Abstract
Hepatitis C virus (HCV) NS5B polymerase is a key target for the development of therapeutic agents aimed at the treatment of HCV infections. Here we report on the identification of novel allosteric inhibitors of HCV NS5B through a combination of structure-based virtual screening, synthesis and structure-activity relationship (SAR) optimization approach. Virtual screening of 260,000 compounds from the ChemBridge database against the tetracyclic indole inhibitor binding pocket of NS5B (allosteric pocket-1, AP-1), sequentially down-sized the library by 4 orders of magnitude to yield 23 candidates. In vitro evaluation of the NS5B inhibitory activity of the in-silico selected compounds resulted in 17% hit rate, identifying two novel chemotypes. Of these, compound 3, bearing the rhodanine scaffold, proved amenable for productive SAR exploration and synthetic modification. As a result, 25 derivatives that exhibited IC50 values ranging from 7.7 to 68.0 μM were developed. Docking analysis of lead compound 28 within the tetracyclic indole- and benzylidene-binding allosteric pockets (AP-1 and AP-3, respectively) of NS5B revealed topological similarities between these two pockets. Compound 28, a novel rhodanine analog with NS5B inhibitory potency in the low micromolar level range may be a promising lead for future development of more potent NS5B inhibitors.
Keywords: NS5B polymerase; Virtual screening; Rhodanine; Imidazocoumarin, SAR
1. Introduction
Hepatitis C virus (HCV), the etiological agent of parenteral non-A, non-B hepatitis, often causes the development of malignant chronic disease, including liver cirrhosis and hepatocellular carcinoma, which frequently ends in liver failure.1-4 An estimated 200 million cases of HCV infections exist worldwide, of which over 4.1 million infections occur in the United States.5 At present, neither a vaccine nor a therapy with effective broad spectrum mode of action against all genotypes of HCV is available.6-8 Current HCV therapy comprising of pegylated interferon α (PEG-IFN-α) in combination with ribavirin has found limited patient compliance due to severe adverse effects.9,10 Therefore, there is a need to develop novel anti-HCV agents with high therapeutic index, reduced side-effects, and a convenient route of administration.
HCV is an enveloped, positive-stranded RNA virus with ~9.6 kb genome that encodes three structural (Core, E1, and E2) and seven nonstructural (p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B) proteins.11-13 Nonstructural protein 5B (NS5B), a 66 kDa RNA-dependent RNA polymerase (RdRp) is an important therapeutic target, since it plays a pivotal role in replicating the HCV RNA genome and the host lacks its functional equivalent.14-16 The availability of several high-resolution three-dimensional crystal structures of NS5B either alone or in complex with ribonucleotide substrates or non-nucleoside inhibitors, has resulted in the identification of five distinct NS5B inhibitor binding pockets to-date.8,17-19 Three of these correspond to NS5B allosteric inhibitor binding pockets: allosteric pocket (AP)-1, -2, and -3.17-20 AP-1 is located on the surface of the thumb domain adjacent to the allosteric GTP-binding site.21,22 Inhibitors identified against this pocket include benzimidazole23, indole24, quinoxaline25, thieno[3,2-b]pyrrole26, and coumestan27 derivatives. AP-2 is located in the thumb domain, next to AP-1.28-30 Chemotypes of AP-2 binding analogs include the thiazolone31, N,N-disubstituted phenylalanine32, thiophene-2-carboxylic acid33, pyranoindole34, dihydropyranone35, and thiazolidin-4-one36 derivatives. AP-3 is located adjacent to the active site and includes inhibitors such as benzothiadiazine37, benzylidene38, proline sulfonamide39, anthranilic acid40, acrylic acid41, and pyrrolidine42 derivatives. Crystallographic analyses of these NS5B-inhibitor complexes in combination with high-throughput-screening (HTS), have uncovered several diverse chemical scaffolds of HCV NS5B non-nucleoside inhibitors (NNIs).17-20 In recent years, virtual screening has been emerging as a complementary approach to HTS in an effort to identify potential leads in the drug discovery process.43
Previously, we have reported on the utility of 3D QSAR methodologies such as the comparative molecular field analysis (CoMFA) and comparative molecular similarity indices analysis (CoMSIA) for investigating and optimizing benzimidazole HCV NS5B polymerase inhibitors which bind to AP-1 of NS5B.44 In another study, we identified 2,3-disubstituted-4-thiazolidinone derivatives, as modest inhibitors of NS5B polymerase, and predicted that they bind to AP-2 of NS5B.36 In the present study, we have employed virtual screening tool to identify novel small molecule NNIs of NS5B and further optimized the 5-benzylidene rhodanine-3-acetic acid hit based on the crystal structure of NS5B in complex with a tetracyclic indole inhibitor (PDB ID: 2dxs).24
2. Results
2.1. Validity of docking and virtual screening approach
As a first step, we validated the ability of the docking algorithm to reproduce the co-crystallized pose of the tetracyclic indole, thiazolone and benzylidene inhibitors within their respective allosteric pockets on NS5B. This yielded a good agreement between the docked and crystal structures as is evident from the root mean square (rms) deviation values of 0.827, 0.332 and 0.111 Å for tetracyclic indole, thiazolone and benzylidene inhibitors, respectively. This confirms the reliability of the Glide docking procedure in reproducing the experimentally observed binding mode for NS5B inhibitors. Therefore, the parameters set for Glide docking appear reasonable to provide meaningful insight into the predicted binding mode for hitherto untested NS5B inhibitors.
We next evaluated whether this methodology was amenable to discriminate between known inhibitors and randomly chosen drug-like molecules from the ChemBridge database. For this we docked the “validation set” into the tetracyclic indole binding site of NS5B, and analyzed the list of the “hit” molecules for the top 10% highest scoring compounds. This investigation yielded 11 of the 20 known NNIs as the top 10% scoring molecules (Supplementary data, Fig. S1). Of these 11 compounds, the most potent benzimidazole analog was identified as the top ranked hit with a VS (virtual screening) GlideScore of -7.07. It was interesting to note that approximately 8 compounds with low micromolar to sub-micromolar NS5B inhibitory activity exhibited a VS GlideScore of ~ -5.00. Since the majority of these known NS5B NNIs seeded in the “validation dataset” as test standards yielded VS GlideScores ranging between -5.0 to -7.0 (Supplementary data, Fig. S1), we set VS GlideScore -5.0 as a threshold parameter to identify diverse hit molecules.
2.2. Virtual screening of compounds against AP-1 of HCV NS5B
A step-wise strategy for virtual screening was employed to identify allosteric inhibitors of NS5B polymerase. Towards this end, a ChemBridge database comprising of 260,000 compounds was filtered according to the criteria described in the Methods section, to generate a new database comprising of 52,000 3D compounds (Fig. 1). Irrespective of whether the ligands were treated as neutral or ionized, did not change the outcome of the screening investigations. However, upon ionization, the ligands exhibited relatively more negative binding energies and their ionizable groups such as –COOH oriented towards the guanidine group of Arg503 as opposed to the backbone of Gly493 as observed in their neutral state. The new database comprising of 52,000 3D compounds was docked in AP-1 of NS5B polymerase, employing the Glide's High Throughput Virtual Screening (HTVS) workflow. This second screening resulted in the elimination of ~98% molecules based on the application of VS GlideScore selection cut-off of -5.0. The remaining 650 compounds were visually inspected based on our previous in-silico44 and wet lab27,36 experience for improbable docking orientations in AP-1 of NS5B. The ensuing compounds were further analyzed for their interactions with the amino acid residues which are important for binding of indole and benzimidazole allosteric inhibitors44, in context of the following parameters: (1) possess at least one hydrophobic group pointing towards hydrophobic pocket 1 (HP-1) formed by residues Val37, Ala393, Ala396, Leu492, and Val494 or hydrophobic pocket 2 (HP-2) formed by residues Leu392, Ala395, Ala396, Ile424, Leu425, His428, and Phe429, (2) form at least one hydrogen bond with His428, Ser431 and Gly493 or ionic interaction with the guanidine group of Arg503, and (3) display structural uniqueness (to avoid redundancy among chemically similar hits) (Fig. 2). This rigorous in-silico screening criteria ultimately filtered out 627 compounds, thus yielding a set of 23 compounds bearing structural diversity and good docking orientations for biological evaluation.
