Abstract
We introduce a stepwise approach for family-based designs for selecting a set of markers in a gene that are independently associated with the disease. The approach is based on testing the effect of a set of markers conditional on another set of markers. Several likelihood-based approaches have been proposed for special cases, but no model-free based tests have been proposed. We propose two types of tests in a family-based framework that are applicable to arbitrary family structures and completely robust to population stratification. We propose methods for ascertained dichotomous traits and unascertained quantitative traits. We first propose a completely model-free extension of the FBAT main genetic effect test. Then, for power issues, we introduce two model-based tests, one for dichotomous traits and one for continuous traits. Lastly, we utilize these tests to analyze a continuous lung function phenotype as a proxy for asthma in the Childhood Asthma Management Program. The methods are implemented in the free R package fbati.
Key Words: Binary trait, Candidate gene analysis, Family-based association tests, FBAT-C, Linkage disequilibrium (LD), Model-based test, Model-free test, Nuclear families, Quantitative trait
Introduction
An important methodological issue in association studies of complex disorders is differentiating between true predisposing etiological allele variants and those that are only in linkage disequilibrium (LD) to those loci [Koeleman et al., 2000; Cordell and Clayton, 2002]. Our objective here is to provide methods for selecting a subset of markers in a gene which explain the disease marker association seen in the entire set. There is a complicated relationship between LD and association as described in detail in Nielsen et al. [2008]; strong LD between markers does not always guarantee redundant association results. However, testing each marker at a time, when there is even one DSL in the region, is likely to result in multiple significant χ2 test statistics due to the LD between the markers. In our example, multiple SNPs in the IL10 gene test as significant for association with lung function. A test is needed that can evaluate the contribution of a marker while allowing for an effect at one or more nearby markers.
Case-control methods are used in genetic studies because they can have higher power than family-based tests, but family-based tests of a main genetic effect can have the advantage of being constructed to be completely robust to population substructure and model-free [Laird and Lange, 2006]. In family-based designs, testing a genetic effect at one marker, while conditioning on another marker, has been partially considered in the literature. Koeleman et al. [2000] propose a likelihood-based approach to test for one marker in the presence of another. In a more general framework, Cordell and Clayton [2002], Cordell [2004], and Cordell et al. [2004] suggest a model-building approach using the retrospective likelihood of the genotype conditional on the trait and parental haplotype distribution to model multiple loci. This model based approach is completely robust to population substructure, but is limited to dichotomous traits and families with both parents genotyped. Dudbridge [2008] extends this model-based likelihood approach to missing parents and to quantitative traits using a normal model, but the results can be biased if the normal model does not hold. In principle, the normal model can be extended to using ascertained traits, unlike the approach we will present for quantitative traits. The approach by Dudbridge [2008] is not completely robust to population stratification when there are missing parents, but in practice it performs well when this assumption is violated. We present here methods that are applicable to arbitrary family structures, are completely robust to population stratification, and do not require distributional assumptions on the traits.
When using a test based on multiple genetic markers, we need to consider the difficulty in reconstructing the parental genotypes. When parents are effectively present, as is required in the test by Cordell and Clayton [2002], Cordell [2004], and Cordell et al. [2004] or available through nuisance parameters as in the test by Dudbridge [2008], the haplotype density and phase resolution is not very difficult to compute, even for larger numbers of markers. However, once there are missing parents, reconstructing the haplotype density and phase resolution is more difficult, and can be computationally infeasible if there are more than a few markers. Thus it is advantageous whenever possible to avoid reconstructing the haplotype density of all of the markers when parents are missing and testing multiple markers. Our approaches are constructed with this thought in mind.
We first propose a model-free test for any trait that is completely robust to population substructure and phenotypic model misspecification, and allows for arbitrary pedigrees. We then propose separate model-based tests for dichotomous and continuous traits based on a linear model. The advantage of the model-based method for dichotomous traits over the previous methodology is its method of handling missing parents. This advantage is also shared by the method for quantitative traits, which has the additional advantage of being less restrictive on the phenotypic model than previous approaches. The model-based tests are still completely robust to population substructure, but not to phenotypic model misspecification. We assess the robustness of the model-based tests to phenotypic model misspecification via simulation. In our tests, we avoid reconstructing the full haplotype density by instead conditioning on the haplotype density of small subsets of the markers, or, preferably, the univariate densities of each of the markers. This results in more informative families, especially when parents are missing. We utilize the proposed test to analyze a lung function phenotype in the Childhood Asthma Management Program (CAMP) study.
Methods
We propose several methods, all of which are completely robust to population stratification. We first propose a model-free test for any trait, and then propose separate tests for dichotomous and continuous traits that assume a log-linear and linear disease model, respectively. The latter tests are more powerful, but not completely robust to phenotypic model misspecification. Suppose that Yij indicates an individual's trait. Suppose the genotypes of the k-th marker for the j-th individual in the i-th family are given by gij,k, and Xij,k = X(gij,k) is some coding of the marker, e.g. additive or genotype. We are interested in testing the set of M markers Xij,m = (Xij,m1 … Xij,mm) conditional on the set of C markers Xij,c = (Xij,c1 … Xij,cC). Suppose that the sufficient statistic for parental mating type at marker a is given by Sa. Let S{a,b} = {Sa, Sb}, i.e. the sufficient statistic for parental mating type at marker a and the sufficient statistic for parental mating type at marker b, and let SH({a,b}) be the sufficient statistic for the haplotype density of the parental mating type of those markers. The distribution of the sufficient statistics for the SNP and haplotype density is given in Rabinowitz and Laird [2000] and Horvath et al. [2004]. All tests proposed are completely robust to population substructure by conditioning on the sufficient statistic for parental mating type, although some will use the haplotype density, and some will use just the SNP density.
