Abstract
The 3-dimensional structure of an allergen defines the accessible parts on the surface of the molecule or epitopes that interact with antibodies. Mapping the antigenic determinants for IgE antibody binding has been pursued through strategies based on the use of overlapping synthetic peptides, recombinant allergenic fragments or unfolded allergens. These approaches led to the identification of mostly linear epitopes and are useful for food allergens that undergo digestion or food processing. For inhaled allergens, conformational epitopes appear to be the primary targets of IgE responses. Knowledge of the molecular structure of allergens alone and in complex with antibodies that interfere with IgE antibody binding is important to understand the immune recognition of B cell-antigenic determinants on allergens and the design of recombinant allergens for immunotherapy. Starting with the molecular cloning and expression of allergens, and with the advent of X-ray crystallography and nuclear magnetic resonance techniques, we have been able to visualize conformational epitopes on allergens.
Key Words: Conformational epitope, Linear epitope, X-ray crystallography, Allergen structure, IgE antibody, Bla g 2
Introduction
The term epitope, coined by Niels Jerne in 1960 [1], refers to an antigenic determinant or part of a molecule recognized by an antibody. In allergic disease, IgE antibodies are produced against specific epitopes from foreign proteins or glycoproteins during sensitization. Subsequently, the specific recognition of these epitopes on the allergen by IgE antibodies bound to effector cells such as basophils and mast cells is essential for the development of the allergic response. Whereas monoclonal antibodies (mAb) only recognize a single epitope, multiple epitopes are involved in an IgE antibody polyclonal response. Since the 1980s, several studies colocalized IgE and mAb-binding epitopes by inhibition assays, but without revealing the specific residues involved in the allergen-antibody interaction [2,3]. During this decade, the first X-ray crystal structures of epitopes were described in an allergen, the hen egg-white lysozyme, although their overlap with IgE antibody-binding sites is unknown [4,5,6]. The identification of the 3-dimensional structure of epitopes involved in allergic disease can be used to better understand the immune response to allergens, including the cross-reactivity among homologous proteins, and for the design of recombinants for immunotherapy. This review addresses strategies that have been used to identify IgE antibody-binding epitopes on allergens, without analyzing carbohydrate or drug epitopes. Special emphasis is given to X-ray crystallographic studies, in particular the cockroach allergen Bla g 2, which revealed the precise structure of allergen epitopes.
Broadening the Concept of Conformational Epitope
Important differences exist between B and T cell epitopes, and their localization in the allergen does not necessarily coincide. Whereas T cell epitopes are only linear and distributed throughout the primary structure of the allergen, B cell epitopes recognized by IgE antibodies are either linear or conformational and are located on the surface of the molecule accessible to antibodies. This review will focus on B cell epitopes.
A linear epitope (also known as continuous or sequential epitope) involves a protein segment of consecutive amino acids recognized by the variable region of an antibody (N-terminus of the Fab fragment), whereas a conformational or discontinuous epitope comprises amino acids that are close in space in the folded molecule, despite being noncontiguous in the amino acid sequence (fig. 1, first column). Unlike linear epitopes, conformational epitopes are dependent on the 3-dimensional structure of the protein. For globular inhaled allergens, conformational epitopes play a very important role in allergenicity [7,8]. For food allergens, some IgE antibody-binding epitopes may have been inaccessible in the properly folded allergenic molecule, but after food processing or digestion, an increase in the number or accessibility of IgE antibody-binding epitopes may occur. For example, the IgE-binding epitopes in the trimeric peanut allergen Ara h 1 are found in areas of interaction between monomers, so the trimeric structure of the allergen may protect IgE-binding epitopes from degradation [9]. However, for food allergens related to the oral allergy syndrome such as Bet v 1-related food allergens, loss of the structural conformation by protein denaturation/digestion leads to a loss of some or all the B cell epitopes (but not the T cell epitopes) [10].
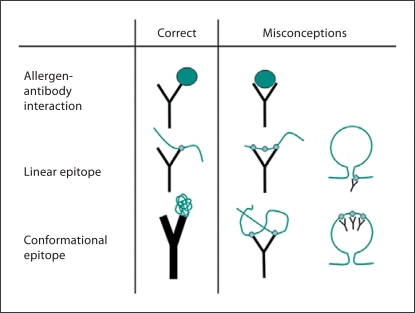
Fig. 1.
Representations of allergen-antibody interactions and linear and conformational epitopes. The allergen and the epitopes are shown as a big cyan sphere and small spheres, respectively. Correct representations and misconceptions are on the left and the right, respectively. The fragment of allergen connecting the 2 Fab of the antibody and containing 3 small spheres is a misinterpretation of a linear epitope (second row, second column).
Since conformational epitopes depend on the 3-dimensional structure of the protein, changes in protein folding may lead to changes in the number of epitopes. Calcium-binding proteins, for example, undergo conformational changes upon calcium binding which may hide or expose IgE epitopes. The cockroach allergen Bla g 6 is a troponin C and has a very flexible molecular structure that extends from a closed conformation in the apo form (calcium-free) to a dumbbell-shaped open molecule with a long α-helix in the calcium-bound form by unbending 90° [11]. Other calcium-binding proteins from plants and fish have been described [12]. The accessibility of epitopes is reduced in the calcium-depleted form, and IgE antibodies will mostly bind the allergen with the conformation that is dominant during sensitization. Similarly, recombinant allergens may fold differently when expressed in different expression systems, and an expression system may lead to the loss of B cell epitopes [13].
Initial B cell epitope mapping studies using peptides designed from partial sequences of allergens allowed the distinction between linear and conformational epitopes. An epitope was linear if the antibody bound to a small peptide or unfolded allergen. The presence of conformational B cell epitopes could be detected by the reduction of the capacity to bind IgE antibodies after loss of the native 3-dimensional structure of the allergen due to denaturation (by heat or reduction of sulfide bonds, for example) or digestion, or by the removal of a propeptide (as described for enzymes like pro-Der p 1) [7,8,14].
Because of the resolution of the X-ray crystal structure of allergen-antibody complexes, the dichotomy between linear and conformational epitopes disappears, and the concept of conformational or discontinuous epitopes becomes broader. Epitopes involve several allergen loops and are therefore conformational, whereas continuous epitopes are part of discontinuous epitopes [15,16,17].
Correct representations of allergen-antibody interactions as well as linear and conformational epitopes are an indication of a correct understanding of the concepts involved (fig. 1, left). However, erroneous representations are often encountered and should be avoided (fig. 1, right). Some misconceptions involve the recognition of 2 epitopes by the 2 Fab of the antibody (fig. 1, second column), or the additional disproportioned representation of allergen loops versus the antibody (fig. 1, third column). Only 1 Fab of the antibody binds to an epitope on the allergen, unless the epitope was repeated and oriented in a way and at an optimal distance for recognition by the 2 Fab of the same antibody.
