Summary
The exon junction complex (EJC) is assembled on spliced mRNAs upstream of exon-exon junctions, and can regulate their subsequent translation, localization, or degradation. We isolated mutations in Drosophila mago nashi (mago), which encodes a core EJC subunit, based on their unexpectedly specific effects on photoreceptor differentiation. Loss of Mago prevents Epidermal growth factor receptor signaling, due to a large reduction in MAPK mRNA levels. MAPK expression also requires the EJC subunits Y14 and eIF4AIII, and EJC-associated splicing factors. Mago depletion does not affect the transcription or stability of MAPK mRNA, but alters its splicing pattern. MAPK expression from an exogenous promoter requires Mago only when the template includes introns. MAPK is the primary functional target of mago in eye development; in cultured cells, Mago knockdown disproportionately affects other large genes located in heterochromatin. These data support a nuclear role for EJC components in splicing a specific subset of introns.
Keywords: MAPK, EGFR, EJC, Mago, heterochromatin, splicing
Introduction
The exon junction complex (EJC) plays an important role in coupling nuclear and cytoplasmic events in gene expression; its recruitment allows nuclear pre-mRNA splicing to influence the subsequent fate of the spliced mRNAs (Tange et al., 2004). The EJC is assembled onto mRNAs during splicing, 20–24 bases upstream of each exon junction (Gehring et al., 2009a). The DEAD-box RNA helicase eIF4AIII is the first subunit to associate with pre-mRNA, through interactions with the intron-binding protein IBP160 (Gehring et al., 2009a; Ideue et al., 2007). eIF4AIII then recruits Magoh (known as Mago in Drosophila) (Boswell et al., 1991; Kataoka et al., 2001) and Y14 (Hachet and Ephrussi, 2001; Kataoka et al., 2000; Le Hir et al., 2000; Mohr et al., 2001), which stabilize eIF4AIII binding by inhibiting its ATPase activity (Andersen et al., 2006; Ballut et al., 2005; Bono et al., 2006). These three subunits constitute the pre-EJC; the fourth core subunit, MLN51 (Barentsz (Btz) in Drosophila) (Degot et al., 2004; van Eeden et al., 2001), is added after export of spliced mRNA to the cytoplasm (Gehring et al., 2009a; Herold et al., 2009). Many accessory proteins transiently interact with this core complex and modulate its function (Tange et al., 2004). The EJC remains bound to cytoplasmic mRNA until it is displaced by the ribosome-associated disassembly factor Pym during the first round of translation (Dostie and Dreyfuss, 2002; Gehring et al., 2009b).
The EJC has been shown to regulate post-transcriptional events that include mRNA localization, translation and degradation. In vertebrate cells, the presence of the EJC on spliced mRNAs increases their translational yield (Nott et al., 2004; Wiegand et al., 2003), in part by recruiting S6 Kinase 1 (Ma et al., 2008). The EJC is best known for its role in nonsense-mediated decay (NMD), a surveillance mechanism that degrades mRNAs containing premature termination codons (PTCs) (Chang et al., 2007). In mammals, NMD is greatly enhanced by the presence of a spliceable intron downstream of a PTC, and is mediated by the EJC and accessory factors that include three up-frameshift (UPF) proteins (Cheng et al., 1994; Stalder and Muhlemann, 2008; Thermann et al., 1998). However, NMD can occur independently of splicing or the EJC in lower organisms such as Drosophila (Gatfield et al., 2003). In Drosophila, the EJC has a role in mRNA localization; all four core EJC components are required to localize oskar mRNA to the posterior pole of the oocyte (Hachet and Ephrussi, 2001; Mohr et al., 2001; Newmark and Boswell, 1994; Palacios et al., 2004; van Eeden et al., 2001).
We isolated mutant alleles of mago based on their specific defects in Epidermal growth factor receptor (EGFR)-dependent processes in eye development. Phosphorylation of Mitogen-activated protein kinase (MAPK) is a critical step in signal transduction downstream of the EGFR and other receptor tyrosine kinases (Katz et al., 2007). Loss of mago strongly reduces the total level of the mRNA encoding Rolled (Rl), the Drosophila Extracellular signal-regulated kinase (ERK)-related MAPK. Y14 and eIF4AIII, the other two subunits of the pre-EJC, also positively regulate MAPK transcript levels, but Btz does not. An intronless MAPK cDNA is independent of mago and can rescue photoreceptor differentiation in mago mutant clones; inclusion of the introns renders it Mago-dependent. Mago does not affect MAPK transcription or mRNA stability, but alters its splicing pattern. MAPK is a large gene located in heterochromatin; a genome-wide survey of Mago-regulated genes found that genes that shared these features were over-represented. Based on these observations, we propose that the pre-EJC is essential to splice a specific set of transcripts that includes the critical signal transduction component MAPK.
Results
mago is required for EGFR signaling in eye and wing development
EGFR signaling plays a critical role in Drosophila eye development. Differentiation of regularly spaced clusters, each containing eight photoreceptor cells, progresses from posterior to anterior across the third instar larval eye imaginal disc, led by an indentation known as the morphogenetic furrow (MF). R8, the first photoreceptor to form in each developing cluster, induces EGFR activation in surrounding cells to promote their differentiation into R1–R7 photoreceptors (Fig. 1A)(Roignant and Treisman, 2009). In a genetic screen for mutations affecting photoreceptor differentiation (Janody et al., 2004), we isolated three alleles of mago nashi (mago) (Fig. 1B). In large clones of mago mutant cells in the eye disc, R8 differentiation, visualized using the marker Senseless (Sens), initiated correctly immediately posterior to the MF; however, few other photoreceptors were recruited (Fig. 1E, F). This phenotype resembles those reported for mutations in components of the EGFR pathway (Halfar et al., 2001; Yang and Baker, 2003).
Figure 1. mago is required for EGFR-dependent processes in the eye and wing discs.
(A) Photoreceptor differentiation proceeds from posterior (P) to anterior (A) across the eye disc. R8 cells differentiate first, immediately posterior to the MF, and produce the ligand Spi, which activates the EGFR in surrounding cells and induces the sequential formation of R1–R7 cells. (B) Sequence comparison of Drosophila Mago and human Magoh. Identical amino acids are in red. The amino acid changes in our three mago alleles are indicated. (C-N) third instar eye imaginal discs with anterior to the left. Photoreceptors are stained with anti-Elav (C, E, G, blue in D, F, H, J, L). R8 is stained with anti-Senseless (red in D, F, H). (C, D) wildtype. (E, F) large mago93D mutant clones generated in a Minute background are marked by the absence of GFP (green in F). R8 cells start to differentiate normally, but differentiation of other photoreceptors is impaired. (G, H) large mago93D mutant clones are marked by the absence of GFP (green in H) in discs expressing p35 in all cells posterior to the MF. Rescue of apoptosis in mago mutant clones does not restore R1–R7 differentiation. Insets show enlargements of the boxed regions; note the loss of photoreceptors other than R8 in (E-H). (I-N) mago93D clones are marked by the absence of GFP (green in J, L, N) and stained with anti-Caspase 3 (I, red in J), anti-Cyclin B (K, red in L), or anti-Pointed P1 (M, magenta in N). The mutant clones show increased apoptosis and increased Cyclin B expression (insets show enlargements of boxed regions), indicating a failure to arrest in G1, and do not express Pnt-P1. (O-R) wing discs expressing GFP (green in P) or containing mago93D clones marked by the absence of GFP (green in R). argos-lacZ expression revealed by anti-β-galactosidase staining (O, Q, magenta in P, R) is absent in mago mutant clones.