Fig. 1.
Virtual screening parameters employed for identifying potential NS5B allosteric inhibitors from the ChemBridge database.
Fig. 2.
Representative key pharmacophoric features employed for visual inspection and selection of compounds against tetracyclic indole inhibitor binding allosteric pocket of NS5B. The pharmacophoric features include hydrophobic groups pointing to HP-1 and/or HP-2, hydrogen bond acceptor or negatively charged groups and the central heterocyclic scaffold to hold these features together.
2.3. In vitro screening of inhibitors
To investigate the inhibitory activity of the virtually screened 23 candidates against NS5B, in vitro NS5B RNA dependent RNA polymerase (RdRp) inhibition assay was carried out as described in Methods section.27,36 The RdRp reaction was performed on poly rA/U12 template-primer (TP), employing recombinant HCV NS5B (genotype 1b) with an N-terminal His-tag and C-terminal 21-amino acid deletion (NS5BCΔ21).27,36 LQB34, a coumestan derivative, previously characterized by us as a NS5B inhibitor was included as an internal reference standard.27 To identify candidates belonging to a wider range of structural scaffolds, preliminary screening was conducted at a concentration of 250 μM for each compound. This analysis yielded four compounds exhibiting ≥50% inhibition of NS5B RdRp activity (Table 1), thus demonstrating a 17% hit rate. Of these, compounds 3 (rhodanine analog) and 4 (imidazocoumarin analog) exhibited IC50 values of 55.2 μM and 60.2 μM, respectively. Further exploration of SAR around imidazocoumarin analog 4 resulted in either inactive or poorly active analogs (Supplementary data, compounds 39-43, Table S1). Thus we pursued compound 3 for further SAR investigations.
Table 1.
Anti-NS5B RdRp activity of compounds 1-4
 | ||||
|---|---|---|---|---|
| Compounda | MW | ClogPb | % inhibition @ 250 μMc | IC50 (μM)d |
| 1 | 283 | 1.73 | 55±2.8 | ~200 |
| 2 | 282 | 3.24 | 48.6±4.9 | ~250 |
| 3 | 347 | 2.31 | 71±7.0 | 55.2±1.10 |
| 4 | 262 | 2.89 | 62.5±7.7 | 60.2±1.17 |
Compounds acquired from ChemBridge were reported to have a purity ≥90% by 1H NMR
ClogP was calculated using QikProp v3.0
Percent inhibition was determined at 250 μM concentration of the indicated compound and represents an average of at least two independent measurements in duplicate. NS5B RdRp activity in the absence of the inhibitor was taken as 100 percent after subtraction of residual background activity.
The IC50 values of compounds 3 and 4 were determined from dose-response curves using 8-12 concentrations of each compound in duplicate in two independent experiments. Curves were fitted to data points using nonlinear regression analysis and IC50 values were interpolated from the resulting curves using GraphPad Prism 3.03 software.
2.4. Development of SAR around rhodanine scaffold
Following the initial rhodanine hit 3, commercially available rhodanine analogs (compounds 5-22) were obtained for exploration of SAR around 3- and 5-position of the rhodanine scaffold, leading to the development of detailed SAR data (Table 2) and identification of potent analog (compound 22, IC50 = 10.6 μM) as described below.
Table 2.
Structure-activity relationship of compound 3 derivatives
 | |||||
|---|---|---|---|---|---|
| Compda | R | Ar | MW | ClogPb | IC50 (μM)c |
| 5 | -CH2COOH | 3-CF3Ph | 347 | 2.31 | 50.9±1.4 |
| 6 | -CH2COOH | 3-BrPh | 358 | 3.02 | 20.2±1.2 |
| 7 | -CH2COOH | 2,4-ClPh | 348 | 3.40 | 17.9±1.2 |
| 8 | -CH2CH2COOH | 2,4-ClPh | 362 | 3.96 | 19.9±1.3 |
| 9 | -CH2CH2SO3H | 2,4-ClPh | 398 | 3.20 | 23.5±1.2 |
| 10 | -CH2CH2CH2COOH | 2,4-ClPh | 376 | 4.31 | 58.0±1.5 |
| 11 | -(CH2)5COOH | 2,4-ClPh | 404 | 5.10 | 16.1±1.3 |
| 12 | -CH2COOH | naphthalene-1-yl | 329 | 3.60 | 61.4±1.3 |
| 13 | -CH(COOH)CH3 | naphthalen-1-yl | 343 | 3.94 | 68.1±1.5 |
| 14 | -CH(COOH)CH(CH3)2 | naphthalen-1-yl | 371 | 4.64 | 16.9±1.2 |
| 15 | -CH(COOH)CH(CH3)2 | 4-FPh | 339 | 4.13 | 42.9±1.3 |
| 16 | -CH(COOH)CH(CH3)2 | 2-ClPh | 355 | 4.18 | 36.1±1.2 |
| 17 | -CH(COOH)CH(CH3)2 | 2,4-ClPh | 390 | 4.73 | 14.1±1.2 |
| 18 | -CH(COOH)CH2CH2CH3 | 2,4-ClPh | 390 | 4.71 | 28.2±1.1 |
| 19 | -CH(COOH)CH2CH2SCH3 | 2,4-ClPh | 422 | 5.26 | 29.2±1.2 |
| 20 | -CH(COOH)CH2CH(CH3)2 | 2,4-ClPh | 404 | 5.28 | 18.9±1.1 |
| 21 | -CH(COOH)CH3 | 2,4-ClPh | 362 | 3.94 | 13.3±1.2 |
| 22 | -CH(COOH)CH2Ph | 2,4-ClPh | 438 | 5.77 | 10.6±1.5 |
| 23 | -CH2COOH | 2-NO2Ph | 324 | 2.10 | 16.2±1.4 |
| 24 | -CH2COOH | 4-NO2Ph | 324 | 2.01 | 21.9±1.3 |
| 25 | -CH2COOH | 4-FPh | 297 | 2.65 | 42.1±1.0 |
| 26 | -CH2COOH | 3-ClPh | 313 | 2.82 | 18.4±1.2 |
| 27 | -CH2COOH | 3,4-ClPh | 348 | 3.39 | 8.4±1.5 |
| 28 | -CH2COOH | 3-phenoxyPh | 371 | 4.26 | 7.7±1.4 |
Compounds 5-8, and 12 acquired from ChemBridge and compounds 9-11 and 13-22 acquired from Sigma were reported to have a purity ≥ 90% by 1H NMR, compounds 23-28 were synthesized in the present study. The key for b and c is the same as that of corresponding b and d keys used in Table 1.
