by mef2 and hdac5. Proc Natl Acad Sci U S A. 2003;100:1711–1716. doi: 10.1073/pnas.0337639100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakagawa Y, Kuwahara K, Takemura G, Akao M, Kato M, Arai Y, Takano M, Harada M, Murakami M, Nakanishi M, Usami S, Yasuno S, Kinoshita H, Fujiwara M, Ueshima K, Nakao K. P300 plays a critical role in maintaining cardiac mitochondrial function and cell survival in postnatal hearts. Circ Res. 2009;105:746–754. doi: 10.1161/CIRCRESAHA.109.206037. [DOI] [PubMed] [Google Scholar]
- 26.Schaeffer PJ, Wende AR, Magee CJ, Neilson JR, Leone TC, Chen F, Kelly DP. Calcineurin and calcium/calmodulin-dependent protein kinase activate distinct metabolic gene regulatory programs in cardiac muscle. J Biol Chem. 2004;279:39593–39603. doi: 10.1074/jbc.M403649200. [DOI] [PubMed] [Google Scholar]
- 27.Handschin C, Rhee J, Lin J, Tarr PT, Spiegelman BM. An autoregulatory loop controls peroxisome proliferator-activated receptor gamma coactivator 1alpha expression in muscle. Proc Natl Acad Sci U S A. 2003;100:7111–7116. doi: 10.1073/pnas.1232352100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jager S, Handschin C, St-Pierre J, Spiegelman BM. Amp-activated protein kinase (ampk) action in skeletal muscle via direct phosphorylation of pgc-1alpha. Proc Natl Acad Sci U S A. 2007;104:12017–12022. doi: 10.1073/pnas.0705070104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nisoli E, Clementi E, Paolucci C, Cozzi V, Tonello C, Sciorati C, Bracale R, Valerio A, Francolini M, Moncada S, Carruba MO. Mitochondrial biogenesis in mammals: The role of endogenous nitric oxide. Science. 2003;299:896–899. doi: 10.1126/science.1079368. [DOI] [PubMed] [Google Scholar]
- 30.Carraway MS, Suliman HB, Jones WS, Chen CW, Babiker A, Piantadosi CA. Erythropoietin activates mitochondrial biogenesis and couples red cell mass to mitochondrial mass in the heart. Circ Res. doi: 10.1161/CIRCRESAHA.109.214353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Borniquel S, Valle I, Cadenas S, Lamas S, Monsalve M. Nitric oxide regulates mitochondrial oxidative stress protection via the transcriptional coactivator pgc-1alpha. Faseb J. 2006;20:1889–1891. doi: 10.1096/fj.05-5189fje. [DOI] [PubMed] [Google Scholar]
- 32.Wu H, Kanatous SB, Thurmond FA, Gallardo T, Isotani E, Bassel-Duby R, Williams RS. Regulation of mitochondrial biogenesis in skeletal muscle by camk. Science. 2002;296:349–352. doi: 10.1126/science.1071163. [DOI] [PubMed] [Google Scholar]
- 33.Puigserver P, Rhee J, Lin J, Wu Z, Yoon JC, Zhang CY, Krauss S, Mootha VK, Lowell BB, Spiegelman BM. Cytokine stimulation of energy expenditure through p38 map kinase activation of ppargamma coactivator-1. Mol Cell. 2001;8:971–982. doi: 10.1016/s1097-2765(01)00390-2. [DOI] [PubMed] [Google Scholar]
- 34.Knutti D, Kralli A. Pgc-1, a versatile coactivator. Trends Endocrinol Metab. 2001;12:360–365. doi: 10.1016/s1043-2760(01)00457-x. [DOI] [PubMed] [Google Scholar]
- 35.Sano M, Tokudome S, Shimizu N, Yoshikawa N, Ogawa C, Shirakawa K, Endo J, Katayama T, Yuasa S, Ieda M, Makino S, Hattori F, Tanaka H, Fukuda K. Intramolecular control of protein stability, subnuclear compartmentalization, and coactivator function of peroxisome proliferator-activated receptor gamma coactivator 1alpha. J Biol Chem. 2007;282:25970–25980. doi: 10.1074/jbc.M703634200. [DOI] [PubMed] [Google Scholar]
- 36.Li X, Monks B, Ge Q, Birnbaum MJ. Akt/pkb regulates hepatic metabolism by directly inhibiting pgc-1alpha transcription coactivator. Nature. 2007;447:1012–1016. doi: 10.1038/nature05861. [DOI] [PubMed] [Google Scholar]
- 37.Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of pgc-1alpha and sirt1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 38.Lerin C, Rodgers JT, Kalume DE, Kim SH, Pandey A, Puigserver P. Gcn5 acetyltransferase complex controls glucose metabolism through transcriptional repression of pgc-1alpha. Cell Metab. 2006;3:429–438. doi: 10.1016/j.cmet.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 39.Gerhart-Hines Z, Rodgers JT, Bare O, Lerin C, Kim SH, Mostoslavsky R, Alt FW, Wu Z, Puigserver P. Metabolic control of muscle mitochondrial function and fatty acid oxidation through sirt1/pgc-1alpha. Embo J. 2007;26:1913–1923. doi: 10.1038/sj.emboj.7601633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Teyssier C, Ma H, Emter R, Kralli A, Stallcup MR. Activation of nuclear receptor coactivator pgc-1alpha by arginine methylation. Genes Dev. 2005;19:1466–1473. doi: 10.1101/gad.1295005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Charest-Marcotte A, Dufour CR, Wilson BJ, Tremblay AM, Eichner LJ, Arlow DH, Mootha VK, Giguere V. The homeobox protein prox1 is a negative modulator of err{alpha}/pgc-1{alpha} bioenergetic functions. Genes Dev. 2010;24:537–542. doi: 10.1101/gad.1871610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Finck BN, Gropler MC, Chen Z, Leone TC, Croce MA, Harris TE, Lawrence JC, Jr., Kelly DP. Lipin 1 is an inducible amplifier of the hepatic pgc-1alpha/pparalpha regulatory pathway. Cell Metab. 2006;4:199–210. doi: 10.1016/j.cmet.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 43.Yang J, Williams RS, Kelly DP. Bcl3 interacts cooperatively with peroxisome proliferator-activated receptor gamma (ppargamma) coactivator 1alpha to coactivate nuclear receptors estrogen-related receptor alpha and pparalpha. Mol Cell Biol. 2009;29:4091–4102. doi: 10.1128/MCB.01669-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fan M, Rhee J, Pierre J, Handschin C, Puigserver P, Lin J, Jaeger S, Erdjument-Bromage H, Tempst P, Spiegelman BM. Suppression of mitochondrial respiration through recruitment of p160 myb binding protein to pgc-1alpha: Modulation by p38 mapk. Genes Dev. 2004;18:278–289. doi: 10.1101/gad.1152204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kelly TJ, Lerin C, Haas W, Gygi SP, Puigserver P. Gcn5-mediated transcriptional control of the metabolic coactivator pgc-1beta through lysine acetylation. J Biol Chem. 2009;284:19945–19952. doi: 10.1074/jbc.M109.015164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin J, Yang R, Tarr PT, Wu PH, Handschin C, Li S, Yang W, Pei L, Uldry M, Tontonoz P, Newgard CB, Spiegelman BM. Hyperlipidemic effects of dietary saturated fats mediated through pgc-1beta coactivation of srebp. Cell. 2005;120:261–273. doi: 10.1016/j.cell.2004.11.043. [DOI] [PubMed] [Google Scholar]