Loss of EGFR signaling also leads to apoptosis in the eye disc (Halfar et al., 2001; Yang and Baker, 2003). mago mutant clones strongly accumulated activated caspases, indicative of apoptosis (Fig. 1I, J). To test whether the lack of photoreceptor differentiation in mago mutant clones was simply a consequence of cell death, we blocked cell death in the eye disc by expressing the anti-apoptotic peptide p35 (Hay et al., 1994). This rescued the loss of R8 cells, but did not restore their ability to recruit additional photoreceptors (Fig. 1G, H). Like known components of the EGFR pathway, mago thus independently controls both photoreceptor differentiation and cell survival. A third function of EGFR signaling in the eye disc is to arrest differentiating photoreceptors in the G1 phase of the cell cycle. In the absence of EGFR signaling, re-entry of these cells into the cell cycle can be visualized by increased expression of Cyclin B, a marker of S and G2 phases (Yang and Baker, 2003). mago mutant clones accumulated Cyclin B in extra cells (Fig. 1K, L), indicating a failure of G1 arrest.
To further confirm a requirement for mago in EGFR signaling, we examined the expression of EGFR target genes. Expression of the transcription factor Pointed P1 (PntP1) is induced by EGFR signaling as photoreceptors initiate their differentiation just posterior to the MF; in mago mutant clones, PntP1 expression was lost (Fig. 1M, N). During wing development, EGFR signaling activates expression of the target gene argos in the wing vein primordia (Fig. 1O, P) (Golembo et al., 1996). argos expression was strongly reduced in mago mutant cells in the wing disc (Fig. 1Q, R). The requirement for mago for EGFR signaling in both eye and wing development suggests that it has a general function in this pathway. Its effect on the EGFR pathway appears quite specific, since the normal pattern of R8 differentiation would be incompatible with a role for mago in signaling by Hedgehog, Notch, or Wingless in the developing eye (Roignant and Treisman, 2009).
Mago acts downstream of Raf and upstream of MAPK activation
To determine the point at which mago acts in the EGFR signaling pathway, we performed epistasis experiments in the eye disc. Spitz (Spi) is the primary ligand that induces EGFR activation in R1–7; activated EGFR feeds into the Ras/MAPK pathway common to other receptor tyrosine kinases (Fig. 2M). The GTP-bound form of Ras activates the protein kinase Raf, initiating a kinase cascade in which Raf phosphorylates Downstream of raf1 (Dsor1 or MEK), which in turn phosphorylates MAPK. Phosphorylated MAPK enters the nucleus and phosphorylates specific transcription factors to regulate target gene expression. We expressed constitutively active forms of these components of the pathway specifically within mago mutant clones. Constitutively secreted Spi (Schweitzer et al., 1995), activated EGFR (Queenan et al., 1997), activated Ras (Karim and Rubin, 1998) and activated Raf (Martin-Blanco et al., 1999) all failed to induce photoreceptor differentiation in mago mutant clones (Fig. 2A-F, I, J), although each induced ectopic photoreceptors when expressed in wild-type cells (Fig. 2G, H) (Miura et al., 2006; Roignant et al., 2006). However, an activated form of MAPK, RolledSEM (Ciapponi et al., 2001), fully rescued the lack of photoreceptors in mago mutant cells (Fig. 2K-L). Similar epistasis experiments in the wing disc, using argos-lacZ to monitor pathway activation, likewise showed that only activated MAPK could induce argos expression in mago mutant cells (Fig. S1). The activity of mago is thus required downstream of Raf activation but upstream of MAPK activation.
Figure 2. mago acts downstream of Raf but upstream of phospho-MAPK in the eye disc.
(A-L) eye discs in which photoreceptors are stained with anti-Elav (A, C, E, G, I, K; magenta in B, D, F, H, J, L). Clones mutant for mago93D and/or expressing UAS transgenes are labeled by GFP expression (green in B, D, F, H, J, L) and indicated by green arrows in A, C, E, G. I, K. Expression of UAS- sSpi (A, B), UAS-EGFRλtop (C, D), UAS-RasV12 (E, F) or UAS-Raf179 (I, J) does not rescue photoreceptor differentiation in mago mutant clones. (G, H) Expression of UAS- Raf179 in wildtype clones leads to excessive photoreceptor differentiation. (K, L) Expression of UAS-RolledSEM induces excessive photoreceptor differentiation in mago93D clones. (M) a simplified diagram of the EGFR signaling pathway. See also Figure S1.
Mago is required to maintain MAPK levels sufficient for signaling
Since mago encodes a subunit of the EJC, we reasoned that it might control the expression of a component of the EGFR pathway. Indeed, we found that the levels of MAPK protein were strongly reduced in mago mutant clones in both the eye and wing discs (Fig. 3A-D). To determine the extent of the reduction, we compared protein extracts from wild-type eye discs expressing GFP in all cells and from eye discs containing large mago mutant clones marked by the absence of GFP. Quantification of MAPK on Western blots relative to GFP and Tubulin, to correct for the proportion of wild-type cells, showed that mago mutant cells expressed MAPK at only 16% of the wild-type level (Fig. 3E).
Figure 3. mago is required to maintain normal MAPK protein levels.
(A-D) Anti-MAPK staining (A, C, magenta in B, D) of eye discs (A, B) or wing discs (C, D) containing mago93D clones marked by the absence of GFP (green in B, D). MAPK protein levels are strongly reduced in mago mutant clones. (E) Western blot using protein extracts derived either from eye discs expressing GFP in all cells (WT) or from eye discs containing large mago93D clones lacking GFP. The ratio of GFP to Tubulin was used to quantify the amount of remaining wildtype tissue in mago93D mutant eye discs. MAPK levels are decreased by 84% when normalized to GFP. (F) MAPK and Tubulin staining of S2R+ cells treated with lacZ or mago dsRNA. MAPK is specifically reduced. (G) D2F cells treated with lacZ or mago dsRNA were incubated with sSpi conditioned media for 0 or 30 min. Protein lysates were blotted with antibodies to Tubulin, MAPK, diphospho-MAPK, MEK, and Mago. (H) S2R+ cells treated with lacZ or mago dsRNA were incubated with 25 µg/ml insulin for 0 or 10 min. Lysates were blotted with antibodies to Tubulin, MAPK and diphospho-MAPK. mago dsRNA reduced MAPK phosphorylation after sSpi or insulin treatment due to a decrease in total MAPK protein.
We next examined whether these results could be generalized to cultured Drosophila S2 and S2R+ cells, in which mago is expressed and can be knocked down by RNA interference (RNAi; Fig. 3G, H). Mago depletion in these cells resulted in a 75% reduction in MAPK protein levels in comparison to Tubulin, visible both on Western blots and in immunohistochemical stainings (Fig. 3F-H). This effect was specific, since MEK levels were not significantly reduced (Fig. 3G). As expected, the loss of MAPK protein strongly reduced the potency of EGFR signaling; MAPK phosphorylation induced by treatment of an S2 cell line that stably expresses the EGFR (D2F; (Schweitzer et al., 1995)) with media conditioned by cells expressing Spi (Miura et al., 2006) was strongly reduced in cells treated with mago RNAi (Fig. 3G). S2R+ cells treated with insulin to activate the endogenous Insulin receptor also showed reduced MAPK phosphorylation and MAPK protein levels when Mago was knocked down (Fig. 3H), supporting a general role for Mago in receptor tyrosine kinase signaling.
The other subunits of the pre-EJC also control MAPK levels
Mago is a subunit of the core EJC, which also includes three other proteins, Y14, Btz and eIF4AIII. To determine whether the effect of Mago on MAPK was due to its function within the EJC, we tested whether these other EJC subunits were also required for EGFR signaling and MAPK expression. Existing alleles of Y14/tsunagi (tsu), which have P element sequences inserted in the 5’UTR (Mohr et al., 2001) (Fig. 4A), did not affect photoreceptor differentiation (Fig. S2A, D). However, these alleles did not abolish Y14 protein expression (Fig. S2B, C, E, F). We therefore generated a Y14 null allele by imprecise excision of the P element tsu1. The tsuΔ18 deletion removed the entire Y14 coding sequence, resulting in the complete absence of detectable Y14 protein (Fig. 4A, Fig. S2H, I). Clones homozygous for tsuΔ18 strongly resembled mago mutant clones, showing both failure to differentiate R1–7 photoreceptors and extensive apoptosis (Fig. 4B-D and Fig. S2G). We prevented cell death in tsuΔ18 mutant clones using a mutation in dark, a gene necessary for apoptosis (Srivastava et al., 2007); as for mago, this allowed R8 survival, but did not rescue recruitment of other photoreceptors (Fig. 4E-G). Depletion of Y14 by RNAi in S2R+ cells also reduced MAPK levels (Fig. 4K). Mago and Y14 thus appear to act together to maintain MAPK protein levels sufficient for EGFR signaling.
