Abstract
Until even only a few years ago, the idea that effective therapies for human mitochondrial disorders resulting from dysfunction of the respiratory chain/oxidative phosphorylation system (OxPhos) could be developed was unimaginable. The obstacles to treating diseases caused by mutations in either mitochondrial DNA (mtDNA) or nuclear DNA (nDNA), and which had the potential to affect nearly every organ system, seemed overwhelming. However, while clinically applicable therapies still remain largely in the future, the landscape has changed dramatically; we can now envision the possibility of treating some of these disorders. Among these are techniques to upregulate mitochondrial biogenesis, to enhance organellar fusion and fission, to “shift heteroplasmy,” and to eliminate the burden of mutant mtDNAs via cytoplasmic transfer.
Introduction
Cellular production of energy, in the form of adenosine triphosphate (ATP), is derived from the oxidation of substrates - mainly carbohydrates and fats. While some ATP is produced in the cytoplasm (e.g. during the conversion of glucose to pyruvate in anaerobic glycolysis), the vast majority is produced under aerobic conditions within the mitochondria, in a process known as oxidative phosphorylation (OxPhos). In broad view, the OxPhos system consists of two elements: (1) the respiratory chain, which takes the chemical energy stored in the food we eat to create a thermodynamic gradient that drives ATP synthesis, and (2) the ATP synthesizing machinery itself, which combines adenosine diphosphate (ADP) and inorganic phosphate (Pi) to generate ATP that is then exported to all parts of the cell.
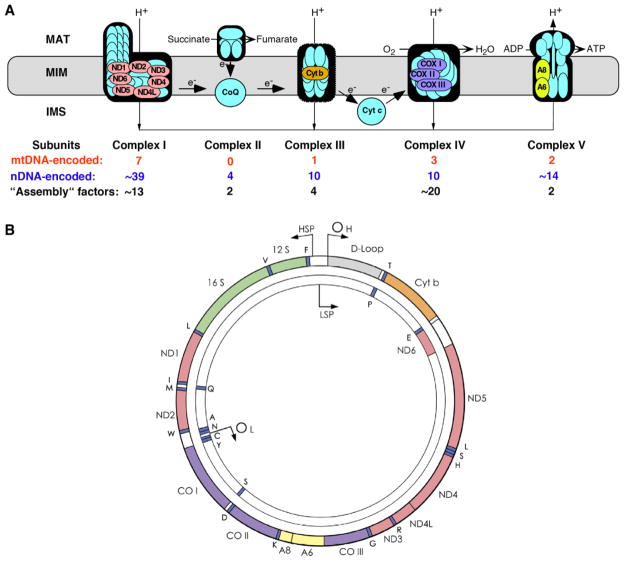
The OxPhos system consists of five multisubunit complexes and two electron carriers (Figure 1A). Reducing equivalents in the form of protons in NADH and FADH2, derived from the oxidation of acetyl-CoA in the tricarboxylic acid cycle, are pumped across the mitochondrial inner membrane (MIM) into the intermembrane space (IMS) through respiratory complexes I, III, and IV to generate a “proton-motive gradient” across the MIM (between the IMS and the matrix). The resulting change in proton-motive force (PMF), which consists of a ΔpH (chemical) component and a ΔΨ (electrical) component, drives the synthesis of ATP from ADP and Pi by complex V (called the FoF1-ATPase). Notably, the OxPhos system is the only cellular “machine” constructed from gene products encoded by both nDNA and mtDNA (Figure 1B). A failure of OxPhos, even a partial one, can have devastating effects; thus it is no surprise that mitochondrial disorders are highly debilitating and often fatal [1].
Figure 1.
The human oxidative phosphorylation system. A. Shown are the four complexes of the respiratory chain (I [NADH dehydrogenase-CoQ reductase], II [succinate dehydrogenase-CoQ reductase], III [CoQ-cytochrome c reductase], and IV [cytochrome c oxidase]), the FoF1-ATPase (complex V), and the two electron carriers, coenzyme Q (CoQ; also called ubiquinone) and cytochrome c (Cyt c). The 13 colored subunits are encoded by mtDNA (see B); the rest are nDNA-encoded. Each of the complexes also require “assembly” factors, all nDNA-encoded, for their synthesis and maintenance. MAT, matrix. B. The 16.6-kb human mitochondrial genome (mtDNA). It encodes 13 polypeptides (A, ATPase; CO, cytochrome oxidase; Cyt b, cytochrome b; ND, NADH dehydrogenase; colors as in A), 2 rRNAs (12S and 16S, in green) and 22 tRNAs (1-letter amino acid code; in blue); the origins of replication (OH, OL) and the promoters of transcription (HSP, LSP) off the “heavy” and “light” strands are also indicated.
Mutations in nDNA genes encoding not only the respiratory chain subunits, but also the large number of proteins required to assemble the individual respiratory complexes, (including incorporation of prosthetic groups), to generate the proper phospholipid milieu of the organelle, and to maintain the integrity of the mtDNA replication, transcription and translation machinery cause classic Mendelian disorders [2,3]. To date, mutations in approximately 70 such genes have been identified [4], and many more will probably be found in the future.
Mutations in mtDNA are usually maternally inherited, but can also arise sporadically [5]. In healthy individuals, mitochondrial genomes, which are present in hundreds or thousands of copies per cell, are genetically identical, a situation known as homoplasmy, but in a typical mtDNA-derived disorder, most, but not all, mitochondrial genomes are mutated, a situation known as heteroplasmy (although patients with homoplasmic mutations have also been described). More than 200 pathogenic mtDNA point mutations and an equal number of large-scale partial deletions have been reported [6]. Although the total copy number of mtDNAs in each cell is under tight control [7], the ratio of normal:mutated mtDNA in daughter cells following cell division can shift due to random drift (a phenomenon known as mitotic segregation) and the clinical phenotype can vary accordingly, both in space (i.e. among cells and tissues) and in time. Thus, the nature of mtDNA as a population of molecules makes both diagnosis and treatment truly difficult. Fortunately, however, with but one exception [8], all pathogenic heteroplasmic mtDNA mutations are functionally recessive, in the sense that it takes a large proportion of mutated genomes above a “threshold” for dysfunction (typically >70–80%) to cause disease; conversely, a small increase in the proportion of normal mtDNAs is sufficient to rescue function, a property that could be exploited therapeutically.
Potential therapeutic strategies
Given that mitochondrial diseases can result from mutations in either mtDNA, whose transmission is governed by the “loose” rules of population genetics, or nuclear DNA, whose transmission is governed by the stricter rules of Mendelian genetics, it is likely that therapeutic strategies will have to be tailored to accomodate the various classes of mutation. Numerous approaches have been discussed in recent reviews [9,10]. These include “enhancement” of respiratory chain function (e.g. supplementation with carnitine, coenzyme Q10 [CoQ10]. thiamine, succinate, folate [11], and methylene blue [12]); removal of noxious metabolites (e.g. lactate [13]); scavenging of free radicals “leaking” from the impaired respiratory chain [14]; treatment of symptoms (e.g. seizures, endocrinopathy, and diabetes); exercise [15]; surgery (e.g. correction of ptosis, or droopy eyelids [16]; cochlear implants for hearing loss [17]; heart transplantation for cardiopathies [18]); and genetic counseling [19]. Notably, therapeutic trials for mitochondrial diseases have not only been generally ineffective, but they have also been inadequately designed, often anecdotal or underpowered. Thus, there is an urgent need in this field for rigorous, double-blinded, placebo controlled studies. Here we focus on recent advances in our understanding of both mtDNA-related and Mendelian mitochondrial disorders that point to rational treatment approaches.