Model-Free Method for Binary and Continuous Traits (FBAT-C Robust)
A model-free test can be constructed similar to that of the FBAT main genetic effect statistic [Rabinowitz and Laird, 2000; Schaid, 1996]. However, instead of basing the test statistic on P(Xij,mk∣Si,mk), we additionally condition on the other markers using the distribution
| (1) |
Conditioning on Yi is not necessary since under the null hypothesis, the distribution does not depend on Y, as shown in the Appendix. Let Tij be the mean centered for quantitative traits or an indicator of disease status for dichotomous traits [Lunetta et al., 2000]. Let
| (2) |
This is essentially the standard FBAT main genetic effect test [Laird et al., 2000; Rabinowitz and Laird, 2000] in the univariate case, except we have replaced E[X∣S] by E[Xij,m∣Si,H(mk,c), gij,c].
The distribution of P(Xij,mk, Xij,c∣Si,H(mk,c)) is given by Horvath et al. [2004], but we do not need to estimate weights for the phases of the offspring in the cases where phase is not completely resolved. It is a very technical detail as we shall explain. We condition on the sufficient statistic for parental mating type and phase resolution to make the test robust to population substructure. It is easiest to think of the sufficient statistic as a partition of the sample space, or a set of possible outcomes consistent with what was observed or can be reconstructed about the parental mating type [Rabinowitz and Laird, 2000; Horvath et al. 2004]. Construction of this set in Horvath et al. [2004] involves finding the set of unphased genotypes that are consistent with all possible phased parental mating types. Horvath et al. [2004] then estimate the phases of the offspring; however, this is not necessary here as the disease model does not depend on phase. A similar but less general conditioning argument is given in Cordell and Clayton [2002].
Let Ui,m = (Ui,m1 … Ui,mM). Then the resulting test statistic ΣjUi,m is similar to the FBAT main genetic effects test in the univariate case, or the multimarker test [Rakovski et al., 2007; Chapman et al., 2003] when multiple markers are being analyzed. However, here we have a more restrictive conditioning set for computing the expected value of Xij,mk. Proof that this has expectation zero under the null hypothesis is provided in the Appendix. The resulting test, using the empirical variance, is given by
which asymptotically follows a χ2 distribution with rank (Σi (Ui,m) (Ui,m)T) degrees of freedom. We will refer to this test as the FBAT-C Robust test.
Model-Based Methods
Despite the attractiveness of the completely robust test, the FBAT-C Robust test is a lot less powerful than a test based on a disease model. Thus we introduce two model-based methods. We will explore how robust the tests are to phenotypic model misspecification by simulation.
Model-Based Method for an Ascertained Binary Trait (FBAT-C Log-Linear)
For a dichotomous trait, we can alternatively base the likelihood on the joint distribution of the markers being analyzed and conditioned on
| (3) |
where ξi = Si,{H({m1,c}), …,H({mM,c}),H(c)}, rather than equation 1. By not conditioning on Xi,c, the resulting likelihood provides a more powerful test than the model-free approach, but one must assume a model for the trait, as we allow Yi to possibly depend on Xi,c.
For a binary trait, suppose that the probability of disease is given by the generalized linear model
| (4) |
Notice that the disease model does not depend upon phase. The intercept αi allows for other individual or family effects. Covariates are not necessary to model, as we will see that they cancel out of the retrospective likelihood proposed below. Our null hypothesis is that βm = 0, i.e. none of the markers in m are in linkage disequilibrium with any disease locus after adjusting for the markers in c. Suppose we additionally assume that we have phenotypic independence of the sibs, so that
| (5) |
Such an assumption is fairly reasonable in this case since all family-specific covariates and any family factors that might be modeled in equation 4 will cancel out of the likelihood in equation 3, along with αi. Using this likelihood, equations 4 and 5, and using the distribution for P(Xi∣Si,H({mk,c})) as described in the model-free test, we can construct a score test of H0: βm = 0. The log link function in equation 4 is necessary for the baseline disease prevalence to cancel out of the formula. This also requires that we only use the affected offspring, i.e. unaffected sibs are used only to reconstruct parental haplotype transmissions. This restriction was not necessary in the model-free test, although being able to use the unaffected siblings will generally not allow to overcome the power gained from assuming a disease model. The first difference between this retrospective likelihood and the one given by Cordell and Clayton [2002] is that this likelihood conditions on the sufficient statistic for parental mating type, and so can be used when there are missing parents. Unlike the extension proposed by Dudbridge [2008], we do not estimate nuisance parameters for the parental genotypes, and so the test is completely robust to population stratification. We also use the likelihood in equation 3 instead of the product of the marginals ΠjP(Xij∣Si,H({m,c}), Yij = 1; β) as done by Cordell and Clayton [2002] and Dudbridge [2008], although when parents are observed under the assumption in equation 5 they are equivalent. Both Cordell and Clayton [2002] and Dudbridge [2008] also use the haplotype density of all of the markers SH({m,c}), whereas we will use a less restrictive set.
To help us define our estimating equations, let
| (6) |
The derivatives of the log-likelihood based on equation 3 gives us the estimating equations ΣiUi,m(βm, βc) and ΣiUi,c(βm, βc). Details and proof that these have expectation zero under the null hypothesis are provided in the Appendix. We estimate the nuisance parameter βc by solving the estimating equation ΣiUi,c(0, βc) = 0. The contribution of the i-th family, adjusted for estimation of the nuisance parameter, is given by
| (7) |
Then the test statistic is given by (ΣiWi)T (ΣiWiWTi)– (ΣiWi). Under weak regularity conditions, this follows a χ2 distribution with degrees of freedom given by rank (ΣiWiWTi)–. We will denote this test by FBAT-C Log-Linear.
If we had instead based the likelihood on P(Xi,mk∣Xi,c, Yi = 1, Si,H({mk,c}); β), the resulting score test is identical to the model-free FBAT-C Robust test. This is because then the likelihood at βm = 0 no longer depends on Yi = 1 or on β. The added power in the FBAT-C Log-Linear test comes from modeling Xij,c rather than conditioning on it.