Figure 4. mago, Y14 and eIF4AIII but not btz are required for MAPK expression and function.
(A) Genomic structure of Y14, showing the coding regions (black), UTRs (white) and the position of the tsu1 P element and tsu5 allele. The tsuΔ18 deletion removes the whole Y14 open reading frame without disrupting the adjacent gene Mys-45. (B-J) Third instar eye discs containing large clones marked by the absence of GFP (green in D, G, J) homozygous for the Y14 null allele tsuΔ18 (B-D); for tsuΔ18, darkN28 (E-G); and for btz2 (H-J). Photoreceptors are stained with anti-Elav (B, E, H; blue in D, G, J) and R8 is stained with anti-Sens (C, F, I; red in D, G, J). Arrows in D, G point to clusters containing only R8. Like mago, Y14 is required independently for both photoreceptor differentiation and cell survival, but btz is not necessary for either. (K) Protein lysates from S2R+ cells treated with the indicated dsRNAs were blotted with antibodies to Tubulin and MAPK. Mago, Y14 and eIF4AIII are required to maintain MAPK levels, but Btz is not. See also Figure S2.
We examined btz function using the likely null allele btz2, a deletion of the N-terminal half of the protein (van Eeden et al., 2001). Surprisingly, we found that photoreceptors differentiated normally in btz mutant clones (Fig. 4H-J). Consistently, RNAi directed against btz in S2R+ cells had no effect on MAPK expression (Fig. 4K), although it was effective at lowering btz levels (Fig. S2M). Because eIF4AIII is the RNA binding component of the EJC, its absence is predicted to abolish all EJC functions. No null allele of eIF4AIII is available, and clones homozygous for the missense allele eIF4AIII19 (Palacios et al., 2004) only weakly affected photoreceptor differentiation (Fig. S2J-L). However, we found that depletion of eIF4AIII by RNAi in S2R+ cells strongly reduced MAPK levels relative to Tubulin (Fig. 4K). The requirement of Mago, Y14 and eIF4AIII, but not Btz, for MAPK regulation suggests that this function is performed by the pre-EJC prior to Btz addition.
MAPK is the primary target of the pre-EJC in photoreceptor differentiation
If the pre-EJC bound upstream of MAPK exon junctions acts directly on MAPK mRNA, then expression of a MAPK cDNA lacking introns should be independent of EJC components. We found that HA-tagged MAPK expressed from a cDNA template in S2R+ cells was unaffected by treatment with mago dsRNA, although endogenous MAPK levels were strongly reduced (Fig. 5A, B). We next asked whether this construct could mediate EGFR signaling in cells lacking EJC subunits in vivo. Expression of UAS-MAPK-HA in mago or Y14 mutant clones in the eye disc largely restored the differentiation of R1–7 photoreceptors, and enabled these mutant cells to respond to activated Ras by differentiating extra photoreceptors (Fig. 5C-H, Fig. S3). Reduction of MAPK levels is thus the primary reason for the photoreceptor defects in mago or Y14 mutant clones.
Figure 5. A MAPK cDNA rescues photoreceptor differentiation in mago mutant cells.
(A) Diagram of the MAPK-HA cDNA construct. (B) Western blot of protein extracts from S2R+ cells transfected with UAS-MAPK cDNA, UAS-GFP and actin-GAL4, and treated with lacZ or mago dsRNA. Expression of endogenous MAPK (lower band), but not the HA-tagged MAPK cDNA (upper band), is reduced in the absence of Mago. (C-H) Eye discs containing mago93D clones alone (C, D) or mago93D clones expressing MAPK cDNA (E-H), positively marked by GFP expression (H, green in D, F) and by anti-HA staining (G, blue in F). Photoreceptors are stained with anti-Elav (C, E; red in D, F). MAPK cDNA restores almost normal photoreceptor differentiation to mago mutant cells. See also Figure S3.
The pre-EJC regulates MAPK post-transcriptionally
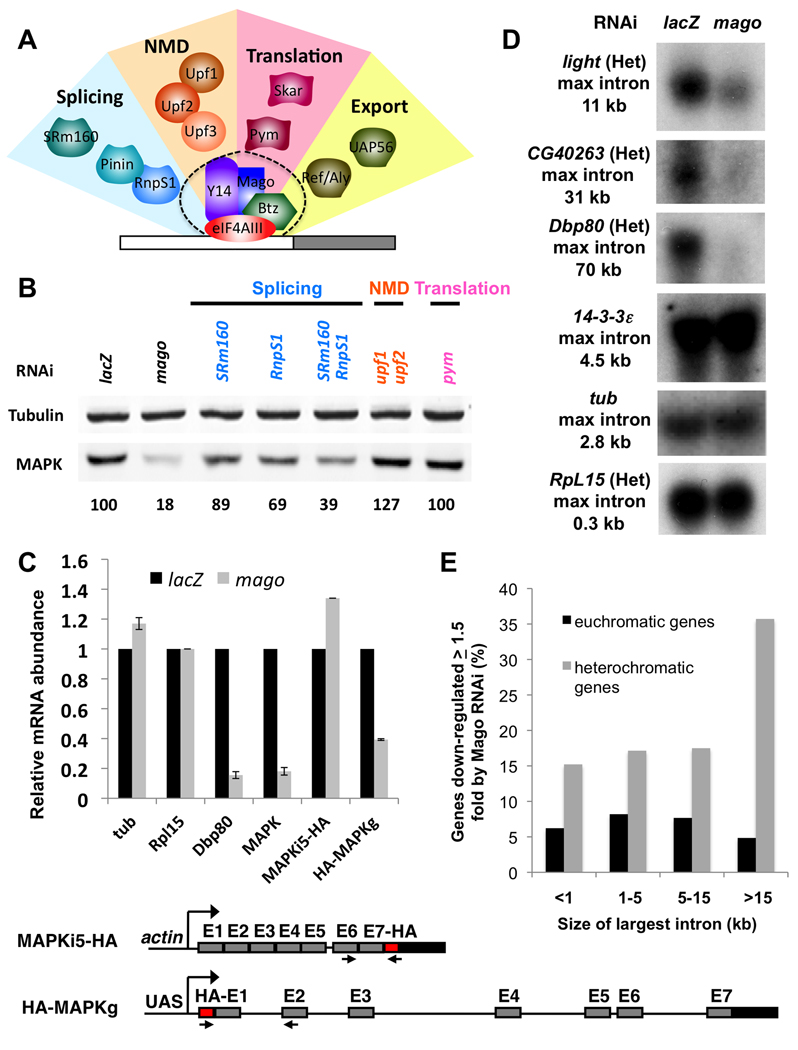
To discriminate whether the pre-EJC acts by affecting the synthesis, stability or translation of MAPK mRNA, we first examined MAPK mRNA levels by Northern blotting in S2R+ cells in which Mago was knocked down by RNAi. We observed a dramatic reduction in MAPK transcript levels (Fig. 6A), indicating that Mago is required for the production or stabilization of mature MAPK mRNA. Using quantitative RT-PCR (qRT-PCR) with primers in the first two exons to measure the levels of MAPK transcripts, we found that Mago depletion caused a 70% reduction in MAPK mRNA, while other mRNAs examined were expressed at normal levels (Fig. 6B, C). This decrease is similar to the 75% reduction in MAPK protein, suggesting that Mago does not significantly affect MAPK translation. Consistent with this interpretation, RNAi directed against the EJC disassembly factor Pym, which enhances translation of EJC-bound transcripts in vertebrates (Diem et al., 2007), did not affect MAPK expression (Fig. 7B). In C. elegans and human stem cells, translation of MAPK transcripts is inhibited by Pumilio family (PUF) proteins bound to their 3’UTRs (Lee et al., 2007). However, knocking down Pumilio (Pum), the only Drosophila PUF protein, did not increase MAPK levels in the presence or absence of Mago (Fig. S4A, B), indicating that the pre-EJC does not act by counteracting Pum-mediated translational repression.