Enhancement of mitochondrial biogenesis
Since the overwhelming majority of proteins in mitochondria are encoded by nuclear DNA, it is not surprising that mutations in a variety of nuclear-encoded structural subunits and assembly factors cause profound defects in mitochondrial function [20]. Moreover, mutations in nuclear-encoded mtDNA-associated proteins, such as DNA polymerase γ [21], cause a variety of mtDNA mutations, including mtDNA depletion [22]. Collectively, these mutations cause severe mitochondrial dysfunction with highly variable clinical presentation and scant therapeutic possibilities.
In recent years, enhancement of overall mitochondrial biogenesis has emerged as an exciting therapeutic prospect. The transcriptional co-activators PGC (peroxisome proliferator-activated receptor γ coactivator)-1α, PGC-1β, and PRC (PGC-related coactivator) have been identified as activators of a signaling cascade controlling the expression of nuclear-encoded mitochondrial OxPhos components [23]. This pathway also regulates mtDNA through modulation of TFAM (transcription factor A, mitochondrial), the mtDNA packaging and transcription factor [24].
Accordingly, upregulation of PGC-1α signaling could increase the total number of mitochondria within cells, thereby restoring mitochondrial bioenergetics to near-normal levels, assuming that the nuclear mutation causing the respiratory deficit leaves at least some residual OxPhos function. Using a muscle-specific knockout of the COX10 assembly factor, Moraes and colleagues show that PGC-1α activation protects muscle from mitochondrial dysfunction: COX10-null mice have only 20–40% of wild-type (WT) mitochondrial function, but transgenic overexpression of PGC-1α in these mice restores full mitochondrial function; importantly, COX10−/− mice treated with bezafibrate, a PGC-1α agonist, have 85% of WT OxPhos activity and their muscle function is improved compared to untreated littermates [25].
These data validate the manipulation of PGC-1α signaling as a way to affect mitochondrial function, making this a candidate therapeutic approach for treating mitochondrial disease. The development of PGC-1α agonists such as bezafibrate [25] and pyrroloquinoline quinone [26] could allow for the development of rational therapeutics for inherited mitochondrial diseases. Looking down the road, enhancing mitochondrial biogenesis might restore mitochondrial function in a variety of other cellular contexts, with the potential to combat mitochondrial dysfunction in common human conditions such as diabetes [27], age-associated sarcopenia [28], and myocardial infarction [29].
We note that the proliferation of mitochondria in response to PGC-1α agonists resembles the mitochondrial proliferation in muscle (“ragged red fibers”, or RRF) that is often found in patients with pathogenic mtDNA mutations [1]. If mitochondrial biogenesis via RRF cannot rescue disease in the first place, why should upregulation of PGC-1α prove effective after the fact? We believe that the two processes, while superficially similar, operate via different mechanisms. PGC-1α stimulates the biogenesis of all mitochondria, whether dysfunctional or not [28], whereas the RRFs in patients, even those who are heteroplasmic, are composed almost exclusively of mitochondria harboring mutated mtDNAs, with essentially no proliferation of mitochondria harboring normal mtDNAs [30,31]. Thus, upregulating PGC-1α function is indeed a reasonable approach to treatment, but clearly we need a better understanding of the role of mitochondrial biogenesis in both pathogenesis and therapy.
Bone marrow transplantation in MNGIE
Mitochondrial neurogastrointestinal encephalomyopathy (MNGIE) is a distinct autosomal recessive multisystemic disease characterized clinically by ptosis and ophthalmoparesis, severe gastrointestinal dysmotility, extreme cachexia, degeneration of peripheral nerves, and brain white matter abnormalities (leukoencephalopathy) leading to death in early to mid-adulthood [32]. MNGIE is caused by loss-of-function mutations in the TYMP gene encoding the cytosolic enzyme thymidine phosphorylase (TPase) [33]. Loss of TPase activity results in dramatic elevations of thymidine and deoxyuridine in blood and tissues [34]. The accumulated nucleosides produce deoxynucleotide pool imbalances, which, in turn, cause instability of mtDNA, manifesting as multiple deletions, depletion, and site-specific point mutations [35].
Based on our knowledge of the pathogenic mechanism of MNGIE, therapies to eliminate the toxic nucleosides should ameliorate or even cure this fatal disease. Single treatments of hemodialysis filtered thymidine from the circulation of two MNGIE patients, but only transiently: the nucleoside re-accumulated to baseline levels in plasma within 24 hours of treatment [36] and chronic hemodialysis for over 1.5 years failed to prevent disease progression and death in one patient [37]. A young woman with MNGIE treated with peritoneal dialysis for three years showed clinical improvements of gastrointestinal symptoms, but strangely, plasma nucleoside levels did not change [38].
TPase enzyme replacement therapy is another promising approach to MNGIE. Towards this goal, a single dose of erythrocyte-encapsulated TPase was administered to a severely affected individual, resulting in partial reductions of thymidine and deoxyuridine in blood and urine; however, the patient died of pneumonia three weeks later. Because platelets contain abundant amounts of TPase, platelets were infused in two MNGIE patients, partially restoring circulating TPase activity and reducing plasma nucleosides [39].
Based on this proof-of-principle study, allogeneic hematopoetic stem cell transplantation (AHSCT) has been performed to restore TPase activity permanently [40]. To date, of the 11 patients who have undergone AHSCT, 5 are alive after successful transplant. All show normalization of blood TPase activity and reductions of plasma thymidine and deoxyuridine to barely-detectable levels; slight clinical improvements were noted after one year. A patient transplanted four and a-half years ago exhibits markedly improved gastrointestinal function; she discontinued parenteral nutrition and gained 3 kilograms; her neuropathy has also improved as assessed clinically and by electrophysiological studies. A prospective clinical trial of AHSCT for MNGIE is being planned.
CoQ supplementation
Two features make CoQ10 extremely popular in the management of patients with mitochondrial disorders: its well-documented safety even at very high doses, and its dual role as a component of the respiratory chain and as a potent antioxidant. There are numerous anecdotal reports of the beneficial effect of CoQ10 in mitochondrial diseases, usually as a component of a “cocktail” that also includes L-carnitine, vitamin B complex, vitamin C, and vitamin K1 [11], but a rigorous placebo-controlled trial is still lacking (and badly needed).