Moving –CF3 group on the 5-benzylidene moiety of 3 (IC50 = 55.2 μM) from para to the meta position (compound 5, IC50 = 50.9 μM) led to a marginal enhancement of inhibitory activity. Bioisosteric replacement of the –CF3 group in compound 5 with –Br group (compound 6, IC50 = 20.2 μM) resulted in a 2.5-fold increase in inhibitory activity. Further improvement in inhibitory activity was obtained when 3-CF3 benzylidene was replaced with 2,4-dichlorobenzylidene moiety (compound 7, IC50 = 17.9 μM). We purchased compounds 8-11, to explore the effect of 3-position substituents on rhodanine ring in the presence of 2,4-dichlorobenzylidene substituent at 5-position of the rhodanine ring. Separating α–carboxyl group from the rhodanine core by ethylene bridge resulted in a marginal decrease in activity (compound 8, IC50 = 19.9 μM). Further decrease in activity was observed when carboxyl group in compound 8 was replaced with bioisosteric sulfonic acid group (compound 9, IC50 = 23.5 μM). Separation of carboxyl group from the rhodanine core by a propylene linker resulted in substantial loss of activity (compound 10, IC50 = 58.0 μM), in contrast the activity improved by 3.6-fold when the pentylene linker was used (compound 11, IC50 = 16.1 μM). Several additional analogs such as: 5-(2,4-dichlorobenzylidene)-3-ethyl-2-thioxothiazolidin-4-one, 2-(4-oxo-2-thioxothiazolidin-3-yl)acetic acid, and 2-(5-benzylidene-4-oxo-2-thioxothiazolidin-3-yl)acetic acid lacking (a) a carboxyl group at N3 substituent, (b) benzylidene moiety at C5-position, and (c) substituents on benzylidene moiety, respectively, were found to be either inactive or poorly active (Supplementary data, compounds 29-34, Table S1). In addition, analogs with electron donating substituents on the benzylidene moiety (Supplementary data, compounds 35 and 36, Table S1) exhibited poor activity, whereas electron withdrawing 3-cyano and 3-carboxy substituents on the benzylidene moiety (Supplementary data, compounds 37 and 38, Table S1) exhibited higher NS5B inhibitory activity.
The 1-naphthylidene substituent at 5-position of the rhodanine ring was detrimental to the NS5B inhibitory activity when the N3-substituent was either an acetic acid (compound 12, IC50 = 61.4 μM) or α-methyl acetic acid (compound 13, IC50 = 68.1 μM), in contrast to a bulkier α-isopropyl acetic acid substituent (compound 14, IC50 = 16.9 μM) which was beneficial. We next obtained compounds 15-17, which similar to compound 14 carried the α-isopropyl acetic acid substituent at 3-position of the rhodanine ring, but harbored a series of substituted benzylidenes in lieu of 1-naphthylidene moiety. While 4-fluorobenzylidene (compound 15, IC50 = 42.9 μM) and 2-chlorobenzylidene (compound 16, IC50 = 36.1 μM) analogs proved to be approximately 2-3 fold less active compared to compound 14, comparable inhibitory activity was observed in the presence of 2,4-dichlorobenzylidene substituent (compound 17, IC50 = 14.1 μM).
Based on these findings, we procured compounds 18-22 bearing 2,4-dichlorobenzylidene substituent at 5-position and a various substituents at 3-position of the rhodanine ring. While α-propylacetic acid (compound 18, IC50 = 28.2 μM) and α-methylthioethyl acetic acid (compound 19, IC50 = 29.2 μM) substituents resulted in two-fold decrease in inhibitory activity, the bulkier α-isobutyl acetic acid analog (compound 20, IC50 = 18.9 μM) and the less bulky α-methyl acetic acid analog (compound 21, IC50 = 13.3 μM) exhibited activities in a comparable range to compound 17. Finally, compound 22 having α-benzyl acetic acid group at 3-position (IC50 = 10.6 μM) proved to be the most potent compound of the commercially available rhodanine series (Table 2).
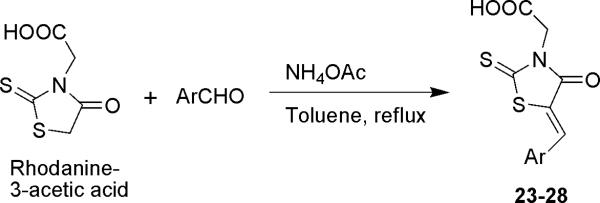
With the SAR information gathered from the in vitro evaluation of commercially available rhodanine analogs against NS5B, we sought to synthesize novel rhodanine analogs in order to identify more potent compounds. We chose rhodanine-3-acetic acid as the scaffold for further SAR exploration aiming at variations at C5-benzylidene substituent. This choice was based on the following rationale: 1) reasonably active compounds 6 (IC50 = 20.2 μM) and 7 (IC50 = 17.9 μM) both bearing rhodanine-3-acetic acid functionality; 2) absence of stereogenic center; 3) easy availability of starting material; and 4) one step synthetic derivatization at C5-position through Knoevenagel condensation leading to target compounds. Addition of arylalkylidene moiety at C5-position of the rhodanine-3-acetic acid was accomplished by a Knoevenagel condensation to afford target compounds 23-28 as shown in Scheme 1. This condensation reaction with aromatic aldehyde provided only the Z isomer, as determined by the chemical shift of the methylene proton ranging from 7.9-8.1 as a singlet.45,46 The synthesized compounds 23-28 were subjected to in vitro NS5B evaluation (Table 2). The 2-nitrobenzylidene analog (compound 23, IC50 = 16.2 μM) exhibited higher activity versus the 4-nitrobenzylidene analog (compound 24, IC50 = 21.9 μM), suggesting that the 2-position of the benzylidene moiety may be favorable for nitro substituent. Replacement of nitro group in compound 24 with fluoro (compound 25, IC50 = 42.1 μM) led to ~2-fold decrease in activity indicating the importance of the size of the electron withdrawing group. The 3-chlorobenzylidene analog (compound 26, IC50 = 18.4 μM) exhibited comparable activity to compound 24. Finally, more than two-fold enhancement of inhibitory activity was obtained when 3-chlorobenzylidene was replaced with either 3,4-dichlorobenzylidene (compound 27, IC50 = 8.4 μM) or 3-phenoxybenzylidene (compound 28, IC50 = 7.7 μM), which led to the identification of the two most potent compounds from the in house synthetic efforts on rhodanine analogs. These data illustrate that carboxyl group at α-position of the N3-substituent and electron withdrawing group bearing benzylidene moiety at C5-position are essential pharmacophoric features of the rhodanine series of compounds for NS5B polymerase inhibitory activity.
Scheme 1.
