Summary
Rv3671c, a putative serine protease, is crucial for persistence of M. tuberculosis in the hostile environment of the phagosome. We show that Rv3671c is required for M. tuberculosis resistance to oxidative stress in addition to its role in protection from acidification. Structural and biochemical analyses demonstrate that the periplasmic domain of Rv3671c is a functional serine protease of the chymotrypsin family and, remarkably, that its activity increases upon oxidation. High-resolution crystal structures of this protease in an active strained state and in an inactive relaxed state reveal that a solvent-exposed disulfide bond controls the protease activity by constraining two distant regions of Rv3671c and stabilizing it in the catalytically active conformation. In vitro biochemical studies confirm that activation of the protease in an oxidative environment is dependent on this reversible disulfide bond. These results suggest that the disulfide bond modulates activity of Rv3671c depending on the oxidative environment in vivo.
Keywords: acid stress, oxidative stress, persistence, serine protease, tuberculosis
Introduction
Mycobacterium tuberculosis presents a major threat to global health. This bacterium has infected one out of every three people worldwide with most individuals harboring M. tuberculosis in the latent, asymptomatic form. A significant fraction of latent infections (~5–10%) are expected to develop into active pulmonary disease (Kumar, 2007). M. tuberculosis evades the onslaught of the immune system by complex resistance mechanisms, which allow the pathogen to persist within the host and to multiply during disease. In macrophages that have been activated with the T cell–derived cytokine IFN-γ, M. tuberculosis is exposed to several potentially bactericidal defenses, including acidic pH (Schaible et al., 1998) and reactive oxygen and nitrogen intermediates (ROI and RNI) (MacMicking et al., 1997). However, M. tuberculosis can protect itself from this acidic environment by active mechanisms of pH homeostasis (Vandal et al., 2008).
Resistance against acidification depends on several proteins including a membrane protein encoded by Rv3671c (Vandal et al., 2008; Vandal et al., 2009). An M. tuberculosis mutant with its Rv3671c gene disrupted by a transposon insertion was hypersensitive to low pH (Vandal et al., 2008). This mutant failed to maintain a near neutral intrabacterial pH and lost viability both when exposed to acidified (pH 4.5) phosphate-citrate buffer or when residing within IFNγ-activated, acidified macrophages. Moreover, virulence of the Rv3671c mutant was severely attenuated in a mouse model, as displayed by its reduced growth during the acute phase of infection and failed persistence during the chronic phase (Vandal et al., 2008). Therefore, the Rv3671c protein might be a suitable target for development of novel agents effective against not only active but also latent tuberculosis.
Rv3671c is predicted to encode a membrane associated serine protease (Cole et al., 1998) that is one of 38 proteases conserved among M. leprae, M. bovis, M. avium paratuberculosis and M. tuberculosis (Ribeiro-Guimaraes and Pessolani, 2007). Secondary structure prediction of its N-terminal domain indicates the presence of four putative transmembrane helices that likely anchor the protease to the cytoplasmic membrane (Sonnhammer et al., 1998). This topology places the C-terminal protease domain of Rv3671c in the periplasm. The presence of a periplasmic space in M. tuberculosis was substantiated through cryo-electron microscopy studies, which revealed the presence of an outer membrane analogous to that in Gram-negative bacteria (Hoffmann et al., 2008; Zuber et al., 2008). The protease domain of Rv3671c contains the catalytic triad composed of His235, Asp264 and Ser343, conserved in the serine protease family (Fig. 1). A Ser343Ala mutant of Rv3671c failed to complement the acid hyper-susceptibility of the Rv3671c transposon mutant. This observation suggests that Rv3671c-mediated proteolysis is required for acid resistance of M. tuberculosis (Vandal et al., 2008).
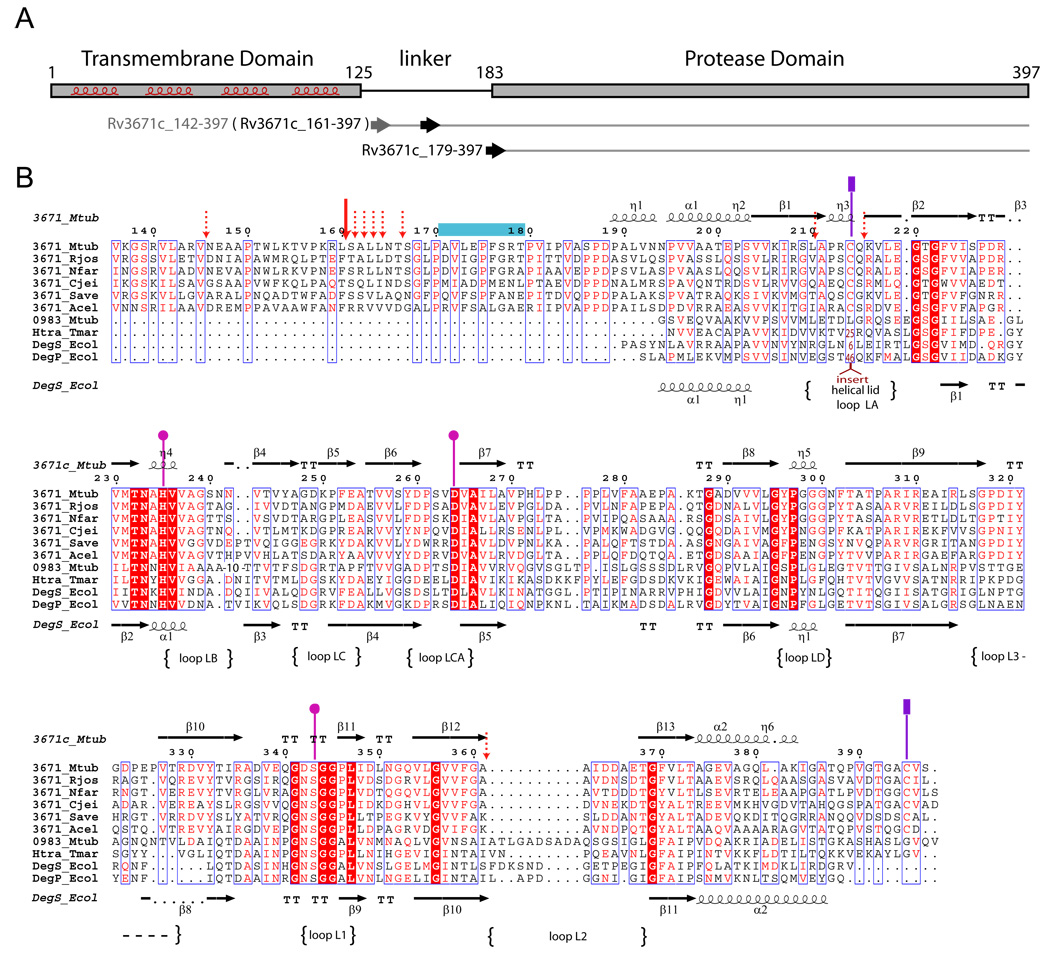
Figure 1.
A. Domain organization of the Rv3671c protein. The predicted transmembrane helices are highlighted. The horizontal arrows indicate the first residues of the recombinant protein constructs (residues 142 and 179) and the first residue (residue 161) of the autocleavage product of Rv3671c_142-397. B. Multiple sequence alignment of Rv3671c homologs from Rhodococcus jostii RHA1 (Rjos), Nocardia farcinica IFM 10152 (Nfar), Corynebacterium jeikeium K411 (Cjei), Streptomyces avermitilis MA-4680 (Save), Acidothermus cellulolyticus 11B (Acel) and four HtrA homologs: DegS and DegP from Escherichia coli K12 (Ecol), Rv0983 from Mycobacterium tuberculosis H37Rv (Mtub), HtrA from T. maritima (Tmar). The residues of the catalytic triad are designated by pink circles. The cysteine residues of the disulfide bond are marked by purple rectangles. The dominant and minor autocleavage sites (as determined by MALDI-MS analysis) are marked by the solid and dashed red arrows, respectively. The peptidic substrate mimic bound to the active site in the crystal is denoted by a blue horizontal bar. The loop names correspond to the serine protease nomenclature (Perona and Craik, 1995).
See also Figure S1.
Here, we demonstrate that Rv3671c is a functional serine protease. We report its crystal structures in active (bound to a substrate mimic) and inactive conformations. This structural and biochemical analysis reveals a labile intramolecular disulfide bond that regulates the proteolytic activity of Rv3671c in vitro, by stabilizing the protease active site in the substrate-bound conformation. We show that Rv3671c not only confers resistance to acidification, but it is also required to protect M. tuberculosis from oxidative stress.
Results
Rv3671c is a serine protease activated by oxidation
A BLAST (Altschul et al., 1990) search for homologous proteins indicates that M. tuberculosis Rv3671c is a putative serine protease of the chymotrypsin clan, with an additional N-terminal domain (Fig. 1). Neither Rv3671c nor its close relatives in phylogenetically related actinobacteria have been characterized. The nearest characterized sequence homologs of Rv3671c is HtrA from Thermotoga maritima (Kim et al., 2003). HtrA (high-temperature requirement A) family of serine proteases also include the well-characterized DegS protein (Sohn et al., 2007; Zeth, 2004). The protease domains of HtrA and Rv3671c exhibit approximately 20% sequence identity. However, HtrA proteases, unlike Rv3671c, contain one or more C-terminal PDZ domains, which regulate their protease activities by binding to regions of unfolded proteins in the periplasm (Krojer et al., 2008a; Schlieker et al., 2004; Sohn et al., 2007; Songyang et al., 1997; Walsh et al., 2003).