There is a group of disorders, however, in which CoQ10 supplementation can be of substantial benefit, in some cases even life-saving: these are the primary CoQ10 deficiency syndromes. Initially described in patients with the triad of mitochondrial and lipid storage myopathy, recurrent myoglobinuria and central nervous system dysfunction (seizures, ataxia and mental retardation) [41], muscle CoQ10 deficiency was later identified in patients with cerebellar ataxia and cerebellar atrophy inconsistently associated with central or peripheral nervous system problems [42]. In 2006 the Hirano group identified the first mutations in genes encoding CoQ10 biosynthetic proteins [43,44]. Remarkably, an infant boy with a COQ2 mutation, a severe mitochondrial encephalomyopathy (including strokes) and renal dysfunction (glomerulonephrosis) returned to normal after CoQ10 administration; preventive CoQ10 supplementation was effective in his younger sister who had incipient renal disease [45].
Although the response to CoQ10 administration appears to vary in different patients, probably depending on the mutant biosynthetic gene, supplementation with high CoQ10 doses should be tried in all patients.
Enhanced organellar fusion/fission
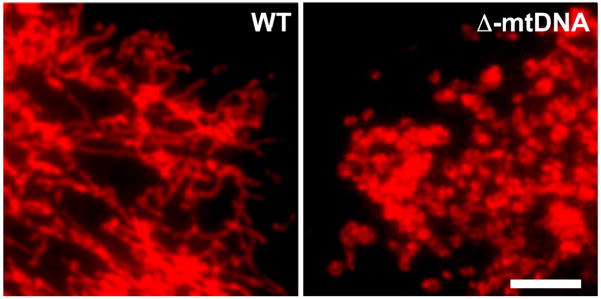
The mitochondrial network maintains a high degree of adaptability in response to various metabolic and environmental stimuli [46], as evidenced by the ability of this network to undergo fission and fusion events and to distribute mtDNA nucleoids (assemblies of mtDNA with its associated proteins) in such a way that, in response to fission, each mitochondrion will contain at least one nucleoid [47]. Accordingly, both genetic and environmental factors that negatively impact mitochondrial function will necessarily impair the responsive adaptability of the mitochondrial network, as demonstrated by the inability of cells carrying large-scale partial deletions of mtDNA (Δ-mtDNAs) to maintain a reticular mitochondrial morphology (Figure 2). While mitochondrial morphology can be affected by non-disease factors, such as metabolic status [48] and position in the cell cyle [49,50], the fragmentation of mitochondria is often a symptom of OxPhos dysfunction [51], because the mere addition of an uncoupler of membrane potential to normal cells is sufficient to convert elongated mitochondria to fragmented ones [52].
Figure 2.
Mitochondrial morphology in different mtDNA backgrounds. Cybrid cells homoplasmic for either WT mtDNA (WT) or a 1.9-kb partially-deleted mtDNA (Δ-mtDNA) were stained with MitoTracker Red and visualized by fluorescence microscopy. Note that WT cells maintain a reticular morphology, suggesting that these cells maintain a united mitochondrial network. Conversely, the cells carrying Δ-mtDNA displayed a uniformly fragmented morphology. Size bar = 10 μM.
If mitochondrial fragmentation is the consequence, rather than the cause, of mitochondrial dysfunction, reversing mitochondrial morphology from fragmented to tubular as a therapeutic strategy might not be viewed as a sensible approach to therapy. However, recent studies of the relationship between mitochondrial fusion/fission and autophagy [53] imply that enhancing mitochondrial fusion could “normalize” the distribution of mutated and WT mtDNAs within the cell, thereby enhancing intra-organellar complementation of function [54], while at the same time allowing the elimination of mitochondria that still contain high levels of mutated mtDNAs via autophagy [53]. Thus, manipulating organellar dynamics using a pharmacological agent might be a promising therapeutic strategy.
Mitochondrial shape in mammals is maintained in a dynamic equilibrium between factors that promote organellar fusion (e.g. mitofusins 1 and 2 [MFN1, MFN2] and optic atrophy protein 1 [OPA1]) and those promoting fission (e.g. mitochondrial fission protein 1 [FIS1] and dynamin-related protein 1 [DRP1]) [55–57]. Genetic experiments show that inhibiting fusion shifts the equilibrium towards fission [58], and vice versa [59]. Thus, adding a pharmacological agent that inhibits fission to cells containing fragmented mitochondria should increase the proportion of tubular mitochondria. In fact, Nunnari’s group recently showed that a compound dubbed mdivi-1 (mitochondrial division inhibitor 1), a quinazolinone whose structure is somewhat reminiscent of a steroid, does exactly that, probably by binding to DRP1 and inhibiting its GTPase activity, thereby preventing its self-assembly into a “ligature” that severs the mitochondrion [60]. More recently, mdivi-1 rescued mitochondrial morphological and functional defects induced by mutations in PINK1 (PTEN-related protein kinase 1) [61], a protein involved in mitochondrial autophagy.
Because a number of neurodegenerative diseases result from aberrant mitochondrial fusion (e.g. dominant optic atrophy due to mutations in OPA1, and Charcot-Marie-Tooth disease type 2A due to mutations in MFN2 [57]), promoting fusion via a fission-inhibiting compound such as mdivi-1 could have therapeutic potential. Using a compound like mdivi-1 for OxPhos diseases is less straightforward, but should be examined. Although agents that act upon the fusion apparatus have not yet been identified, data from mdivi-1 are encouraging. Thus, a drug-based approach to shift the balance towards more elongated mitochondria is an exciting prospect.
Modulation of aberrant calcium homeostasis
One feature shared by many mitochondrial diseases is aberrant calcium homeostasis. Because calcium (Ca2+) is such an important factor in regulating cellular behavior, strategies to normalize Ca2+ distribution, even though they are fundamentally “symptomatic” in nature, might have highly salutary effects in patients. In this regard, pathogenic mutations in mtDNA, causing MERRF (myoclonus epilepsy with ragged-red fibers [1]) [62] and in nDNA, causing Leigh syndrome associated with complex I deficiency [63], both result in loss of the ability of mitochondria to take up and buffer cytosolic Ca2+. Notably, in both cases, treating the patient cells with CGP37157, a mitochondrial calcium channel blocker, ameliorates the defect [62,63]. The resetting of calcium homeostasis in two respiratory chain disorders with vastly different etiologies warrants follow-up, and perhaps even a clinical trial.
Heteroplasmic shifting
The relative mtDNA mutation load within a given heteroplasmic cell determines whether the cell has effective mitochondrial function (the “threshold effect”). Importantly, cellular heteroplasmy can drift, both in space and in time [64], and the accumulation of mtDNA mutations, which varies among tissues [65], may also depend on the mode of mtDNA replication in that tissue [66]. Thus, increasing the proportion of mtDNAs while concomitantly reducing the proportion of mutant mtDNAs (“heteroplasmic shifting”) is possible [67], and existing cellular pathways might provide the means to implement this shift. Moreover, the packaging of mtDNA into protein-DNA assemblies called nucleoids provides an underlying genetic basis for heteroplasmic shifting, as nucleoids do not frequently exchange mtDNAs [54].