Preparation of compounds 23-28
2.5. Docking analysis
We have identified a novel rhodanine analog (compound 3) as a “hit” molecule by virtual screening against AP-1 of NS5B. To gain a better insight into the potential binding mode of the optimized derivatives of this initial hit, we docked compounds 27 and 28 into each of the three reported HCV NS5B NNI binding sites represented by AP-124, AP-231 and AP-338 (Table 3). For comparison, we also docked two structurally related compounds, Amgen 5o (AP-3 bound inhibitor)38 and Valeant 6 (AP-2 bound inhibitor)31 within AP-1, AP-2 and AP-3 of NS5B (Table 3). Since the ionized state of the ligands at physiological pH is known to influence their binding modes and energetics of binding at the target sites, we have used the ionized form of these compounds for docking experiments.43 Consistent with their experimental binding modes, Amgen 5o exhibited tighter binding in AP-3 versus AP-1 and AP-2 of NS5B, while Valeant 6 bound much tighter in AP-2 compared to AP-1 or AP-3, as deduced from their relative XPGlideScore values (Table 3). Surprisingly compound 28 appeared to bind more favorably in AP-3 (XP-GlideScore = -6.37) versus AP-1 (XP-GlideScore = -5.14), whereas AP-2 (XP-GlideScore = -4.66) appeared to be the least favored. A similar binding trend was observed with compound 27. This observation was rather intriguing given that compounds 27 and 28 were derived from compound 3, our “hit” molecule identified through screening for AP-1 binding molecules. This aroused our curiosity to examine the binding energies for compound 3 and compound 22, the most potent compound of the commercially available series, within each of the three allosteric pockets of NS5B. Consistent with our screening analysis, both these compounds exhibited a better fit in AP-1 versus the other two pockets (Table 3). It may be noted that Amgen 5o (IC50 = 0.2 μM), although several-fold more potent than compound 28 (IC50 = 7.7 μM), exhibited weaker binding energetics compared to compound 28 in AP-3 (Table 3). This discrepancy is not surprising in view of the observation that Amgen 5o forms a covalent bond with Cys366 in the NS5B co-crystal structure.38 Consistent with this parameter, when the Amgen 5o bound structure (C5-benzylidene moiety linked to the rhodanine ring through a single bond) was docked into AP-3 it yielded XP-GlideScore value of -7.94 (through space interaction, data not shown).
Table 3.
Binding energies of compounds 3, 22, 27, 28, Amgen 5o and Valeant 6 in their ionized state within each of the allosteric pockets of NS5B
 | ||||
|---|---|---|---|---|
| Compounds | XP-GlideScore (kcal/mol) |
IC50 (μM) | ||
| AP-1 | AP-2 | AP-3 | ||
| 3 | -6.00 | -5.92 | -5.46 | 55.2 |
| 22 | -5.50 | -4.79 | -3.70 | 10.6 |
| 27 | -5.29 | -5.18 | -8.00 | 8.4 |
| 28 | -5.14 | -4.66 | -6.37 | 7.7 |
| Amgen 5o | -3.80 | -3.30 | -4.79 | 0.2 |
| Valeant 6 | -3.49 | -7.54 | -7.10 | 3.0 |
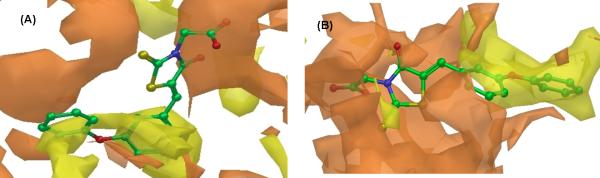
Further analysis of the Macromodel hydrophilic (orange color)/hydrophobic (yellow color) surface area representation of AP-1 (Fig. 3A) and AP-3 (Fig. 3B) of NS5B upon docking of compound 28 indicates topological similarities between these two pockets. For example, the 3-phenoxybenzylidene moiety of compound 28 appears to be completely engulfed within the yellow hydrophobic surface area in both AP-1 and AP-3 of NS5B. The rhodanine acetic acid moiety is surrounded by hydrophilic surface area in both AP-1 and AP-3 of NS5B. This finding may thus explain why compound 28 binds within both pockets, albeit with much stronger interaction in AP-3 versus AP-1, as indicated by its corresponding XP-GlideScores within each of these pockets (Table 3).
Fig. 3.
Macromodel hydrophilic (orange color) and hydrophobic (yellow color) surface representations of AP-1 (panel A) and AP-3 (panel B) of NS5B in the presence of compound 28.
As shown in Fig. 4A, compound 28 appears to span the entire AP-1 pocket (i.e., hydrophobic contacts with HP-1 and HP-2 and ionic interaction with Arg503). The solvent-exposed acetic acid group at N3-position of the rhodanine ring forms ionic interaction with the guanidine group of Arg503 (-COO---H2N-Arg503, 1.71 Å). A hydrogen bond was seen between the C4 carbonyl oxygen atom of the rhodanine ring and the guanidine group of Arg503 (-C=O---H2N-Arg503, 1.82 Å, 151.6°). The rhodanine ring is stabilized by the side chains of Pro495 and Trp500. The phenyl ring of the benzylidene group at C5-substituent forms hydrophobic contacts with several residues present in HP-2 such as Leu392, Ala395, Ala396, Ile424, Leu425, His428, Phe429 and Val494 whereas the terminal phenyl ring is stabilized by hydrophobic interactions with residues from HP-1 such as Val37, Ala393, Ala396, Leu492 and Val494.
Fig. 4.
XP-Glide predicted binding mode of compound 28 within AP-1 (panel A) and AP-3 (panel B) of NS5B. Important amino acids are depicted as sticks with the atoms colored as carbon – green, hydrogen – white, nitrogen – blue, oxygen – red and sulfur – yellow) whereas the inhibitor is shown as ball and stick model with the same color scheme as above except carbon atoms are represented in orange. Dotted black lines indicate either hydrogen bonding or ionic interaction.
Fig. 4B shows the binding mode of compound 28 within AP-3 of NS5B. The carboxyl group forms ionic interaction with the ε-amino group of Lys141 (-COO---H3N-Lys141, 1.79 Å). In addition, the carboxyl oxygen atom forms water1307 mediated hydrogen bonding interaction with the side chain of Glu143 and water1249 (-COO---H2O1307, 1.69 Å, 159.6°; 1307OH2---OOC-Glu143, 1.88 Å, 174.4°; 1307H2O---H2O1249, 2.12 Å, 166.6°). We hypothesize that a salt-bridge interaction between the ε-amino group of Lys141 and the carboxylate group of Glu143 might have destabilized due to the formation of salt-bridge interaction between the ε-amino group of Lys141 and the carboxylate group of compound 28. The C4 carbonyl oxygen atom of the rhodanine ring forms water mediated hydrogen bonding interactions with the backbone atoms of Gln446 and Gly449 (-C=O---H2O1432, 2.24 Å, 129.7°; 1432OH2---O=C-Gln446, 2.36 Å, 169.9°; 1432H2O---HN-Gly449, 2.13 Å, 169.3°). The rhodanine ring is stabilized by the side chains of Arg158, Gln446 and Ser556. The phenyl ring of the benzylidene group at C5-substituent forms hydrophobic contacts with Phe193, Cys366 and Tyr448, whereas the terminal phenyl ring is extensively stabilized by hydrophobic interactions with residues Leu384, Met414, Tyr415 and Tyr448. Apart from hydrophobic interactions, the terminal phenyl ring is also stabilized through π-cation type interaction with the guanidine group of Arg200. These models thus provide insight into the binding of compound 28 in AP-1 and AP-3 of NS5B.
3. Discussion
In this study, we have employed the virtual screening approach in an effort to identify and further design new inhibitors of HCV NS5B, a validated target for antiviral therapy against HCV. A number of previous studies have reported the utility of this strategy towards identifying novel lead molecules targeting HCV NS5B.47-51 These screening campaigns have explored several commercially available diversity set chemical libraries such as the 3D Pharmo DBTM (Equispharm, Korea), National Cancer Institute (NCI), LeadQuest, Maybridge, French National Library and Prestwick through pharmacophore or docking-based virtual screening approach with equal success.47-51 Notably, these studies have highlighted the cost-effectiveness of this strategy towards identifying potential leads by virtue of downsizing the library by several orders of magnitude. In our studies, we have explored 260,000 molecules from the ChemBridge compound library through virtual screening against the tetracyclic indole inhibitor binding pocket (AP-1) of NS5B24, and identified 23 candidates (Fig. 1), thus downsizing the library by 4 orders of magnitude, consistent with afore-mentioned reports.