Allergenicity Predictions
Many algorithms have been developed to predict protein antigenicity and identify sequential epitopes based on intrinsic amino acid properties and properties that are a function of the structural context in the molecule (hydrophilicity, antigenicity, segmental mobility, flexibility and accessibility) [18]. The accuracy of these algorithms ranges from 35 to 75%. Allergenicity and antigenicity predictions differ in the fact that the first one is mainly restricted to the identification of epitopes involved in IgE antibody binding. Due to the interest in the production of safe improved crops, scientists have focused on the prediction of allergenicity in engineered foods. One of the criteria defined by the FAO/WHO Codex alimentarius guidelines for allergenicity prediction in transgenic proteins has been based on screening for identities of at least 6 contiguous amino acids shared with allergenic proteins, as potential linear IgE antibody-binding epitopes [19]. Recently, efforts have focused on incorporating structural information into the predictions. Three conformational epitope prediction servers exist that require the availability of the 3-dimensional structure of the protein [18,20,21]. Epitope predictions can be done with the structure of more than 50 allergens actually available at the Protein Data Bank (http://www.rcsb.org) (table 1). An in silico method using an epitope motif database containing 1,013 antibody-binding motifs was developed to predict antigenic epitopes in proteins in a structural context [22]. The server SDAP allows the prediction of cross-reactivity and allergenicity by using allergen sequences, known 3-dimensional structures of allergens, models of allergens with unknown structures and IgE epitopes and binding data from the literature [23]. However, since most of the epitopes described in the literature are linear, there is a way to go before being able to predict allergenicity in a molecule based on known conformational epitopes from allergens. Therefore, empirical strategies are nowadays the most accurate way to identify amino acids involved in IgE antibody binding.
Table 1.
Tertiary structures of allergens
| Allergen | Species of origin | Function/structure | X-ray crystal structure | NMR structure | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Inhaled – indoors | |||||||||||
| Bla g 2 | Blattella germanica | inactive aspartic protease | 1YG9* | 2NR6*,1 | |||||||
| Bla g 4 | Blattella germanica | lipocalin | 3EBK | ||||||||
| Blo t 5 | Blomia tropicalis | unknown | 2JMH | ||||||||
| Bos d 2 | Bos domesticus | lipocalin | 1BJ7 | ||||||||
| Der f 1 | Dermatophagoides farinae | cysteine protease | 3D6S | ||||||||
| Der f 2 | Dermatophagoides farinae | lipid-binding protein | 1XWV | 2F08 | 1AHK | 1AHM | 1WRF | ||||
| Der f 13 | Dermatophagoides farinae | fatty acid-binding protein | 2A0A | ||||||||
| Der p 1 | Dermatophagoides pteronyssinus | cysteine protease | 1XKG | 2AS8* | 3F5V | ||||||
| Der p 2 | Dermatophagoides pteronyssinus | lipid-binding protein | 1KTJ* | 1A9V* | |||||||
| Fel d 1 | Felis domesticus | uteroglobin | 1PUO* | 1ZKR* | 2EJN* | ||||||
| Mus m 1 | Mus musculus | lipocalin | 1MUP | ||||||||
| Per a 4 | Periplaneta americana | lipocalin | 3EBW | ||||||||
| Rat n 1 | Rattus norvegicus | lipocalin | 2A2G | 2A2U | |||||||
| Inhaled – outdoors | |||||||||||
| Amb t 5 | Ambrosia trifida | unknown | 1BBG | 2BBG | 3BBG | ||||||
| Ara t 8 | Arabidopsis thaliana | profilin | 1A0K | 3NUL | |||||||
| Bet v 1 | Betula verrucosa | pathogenesis-related protein | 1BV1 | 1QMR* | 1FM4 | 1LLT* | 1FSK1 | 1BTV | 1B6F | ||
| Bet v 2 | Betula verrucosa | profilin | 1CQA | ||||||||
| Bet v 4 | Betula verrucosa | Ca-binding protein | 1H4B | ||||||||
| Che a 3 | Chenopodium album | polcalcin | 2OPO | ||||||||
| Chi t 1 | Chironomus thummi thummi | hemoglobin | 1ECO | ||||||||
| Equ c 1 | Equus caballus | lipocalin | 1EW3 | ||||||||
| Hev b 6 | Hevea brasiliensis | hevein precursor | 1Q9B | 1WKX | 1T0W | 1HEV | |||||
| Hev b 8 | Hevea brasiliensis | latex profilin | 1G5U | ||||||||
| Jun a 1 | Juniperus ashei | pectate lyase | 1PXZ | ||||||||
| Ole e 6 | Olea europaea | unknown | 1SS3 | ||||||||
| Ole e 9 | Olea europaea | β-1,3-glucanase (C-terminus) | 2JON | ||||||||
| Phl p 1 | Phleum pratense | expansin | 1N10 | ||||||||
| Phl p 2 | Phleum pratense | grass group II/III | 1WHO | 1WHP | 2VXQ1 | 1BMW | |||||
| Phl p 3 | Phleum pratense | grass group II/III | 2JNZ | ||||||||
| Phl p 5 | Phleum pratense | unknown | 1L3P | ||||||||
| Phl p 6 | Phleum pratense | unknown | 1NLX | ||||||||
| Phl p 7 | Phleum pratense | Ca-binding protein | 1K9U | ||||||||
| Zea m 1 | Zea mays | β-expansin | 2HCZ | ||||||||
| Ingested – food | |||||||||||
| Api g 1 | Apium graveolens | pathogenesis-related protein | 2BK0 | ||||||||
| Ara h 3 | Arachis hypogaea | cupin (11S globulin, glycinin) | 3C3V | ||||||||
| Ara h 6 | Arachis hypogaea | conglutin (2S albumin) | 1W2Q | ||||||||
| Bos d 4 | Bos domesticus | α-lactalbumin | 1F6R | 1F6S | 1HFZ | 2G4N | |||||
| Bos d 5 | Bos domesticus | β-lactoglobulin | 1GX8 | 1GX9 | 2AKQ | 1BSO | 1UZ2* | 2R561 | |||
| Gal d 2 | Gallus domesticus | ovalbumin | 1JTI* | 1OVA | 1UHG* | ||||||
| Gal d 3 | Gallus domesticus | ovotransferrin | 1RYX | 2D3I | 1OVT | 1AIV | 1TFA | 1IEJ | |||
| Gal d 4 | Gallus domesticus | lysozyme2 | 1LYZ | 1H6M* | 1YQV1 | 1FDL1 | 1MLC1 | 3HFM1 | 1GXV | 1GXX | |
| Mus a x | Musa acuminata | banana glucanase | 2CYG | ||||||||
| Pru av 2 | Prunus avium | thaumatin-like protein | 2AHN | ||||||||
| Pru p 3 | Prunus persica | nonspecific lipid transfer protein | 2ALG | 2B5S | |||||||
| Ric c 3 | Ricinus communis | 2S albumin storage protein | 1PSY | ||||||||
| Injected | |||||||||||
| Api m 1 | Apis mellifera | phospholipase A2 | 1POC | ||||||||
| Api m 2 | Apis mellifera | hyaluronidase | 1FCQ | 1FCU | 1FCV | 2J881 | |||||
| Api m 4 | Apis mellifera | mellitin | 2MLT | 1BH1* | |||||||
| Sol i 3 | Solenopsis invicta | unknown | 2VZN | ||||||||
| Ves v 2 | Vespula vulgaris | hyaluronidase | 2ATM | ||||||||
| Ves v 5 | Vespula vulgaris | antigen 5 | 1QNX | ||||||||
| Through skin | |||||||||||
| Mala s 1 | Malassezia sympodialis | β-propeller fold | 2P9W | ||||||||
| Mala s 6 | Malassezia sympodialis | cyclophilin | 2CFE | ||||||||
| Mala s 13 | Malassezia sympodialis | thioredoxin | 2J23 | ||||||||
Modified or mutated molecule.