- 47.Arany Z. Pgc-1 coactivators and skeletal muscle adaptations in health and disease. Curr Opin Genet Dev. 2008;18:426–434. doi: 10.1016/j.gde.2008.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leick L, Hellsten Y, Fentz J, Lyngby SS, Wojtaszewski JF, Hidalgo J, Pilegaard H. Pgc-1alpha mediates exercise-induced skeletal muscle vegf expression in mice. American journal of physiology. 2009;297:E92–103. doi: 10.1152/ajpendo.00076.2009. [DOI] [PubMed] [Google Scholar]
- 49.Pierre J, Drori S, Uldry M, Silvaggi JM, Rhee J, Jager S, Handschin C, Zheng K, Lin J, Yang W, Simon DK, Bachoo R, Spiegelman BM. Suppression of reactive oxygen species and neurodegeneration by the pgc-1 transcriptional coactivators. Cell. 2006;127:397–408. doi: 10.1016/j.cell.2006.09.024. [DOI] [PubMed] [Google Scholar]
- 50.Banke NH, Wende AR, Leone TC, O'Donnell JM, Abel ED, Kelly DP, Lewandowski ED. Preferential oxidation of triacylglyceride-derived fatty acids in heart is augmented by the nuclear receptor ppar{alpha} Circ Res. 2010 doi: 10.1161/CIRCRESAHA.110.221713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wende AR, Huss JM, Schaeffer PJ, Giguere V, Kelly DP. Pgc-1alpha coactivates pdk4 gene expression via the orphan nuclear receptor erralpha: A mechanism for transcriptional control of muscle glucose metabolism. Mol Cell Biol. 2005;25:10684–10694. doi: 10.1128/MCB.25.24.10684-10694.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Russell LK, Mansfield CM, Lehman JJ, Kovacs A, Courtois M, Saffitz JE, Medeiros DM, Valencik ML, McDonald JA, Kelly DP. Cardiac-specific induction of the transcriptional coactivator peroxisome proliferator-activated receptor gamma coactivator-1alpha promotes mitochondrial biogenesis and reversible cardiomyopathy in a developmental stage-dependent manner. Circ Res. 2004;94:525–533. doi: 10.1161/01.RES.0000117088.36577.EB. [DOI] [PubMed] [Google Scholar]
- 53.Liang H, Balas B, Tantiwong P, Dube J, Goodpaster BH, O'Doherty RM, DeFronzo RA, Richardson A, Musi N, Ward WF. Whole body overexpression of pgc-1alpha has opposite effects on hepatic and muscle insulin sensitivity. American journal of physiology. 2009;296:E945–954. doi: 10.1152/ajpendo.90292.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kelly DP, Scarpulla RC. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes Dev. 2004;18:357–368. doi: 10.1101/gad.1177604. [DOI] [PubMed] [Google Scholar]
- 55.Scarpulla RC. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol Rev. 2008;88:611–638. doi: 10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- 56.Hock MB, Kralli A. Transcriptional control of mitochondrial biogenesis and function. Annu Rev Physiol. 2009;71:177–203. doi: 10.1146/annurev.physiol.010908.163119. [DOI] [PubMed] [Google Scholar]
- 57.Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla RC, Spiegelman BM. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator pgc-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 58.Mootha VK, Handschin C, Arlow D, Xie X, Pierre J, Sihag S, Yang W, Altshuler D, Puigserver P, Patterson N, Willy PJ, Schulman IG, Heyman RA, Lander ES, Spiegelman BM. Erralpha and gabpa/b specify pgc-1alpha-dependent oxidative phosphorylation gene expression that is altered in diabetic muscle. Proc Natl Acad Sci U S A. 2004;101:6570–6575. doi: 10.1073/pnas.0401401101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Baar K, Song Z, Semenkovich CF, Jones TE, Han DH, Nolte LA, Ojuka EO, Chen M, Holloszy JO. Skeletal muscle overexpression of nuclear respiratory factor 1 increases glucose transport capacity. FASEB J. 2003;17:1666–1673. doi: 10.1096/fj.03-0049com. [DOI] [PubMed] [Google Scholar]
- 60.Tremblay AM, Giguere V. The nr3b subgroup: An overrview. Nucl Recept Signal. 2007;5:e009. doi: 10.1621/nrs.05009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Giguere V. Transcriptional control of energy homeostasis by the estrogen-related receptors. Endocr Rev. 2008;29:677–696. doi: 10.1210/er.2008-0017. [DOI] [PubMed] [Google Scholar]
- 62.Villena JA, Kralli A. Erralpha: A metabolic function for the oldest orphan. Trends Endocrinol Metab. 2008;19:269–276. doi: 10.1016/j.tem.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huss JM, Torra IP, Staels B, Giguere V, Kelly DP. Estrogen-related receptor alpha directs peroxisome proliferator-activated receptor alpha signaling in the transcriptional control of energy metabolism in cardiac and skeletal muscle. Mol Cell Biol. 2004;24:9079–9091. doi: 10.1128/MCB.24.20.9079-9091.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huss JM, Kopp RP, Kelly DP. Peroxisome proliferator-activated receptor coactivator-1alpha (pgc-1alpha) coactivates the cardiac-enriched nuclear receptors estrogen-related receptor-alpha and -gamma. Identification of novel leucine-rich interaction motif within pgc-1alpha. J Biol Chem. 2002;277:40265–40274. doi: 10.1074/jbc.M206324200. [DOI] [PubMed] [Google Scholar]
- 65.Schreiber SN, Knutti D, Brogli K, Uhlmann T, Kralli A. The transcriptional coactivator pgc-1 regulates the expression and activity of the orphan nuclear receptor estrogen-related receptor alpha (erralpha) J Biol Chem. 2003;278:9013–9018. doi: 10.1074/jbc.M212923200. [DOI] [PubMed] [Google Scholar]
- 66.Dufour CR, Wilson BJ, Huss JM, Kelly DP, Alaynick WA, Downes M, Evans RM, Blanchette M, Giguere V. Genome-wide orchestration of cardiac functions by the orphan nuclear receptors erralpha and gamma. Cell Metab. 2007;5:345–356. doi: 10.1016/j.cmet.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 67.Rangwala SM, Wang X, Calvo JA, Lindsley L, Zhang Y, Deyneko G, Beaulieu V, Gao J, Turner G, Markovits J. Estrogen-related receptor {gamma} is a key regulator of muscle mitochondrial activity and oxidative capacity. J Biol Chem. 2010 doi: 10.1074/jbc.M110.125401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Alaynick WA, Kondo RP, Xie W, He W, Dufour CR, Downes M, Jonker JW, Giles W, Naviaux RK, Giguere V, Evans RM. Errgamma directs and maintains the transition to oxidative metabolism in the postnatal heart. Cell Metab. 2007;6:13–24. doi: 10.1016/j.cmet.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 69.Alaynick WA, Way JM, Wilson SA, Benson WG, Pei L, Downes M, Yu R, Jonker JW, Holt JA, Rajpal DK, Li H, Stuart J, McPherson R, Remlinger KS, Chang CY, McDonnell DP, Evans RM, Billin AN. Errgamma regulates cardiac, gastric, and renal potassium homeostasis. Mol Endocrinol. 2010;24:299–309. doi: 10.1210/me.2009-0114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Luo J, Sladek R, Bader JA, Matthyssen A, Rossant J, Giguere V. Placental abnormalities in mouse embryos lacking the orphan nuclear receptor err-beta. Nature. 1997;388:778–782. doi: 10.1038/42022. [DOI] [PubMed] [Google Scholar]