Amongst the two novel chemotypes identified upon in vitro RdRp screening of the in-silico selected 23 compounds (Table 1), pursuit of SAR around the imidazocoumarin scaffold (compound 4) had to be abandoned as it yielded either inactive or poorly active analogs (Supplementary data, compounds 39-43, Table S1). In contrast, compound 3, bearing the rhodanine scaffold proved to be a suitable “hit” for development of more potent novel analogs, through SAR optimization and chemical synthesis (Table 2). Previous studies have reported the IC50 values for lead molecules identified through virtual screening, ranging from 20 μM for the 3D Pharmo DBTM set51 to 1 μM for compounds originating from the French National Library/Prestwick commercial library and the NCI library put together.50 Our “hit” compound 3, though it exhibited a higher IC50 value of 55 μM, a systematic SAR exploration and one step synthetic derivatization around a selected parent heterocyclic core of compound 7, ultimately yielded compound 28 (IC50 = 7.7 μM), the most potent NS5B inhibitor of this series. These findings clearly illustrate the applicability of the virtual screening protocol to develop novel analogs with a clear improvement in their biological activity.
Our two lead molecules, compounds 27 and 28, share some structural features with previously reported NS5B polymerase inhibitors from Amgen38 and Valeant.31 These may be compared and contrasted here as follows. For instance, Amgen 5o possesses an arylsulfonamide group at the N3-position as opposed to (un)branched carboxyalkyl groups present in our compounds (Table 3). Valeant 6 on the other hand harbors a thiazolone core in lieu of the 4-oxo-2-thioxothiazolidine core present in our compounds and in addition, bears 4-fluorophenylglycine at 2-position of the thiazolone ring instead of (un)branched carboxyalkyl moiety at N3-position as seen in our compounds (Table 3). Thus, our lead molecules appear to be more closely related to Amgen 5o, an AP-3 bound inhibitor38, an aspect consistent with their predicted binding mode in AP-3 of NS5B (Table 3). However, in the absence of a co-crystal structure for these compounds, one cannot rule out their binding in AP-1, given that these analogs were originally identified through screening against this pocket. We speculate that topological similarities between AP-1 and AP-3 of NS5B upon docking of compound 28 as indicated by the Macromodel hydrophilic/hydrophobic surface area representation (Fig. 3) may potentially account for the identification of our leads through AP-1 of NS5B.
Given the feasibility that these compounds may bind either AP-1 or AP-3 of NS5B, we have examined the binding characteristic of compound 28 within both these pockets, in order to gain insight for future design and development of more potent novel NS5B inhibitors. As shown in Fig. 4A, the phenyl ring of the benzylidene group at C5-substituent of compound 28 bound to HP-2 in AP-1 offers room for further modifications with small hydrophobic groups such as methyl and halogens at 4, 5 and 6 positions. Similarly, the terminal phenyl ring of compound 28 bound to HP-1 can tolerate substituents such as methyl and halogens. The α-carbon atom of the substituents at the N3-position of the rhodanine ring, being solvent-exposed, may be branched with a variety of amino acid side chains with appropriate stereochemistry to optimize the pharmacokinetic profile of this series of compounds.
Analysis of the XP-Glide predicted binding pose of compound 28 in AP-3 (Fig. 4B), suggests that 4, 5 and 6 position of the phenyl ring of the C5-benzylidene substituent can be substituted with small hydrophobic groups. Similarly, branching at the methine group of the C5-benzylidene substituent with small hydrophobic groups such as methyl, ethyl, or cyclopropyl may be well tolerated without compromising the inhibitory potency of the parent compounds. Further, the terminal phenyl ring can accommodate small groups such as methyl and halogens at ortho, meta and para positions. The vacant region around α-carbon of N3-substituent similar to that found in AP-1, provides further opportunity for optimizing the amino acid side chains in terms of bulk, hydrophobicity, charge, polarity and also the stereochemistry of the resulting chiral center.
4. Conclusions
The ChemBridge database consisting of 260,000 compounds was screened against the tetracyclic indole inhibitor binding allosteric pocket (AP-1) of NS5B to identify novel inhibitors. Thus, a total of 25 inhibitors belonging to the rhodanine scaffold with IC50 values in the range of 7.7 to 68.0 μM were identified through a combined use of virtual screening, SAR analysis, synthesis and biological evaluation. Current efforts are underway to establish the experimental binding mode of compound 28 with NS5B. In the interim the docking models of compound 28 within AP-1 and AP-3 of NS5B may serve as a guide for future NS5B inhibitor development endeavor.
5. Materials and Methods
5.1. Virtual Screening
A ChemBridge database comprising of a) the first 220,000 compounds from the 430,000 compounds present in Express Pick, February 2008, b) 30,000 compounds from MW set, and c) 10,000 compounds from Kinase set (ChemBridge Corporation) was initially prefiltered according to Lipinski's “Rule of Five”52, use of no tautomers, removal of metals from salts and one stereoisomer per ligand. Since previous studies have treated the ligands as either neutral or ionized during virtual screening experiments53, in this study we treated ligands independently as neutral and ionized. These filtering criteria resulted in filtering out ~80% of the compounds from the original 2D databases to generate 52,000 3D molecules using LigPrep (Schrodinger, LLC: New York), a ligand preparation software which generates a minimized conformation of each ligand.
Crystal structures of NS5B polymerase in complex with tetracyclic indole (PDB ID: 2DXS)24, thiazolone (PDB ID: 2I1R)31 and benzylidene (PDB ID: 2AX1)38 inhibitors, representing AP-1, AP-2 and AP-3 pockets, respectively, were used in the present study. AP-1 was used to perform the entire virtual screening operation, while AP-2 and AP-3 were employed as controls to evaluate the comparative binding potential of the newly identified analogs. The protonation states of residues in the allosteric site were adjusted to the dominant ionic forms at pH 7.4. Water molecules of crystallization were removed from all complexes except 2AX1, where the bound ligand was stabilized through hydrogen bonding with water molecule. The resultant protein was minimized for docking using the protein preparation facility implemented in Maestro v8.0 and Impact program v4.5 (Schrodinger, LLC: New York).
To validate the accuracy of our docking approach, we determined ‘the lowest energy pose’ (binding conformation) predicted by the XP-GlideScore function, versus the experimental binding mode as determined by X-ray crystallography. Towards this end, we removed the aforementioned crystallographic bound inhibitors from their binding sites, minimized and then re-docked them into their respective binding site on HCV NS5B polymerase. To establish the specificity of the Glide (Grid-based ligand docking with energetics) methodology (Schrodinger, LLC: New York) in discriminating known inhibitors from the randomly chosen drug-like molecules taken from ChemBridge database, the “validation dataset” comprising of 1000 molecules, of which 20 were known NS5B NNIs belonging to benzimidazole23, indole24 and quinoxaline25 series, were docked into the tetracyclic indole binding AP-1 of NS5B.
A flowchart depicting the various steps of virtual screening (VS), including database filtration and subsequent high throughput docking (default settings) studies is shown in Fig. 1.