Allergen in complex with an antibody fragment.
Structures selected from multiple Protein Data Bank entries.
Prediction of Cross-Reactive Epitopes
Prediction of cross-reactive epitopes can be achieved by identifying amino acid similarities on the molecular surface of cross-reactive allergens. Allergens are recognized as foreign by the human body, but in many cases they also share homology with endogenous proteins. Proteins with a sequence identity to a human homologue above approximately 62% have been reported to be rarely allergenic [24]. To further predict allergenic epitopes, cross-reactive allergens can be compared to homologous endogenous proteins that are not allergens, and areas in common can be in most cases excluded as potential IgE antibody-binding epitopes. Differences at the level of molecular surface between allergens and human proteins may be relevant for the development of allergic disease.
Strategies for B Cell Epitope Mapping: Advantages, Disadvantages and Pitfalls
Strategies Not Based on the 3-Dimensional Structure of the Allergen
Several strategies have been developed for the identification of B cell epitopes on allergens. Most of the mapping techniques are based on IgE antibody recognition of: (1) overlapping synthetic peptides derived from the linear sequence of the allergen, (2) small allergen fragments expressed as recombinant proteins or resulting from allergen digestion, or (3) unfolded allergens by immunoblot. These mapping techniques are especially useful for food allergens which have mainly linear epitopes after food processing or digestion. As a result of mapping using these techniques, an association between the responses to linear epitopes in foods and the severity of the allergic disease has been described for some foods. The specificity of IgE antibodies to sequential epitopes of hen's egg ovomucoid and cow's milk proteins has been reported to be a useful marker of persistence to the respective food allergies [25,26]. The IgE epitope diversity (i.e., number of epitopes recognized) has also been correlated with peanut allergy severity [27]. However, the outcome of these mapping approaches is limited to the identification of linear epitopes. For globular allergens such as inhaled allergens (which are not subjected to denaturation or digestion) or food allergens involved in oral allergy syndrome, these techniques have the important limitation of not being able to identify conformational epitopes [28]. In these cases, approaches based on the 3-dimensional structure of the protein are preferable.
Strategies Based on the 3-Dimensional Structure of the Allergen
One strategy for B cell epitope mapping involves the identification of mimotopes or residues resembling the epitope of an allergen by phage display technology. This technique selects phage-displayed peptides that are recognized by an antibody and uses an algorithm or the physicochemical characteristics of the amino acids from the selected peptide to localize the putative epitope on the allergen surface [29]. Mimotopes for Bet v 1, Bla g 2, Der p 1, Der p 2, Lol p 1, Pru p 3 and Phl p 5a have been identified by phage display technology. Although the mimotopes are not necessarily identical to the allergenic epitope, this technique estimates the location of residues involved in antibody binding on the allergen.
Another strategy for B cell epitope mapping is the selection of surface-exposed charged residues, based on the knowledge of the 3-dimensional structure of the allergen, followed by site-directed mutagenesis studies to identify low IgE antibody-binding mutants [30,31]. However, this technique may, in some cases, require the expression of a large number of mutants [30]. This number can be significantly reduced if epitopes from antibodies overlapping with IgE antibody binding are first identified by X-ray crystallography or nuclear magnetic resonance (NMR). Subsequently, mutants with substitutions of residues involved in the epitope can be selectively expressed and tested for IgE antibody binding to identify important amino acids for the allergen-antibody interaction [32]. A reduction or lack of IgE antibody binding could be either due to changes in the 3-dimensional structure of the allergen related to the substitution or to a single amino acid change in a mutant that folds like the wild-type allergen. Therefore, an analysis of the overall fold of the expressed mutants is required for a correct interpretation of the mutational studies. For this purpose, techniques such as CD spectra, combined, if possible, with the assessment of allergen binding to specific mAb nonoverlapping with the one under study, should be performed.
Several NMR techniques have been applied to allergen epitope mapping. The main limitation of NMR is the relatively small molecular size of the proteins to be analyzed (up to 25 kDa for high-quality de novo structures). Epitopes have been identified in Der p 2 by using the whole mAb to protect the epitope from hydrogen exchange, followed by the NMR analysis of the allergen alone [33]. Transverse relaxation-optimized spectroscopy NMR was used to perform chemical shift mapping to solve the structure of Blo t 5 with the Fab’ of a mAb. A discontinuous epitope overlapping with human IgE epitope(s) was identified [34]. Finally, NMR differential exchange broadening experiments were performed for complexes of intact mAb and Der f 2, and 2 conformational nonoverlapping epitopes were identified on the allergen [35].
The resolution of the X-ray crystal structure of allergen-antibody complexes is a precise way to identify allergen epitopes in proteins with a wider range of molecular weights than can be analyzed by NMR. The main limitation of this approach for B cell epitope mapping is the lack of natural IgE mAb in milligram amount required for X-ray crystallography studies. The IgE antibody response is polyclonal, and the concentration in sera is low (∼0.05%) compared to IgG antibodies. Therefore, either IgG mAb overlapping with IgE or IgE mAb from combinatorial libraries (obtained in vitro from a random combination of heavy and light chains of antibodies) can be obtained in high amounts for crystallization and used as surrogates for IgE antibodies. All the allergen-antibody complexes solved by X-ray crystallography were obtained using Fab or Fab’ fragments of murine mAb or Fab fragments derived from phage surface-displayed combinatorial (human) antibody Fab libraries. Several considerations need to be taken into account when interpreting these studies. First, reduction of human IgE antibody binding by murine mAb indicates that these murine antibodies are competing with the natural human polyclonal serum IgE for binding to the epitope, but does not necessarily demonstrate that the epitopes are identical. The epitopes could be overlapping. Second, Fab fragments derived from phage display technology result from a random combination of heavy and light chains. During the selection of Fabs, the probability that the combination of heavy and light chains with the highest affinity for the allergen is selected is high, and this combination might not correspond to the pairings which in nature are evolutionarily selected. Therefore, although both approaches are not suitable to exactly define naturally occurring human IgE-binding epitopes, they are useful to identify important amino acids involved in the IgE-allergen interaction when combined with mutational analysis.
Epitopes Defined by X-Ray Crystallography
With the development of the field of structural biology, the resolution of the structure of protein-antibody complexes revealed the structure of antigenic epitopes and the corresponding paratopes on the antibody. In the 1980s, the structure of 5 protein-antibody complexes was solved: 3 Fab antibody fragments with lysozyme from chicken egg-white, which is an allergen, and 2 complexes of Fab with neuraminidase from influenza virus [4,5,6,15]. The 5 structures defined conformational epitopes occupying large areas of 15–22 amino acids on several surface loops, with a buried surface on the antigen of 650–900 Å2. There were 75–120 hydrogen bonds between the antibody and the antigen as well as salt links and hydrophobic interactions. Energy calculations suggested that from the 15–22 residues in the antigen in contact with a similar number of residues on the antibody paratope, only 5–6 contributed most of the binding energy [15].