In practice, we would code each of the markers Xij,ck using indicator variables for each genotype to prevent model misspecification, i.e. Xij,ck = (Igij,ck = AAIgij,ck = Aa). In contrast, the coding of Xij,mk does not affect the validity of the test. However, using either the additive coding (i.e. the number of disease alleles) or indicator variables for genotype for Xij,mk is necessary when we are testing multiple markers conditional on another marker. With the additive coding and indicator variable coding in the bi-allelic case, the value of the test statistic does not depend on which allele is coded as the disease allele. This is not the case for a dominant or recessive coding. Using indicator variables would be most appropriate in the case of a dominant or recessive gene.
Linear Model for an Unascertained Continuous Trait (FBAT-C Linear)
We take a slightly different approach for quantitative traits, assuming a model only for the mean of the trait. Assume that the mean of the trait follows a linear model
| (8) |
We additionally assume only that Mendel's laws hold and make no distributional assumptions on the error, rather than normal error as in Dudbridge [2008]. The term μij = μij(Si,{m,c}) encodes the dependence of the trait on the parental mating type. We leave this term unspecified, so that the test is completely robust to population stratification.
Let ΔXij,mk = Xij,mk – E(Xij,mk∣Si,mk), and ΔXij,m = (ΔXij,m1 … ΔXij,mM). The parameters βm and βc could be estimated using G-Estimation [Robins et al., 1992] as in Vansteelandt et al. [2008]. However, here we are instead interested in testing βm, treating βc as a nuisance parameter; that is, here we are coding each allele instead of grouping them as a single haplotype. Since we also assume no haplotype effect, we use a slightly different parental mating type conditioning set than Vansteelandt et al. [2008], and a very similar G-Estimator. Define the residual phenotype
To define the estimating equations, similar to before, we let
| (9) |
with Ui,m and Ui,c defined as before in equation 6. Then our estimating equations can be given by ΣiUi,m and ΣiUi,c. Note that these equations do not depend on the haplotype density at all, and so will have more informative families than the other proposed methods. The estimating equation has expectation 0 (see Appendix) for every choice of μij. Thus the choice of μij will not affect the validity of the test, only the power. A powerful two-step approach for estimating μij is as follows. In the first step, we estimate μij by the sample mean of the trait, possibly after adjustment for covariates. We then estimate βc using the data. In the second step, we let μij be the predicted value of the residual eij(βm, βc) from each strata of parental mating type. Misspecification of this second step will not invalidate the approach, but may at worst lead to some loss in power.
Construction of the test statistic then proceeds as in that of the FBAT-C Log-Linear test. The nuisance parameter is solved as before under the null hypothesis from the equation Σi,jUij,c(0, βc) = 0, which has the following closed form estimate
Intuitively, the estimating equation for the nuisance parameter βc is very similar to the FBAT main genetic effects test. Let the contribution of the i-th family, adjusted for estimating the nuisance parameter, be given by
Then the test statistic is given by (ΣiWi)T (ΣiWiWTi)– (ΣiWi). Under weak regularity conditions, this follows a χ2 distribution with rank (ΣiWiWTi)– degrees of freedom, as shown in the Appendix. This test statistic is intuitively similar to a test constructed by first regressing the effect of the marker being conditioned on the trait, and second using this residual as a new trait in the standard FBAT main genetics effect test of the marker being analyzed; however, the G-Estimation approach uses Mendel's laws and evaluates the contributions of all markers simultaneously. Additionally, if we were to avoid modeling Xc in equation 8, and use S{H(m1,c), … H(mM,c)}, then the G-Estimation would yield the FBAT-C Robust test.
Stepwise Strategy
Each test described above (FBAT-C Robust, Log-Linear, and Linear) can be applied using a stepwise approach to determine a set of markers that best explain the association with the disease. The stepwise approach is intended to be used after a significant result has been found from a multimarker test [Chapman et al., 2003; Rakovski et al., 2007]. The stepwise approach begins with a univariate analysis of each marker, with the FBAT main genetic effects test by Rabinowitz and Laird [2000], choosing the most significant marker. Then the FBAT-C test can be applied by conditioning on the markers from the previous step, and testing each of the other markers. A step-down approach is then applied to ensure all of the markers are really necessary.
Simulations
Power
In all plots and tables displayed here, unless otherwise noted, we ran 10,000 simulations with an empirical type I error rate of 0.05. Each consisted of 500 families, of different structures as will be indicated. Results are displayed under an additive disease model.
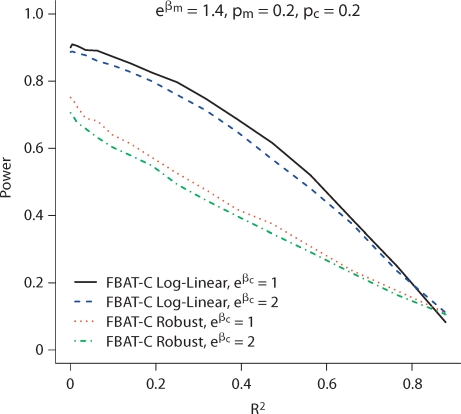
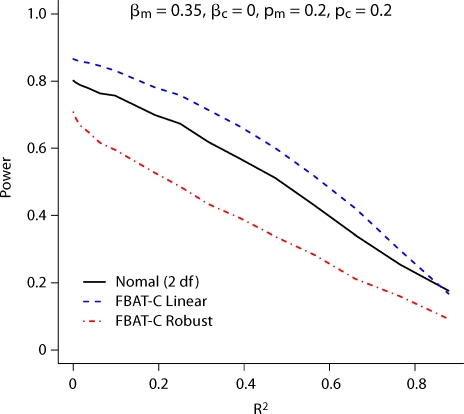
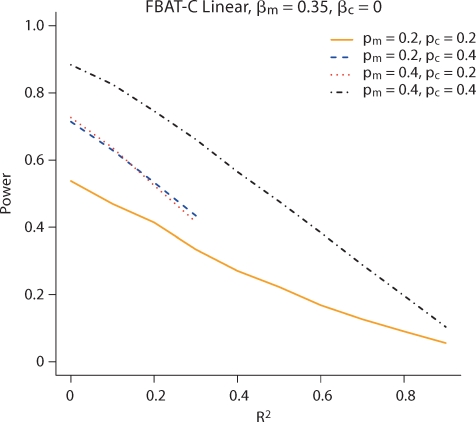
We first compare the power of the model-based FBAT-C Log-Linear and FBAT-C Linear tests to the FBAT-C Robust test, and the other approaches proposed in the literature. For power, we simulate the data under the models given in equation 4 for dichotomous traits and equation 8 with standard normal error for continuous traits. Figure 1 and 2 show that the model-based methods can be substantially more powerful than the robust method for dichotomous and continuous traits, respectively. They also show that the test will not have much if any power if the markers being analyzed are too highly correlated (R2, rather than D′) with the markers being conditioned on, depending on the sample size and magnitude of the effect size. If the markers are perfectly correlated, then they are indistinguishable. Results in this plot are shown for allele frequencies of 0.2 for both alleles, but the relative power difference between the tests does not change much as a function of allele frequency. We vary the allele frequencies in figure 3; both the allele frequency of the marker being tested and that being conditioned on affect the power of the disease. In figure 1, the FBAT-C Log-Linear test has the same power as the likelihood ratio test introduced by Cordell and Clayton [2002], since for trios the FBAT-C Log-Linear test is a score test of the same likelihood. The FBAT-C Linear model is more powerful than the normal model implemented in the software of Dudbridge [2008], largely because the latter is a two degree of freedom test while the former is a one degree of freedom test. Results are similar for discordant sibpairs; the nuisance parameters for parents do not increase the power of the test much, if at all (results shown in online supplementary figures 1 and 2, www.karger.com/doi/10.1159/000264447).