Figure 6. Mago affects MAPK mRNA levels post-transcriptionally.
(A) MAPK and Ribosomal protein L32 (RpL32) mRNA detected by Northern blotting in S2R+ cells treated with lacZ or mago dsRNA. (B) Transcript levels in S2R+ cells treated with the control dsRNA aveugle (ave), an upstream component of the EGFR pathway, or with mago dsRNA were measured by qRT-PCR. mago RNAi reduced MAPK mRNA levels by 70%, but did not affect mRNAs encoding other EGFR pathway components. (C) Diagram showing the 7 exons and 6 introns of the 50 kb MAPK gene. Primers spanning each exon-exon junction were used to detect mRNA levels by qRT-PCR in lacZ or mago dsRNA-treated S2R+ cells. (D) MAPK pre-mRNA levels in S2R+ cells treated with lacZ or mago dsRNA were assessed by qRT-PCR using primers spanning the exon-intron junctions. Pre-mRNA levels are reduced in some regions of the MAPK gene and increased in others in Mago-depleted cells, suggesting that Mago does not affect MAPK transcription. For C and D, the mean of 5 experiments is shown; error bars indicate standard deviations. β-tubulin (tub), RpL15 and Histone H3 (His3) were used as controls. Signals detected in the absence of reverse transcriptase are also plotted (lacZ-no RT, mago-no RT) but are negligible on the scale of these graphs. (E) RT-PCR using primers in exons 3 and 7 amplified a smaller product in cells treated with mago, but not lacZ, dsRNA (arrow). The structure of this product is diagrammed below. See also Figure S4 and Table S2.
Figure 7. The pre-EJC may facilitate splicing of large introns in heterochromatic genes.
(A) Proteins associated with the core EJC and their known functions. (B) MAPK and Tubulin levels detected by Western blotting of lysates from S2R+ cells treated with the indicated dsRNAs. Quantification of MAPK levels relative to the lacZ control is shown below the blot. Knocking down the splicing factors SRm160 and RnpS1, especially in combination, reduces MAPK levels. (C) qRT-PCR was used to measure mRNA transcribed from the UAS-HA-MAPK genomic construct shown below (HA-MAPKg, primers in the HA sequence and exon 2) and from actin-MAPKi5-HA (primers in exon 6 and the HA sequence) in S2R+ cells treated with lacZ or mago dsRNA. Controls were tub, RpL15, Dbp80, and endogenous MAPK (primers in exons 2 and 3), and HA-MAPKg levels were normalized to transcripts from cotransfected UAS-GFP, also driven by actin-GAL4. Mago depletion reduces mRNA expressed from the genomic construct. (D) Northern blots of RNA from S2R+ cells treated with lacZ or mago dsRNA. Expression of the large heterochromatic (Het) genes light, CG40263 and Dbp80 is reduced by mago RNAi, but expression of the small genes tub, 14-3-3 ε and RpL 15 is unaffected. (E) Genes downregulated ≥1.5 fold in mago dsRNA-treated S2R+ cells broken down by location in euchromatin or heterochromatin and by the size of their largest introns are shown as a percentage of the total number of genes in each category that are expressed in control lacZ dsRNA-treated cells. Heterochromatic genes with introns larger than 15 kb are the most likely to be dependent on Mago. See also Figure S5 and Table S1.
The primary step regulated by the pre-EJC is thus likely to be the transcription, processing or degradation of MAPK mRNA. To assess MAPK mRNA stability, we treated cells with actinomycin D to block transcription and measured the levels of MAPK transcripts over time. The stability of MAPK mRNA relative to Ribosomal protein L15 (RpL15) mRNA was only slightly decreased by Mago RNAi treatment (Fig. S4C), making it unlikely to account for the large reduction in total MAPK mRNA levels. A reduction in transcriptional initiation would be expected to reduce all regions of the MAPK mRNA to the same extent; however, both qRT-PCR and deep sequencing of mRNA showed that transcripts of 5’ exons were decreased more than 3’ exons (Fig. 6C, Fig. S4D). To further evaluate MAPK transcription, we examined the expression level of the pre-mRNA by qRT-PCR, using primers to amplify intron-exon junctions. We found that while some regions of the MAPK pre-mRNA were reduced in Mago-depleted cells, others were increased (Fig. 6D), arguing against an effect of Mago on MAPK transcription. The variability in both mRNA and pre-mRNA levels over the length of the gene is suggestive of defects in splicing. Consistent with this model, we detected an abnormal MAPK splice product in Mago-depleted, but not control cells. Sequencing of this product revealed that it results from splicing of a cryptic 5’ splice site 7 bases into exon 4 directly to exon 7, skipping exons 5 and 6 (Fig. 6E). Although splicing defects could result in frameshifts that would lead to NMD, we did not observe further stabilization of MAPK pre-mRNA when we knocked down both Mago and the NMD factor Upf1 (Fig. S5A); abnormal splice products may thus be degraded within the nucleus.
MAPK expression requires splicing-related EJC accessory factors
The core EJC has been reported to associate with accessory factors involved in splicing, NMD, translation, and mRNA export (Fig. 7A) (Diem et al., 2007; Le Hir et al., 2001; Le Hir et al., 2000; Li et al., 2003; Ma et al., 2008). To investigate which of these processes are involved in regulation of MAPK by the pre-EJC, we knocked down a representative set of factors in S2R+ cells. Depletion of the NMD factors Upf1 and Upf2 (Chang et al., 2007) had no effect on MAPK (Fig. 7B), as expected since NMD in Drosophila does not require the EJC (Gatfield et al., 2003). However, depletion of the EJC-associated splicing factors SRm160 and RnpS1 (Reichert et al., 2002; Trembley et al., 2005) reduced MAPK levels relative to Tubulin, especially when both were knocked down simultaneously (Fig. 7B), supporting a role for the EJC in splicing the MAPK transcript. Our data probably underestimate the effect of these splicing factors on MAPK, since SRm160 was only partially depleted (Fig. S5B). Depletion of RnpS1 by transgenic RNAi in vivo in the wing imaginal disc also reduced MAPK levels (Fig. S5C, D).
If the pre-EJC is required for MAPK splicing, it should affect the expression of a MAPK construct that contains the endogenous introns but is expressed from an exogenous promoter. We used recombineering to place the MAPK genomic region, including all its introns, downstream of a UAS promoter and HA tag. Expression of HA-MAPK from this construct driven by actin-GAL4 in S2R+ cells was strongly reduced by mago RNAi (Fig. 7C, Fig. S5E). In contrast, an actin promoter-driven HA-MAPK cDNA construct containing only the smallest intron, I5, was not affected by Mago depletion (Fig. 7C, Fig. S5E), arguing that one or more of the larger MAPK introns confers the requirement for the pre-EJC.
Mago promotes the expression of heterochromatic genes with large introns
The MAPK gene has two unusual features: it contains introns of up to 25 kb, much longer than the average size of 1.4 kb for Drosophila (Yu et al., 2002) (Fig. 6C), and it is expressed despite its location within a region of constitutive heterochromatin (Eberl et al., 1993). We investigated whether other genes that shared these features showed a similar requirement for the pre-EJC. We found that transcript levels of several other genes with large introns located in heterochromatin were strongly reduced in cells depleted for Mago, while small genes in heterochromatin or euchromatin were unaffected (Fig. 7D). To extend these results, we carried out a genome-wide survey of genes affected by Mago depletion by deep sequencing of mRNA isolated from control or mago dsRNA-treated S2R+ cells. We found that genes located in heterochromatin were over-represented among those showing reduced expression after Mago knockdown. Expression of 18.5% of the heterochromatic genes detected in these cells (43/232), but only 6.6% of the euchromatic genes (505/7638), was reduced by at least 1.5 fold in comparison to cells treated with lacZ RNAi (Fig. 7E, Table S1). Among heterochromatic genes, those with introns larger than 15 kb were twice as likely to be affected by mago RNAi as genes with smaller or no introns, while regulation showed no correlation with intron size for euchromatic genes (Fig. 7E, Table S1).