Since the publication of the crystallographic structures of lysozyme-antibody complexes, 5 additional allergen-antibody complexes have been reported. The allergens are birch pollen allergen Bet v 1, bee venom allergen Api m 2 (hyaluronidase), cockroach allergen Bla g 2, β-lactoglobulin from bovine milk and timothy grass pollen allergen Phl p 2 [36,37,38,39,40]. The first 3 are complexes with Fab fragments of murine IgG1 antibodies, and the last 2 are complexes with human IgE-derived Fab fragments. The 3 murine IgG1 antibodies inhibited IgE antibody binding to the allergen, suggesting that at least a partial overlap of the IgG and IgE epitopes occurs. The surfaces reported to be involved in the interaction are comparable: the buried areas on the allergen are 931 Å2 for Bet v 1, 870 Å2 for Bla g 2, 890 Å2 for β-lactoglobulin and 855 Å2 for Phl p 2, and the total contact surface area for the Api m 2 complex interface is 1,274 Å2. All the epitopes are conformational, despite the dominance of a long loop in the Bet v 1 (residues Glu42-Thr52), Api m 2 (Arg138, His141-Arg148) and Bla g 2 (Lys60-Asn70) epitopes. Interestingly, this loop, involving several contiguous amino acids, is comparable to a linear epitope and represents an important part (∼60–80%) of the interaction [36,37,38]. Yet, there is evidence that even the conformation of this linear loop can be important [37]. The epitopes defined by these 3 IgG antibodies have an overall convex or protruding shape that complements the shape of the paratope on the antibody. In contrast, the 2 known epitopes in the complexes with IgE-derived Fab involve β-sheets and are planar. An analysis of epitope shapes in X-ray crystal structures of immunocomplexes showed that most of the epitopes are convex (68%) and a minority are concave (23%) or planar (9%) [39]. However, there is not yet enough evidence that a flat surface is a common feature of IgE antibody-binding epitopes.
The conclusions from the analysis of the X-ray crystal structures of antigen-antibody complexes also apply to the allergen-antibody complexes: (1) complementarity in shape and charge between paratope and epitope are important for binding, (2) only a small fraction of the total complementarity-determining region (CDR) surface is involved in the antigen interaction, (3) the recognition sites are plastic and conformational changes may occur in allergen and/or antibody upon complex formation, (4) side chains contribute a major part of the interaction, (5) hydrogen bonds, van der Waals interactions and salt bridges provide stability to the complex, (6) solvent molecules may also contribute to the interaction, and (7) amino acids with aromatic rings (Phe, Trp) play a significant role in binding [41]. These very hydrophobic amino acids, which are mostly buried in water-soluble molecules, are unusually frequent in CDRs. It has been postulated that the exposure of aromatics, made possible by the strength of the bilayer β-sheet structure of the antibody, enhances the capacity for binding proteins by making the CDR surface ‘sticky’ [41]. Additionally, some of the hydrophobic electron-rich aromatic residues (Phe, Tyr, Trp) interact with a cationic side chain (Arg or Lys) and establish cation-π interactions. These noncovalent bonds have been reported to be important for molecular recognition, including antibody-protein interactions [42,43]. The energy involved in cation-π interactions is of the same order of magnitude as hydrogen bonds or salt bridges [43]. Although cation-π interactions are not present in all protein-antibody interactions, at least 1–3 cation-π interactions were observed in the antibody complexes with Api m 2, Bla g 2, β-lactoglobulin, Phl p 2 and lysozyme (1yqv, 1mlc, 3hfm, 1jhl).
Information about the structural basis of cross-reactivity can also be inferred from the analysis of some of the complexes and the comparison of homologous allergens. The Phl p 2-antibody complex showed that a β-sheet epitope is highly conserved (7 residues) among group 2/3 grass pollen allergens, whereas only 3 residues were conserved with the homologous C-terminus of Phl p 1. These studies supported the extensive cross-reactivity between group 2 and 3 grass allergens and the lack of relevant cross-reactivity between group 2/3 and group 1 allergens [40]. Similarly, the Bet v 1-antibody complex revealed a dominant IgE antibody-binding epitope conserved among major homologous allergens within the Fagales order.
Recently, 2 more allergen-Fab antibody structures have been solved [44,45]. The structure of a complex of Der f 1 with an Fab of a mAb that cross-reacts with Der p 1, together with the structures of both mite group 1 allergens, provides the structural basis for the cross-reactivity between Der p 1 and Der f 1 mite allergens [44,46]. An additional structure of Bla g 2 in complex with an antibody (4C3 mAb) nonoverlapping with the one (7C11 mAb) previously crystallized with the allergen has also been solved. The antibodies 7C11 and 4C3 bind to the 2 opposite lobes of the allergen. The Bla g 2-7C11 interaction involves amino acid residues, whereas carbohydrates, in addition to amino acid residues, are involved in the Bla g 2-4C3 interaction [38,45].
X-Ray Crystal Structures of Bla g 2 Alone and in Complex with an mAb Overlapping with an IgE Epitope
Bla g 2 is one of the main inhaled allergens associated with cockroach allergy and shows the highest prevalence of sensitization among known cockroach allergens (50–70%) [47,48]. Bla g 2 is homologous to the family of aspartic protease enzymes (∼28% identity to pepsin). However, important substitutions in the catalytic site render it inactive. Bla g 2 is related to the pregnancy-associated glycoprotein secreted by ruminants during pregnancy [49,50]. The X-ray crystal structure confirmed that Bla g 2 has a 3-dimensional structure typical of aspartic proteases, with a bilobal shape originated from gene duplication. Each lobe shares a similar overall folding despite little similarity at the level of the primary structure [51]. Unusual features of Bla g 2 for an aspartic protease are: (1) lack of standard aspartic protease activity, (2) the presence of a zinc ion bound to the molecule, and (3) the presence of 5 instead of the usual 2 or 3 disulfide bonds. The stability conferred to the molecule by the last 2 structural features may contribute to the persistence of Bla g 2 in the environment and its relevance for a continuous exposure associated with the development of asthma.
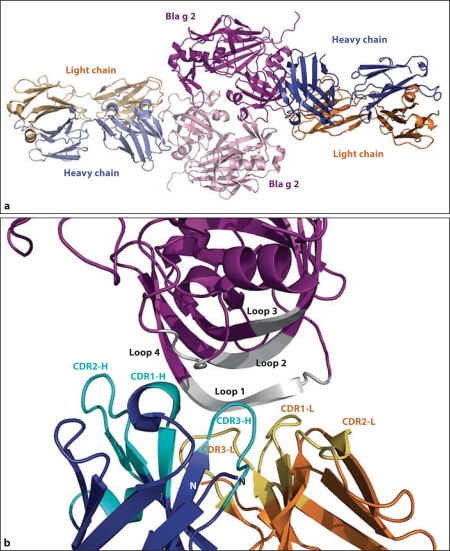
To identify antigenic determinants on Bla g 2, the allergen was cocrystallized with an Fab’ fragment of an mAb (7C11) that interferes with IgE antibody binding. The X-ray crystal structure of the allergen-antibody complex revealed that Bla g 2 forms a dimer, and each monomer interacts with 1 Fab’ molecule, forming a complex of 6 molecules (fig. 2a). There are 2 main interactions in the complex that were analyzed by site-directed mutagenesis: (1) between allergen and antibody and (2) between Bla g 2 monomers.