When deprived of glucose and treated with ketogenic media supplements, heteroplasmic cytoplasmic hybrid (cybrid) cells harboring a large-scale partial deletion of mtDNA shifted their heteroplasmy below threshold and recovered mitochondrial function [68]. While the mechanism by which this shift occurred is unclear, these results are consistent with selective degradation of defective mitochondria (i.e. mitochondrial autophagy or mitophagy) as the mediator of the shift. Autophagy is the sequestration of cellular components within autophagosomes, followed by lysosomal fusion to degrade the sequestered materials. Autophagosomes target dysfunctional mitochondria within the cell, performing a mitochondrial “quality control” function [69]. Parkin, an E3 ubiquitin ligase, is selectively recruited to functionally impaired mitochondria [70] in a PINK1-dependent manner [71], suggesting that autophagy might degrade mitochondria carrying mutant mtDNAs, thereby providing a therapeutic strategy that relies on an intrinsic cellular pathway. Suomalainen’s group recently used a ketogenic diet to rescue metabolic function in a mouse model of late-onset mitochondrial myopathy due to deletions of mtDNA, but interestingly, the metabolic improvement was not accompanied by any obvious reduction in mutation load [72]. This discrepancy may be explained, however, by the possibility that instead of reducing mutation load, the ketogenic stress “normalized” the distribution of the mutation so that cells contained more mitochondria below the threshold for dysfunction [54].
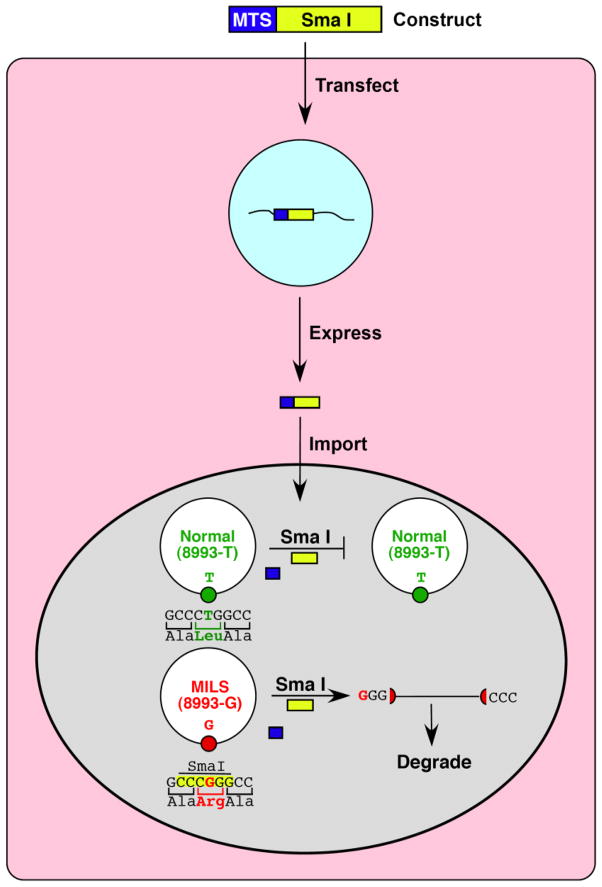
Heteroplasmic shifting can also be accomplished genetically. For example, the T to G mutation at nt-8993 in the MTATP6 gene [73] that causes MILS (maternally-inherited Leigh syndrome) and/or NARP (neuropathy, ataxia, and retinitis pigmentosa) creates a novel and unique SmaI site in human mtDNA. Since WT-mtDNA has no SmaI sites, if functioning SmaI could be targeted to mitochondria in cells from heteroplasmic NARP/MILS patients, it would be possible to selectively cleave (and thereby destroy) the mutated mtDNAs while leaving the WT-mtDNAs intact (Figure 3). In 2002, Tanaka and colleagues demonstrated that such an approach is possible [74]. Although this strategy would only work for a mutation that generates a unique restriction site absent in WT-mtDNA, the precedent for importation of a nuclease into mitochondria has been set.
Figure 3.
Mitochondrial importation of a restriction endonuclease to cleave mutated mtDNAs selectively. The example shown is of importation of SmaI (yellow rectangle) to eliminate the T&G mutation at mtDNA position 8993 (Leu&Arg mutation at aa-156 in ATP6) associated with NARP/MILS. Following transfection of the construct into the cell (pink), mitochondrially-targeted SmaI expressed from the nucleus (blue) is targeted to the organelle (gray) via an N-terminal mitochondrial targeting sequence (MTS; dark blue rectangle) that is cleaved from the precursor polypeptide following import. Only mutated mtDNAs are cleaved and then degraded, leaving the normal mtDNAs intact.
A more generalizable approach would be to import a restriction endonuclease that selectively targets any mutated mtDNA. The technology for such a strategy is taking shape and is based on the exquisite DNA-binding selectivity of zinc (Zn) fingers (ZFs), so-called because they contain histidine and cysteine residues that bind Zn; the Zn-ligated domain of the polypeptide binds DNA in a highly sequence-specific manner [75]. ZFs, which are components of many transcription factors, can be modified for use as selective DNA binding and cleaving agents. In particular, appending a restriction endonuclease to a ZF creates a zinc-finger nuclease (ZFN) that behaves identically to a restriction endonuclease but with a DNA binding specificity that can be tailored to cleave almost any nuclear DNA sequence [76]. Although creating a mitochondrially-targeted ZFN is the obvious next step, this is technically challenging. However, Minzcuk et al. recently targeted both a ZF-DNA methylase [77] and a ZFN [78] to mitochondria. This selective mtDNA targeting approach is strengthened by the recent identification of transcription activator-like (TAL) effector polypeptides in plants that also bind DNA selectively [79], and could also be modified to target mtDNA. In all of these cases, the heteroplasmic shifting may be most effective in tissues that are especially active in recombination, such as the heart [80,81].
These genetic approaches for shifting heteroplasmy have great promise for “personalized mitochondrial medicine,” in which diseases resulting from specific mtDNA mutations can be targeted selectively. Moreover, disposing of the genetic construct after use is also plausible (e.g. by providing a second endonuclease that cleaves the plasmid itself), which could prevent the plasmid from wreaking any havoc once the “mito-ZFN” or “mito-TAL” has finished its work.