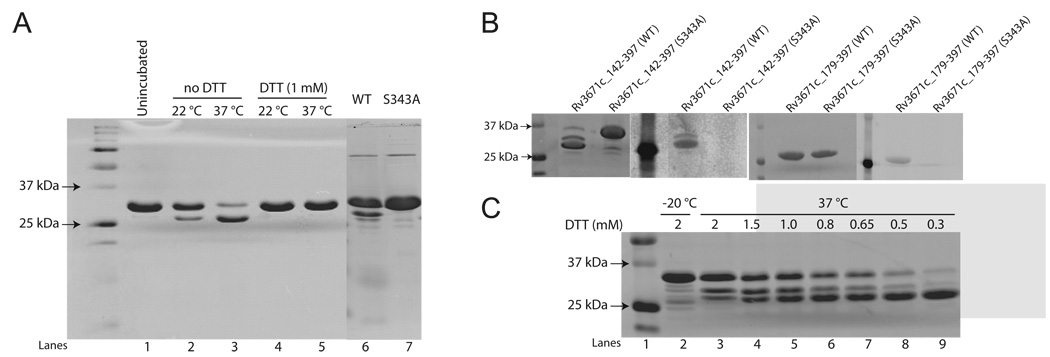
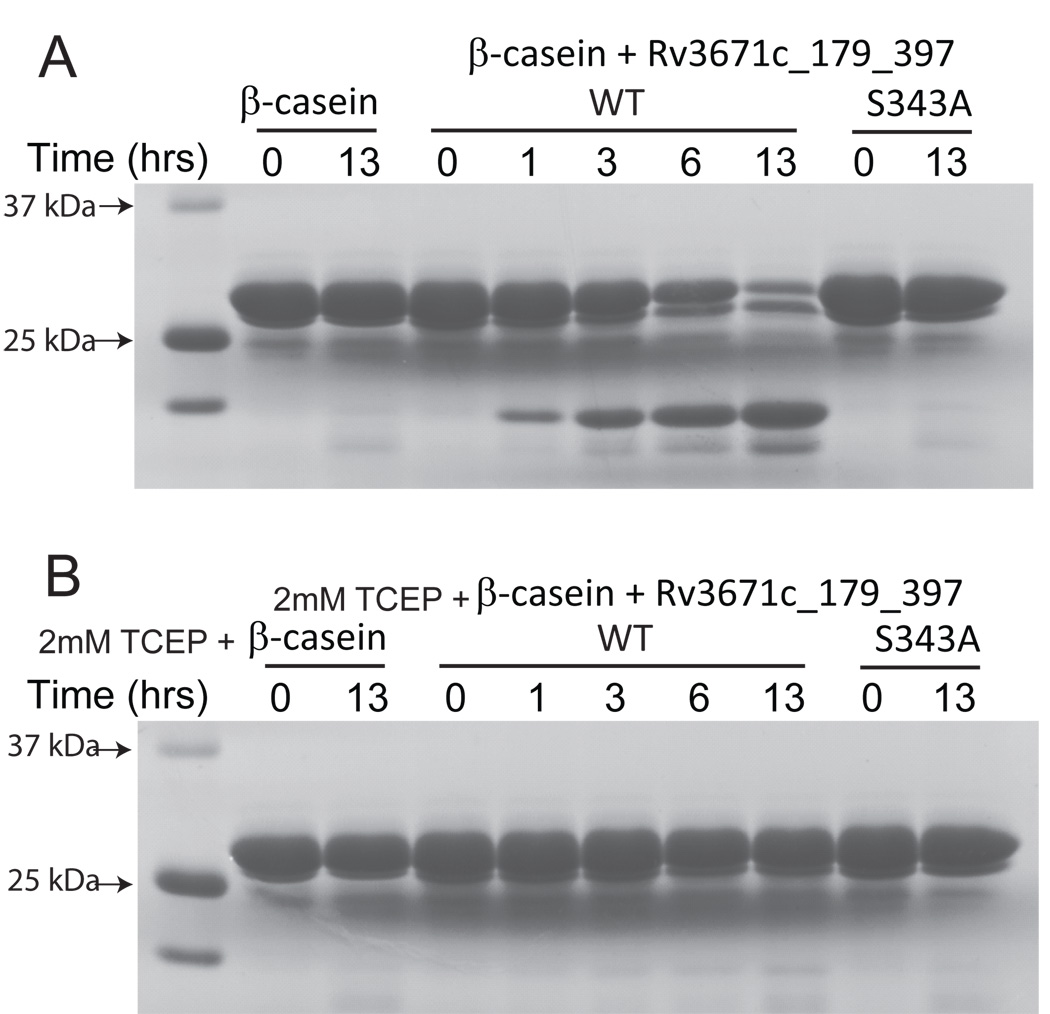
In order to understand the function and regulation of Rv3671c protease, we pursued expression in E. coli of both the full-length protein and the protease domain. Overexpression of the full-length protein was toxic to E. coli (not shown). In contrast, a domain (residues 142–397; Rv3671c_142-397) lacking the predicted transmembrane region was overexpressed successfully and purified in a soluble form (Fig. 2A). Rv3671c_142-397 is a monomer in solution, judged by its retention time in size-exclusion chromatography (Fig. S1A). It exhibited autoproteolytic activity, cleaving itself near its N-terminus (Fig. 2A). Protein identification by liquid chromatography-mass spectrometry (LC/MS/MS) revealed that autocleavage occurred after Leu160, between sequences PKRL and SALL (Fig. 1B). In addition to this dominant cleavage, minor autoproteolysis occurred at other sites, mostly in the vicinity of Leu160 (after residues Val145, Ser161, Ala162, Leu163, Leu164, Thr166) and at Ala361 (Fig. 1B). The mutant of the predicted catalytic residue, Ser343Ala, did not exhibit any autocleavage activity (Fig 2A, lane 7) confirming that the cleavage is auto-proteolytic and not due to a contaminating protease. Autoproteolysis of the wild-type protein was inhibited by PMSF, a known inhibitor of serine proteases (Fig. S1B). Moreover, the wild-type protein but not the Ser343Ala mutant reacted strongly with fluorophosphonate (FP)-TAMRA, a serine hydrolase-directed probe (Jessani et al., 2002; Patricelli et al., 2001) (Fig. 2B). Taken together, these data conclusively demonstrate that Rv3671c encodes a functional serine protease. Remarkably, the autocleavage activity of Rv3671c was significant only when no reducing agent was present in buffers. Since reducing agent was present during purification, the autoproteolytic activity of the purified protein became detectable only upon storage at 4 °C as the reducing agent degraded, or upon eliminating the reducing agent by buffer exchange. We observed concentration-dependent inhibition of autoproteolysis in such sample by reducing agents (DTT, β-mercaptoethanol, and TCEP) (Fig. 2C; inhibition by DTT shown as representative), indicating reversibility of the oxidation-dependent activation. Autocleavage of the protease domain did not occur during overexpression likely due to the reducing nature of the E. coli cytoplasm. In addition, we tested whether Rv3671c is able to cleave an independent substrate. Rv3671c displayed multiple turnover cleavage of β-casein and, similarly to autocleavage, this trans-cleavage activity was efficiently inhibited by a reducing agent (Fig. 3).
Figure 2.
Autocleavage activity of Rv3671c protease. A. A coomassie-stained 15% SDS-PAGE gel demonstrates the autocleavage activity of wild-type Rv3671c (residues 142 to 397) in the absence but not in the presence of 1 mM DTT. The S343A Rv3671c mutant (lane 7) does not exhibit autoproteolytic activity. B. SDS-PAGE showing covalent binding of FP-TAMRA to wild-type Rv3671c and not to the Ser343Ala mutant. 3 µg Rv3671c_142-397 (panels 1,2) and Rv3671c_179-397 (panels 3,4) and the respective S343A mutant proteins were incubated in the presence of 2 µM FP-TAMRA for 30 min and then separated on a 15% SDS PAGE. Panels 1 and 3 show the coomassie-stained gels and panels 2 and 4 show the fluorescent images. C. The inhibition of autoproteolysis of Rv3671c by DTT (data for other reducing agents are not shown). Wild-type Rv3671c_142-397 (2 µg) was incubated with the indicated amounts of DTT at 37 °C for 14 hrs and then separated on a 15% SDS-PAGE.
Figure 3.
Multiple turnover cleavage of β-casein. 10 µg β-casein were incubated for the indicated times with 1 µg Rv3671c_179-397 in the absence (A) and in the presence (B) of 2 mM TCEP. The Ser343Ala mutant of Rv3671c displayed no cleavage activity.
The structure of the Rv3671c protease
The dominant autocleavage product (residues 161–397) of Rv3671c_142-397 protease domain was crystallized in the absence of any reducing agent. No crystals were obtained if the crystallization solution contained a reducing agent. The crystal structure, determined and refined to 1.3 Ǻ resolution (Table I), reveals a monomeric protease with a chymotrypsin fold (Fig. 4A) consistent with its monomeric state in solution. As in other serine proteases, two β-barrel subdomains, each comprised of six antiparallel beta strands, contribute the catalytic triad of residues, His235 and Asp264 by one subdomain and Ser343 by the other.
Table I.