- 71.Larsson NG, Barsh GS, Clayton DA. Structure and chromosomal localization of the mouse mitochondrial transcription factor a gene (tfam) Mamm Genome. 1997;8:139–140. doi: 10.1007/s003359900373. [DOI] [PubMed] [Google Scholar]
- 72.Falkenberg M, Gaspari M, Rantanen A, Trifunovic A, Larsson NG, Gustafsson CM. Mitochondrial transcription factors b1 and b2 activate transcription of human mtdna. Nat Genet. 2002;31:289–294. doi: 10.1038/ng909. [DOI] [PubMed] [Google Scholar]
- 73.Virbasius JV, Scarpulla RC. Activation of the human mitochondrial transcription factor a gene by nuclear respiratory factors: A potential regulatory link between nuclear and mitochondrial gene expression in organelle biogenesis. Proc Natl Acad Sci U S A. 1994;91:1309–1313. doi: 10.1073/pnas.91.4.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gleyzer N, Vercauteren K, Scarpulla RC. Control of mitochondrial transcription specificity factors (tfb1m and tfb2m) by nuclear respiratory factors (nrf-1 and nrf-2) and pgc-1 family coactivators. Mol Cell Biol. 2005;25:1354–1366. doi: 10.1128/MCB.25.4.1354-1366.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Parisi MA, Xu B, Clayton DA. A human mitochondrial transcriptional activator can functionally replace a yeast mitochondrial hmg-box protein both in vivo and in vitro. Mol Cell Biol. 1993;13:1951–1961. doi: 10.1128/mcb.13.3.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Larsson NG, Wang J, Wilhelmsson H, Oldfors A, Rustin P, Lewandoski M, Barsh GS, Clayton DA. Mitochondrial transcription factor a is necessary for mtdna maintenance and embryogenesis in mice. Nat Genet. 1998;18:231–236. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- 77.Li H, Wang J, Wilhelmsson H, Hansson A, Thoren P, Duffy J, Rustin P, Larsson NG. Genetic modification of survival in tissue-specific knockout mice with mitochondrial cardiomyopathy. Proc Natl Acad Sci U S A. 2000;97:3467–3472. doi: 10.1073/pnas.97.7.3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang J, Wilhelmsson H, Graff C, Li H, Oldfors A, Rustin P, Bruning JC, Kahn CR, Clayton DA, Barsh GS, Thoren P, Larsson NG. Dilated cardiomyopathy and atrioventricular conduction blocks induced by heart-specific inactivation of mitochondrial DNA gene expression. Nat Genet. 1999;21:133–137. doi: 10.1038/5089. [DOI] [PubMed] [Google Scholar]
- 79.Ikeuchi M, Matsusaka H, Kang D, Matsushima S, Ide T, Kubota T, Fujiwara T, Hamasaki N, Takeshita A, Sunagawa K, Tsutsui H. Overexpression of mitochondrial transcription factor a ameliorates mitochondrial deficiencies and cardiac failure after myocardial infarction. Circulation. 2005;112:683–690. doi: 10.1161/CIRCULATIONAHA.104.524835. [DOI] [PubMed] [Google Scholar]
- 80.Lehmann JM, Moore LB, Smith-Oliver TA, Wilkison WO, Willson TM, Kliewer SA. An antidiabetic thiazolidinedione is a high affinity ligand for peroxisome proliferator-activated receptor gamma (ppar gamma) J Biol Chem. 1995;270:12953–12956. doi: 10.1074/jbc.270.22.12953. [DOI] [PubMed] [Google Scholar]
- 81.Burkart EM, Sambandam N, Han X, Gross RW, Courtois M, Gierasch CM, Shoghi K, Welch MJ, Kelly DP. Nuclear receptors pparbeta/delta and pparalpha direct distinct metabolic regulatory programs in the mouse heart. J Clin Invest. 2007;117:3930–3939. doi: 10.1172/JCI32578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Duncan JG, Bharadwaj KG, Fong JL, Mitra R, Sambandam N, Courtois MR, Lavine KJ, Goldberg IJ, Kelly DP. Rescue of cardiomyopathy in peroxisome proliferator-activated receptor-alpha transgenic mice by deletion of lipoprotein lipase identifies sources of cardiac lipids and peroxisome proliferator-activated receptor-alpha activators. Circulation. 2010;121:426–435. doi: 10.1161/CIRCULATIONAHA.109.888735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Leone TC, Weinheimer CJ, Kelly DP. A critical role for the peroxisome proliferator-activated receptor alpha (pparalpha) in the cellular fasting response: The pparalpha-null mouse as a model of fatty acid oxidation disorders. Proc Natl Acad Sci U S A. 1999;96:7473–7478. doi: 10.1073/pnas.96.13.7473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Campbell FM, Kozak R, Wagner A, Altarejos JY, Dyck JR, Belke DD, Severson DL, Kelly DP, Lopaschuk GD. A role for peroxisome proliferator-activated receptor alpha (pparalpha) in the control of cardiac malonyl-coa levels: Reduced fatty acid oxidation rates and increased glucose oxidation rates in the hearts of mice lacking pparalpha are associated with higher concentrations of malonyl-coa and reduced expression of malonyl-coa decarboxylase. J Biol Chem. 2002;277:4098–4103. doi: 10.1074/jbc.M106054200. [DOI] [PubMed] [Google Scholar]
- 85.Hafstad AD, Khalid AM, Hagve M, Lund T, Larsen TS, Severson DL, Clarke K, Berge RK, Aasum E. Cardiac peroxisome proliferator-activated receptor-alpha activation causes increased fatty acid oxidation, reducing efficiency and post-ischaemic functional loss. Cardiovasc Res. 2009;83:519–526. doi: 10.1093/cvr/cvp132. [DOI] [PubMed] [Google Scholar]