Some large introns are spliced by a recursive mechanism that relies on cryptic splice sites located within the intron (Burnette et al., 2005); however, no consensus recursive splice site is present in the MAPK gene (A. Javier Lopez, pers. comm.). Of the 84 genes with predicted recursive splice sites (Burnette et al., 2005) that are expressed in S2R+ cells, only 4 (5%) were down-regulated at least 1.5 fold by Mago depletion. Since 7% of all expressed genes were down-regulated to this extent, recursively spliced genes are under-represented among Mago targets. It is possible that the pre-EJC specifically facilitates the splicing of large introns that cannot be subdivided by recursive splice sites, which may be more common in heterochromatic genes (Smith et al., 2007). Alternatively, other features of the affected introns, such as the strength of their splice sites, the presence of repetitive sequences (Dimitri et al., 2003), or the chromatin structure of the DNA template, may contribute to determining the requirement for the pre-EJC.
Discussion
The EJC is thought to bind to all spliced mRNAs independently of their sequence (Le Hir et al., 2000), allowing them to be distinguished from unspliced transcripts in the cytoplasm. Despite these very general binding properties, we find that loss of core EJC subunits causes surprisingly specific defects. Our investigation of the basis for the effect of EJC subunits on one target gene, MAPK, has revealed a function of the pre-EJC during the splicing process.
Specificity of EJC function
Our genome-wide expression analysis found that loss of Mago reduces the transcript levels of only 7% of the genes expressed in S2R+ cells by 1.5 fold or more. The number of genes directly regulated by the pre-EJC is likely to be much smaller, since transcript levels were measured after an extensive period of RNAi treatment that was necessary to eliminate the Mago protein. The ability of MAPK to rescue photoreceptor differentiation in mago mutant clones also suggests that many genes are down-regulated as an indirect consequence of loss of MAPK. Similarly, many of the defects of mouse neuroepithelial stem cells heterozygous for Magoh are rescued by restoring the expression of a single gene, Lis1 (Silver et al., 2010). Cytoplasmic functions of the EJC also show specificity; for instance, the EJC is required to localize oskar mRNA to the posterior of the oocyte, but has no effect on the subcellular localization of other spliced mRNAs such as bicoid or gurken (Hachet and Ephrussi, 2001; Newmark and Boswell, 1994; Newmark et al., 1997). This functional specificity might indicate that EJC components are in fact assembled on only a subset of spliced transcripts. Indeed, only the first intron in the oskar transcript contributes to its localization by the EJC (Hachet and Ephrussi, 2004). However, experiments in vertebrate and Drosophila cells have found no specific requirement for EJC assembly other than an upstream exon at least 20 bases long (Herold et al., 2009; Ideue et al., 2007; Le Hir et al., 2001; Tange et al., 2004). Localization of EJC components to particular cytoplasmic regions in Drosophila oocytes (Hachet and Ephrussi, 2001; Mohr et al., 2001) and mammalian neurons (Giorgi et al., 2007) may simply represent their selective retention on mRNAs that are translationally repressed (Dostie and Dreyfuss, 2002; Gehring et al., 2009b).
The importance of MAPK for receptor tyrosine kinase signaling has led to the evolution of multiple mechanisms to regulate its expression as well as its phosphorylation (Lee et al., 2007; Nykamp et al., 2008). Other vital targets for the pre-EJC may be found in the ovary. mago and Y14, but not btz, are required early in oogenesis for germline stem cell differentiation and oocyte specification (Parma et al., 2007; van Eeden et al., 2001). Since germline inactivation of the Ras pathway has no effect on oogenesis (Hou et al., 1995), these functions of Mago and Y14 may reflect a requirement for the pre-EJC to splice transcripts other than MAPK.
A role for the pre-EJC in splicing
The EJC has been shown to act on previously spliced mRNAs in the cytoplasm to increase their translation, direct their subcellular localization, or target them for degradation if they contain premature stop codons (Tange et al., 2004). However, none of these mechanisms could explain the strong reduction of MAPK mRNA levels in the absence of pre-EJC subunits. We have provided several lines of evidence suggesting that the pre-EJC facilitates splicing of a specific subset of introns, including at least one present in the MAPK pre-mRNA. First, MAPK is not an indirect transcriptional target of the pre-EJC, because MAPK pre-mRNA is not uniformly reduced in the absence of mago, and Mago is required for the expression of a MAPK genomic construct driven by a heterologous promoter. Second, the EJC-associated splicing factors RnpS1 and SRm160 (Reichert et al., 2002) contribute to maintaining normal MAPK levels, while Btz, the only core EJC subunit absent from the spliceosomal complex (Gehring et al., 2009a), is dispensable for MAPK expression. Third, an abnormally spliced MAPK product is detected in Mago-depleted cells. Finally, heterochromatic genes with large introns show an increased propensity for regulation by Mago. Previous experiments did not detect any positive function for the EJC in splicing; however, they were performed in vitro using short introns with strong splice sites (Zhang and Krainer, 2007), and would therefore have missed a function specific to one class of introns.
It will be interesting to determine what features of introns make their splicing dependent on the pre-EJC. Our genome-wide analysis points to heterochromatic location and intron size as two characteristics that are likely to be important. Unlike mammalian genomes, the Drosophila genome contains primarily short introns (Yu et al., 2002). Large introns are most common in heterochromatic genes such as MAPK, where they are rich in repetitive DNA composed of transposons, retrotransposons and satellite sequences (Dimitri et al., 2003). Production of endo-siRNAs from such repetitive elements (Ghildiyal et al., 2008) or the presence of splice sites within these elements (Ponicsan et al., 2010) could interfere with the splicing of the introns they occupy. Chromatin structure might also directly influence splicing, as suggested by recent studies showing differences in nucleosome occupancy and histone modifications between exons and introns, and recruitment of splicing regulators by chromatin-binding proteins (Schwartz and Ast, 2010; Luco et al., 2010).
Recognition of splice sites over long distances poses a challenge to the splicing machinery. Splice sites for large introns are initially identified by an exon definition mechanism (Fox-Walsh et al., 2005). The pre-EJC, which is assembled upstream of the 5’ splice site during splicing (Gehring et al., 2009a), might interact with other factors across the exon to facilitate recognition of the upstream 3’ splice site. Perhaps pre-EJC complexes deposited upstream of introns that can be easily detected due to their small size, strong splice sites, or other features, contribute to the subsequent recognition of neighboring introns. Alternatively, since the pre-EJC is assembled prior to exon ligation (Gehring et al., 2009a), it might act during its own recruitment into the spliceosome to promote the second step of splicing. We cannot distinguish these alternatives at present, since our measurements of 5’ and 3’ splice junctions in the MAPK pre-mRNA were made at steady state and thus reflect the balance between transcription, splicing and degradation. The presence of recursive splice sites that allow large introns to be spliced in multiple steps (Burnette et al., 2005) makes genes less likely to require the EJC. Interestingly, recursive splice sites are much less common in vertebrate introns than in Drosophila (Shepard et al., 2009), suggesting that the EJC-dependent mechanism might be more widely used in higher organisms. Our data challenge the view that the EJC acts only as a marker that affects post-splicing events, and suggest that this complex also functions within the nucleus to process a specific set of transcripts.
Experimental Procedures
Drosophila genetics
Three lethal alleles of mago were isolated in a mosaic screen for genes required for photoreceptor differentiation (Janody et al., 2004). The mutations were mapped to the mago genomic region by meiotic recombination with P(w+) elements (Zhai et al., 2003). The coding region of mago was amplified by PCR from homozygous mutant larvae and sequenced. Two alleles altered conserved amino acids (mago93D, S39F; mago115F, E21K) and one introduced a premature stop codon (mago69L, R121@). These alleles failed to complement the previously described strong allele mago3 (Boswell et al., 1991), which also showed the same eye disc phenotype. To generate a Y14 null allele, we mobilized the P(w+) element P{EP}tsuEP567 (tsu1) (Flybase) by crossing it to the transposase stock A2–3, CyO; TM3/T(2;3)apterousxa. We generated 89 independent excision lines, identified deletions by PCR using primers flanking the P-element, and determined the breakpoints by sequencing the PCR products. tsuΔ18 is a 711 bp deficiency that removes the entire Y14 coding sequence.