Data collection and refinement statistics for structures of Rv3671c.
| Rv3671c_161-397 | Rv3671c_179-397 | Rv3671c_179-397 S343A | |
|---|---|---|---|
| Data collection | |||
| Space group | P3121 | C2 | P1 |
| Monomers/a.u. | 1 | 2 | 2 |
| Cell dimensions | |||
| a, b, c (Å) | 64.7, 64.7, 98.7 | 119.1, 43.7, 71.5 | 42.6, 44.2, 53.4 |
| α, β, γ (°) | 90, 90, 120 | 90, 104.8, 90 | 101.7, 97.6, 105.1 |
| Resolution (Å) | 50.0-1.3 (1.35-1.3)a | 50.00-1.75 (1.81-1.75) | 50-2.1 (2.14-2.10) |
| Rmerge (%) | 6.7 (62.0) | 5.6 (31.5) | 6.4 (21.5) |
| I/σI | 24.7 (3.2) | 44.6 (3.4) | 22.6 (3.8) |
| Completeness | 99.5 (100) | 96.5 (69.9) | 95.0 (84.3) |
| Redundancy | 10.1 (6.7) | 6.9 (4.1) | 2.1 (1.5) |
| Refinement | |||
| Resolution (Å) | 30.0-1.3 (1.33-30) | 40.00-1.75 (1.80-1.75) | 40.0-2.1 (2.16-2.10) |
| No. reflections | 56042 | 32887 | 18853 |
| Rwork, % | 16.4 (23.8) | 18.8 (25.5) | 22.7 (26.5) |
| Rfree, % | 19.9 (26.5) | 23.9 (28.9) | 27.6 (30.5) |
| No. atoms/a.u. | 1999 | 3280 | 2984 |
| Average B-factors | |||
| Protein | 16.5b | 16c | 29.8c |
| Water | 36.2b | 41c | 44.5c |
| r.m.s. deviations | |||
| Bond length (Å) | 0.01 | 0.009 | 0.006 |
| Bond angle (°) | 1.454 | 1.184 | 0.926 |
| Ramachandran plotd: | |||
| % residues in the regions: | |||
| Most favorable | 89.9 | 90.8 | 88.7 |
| Additional allowed | 10.1 | 9.2 | 11.0 |
| Generously allowed | 0 | 0 | 0.3 |
| Disallowed | 0 | 0 | 0 |
The values for the highest-resolution shell are given in parentheses.
Anisotropic B-factor refinement
The B-factor values are exaggerated by TLS refinement.
PROCHECK output.
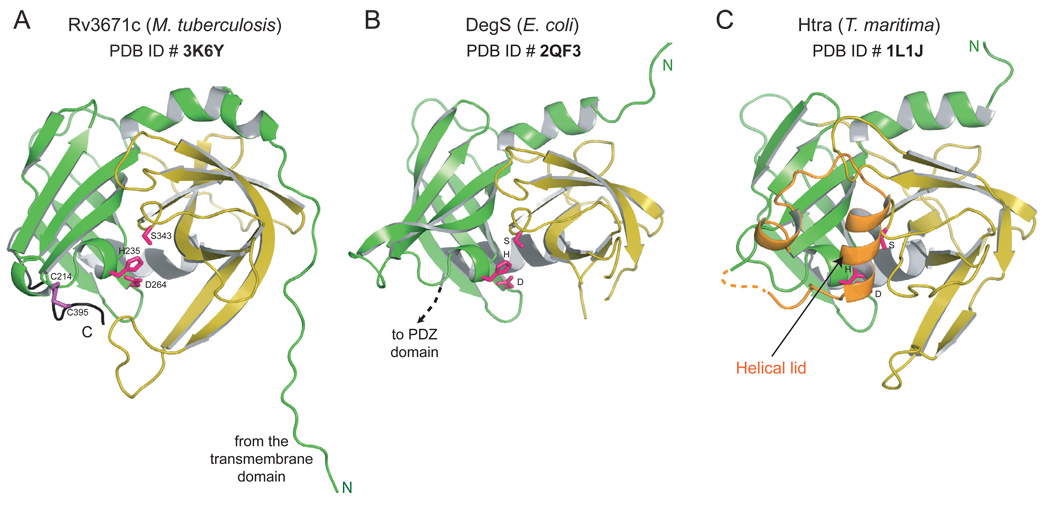
Figure 4.
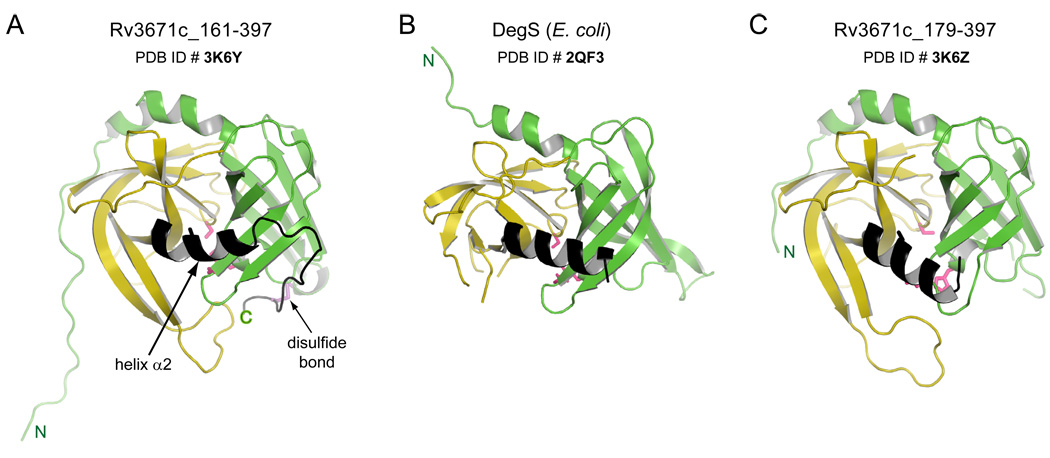
Structure of Rv3671c. Cartoon representations of A. the structure of Rv3671c B. DegS from E. coli (PDB ID: 2QF3) (Sohn et al., 2007) and C. HtrA from T. maritima (PDB ID: 1L1J) (Kim et al., 2003) are shown in similar orientation. The N- and C-terminal β-barrel subdomains are colored green and yellow, respectively. The catalytic residues and the disulfide bond forming cysteines of Rv3671c are shown as pink and purple sticks.
See also Figure S2.
Even though the β-barrel core folds of Rv3671c are most similar to the protease domains of DegS (Sohn et al., 2007, 2009; Zeth, 2004) (Fig. 4B) and HtrA from T. maritima (Kim et al., 2003) (Fig. 4C), Rv3671c differs from these proteins in several important respects. The homotrimeric T. maritima HtrA as well as E. coli DegP proteins contain an insertion in the protease fold (called the helical lid in T. maritima HtrA and LA loop in DegP; Fig. 4C and Fig. S2, respectively; also designated in Fig. 1). The helical lid of T. maritima HtrA is thought to block the active site of the same monomer (Kim et al., 2003; Kim et al., 2008) whereas the LA loop of one monomer of DegP functions by blocking the active site of another monomer in a higher-order assembly (Krojer et al., 2008b), in both cases resulting in inactivation of the protease. Rv3671c and DegS lack such an insertion. Another prominent distinguishing feature of Rv3671c from all HtrA proteins is its extreme C-terminal region (residues 384–397; colored black in Figs. 4A and 5A) that lacks secondary structure and wraps around the N-terminal β-barrel subdomain (residues 161–280; colored green in Figs. 4A and 5A). Strikingly, Cys395, the third residue from the C-terminus, forms a disulfide bond with Cys214 of the N-terminal subdomain. These two Cys residues are conserved in the Rv3671c family but not present in HtrA (Fig. 1B). Another noticeable feature of the Rv3671c structure is a kink in helix α2 (residues 374–383) preceding Cys395 (Fig. 5A) that marks a junction between an α-helix and a 310-helix. Helical kinks commonly occur at glycine residues (Rigoutsos et al., 2003); therefore, it is intriguing that this kink is located at Val377, a residue conserved in the Rv3671c family, and not at a nearby non-conserved Gly379. This kink might reflect a strain imposed by the Cys214-Cys395 disulfide bond since the homologous helix that precedes the PDZ domain in the HtrA family is straight (Fig. 5B).
Figure 5.
Cartoon representation of the back views of A. Rv3671c_161-397 (active state) B. DegS from E. coli and C. Rv3671c_179-397 (inactive state) are shown in similar orientations. The kink in helix α2 for Rv3671c_161-397 is indicated by the arrow. Corresponding α2 helices in all panels are colored in black.