- 86.Finck BN, Lehman JJ, Leone TC, Welch MJ, Bennett MJ, Kovacs A, Han X, Gross RW, Kozak R, Lopaschuk GD, Kelly DP. The cardiac phenotype induced by pparalpha overexpression mimics that caused by diabetes mellitus. J Clin Invest. 2002;109:121–130. doi: 10.1172/JCI14080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Finck BN, Han X, Courtois M, Aimond F, Nerbonne JM, Kovacs A, Gross RW, Kelly DP. A critical role for pparalpha-mediated lipotoxicity in the pathogenesis of diabetic cardiomyopathy: Modulation by dietary fat content. Proc Natl Acad Sci U S A. 2003;100:1226–1231. doi: 10.1073/pnas.0336724100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yang J, Sambandam N, Han X, Gross RW, Courtois M, Kovacs A, Febbraio M, Finck BN, Kelly DP. Cd36 deficiency rescues lipotoxic cardiomyopathy. Circ Res. 2007;100:1208–1217. doi: 10.1161/01.RES.0000264104.25265.b6. [DOI] [PubMed] [Google Scholar]
- 89.Cheng L, Ding G, Qin Q, Xiao Y, Woods D, Chen YE, Yang Q. Peroxisome proliferator-activated receptor delta activates fatty acid oxidation in cultured neonatal and adult cardiomyocytes. Biochem Biophys Res Commun. 2004;313:277–286. doi: 10.1016/j.bbrc.2003.11.127. [DOI] [PubMed] [Google Scholar]
- 90.Cheng L, Ding G, Qin Q, Huang Y, Lewis W, He N, Evans RM, Schneider MD, Brako FA, Xiao Y, Chen YE, Yang Q. Cardiomyocyte-restricted peroxisome proliferator-activated receptor-delta deletion perturbs myocardial fatty acid oxidation and leads to cardiomyopathy. Nat Med. 2004;10:1245–1250. doi: 10.1038/nm1116. [DOI] [PubMed] [Google Scholar]
- 91.Duan SZ, Ivashchenko CY, Russell MW, Milstone DS, Mortensen RM. Cardiomyocyte-specific knockout and agonist of peroxisome proliferator-activated receptor-gamma both induce cardiac hypertrophy in mice. Circ Res. 2005;97:372–379. doi: 10.1161/01.RES.0000179226.34112.6d. [DOI] [PubMed] [Google Scholar]
- 92.Son NH, Park TS, Yamashita H, Yokoyama M, Huggins LA, Okajima K, Homma S, Szabolcs MJ, Huang LS, Goldberg IJ. Cardiomyocyte expression of ppargamma leads to cardiac dysfunction in mice. J Clin Invest. 2007;117:2791–2801. doi: 10.1172/JCI30335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Arany Z, Foo SY, Ma Y, Ruas JL, Bommi-Reddy A, Girnun G, Cooper M, Laznik D, Chinsomboon J, Rangwala SM, Baek KH, Rosenzweig A, Spiegelman BM. Hif-independent regulation of vegf and angiogenesis by the transcriptional coactivator pgc-1alpha. Nature. 2008;451:1008–1012. doi: 10.1038/nature06613. [DOI] [PubMed] [Google Scholar]
- 94.Bloor CM. Angiogenesis during exercise and training. Angiogenesis. 2005;8:263–271. doi: 10.1007/s10456-005-9013-x. [DOI] [PubMed] [Google Scholar]
- 95.Baar K, Wende AR, Jones TE, Marison M, Nolte LA, Chen M, Kelly DP, Holloszy JO. Adaptations of skeletal muscle to exercise: Rapid increase in the transcriptional coactivator pgc-1. Faseb J. 2002;16:1879–1886. doi: 10.1096/fj.02-0367com. [DOI] [PubMed] [Google Scholar]
- 96.Norrbom J, Sundberg CJ, Ameln H, Kraus WE, Jansson E, Gustafsson T. Pgc-1alpha mrna expression is influenced by metabolic perturbation in exercising human skeletal muscle. J Appl Physiol. 2004;96:189–194. doi: 10.1152/japplphysiol.00765.2003. [DOI] [PubMed] [Google Scholar]
- 97.Koves TR, Li P, An J, Akimoto T, Slentz D, Ilkayeva O, Dohm GL, Yan Z, Newgard CB, Muoio DM. Peroxisome proliferator-activated receptor-gamma co-activator 1alpha-mediated metabolic remodeling of skeletal myocytes mimics exercise training and reverses lipid-induced mitochondrial inefficiency. J Biol Chem. 2005;280:33588–33598. doi: 10.1074/jbc.M507621200. [DOI] [PubMed] [Google Scholar]
- 98.Cluberton LJ, McGee SL, Murphy RM, Hargreaves M. Effect of carbohydrate ingestion on exercise-induced alterations in metabolic gene expression. J Appl Physiol. 2005;99:1359–1363. doi: 10.1152/japplphysiol.00197.2005. [DOI] [PubMed] [Google Scholar]
- 99.Geng T, Li P, Okutsu M, Yin X, Kwek J, Zhang M, Yan Z. Pgc-1alpha plays a functional role in exercise-induced mitochondrial biogenesis and angiogenesis but not fiber-type transformation in mouse skeletal muscle. Am J Physiol Cell Physiol. 2010;298:C572–579. doi: 10.1152/ajpcell.00481.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Heineke J, Auger-Messier M, Xu J, Oka T, Sargent MA, York A, Klevitsky R, Vaikunth S, Duncan SA, Aronow BJ, Robbins J, Cromblehol TM, Molkentin JD. Cardiomyocyte gata4 functions as a stress-responsive regulator of angiogenesis in the murine heart. J Clin Invest. 2007;117:3198–3210. doi: 10.1172/JCI32573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sano M, Minamino T, Toko H, Miyauchi H, Orimo M, Qin Y, Akazawa H, Tateno K, Kayama Y, Harada M, Shimizu I, Asahara T, Hamada H, Tomita S, Molkentin JD, Zou Y, Komuro I. P53-induced inhibition of hif-1 causes cardiac dysfunction during pressure overload. Nature. 2007;446:444–448. doi: 10.1038/nature05602. [DOI] [PubMed] [Google Scholar]
- 102.Izumiya Y, Shiojima I, Sato K, Sawyer DB, Colucci WS, Walsh K. Vascular endothelial growth factor blockade promotes the transition from compensatory cardiac hypertrophy to failure in response to pressure overload. Hypertension. 2006;47:887–893. doi: 10.1161/01.HYP.0000215207.54689.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lai L, Leone TC, Zechner C, Schaeffer PJ, Kelly SM, Flanagan DP, Medeiros DM, Kovacs A, Kelly DP. Transcriptional coactivators pgc-1alpha and pgc-lbeta control overlapping programs required for perinatal maturation of the heart. Genes Dev. 2008;22:1948–1961. doi: 10.1101/gad.1661708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pierre J, Lin J, Krauss S, Tarr PT, Yang R, Newgard CB, Spiegelman BM. Bioenergetic analysis of peroxisome proliferator-activated receptor gamma coactivators 1alpha and 1beta (pgc-1alpha and pgc-1beta) in muscle cells. J Biol Chem. 2003;278:26597–26603. doi: 10.1074/jbc.M301850200. [DOI] [PubMed] [Google Scholar]
- 105.Vercauteren K, Gleyzer N, Scarpulla RC. Short hairpin rna-mediated silencing of prc (pgc-1-related coactivator) results in a severe respiratory chain deficiency associated with the proliferation of aberrant mitochondria. J Biol Chem. 2009;284:2307–2319. doi: 10.1074/jbc.M806434200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wallace DC. Mitochondrial defects in cardiomyopathy and neuromuscular disease. Am Heart J. 2000;139:S70–85. doi: 10.1067/mhj.2000.103934. [DOI] [PubMed] [Google Scholar]
- 107.DiMauro S, Schon EA. Mitochondrial respiratory-chain diseases. N Engl J Med. 2003;348:2656–2668. doi: 10.1056/NEJMra022567. [DOI] [PubMed] [Google Scholar]
- 108.DiMauro S, Schon EA. Mitochondrial DNA mutations in human disease. Am J Med Genet. 2001;106:18–26. doi: 10.1002/ajmg.1392. [DOI] [PubMed] [Google Scholar]