Immunohistochemistry and Western blot analysis
Antibody staining of eye and wing imaginal discs was performed as described (Roignant et al., 2006). Secondary antibodies were from Jackson Immunoresearch; FITC, TRITC or Cy5 conjugates were used at 1:200 and Alexa 488 conjugates at 1:1000. Images were captured on a Leica TCS NT or a Zeiss LSM 510 confocal microscope. Western blots were performed as described (Miura et al., 2006). To make protein extract from eye-antennal discs, 100 discs of each genotype were lysed in ice-cold lysis buffer (50mM Tris-HCl pH 8.0, 150mM NaCl, 1% Triton X-100, protease inhibitor cocktail (Roche), 5mM EDTA, 5mM NaF, 1mM Na3VO4, 0.1% SDS, 0.5% sodium deoxycholate).
Cell culture and RNAi
S2R+ cells were maintained in Schneider’s medium supplemented with 10% fetal calf serum; D2F cells (Schweitzer et al., 1995) were additionally supplemented with 150 µg/ml G418. Double-stranded RNAs (dsRNAs) were generated using the MEGAscript T7 and T3 kit (Ambion) as described (Roignant et al., 2006). S2R+ cells (106 cells/well) were treated with 15 µg dsRNA. Cells were transfected using Effectene (Qiagen) 3 days after dsRNA incubation and returned to medium containing 15 µg dsRNA. Cells were harvested after 7 days and lysed in ice-cold lysis buffer. MAPK phosphorylation was induced in S2R+ cells by incubation in PBS containing 25 µg/ml bovine insulin (Sigma) for 10 min. In D2F cells, MAPK phosphorylation was induced by incubation in sSpiCS conditioned medium for 30 min (Miura et al., 2006), following EGFR induction for 3 hr with 200 µM Cu2SO4.
Measurement of RNA levels
Total RNA was extracted from cells using TRIzol (Invitrogen) and treated with RQ1 RNase-Free DNase (Promega). Reverse transcription was performed from 2 µg of total RNA using M-MLV Reverse Transcriptase (Promega). The exponential phases of PCR reactions were determined on 18–23 cycles to allow semiquantitative comparisons of cDNAs. For qRT-PCR analysis, cDNA was amplified using Power SYBR green and a real-time PCR ABI 7900HT Sequence Detection Systems machine (Applied Biosystems). The relative abundance of transcripts was calculated as described (Carrera et al., 2008). All experiments were repeated at least three times and the data are presented as the mean +/− standard deviation. Primer sequences are given in Table S2. For Northern blots, 10 µg of total RNA were denatured for 30 min at 55°C in glyoxal loading buffer and separated on a 1% agarose gel. RNA was transferred to a Hybond-XL membrane (Amersham), UV crosslinked and probed with PCR fragments (500 bp-1 kb) radioactively labeled with [32P]-dCTP. Membranes were exposed to X-ray film for 24–72 h at − 80°C.
Deep sequencing of mRNA
S2R+ cells were treated with 15 µg lacZ or mago dsRNAs for 3 days and placed in fresh medium with 15 µg dsRNA for another 3 days. Total RNA was extracted using TRIzol, cleaned using RNeasy Mini Kit (Qiagen) and treated with RQ1 RNase-Free DNase (Promega). Isolation of Poly(A)+ mRNA, RT reactions, and purification of the cDNA templates were performed following the mRNA-Seq Sample Preparation kit protocol from Illumina. Each cDNA sample was uploaded onto one lane of a flow cell and sequenced in a 54-nucleotide single-end run on an Illumina Genome Analyser II.
Raw images were analyzed by Illumina RTA version 1.6 using Phix control lane for estimating base calling parameters. Reads were generated and aligned to the D. melanogaster genome (dm3) and exon-exon splice junction database (prepared using a UCSC annotation database downloaded on 4/23/10) by Illumina CASAVA version 1.6 using default filtering parameters. The first and last two nucleotides were trimmed out. In total, 22,537,621 and 21,130,945 50bp reads for LacZ and Mago samples, respectively, were sequenced. 46% of LacZ and 44% of Mago reads had eligible alignments after filtering for contaminants, repeats, and reads aligned to multiple genes. We normalized gene counts using the RPKM method (reads per kilobase of transcript per million mapped sequence reads)(Mortazavi et al., 2008), and calculated the fold-change of RPKM between the LacZ and Mago samples. The data have been deposited in the GenBank Short Read Archive (accession number SRA022032.3). A list of genes in heterochromatin was obtained from Madeline Crosby (Flybase).
Supplementary Material
Acknowledgements
We thank Robert Boswell, Barry Dickson, Anne Ephrussi, Ruth Lehmann, James Skeath, Daniel St Johnston, Marc Therrien, the Developmental Studies Hybridoma Bank, the Drosophila Genomics Resource Center and the Bloomington Drosophila stock center for reagents. We also thank Antonio-Javier Lopez for searching for recursive splice sites in MAPK pre-mRNA, and Madeline Crosby for providing a list of heterochromatic genes. We are grateful to the NYU Cancer Institute Genomics Facility and Laura Hogan for assistance with qRT-PCR and deep sequencing, and to Stuart Brown and Zuojian Tang for bioinformatics analysis. The manuscript was improved by the critical comments of Sergio Astigarraga, Inés Carrrera, Kerstin Hofmeyer, Kevin Legent, Sylvie Ozon Rickman, Hyung Don Ryoo, Josie Steinhauer, Andrea Zamparini and Jiri Zavadil. This work was supported by the National Institutes of Health (grant EY13777 to J.E.T.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andersen CB, Ballut L, Johansen JS, Chamieh H, Nielsen KH, Oliveira CL, Pedersen JS, Seraphin B, Le Hir H, Andersen GR. Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science. 2006;313:1968–1972. doi: 10.1126/science.1131981. [DOI] [PubMed] [Google Scholar]
- Ballut L, Marchadier B, Baguet A, Tomasetto C, Seraphin B, Le Hir H. The exon junction core complex is locked onto RNA by inhibition of eIF4AIII ATPase activity. Nat Struct Mol Biol. 2005;12:861–869. doi: 10.1038/nsmb990. [DOI] [PubMed] [Google Scholar]
- Bono F, Ebert J, Lorentzen E, Conti E. The crystal structure of the exon junction complex reveals how it maintains a stable grip on mRNA. Cell. 2006;126:713–725. doi: 10.1016/j.cell.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Boswell RE, Prout ME, Steichen JC. Mutations in a newly identified Drosophila melanogaster gene, mago nashi, disrupt germ cell formation and result in the formation of mirror-image symmetrical double abdomen embryos. Development. 1991;113:373–384. doi: 10.1242/dev.113.1.373. [DOI] [PubMed] [Google Scholar]
- Burnette JM, Miyamoto-Sato E, Schaub MA, Conklin J, Lopez AJ. Subdivision of large introns in Drosophila by recursive splicing at nonexonic elements. Genetics. 2005;170:661–674. doi: 10.1534/genetics.104.039701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrera I, Zavadil J, Treisman JE. Two subunits specific to the PBAP chromatin remodeling complex have distinct and redundant functions during Drosophila development. Mol Cell Biol. 2008;28:5238–5250. doi: 10.1128/MCB.00747-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- Cheng J, Belgrader P, Zhou X, Maquat LE. Introns are cis effectors of the nonsense-codon-mediated reduction in nuclear mRNA abundance. Mol Cell Biol. 1994;14:6317–6325. doi: 10.1128/mcb.14.9.6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciapponi L, Jackson DB, Mlodzik M, Bohmann D. Drosophila Fos mediates ERK and JNK signals via distinct phosphorylation sites. Genes Dev. 2001;15:1540–1553. doi: 10.1101/gad.886301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degot S, Le Hir H, Alpy F, Kedinger V, Stoll I, Wendling C, Seraphin B, Rio MC, Tomasetto C. Association of the breast cancer protein MLN51 with the exon junction complex via its speckle localizer and RNA binding module. J Biol Chem. 2004;279:33702–33715. doi: 10.1074/jbc.M402754200. [DOI] [PubMed] [Google Scholar]