Proteolytic activity of Rv3671c is enhanced in the presence of the Cys214-Cys395 disulfide
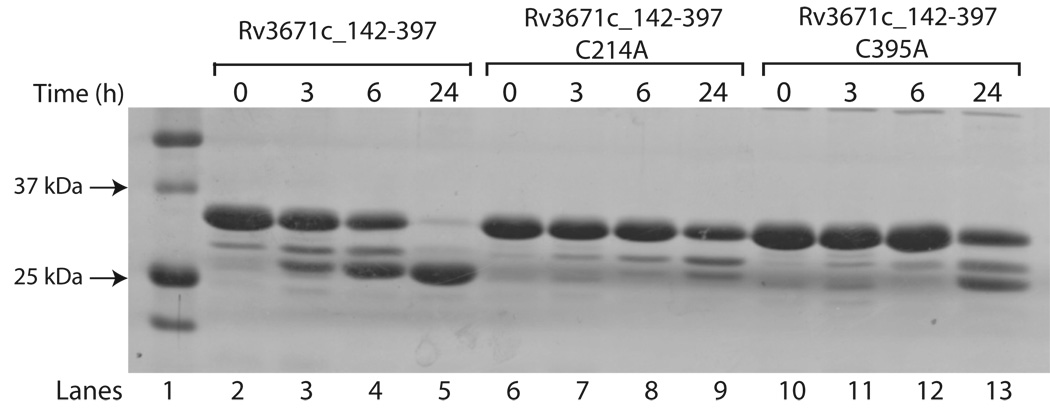
Rv3671c protease is inactivated in the presence of reducing agents. In order to test directly whether this inactivation is a result of breaking the disulfide bond, we measured proteolytic activity of Cys214Ala and Cys395Ala mutants. Either mutation resulted in a significant decrease of protease activity (Fig. 6). The Cys214-Cys395 disulfide linkage holds the protease in a strained state and appears to be required for its optimal activity. The considerable loss of activity in the presence of a reducing agent or as result of a Cys mutation is most consistent with a conformational equilibrium of the protease between active and inactive states in the absence of the disulfide bond. This equilibrium must be shifted to the active state upon disulfide bond formation. The disulfide bond formation itself is reversible as the reduced bond can be reformed upon oxidation (Fig. S3).
Figure 6.
Auto-cleavage of C214A and C395A mutants of Rv3671c. Autoproteolysis of C214A and C395A mutants of Rv3671c is severely impaired compared to the wild-type. The proteases were incubated at 37 °C for the indicated times and then separated on a 15% SDS-PAGE.
See also Figure S3.
The active site of Rv3671c
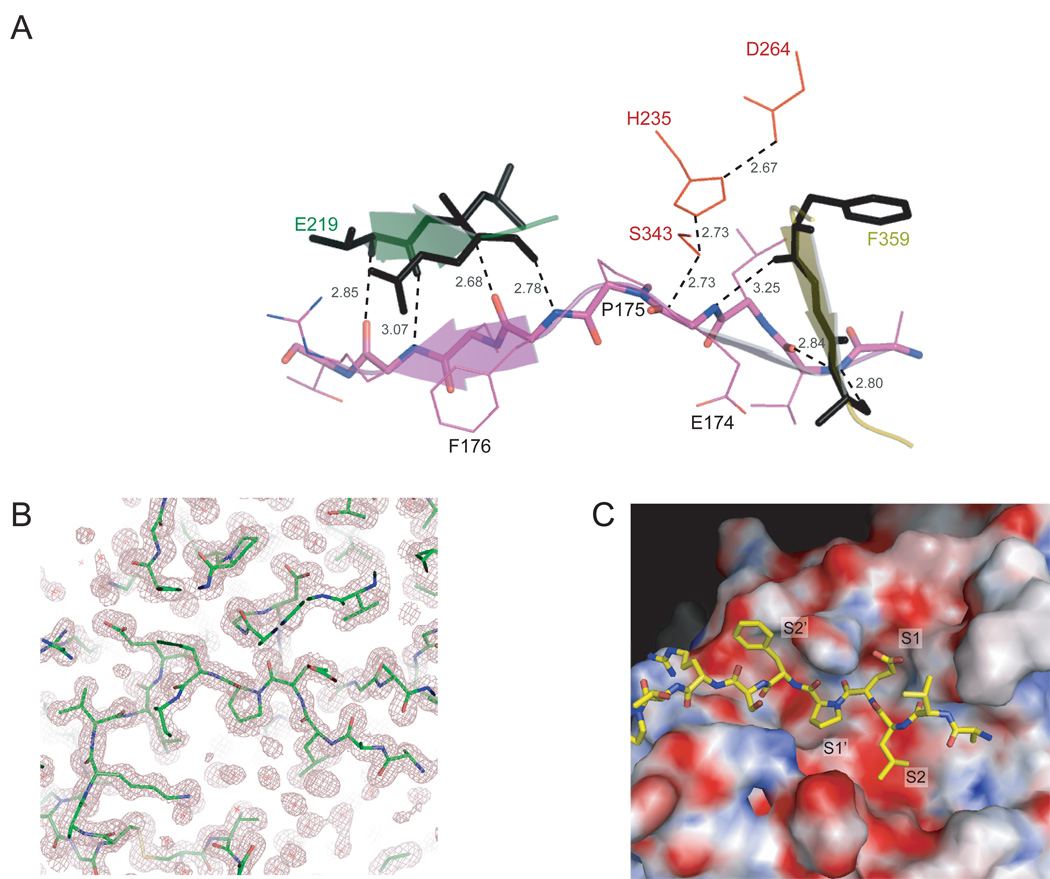
The active site of each Rv3671c monomer is bound to a peptidic stretch AVLEPFSRT (residues 171 to 179) provided by its crystal-packing neighbor. This peptide belongs to the linker region (residues 125 to 183) that connects the protease with the predicted transmembrane domain (Fig. 1B). Residues 161–183 of the linker region lack secondary structure for this protein construct. The mutual disposition of residues within the catalytic triad (His235, Asp264, Ser343) and the backbone of the bound peptide near the scissile bond match very well those in numerous structures of other serine proteases. These include classical structures of active trypsin and chymotrypsin (Birktoft and Blow, 1972; Huber et al., 1974), recently reported ultra-high resolution structures of serine proteases in the active state (Fuhrmann et al., 2004; Radisky et al., 2006) as well as the structure of the active state of DegS (Sohn et al., 2007). The P1 and P1’ positions of the active site (the nomenclature follows the convention in proteases (Schechter and Berger, 1967)) are occupied by residues Glu174 and Pro175, respectively (Fig. 7A, B). The amide bond between these residues is properly positioned for the nucleophilic attack but remains uncleaved in the structure. Serine proteases usually disfavor cleavage immediately before and after a proline (Bromme et al., 1986; Markert et al., 2003), presumably due to insufficient peptide backbone flexibility at the scissile bond. The distance between the Oγ atom of Ser343 and the carbonyl carbon of the Pro in the P1’ position is 2.73 Å thus representing the conformation of a binary complex prior to the nucleophilic attack by Ser343. The distance between the Nε2 atom of His235 and the Oγ atom of Ser343 is also 2.73 Å and that between the Oδ1 of Asp264 and the Nδ1 of His235 is 2.67 Å (Fig. 7A), indicating strong hydrogen bonds. The latter distance approaches that of a low-barrier hydrogen bond (LBHB; < 2.55 Å) predicted to be relevant for a transition state (Cleland and Kreevoy, 1994). However the hydrogen atom between the Oδ1 the Nδ1 is likely to be somewhat shifted towards the Nδ1 of His235 as observed directly for a very similar Oδ1-Nδ1 distance (2.68 Å) in the 0.98 Å-resolution structure of proteinase K from Tritirachium album limber (Betzel et al., 2001) and in the ultra-high 0.78 Å-resolution structure of subtilisin from Bacillus lentus (Kuhn et al., 1998), both also determined at acidic pH. In these structures, due to this hydrogen sharing, the catalytic Asp is thought to be only partially negatively charged or uncharged. Thus His235 appears to be positioned appropriately to serve as the general base. The negative charge on the carbonyl oxygen (the oxyanion) is likely stabilized by the main chain NH groups of conserved Gly341 and Ser343 (Fig. 7B). Indeed, the respective interatomic distances of 2.71 Å and 3.12 Å are consistent with hydrogen bond formation typical for a functional oxyanion hole. Therefore, this Rv3671c structure provides a high-resolution view of a serine protease in an active state bound to an uncleaved peptide substrate mimic.
Figure 7.
The active site of Rv3671c protease with a bound peptide. A. A mixed cartoon-stick view of the peptidic substrate mimic (residues 171–179 of the N-terminal region) bound in the catalytic pocket of Rv3671c protease. The residues of the catalytic triad are shown in red. The peptide backbone forms the β-sheet extensions (shown by the arrows) of the N-terminal and the C-terminal β-barrel subdomains. The hydrogen bonds to the peptide backbone and bonds relevant for catalysis are shown by the dashed lines, together with their distances in Ǻ. B. 2Fo-Fc electron density map (contoured at 1σ) of the active site demonstrates the bound peptide and the oxyanion hole. C. Surface electrostatic potential of the active site. Positively charged, negatively charged and hydrophobic regions are shown in blue, red and white, respectively.
The peptide substrate mimic bound to the active site of Rv3671c pinpoints the topography of the substrate-binding pockets (Fig. 7B). This peptide extends on both sides of the potential scissile bond, spanning residues from P4 to P5’ positions, representing a rare view of the peptidic part both downstream and upstream of the cleavage site within one structure (Fig. 7B). The region N-terminal to the scissile bond (residues Ala171 and Val172 of the substrate mimic) is hydrogen bonded to strand β12 of the C-terminal β-barrel subdomain and the region C-terminal to the scissile bond (residues Phe176, Ser177, Arg178) is bonded to strand β2 of the N-terminal β-barrel subdomain. Through these interactions, the backbone of the peptide substrate mimic extends the β-sheets of the core β-barrel sub-domains on both sides of the potential scissile bond (Fig. 7A).