Fig. 1.
Power for trios with a dichotomous trait of the test of Xm conditional on Xc. R2 is between the markers Xm and Xc, where Xm is the DSL.
Fig. 2.
Power for trios of the test of Xm conditional on Xc for a continuous trait.
Fig. 3.
Power of the test of Xm conditional on Xc for a continuous trait using FBAT-C Linear for different allele frequencies. The two power curves, where pm ≠ pc, cannot be perfectly correlated (binary data), and are shown only as far as they can be correlated.
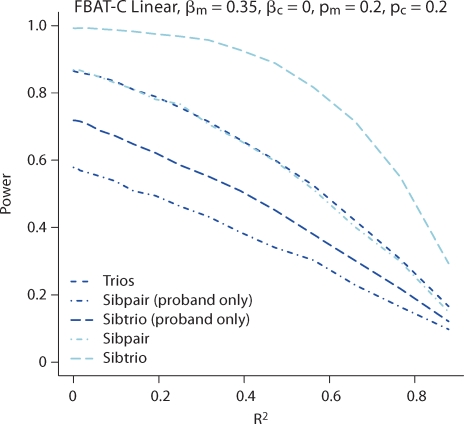
Next, we explore the power of different family structures in figure 4; results are similar to that of the usual FBAT main genetic effect test. Under a quantitative trait, we compare trios, sibships with two offspring and no parents (sibpairs), and sibships with three offspring and no parents (sibtrios). For the sibpairs and sibtrios, when parents are missing, we then consider the case when only one offspring in the sibship is phenotyped, which would be more common in a dichotomous trait. In this case trios are more powerful than sibtrios, which are more powerful than sibships. This relationship is preserved no matter what the allele frequency of the other model parameters are. However, with a quantitative trait, often more offspring are phenotyped, so we then compare to the cases when all of the offspring are phenotyped. When all offspring are phenotyped, the sibpair is about as powerful as trios, and the sibtrio is the most powerful.
Fig. 4.
Power of the test of Xm conditional on Xc for a continuous trait using FBAT-C Linear. For the sibpair and sibtrio simulations, those indicated with ‘proband only’ have only the proband phenotyped; the others have phenotyped all offspring.
Robustness
Secondly, we test the robustness of the two model-based tests. The FBAT-C Robust test always preserves the type I error rate, so we focus here on the performance of the model-based tests. The results we display for all of the robustness tests are all done with an allele frequency of 0.2. Other allele frequencies were simulated, but the main parameter that inflated the type I error was the correlation between the markers. In the cases where the type I error was inflated, it was generally more inflated for more highly correlated markers, and for higher βc values, unless otherwise specified. Thus most charts focus on showing a range of R2 values or just more extreme R2 values when other parameters are of interest, and reasonable βc values. We begin by considering the robustness of the FBAT-C Log-Linear method. One potential misspecification is if the scale of the model is wrong, for example if the link function in equation 4 should instead be on a logistic scale, although they should be similar for a rare enough disease. To investigate the robustness of the test, we simulated data under the logistic model. In the results in online supplementary table 1, the model-based test is generally overly conservative, but preserves the nominal type I error rate. This does not generally translate into more power for the FBAT-C test than the FBAT-C Robust test under the misspecified case, except for the cases when the markers are very highly correlated, and there is low power of the test anyway (results not shown).
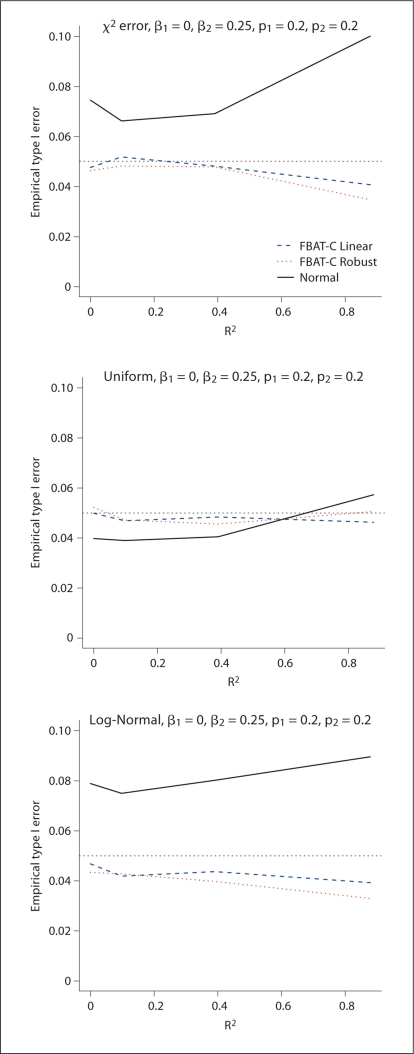
Next, we tested the robustness of the model-based tests for continuous traits to the disease model. We modeled the mean as in equation 8 with a uniform and χ2 variance distribution. Figure 5 shows that FBAT-C Linear performs as expected and desired, while the approach based on a normal likelihood [Dudbridge, 2008] has an inflated type I error. Second, we simulated what would happen if the mean was really specified by a log-normal distribution, to test what would happen when the linear model did not hold for the mean. We see very little departure from the nominal type I error rate in figure 5 for the FBAT-C Linear test. Third, we tested whether phenotypic correlation amongst the siblings would bias the test. Even under strong phenotypic correlation, the test shows little if any departure from the nominal type I error rate (results not shown). Last, we found that the approach by Dudbridge [2008] did not inflate the type I error rate much, if at all, for discordant sibpairs (results shown in the supplementary material).