- 109.Ingwall JS. Mann D, editor. Energetic basis for heart failure. Heart failure: A companion to braunwald's heart disease. 2002 [Google Scholar]
- 110.Ingwall JS. Atp and the heart. Kluwer Academic Publishers; Boston: 2003. [Google Scholar]
- 111.Ingwall JS, Weiss RG. Is the failing heart energy starved? On using chemical energy to support cardiac function. Circ Res. 2004;95:135–145. doi: 10.1161/01.RES.0000137170.41939.d9. [DOI] [PubMed] [Google Scholar]
- 112.Lebrecht D, Walker UA. Role of mtdna lesions in anthracycline cardiotoxicity. Cardiovasc Toxicol. 2007;7:108–113. doi: 10.1007/s12012-007-0009-1. [DOI] [PubMed] [Google Scholar]
- 113.Pastores GM, Santorelli FM, Shanske S, Gelb BD, Fyfe B, Wolfe D, Willner JP. Leigh syndrome and hypertrophic cardiomyopathy in an infant with a mitochondrial DNA point mutation (t8993g) Am J Med Genet. 1994;50:265–271. doi: 10.1002/ajmg.1320500310. [DOI] [PubMed] [Google Scholar]
- 114.Andreu AL, Checcarelli N, Iwata S, Shanske S, DiMauro S. A missense mutation in the mitochondrial cytochrome b gene in a revisited case with histiocytoid cardiomyopathy. Pediatr Res. 2000;48:311–314. doi: 10.1203/00006450-200009000-00008. [DOI] [PubMed] [Google Scholar]
- 115.Palmieri L, Alberio S, Pisano I, Lodi T, Meznaric-Petrusa M, Zidar J, Santoro A, Scarcia P, Fontanesi F, Lamantea E, Ferrero I, Zeviani M. Complete loss-of-function of the heart/muscle-specific adenine nucleotide translocator is associated with mitochondrial myopathy and cardiomyopathy. Hum Mol Genet. 2005;14:3079–3088. doi: 10.1093/hmg/ddi341. [DOI] [PubMed] [Google Scholar]
- 116.Kelly DP, Strauss AW. Inherited cardiomyopathies. N Engl J Med. 1994;330:913–919. doi: 10.1056/NEJM199403313301308. [DOI] [PubMed] [Google Scholar]
- 117.Loeffen J, Elpeleg O, Smeitink J, Smeets R, Stockler-Ipsiroglu S, Mandel H, Sengers R, Trijbels F, van den Heuvel L. Mutations in the complex i ndufs2 gene of patients with cardiomyopathy and encephalomyopathy. Ann Neurol. 2001;49:195–201. doi: 10.1002/1531-8249(20010201)49:2<195::aid-ana39>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 118.Marin-Garcia J, Goldenthal MJ. Mitochondrial centrality in heart failure. Heart Fail Rev. 2008;13:137–150. doi: 10.1007/s10741-007-9079-1. [DOI] [PubMed] [Google Scholar]
- 119.Antonicka H, Mattman A, Carlson CG, Glerum DM, Hoffbuhr KC, Leary SC, Kennaway NG, Shoubridge EA. Mutations in cox15 produce a defect in the mitochondrial heme biosynthetic pathway, causing early-onset fatal hypertrophic cardiomyopathy. Am J Hum Genet. 2003;72:101–114. doi: 10.1086/345489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bennett MJ, Rinaldo P, Strauss AW. Inborn errors of mitochondrial fatty acid oxidation. Crit Rev Clin Lab Sci. 2000;37:1–44. doi: 10.1080/10408360091174169. [DOI] [PubMed] [Google Scholar]
- 121.Strauss AW, Powell CK, Hale DE, Anderson MM, Ahuja A, Brackett JC, Sims HF. Molecular basis of human mitochondrial very-long-chain acyl-coa dehydrogenase deficiency causing cardiomyopathy and sudden death in childhood. Proc Natl Acad Sci U S A. 1995;92:10496–10500. doi: 10.1073/pnas.92.23.10496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hale DE, Batshaw ML, Coates PM, Frerman FE, Goodman SI, Singh I, Stanley CA. Long-chain acyl coenzyme a dehydrogenase deficiency: An inherited cause of nonketotic hypoglycemia. Pediatr Res. 1985;19:666–671. doi: 10.1203/00006450-198507000-00006. [DOI] [PubMed] [Google Scholar]
- 123.Ibdah JA, Paul H, Zhao Y, Binford S, Salleng K, Cline M, Matern D, Bennett MJ, Rinaldo P, Strauss AW. Lack of mitochondrial trifunctional protein in mice causes neonatal hypoglycemia and sudden death. J Clin Invest. 2001;107:1403–1409. doi: 10.1172/JCI12590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Rocchiccioli F, Wanders RJ, Aubourg P, Vianey-Liaud C, Ijlst L, Fabre M, Cartier N, Bougneres PF. Deficiency of long-chain 3-hydroxyacyl-coa dehydrogenase: A cause of lethal myopathy and cardiomyopathy in early childhood. Pediatr Res. 1990;28:657–662. doi: 10.1203/00006450-199012000-00023. [DOI] [PubMed] [Google Scholar]
- 125.Graham BH, Waymire KG, Cottrell B, Trounce IA, MacGregor GR, Wallace DC. A mouse model for mitochondrial myopathy and cardiomyopathy resulting from a deficiency in the heart/muscle isoform of the adenine nucleotide translocator. Nat Genet. 1997;16:226–234. doi: 10.1038/ng0797-226. [DOI] [PubMed] [Google Scholar]
- 126.Nahrendorf M, Spindler M, Hu K, Bauer L, Ritter O, Nordbeck P, Quaschning T, Hiller KH, Wallis J, Ertl G, Bauer WR, Neubauer S. Creatine kinase knockout mice show left ventricular hypertrophy and dilatation, but unaltered remodeling post-myocardial infarction. Cardiovasc Res. 2005;65:419–427. doi: 10.1016/j.cardiores.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 127.Li Y, Huang TT, Carlson EJ, Melov S, Ursell PC, Olson JL, Noble LJ, Yoshimura MP, Berger C, Chan PH, Wallace DC, Epstein CJ. Dilated cardiomyopathy and neonatal lethality in mutant mice lacking manganese superoxide dismutase. Nat Genet. 1995;11:376–381. doi: 10.1038/ng1295-376. [DOI] [PubMed] [Google Scholar]
- 128.Lewis W, Day BJ, Kohler JJ, Hosseini SH, Chan SS, Green EC, Haase CP, Keebaugh ES, Long R, Ludaway T, Russ R, Steltzer J, Tioleco N, Santoianni R, Copeland WC. Decreased mtdna, oxidative stress, cardiomyopathy, and death from transgenic cardiac targeted human mutant polymerase gamma. Lab Invest. 2007;87:326–335. doi: 10.1038/labinvest.3700523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kurtz DM, Rinaldo P, Rhead WJ, Tian L, Millington DS, Vockley J, Hamm DA, Brix AE, Lindsey JR, Pinkert CA, O'Brien WE, Wood PA. Targeted disruption of mouse long-chain acyl-coa dehydrogenase gene reveals crucial roles for fatty acid oxidation. Proc Natl Acad Sci U S A. 1998;95:15592–15597. doi: 10.1073/pnas.95.26.15592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Exil VJ, Roberts RL, Sims H, McLaughlin JE, Malkin RA, Gardner CD, Ni G, Rottman JN, Strauss AW. Very-long-chain acyl-coenzyme a dehydrogenase deficiency in mice. Circ Res. 2003;93:448–455. doi: 10.1161/01.RES.0000088786.19197.E4. [DOI] [PubMed] [Google Scholar]