The cavity size and the hydrophobic nature of S1 pocket (Fig. 7C) appear most consistent with housing a small or flexible hydrophobic side chain such as Leu, Val or Ala, but not bulkier Trp, Tyr or Phe residues, as found in elastase (Bode et al., 1986; Meyer et al., 1986). The dominant autocleavage observed after Leu160 and several minor cleavage sites occurring after Leu or Ala residues are in agreement with this pocket’s structure. The Glu residue of the peptide interacting with the fairly shallow S1 pocket is in a bent conformation such that its aliphatic part (Cβ-Cδ) contacts with the Cγ of conserved Val338 and with the Cβ atom of Ala361 whereas its carboxyl moiety is solvent-exposed. The S2 pocket is deep and negatively charged (at neutral pH) at the bottom (Fig. 7C) due to the catalytic Asp264. Thus this pocket is most consistent with a Lys or an Arg at the P2 position. The aliphatic part of such Lys or Arg would interact with the conserved Ile320 and Val396. Indeed, Arg159 is at the P2 position of the dominant cleavage site (Fig. 1). However, at the low pH of the crystallization solution (5.25), the negative charge of Asp264 must be largely neutralized by the strong salt bridge with the positively charged His235 (the Oδ1-Nδ1 distance is 2.67 Å). In the structure, this pocket is partially occupied by a Leu that is stabilized by hydrophobic interactions with Ile320 and Val396. The rest of the pocket is filled with water molecules, one of which makes a hydrogen bond with Asp264 (not shown). The residue at the P3 position is highly solvent-exposed and does not appear to make any specific contacts. The S4 pocket is shallow and entirely hydrophobic due to Leu315, Phe 359 and Phe370 and appears perfectly suited for a small hydrophobic residue such as Ala or Val. In the structure it is occupied by an Ala residue. The P1’, P2’ and P3’ residues (Pro, Phe and Ser) face the solvent and make limited contacts with the hydrophobic/weakly polar S2’/S3’ pockets. The residues in the P4’ and P5’ positions are located in shallow protein pockets and are even more highly solvent exposed (Fig. 7C). Therefore, there may not be specificity rules for the P1’ – P5’ positions. In summary, this structure suggests relaxed specificity with several specificity requirements for P1, P2 and P4 positions.
The structures of Rv3671c in an inactive conformation
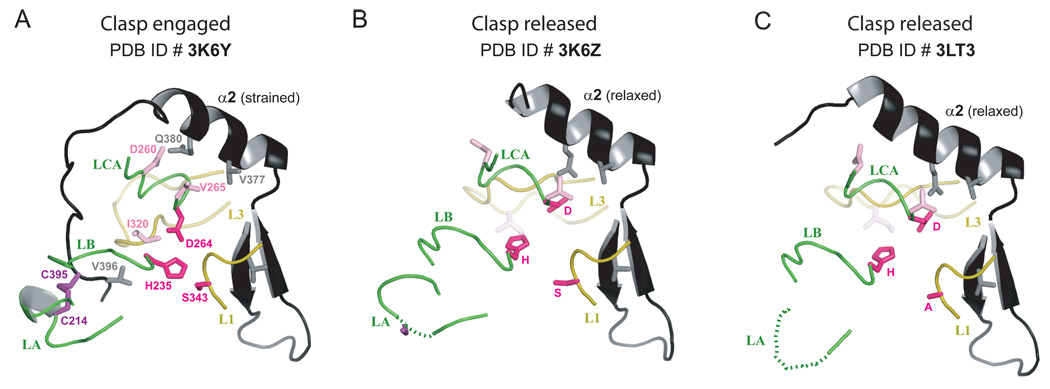
To investigate potential conformational plasticity of the active site pocket, we carried out crystallization trials of an Rv3671c construct that is further truncated at the N-terminus (residues 179–397; Rv3671c_179-397). This construct lacks the region mimicking a bound substrate in our earlier structure. Rv3671c_179-397 crystallized in a different crystal form (Table I). Its structure (Fig. 5C) was determined by molecular replacement using a pruned version of the Rv3671c_161-397 structure as a search model. Notably, two regions were not observed in the electron density map due to disorder: 1) loop LA (Fig. 1; residues 210–217) containing Cys214 and 2) residues 387–397 at the C-terminal region containing Cys395 (Fig. 8B). SDS-PAGE and MALDI-TOF analysis of the preparation of Rv3671c_179-397 used for crystallization revealed that this protein was proteolysed at multiple sites in loop LA (data not shown), explaining the absence of electron density in this region. No proteolysis in the disordered C-terminal region was observed. An incision in loop LA near Cys214 likely relieves the constraint on Cys395 and, therefore, must have a similar effect on the protein conformation to that produced by breaking the disulfide bond. Releasing Cys395 resulted in the increased conformational freedom of the C-terminus and in relaxing the strained active conformation of helix α2 observed in the previous structure (Fig. 5A, C). Helix α2 became straight and fully α-helical. Comparison of the strained and relaxed conformation of Rv3671c demonstrates that the C-terminal region of this protein acts as a clasp by embracing regions containing residues of the catalytic triad (Fig. 8A). Thus, the effects of proteolysis and, by inference, the breaking of the disulfide bond, are expected to be transmitted to the active site. We observed conformational changes of several protein loops (Fig. 8B) and, most notably, disarrangement of all three catalytic triad residues from their optimal positions seen in the previous structure (Fig. 8A). Such mispositioning of the active site residues signifies an inactive state of the protease. Because the inactive conformation of the protease active site is unoccupied, closing-opening of the clasp and the coupled conformational changes of the loops including the order-disorder transitions of residues 210–219 and the C-terminus (residues 387–397), suggest a conformational equilibrium that is shifted towards the inactive state in the absence of the bound substrate when the disulfide bond is reduced. The likely role of the disulfide bond is thus to stabilize the active conformation of the protease.
Figure 8.
Cartoon representations of the two conformations of the clasp (in black) in the three Rv3671c structures. Protein loops (loop LA: residues 210–218, loop LB: residues 235–242, loop LCA: residues 258–265, loop L3: residues 316–328 and loop L1: residues 342–346) and the catalytic triad residues are highlighted, demonstrating their concerted conformational changes. A. The active (strained) state of the protease with the clasp engaged, as observed in the structure of Rv3671c_161-397. B,C. The inactive (relaxed) state of the protease with the clasp released, as observed in the structures of Rv3671c_179-397 (panel B) and its Ser343Ala mutant (panel C).
Because it is possible that the inactive state could be an artifact of proteolysis unrelated to substrate binding or formation of the disulfide, we determined a crystal structure of the proteolytically inactive Ser343Ala mutant of Rv3671c_179-397. This protein crystallized in yet a different crystal form from those of the two above structures. Despite the lack of self-cleavage, the conformation of Rv3671c_179-397_S343A very closely resembles the inactive conformation of the wild-type Rv3671c_179-397 (Fig. 8C). Similar disorder of loop LA and the C-terminal region is observed. As seen in the structure of Rv3671c_179-397, the disulfide bond is absent in the electron density of Rv3671c_179-397_S343A. In both structures, straightening of the C-terminal helix α2 prevents Cys395 from approaching Cys214 for disulfide bond formation, even if the disordered C-terminal stretch (residues 389–395) could adopt a fully extended conformation. Therefore, the disulfide bond must be absent in this conformation. Consistent with this structural observation, Rv3671c_179-397 Ser343Ala protein does not readily form a disulfide upon oxidation (Fig. S3). Therefore, the inactive conformation is favored in the reduced form of the protease, in the substrate-unbound state. This structure provides strong evidence that the conformational state of the protease is tightly coupled to the state of the disulfide. In the inactive conformation the protease active site is not occupied by a peptide. Therefore, closing of the clasp and the coupled conformational changes of the loops (Fig. 8) including the disorder-order transitions of residue ranges 210–219 and 387–397, support our model of the disulfide bond stabilizing the active site in the active conformation required for substrate binding and catalysis.
Structural details of the conformational transition from the inactive to the active state
The structures of Rv3671c in the inactive and the active states (Fig. 8) demonstrate that the effect of the clasp closure is propagated to the active site region through direct contacts between the C-terminal region (residues 389–397) and the protease loops. Notably, this region of the closed clasp interacts with loop L3, which in turn interacts with loop L2, resulting in dramatic movements of these loops (Fig. 8). Loop L2 sets up the S1 pocket by positioning its Ala361. Loop L3 contacts the short β-hairpin that contains the catalytic Asp264. In addition, the C-terminal region appears to push loop LB (238–242), which likely repositions the catalytic His235, located in the immediate vicinity.
Our data suggest that these conformational changes represent an equilibrium process when the disulfide bond is reduced. The disulfide bond formation shifts this equilibrium towards the active state. Substrate binding likely also shifts the equilibrium to the active state. Under oxidative conditions, substrate binding and turnover are expected to promote the disulfide bond formation. Consistently with this model, we observe that the formation of the disulfide bond occurs more readily in the wild type Rv3671c than it does in its Ser343Ala mutant (Fig. S3). Even though the mechanism of DegS activation is different in that it involves the relief of protease inhibition by the PDZ domain (absent in Rv3671c) upon its binding to a regulatory OMP peptide (Sohn et al., 2007, 2009; Sohn and Sauer, 2009), DegS is also activated by binding to its substrate, RseA (Sohn and Sauer, 2009). In addition, in the PDZ-containing HtrA proteins, loops L2 and L3 also contain major regulatory structural determinants.