Fig. 5.
FBAT-C Linear validity results for trios. The plots of the χ2 and uniform distribution follow a linear model, but have nonnormal variance. The log-normal model follows a non-linear model.
In summary the model-based method for a dichotomous test often preserves the type I error rate, but is over-conservative when the log-linear conditional mean model is misspecified. The model-based method for continuous traits also performs well.
Stepwise Strategy
Lastly we test how well our overall strategy performs. We apply this for our FBAT-C Linear test and our FBAT-C Robust test. We additionally consider one other approach, where all markers are univariately tested, and those with a significant p value after Bonferroni correction are chosen; we denote this approach the Bonferroni approach. To simulate data to test the strategy, we use the haplotype frequency from the dataset (table 1) in our application with 682 trios. We consider validity in this case to be when the approach finds a marker (or markers), but there is no association, i.e. no markers should be found because there is no DSL. Recall that the FBAT-C approaches all begin with a multimarker test, and so should be conservative in this case. When testing the validity of the test, the FBAT-C Linear and FBAT-C Robust have an empirical type I error rate of 0.018, and the Bonferroni approach has an empirical type I error rate of 0.032 (500 simulations, approximate SE 0.01). We then simulated three different cases to test how well the approach works if there really was a DSL, or two DSLs, in the set of typed markers. Lastly we looked at how well the approach would work if a DSL was untyped; we took the two most highly correlated SNPs from our dataset (correlation 0.93), using one as the untyped DSL, and saw how often the other correlated marker was chosen. Results for a continuous trait are shown in table 2, and are similar for a dichotomous trait (results not shown). With one DSL, the FBAT-C Linear and FBAT-C Robust tests perform similarly, as the initial univariate step is the same. However, when there is more than one DSL, then the FBAT-C Linear approach performs better than the FBAT-C Robust test. The Bonferroni approach generally does better for the case of picking up only non-DSL markers, but has a lot more cases where it picks up only one of the DSLs or one of the DSLs and additional markers (the ‘at least one DSL’ column), as would be expected.
Table 1.
R2 (D') of markers in the CAMP dataset
| rs3024509 | rs3024492 | rs3024496 | rs1800896 | rs1800872 | rs1800871 | |
|---|---|---|---|---|---|---|
| rs3024509 | 1 | |||||
| rs3024492 | 0.02 (1.00) | 1 | ||||
| rs3024496 | 0.07 (0.96) | 0.33 (0.99) | 1 | |||
| rs1800896 | 0.07 (0.96) | 0.33 (0.99) | 0.90 (0.95) | 1 | ||
| rsl800872 | 0.02 (0.95) | 0.10 (0.98) | 0.30 (0.96) | 0.31 (0.99) | 1 | |
| rs1800871 | 0.02 (0.88) | 0.10 (0.94) | 0.29 (0.93) | 0.29 (0.95) | 0.93 (0.98) | 1 |
Table 2.
. Empirical results for different strategies using the haplotype frequencies from the CAMP data with 682 trios
| Strategy | β | FBAT-C Linear |
FBAT-C Robust |
Bonferroni |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| exactly all DSLs | at least one DSL | only non-DSL markers | exactly all DSLs | at least one DSL | only non-DSL markers | exactly all DSLs | at least one DSL | only non-DSL markers | ||
| 1 DSLa | 0.25 | 0.396 | 0.032 | 0.150 | 0.392 | 0.112 | 0.150 | 0.238 | 0.028 | 0.040 |
| 0.5 | 0.832 | 0.082 | 0.084 | 0.814 | 0.194 | 0.104 | 0.118 | 0.078 | 0.000 | |
| 2 DSLa | 0.25 | 0.242 | 0.470 | 0.052 | 0.116 | 0.540 | 0.084 | 0.026 | 0.646 | 0.000 |
| 0.5 | 0.672 | 0.300 | 0.000 | 0.450 | 0.362 | 0.028 | 0.088 | 0.794 | 0.004 | |
| Untyped DSLb | 0.25 | 0.372 | 0.058 | 0.064 | 0.354 | 0.056 | 0.066 | 0.352 | 0.166 | 0.046 |
| 0.5 | 0.834 | 0.162 | 0.002 | 0.846 | 0.132 | 0.016 | 0.254 | 0.744 | 0.002 | |
When the true model is one DSL, the column ‘at least one DSL’ indicates the case when the DSL is found and additional non-DSL markers are found. When the true model is two DSLs the column ‘at least one DSL’ included the case when only one of the two DSL markers is found, with or without any additional markers. The columns for each strategy do not sum to 1, as the case when no markers are found is not included in the table.
For 1 and 2 DSLs, random markers were chosen as the DSL(s).
For the untyped DSL, rs1800872 was chosen as the DSL, but was omitted from the marker set, and rs1800871 was considered to be ‘the true DSL’ because of the high correlation (0.93). Based on 500 simulations; approximate SE <0.01.