5.2. Chemistry
Melting points (mp) of the synthesized compounds were determined on a Thomas-Hoover capillary melting point apparatus and are uncorrected. 2-nitrobenzaldehyde and 4-nitrobenzaldehyde were obtained from Sigma-Aldrich (Saint Louis, MO, USA); rhodanine-3-acetic acid, 3-chlorobenzaldehyde, 4-fluorobenzaldehyde, 3,4-dichlorobenzaldehye and 3-phenoxybenzaldehyde were obtained from VWR (Bridgeport, NJ, USA) and TCI America (Portland, OR, USA) and were used as received. All compounds were checked for their homogeneity by TLC using silica as stationary phase. The 1H NMR spectra were recorded on a Bruker 400 Avance DPX spectrometer outfitted with a z-axis gradient probe. The chemical shifts are reported as parts per million (δ ppm) downfield from tetramethylsilane (TMS) as an internal standard. Data are reported as follows: chemical shift, multiplicity (s) singlet, (d) doublet, (t) triplet, (m) multiplet. The C, H, and N analyses were performed by Atlantic Microlabs, Inc., (Norcross, GA, USA) and the observed values were within ±0.4% of calculated values.
5.2.1. General Procedure for the preparation of 5-benzylidene rhodanine-3-acetic acid derivatives
To a three neck reaction flask equipped with reflux condenser, rhodanine-3-acetic acid (0.3 g, 1.57 mmol) in 6 mL anhydrous toluene was added, followed by addition of ammonium acetate (1.60-3.20 mmol) and 1.57 mmol of the corresponding aromatic aldehydes. The reaction mixture was refluxed for 1-2 h under inert condition. The reaction was monitored by TLC. The reaction mixture was evaporated under vacuum, and the residue was dissolved in water. Aqueous solution was then acidified with conc. HCl and precipitated product was extracted into ethyl acetate, dried over sodium sulfate and evaporated under vacuum to obtain the purified target compounds.
5.2.2. 2-(5-(2-nitrobenzylidene)-4-oxo-2-thioxothiazolidin-3-yl)acetic acid (23)
Brown solid. Yield = 0.37 g, (72%), mp 201-204 °C; Rf = 0.74 (DCM:MeOH 80:20), 1H NMR (DMSO-d6) δ 4.76 (2H, s), 7.78 (2H, d, J = 7.48 Hz), 7.92 (1H, t, J = 7.58 Hz), 8.15 (1H, s), 8.25 (1H, d, J = 8.36 Hz) 13.49 (1H, s). Anal. Calcd. for C12H8N2O5S2: C, 44.44; H, 2.47; N, 8.64. Found: C, 44.65; H, 2.49; N, 8.44.
5.2.3. 2-(5-(4-nitrobenzylidene)-4-oxo-2-thioxothiazolidin-3-yl)acetic acid (24)
Yellow solid. Yield = 0.39 g, (76%), mp 259-262 °C, (lit. 240-243 °C)54; Rf = 0.52 (DCM:MeOH 80:20), 1H NMR (DMSO-d6) δ 4.77 (2H, s), 7.95 (2H, d, J = 8.76 Hz), 8.03 (1H, s), 8.37 (2H, d, J = 8.76 Hz), 13.54 (1H, s). Anal. Calcd. for C12H8N2O5S2: C, 44.44; H, 2.47; N, 8.64. Found: C, 44.21; H, 2.33; N, 8.44.
5.2.4. 2-(5-(4-fluorobenzylidene)-4-oxo-2-thioxothiazolidin-3-yl)acetic acid (25)
Yellow solid. Yield = 0.33 g, (72%), mp 256-259 °C; Rf = 0.54 (DCM:MeOH 80:20),1H NMR (DMSO-d6) δ 4.75 (2H, s), 7.41 (2H, d, J = 8.76 Hz), 7.75 (2H, d, J = 5.52 Hz), 7.93 (1H, s), 13.50 (1H, s). Anal. Calcd. for C12H8FNO3S2: C, 48.48; H, 2.71; N, 4.71. Found: C, 48.57; H, 2.67; N, 4.73.
5.2.5. 2-(5-(3-chlorobenzylidene)-4-oxo-2-thioxothiazolidin-3-yl)acetic acid (26)
Yellow solid. Yield = 0.36 g, (73%), mp 220-223 °C; Rf = 0.62 (DCM:MeOH 80:20), 1H NMR (DMSO-d6) δ 4.75 (2H, s), 7.61 (3H, s), 7.79 (1H, s), 7.91 (1H, s), 13.50 (1H, s). Anal. Calcd. for C12H8ClNO3S2: C, 45.93; H, 2.55; N, 4.47. Found: C, 45.97; H, 2.43; N, 4.50.
5.2.6. 2-(5-(3,4-dichlorobenzylidene)-4-oxo-2-thioxothiazolidin-3-yl)acetic acid (27)
Yellow solid. Yield = 0.47 g, (83%), mp 263-266 °C; Rf = 0.35 (DCM:MeOH 80:20), 1H NMR (DMSO-d6) δ 4.74 (2H, s), 7.61 (1H, d, J = 8.80 Hz), 7.83 (1H, d, J = 8.44 Hz), 7.90 (1H, s), 8.00 (1H, s), 13.51 (1H, s). Anal. Calcd. for C12H7Cl2NO3S2: C, 41.39; H, 2.03; N, 4.02. Found: C, 41.48; H, 1.96; N, 4.01.
5.2.7. 2-(5-(3-phenoxybenzylidene)-4-oxo-2-thioxothiazolidin-3-yl)acetic acid (28)
Brown solid. Yield = 0.45 g, (87%), mp 178-180 °C; Rf = 0.62 (DCM:MeOH 80:20), 1H NMR (DMSO-d6) δ 4.73 (2H, s), 7.10 (2H, d, J = 8.52 Hz), 7.22 (2H, m), 7.28 (1H, s), 7.45 (3H, t, J = 7.68 Hz), 7.58 (1H, t, J = 7.98 Hz), 7.90 (1H, s), 13.46 (1H, s). Anal. Calcd. for C18H13NO4S2. 1/2 H2O: C, 56.83; H, 3.71; N, 3.68. Found: C, 57.10; H, 3.75; N, 3.72.
5.3. Determination of the inhibition constant
The compounds were dissolved in dimethylsulfoxide (DMSO) as a 10 mM stock solution and stored at -20 °C for no more than 1 week. Serial dilutions were made in DMSO immediately prior to the assay. Recombinant NS5BCΔ21 protein was purified from the plasmid pThNS5BCΔ21 expressed in Escherichia coli DH5α, by Ni-NTA column chromatography as described previously.27,36 NS5BCΔ21 enzyme stocks were stored in aliquots at -80 °C in buffer containing 50 mM Tris–HCl [pH 8.0], 100 mM NaCl, 5 mM MgCl2 and 50% glycerol. The effect of the compounds on the RdRp activity of NS5BCΔ21 was evaluated by the standard primer-dependent elongation assays as previously described.27,36 Briefly, enzymatic reaction mixtures (25 μL) containing 20 mM Tris-HCl (pH 7.0), 100 mM NaCl, 100 mM sodium glutamate, 0.01% BSA, 0.01% Tween-20, 5% glycerol, 20 U/mL of RNase Out, 0.5 μM of poly rA/U12, 25 μM UTP, 2-5 μCi [α-32P]UTP, 300-500 ng of NS5BCΔ21 and 0.5 mM MnCl2 in the presence or absence of the inhibitor were incubated for 1 h at 30°C. Reactions were terminated by the addition of ice cold 5% (v/v) trichloroacetic acid (TCA) containing 0.5 mM pyrophosphate. Precipitated RNA products were evaluated by GF-B filter binding assay and quantified on a liquid scintillation counter (Packard). RNA synthesis in the absence of the inhibitor was set at 100% (control), and that in the presence of the inhibitor was expressed relative to this control. The concentration of the compounds inhibiting 50% of NS5B RdRp activity (IC50) were calculated from the inhibition curve as a function of inhibitor concentrations employing GraphPad Prism 3.03 software (GraphPad Software, Inc., San Diego, CA).