The M. tuberculosis Rv3671c mutant is hypersensitive to oxidative damage
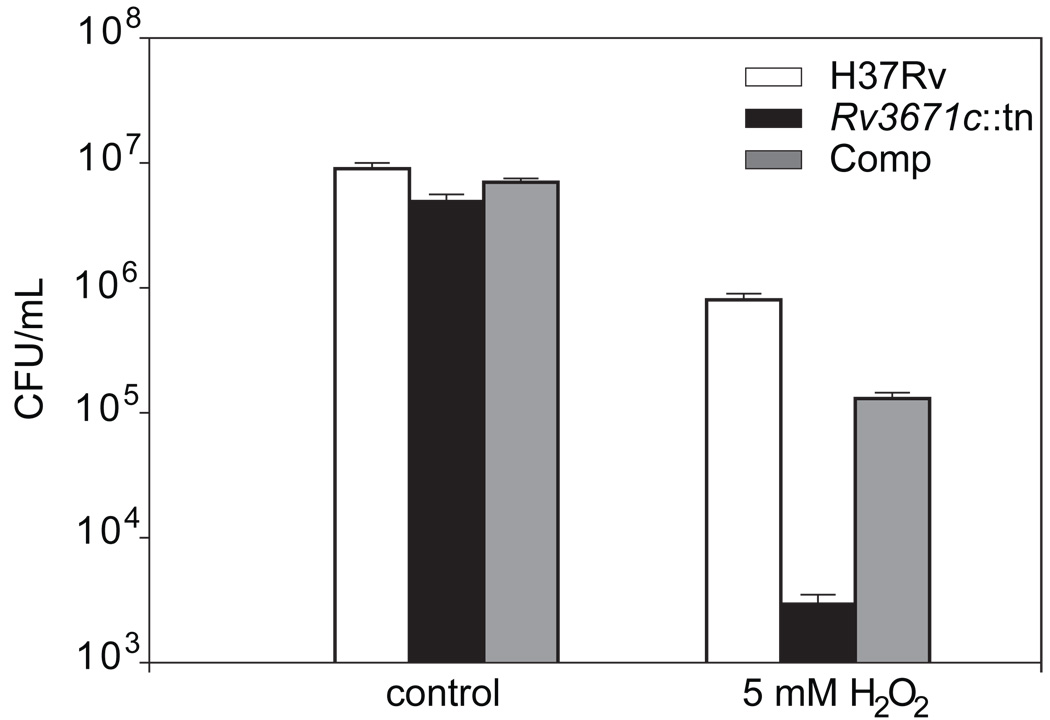
The biochemical and structural data demonstrated that the purified Rv3671c protease is selectively active in an oxidative environment. This suggests that Rv3671c might be important during exposure to oxidative stress. To test whether the Rv3671c transposon mutant displays increased sensitivity to oxidative stress, we exposed the wild-type, the mutant and the Rv3671c-complemented strains to H2O2 at pH 7. Viability of the Rv3671c mutant was reduced over 100-fold compared to that of wild-type M. tuberculosis upon exposure to H2O2 (Fig. 9). When complemented with a copy of the wild-type gene, the Rv3671c mutant survived similarly to wild-type M. tuberculosis. These results indicate that Rv3671c protects M. tuberculosis not only against acidification but also against oxidative stress.
Figure 9.
Hypersensitivity of the Rv3671c mutant to oxidative damage. The CFU count of wild type M. tuberculosis, the Rv3671c transposon mutant and complemented mutant was determined in the absence of H2O2 (control) or after incubation with 5 mM H2O2 for 2 hr. In each case, mean ± standard deviation values are calculated from triplicate independent cultures.
Discussion
The Rv3671c protein protects M. tuberculosis against both acidic and oxidative stress encountered in the macrophage phagosome. We demonstrate that Rv3671c is proteolytically active and this activity depends on its active site residue, Ser343. Earlier we showed that the Ser343Ala mutant of Rv3671c failed to rescue the M. tuberculosis Rv3671c transposon mutant from its hypersensitivity to acidic stress (Vandal et al., 2008). Taken together, these observations indicate that the proteolytic activity of Rv3671c is indispensable for protection against acid. Strikingly, the activity of Rv3671c in vitro increases significantly upon oxidation which promotes formation of a disulfide bond between two conserved solvent-exposed Cys residues. E. coli HtrA, a protein homologous to Rv3671c involved in bacterial heat shock response, contains a disulfide bond that appears to be important for stability of its tertiary structure and the oligomerization state (Skorko-Glonek et al., 2003). Formation of the disulfides in this and other bacterial proteins is commonly mediated by DsbA, whose oxidizing potential is regenerated by DsbB (Nakamoto and Bardwell, 2004). Even though M. tuberculosis lacks a homolog of DsbB, it contains a homolog of eukaryotic VKOR, which has been shown to restore disulfide bond formation in DsbB-deficient E. coli and proposed to have DsbB functions in bacteria lacking DsbB (Dutton et al., 2008). Formation of disulfide bonds in proteins is thought to be important for stabilizing their native globular fold. Once formed, the disulfide bonds are constrained by other elements of the protein structure and thus unreactive. Classical protein folding studies demonstrate that disulfide bonds in trypsin are critical for kinetic control of refolding as well as for structural stability (Epstein and Anfinsen, 1962; Liener, 1957), whereas in the active trypsin, none of these six disulfide bonds can be reduced with DTT at concentrations up to 10 mM (Sondack and Light, 1971). Rv3671c is different in that its overall fold appears to be independent of the integrity of the highly labile and reversible disulfide bond whereas the proteolytic activity is strongly enhanced by its presence. Indeed, the Cys214Ala and Cys395Ala mutants of Rv3671c behave similarly to the wild-type protein (with or without reducing agent) on the size exclusion column (data not shown) and retain some proteolytic activity indicating that the protein fold is not globally destabilized upon disulfide bond reduction. Moreover, the global fold of Rv3671c is essentially the same in the three structures reported here. One of these structures represents the substrate-bound, active state of the protease containing the disulfide bond and the other two structures represent the unbound, inactive state, in which this bond is not present. It remains to be determined whether the formation of this disulfide bond in vivo is constitutive, regulated in response to stress (oxidation and acidification) or regulated by a protein factor such as VKOR.
HtrA proteases play an important role in protecting E. coli and other bacteria from environmental and cellular stress (Clausen et al., 2002). E. coli DegS and DegP proteases are involved in stress response through their allosteric activation upon binding of unfolded outer membrane proteins to their PDZ domains and upon substrate binding to the protease (Krojer et al., 2008b; Schlieker et al., 2004; Sohn et al., 2007; Sohn and Sauer, 2009; Walsh et al., 2003). M. tuberculosis contains three HtrA proteases (Cole et al., 1998). Trimeric HtrA2 is required for full virulence of M. tuberculosis in mice and can function both as a protease and a chaperone. A crystal structure of M. tuberculosis HtrA2 (Mohamedmohaideen et al., 2008) revealed that unlike DegP, but similarly to DegS and Rv3671c, it lacks the LA loop. The activity of M. tuberculosis HtrA2 is probably controlled through peptide binding to the PDZ domain, similarly to DegS, however the structural details of this control may be distinct (Mohamedmohaideen et al., 2008). Unlike HtrA, Rv3671c does not form oligomers and likely does not have a chaperone function. In addition, Rv3671c lacks a PDZ domain and does not contain an LA loop similar to that of DegP. Thus, Rv3671c’s mechanism of activation differs from those of known HtrA proteins. Indeed, this study demonstrates that distinct conserved features unique to the Rv3671c family that are responsible conformational changes that are coupled to its transition from the inactive to the active state. However, it is likely that substrates act as activators of Rv3671c by stabilizing its active state, similarly to substrate activation of DegS (Sohn and Sauer, 2009) and probably other proteins in the broad HtrA family. Rv3671c exhibits autoproteolytic activity and the biological significance of this activity is currently under investigation. It is tempting to speculate that autocleavage releases the protease from its membrane tether to the periplasm in vivo.
Based on our structural studies, we propose that the redox-sensitive disulfide bond increases Rv3671c’s proteolytic activity by stabilizing the protease in the conformation in which the active site residues are properly positioned for substrate binding and catalysis. This conformational control is indirect in that it is exerted by a disulfide tethering the two β-barrel subdomains. The tether constrains in a clasp-like fashion several β-sheet and loop regions, including those containing active site residues. This active conformation of Rv3671c is strained as manifested by the bent helix α2. In contrast, in the inactive conformation, where the tether appears to be absent and the clasp disengaged, the C-terminal region (residues 384–397; a part of the clasp) is disordered and several regions of the protein close to the active site adopt a different conformation. The effects of these conformational changes propagate to the active site, where the catalytic triad residues are severely misaligned from their optimal positions. The helix that was bent is now straight, which signifies that the inactive state of Rv3671c is relaxed. Helical bends are associated with ligand binding or conformational transitions in other systems (Ivanov et al., 2007; Sreekanth et al., 2008).
Our structural and biochemical observations suggest that when the Cys thiols of Rv3671c are reduced the protease favors the inactive state, potentially to minimize degradation of other functionally active proteins. Changes in the oxidative state of the periplasm are therefore predicted to modulate the proteolytic activity of this protease.
The mechanism by which Rv3671c protects M. tuberculosis against acid and oxidative damage remains to be identified; however the homology of Rv3671c to HtrA might provide clues. HtrA and DegP are thought to degrade excess, misfolded or heat-denatured proteins in the periplasm of Gram-negative bacteria (Kim and Kim, 2005; Krojer et al., 2008a). Rv3671c may function similarly in degrading proteins unfolded due to acid and oxidative stresses. Consistent with this model, Rv3671c is able to degrade an unrelated protein (Fig. 3). We cannot exclude that, instead of being directly involved in a stress response, Rv3671c could be essential for maintenance of the cell wall integrity and, once deleted, renders the bacteria unable to maintain their internal pH. In addition to Rv3671c, we recently identified several other genes, each of which is required for protecting M. tuberculosis from both acid and oxidative stress (Vandal et al., 2009). Therefore it appears that defense against acid and oxidative stress can be achieved through a common pathway. For example, remodeling ion channels, proton pumps or membrane lipids could each cause reduced permeability of the membrane to both protons and reactive oxygen species and provide defense against both stresses. Finding specific molecular targets of the Rv3671c protease is a subject of future inquiry, which will shed light into this specific mechanism of protection of M. tuberculosis from acid and oxidative stress.