Application to SNPs in the IL10 Gene
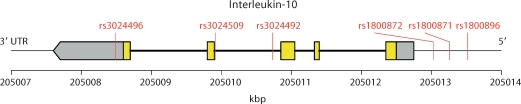
We apply our test to six SNPs in the Interleukin-10 (IL10) gene and its promoter regions in the Childhood Asthma Management Program (CAMP), as previously described in Lyon et al. [2004]. The SNPs are shown in figure 6. We use the continuous measurement of post-bronchodilator forced expiratory volume (FEV) as a proxy for lung function, adjusted for age, height, gender, weight, and race [CAMP, 1999]. There are 682 phenotyped offspring, with almost all having parental genotype information. The multimarker test [Rakovski et al., 2007] of these six markers has a p value of 0.0131, and the correlation matrix of the markers is shown in table 1.
Fig. 6.
The SNPs in the IL10 gene of the CAMP dataset.
We apply our FBAT-C Linear test, our FBAT-C Robust tests, and the normal model proposed by Dudbridge [2008] to the dataset. For illustrative purposes, we also dichotomize the trait by its median, and apply our FBAT-C Log-Linear and FBAT-C Robust tests. To demonstrate the test, we apply it in a stepwise fashion, as in stepwise regression as described above. We begin by choosing the most significant marker from the univariate FBAT main genetic effects test, which is the same as the univariate marker rs3024496 chosen by the normal model approach (table 3). The results of the stepwise approach are shown in table 4. The strategy using the FBAT-C Linear approach, the most powerful of our proposed approaches, begins with rs3024496, and then also chooses rs1800872. These two markers are not very strongly correlated (R2 = 0.30, table 1). Results of the haplotype test Horvath et al. [2004] are given in table 5, showing that there may be an interaction between these two markers.
Table 3.
Univariate results of the CAMP dataset
| Marker | Allele frequency | Quantitative trait
|
Dichotomized trait |
||||
|---|---|---|---|---|---|---|---|
| # Inf | FBAT | normal | # Inf | FBAT | |||
| rs3024509 | 0.057 | 127 | 0.6462 | 0.7033 | 64 | 0.3390 | |
| rs3024492 | 0.222 | 341 | 0.0290 | 0.0310 | 181 | 0.8946 | |
| rs3024496 | 0.457 | 446 | 0.0019 | 0.0032 | 235 | 0.0106 | |
| rs1800896 | 0.455 | 395 | 0.0751 | 0.0278 | 209 | 0.0351 | |
| rs1800872 | 0.280 | 365 | 0.7894 | 0.8714 | 196 | 0.3778 | |
| rs1800871 | 0.284 | 378 | 0.6233 | 0.7812 | 199 | 0.3266 | |
Results are first given for the quantitative trait; p values are given using the FBAT main genetic effects test (FBAT), and using the normal model (normal) as proposed by Dudbridge [2008]. Then results of the median-dichotomized trait are presented using the FBAT main genetic effects test. The number of informative families, i.e. the number of families with non-zero contributions to the test statistic, is given in the ‘# Inf column.
Table 4.
Stepwise results using the FBAT-C Linear and FBAT-C Robust approaches presented in the paper
| Step | Marker analyzed | Marker condition | Quantitative trait |
Dichotomized trait |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| FBAT-C Linear |
FBAT-C Robust |
normal model |
FBAT-C Log-Linear |
FBAT-C Robust |
|||||||
| # Inf | p value | # Inf | p value | p value | # Inf | p value | # Inf | p value | |||
| Up | rs3024509 | rs3024496 | 121|(121,315) | 0.7996 | 71 | 0.3509 | 0.9673 | 60|(136,235) | 0.6535 | 40 | 0.5271 |
| rs3024492 | 299|(299,314) | 0.6139 | 122 | 0.3276 | 0.8030 | 160|(136,234) | 0.1631 | 67 | 0.3352 | ||
| rs1800896 | 341|(341,282) | 0.2327 | 16 | - | 0.6274 | 176|(127,214) | 0.6099 | 9 | - | ||
| rs1800872 | 334|(334,282) | 0.0490 | 158 | 0.1003 | 0.0910 | 177|(126,213) | 0.5330 | 92 | 0.3820 | ||
| rs1800871 | 331|(331,286) | 0.0951 | 154 | 0.0701 | 0.1146 | 175|(124,216) | 0.4023 | 86 | 0.1676 | ||
| Up | rs3024509 | rs3024496, rs1800872 | 108|(108,282), (401,329) | 0.6867 | |||||||
| rs3024492 | 266|(266,281), (400,328) | 0.4493 | |||||||||
| rs1800896 | 339|(339,281), (400,328) | 0.8362 | |||||||||
| rs1800871 | rs1800872 | 304|(304,259), (369,300) | 0.2178 | ||||||||
| Down | rs3024496 | rs1800872 | 3461(346,329) | 0.0038 | |||||||
| rs1800872 | rs3024496 | 334|(334,282) | 0.0490 | ||||||||
Results are also presented using a normal model as proposed by Dudbridge [2008]. The number of informative families, i.e. the number of families with non-zero contributions, is given in the ‘# Inf column. In the ‘# Inf column, the first number corresponds to the analysis allele (under an additive coding), and the subsequent pairs of numbers correspond to the number of informative families for estimating each nuisance parameter (under a codominant coding). The results of each test are shown only as far as the approach would have gone, and if there were ≥20 informative families.
Table 5.
Haplotype results of the CAMP data at the SNPs rs3024496 and rs1800872, respectively
| Haplotype | Frequency | # Inf | p value | Transmission |
|---|---|---|---|---|
| C C | 0.454 | 405 | 0.0151 | + |
| T C | 0.271 | 379 | 0.0061 | − |
| T A | 0.271 | 363 | 0.9490 | − |
| C A | 0.004 | 52 | 0.0623 | + |
For transmission, ‘+’ indicates rarer allele overtransmitted, and ‘–’ indicates undertransmitted.