Fig. 2.
a Ribbon representation of the 3-dimensional structure of the complex between Bla g 2 (purple) and the heavy (blue) and light (orange) chains of the Fab’ fragment of the 7C11 mAb. b Conformational epitope of 7C11 mAb on Bla g 2. The 4 loops involved in the epitope are colored in light grey and the complementarity-determining regions of the heavy and light chains are shown in cyan and yellow, respectively. The N-termini of the heavy and light chains of the mAb are indicated by N.
Identifying Antigenic Determinants on the Cockroach Allergen Bla g 2
The Bla g 2-7C11 mAb interaction defined a large epitope on the surface of the N-terminal domain of the allergen with average contact areas between both molecules of 870 Å2. The epitope includes 4 loops (the predominant loop 1, residues 60–70; loop 2, residues 83–86; loop 3, residues 98–100, and loop 4, residues 129–132), interacting with all 6 CDRs of the Fab’ (fig. 2; table 2). The allergen did not suffer major conformational changes upon antibody binding, the largest one occurring in the area in the vicinity of Asp68a, which is the only residue interacting with both the heavy and the light chain of the antibody.
Table 2.
Main residues involved in the Bla g 2-7C11 mAb interaction
| Bla g 2 residue | 7C11 mAb residue/antibody loop | ||
|---|---|---|---|
| Loop 1 | 60–70 | ||
| K65 | Y531 | L2 | |
| Y66 | S30 | L1 | |
| I67 | W99 | H3 | |
| I67 | R101 | H3 | |
| D68a | H35 | H1 | |
| D68a | Q96 | L3 | |
| G69 | H91 | L3 | |
| N70 | R59 | H2 | |
| Loop 2 | 83–86 | ||
| R83 | Y921 | L3 | |
| E86 | R101 | H3 | |
| Loop 3 | 98–100 | ||
| Q98 | R101 | H3 | |
| D100 | R101 | H3 | |
| Loop 4 | 129–132 | ||
| K132 (monomer 1) | K30 | H1 | |
| K132 (monomer 1) | D52 | H2 | |
| K132 (monomer 2) | Y331 | H1 | |
Cation-π interaction.
Two thirds of the amino acids involved in the Bla g 2-7C11 mAb interaction belong to the continuous part of the epitope of loop 1, which includes 9 consecutive residues, plus Arg83 from loop 2 (fig. 2b). Most of the interactions are polar, involving hydrogen bonds and ion pairs. The involvement of a large continuous segment of the allergen for the largest contact with the antibody has also been observed in the complexes with Bet v 1 and Api m 2. This observation agrees with the concept of linear epitopes as part of discontinuous epitopes. The additional shorter loops 2 and 3 are supported by main chain interactions forming a short β-sheet, recognized by the antibody in a similar way to the ones described for the β-lactoglobulin and Phl p 2 complexes with IgE-Fab. However, the extent of the β-sheets involved in these last 2 interactions seemed to be larger, involving 4 stranded antiparallel β-sheets, and the epitopes were mainly located in secondary structure elements, a feature not observed in 82 structures of protein-antibody complexes [39].
Interestingly, there were 3 cation-π interactions between 3 pairs of residues involving Lys65, Arg83 and Lys132 from Bla g 2, and Tyr53 (L2), Tyr92 (L3) and Tyr33 (H1) in the antibody, respectively (table 2). The cation-π interactions involving Lys65 and Arg 83 are close to the predominant loop 1, which shows the major conformational change upon antibody binding. Therefore, we proposed that cation-π interactions are important for the long-range recognition of the epitopes that show conformational changes by an ‘induced fit’ mechanism. The allergen-antibody recognition involving the loop that includes Lys132 occurs through a ‘lock-and-key’ mechanism. Site-directed mutagenesis of K132 to alanine strongly affected the mAb 7C11 and IgE antibody binding, without significantly changing the allergen 3-dimensional structure [52]. These results indicate that Lys132 is a major contributor to the mAb epitope and that this residue is involved in a major IgE antibody-binding epitope on Bla g 2. The mAb 7C11 inhibits IgE antibody binding up to ∼40%, which represents a significant contribution of the mAb epitope (and specifically Lys132) to IgE antibody binding. This result further supports the importance of the 7C11 mAb epitope on IgE antibody binding, especially considering that the 7C11 mAb epitope represents a very small percentage of the total allergen surface (∼6% calculated from the ∼870 Å2 covered by the antibody vs. the total allergen surface which is 15,061 Å2). It is likely that IgE antibodies also bind to the C-terminal lobe of Bla g 2. However, an equivalent IgE epitope could not be identified by site-directed mutagenesis of the region in the C-terminus structurally equivalent to the 7C11 mAb epitope [53]. A similar outcome has been reported by mutagenesis of a dominant Bet v 1 epitope and a Der f 13 epitope [32,54]. The mutated Der f 13 was able to induce PBMC proliferation comparable to the wild-type allergen and shift the secretion of cytokines from a Th2 to a Th1 pattern. Mutagenesis studies based on the known structure of IgE antibody-binding epitopes will provide the basis to design recombinant hypoallergenic forms of immunotherapy.
Relevance of Oligomerization for Allergic Disease
The number of IgE epitopes on an allergen molecule (as well as the concentration and affinity of allergen-specific antibodies) determines the extent of degranulation [55,56]. Therefore, a dimeric or oligomeric form of an allergen may favor cross-linking versus the monomer due to an effective increase of epitopes. Evidence in favor of the contribution of oligomerization to allergenicity has been reported for the cockroach allergen Bla g 2, which is a dimeric allergen in solution and in the crystallized complex with Fab’ fragments of the mAb 7C11 (fig. 2a). In the X-ray crystal structure of the allergen-antibody complex, the Bla g 2 dimer is stabilized by a quasi- four-helical bundle comprised of 1 short α-helix and 1 helical turn from each monomer. The dimer interface of 1,139 Å2 per molecule is above the consensus cutoff value (∼850 Å2) that allows distinction between real physiological contacts and contacts caused by crystal packing [57]. Two mutants of Bla g 2 with substitutions of residues involved in the dimer interface, including hydrophobic interactions, were expressed in Pichia pastoris and purified. The dimeric wild-type Bla g 2 induced significantly more β-hexosaminidase release from sensitized mast cells than the monomeric mutant, suggesting a certain role of oligomerization in allergenicity of Bla g 2 [38].
Oligomerization is a common phenomenon among allergens and has often been observed in the X-ray crystal structures of allergens or in solution [58,59,60]. Since oligomerization in X-ray crystal structures does not always reflect the oligomeric state in physiological conditions, techniques such as size exclusion chromatography or light scattering can be used to assess oligomerization of allergens in solution. More experimental evidence is needed to assess the extent of the contribution of allergen oligomerization to allergenicity. This effect would not necessarily always occur in artificially oligomerized recombinant allergens. Oligomers engineered artificially from a wild type that is not an oligomer may have reduced allergenicity if misfolding affects the conformation of IgE epitopes or if some epitopes are hidden in the oligomeric interface [61,62].
Final Note
IgE antibody-binding epitopes were initially localized by indirect mapping techniques that led to the identification of mainly linear epitopes. With the development of structural biology techniques such as X-ray crystallography and NMR, the specific epitopes involved in the allergen-antibody interaction can be identified. These structural studies will contribute to understanding intrinsic properties of proteins related to allergenicity.