The dramatic defect in virulence of the M. tuberculosis lacking Rv3671c suggests that inhibitors targeting this protease might have chemotherapeutic potential. Our structural and biochemical studies provide a foundation for further mechanistic studies of Rv3671c that could facilitate structure-assisted design of inhibitors for this potential drug target.
Materials and Methods
Strains, media and measurement of sensitivity to hydrogen peroxide
M. tuberculosis strains were grown at 37°C in a humidified incubator with 5% C02 in Middlebrook 7H9 medium (Difco) containing 0.2% glycerol, 0.5% BSA, 0.2% dextrose, 0.085% NaCl and 0.05% Tween-80, or on Middlebrook 7H10 agar (Difco) containing 10% oleic acid-albumin-dextrose-catalase (OADC) (Becton Dickinson). Sensitivity to hydrogen peroxide was determined as reported (Vandal et al., 2009).
Expression and purification of Rv3671c
The truncated forms of Rv3671c, Rv3671c_142-397 and Rv3671c_179-397 were cloned into pET19bpps (Tsodikov et al., 2007) and pEt28a, respectively, yielding constructs bearing a polyhistidine tag. The S343A mutation was introduced into both constructs by PCR. All proteins were overexpressed in E. coli BL21 (DE3) and purified by using Ni2+ and size exclusion chromatography. The details of cloning, expression and purification can be found in Supplementary Information.
FP-TAMRA binding
Flourophosphonate-tamra (FP-TAMRA) was kindly provided by Dr. Benjamin Cravatt. 3 µg purified protease and 2 µM FP-tamra were combined in 20 mM Tris-HCl pH 7.4. Samples were incubated in the dark for 30 minutes at room temperature, boiled in SDS-containing sample buffer, resolved on a 15% SDS-PAGE and visualized using Coomassie stain and a fluorescent scanner (Odyssey Infrared Imaging System, LI-COR Biosciences).
Proteolysis assays
Autoproteolysis
Purified proteins at final concentration of 8 µM were incubated at 37°C in 20 mM Tris-HCl buffer pH 7.4. Aliquots were removed at indicated time points, boiled in SDS-containing sample buffer, resolved on a 15% SDS-PAGE and visualized using Coomassie stain. In the DTT titration experiment, the indicated amounts of DTT were added prior to incubation and the samples were incubated overnight. In the PMSF titration experiment, the indicated amounts of PMSF were added; the samples were incubated at 25 °C for 15 minutes, and then at 37 °C overnight.
Casein proteolysis
10 µg β-casein (Sigma) was incubated at 37 °C with 1 µg Rv3671c_179-397 in 50 mM Tris-HCl buffer pH 7.4 with and without 2 mM tris(2-carboxyethyl) phosphine (TCEP). Aliquots were removed and boiled in SDS-containing sample buffer at indicated time points, resolved on a 15% SDS-PAGE and visualized using Coomassie stain.
Maldi-MS Analysis
The gel bands corresponding to proteolytic products were excised, reduced with 10 mM DTT and alkylated with 55 mM iodoacetamide, and then digested with trypsin at 37°C overnight. The digestion products were analyzed by MALDI-TOF with a PerSeptive MALDI-TOF DE-STR mass spectrometer (Applied Biosystems) and LC-MS/MS analysis with the LTQ-Orbitrap mass spectrometer (Thermo). The masses of peptide ions from each gel band were analyzed and compared with the tryptic peptides from the whole length protein to determine the cleavage sites. The cleavage sites were also determined from MS/MS spectra generated from LC-MS/MS analysis by database searching and manual interpretation.
Crystallization, data collection and structure determination
All proteins were crystallized by vapor diffusion at 22 °C. Crystallization and cryoprotection is described in detail in the Supplementary Information. X-ray diffraction data were collected at the LS-CAT beamline at the Advanced Photon Source at the Argonne National laboratory at 100 K and processed using program HKL2000 (Otwinowski, 1997). The structures of all Rv3671c protease variants were determined by molecular replacement by using program Phaser (McCoy et al., 2007) as described in detail for each variant in Supplementary Information. The observed conformational differences between the structure of Rv3671c_161-397 and that of Rv3671c_179-397 are not caused by the different crystallization pH per se as the protease displays significant activity over a wide range of pH from 4.5 to 8.8 (Fig. S4). The structures were refined and rebuilt iteratively by using programs REFMAC (Murshudov et al., 1997) and COOT (Emsley and Cowtan, 2004), respectively. The data collection and refinement statistics are given in Table I. The structures of Rv3671c_161-397, Rv3671c_179-397 and Rv3671c_179-397 Ser343Ala were deposited into the Protein Data Bank (PDB ID’s are 3K6Y, 3K6Z and 3LT3, respectively).
Supplementary Material
Acknowledgements
We thank Eli Eisman for technical assistance with crystallization of Rv3671c_161-397, Dirk Schnappinger, Carl Nathan and Evette Radisky for helpful comments and Spencer Anderson and the staff of sector LS-CAT of the Advanced Photon Source at the Argonne National Laboratory for assistance with the collection of the diffraction data. This work was funded by NIH ROI AI081725 (S.E.). The Department of Microbiology and Immunology acknowledges the support of the William Randolph Hearst Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Betzel C, Gourinath S, Kumar P, Kaur P, Perbandt M, Eschenburg S, Singh TP. Structure of a serine protease proteinase K from Tritirachium album limber at 0.98 A resolution. Biochemistry. 2001;40:3080–3088. doi: 10.1021/bi002538n. [DOI] [PubMed] [Google Scholar]
- Birktoft JJ, Blow DM. Structure of crystalline -chymotrypsin. V. The atomic structure of tosyl- -chymotrypsin at 2 A resolution. J Mol Biol. 1972;68:187–240. doi: 10.1016/0022-2836(72)90210-0. [DOI] [PubMed] [Google Scholar]
- Bode W, Wei AZ, Huber R, Meyer E, Travis J, Neumann S. X-ray crystal structure of the complex of human leukocyte elastase (PMN elastase) and the third domain of the turkey ovomucoid inhibitor. Embo J. 1986;5:2453–2458. doi: 10.1002/j.1460-2075.1986.tb04521.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromme D, Peters K, Fink S, Fittkau S. Enzyme-substrate interactions in the hydrolysis of peptide substrates by thermitase, subtilisin BPN', and proteinase K. Arch Biochem Biophys. 1986;244:439–446. doi: 10.1016/0003-9861(86)90611-9. [DOI] [PubMed] [Google Scholar]
- Clausen T, Southan C, Ehrmann M. The HtrA family of proteases: implications for protein composition and cell fate. Mol Cell. 2002;10:443–455. doi: 10.1016/s1097-2765(02)00658-5. [DOI] [PubMed] [Google Scholar]
- Cleland WW, Kreevoy MM. Low-barrier hydrogen bonds and enzymic catalysis. Science. 1994;264:1887–1890. doi: 10.1126/science.8009219. [DOI] [PubMed] [Google Scholar]
- Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, 3rd, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- Dutton RJ, Boyd D, Berkmen M, Beckwith J. Bacterial species exhibit diversity in their mechanisms and capacity for protein disulfide bond formation. Proc Natl Acad Sci U S A. 2008;105:11933–11938. doi: 10.1073/pnas.0804621105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Epstein CJ, Anfinsen CB. The reversible reduction of disulfide bonds in trypsin and ribonuclease coupled to carboxymethyl cellulose. J Biol Chem. 1962;237:2175–2179. [PubMed] [Google Scholar]
- Fuhrmann CN, Kelch BA, Ota N, Agard DA. The 0.83 A resolution crystal structure of alpha-lytic protease reveals the detailed structure of the active site and identifies a source of conformational strain. J Mol Biol. 2004;338:999–1013. doi: 10.1016/j.jmb.2004.03.018. [DOI] [PubMed] [Google Scholar]
- Hoffmann C, Leis A, Niederweis M, Plitzko JM, Engelhardt H. Disclosure of the mycobacterial outer membrane: cryo-electron tomography and vitreous sections reveal the lipid bilayer structure. Proc Natl Acad Sci U S A. 2008;105:3963–3967. doi: 10.1073/pnas.0709530105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber R, Kukla D, Bode W, Schwager P, Bartels K, Deisenhofer J, Steigemann W. Structure of the complex formed by bovine trypsin and bovine pancreatic trypsin inhibitor. II. Crystallographic refinement at 1.9 A resolution. J Mol Biol. 1974;89:73–101. doi: 10.1016/0022-2836(74)90163-6. [DOI] [PubMed] [Google Scholar]