- 131.Neubauer S. The failing heart--an engine out of fuel. N Engl J Med. 2007;356:1140–1151. doi: 10.1056/NEJMra063052. [DOI] [PubMed] [Google Scholar]
- 132.Starling RC, Hammer DF, Altschuld RA. Human myocardial atp content and in vivo contractile function. Mol Cell Biochem. 1998;180:171–177. [PubMed] [Google Scholar]
- 133.Beer M, Seyfarth T, Sandstede J, Landschutz W, Lipke C, Kostler H, von Kienlin M, Harre K, Hahn D, Neubauer S. Absolute concentrations of high-energy phosphate metabolites in normal, hypertrophied, and failing human myocardium measured noninvasively with (31)p-sloop magnetic resonance spectroscopy. J Am Coll Cardiol. 2002;40:1267–1274. doi: 10.1016/s0735-1097(02)02160-5. [DOI] [PubMed] [Google Scholar]
- 134.Weiss RG, Gerstenblith G, Bottomley PA. Atp flux through creatine kinase in the normal, stressed, and failing human heart. Proc Natl Acad Sci U S A. 2005;102:808–813. doi: 10.1073/pnas.0408962102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Neubauer S, Horn M, Cramer M, Harre K, Newell JB, Peters W, Pabst T, Ertl G, Hahn D, Ingwall JS, Kochsiek K. Myocardial phosphocreatine-to-atp ratio is a predictor of mortality in patients with dilated cardiomyopathy. Circulation. 1997;96:2190–2196. doi: 10.1161/01.cir.96.7.2190. [DOI] [PubMed] [Google Scholar]
- 136.Ingwall JS. Energy metabolism in heart failure and remodelling. Cardiovasc Res. 2009;81:412–419. doi: 10.1093/cvr/cvn301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.de las Fuentes L, Herrero P, Peterson LR, Kelly DP, Gropler RJ, Davila-Roman VG. Myocardial fatty acid metabolism: Independent predictor of left ventricular mass in hypertensive heart disease. Hypertension. 2003;41:83–87. doi: 10.1161/01.hyp.0000047668.48494.39. [DOI] [PubMed] [Google Scholar]
- 138.de las Fuentes L, Soto PF, Cupps BP, Pasque MK, Herrero P, Gropler RJ, Waggoner AD, Davila-Roman VG. Hypertensive left ventricular hypertrophy is associated with abnormal myocardial fatty acid metabolism and myocardial efficiency. J Nucl Cardiol. 2006;13:369–377. doi: 10.1016/j.nuclcard.2006.01.021. [DOI] [PubMed] [Google Scholar]
- 139.Laine H, Katoh C, Luotolahti M, Yki-Jarvinen H, Kantola I, Jula A, Takala TO, Ruotsalainen U, Iida H, Haaparanta M, Nuutila P, Knuuti J. Myocardial oxygen consumption is unchanged but efficiency is reduced in patients with essential hypertension and left ventricular hypertrophy. Circulation. 1999;100:2425–2430. doi: 10.1161/01.cir.100.24.2425. [DOI] [PubMed] [Google Scholar]
- 140.Sorokina N, O'Donnell JM, McKinney RD, Pound KM, Woldegiorgis G, LaNoue KF, Ballal K, Taegtmeyer H, Buttrick PM, Lewandowski ED. Recruitment of compensatory pathways to sustain oxidative flux with reduced carnitine palmitoyltransferase i activity characterizes inefficiency in energy metabolism in hypertrophied hearts. Circulation. 2007;115:2033–2041. doi: 10.1161/CIRCULATIONAHA.106.668665. [DOI] [PubMed] [Google Scholar]
- 141.Stanley WC, Recchia FA, Lopaschuk GD. Myocardial substrate metabolism in the normal and failing heart. Physiol Rev. 2005;85:1093–1129. doi: 10.1152/physrev.00006.2004. [DOI] [PubMed] [Google Scholar]
- 142.Ide T, Tsutsui H, Hayashidani S, Kang D, Suematsu N, Nakamura K, Utsumi H, Hamasaki N, Takeshita A. Mitochondrial DNA damage and dysfunction associated with oxidative stress in failing hearts after myocardial infarction. Circ Res. 2001;88:529–535. doi: 10.1161/01.res.88.5.529. [DOI] [PubMed] [Google Scholar]
- 143.Tsutsui H. Mitochondrial oxidative stress and heart failure. Intern Med. 2006;45:809–813. doi: 10.2169/internalmedicine.45.1765. [DOI] [PubMed] [Google Scholar]
- 144.L'Ecuyer T, Sanjeev S, Thomas R, Novak R, Das L, Campbell W, Heide RV. DNA damage is an early event in doxorubicin-induced cardiac myocyte death. Am J Physiol Heart Circ Physiol. 2006;291:H1273–1280. doi: 10.1152/ajpheart.00738.2005. [DOI] [PubMed] [Google Scholar]
- 145.Lewis W. Defective mitochondrial DNA replication and nrtis: Pathophysiological implications in aids cardiomyopathy. Am J Physiol Heart Circ Physiol. 2003;284:H1–9. doi: 10.1152/ajpheart.00814.2002. [DOI] [PubMed] [Google Scholar]
- 146.Frerichs FC, Dingemans KP, Brinkman K. Cardiomyopathy with mitochondrial damage associated with nucleoside reverse-transcriptase inhibitors. N Engl J Med. 2002;347:1895–1896. doi: 10.1056/NEJM200212053472320. [DOI] [PubMed] [Google Scholar]
- 147.Karamanlidis G, Nascimben L, Couper GS, Shekar PS, del Monte F, Tian R. Defective DNA replication impairs mitochondrial biogenesis in human failing hearts. Circ Res. 2010;106:1541–1548. doi: 10.1161/CIRCRESAHA.109.212753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Lehman JJ, Kelly DP. Gene regulatory mechanisms governing energy metabolism during cardiac hypertrophic growth. Heart Fail Rev. 2002;7:175–185. doi: 10.1023/a:1015332726303. [DOI] [PubMed] [Google Scholar]
- 149.Lehman JJ, Kelly DP. Transcriptional activation of energy metabolic switches in the developing and hypertrophied heart. Clin Exp Pharmacol Physiol. 2002;29:339–345. doi: 10.1046/j.1440-1681.2002.03655.x. [DOI] [PubMed] [Google Scholar]
- 150.Huss JM, Kelly DP. Nuclear receptor signaling and cardiac energetics. Circ Res. 2004;95:568–578. doi: 10.1161/01.RES.0000141774.29937.e3. [DOI] [PubMed] [Google Scholar]
- 151.Sihag S, Cresci S, Li AY, Sucharov CC, Lehman JJ. Pgc-1alpha and erralpha target gene downregulation is a signature of the failing human heart. J Mol Cell Cardiol. 2009;46:201–212. doi: 10.1016/j.yjmcc.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Sano M, Schneider MD. Cyclin-dependent kinase-9: An rnapii kinase at the nexus of cardiac growth and death cascades. Circ Res. 2004;95:867–876. doi: 10.1161/01.RES.0000146675.88354.04. [DOI] [PubMed] [Google Scholar]
- 153.Arany Z, Novikov M, Chin S, Ma Y, Rosenzweig A, Spiegelman BM. Transverse aortic constriction leads to accelerated heart failure in mice lacking ppar-gamma coactivator 1alpha. Proc Natl Acad Sci U S A. 2006;103:10086–10091. doi: 10.1073/pnas.0603615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Huss JM, Imahashi K, Dufour CR, Weinheimer CJ, Courtois M, Kovacs A, Giguere V, Murphy E, Kelly DP. The nuclear receptor erralpha is required for the bioenergetic and functional adaptation to cardiac pressure overload. Cell Metab. 2007;6:25–37. doi: 10.1016/j.cmet.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 155.Palomer X, Alvarez-Guardia D, Rodriguez-Calvo R, Coll T, Laguna JC, Davidson MM, Chan TO, Feldman AM, Vazquez-Carrera M. Tnf-alpha reduces pgc-1alpha expression through nf-kappab and p38 mapk leading to increased glucose oxidation in a human cardiac cell model. Cardiovasc Res. 2009;81:703–712. doi: 10.1093/cvr/cvn327. [DOI] [PubMed] [Google Scholar]