Supplementary Material
Acknowledgements
This work was supported by the National Institute of Health research grant DK066837 to NKB. TTT was supported by start-up funds and resources from the College of Pharmacy of St. John's University. We thank Dr. Santosh Khedkar from Harvard Medical School, Dr. V. Hariprasad from Huntsman Cancer Institute and Dr. Narayanan Ramasubbu from University of Medicine and Dentistry of New Jersey for many helpful suggestions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary data
Supplementary data associated with this article can be found in the online version, at .........................
References
- 1.Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. Science. 1989;244:359. doi: 10.1016/s0168-8278(02)00051-x. [DOI] [PubMed] [Google Scholar]
- 2.Alter MJ, Margolis HS, Krawczynski K, Judson FN, Mares A, Alexander WJ, Hu PY, Miller JK, Gerber MA, Sampliner RE, et al. The New England J. Med. 1992;327:1899. doi: 10.1056/NEJM199212313272702. [DOI] [PubMed] [Google Scholar]
- 3.Alter MJ. Infect. Agents Dis. 1993;2:155. [PubMed] [Google Scholar]
- 4.Di Bisceglie AM, Order SE, Klein JL, Waggoner JG, Sjogren MH, Kuo G, Houghton M, Choo QL, Hoofnagle JH. Am. J. Gastroenterol. 1991;86:335. [PubMed] [Google Scholar]
- 5.Alter MJ. World J. Gastroenterol. 2007;13:2436. doi: 10.3748/wjg.v13.i17.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang QM, Heinz BA. Prog. Drug Res. 2000;55:1. doi: 10.1007/978-3-0348-8385-6_1. [DOI] [PubMed] [Google Scholar]
- 7.Abrignani S, Houghton M, Hsu HH. J. Hepatol. 1999;31(Suppl 1):259. doi: 10.1016/s0168-8278(99)80413-9. [DOI] [PubMed] [Google Scholar]
- 8.Huang Z, Murray MG, Secrist JA., 3rd Antiviral Res. 2006;71:351. doi: 10.1016/j.antiviral.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 9.Ferenci P, Fried MW, Shiffman ML, Smith CI, Marinos G, Goncales FL, Jr., Haussinger D, Diago M, Carosi G, Dhumeaux D, Craxi A, Chaneac M, Reddy KR. J. Hepatol. 2005;43:425. doi: 10.1016/j.jhep.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 10.Arataki K, Kumada H, Toyota K, Ohishi W, Takahashi S, Tazuma S, Chayama K. Intervirology. 2006;49:352. doi: 10.1159/000095155. [DOI] [PubMed] [Google Scholar]
- 11.Bartenschlager R, Ahlborn-Laake L, Mous J, Jacobsen H. J. Virol. 1994;68:5045. doi: 10.1128/jvi.68.8.5045-5055.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reed KE, Rice CM. Curr. Top. Microbiol. Immunol. 2000;242:55. doi: 10.1007/978-3-642-59605-6_4. [DOI] [PubMed] [Google Scholar]
- 13.Tanji Y, Hijikata M, Hirowatari Y, Shimotohno K. J. Virol. 1994;68:8418. doi: 10.1128/jvi.68.12.8418-8422.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Behrens SE, Tomei L, De Francesco R. Embo. J. 1996;15:12. [PMC free article] [PubMed] [Google Scholar]
- 15.Moradpour D, Penin F, Rice CM. Nat. Rev. Microbiol. 2007;5:453. doi: 10.1038/nrmicro1645. [DOI] [PubMed] [Google Scholar]
- 16.Wang QM, Heinz BA. Prog. Drug Res. 2001 Spec No, 79. [Google Scholar]
- 17.Soriano V, Peters MG, Zeuzem S. Clin. Infect. Dis. 2009;48:313. doi: 10.1086/595848. [DOI] [PubMed] [Google Scholar]
- 18.Beaulieu PL. Curr. Opin. Investig. Drugs. 2007;8:614. [PubMed] [Google Scholar]
- 19.Koch U, Narjes F. Curr. Topics Med. Chem. 2007;7:1302. doi: 10.2174/156802607781212211. [DOI] [PubMed] [Google Scholar]
- 20.Talele TT. Curr. Bioactive Compounds. 2008;4:86. [Google Scholar]
- 21.Bressanelli S, Tomei L, Rey FA, De Francesco R. J. Virol. 2002;76:3482. doi: 10.1128/JVI.76.7.3482-3492.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Di Marco S, Volpari C, Tomei L, Altamura S, Harper S, Narjes F, Koch U, Rowley M, De Francesco R, Migliaccio G, Carfi A. J. Biol. Chem. 2005;280:29765. doi: 10.1074/jbc.M505423200. [DOI] [PubMed] [Google Scholar]
- 23.Beaulieu PL, Bos M, Bousquet Y, DeRoy P, Fazal G, Gauthier J, Gillard J, Goulet S, McKercher G, Poupart MA, Valois S, Kukolj G. Bioorg. Med. Chem. Lett. 2004;14:967. doi: 10.1016/j.bmcl.2003.12.032. [DOI] [PubMed] [Google Scholar]
- 24.Ikegashira K, Oka T, Hirashima S, Noji S, Yamanaka H, Hara Y, Adachi T, Tsuruha J, Doi S, Hase Y, Noguchi T, Ando I, Ogura N, Ikeda S, Hashimoto H. J. Med. Chem. 2006;49:6950. doi: 10.1021/jm0610245. [DOI] [PubMed] [Google Scholar]
- 25.Rong F, Chow S, Yan S, Larson G, Hong Z, Wu J. Bioorg. Med. Chem. Lett. 2007;17:1663. doi: 10.1016/j.bmcl.2006.12.103. [DOI] [PubMed] [Google Scholar]
- 26.Ontoria JM, Martin Hernando JI, Malancona S, Attenni B, Stansfield I, Conte I, Ercolani C, Habermann J, Ponzi S, Di Filippo M, Koch U, Rowley M, Narjes F. Bioorg. Med. Chem. Lett. 2006;16:4026. doi: 10.1016/j.bmcl.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 27.Kaushik-Basu N, Bopda-Waffo A, Talele TT, Basu A, Costa PR, da Silva AJ, Sarafianos SG, Noel F. Nucleic Acids Res. 2008;36:1482. doi: 10.1093/nar/gkm1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Biswal BK, Cherney MM, Wang M, Chan L, Yannopoulos CG, Bilimoria D, Nicolas O, Bedard J, James MN. J. Biol. Chem. 2005;280:18202. doi: 10.1074/jbc.M413410200. [DOI] [PubMed] [Google Scholar]