Cytoplasmic transfer
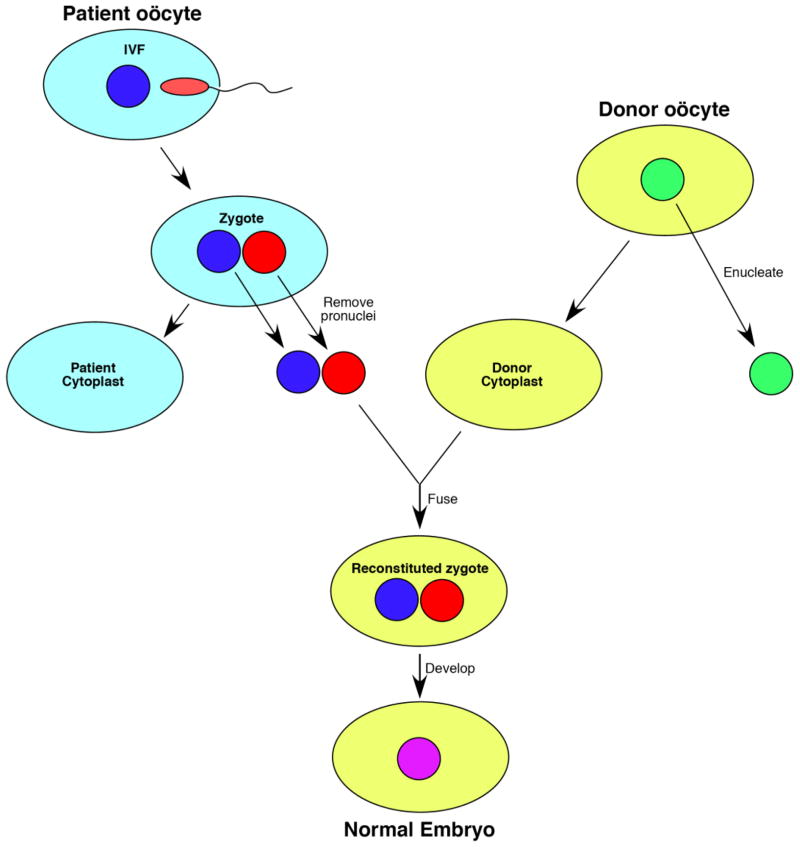
The best gene therapy for a maternally-inherited disease is one in which the mtDNA mutation is eliminated forever. One way to accomplish this, at least in principle, is to eliminate the offending mtDNAs by transferring an in vitro-fertilized nucleus from the ooplasm of a woman carrying a mtDNA mutation (e.g. MELAS [mitochondrial encephalomyopathy, lactic acidosis and stroke-like episodes]) to an enucleated oocyte from a normal donor; the embryo, which contains a normal nucleus from the MELAS mother (and father) and normal mtDNA from the cytoplasmic donor, could be implanted into the mother’s uterus (Figure 4). This approach would result in a mitochondrially normal child carrying the nuclear traits of both parents. The feasibility of cytoplasmic transfer has now been demonstrated by the Newcastle group in the United Kingdom [82], and a variant of this approach was recently used in the United States to produce two “transmitochondrial” rhesus monkeys [83]. Of course, the ethics of cytoplasmic transfer would need to be addressed before this type of germline gene therapy can be undertaken [84].
Figure 4.
One approach to cytoplasmic transfer [82]. Following in vitro-fertilization, the haploid parental pronuclei (i.e. the normal pronucleus from the ooplasm of a woman carrying a mtDNA mutation [blue] and the normal pronucleus from the father [red]) are transferred to an enucleated donor oocyte containing normal mtDNAs (yellow); following fusion of the pronuclei, the embryo now contains a normal diploid nucleus from the parents (purple) and normal mtDNA from the cytoplasmic donor, and can be implanted into the mother’s uterus.
Concluding remarks
Among the new approaches discussed here, some, like the modulation of Ca2+ homeostasis, could be undertaken soon, with clinical trials that could assess effectiveness relatively quickly. Enhancement of mitochondrial biogenesis, especially for nDNA mutations, will likely take much longer, as the long-term use of pharmacological agents to upregulate the number of organelles (both functional and dysfunctional) will likely have untoward side effects With respect to mtDNA mutations, the most exciting prospects for therapy lie in the strategies to shift mtDNA heteroplasmy. Manipulating heteroplasmy using pharmacological approaches could be tried right now, but the readouts, both clinical and genetic, will be difficult to assess, as a perceived improvement in metabolic function may not be related to an obvious drop in the mutation load [72]. In the long run, eliminating mutated mtDNAs permanently using ZFN-like technology is even more exciting, but of course, all the issues that have plagued the gene therapy field (e.g. delivery to target tissues, immunological reactions, deleterious gene integration events) will need to be addressed. One can also envision “combination” therapies based on these ideas, employing approaches that, for example, reduce mutant load (e.g. via an “acute” course of ZFN therapy), homogenize the distribution of the mutation (e.g. via enhanced organellar fusion), and upregulate normal mitochondrial function (e.g. via autophagy or Ca2+ remediation). The choice of treatment will likely vary depending on the mutation. For example, shifting strategies may work best on mtDNA mutations affecting protein synthesis (e.g. in tRNAs), which usually cause RRFs, whereas strategies to upregulate organellar biogenesis may work best for mutations in OxPhos structural subunits, where RRFs are rarer.
The potential to develop the therapeutic strategies described above has ramifications that extend beyond the treatment of “authentic” mitochondrial OxPhos diseases. For example, there is growing evidence that the accumulation of somatic mutations in mtDNA play a role in the normal aging process [85], in Parkinson disease [86], and perhaps even in cancer [87]. There is also growing evidence for a mitochondrial connection in neurodegenerative diseases in which altered mitochondrial dynamics and mitophagy might play significant pathogenic roles. This is true not only in the case of Parkinson disease, where the evidence for altered mitophagy is particularly strong [70], but also in amyotrophic lateral sclerosis, where mitochondrial function and axonal transport are altered [88], and in Alzheimer disease, where recent evidence points to altered communication between the endoplasmic reticulum and mitochondria in the pathogenesis of the disease [89,90]. Thus, the strategies described above, designed to help a few thousand patients, might eventually be used to treat millions.
Acknowledgments
This review was supported by grants from the National Institutes of Health (NS11766 and HD32062), the Muscular Dystrophy Association, the Ellison Medical Foundation, the Alzheimer Drug Discovery Foundation, and the Marriott Mitochondrial Disorder Clinical Research Fund (MMDCRF).