- Diem MD, Chan CC, Younis I, Dreyfuss G. PYM binds the cytoplasmic exon-junction complex and ribosomes to enhance translation of spliced mRNAs. Nat Struct Mol Biol. 2007;14:1173–1179. doi: 10.1038/nsmb1321. [DOI] [PubMed] [Google Scholar]
- Dimitri P, Junakovic N, Arca B. Colonization of heterochromatic genes by transposable elements in Drosophila. Mol Biol Evol. 2003;20:503–512. doi: 10.1093/molbev/msg048. [DOI] [PubMed] [Google Scholar]
- Dostie J, Dreyfuss G. Translation is required to remove Y14 from mRNAs in the cytoplasm. Curr Biol. 2002;12:1060–1067. doi: 10.1016/s0960-9822(02)00902-8. [DOI] [PubMed] [Google Scholar]
- Eberl DF, Duyf BJ, Hilliker AJ. The role of heterochromatin in the expression of a heterochromatic gene, the rolled locus of Drosophila melanogaster. Genetics. 1993;134:277–292. doi: 10.1093/genetics/134.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox-Walsh KL, Dou Y, Lam BJ, Hung SP, Baldi PF, Hertel KJ. The architecture of pre-mRNAs affects mechanisms of splice-site pairing. Proc Natl Acad Sci U S A. 2005;102:16176–16181. doi: 10.1073/pnas.0508489102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatfield D, Unterholzner L, Ciccarelli FD, Bork P, Izaurralde E. Nonsense-mediated mRNA decay in Drosophila: at the intersection of the yeast and mammalian pathways. EMBO J. 2003;22:3960–3970. doi: 10.1093/emboj/cdg371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring NH, Lamprinaki S, Hentze MW, Kulozik AE. The hierarchy of exon-junction complex assembly by the spliceosome explains key features of mammalian nonsense-mediated mRNA decay. PLoS Biol. 2009a;7:e1000120. doi: 10.1371/journal.pbio.1000120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring NH, Lamprinaki S, Kulozik AE, Hentze MW. Disassembly of exon junction complexes by PYM. Cell. 2009b;137:536–548. doi: 10.1016/j.cell.2009.02.042. [DOI] [PubMed] [Google Scholar]
- Ghildiyal M, Seitz H, Horwich MD, Li C, Du T, Lee S, Xu J, Kittler EL, Zapp ML, Weng Z, Zamore PD. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science. 2008;320:1077–1081. doi: 10.1126/science.1157396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgi C, Yeo GW, Stone ME, Katz DB, Burge C, Turrigiano G, Moore MJ. The EJC factor eIF4AIII modulates synaptic strength and neuronal protein expression. Cell. 2007;130:179–191. doi: 10.1016/j.cell.2007.05.028. [DOI] [PubMed] [Google Scholar]
- Golembo M, Schweitzer R, Freeman M, Shilo BZ. argos transcription is induced by the Drosophila EGF receptor pathway to form an inhibitory feedback loop. Development. 1996;122:223–230. doi: 10.1242/dev.122.1.223. [DOI] [PubMed] [Google Scholar]
- Hachet O, Ephrussi A. Drosophila Y14 shuttles to the posterior of the oocyte and is required for oskar mRNA transport. Curr Biol. 2001;11:1666–1674. doi: 10.1016/s0960-9822(01)00508-5. [DOI] [PubMed] [Google Scholar]
- Hachet O, Ephrussi A. Splicing of oskar RNA in the nucleus is coupled to its cytoplasmic localization. Nature. 2004;428:959–963. doi: 10.1038/nature02521. [DOI] [PubMed] [Google Scholar]
- Halfar K, Rommel C, Stocker H, Hafen E. Ras controls growth, survival and differentiation in the Drosophila eye by different thresholds of MAP kinase activity. Development. 2001;128:1687–1696. doi: 10.1242/dev.128.9.1687. [DOI] [PubMed] [Google Scholar]
- Hay BA, Wolff T, Rubin GM. Expression of baculovirus P35 prevents cell death in Drosophila. Development. 1994;120:2121–2129. doi: 10.1242/dev.120.8.2121. [DOI] [PubMed] [Google Scholar]
- Herold N, Will CL, Wolf E, Kastner B, Urlaub H, Luhrmann R. Conservation of the protein composition and electron microscopy structure of Drosophila melanogaster and human spliceosomal complexes. Mol Cell Biol. 2009;29:281–301. doi: 10.1128/MCB.01415-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou XS, Chou TB, Melnick MB, Perrimon N. The Torso receptor tyrosine kinase can activate Raf in a Ras-independent pathway. Cell. 1995;81:63–71. doi: 10.1016/0092-8674(95)90371-2. [DOI] [PubMed] [Google Scholar]
- Ideue T, Sasaki YT, Hagiwara M, Hirose T. Introns play an essential role in splicing-dependent formation of the exon junction complex. Genes Dev. 2007;21:1993–1998. doi: 10.1101/gad.1557907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janody F, Lee JD, Jahren N, Hazelett DJ, Benlali A, Miura GI, Draskovic I, Treisman JE. A mosaic genetic screen reveals distinct roles for trithorax and Polycomb group genes in Drosophila eye development. Genetics. 2004;166:187–200. doi: 10.1534/genetics.166.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim FD, Rubin GM. Ectopic expression of activated Ras1 induces hyperplastic growth and increased cell death in Drosophila imaginal tissues. Development. 1998;125:1–9. doi: 10.1242/dev.125.1.1. [DOI] [PubMed] [Google Scholar]
- Kataoka N, Diem MD, Kim VN, Yong J, Dreyfuss G. Magoh, a human homolog of Drosophila Mago nashi protein, is a component of the splicing-dependent exon-exon junction complex. EMBO J. 2001;20:6424–6433. doi: 10.1093/emboj/20.22.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka N, Yong J, Kim VN, Velazquez F, Perkinson RA, Wang F, Dreyfuss G. Pre-mRNA splicing imprints mRNA in the nucleus with a novel RNA-binding protein that persists in the cytoplasm. Mol Cell. 2000;6:673–682. doi: 10.1016/s1097-2765(00)00065-4. [DOI] [PubMed] [Google Scholar]
- Katz M, Amit I, Yarden Y. Regulation of MAPKs by growth factors and receptor tyrosine kinases. Biochim Biophys Acta. 2007;1773:1161–1176. doi: 10.1016/j.bbamcr.2007.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Gatfield D, Izaurralde E, Moore MJ. The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J. 2001;20:4987–4997. doi: 10.1093/emboj/20.17.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Izaurralde E, Maquat LE, Moore MJ. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon-exon junctions. Embo J. 2000;19:6860–6869. doi: 10.1093/emboj/19.24.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MH, Hook B, Pan G, Kershner AM, Merritt C, Seydoux G, Thomson JA, Wickens M, Kimble J. Conserved regulation of MAP kinase expression by PUF RNA-binding proteins. PLoS Genet. 2007;3:e233. doi: 10.1371/journal.pgen.0030233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Lin RI, Lai MC, Ouyang P, Tarn WY. Nuclear Pnn/DRS protein binds to spliced mRNPs and participates in mRNA processing and export via interaction with RNPS1. Mol Cell Biol. 2003;23:7363–7376. doi: 10.1128/MCB.23.20.7363-7376.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luco RF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith OM, Misteli T. Regulation of alternative splicing by histone modifications. Science. 2010;327:996–1000. doi: 10.1126/science.1184208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma XM, Yoon SO, Richardson CJ, Julich K, Blenis J. SKAR links pre-mRNA splicing to mTOR/S6K1-mediated enhanced translation efficiency of spliced mRNAs. Cell. 2008;133:303–313. doi: 10.1016/j.cell.2008.02.031. [DOI] [PubMed] [Google Scholar]