- 156.Garnier A, Fortin D, Delomenie C, Momken I, Veksler V, Ventura-Clapier R. Depressed mitochondrial transcription factors and oxidative capacity in rat failing cardiac and skeletal muscles. J Physiol. 2003;551:491–501. doi: 10.1113/jphysiol.2003.045104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Lin J, Wu PH, Tarr PT, Lindenberg KS, Pierre J, St, Zhang CY, Mootha VK, Jager S, Vianna CR, Reznick RM, Cui L, Manieri M, Donovan MX, Wu Z, Cooper MP, Fan MC, Rohas LM, Zavacki AM, Cinti S, Shulman GI, Lowell BB, Krainc D, Spiegelman BM. Defects in adaptive energy metabolism with cns-linked hyperactivity in pgc-1alpha null mice. Cell. 2004;119:121–135. doi: 10.1016/j.cell.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 158.Arany Z, He H, Lin J, Hoyer K, Handschin C, Toka O, Ahmad F, Matsui T, Chin S, Wu PH, Rybkin II, Shelton JM, Manieri M, Cinti S, Schoen FJ, Bassel-Duby R, Rosenzweig A, Ingwall JS, Spiegelman BM. Transcriptional coactivator pgc-1 alpha controls the energy state and contractile function of cardiac muscle. Cell Metab. 2005;1:259–271. doi: 10.1016/j.cmet.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 159.Leone T, Lehman J, Finck B, Schaeffer PJ, W A, Boudina S, Courtois M, Wozniak DF, Sambandam N, Bernal-Mizrachi C, Chen Z, Holloszy JO, Medeiros DM, Schmidt RE, Saffitz JE, Abel ED, Semenkovich CF, Kelly DP. Pgc-1alpha deficiency causes multi-system energy metabolic derangements: Muscle dysfunction, abnormal weight control and hepatic steatosis. PLOS. 2005;3:e101. doi: 10.1371/journal.pbio.0030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Zhang Y, Huypens P, Adamson AW, Chang JS, Henagan TM, Boudreau A, Lenard NR, Burk D, Klein J, Perwitz N, Shin J, Fasshauer M, Kralli A, Gettys TW. Alternative mrna splicing produces a novel biologically active short isoform of pgc-1alpha. J Biol Chem. 2009;284:32813–32826. doi: 10.1074/jbc.M109.037556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Chang JS, Huypens P, Zhang Y, Black C, Kralli A, Gettys TW. Regulation of nt-pgc-1alpha subcellular localization and function by protein kinase a-dependent modulation of nuclear export by crm1. J Biol Chem. 2010;285:18039–18050. doi: 10.1074/jbc.M109.083121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Lehman JJ, Boudina S, Banke NH, Sambandam N, Han X, Young DM, Leone TC, Gross RW, Lewandowski ED, Abel ED, Kelly DP. The transcriptional coactivator pgc-1alpha is essential for maximal and efficient cardiac mitochondrial fatty acid oxidation and lipid homeostasis. Am J Physiol Heart Circ Physiol. 2008;295:H185–196. doi: 10.1152/ajpheart.00081.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Lelliott CJ, Medina-Gomez G, Petrovic N, Kis A, Feldmann HM, Bjursell M, Parker N, Curtis K, Campbell M, Hu P, Zhang D, Litwin SE, Zaha VG, Fountain KT, Boudina S, Jimenez-Linan M, Blount M, Lopez M, Meirhaeghe A, Bohlooly YM, Storlien L, Stromstedt M, Snaith M, Oresic M, Abel ED, Cannon B, Vidal-Puig A. Ablation of pgc-1beta results in defective mitochondrial activity, thermogenesis, hepatic function, and cardiac performance. PLoS Biol. 2006;4:e369. doi: 10.1371/journal.pbio.0040369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Sonoda J, Mehl IR, Chong LW, Nofsinger RR, Evans RM. Pgc-1beta controls mitochondrial metabolism to modulate circadian activity, adaptive thermogenesis, and hepatic steatosis. Proc Natl Acad Sci U S A. 2007;104:5223–5228. doi: 10.1073/pnas.0611623104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Vianna CR, Huntgeburth M, Coppari R, Choi CS, Lin J, Krauss S, Barbatelli G, Tzameli I, Kim YB, Cinti S, Shulman GI, Spiegelman BM, Lowell BB. Hypomorphic mutation of pgc-1beta causes mitochondrial dysfunction and liver insulin resistance. Cell Metab. 2006;4:453–464. doi: 10.1016/j.cmet.2006.11.00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.McClure TD, Young ME, Taegtmeyer H, Ning XH, Buroker NE, Lopez-Guisa J, Portman MA. Thyroid hormone interacts with pparalpha and pgc-1 during mitochondrial maturation in sheep heart. Am J Physiol Heart Circ Physiol. 2005;289:H2258–2264. doi: 10.1152/ajpheart.00473.2005. [DOI] [PubMed] [Google Scholar]
- 167.Portman MA, Xiao Y, Qian K, Tucker RL, Parish SM, Ning XH. Thyroid hormone coordinates respiratory control maturation and adenine nucleotide translocator expression in heart in vivo. Circulation. 2000;102:1323–1329. doi: 10.1161/01.cir.102.11.1323. [DOI] [PubMed] [Google Scholar]
- 168.Sadana P, Zhang Y, Song S, Cook GA, Elam MB, Park EA. Regulation of carnitine palmitoyltransferase i (cpt-ialpha) gene expression by the peroxisome proliferator activated receptor gamma coactivator (pgc-1) isoforms. Mol Cell Endocrinol. 2007;267:6–16. doi: 10.1016/j.mce.2006.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Davila-Roman VG, Vedala G, Herrero P, de las Fuentes L, Rogers JG, Kelly DP, Gropler RJ. Altered myocardial fatty acid and glucose metabolism in idiopathic dilated cardiomyopathy. J Am Coll Cardiol. 2002;40:271–277. doi: 10.1016/s0735-1097(02)01967-8. [DOI] [PubMed] [Google Scholar]
- 170.Meigs JB, Hu FB, Rifai N, Manson JE. Biomarkers of endothelial dysfunction and risk of type 2 diabetes mellitus. JAMA. 2004;291:1978–1986. doi: 10.1001/jama.291.16.1978. [DOI] [PubMed] [Google Scholar]
- 171.Ohara Y, Peterson TE, Harrison DG. Hypercholesterolemia increases endothelial superoxide anion production. J Clin Invest. 1993;91:2546–2551. doi: 10.1172/JCI116491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Won JC, Park JY, Kim YM, Koh EH, Seol S, Jeon BH, Han J, Kim JR, Park TS, Choi CS, Lee WJ, Kim MS, Lee IK, Youn JH, Lee KU. Peroxisome proliferator-activated receptor-gamma coactivator 1-alpha overexpression prevents endothelial apoptosis by increasing atp/adp translocase activity. Arterioscler Thromb Vasc Biol. 2010;30:290–297. doi: 10.1161/ATVBAHA.109.198721. [DOI] [PubMed] [Google Scholar]