Glossary
- AHSCT
Allogeneic hematopoetic stem cell transplantation. Transplantation of hematopoetic stem cells from a donor to a recipient
- COX
Cytochrome c oxidase. Also known as complex IV of the respiratory chain
- Cybrid
Cytoplasmic hybrid cell, derived from the fusion of a cell lacking mtDNA but still containing mitochondria (called a ρ0 cell) and a cytoplast (an enucleated cell) containing mitochondria, typically from a heteroplasmic patient harboring a pathogenic mtDNA mutation. Cybrids are often used to study genotype-phenotype relationships in tissue culture
- Heteroplasmy
The situation in which two or more mtDNA genotypes coexist in the system under discussion (e.g. a cell, tissue, or organism)
- Homoplasmy
The situation in which all mtDNAs in the system under discussion (e.g. a cell, tissue, or organism) are genetically identical
- MELAS
Mitochondrial encephalomyopathy, lactic acidosis and stroke-like episodes. A maternally-inherited mitochondrial disorder usually due to an A to G mutation at mtDNA position 3243 in the tRNALue(UUR) gene. Patients have respiratory chain dysfunction, especially complex I deficiency. It is one of the most common maternally-inherited mitochondrial disorders
- MERRF
Myoclonus epilepsy with ragged-red fibers. A maternally-inherited mitochondrial disorder usually due to an A to G mutation at mtDNA position 8344 in the tRNALys gene. Patients have respiratory chain dysfunction, myoclonic jerks, and ataxia
- Leigh syndrome
A subacute necrotizing encephalopathy that manifests as psychomotor regression, typically affecting infants or young children. Brain MRI scans reveal characteristic lesions in the basal ganglia and brain stem. Inheritance pattern can be maternal, autosomal recessive, or X-linked
- RRF
Ragged-red fibers. A morphological feature in muscle indicating massive mitochondrial proliferation, almost always of mitochondria containing high levels of mutated mtDNAs. So named due to the red-purple appearance of such fibers upon staining with modified Gomori trichrome
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.DiMauro S, Schon EA. Mitochondrial respiratory-chain diseases. N Engl J Med. 2003;348:2656–2668. doi: 10.1056/NEJMra022567. [DOI] [PubMed] [Google Scholar]
- 2.Zeviani M, Di Donato S. Mitochondrial disorders. Brain. 2004;127:2153–2172. doi: 10.1093/brain/awh259. [DOI] [PubMed] [Google Scholar]
- 3.Jacobs HT, Turnbull DM. Nuclear genes and mitochondrial translation: a new class of genetic disease. Trends Genet. 2005;21:312–314. doi: 10.1016/j.tig.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 4.Spinazzola A, Zeviani M. Disorders from perturbations of nuclear-mitochondrial intergenomic cross-talk. J Int Med. 2009;265:174–192. doi: 10.1111/j.1365-2796.2008.02059.x. [DOI] [PubMed] [Google Scholar]
- 5.DiMauro S, Andreu AL. Mutations in mitochondrial DNA as a cause of exercise intolerance. Ann Med. 2001;33:472–476. doi: 10.3109/07853890109002096. [DOI] [PubMed] [Google Scholar]
- 6.Chinnery PF, et al. Risk of developing a mitochondrial DNA deletion disorder. Lancet. 2004;364:592–596. doi: 10.1016/S0140-6736(04)16851-7. [DOI] [PubMed] [Google Scholar]
- 7.Tang Y, et al. Rearrangements of human mitochondrial DNA (mtDNA): new insights into the regulation of mtDNA copy number and gene expression. Mol Biol Cell. 2000;11:1471–1485. doi: 10.1091/mbc.11.4.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sacconi S, et al. A functionally dominant mitochondrial DNA mutation. Hum Mol Genet. 2008;17:1840–1820. doi: 10.1093/hmg/ddn073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DiMauro S, et al. Approaches to the treatment of mitochondrial diseases. Muscle Nerve. 2006;34:265–283. doi: 10.1002/mus.20598. [DOI] [PubMed] [Google Scholar]
- 10.Kerr DS. Treatment of mitochondrial electron transport chain disorders: A review of clinical trials over the past decade. Mol Genet Metab. 2010;99:246–255. doi: 10.1016/j.ymgme.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 11.Marriage BJ, et al. Cofactor treatment improves ATP synthetic capacity in patients with oxidative phosphorylation disorders. Mol Genet Metab. 2004;81:263–272. doi: 10.1016/j.ymgme.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 12.Atamna H, et al. Methylene blue delays cellular senescence and enhances key mitochondrial biochemical pathways. FASEB J. 2008;22:703–712. doi: 10.1096/fj.07-9610com. [DOI] [PubMed] [Google Scholar]
- 13.Kaufmann P, et al. Dichloroacetate causes toxic neuropathy in MELAS: a randomized, controlled clinical trial. Neurology. 2006;66:324–330. doi: 10.1212/01.wnl.0000196641.05913.27. [DOI] [PubMed] [Google Scholar]
- 14.Smith RA, et al. Mitochondria-targeted antioxidants in the treatment of disease. Ann NY Acad Sci. 2008;1147:105–111. doi: 10.1196/annals.1427.003. [DOI] [PubMed] [Google Scholar]
- 15.Murphy JL, et al. Resistance training in patients with single, large-scale deletions of mitochondrial DNA. Brain. 2008;131:2832–2840. doi: 10.1093/brain/awn252. [DOI] [PubMed] [Google Scholar]
- 16.Edmonds JL. Surgical and anesthetic management of patients with mitochondrial dysfunction. Mitochondrion. 2004;4:543–548. doi: 10.1016/j.mito.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 17.Sinnathuray AR, et al. A review of cochlear implantation in mitochondrial sensorineural hearing loss. Otol Neurotol. 2003;24:418–426. doi: 10.1097/00129492-200305000-00012. [DOI] [PubMed] [Google Scholar]
- 18.Bonnet D, et al. Heart transplantation in children with mitochondrial cardiomyopathy. Heart. 2001;86:570–573. doi: 10.1136/heart.86.5.570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steffann J, et al. Analysis of mtDNA variant segregation during early human embryonic development: a tool for successful NARP preimplantation diagnosis. J Med Genet. 2006;43:244–247. doi: 10.1136/jmg.2005.032326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pecina P, et al. Genetic defects of cytochrome c oxidase assembly. Physiol Res. 2004;53(Suppl 1):S213–S223. [PubMed] [Google Scholar]
- 21.Filosto M, et al. Clinical and genetic heterogeneity in progressive external ophthalmoplegia due to mutations in polymerase gamma. Arch Neurol. 2003;60:1279–1284. doi: 10.1001/archneur.60.9.1279. [DOI] [PubMed] [Google Scholar]
- 22.Davidzon G, et al. POLG mutations and Alpers syndrome. Ann Neurol. 2005;57:921–923. doi: 10.1002/ana.20498. [DOI] [PubMed] [Google Scholar]
- 23.Scarpulla RC. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol Rev. 2008;88:611–638. doi: 10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- 24.Ekstrand MI, et al. Mitochondrial transcription factor A regulates mtDNA copy number in mammals. Hum Mol Genet. 2004;13:935–944. doi: 10.1093/hmg/ddh109. [DOI] [PubMed] [Google Scholar]