Acknowledgements
I would like to thank Dr. Alla Gustchina, Dr. Alex Wlodawer and Mi Li at the National Cancer Institute for the collaborative structural studies on the cockroach allergen Bla g 2, and Dr. Martin Chapman for his support. The Bla g 2 project described was supported by Award Number R01AI077653 from the National Institute of Allergy and Infectious Diseases. The content is solely the responsibility of the author and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health.
References
- 1.Jerne NK. Immunological speculations. Ann Rev Microbiol. 1960;14:341–358. doi: 10.1146/annurev.mi.14.100160.002013. [DOI] [PubMed] [Google Scholar]
- 2.Olson JR, Klapper DG. Two major human allergenic sites on ragweed pollen allergen antigen E identified by using monoclonal antibodies. J Immunol. 1986;136:2109–2115. [PubMed] [Google Scholar]
- 3.Chapman MD, Heymann PW, Platts-Mills TA. Epitope mapping of two major inhalant allergens, Der p I and Der f I, from mites of the genus Dermatophagoides. J Immunol. 1987;139:1479–1484. [PubMed] [Google Scholar]
- 4.Amit AG, Mariuzza RA, Phillips SE, Poljak RJ. Three-dimensional structure of an antigen-antibody complex at 2.8 A resolution. Science. 1986;233:747–753. doi: 10.1126/science.2426778. [DOI] [PubMed] [Google Scholar]
- 5.Sheriff S, Silverton EW, Padlan EA, Cohen GH, Smith-Gill SJ, Finzel BC, Davies DR. Three-dimensional structure of an antibody-antigen complex. Proc Natl Acad Sci USA. 1987;84:8075–8079. doi: 10.1073/pnas.84.22.8075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Padlan EA, Silverton EW, Sheriff S, Cohen GH, Smith-Gill SJ, Davies DR. Structure of an antibody-antigen complex: crystal structure of the HyHEL-10 Fab-lysozyme complex. Proc Natl Acad Sci USA. 1989;86:5938–5942. doi: 10.1073/pnas.86.15.5938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lombardero M, Heymann PW, Platts-Mills TA, Fox JW, Chapman MD. Conformational stability of B cell epitopes on group I and group II Dermatophagoides spp. allergens. Effect of thermal and chemical denaturation on the binding of murine IgG and human IgE antibodies. J Immunol. 1990;144:1353–1360. [PubMed] [Google Scholar]
- 8.Varshney S, Goldblum RM, Kearney C, Watanabe M, Midoro-Horiuti T. Major mountain cedar allergen, Jun a 1, contains conformational as well as linear IgE epitopes. Mol Immunol. 2007;44:2781–2785. doi: 10.1016/j.molimm.2005.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maleki SJ, Kopper RA, Shin DS, Park CW, Compadre CM, Sampson H, Burks AW, Bannon GA. Structure of the major peanut allergen Ara h 1 may protect IgE-binding epitopes from degradation. J Immunol. 2000;164:5844–5849. doi: 10.4049/jimmunol.164.11.5844. [DOI] [PubMed] [Google Scholar]
- 10.Schimek EM, Zwolfer B, Briza P, Jahn-Schmid B, Vogel L, Vieths S, Ebner C, Bohle B. Gastrointestinal digestion of Bet v 1-homologous food allergens destroys their mediator-releasing, but not T cell-activating, capacity. J Allergy Clin Immunol. 2005;116:1327–1333. doi: 10.1016/j.jaci.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 11.Hindley J, Wünschmann S, Satinover SM, Woodfolk JA, Chew FT, Chapman MD, Pomés A. Bla g 6: a troponin C allergen from Blattella germanica with IgE binding calcium dependence. J Allergy Clin Immunol. 2006;117:1389–1395. doi: 10.1016/j.jaci.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 12.Valenta R, Hayek B, Seiberler S, Bugajska-Schretter A, Niederberger V, Twardosz A, Natter S, Vangelista L, Pastore A, Spitzauer S, Kraft D. Calcium-binding allergens: from plants to man. Int Arch Allergy Immunol. 1998;117:160–166. doi: 10.1159/000024005. [DOI] [PubMed] [Google Scholar]
- 13.Ball T, Edstrom W, Mauch L, Schmitt J, Leistler B, Fiebig H, Sperr WR, Hauswirth AW, Valent P, Kraft D, Almo SC, Valenta R. Gain of structure and IgE epitopes by eukaryotic expression of the major timothy grass pollen allergen, Phl p 1. FEBS J. 2005;272:217–227. doi: 10.1111/j.1432-1033.2004.04403.x. [DOI] [PubMed] [Google Scholar]
- 14.Takai T, Kato T, Yasueda H, Okumura K, Ogawa H. Analysis of the structure and allergenicity of recombinant pro- and mature Der p 1 and Der f 1: major conformational IgE epitopes blocked by prodomains. J Allergy Clin Immunol. 2005;115:555–563. doi: 10.1016/j.jaci.2004.11.024. [DOI] [PubMed] [Google Scholar]
- 15.Laver WG, Air GM, Webster RG, Smith-Gill SJ. Epitopes on protein antigens: misconceptions and realities. Cell. 1990;61:553–556. doi: 10.1016/0092-8674(90)90464-p. [DOI] [PubMed] [Google Scholar]
- 16.Aalberse RC. Structural biology of allergens. J Allergy Clin Immunol. 2000;106:228–238. doi: 10.1067/mai.2000.108434. [DOI] [PubMed] [Google Scholar]
- 17.Schramm G, Bufe A, Petersen A, Haas H, Merget R, Schlaak M, Becker WM. Discontinuous IgE-binding epitopes contain multiple continuous epitope regions: results of an epitope mapping on recombinant Hol l 5, a major allergen from velvet grass pollen. Clin Exp Allergy. 2001;31:331–341. doi: 10.1046/j.1365-2222.2001.01049.x. [DOI] [PubMed] [Google Scholar]
- 18.Kulkarni-Kale U, Bhosle S, Kolaskar AS. CEP: a conformational epitope prediction server. Nucleic Acids Res. 2005;33:W168–W171. doi: 10.1093/nar/gki460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fiers MW, Kleter GA, Nijland H, Peijnenburg AA, Nap JP, van Ham RC. Allermatch, a webtool for the prediction of potential allergenicity according to current FAO/WHO Codex alimentarius guidelines. BMC Bioinformatics. 2004;5:133. doi: 10.1186/1471-2105-5-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haste AP, Nielsen M, Lund O. Prediction of residues in discontinuous B-cell epitopes using protein 3D structures. Protein Sci. 2006;15:2558–2567. doi: 10.1110/ps.062405906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ponomarenko J, Bui HH, Li W, Fusseder N, Bourne PE, Sette A, Peters B. ElliPro: a new structure-based tool for the prediction of antibody epitopes. BMC Bioinformatics. 2008;9:514. doi: 10.1186/1471-2105-9-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Batori V, Friis EP, Nielsen H, Roggen EL. An in silico method using an epitope motif database for predicting the location of antigenic determinants on proteins in a structural context. J Mol Recognit. 2006;19:21–29. doi: 10.1002/jmr.752. [DOI] [PubMed] [Google Scholar]