- Martin-Blanco E, Roch F, Noll E, Baonza A, Duffy JB, Perrimon N. A temporal switch in DER signaling controls the specification and differentiation of veins and interveins in the Drosophila wing. Development. 1999;126:5739–5747. doi: 10.1242/dev.126.24.5739. [DOI] [PubMed] [Google Scholar]
- Miura GI, Buglino J, Alvarado D, Lemmon MA, Resh MD, Treisman JE. Palmitoylation of the EGFR ligand Spitz by Rasp increases Spitz activity by restricting its diffusion. Dev Cell. 2006;10:167–176. doi: 10.1016/j.devcel.2005.11.017. [DOI] [PubMed] [Google Scholar]
- Mohr SE, Dillon ST, Boswell RE. The RNA-binding protein Tsunagi interacts with Mago Nashi to establish polarity and localize oskar mRNA during Drosophila oogenesis. Genes Dev. 2001;15:2886–2899. doi: 10.1101/gad.927001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Newmark PA, Boswell RE. The mago nashi locus encodes an essential product required for germ plasm assembly in Drosophila. Development. 1994;120:1303–1313. doi: 10.1242/dev.120.5.1303. [DOI] [PubMed] [Google Scholar]
- Newmark PA, Mohr SE, Gong L, Boswell RE. mago nashi mediates the posterior follicle cell-to-oocyte signal to organize axis formation in Drosophila. Development. 1997;124:3197–3207. doi: 10.1242/dev.124.16.3197. [DOI] [PubMed] [Google Scholar]
- Nott A, Le Hir H, Moore MJ. Splicing enhances translation in mammalian cells: an additional function of the exon junction complex. Genes Dev. 2004;18:210–222. doi: 10.1101/gad.1163204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nykamp K, Lee MH, Kimble J. C. elegans La-related protein, LARP-1, localizes to germline P bodies and attenuates Ras-MAPK signaling during oogenesis. RNA. 2008;14:1378–1389. doi: 10.1261/rna.1066008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios IM, Gatfield D, St Johnston D, Izaurralde E. An eIF4AIII-containing complex required for mRNA localization and nonsense-mediated mRNA decay. Nature. 2004;427:753–757. doi: 10.1038/nature02351. [DOI] [PubMed] [Google Scholar]
- Parma DH, Bennett PE, Jr, Boswell RE. Mago Nashi and Tsunagi/Y14, respectively, regulate Drosophila germline stem cell differentiation and oocyte specification. Dev Biol. 2007;308:507–519. doi: 10.1016/j.ydbio.2007.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponicsan SL, Kugel JF, Goodrich JA. Genomic gems: SINE RNAs regulate mRNA production. Curr Opin Genet Dev. 2010;20:149–155. doi: 10.1016/j.gde.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queenan AM, Ghabrial A, Schupbach T. Ectopic activation of torpedo/Egfr, a Drosophila receptor tyrosine kinase, dorsalizes both the eggshell and the embryo. Development. 1997;124:3871–3880. doi: 10.1242/dev.124.19.3871. [DOI] [PubMed] [Google Scholar]
- Reichert VL, Le Hir H, Jurica MS, Moore MJ. 5′ exon interactions within the human spliceosome establish a framework for exon junction complex structure and assembly. Genes Dev. 2002;16:2778–2791. doi: 10.1101/gad.1030602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roignant JY, Hamel S, Janody F, Treisman JE. The novel SAM domain protein Aveugle is required for Raf activation in the Drosophila EGF receptor signaling pathway. Genes Dev. 2006;20:795–806. doi: 10.1101/gad.1390506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roignant JY, Treisman JE. Pattern formation in the Drosophila eye disc. Int J Dev Biol. 2009;53:795–804. doi: 10.1387/ijdb.072483jr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Ast G. Chromatin density and splicing destiny: on the cross-talk between chromatin structure and splicing. EMBO J. 2010;29:1629–1636. doi: 10.1038/emboj.2010.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweitzer R, Shaharabany M, Seger R, Shilo BZ. Secreted Spitz triggers the DER signaling pathway and is a limiting component in embryonic ventral ectoderm determination. Genes Dev. 1995;9:1518–1529. doi: 10.1101/gad.9.12.1518. [DOI] [PubMed] [Google Scholar]
- Shepard S, McCreary M, Fedorov A. The peculiarities of large intron splicing in animals. PLoS One. 2009;4:e7853. doi: 10.1371/journal.pone.0007853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver DL, Watkins-Chow DE, Schreck KC, Pierfelice TJ, Larson DM, Burnetti AJ, Liaw HJ, Myung K, Walsh CA, Gaiano N, et al. The exon junction complex component Magoh controls brain size by regulating neural stem cell division. Nat Neurosci. 2010;13:551–558. doi: 10.1038/nn.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CD, Shu S, Mungall CJ, Karpen GH. The Release 5.1 annotation of Drosophila melanogaster heterochromatin. Science. 2007;316:1586–1591. doi: 10.1126/science.1139815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava M, Scherr H, Lackey M, Xu D, Chen Z, Lu J, Bergmann A. ARK, the Apaf-1 related killer in Drosophila, requires diverse domains for its apoptotic activity. Cell Death Differ. 2007;14:92–102. doi: 10.1038/sj.cdd.4401931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalder L, Muhlemann O. The meaning of nonsense. Trends Cell Biol. 2008;18:315–321. doi: 10.1016/j.tcb.2008.04.005. [DOI] [PubMed] [Google Scholar]
- Tange TO, Nott A, Moore MJ. The ever-increasing complexities of the exon junction complex. Curr Opin Cell Biol. 2004;16:279–284. doi: 10.1016/j.ceb.2004.03.012. [DOI] [PubMed] [Google Scholar]
- Thermann R, Neu-Yilik G, Deters A, Frede U, Wehr K, Hagemeier C, Hentze MW, Kulozik AE. Binary specification of nonsense codons by splicing and cytoplasmic translation. EMBO J. 1998;17:3484–3494. doi: 10.1093/emboj/17.12.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trembley JH, Tatsumi S, Sakashita E, Loyer P, Slaughter CA, Suzuki H, Endo H, Kidd VJ, Mayeda A. Activation of pre-mRNA splicing by human RNPS1 is regulated by CK2 phosphorylation. Mol Cell Biol. 2005;25:1446–1457. doi: 10.1128/MCB.25.4.1446-1457.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Eeden FJ, Palacios IM, Petronczki M, Weston MJ, St Johnston D. Barentsz is essential for the posterior localization of oskar mRNA and colocalizes with it to the posterior pole. J Cell Biol. 2001;154:511–523. doi: 10.1083/jcb.200105056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegand HL, Lu S, Cullen BR. Exon junction complexes mediate the enhancing effect of splicing on mRNA expression. Proc Natl Acad Sci U S A. 2003;100:11327–11332. doi: 10.1073/pnas.1934877100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Baker NE. Cell cycle withdrawal, progression, and cell survival regulation by EGFR and its effectors in the differentiating Drosophila eye. Dev Cell. 2003;4:359–369. doi: 10.1016/s1534-5807(03)00059-5. [DOI] [PubMed] [Google Scholar]
- Yu J, Yang Z, Kibukawa M, Paddock M, Passey DA, Wong GK. Minimal introns are not "junk". Genome Res. 2002;12:1185–1189. doi: 10.1101/gr.224602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai RG, Hiesinger PR, Koh TW, Verstreken P, Schulze KL, Cao Y, Jafar-Nejad H, Norga KK, Pan H, Bayat V, et al. Mapping Drosophila mutations with molecularly defined P element insertions. Proc Natl Acad Sci U S A. 2003;100:10860–10865. doi: 10.1073/pnas.1832753100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Krainer AR. Splicing remodels messenger ribonucleoprotein architecture via eIF4A3-dependent and -independent recruitment of exon junction complex components. Proc Natl Acad Sci U S A. 2007;104:11574–11579. doi: 10.1073/pnas.0704946104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.