- 25.Wenz T, et al. Activation of the PPAR/PGC-1α pathway prevents a bioenergetic deficit and effectively improves a mitochondrial myopathy phenotype. Cell Metab. 2008;8:249–256. doi: 10.1016/j.cmet.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Chowanadisai W, et al. Pyrroloquinoline quinone stimulates mitochondrial biogenesis through cAMP response element-binding protein phosphorylation and increased PGC-1α expression. J Biol Chem. 2010;285:142–152. doi: 10.1074/jbc.M109.030130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mootha VK, et al. Errα and Gabpa/b specify PGC-1α-dependent oxidative phosphorylation gene expression that is altered in diabetic muscle. Proc Natl Acad Sci USA. 2004;101:6570–6575. doi: 10.1073/pnas.0401401101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wenz T, et al. Increased muscle PGC-1α expression protects from sarcopenia and metabolic disease during aging. Proc Natl Acad Sci USA. 2009;106:20405–20410. doi: 10.1073/pnas.0911570106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Ikeuchi M, et al. Overexpression of mitochondrial transcription factor a ameliorates mitochondrial deficiencies and cardiac failure after myocardial infarction. Circulation. 2005;112:683–690. doi: 10.1161/CIRCULATIONAHA.104.524835. [DOI] [PubMed] [Google Scholar]
- 30.Mita S, et al. Detection of “deleted” mitochondrial genomes in cytochrome-c oxidase-deficient muscle fibers of a patient with Kearns-Sayre syndrome. Proc Natl Acad Sci USA. 1989;86:9509–9513. doi: 10.1073/pnas.86.23.9509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moraes CT, et al. Molecular analysis of the muscle pathology associated with mitochondrial DNA deletions. Nature Genet. 1992;1:359–367. doi: 10.1038/ng0892-359. [DOI] [PubMed] [Google Scholar]
- 32.Hirano M, et al. Mitochondrial neurogastrointestinal encephalomyopathy (MNGIE): clinical, biochemical, and genetic features of an autosomal recessive mitochondrial disorder. Neurology. 1994;44:721–727. doi: 10.1212/wnl.44.4.721. [DOI] [PubMed] [Google Scholar]
- 33.Nishino I, et al. Thymidine phosphorylase gene mutations in MNGIE, a human mitochondrial disorder. Science. 1999;283:689–692. doi: 10.1126/science.283.5402.689. [DOI] [PubMed] [Google Scholar]
- 34.Valentino ML, et al. Thymidine and deoxyuridine accumulate in tissues of patients with mitochondrial neurogastrointestinal encephalomyopathy (MNGIE) FEBS Lett. 2007;581:3410–3414. doi: 10.1016/j.febslet.2007.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lopez LC, et al. Unbalanced deoxynucleotide pools cause mitochondrial DNA instability in thymidine phosphorylase-deficient mice. Hum Mol Genet. 2009;18:714–722. doi: 10.1093/hmg/ddn401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spinazzola A, et al. Altered thymidine metabolism due to defects of thymidine phosphorylase. J Biol Chem. 2002;277:4128–4133. doi: 10.1074/jbc.M111028200. [DOI] [PubMed] [Google Scholar]
- 37.la Marca G, et al. Pre- and post-dialysis quantitative dosage of thymidine in urine and plasma of a MNGIE patient by using HPLC-ESI-MS/MS. J Mass Spectrom. 2006;41:586–592. doi: 10.1002/jms.1013. [DOI] [PubMed] [Google Scholar]
- 38.Yavuz H, et al. Treatment of mitochondrial neurogastrointestinal encephalomyopathy with dialysis. Arch, Neurol. 2007;64:435–438. doi: 10.1001/archneur.64.3.435. [DOI] [PubMed] [Google Scholar]
- 39.Lara MC, et al. Infusion of platelets transiently reduces nucleoside overload in MNGIE. Neurology. 2006;67:1461–1463. doi: 10.1212/01.wnl.0000239824.95411.52. [DOI] [PubMed] [Google Scholar]
- 40.Schüpbach M, et al. Allogeneic hematopoietic stem cell transplantation for mitochondrial neurogastrointestinal encephalomyopathy (MNGIE) Neurology. 2009;73:332. doi: 10.1093/brain/awv226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ogasahara S, et al. Muscle coenzyme Q deficiency in familial mitochondrial encephalomyopathy. Proc Natl Acad Sci USA. 1989;86:2379–2382. doi: 10.1073/pnas.86.7.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gironi M, et al. Late-onset cerebellar ataxia with hypogonadism and muscle coenzyme Q10 deficiency. Neurology. 2004;62:818–820. doi: 10.1212/01.wnl.0000113719.67643.b7. [DOI] [PubMed] [Google Scholar]
- 43.Lopez LC, et al. Leigh syndrome with nephropathy and CoQ10 deficiency due to decaprenyl diphosphate synthase subunit 2 (PDSS2) mutations. Am J Hum genet. 2006;79:1125–1129. doi: 10.1086/510023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Quinzii C, et al. A mutation in para-hydroxybenzoate-polyprenyl transferase (COQ2) causes primary coenzyme Q10 deficiency. Am J Hum Genet. 2006;78:345–349. doi: 10.1086/500092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Montini G, et al. Early coenzyme Q10 supplementation in primary coenzyme Q10 deficiency. N Engl J Med. 2008;358:2849–2850. doi: 10.1056/NEJMc0800582. [DOI] [PubMed] [Google Scholar]
- 46.Rossignol R, et al. Energy substrate modulates mitochondrial structure and oxidative capacity in cancer cells. Cancer Res. 2004;64:985–993. doi: 10.1158/0008-5472.can-03-1101. [DOI] [PubMed] [Google Scholar]
- 47.Margineantu DH, et al. Cell cycle dependent morphology changes and associated mitochondrial DNA redistribution in mitochondria of human cell lines. Mitochondrion. 2002;1:425–435. doi: 10.1016/s1567-7249(02)00006-5. [DOI] [PubMed] [Google Scholar]
- 48.Park KS, et al. Selective actions of mitochondrial fission/fusion genes on metabolism-secretion coupling in insulin-releasing cells. J Biol Chem. 2008;283:33347–33356. doi: 10.1074/jbc.M806251200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao J, et al. The novel conserved mitochondrial inner-membrane protein MTGM regulates mitochondrial morphology and cell proliferation. J Cell Sci. 2009;122:2252–2262. doi: 10.1242/jcs.038513. [DOI] [PubMed] [Google Scholar]
- 50.Jourdain I, et al. The dynamin related protein Dnm1 fragments mitochondria in a microtubule-dependent manner during the fission yeast cell cycle. Cell Motil Cytoskel. 2009;66:509–523. doi: 10.1002/cm.20351. [DOI] [PubMed] [Google Scholar]
- 51.Willems PH, et al. Mitochondrial dynamics in human NADH:ubiquinone oxidoreductase deficiency. Int J Biochem Cell Biol. 2009;41:1773–1782. doi: 10.1016/j.biocel.2009.01.012. [DOI] [PubMed] [Google Scholar]
- 52.Legros F, et al. Organization and dynamics of human mitochondrial DNA. J Cell Sci. 2004;117:2653–2662. doi: 10.1242/jcs.01134. [DOI] [PubMed] [Google Scholar]