- 23.Ivanciuc O, Mathura V, Midoro-Horiuti T, Braun W, Goldblum RM, Schein CH. Detecting potential IgE-reactive sites on food proteins using a sequence and structure database, SDAP-food. J Agric Food Chem. 2003;51:4830–4837. doi: 10.1021/jf034218r. [DOI] [PubMed] [Google Scholar]
- 24.Jenkins JA, Breiteneder H, Mills EN. Evolutionary distance from human homologs reflects allergenicity of animal food proteins. J Allergy Clin Immunol. 2007;120:1399–1405. doi: 10.1016/j.jaci.2007.08.019. [DOI] [PubMed] [Google Scholar]
- 25.Beyer K, Jarvinen KM, Bardina L, Mishoe M, Turjanmaa K, Niggemann B, Ahlstedt S, Venemalm L, Sampson HA. IgE-binding peptides coupled to a commercial matrix as a diagnostic instrument for persistent cow's milk allergy. J Allergy Clin Immunol. 2005;116:704–705. doi: 10.1016/j.jaci.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 26.Jarvinen KM, Beyer K, Vila L, Bardina L, Mishoe M, Sampson HA. Specificity of IgE antibodies to sequential epitopes of hen's egg ovomucoid as a marker for persistence of egg allergy. Allergy. 2007;62:758–765. doi: 10.1111/j.1398-9995.2007.01332.x. [DOI] [PubMed] [Google Scholar]
- 27.Flinterman AE, Knol EF, Lencer DA, Bardina L, den Hartog Jager CF, Lin J, Pasmans SG, Bruijnzeel-Koomen CA, Sampson HA, van Hoffen E, Shreffler WG. Peanut epitopes for IgE and IgG4 in peanut-sensitized children in relation to severity of peanut allergy. J Allergy Clin Immunol. 2008;121:737–743. doi: 10.1016/j.jaci.2007.11.039. [DOI] [PubMed] [Google Scholar]
- 28.Collins SP, Ball G, Vonarx E, Hosking C, Shelton M, Hill D, Howden ME. Absence of continuous epitopes in the house dust mite major allergens Der p I from Dermatophagoides pteronyssinus and Der f I from Dermatophagoides farinae. Clin Exp Allergy. 1996;26:36–42. doi: 10.1111/j.1365-2222.1996.tb00054.x. [DOI] [PubMed] [Google Scholar]
- 29.Davies JM, O'Hehir RE, Suphioglu C. Use of phage display technology to investigate allergen-antibody interactions. J Allergy Clin Immunol. 2000;105:1085–1092. doi: 10.1067/mai.2000.107040. [DOI] [PubMed] [Google Scholar]
- 30.Chan SL, Ong TC, Gao YF, Tiong YS, Wang DY, Chew FT, Mok YK. Nuclear magnetic resonance structure and IgE epitopes of Blo t 5, a major dust mite allergen. J Immunol. 2008;181:2586–2596. doi: 10.4049/jimmunol.181.4.2586. [DOI] [PubMed] [Google Scholar]
- 31.Garcia-Casado G, Pacios LF, Diaz-Perales A, Sanchez-Monge R, Lombardero M, Garcia-Selles FJ, Polo F, Barber D, Salcedo G. Identification of IgE-binding epitopes of the major peach allergen Pru p 3. J Allergy Clin Immunol. 2003;112:599–605. doi: 10.1016/s0091-6749(03)01605-1. [DOI] [PubMed] [Google Scholar]
- 32.Chan SL, Ong ST, Ong SY, Chew FT, Mok YK. Nuclear magnetic resonance structure-based epitope mapping and modulation of dust mite group 13 allergen as a hypoallergen. J Immunol. 2006;176:4852–4860. doi: 10.4049/jimmunol.176.8.4852. [DOI] [PubMed] [Google Scholar]
- 33.Mueller GA, Smith AM, Chapman MD, Rule GS, Benjamin DC. Hydrogen exchange nuclear magnetic resonance spectroscopy mapping of antibody epitopes on the house dust mite allergen Der p 2. J Biol Chem. 2001;276:9359–9365. doi: 10.1074/jbc.M010812200. [DOI] [PubMed] [Google Scholar]
- 34.Naik MT, Chang CF, Kuo IC, Kung CC, Yi FC, Chua KY, Huang TH. Roles of structure and structural dynamics in the antibody recognition of the allergen proteins: an NMR study on Blomia tropicalis major allergen. Structure. 2008;16:125–136. doi: 10.1016/j.str.2007.10.022. [DOI] [PubMed] [Google Scholar]
- 35.Ichikawa S, Takai T, Inoue T, Yuuki T, Okumura Y, Ogura K, Inagaki F, Hatanaka H. NMR study on the major mite allergen Der f 2: its refined tertiary structure, epitopes for monoclonal antibodies and characteristics shared by ML protein group members. J Biochem (Tokyo) 2005;137:255–263. doi: 10.1093/jb/mvi039. [DOI] [PubMed] [Google Scholar]
- 36.Mirza O, Henriksen A, Ipsen H, Larsen JN, Wissenbach M, Spangfort MD, Gajhede M. Dominant epitopes and allergic cross-reactivity: complex formation between a Fab fragment of a monoclonal murine IgG antibody and the major allergen from birch pollen Bet v 1. J Immunol. 2000;165:331–338. doi: 10.4049/jimmunol.165.1.331. [DOI] [PubMed] [Google Scholar]
- 37.Padavattan S, Schirmer T, Schmidt M, Akdis C, Valenta R, Mittermann I, Soldatova L, Slater J, Mueller U, Markovic-Housley Z. Identification of a B-cell epitope of hyaluronidase, a major bee venom allergen, from its crystal structure in complex with a specific Fab. J Mol Biol. 2007;368:742–752. doi: 10.1016/j.jmb.2007.02.036. [DOI] [PubMed] [Google Scholar]
- 38.Li M, Gustchina A, Alexandratos J, Wlodawer A, Wunschmann S, Kepley CL, Chapman MD, Pomés A. Crystal structure of a dimerized cockroach allergen Bla g 2 complexed with a monoclonal antibody. J Biol Chem. 2008;283:22806–22814. doi: 10.1074/jbc.M800937200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Niemi M, Jylha S, Laukkanen ML, Soderlund H, Makinen-Kiljunen S, Kallio JM, Hakulinen N, Haahtela T, Takkinen K, Rouvinen J. Molecular interactions between a recombinant IgE antibody and the beta-lactoglobulin allergen. Structure. 2007;15:1413–1421. doi: 10.1016/j.str.2007.09.012. [DOI] [PubMed] [Google Scholar]
- 40.Padavattan S, Flicker S, Schirmer T, Madritsch C, Randow S, Reese G, Vieths S, Lupinek C, Ebner C, Valenta R, Markovic-Housley Z. High-affinity IgE recognition of a conformational epitope of the major respiratory allergen Phl p 2 as revealed by X-ray crystallography. J Immunol. 2009;182:2141–2151. doi: 10.4049/jimmunol.0803018. [DOI] [PubMed] [Google Scholar]
- 41.Padlan EA. X-ray crystallography of antibodies. Adv Protein Chem. 1996;49:57–133. doi: 10.1016/s0065-3233(08)60488-x. [DOI] [PubMed] [Google Scholar]