- 173.Valle I, Alvarez-Barrientos A, Arza E, Lamas S, Monsalve M. Pgc-1alpha regulates the mitochondrial antioxidant defense system in vascular endothelial cells. Cardiovasc Res. 2005;66:562–573. doi: 10.1016/j.cardiores.2005.01.026. [DOI] [PubMed] [Google Scholar]
- 174.Olmos Y, Valle I, Borniquel S, Tierrez A, Soria E, Lamas S, Monsalve M. Mutual dependence of foxo3a and pgc-1alpha in the induction of oxidative stress genes. J Biol Chem. 2009;284:14476–14484. doi: 10.1074/jbc.M807397200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Schulz E, Dopheide J, Schuhmacher S, Thomas SR, Chen K, Daiber A, Wenzel P, Munzel T, Keaney JF., Jr. Suppression of the jnk pathway by induction of a metabolic stress response prevents vascular injury and dysfunction. Circulation. 2008;118:1347–1357. doi: 10.1161/CIRCULATIONAHA.108.784298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Kim HJ, Park KG, Yoo EK, Kim YH, Kim YN, Kim HS, Kim HT, Park JY, Lee KU, Jang WG, Kim JG, Kim BW, Lee IK. Effects of pgc-1alpha on tnf-alpha-induced mcp-1 and vcam-1 expression and nf-kappab activation in human aortic smooth muscle and endothelial cells. Antioxid Redox Signal. 2007;9:301–307. doi: 10.1089/ars.2006.1456. [DOI] [PubMed] [Google Scholar]
- 177.Chen Z, Peng IC, Cui X, Li YS, Chien S, Shyy JY. Shear stress, sirt1, and vascular homeostasis. Proc Natl Acad Sci U S A. 2010;107:10268–10273. doi: 10.1073/pnas.1003833107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Xiong S, Salazar G, San Martin A, Ahmad M, Patrushev N, Hilenski L, Nazarewicz RR, Ma M, Ushio-Fukai M, Alexander RW. Pgc-1 alpha serine 570 phosphorylation and gcn5-mediated acetylation by angiotensin ii drive catalase down-regulation and vascular hypertrophy. J Biol Chem. 2010;285:2474–2487. doi: 10.1074/jbc.M109.065235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Qu A, Jiang C, Xu M, Zhang Y, Zhu Y, Xu Q, Zhang C, Wang X. Pgc-1alpha attenuates neointimal formation via inhibition of vascular smooth muscle cell migration in the injured rat carotid artery. Am J Physiol Cell Physiol. 2009;297:C645–653. doi: 10.1152/ajpcell.00469.2008. [DOI] [PubMed] [Google Scholar]
- 180.Chawla A. Control of macrophage activation and function by ppars. Circ Res. 2010;106:1559–1569. doi: 10.1161/CIRCRESAHA.110.216523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Sebastiani M, Giordano C, Nediani C, Travaglini C, Borchi E, Zani M, Feccia M, Mancini M, Petrozza V, Cossarizza A, Gallo P, Taylor RW, d'Amati G. Induction of mitochondrial biogenesis is a maladaptive mechanism in mitochondrial cardiomyopathies. J Am Coll Cardiol. 2007;50:1362–1369. doi: 10.1016/j.jacc.2007.06.035. [DOI] [PubMed] [Google Scholar]
- 182.Garnier A, Zoll J, Fortin D, N'Guessan B, Lefebvre F, Geny B, Mettauer B, Veksler V, Ventura-Clapier R. Control by circulating factors of mitochondrial function and transcription cascade in heart failure: A role for endothelin-1 and angiotensin ii. Circ Heart Fail. 2009;2:342–350. doi: 10.1161/CIRCHEARTFAILURE.108.812099. [DOI] [PubMed] [Google Scholar]
- 183.Rohas LM, St-Pierre J, Uldry M, Jager S, Handschin C, Spiegelman BM. A fundamental system of cellular energy homeostasis regulated by pgc-1alpha. Proc Natl Acad Sci U S A. 2007;104:7933–7938. doi: 10.1073/pnas.0702683104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Duncan JG, Fong JL, Medeiros DM, Finck BN, Kelly DP. Insulin-resistant heart exhibits a mitochondrial biogenic response driven by the peroxisome proliferator-activated receptor-alpha/pgc-1alpha gene regulatory pathway. Circulation. 2007;115:909–917. doi: 10.1161/CIRCULATIONAHA.106.662296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Boudina S, Sena S, Theobald H, Sheng X, Wright JJ, Hu XX, Aziz S, Johnson JI, Bugger H, Zaha VG, Abel ED. Mitochondrial energetics in the heart in obesity-related diabetes: Direct evidence for increased uncoupled respiration and activation of uncoupling proteins. Diabetes. 2007;56:2457–2466. doi: 10.2337/db07-0481. [DOI] [PubMed] [Google Scholar]
- 186.Boudina S, Abel ED. Diabetic cardiomyopathy revisited. Circulation. 2007;115:3213–3223. doi: 10.1161/CIRCULATIONAHA.106.679597. [DOI] [PubMed] [Google Scholar]
- 187.Boudina S, Sena S, O'Neill BT, Tathireddy P, Young ME, Abel ED. Reduced mitochondrial oxidative capacity and increased mitochondrial uncoupling impair myocardial energetics in obesity. Circulation. 2005;112:2686–2695. doi: 10.1161/CIRCULATIONAHA.105.554360. [DOI] [PubMed] [Google Scholar]
- 188.Ahuja P, Zhao P, Angelis E, Ruan H, Korge P, Olson A, Wang Y, Jin ES, Jeffrey FM, Portman M, Maclellan WR. Myc controls transcriptional regulation of cardiac metabolism and mitochondrial biogenesis in response to pathological stress in mice. J Clin Invest. 2010;120:1494–1505. doi: 10.1172/JCI38331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Boudina S, Abel ED. Diabetic cardiomyopathy, causes and effects. Rev Endocr Metab Disord. 2010;11:31–39. doi: 10.1007/s11154-010-9131-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Willy PJ, Murray IR, Qian J, Busch BB, Stevens WC, Jr., Martin R, Mohan R, Zhou S, Ordentlich P, Wei P, Sapp DW, Horlick RA, Heyman RA, Schulman IG. Regulation of ppargamma coactivator 1alpha (pgc-1alpha) signaling by an estrogen-related receptor alpha (erralpha) ligand. Proc Natl Acad Sci U S A. 2004;101:8912–8917. doi: 10.1073/pnas.0401420101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Arany Z, Wagner BK, Ma Y, Chinsomboon J, Laznik D, Spiegelman BM. Gene expression-based screening identifies microtubule inhibitors as inducers of pgc-1alpha and oxidative phosphorylation. Proc Natl Acad Sci U S A. 2008;105:4721–4726. doi: 10.1073/pnas.0800979105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Luptak I, Balschi JA, Xing Y, Leone TC, Kelly DP, Tian R. Decreased contractile and metabolic reserve in peroxisome proliferator-activated receptor-alpha-null hearts can be rescued by increasing glucose transport and utilization. Circulation. 2005;112:2339–2346. doi: 10.1161/CIRCULATIONAHA.105.534594. [DOI] [PubMed] [Google Scholar]