- 53.Wikstrom JD, et al. What can mitochondrial heterogeneity tell us about mitochondrial dynamics and autophagy? Int J Biochem Cell Biol. 2009;41:1914–1927. doi: 10.1016/j.biocel.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 54.Gilkerson RW, et al. Mitochondrial nucleoids maintain genetic autonomy but allow for functional complementation. J Cell Biol. 2008;181:1117–1128. doi: 10.1083/jcb.200712101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hermann GJ, et al. Mitochondrial fusion in yeast requires the transmembrane GTPase Fzo1p. J Cell Biol. 1998;143:359–373. doi: 10.1083/jcb.143.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chan DC. Mitochondria: dynamic organelles in disease, aging, and development. Cell. 2006;125:1241–1252. doi: 10.1016/j.cell.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 57.Chen H, Chan DC. Mitochondrial dynamics - fusion, fission, movement, and mitophagy - in neurodegenerative diseases. Hum Mol Genet. 2009;18:R169–R176. doi: 10.1093/hmg/ddp326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen H, et al. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smirnova E, et al. Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol Biol Cell. 2001;12:2245–2256. doi: 10.1091/mbc.12.8.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cassidy-Stone A, et al. Chemical inhibition of the mitochondrial division dynamin reveals its role in Bax/Bak-dependent mitochondrial outer membrane permeabilization. Dev Cell. 2008;14:193–204. doi: 10.1016/j.devcel.2007.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cui M, et al. Perturbations in mitochondrial dynamics induced by human mutant PINK1 can be rescued by the mitochondrial division inhibitor mdivi-1. J Biol Chem. 2010;285:11740–11752. doi: 10.1074/jbc.M109.066662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brini M, et al. A calcium signaling defect in the pathogenesis of a mitochondrial DNA inherited oxidative phosphorylation deficiency. Nature Med. 1999;5:951–954. doi: 10.1038/11396. [DOI] [PubMed] [Google Scholar]
- 63.Visch HJ, et al. Inhibition of mitochondrial Na+-Ca2+ exchange restores agonist-induced ATP production and Ca2+ handling in human complex I deficiency. J Biol Chem. 2004;279:40328–40336. doi: 10.1074/jbc.M408068200. [DOI] [PubMed] [Google Scholar]
- 64.Jenuth JP, et al. Random genetic drift in the female germline explains the rapid segregation of mammalian mitochondrial DNA. Nat Genet. 1996;14:146–151. doi: 10.1038/ng1096-146. [DOI] [PubMed] [Google Scholar]
- 65.Melov S, et al. Multi-organ characterization of mitochondrial genomic rearrangements in ad libitum and caloric restricted mice show striking somatic mitochondrial DNA rearrangements with age. Nucl Acids Res. 1997;25:974–982. doi: 10.1093/nar/25.5.974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pohjoismaki JL, et al. Mammalian mitochondrial DNA replication intermediates are essentially duplex but contain extensive tracts of RNA/DNA hybrid. J Mol Biol. 2010;397:1144–1155. doi: 10.1016/j.jmb.2010.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Manfredi G, et al. Oligomycin induces a decrease in the cellular content of a pathogenic mutation in the human mitochondrial ATPase 6 gene. J Biol Chem. 1999;274:9386–9391. doi: 10.1074/jbc.274.14.9386. [DOI] [PubMed] [Google Scholar]
- 68.Santra S, et al. Ketogenic treatment reduces deleted mitochondrial DNAs in cultured human cells. Ann Neurol. 2004;56:662–669. doi: 10.1002/ana.20240. [DOI] [PubMed] [Google Scholar]
- 69.Twig G, et al. Mitochondrial fusion, fission and autophagy as a quality control axis: the bioenergetic view. Biochim Biophys Acta. 2008;1777:1092–1097. doi: 10.1016/j.bbabio.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Narendra D, et al. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vives-Bauza C, et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc Natl Acad Sci USA. 107:378–383. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ahola-Erkkilä S, et al. Ketogenic diet slows down mitochondrial myopathy progression in mice. Hum Mol Genet. 2010 doi: 10.1093/hmg/ddq076. [DOI] [PubMed] [Google Scholar]
- 73.Holt IJ, et al. A new mitochondrial disease associated with mitochondrial DNA heteroplasmy. Am J Hum Genet. 1990;46:428–433. [PMC free article] [PubMed] [Google Scholar]
- 74.Tanaka M, et al. Gene therapy for mitochondrial disease by delivering restriction endonuclease SmaI into mitochondria. J Biomed Sci. 2002;9:534–541. doi: 10.1159/000064726. [DOI] [PubMed] [Google Scholar]
- 75.Urnov FD, Rebar EJ. Designed transcription factors as tools for therapeutics and functional genomics. Biochem Pharmacol. 2002;64:919–923. doi: 10.1016/s0006-2952(02)01150-4. [DOI] [PubMed] [Google Scholar]
- 76.Porteus MH, Carroll D. Gene targeting using zinc finger nucleases. Nature Biotechnol. 2005;23:967–973. doi: 10.1038/nbt1125. [DOI] [PubMed] [Google Scholar]
- 77.Minczuk M, et al. Sequence-specific modification of mitochondrial DNA using a chimeric zinc finger methylase. Proc Natl Acad Sci USA. 2006;103:19689–19694. doi: 10.1073/pnas.0609502103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Minczuk M, et al. Development of a single-chain, quasi-dimeric zinc-finger nuclease for the selective degradation of mutated human mitochondrial DNA. Nucl Acids Res. 2008;36:3926–3938. doi: 10.1093/nar/gkn313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Boch J, et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- 80.Fromenty B, et al. High proportions of mtDNA duplications in patients with Kearns-Sayre syndrome occur in the heart. Am J Med Genet. 1997;71:443–452. doi: 10.1002/(sici)1096-8628(19970905)71:4<443::aid-ajmg14>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 81.Pohjoismaki JL, et al. Human heart mitochondrial DNA is organized in complex catenated networks containing abundant four-way junctions and replication forks. J Biol Chem. 2009;284:21446–21457. doi: 10.1074/jbc.M109.016600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Craven L, et al. Pronuclear transfer in human embryos to prevent transmission of mitochondrial DNA disease. Nature. 2010 doi: 10.1038/nature08958. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tachibana M, et al. Mitochondrial gene replacement in primate offspring and embryonic stem cells. Nature. 2009;461:367–372. doi: 10.1038/nature08368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rubenstein DS, et al. Germ-line therapy to cure mitochondrial disease: protocol and ethics of in vitro ovum nuclear transplantation. Camb Q Healthc Ethics. 1995;4:316–339. doi: 10.1017/s0963180100006071. [DOI] [PubMed] [Google Scholar]
- 85.Edgar D, et al. Random point mutations with major effects on protein-coding genes are the driving force behind premature aging in mtDNA mutator mice. Cell Metab. 2009;10:131–138. doi: 10.1016/j.cmet.2009.06.010. [DOI] [PubMed] [Google Scholar]
- 86.Bender A, et al. High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease. Nature Genet. 2006;38:515–517. doi: 10.1038/ng1769. [DOI] [PubMed] [Google Scholar]
- 87.Maximo V, et al. Mitochondria and cancer. Virchows Arch. 2009;454:481–495. doi: 10.1007/s00428-009-0766-2. [DOI] [PubMed] [Google Scholar]
- 88.Magrane J, Manfredi G. Mitochondrial function, morphology, and axonal transport in amyotrophic lateral sclerosis. Antioxid Redox Signal. 2009;11:1615–1626. doi: 10.1089/ars.2009.2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Area-Gomez E, et al. Presenilins are enriched in endoplasmic reticulum membranes associated with mitochondria. Am J Pathol. 2009;175:1810–1816. doi: 10.2353/ajpath.2009.090219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schon EA, Area-Gomez E. Is Alzheimer disease a disorder of mitochondria-associated membranes? J Alzheimers Dis. 2010 doi: 10.3233/JAD-2010-100495. in press. [DOI] [PubMed] [Google Scholar]