Discussion
We began by presenting the FBAT-C Robust test, the test that is totally robust to model misspecification. The FBAT-C Robust test uses the haplotype density of all of the markers being tested and each marker being conditioned on, and has very few informative families, especially if there are lots of markers. We then introduced a disease model for a more powerful approach that generally behaves well under model misspecification. For ascertained dichotomous traits, we introduced the FBAT-C Log-Linear test. The FBAT-C Log-Linear test uses the same haplotype density at the FBAT-C Robust test, but gains an extra power boost from the disease model. Instead of conditioning on the other markers, they are jointly modeled. Finally we introduced the FBAT-C Linear test for quantitative traits. The FBAT-C Linear test does not require normal errors, and does not even need the haploytpe distribution, getting a further power boost. In our tests, we generally avoided reconstructing the haplotype density of all of the markers, and thus also did not include it in our disease model. The disadvantage of this approach is that one can no longer determine whether there is a haplotype effect of the markers, or if the markers are individually contributing to the risk of the disease. However, one can follow-up with a haplotype test, as we did in the application, to determine this. Additionally, the FBAT-C Log-Linear approach could be modified to test for haplotype effects by conditioning on the sufficient statistic for parental mating type of all of the markers rather than the set of pairwise sufficient statistics.
In our application of the FBAT-C approach to the CAMP dataset, we used a stepwise approach to choose the best set of markers to explain the variation in the trait. We assessed how well this approach worked via simulation using our dataset, comparing it to the standard univariate Bonferroni correction approach. The stepwise approach generally performs better at finding the true set of DSLs, at the cost of finding a set of markers that do not contain the DSL more often.
The software is available in the R [R Development Core Team, 2008] package fbati, available from http://cran.r-project.org/. It uses the package fgui for the graphical interface [Hoffmann and Laird, 2009], and the data loading routines of pbatR [Hoffmann and Lange, 2006]. It requires the free FBAT program [Laird et al., 2000] to run. The data format is as described in pbatR.
Supplementary Material
Supplementary Material
Acknowledgements
T.J.H. was funded by MH17119 and ES007142. N.M.L. and C.L. were funded by R01-MH059532. S.V. was supported in part by the IAP Research grant nr. P06/03 from the Belgian Government (Belgian Science Policy).
We thank all of the families for their invaluable participation in the Childhood Asthma Management Study (CAMP). We acknowledge the CAMP investigators and research team, supported by the National Heart, Lung and Blood Institute (NHLBI), National Institutes of Health, for the collection of the CAMP Genetic Ancillary Study data. All work on data collected from the CAMP Genetic Ancillary Study was conducted at the Channing Laboratory of the Brigham and Women's Hospital under appropriate policies and human subject's protections for these two studies. The CAMP Genetics Ancillary Study is supported by U01 HL075419, U01 HL65899, P01 HL083069, R01 HL086601 from the NHLBI.
Appendix
A. Expectation of Model-Free Test Statistic
Here we prove that under H0 equation 9 has expectation 0. For simplicity, we drop the i, j indices. We have that
and hence it suffices to show that
And hence it suffices to show that
| (10) |
We have that
Now, the null hypothesis implies that Y · gm∣gc, SH({m,c}), hence
This verifies equation 10, and so under H0 equation 2 has expectation 0.
Appendix
B. FBAT-C Log-Linear Supplemental Material
B.1 FBAT-C Derivation
The joint retrospective likelihood is given by
| (11) |
and so the log-likelihood contribution of each individual is
We then have that, for each individual marker, the derivative of the log-likelihood evaluated at βm = 0, and with the conditioning set simplified (which we will show we can do in B.2), is given by
where
B.2 Unbiasedness of Estimating Equations
Next, we show that we can simplify the conditioning set for the markers, that the estimating equation ΣiUi has expectation zero under the null hypothesis. We have that
For simplicity, we drop the i indices. First we show that
It is sufficient to show that
Suppose parental genotypes are known, and phase is known of all of the markers. Then this is true by Mendel's laws. If phase is not known, but parents are still present, then we integrate over the phases of the parents and the offspring. We have
| (12) |
where the equality from the first to the second line again follows by Mendel's laws. Thus the result follows when parents are present. Finally suppose that one or both parents are missing. Then we again have equation 12. Now, each combination of the marker being analyzed and conditioned on can be thought of as a single multiallelic marker. Then the equality of P(Xc∣phase, ξ) = P(Xc, ξ) follows from Mendelian transmissions by the arguments in Rabinowitz and Laird [2000] using the sufficient statistic for a multiallelic marker. Similarly, we have that
and so the estimating equations are unbiased.
B.3 Calculations for Wi
Now, when adjusting for the nuisance parameter, we need the following additional derivatives, evaluated under βm = 0. Let
where we define M⊗0 = 1, M⊗1 = M, and M⊗2 = MMT. Then we have that
Appendix
C. Distribution of Wi
From a Taylor series expansion, we have that
Hence
| (13) |
| (14) |
It now follows from the central limit theorem that Wi (equation 7) follows an asymptotically multivariate normal distribution, with variance that can be consistently estimated by the right hand side of expression 13, replacing βc with βcirc;c.
Appendix
D. Unbiasedness of Quantitative Trait Estimating Equations for FBAT-C Linear
Here we show that equation 8 has expectation zero for any μij, similar to section B.2. As an intermediate step, first we show that f(gm∣Sm, Sc) = f(gm∣Sm). Suppose we have both parents, and phase is resolvable, then the transmissions follow Mendel's laws, and the equation holds. If the parents are present, and phase is not resolvable, then we integrate over phases of the parents and offspring. We have
| (15) |
where f(gm∣Sm, Sc, phase) = f(gm∣Sm) follows from Mendel's laws. The result then follows when parents are present. Finally, suppose that one or both parents are missing. Then we again have equation 15. Now, each combination of the marker being analyzed and conditioned on can then be thought of as a single multiallelic marker. The equality f(gm∣Sm, Sc, phase) = f(gm∣Sm) then follows from Mendelian transmissions by the arguments in Rabinowitz and Laird [2000] using the sufficient statistic for a multiallelic marker.
With this result, showing that equation 8 has expectation zero proceeds similarly to that in Vansteelandt et al. [2008].
where we use that E(ΔXij,m∣Sm, Sc) = E(ΔXij,m∣Sm) from our results above.