- Ivanov D, Tsodikov OV, Kasanov J, Ellenberger T, Wagner G, Collins T. Domain-swapped dimerization of the HIV-1 capsid C-terminal domain. Proc Natl Acad Sci U S A. 2007;104:4353–4358. doi: 10.1073/pnas.0609477104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessani N, Liu Y, Humphrey M, Cravatt BF. Enzyme activity profiles of the secreted and membrane proteome that depict cancer cell invasiveness. Proc Natl Acad Sci U S A. 2002;99:10335–10340. doi: 10.1073/pnas.162187599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DY, Kim DR, Ha SC, Lokanath NK, Lee CJ, Hwang HY, Kim KK. Crystal structure of the protease domain of a heat-shock protein HtrA from Thermotoga maritima. J Biol Chem. 2003;278:6543–6551. doi: 10.1074/jbc.M208148200. [DOI] [PubMed] [Google Scholar]
- Kim DY, Kim KK. Structure and function of HtrA family proteins, the key players in protein quality control. J Biochem Mol Biol. 2005;38:266–274. doi: 10.5483/bmbrep.2005.38.3.266. [DOI] [PubMed] [Google Scholar]
- Kim DY, Kwon E, Shin YK, Kweon DH, Kim KK. The mechanism of temperature-induced bacterial HtrA activation. J Mol Biol. 2008;377:410–420. doi: 10.1016/j.jmb.2007.12.078. [DOI] [PubMed] [Google Scholar]
- Krojer T, Pangerl K, Kurt J, Sawa J, Stingl C, Mechtler K, Huber R, Ehrmann M, Clausen T. Interplay of PDZ and protease domain of DegP ensures efficient elimination of misfolded proteins. Proc Natl Acad Sci U S A. 2008a;105:7702–7707. doi: 10.1073/pnas.0803392105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krojer T, Sawa J, Schafer E, Saibil HR, Ehrmann M, Clausen T. Structural basis for the regulated protease and chaperone function of DegP. Nature. 2008b;453:885–890. doi: 10.1038/nature07004. [DOI] [PubMed] [Google Scholar]
- Kuhn P, Knapp M, Soltis SM, Ganshaw G, Thoene M, Bott R. The 0.78 A structure of a serine protease: Bacillus lentus subtilisin. Biochemistry. 1998;37:13446–13452. doi: 10.1021/bi9813983. [DOI] [PubMed] [Google Scholar]
- Kumar V, Abbas AK, Fausto N, Mitchell RN. Robbins Basic Pathology. 8th ed. Saunders Elsevier; 2007. pp. 516–522. [Google Scholar]
- Liener IE. The essentiality of the disulfide linkages in trypsin. J Biol Chem. 1957;225:1061–1069. [PubMed] [Google Scholar]
- MacMicking J, Xie QW, Nathan C. Nitric oxide and macrophage function. Annu Rev Immunol. 1997;15:323–350. doi: 10.1146/annurev.immunol.15.1.323. [DOI] [PubMed] [Google Scholar]
- Markert Y, Koditz J, Ulbrich-Hofmann R, Arnold U. Proline versus charge concept for protein stabilization against proteolytic attack. Protein Eng. 2003;16:1041–1046. doi: 10.1093/protein/gzg136. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer EF, Jr, Radhakrishnan R, Cole GM, Presta LG. Structure of the product complex of acetyl-Ala-Pro-Ala with porcine pancreatic elastase at 1.65 A resolution. J Mol Biol. 1986;189:533–539. doi: 10.1016/0022-2836(86)90322-0. [DOI] [PubMed] [Google Scholar]
- Mohamedmohaideen NN, Palaninathan SK, Morin PM, Williams BJ, Braunstein M, Tichy SE, Locker J, Russell DH, Jacobs WR, Jr, Sacchettini JC. Structure and function of the virulence-associated high-temperature requirement A of Mycobacterium tuberculosis. Biochemistry. 2008;47:6092–6102. doi: 10.1021/bi701929m. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Nakamoto H, Bardwell JC. Catalysis of disulfide bond formation and isomerization in the Escherichia coli periplasm. Biochim Biophys Acta. 2004;1694:111–119. doi: 10.1016/j.bbamcr.2004.02.012. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Patricelli MP, Giang DK, Stamp LM, Burbaum JJ. Direct visualization of serine hydrolase activities in complex proteomes using fluorescent active site-directed probes. Proteomics. 2001;1:1067–1071. doi: 10.1002/1615-9861(200109)1:9<1067::AID-PROT1067>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Perona JJ, Craik CS. Structural basis of substrate specificity in the serine proteases. Protein Sci. 1995;4:337–360. doi: 10.1002/pro.5560040301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radisky ES, Lee JM, Lu CJ, Koshland DE., Jr Insights into the serine protease mechanism from atomic resolution structures of trypsin reaction intermediates. Proc Natl Acad Sci U S A. 2006;103:6835–6840. doi: 10.1073/pnas.0601910103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro-Guimaraes ML, Pessolani MC. Comparative genomics of mycobacterial proteases. Microb Pathog. 2007;43:173–178. doi: 10.1016/j.micpath.2007.05.010. [DOI] [PubMed] [Google Scholar]
- Rigoutsos I, Riek P, Graham RM, Novotny J. Structural details (kinks and non-alpha conformations) in transmembrane helices are intrahelically determined and can be predicted by sequence pattern descriptors. Nucleic Acids Res. 2003;31:4625–4631. doi: 10.1093/nar/gkg639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaible UE, Sturgill-Koszycki S, Schlesinger PH, Russell DG. Cytokine activation leads to acidification and increases maturation of Mycobacterium avium-containing phagosomes in murine macrophages. J Immunol. 1998;160:1290–1296. [PubMed] [Google Scholar]
- Schechter I, Berger A. On the size of the active site in proteases. I. Papain. Biochem Biophys Res Commun. 1967;27:157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- Schlieker C, Mogk A, Bukau B. A PDZ switch for a cellular stress response. Cell. 2004;117:417–419. doi: 10.1016/s0092-8674(04)00453-2. [DOI] [PubMed] [Google Scholar]
- Skorko-Glonek J, Zurawa D, Tanfani F, Scire A, Wawrzynow A, Narkiewicz J, Bertoli E, Lipinska B. The N-terminal region of HtrA heat shock protease from Escherichia coli is essential for stabilization of HtrA primary structure and maintaining of its oligomeric structure. Biochim Biophys Acta. 2003;1649:171–182. doi: 10.1016/s1570-9639(03)00170-5. [DOI] [PubMed] [Google Scholar]
- Sohn J, Grant RA, Sauer RT. Allosteric activation of DegS, a stress sensor PDZ protease. Cell. 2007;131:572–583. doi: 10.1016/j.cell.2007.08.044. [DOI] [PubMed] [Google Scholar]
- Sohn J, Grant RA, Sauer RT. OMP peptides activate the DegS stress-sensor protease by a relief of inhibition mechanism. Structure. 2009;17:1411–1421. doi: 10.1016/j.str.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn J, Sauer RT. OMP peptides modulate the activity of DegS protease by differential binding to active and inactive conformations. Mol Cell. 2009;33:64–74. doi: 10.1016/j.molcel.2008.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sondack DL, Light A. Comparative studies on the modification of specific disulfide bonds of trypsinogen and chymotrypsinogen. J Biol Chem. 1971;246:1630–1637. [PubMed] [Google Scholar]
- Songyang Z, Fanning AS, Fu C, Xu J, Marfatia SM, Chishti AH, Crompton A, Chan AC, Anderson JM, Cantley LC. Recognition of unique carboxyl-terminal motifs by distinct PDZ domains. Science. 1997;275:73–77. doi: 10.1126/science.275.5296.73. [DOI] [PubMed] [Google Scholar]
- Sonnhammer EL, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol. 1998;6:175–182. [PubMed] [Google Scholar]
- Sreekanth R, Pattabhi V, Rajan SS. Characterization of alpha helices interacting with nucleic acids. Comput Biol Chem. 2008;32:378–381. doi: 10.1016/j.compbiolchem.2008.06.002. [DOI] [PubMed] [Google Scholar]
- Tsodikov OV, Ivanov D, Orelli B, Staresincic L, Shoshani I, Oberman R, Scharer OD, Wagner G, Ellenberger T. Structural basis for the recruitment of ERCC1-XPF to nucleotide excision repair complexes by XPA. Embo J. 2007;26:4768–4776. doi: 10.1038/sj.emboj.7601894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandal OH, Pierini LM, Schnappinger D, Nathan CF, Ehrt S. A membrane protein preserves intrabacterial pH in intraphagosomal Mycobacterium tuberculosis. Nat Med. 2008;14:849–854. doi: 10.1038/nmXXXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandal OH, Roberts JA, Odaira T, Schnappinger D, Nathan CF, Ehrt S. Acid-susceptible mutants of Mycobacterium tuberculosis share hypersusceptibility to cell wall and oxidative stress and to the host environment. J Bacteriol. 2009;191:625–631. doi: 10.1128/JB.00932-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh NP, Alba BM, Bose B, Gross CA, Sauer RT. OMP peptide signals initiate the envelope-stress response by activating DegS protease via relief of inhibition mediated by its PDZ domain. Cell. 2003;113:61–71. doi: 10.1016/s0092-8674(03)00203-4. [DOI] [PubMed] [Google Scholar]
- Zeth K. Structural analysis of DegS, a stress sensor of the bacterial periplasm. FEBS Lett. 2004;569:351–358. doi: 10.1016/j.febslet.2004.06.012. [DOI] [PubMed] [Google Scholar]
- Zuber B, Chami M, Houssin C, Dubochet J, Griffiths G, Daffe M. Direct visualization of the outer membrane of mycobacteria and corynebacteria in their native state. J Bacteriol. 2008;190:5672–5680. doi: 10.1128/JB.01919-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.