- 42.Livesay DR, Subramaniam S. Conserved sequence and structure association motifs in antibody-protein and antibody-hapten complexes. Protein Eng Des Sel. 2004;17:463–472. doi: 10.1093/protein/gzh058. [DOI] [PubMed] [Google Scholar]
- 43.Gallivan JP, Dougherty DA. Cation-pi interactions in structural biology. Proc Natl Acad Sci USA. 1999;96:9459–9464. doi: 10.1073/pnas.96.17.9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chruszcz M, Chapman MD, Stura EA, Vailes LD, Saint-Remy J, Minor W, Pomés A. Crystal structure of Der f 1, Der p 1 and a Der f 1 allergen-antibody complex: localization of antibody binding epitopes. J Allergy Clin Immunol. 2009;123:S209. [Google Scholar]
- 45.Pomés A, Li M, Glesner J, Wünschmann S, King EM, Chapman MD, Wlodawer A, Gustchina A. The X-ray crystal structure of two complexes of the cockroach allergen Bla g 2 with fragments of monoclonal antibodies defines two non-overlapping epitopes. J Allergy Clin Immunol. 2009;123:S229. [:] [Google Scholar]
- 46.Chruszcz M, Chapman MD, Vailes LD, Stura EA, Saint-Remy JM, Minor W, Pomés A. Crystal structures of mite allergens Der f 1 and Der p 1 reveal differences in surface-exposed residues that may influence antibody binding. J Mol Biol. 2009;386:520–530. doi: 10.1016/j.jmb.2008.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arruda LK, Vailes LD, Mann BJ, Shannon J, Fox JW, Vedvick TS, Hayden ML, Chapman MD. Molecular cloning of a major cockroach (Blattella germanica) allergen, Bla g 2. Sequence homology to the aspartic proteases. J Biol Chem. 1995;270:19563–19568. doi: 10.1074/jbc.270.33.19563. [DOI] [PubMed] [Google Scholar]
- 48.Satinover SM, Reefer AJ, Pomés A, Chapman MD, Platts-Mills TA, Woodfolk JA. Specific IgE and IgG antibody-binding patterns to recombinant cockroach allergens. J Allergy Clin Immunol. 2005;115:803–809. doi: 10.1016/j.jaci.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 49.Pomés A, Chapman MD, Vailes LD, Blundell TL, Dhanaraj V. Cockroach allergen Bla g 2: structure, function, and implications for allergic sensitization. Am J Respir Crit Care Med. 2002;165:391–397. doi: 10.1164/ajrccm.165.3.2104027. [DOI] [PubMed] [Google Scholar]
- 50.Wünschmann S, Gustchina A, Chapman MD, Pomés A. Cockroach allergen Bla g 2: an unusual aspartic proteinase. J Allergy Clin Immunol. 2005;116:140–145. doi: 10.1016/j.jaci.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 51.Gustchina A, Li M, Wünschmann S, Chapman MD, Pomés A, Wlodawer A. Crystal structure of cockroach allergen Bla g 2, an unusual zinc binding aspartic protease with a novel mode of self-inhibition. J Mol Biol. 2005;348:433–444. doi: 10.1016/j.jmb.2005.02.062. [DOI] [PubMed] [Google Scholar]
- 52.Wünschmann S, Li M, Gustchina A, Wlodawer A, Chapman MD, Pomés A. Mapping of antigenic determinants on Bla g 2 surface. J Allergy Clin Immunol. 2007;119:S105. [Google Scholar]
- 53.Stohr L, Wünschmann S, Chapman MD, Li M, Wlodawer A, Gustchina A, Pomés A. Does structural symmetry of cockroach allergen Bla g 2 determine symmetry of antigenic determinants for IgE antibody binding? J Allergy Clin Immunol. 2009;121:S213. [:] [Google Scholar]
- 54.Spangfort MD, Mirza O, Ipsen H, Van Neerven RJ, Gajhede M, Larsen JN. Dominating IgE-binding epitope of Bet v 1, the major allergen of birch pollen, characterized by X-ray crystallography and site-directed mutagenesis. J Immunol. 2003;171:3084–3090. doi: 10.4049/jimmunol.171.6.3084. [DOI] [PubMed] [Google Scholar]
- 55.Gieras A, Focke-Tejkl M, Ball T, Verdino P, Hartl A, Thalhamer J, Valenta R. Molecular determinants of allergen-induced effector cell degranulation. J Allergy Clin Immunol. 2007;119:384–390. doi: 10.1016/j.jaci.2006.09.034. [DOI] [PubMed] [Google Scholar]
- 56.Christensen LH, Holm J, Lund G, Riise E, Lund K. Several distinct properties of the IgE repertoire determine effector cell degranulation in response to allergen challenge. J Allergy Clin Immunol. 2008;122:298–304. doi: 10.1016/j.jaci.2008.05.026. [DOI] [PubMed] [Google Scholar]
- 57.Ponstingl H, Henrick K, Thornton JM. Discriminating between homodimeric and monomeric proteins in the crystalline state. Proteins. 2000;41:47–57. doi: 10.1002/1097-0134(20001001)41:1<47::aid-prot80>3.3.co;2-#. [DOI] [PubMed] [Google Scholar]
- 58.Lascombe MB, Gregoire C, Poncet P, Tavares GA, Rosinski-Chupin I, Rabillon J, Goubran-Botros H, Mazie JC, David B, Alzari PM. Crystal structure of the allergen Equ c 1. A dimeric lipocalin with restricted IgE-reactive epitopes. J Biol Chem. 2000;275:21572–21577. doi: 10.1074/jbc.M002854200. [DOI] [PubMed] [Google Scholar]
- 59.Scholl I, Kalkura N, Shedziankova Y, Bergmann A, Verdino P, Knittelfelder R, Kopp T, Hantusch B, Betzel C, Dierks K, Scheiner O, Boltz-Nitulescu G, Keller W, Jensen-Jarolim E. Dimerization of the major birch pollen allergen Bet v 1 is important for its in vivo IgE-crosslinking potential in mice. J Immunol. 2005;175:6645–6650. doi: 10.4049/jimmunol.175.10.6645. [DOI] [PubMed] [Google Scholar]
- 60.Kaiser L, Velickovic TC, Badia-Martinez D, Adedoyin J, Thunberg S, Hallen D, Berndt K, Gronlund H, Gafvelin G, van Hage M, Achour A. Structural characterization of the tetrameric form of the major cat allergen Fel d 1. J Mol Biol. 2007;370:714–727. doi: 10.1016/j.jmb.2007.04.074. [DOI] [PubMed] [Google Scholar]
- 61.Vrtala S, Hirtenlehner K, Susani M, Akdis M, Kussebi F, Akdis CA, Blaser K, Hufnagl P, Binder BR, Politou A, Pastore A, Vangelista L, Sperr WR, Semper H, Valent P, Ebner C, Kraft D, Valenta R. Genetic engineering of a hypoallergenic trimer of the major birch pollen allergen Bet v 1. FASEB J. 2001;15:2045–2047. doi: 10.1096/fj.00-0767fje. [DOI] [PubMed] [Google Scholar]
- 62.Reese G, Ballmer-Weber BK, Wangorsch A, Randow S, Vieths S. Allergenicity and antigenicity of wild-type and mutant, monomeric, and dimeric carrot major allergen Dau c 1: destruction of conformation, not oligomerization, is the roadmap to save allergen vaccines. J Allergy Clin Immunol. 2007;119:944–951. doi: 10.1016/j.jaci.2006.11.699. [DOI] [PubMed] [Google Scholar]