- 29.Love RA, Parge HE, Yu X, Hickey MJ, Diehl W, Gao J, Wriggers H, Ekker A, Wang L, Thomson JA, Dragovich PS, Fuhrman SA. J. Virol. 2003;77:7575. doi: 10.1128/JVI.77.13.7575-7581.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang M, Ng KK, Cherney MM, Chan L, Yannopoulos CG, Bedard J, Morin N, Nguyen-Ba N, Alaoui-Ismaili MH, Bethell RC, James MN. J. Biol. Chem. 2003;278:9489. doi: 10.1074/jbc.M209397200. [DOI] [PubMed] [Google Scholar]
- 31.Yan S, Appleby T, Larson G, Wu JZ, Hamatake R, Hong Z, Yao N. Bioorg. Med. Chem. Lett. 2006;16:5888. doi: 10.1016/j.bmcl.2006.08.056. [DOI] [PubMed] [Google Scholar]
- 32.Chan L, Reddy TJ, Proulx M, Das SK, Pereira O, Wang W, Siddiqui A, Yannopoulos CG, Poisson C, Turcotte N, Drouin A, Alaoui-Ismaili MH, Bethell R, Hamel M, L'Heureux L, Bilimoria D, Nguyen-Ba N. J. Med. Chem. 2003;46:1283. doi: 10.1021/jm0340400. [DOI] [PubMed] [Google Scholar]
- 33.Chan L, Pereira O, Reddy TJ, Das SK, Poisson C, Courchesne M, Proulx M, Siddiqui A, Yannopoulos CG, Nguyen-Ba N, Roy C, Nasturica D, Moinet C, Bethell R, Hamel M, L'Heureux L, David M, Nicolas O, Courtemanche-Asselin P, Brunette S, Bilimoria D, Bedard J. Bioorg. Med. Chem. Lett. 2004;14:797. doi: 10.1016/j.bmcl.2003.10.068. [DOI] [PubMed] [Google Scholar]
- 34.Gopalsamy A, Lim K, Ciszewski G, Park K, Ellingboe JW, Bloom J, Insaf S, Upeslacis J, Mansour TS, Krishnamurthy G, Damarla M, Pyatski Y, Ho D, Howe AY, Orlowski M, Feld B, O'Connell J. J. Med. Chem. 2004;47:6603. doi: 10.1021/jm0401255. [DOI] [PubMed] [Google Scholar]
- 35.Li H, Tatlock J, Linton A, Gonzalez J, Borchardt A, Dragovich P, Jewell T, Prins T, Zhou R, Blazel J, Parge H, Love R, Hickey M, Doan C, Shi S, Duggal R, Lewis C, Fuhrman S. Bioorg. Med. Chem. Lett. 2006;16:4834. doi: 10.1016/j.bmcl.2006.06.065. [DOI] [PubMed] [Google Scholar]
- 36.Kaushik-Basu N, Bopda-Waffo A, Talele TT, Basu A, Chen Y, Kucukguzel SG. Front Biosci. 2008;13:3857. doi: 10.2741/2974. [DOI] [PubMed] [Google Scholar]
- 37.Tedesco R, Shaw AN, Bambal R, Chai D, Concha NO, Darcy MG, Dhanak D, Fitch DM, Gates A, Gerhardt WG, Halegoua DL, Han C, Hofmann GA, Johnston VK, Kaura AC, Liu N, Keenan RM, Lin-Goerke J, Sarisky RT, Wiggall KJ, Zimmerman MN, Duffy KJ. J. Med. Chem. 2006;49:971. doi: 10.1021/jm050855s. [DOI] [PubMed] [Google Scholar]
- 38.Powers JP, Piper DE, Li Y, Mayorga V, Anzola J, Chen JM, Jaen JC, Lee G, Liu J, Peterson MG, Tonn GR, Ye Q, Walker NP, Wang Z. J. Med. Chem. 2006;49:1034. doi: 10.1021/jm050859x. [DOI] [PubMed] [Google Scholar]
- 39.Gopalsamy A, Shi M, Ciszewski G, Park K, Ellingboe JW, Orlowski M, Feld B, Howe AY. Bioorg. Med. Chem. Lett. 2006;16:2532. doi: 10.1016/j.bmcl.2006.01.105. [DOI] [PubMed] [Google Scholar]
- 40.Nittoli T, Curran K, Insaf S, DiGrandi M, Orlowski M, Chopra R, Agarwal A, Howe AY, Prashad A, Floyd MB, Johnson B, Sutherland A, Wheless K, Feld B, O'Connell J, Mansour TS, Bloom J. J. Med. Chem. 2007;50:2108. doi: 10.1021/jm061428x. [DOI] [PubMed] [Google Scholar]
- 41.Pfefferkorn JA, Greene ML, Nugent RA, Gross RJ, Mitchell MA, Finzel BC, Harris MS, Wells PA, Shelly JA, Anstadt RA, Kilkuskie RE, Kopta LA, Schwende FJ. Bioorg. Med. Chem. Lett. 2005;15:2481. doi: 10.1016/j.bmcl.2005.03.066. [DOI] [PubMed] [Google Scholar]
- 42.Burton G, Ku TW, Carr TJ, Kiesow T, Sarisky RT, Lin-Goerke J, Hofmann GA, Slater MJ, Haigh D, Dhanak D, Johnson VK, Parry NR, Thommes P. Bioorg. Med. Chem. Lett. 2007;17:1930. doi: 10.1016/j.bmcl.2007.01.034. [DOI] [PubMed] [Google Scholar]
- 43.Lyne PD. Drug Discovery Today. 2002;7:1047. doi: 10.1016/s1359-6446(02)02483-2. [DOI] [PubMed] [Google Scholar]
- 44.Patel PD, Patel MR, Kaushik-Basu N, Talele TT. J. Chem. Inf. Model. 2008;48:42. doi: 10.1021/ci700266z. [DOI] [PubMed] [Google Scholar]
- 45.Ohishi Y, Mukai T, Nagahara M, Yajima M, Kajikawa N, Miyahara K, Takano T. Chem. Pharm. Bull. (Tokyo) 1990;38:1911. doi: 10.1248/cpb.38.1911. [DOI] [PubMed] [Google Scholar]
- 46.Momose Y, Meguro K, Ikeda H, Hatanaka C, Oi S, Sohda T. Chem. Pharm. Bull. (Tokyo) 1991;39:1440. doi: 10.1248/cpb.39.1440. [DOI] [PubMed] [Google Scholar]
- 47.Melagraki G, Afantitis A, Sarimveis H, Koutentis PA, Markopoulos J, Igglessi-Markopoulou O. Bioorg. Med. Chem. 2007;15:7237. doi: 10.1016/j.bmc.2007.08.036. [DOI] [PubMed] [Google Scholar]
- 48.Kim J, Kim KS, Kim DE, Chong Y. Chem. Biol. Drug Design. 2008;72:585. doi: 10.1111/j.1747-0285.2008.00730.x. [DOI] [PubMed] [Google Scholar]
- 49.Betzi S, Eydoux C, Bussetta C, Blemont M, Leyssen P, Debarnot C, Ben-Rahou M, Haiech J, Hibert M, Gueritte F, Grierson DS, Romette JL, Guillemot JC, Neyts J, Alvarez K, Morelli X, Dutartre H, Canard B. Antiviral Res. 2009;84:48. doi: 10.1016/j.antiviral.2009.06.009. [DOI] [PubMed] [Google Scholar]
- 50.Li T, Froeyen M, Herdewijn P. J. Mol. Model. 2009;16:49. doi: 10.1007/s00894-009-0519-9. [DOI] [PubMed] [Google Scholar]
- 51.Ryu K, Kim ND, Choi SI, Han CK, Yoon JH, No KT, Kim KH, Seong BL. Bioorg. Med. Chem. 2009;17:2975. doi: 10.1016/j.bmc.2009.03.024. [DOI] [PubMed] [Google Scholar]
- 52.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Adv. Drug Deliv. Rev. 2001;46:3. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 53.Louise-May S, Yang W, Nie X, Liu D, Deshpande MS, Phadke AS, Huang M, Agarwal A. Bioorg. Med. Chem. Lett. 2007;17:3905. doi: 10.1016/j.bmcl.2007.04.103. [DOI] [PubMed] [Google Scholar]
- 54.Zhou JF, Song YZ, Zhu FX, Zhu YL. Syn. Comm. 2006;36:3297. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.