References
- CAMP The childhood asthma management program (camp): design, rationale, and methods. childhood asthma management program research group. Control Clin Trials. 1999;20:91–120. [PubMed] [Google Scholar]
- Chapman JM, Cooper JD, Todd JA, Clayton DG. Detecting disease associations due to linkage disequilibrium using haplotype tags: a class of tests and the determinants of statistical power. Hum Hered. 2003;56:18–31. doi: 10.1159/000073729. URL http://dx.doi.org/10.1159/000073729. [DOI] [PubMed] [Google Scholar]
- Cordell HJ. Properties of case/pseudocontrol analysis for genetic association studies: Effects of recombination, ascertainment, and multiple affected offspring. Genet Epidemiol. 2004;26:186–205. doi: 10.1002/gepi.10306. URL http://dx.doi.org/ 10.1002/gepi.10306. [DOI] [PubMed] [Google Scholar]
- Cordell HJ, Clayton DG. A unified stepwise regression procedure for evaluating the relative effects of polymorphisms within a gene using case/control or family data: application to hla in type 1 diabetes. Am J Hum Genet. 2002;70:124–141. doi: 10.1086/338007. URL http://dx.doi.org/10.1086/338007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordell HJ, Barratt BJ, Clayton DG. Case/pseudocontrol analysis in genetic association studies: A unified framework for detection of genotype and haplotype associations, gene-gene and gene-environment interactions, and parent-of-origin effects. Genet Epidemiol. 2004;26:167–185. doi: 10.1002/gepi.10307. URL http://dx.doi.org/10.1002/gepi.10307. [DOI] [PubMed] [Google Scholar]
- Dudbridge F. Likelihood-based association analysis for nuclear families and unrelated subjects with missing genotype data. Hum Hered. 2008;66:87–98. doi: 10.1159/000119108. URL http://dx.doi.org/10.1159/000119108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann TJ, Laird NM. fgui: A method for automatically creating graphical user interfaces for command-line R packages. Journal of Statistical Software. 2009;30:1–14. doi: 10.18637/jss.v030.i02. URL http://www.jstatsoft.org/v30/i02/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann T, Lange C. P2BAT: A massive parallel implementation of PBAT for genome-wide association studies in R. Bioinformatics. 2006;22:3103–3105. doi: 10.1093/bioinformatics/btl507. URL http://dx.doi.org/ 10.1093/bioinformatics/btl507. [DOI] [PubMed] [Google Scholar]
- Horvath S, Xu X, Lake SL, Silverman EK, Weiss ST, Laird NM. Family-based tests for associating haplotypes with general phenotype data: application to asthma genetics. Genet Epidemiol. 2004;26:61–69. doi: 10.1002/gepi.10295. URL http://dx. doi.org/10.1002/gepi.10295. [DOI] [PubMed] [Google Scholar]
- Koeleman BP, Dudbridge F, Cordell HJ, Todd JA. Adaptation of the extended transmission/disequilibrium test to distinguish disease associations of multiple loci: the conditional extended transmission/disequilibrium test. Ann Hum Genet. 2000;64:207–213. doi: 10.1017/S0003480000008095. URL http://dx.doi.org/doi:10.1017/S0003480000008095. [DOI] [PubMed] [Google Scholar]
- Laird NM, Horvath S, Xu X. Implementing a unified approach to family-based tests of association. Genet Epidemiol. 2000;19(suppl 1):S36–S42. doi: 10.1002/1098-2272(2000)19:1+<::AID-GEPI6>3.0.CO;2-M. URL http://dx.doi.org/3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Laird NM, Lange C. Family-based designs in the age of large-scale gene-association studies. Nat Rev Genet. 2006;7:385–394. doi: 10.1038/nrg1839. URL http://dx.doi.org/10.1038/nrg1839. [DOI] [PubMed] [Google Scholar]
- Lunetta KL, Faraone SV, Biederman J, Laird NM. Family-based tests of association and linkage that use unaffected sibs, covariates, and interactions. Am J Hum Genet. 2000;66:605–614. doi: 10.1086/302782. URL http://dx.doi.org/10.1086/ 302782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon H, Lange C, Lake S, Silverman EK, Randolph AG, Kwiatkowski D, Raby BA, Lazarus R, Weiland KM, Laird N, Weiss ST. Il10 gene polymorphisms are associated with asthma phenotypes in children. Genet Epidemiol. 2004;26:155–165. doi: 10.1002/gepi.10298. URL http://dx.doi.org/ 10.1002/gepi.10298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen DM, Suchindran S, Smith CP. Does strong linkage disequilibrium guarantee redundant association results? Genet Epidemiol. 2008;32:546–552. doi: 10.1002/gepi.20328. URL http://dx.doi.org/10.1002/gepi.20328. [DOI] [PubMed] [Google Scholar]
- Rabinowitz D, Laird N. A unified approach to adjusting association tests for population admixture with arbitrary pedigree structure and arbitrary missing marker information. Hum Hered. 2000;50:211–223. doi: 10.1159/000022918. [DOI] [PubMed] [Google Scholar]
- Rakovski CS, Xu X, Lazarus R, Blacker D, Laird NM. A new multimarker test for family-based association studies. Genet Epidemiol. 2007;31:9–17. doi: 10.1002/gepi.20186. URL http://dx.doi.org/10.1002/gepi.20186. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria, 2008. ISBN 3–900051–07–0.
- Robins JM, Mark SD, Newey WK. Estimating exposure effects by modelling the expectation of exposure conditional on confounders. Biometrics. 1992;48:479–495. [PubMed] [Google Scholar]
- Schaid DJ. General score tests for associations of genetic markers with disease using cases and their parents. Genet Epidemiol. 1996;13:423–449. doi: 10.1002/(SICI)1098-2272(1996)13:5<423::AID-GEPI1>3.0.CO;2-3. URL http://dx.doi.org/gt;3.0.CO;2–3. [DOI] [PubMed] [Google Scholar]
- Vansteelandt S, Demeo DL, Lasky-Su J, Smoller JW, Murphy AJ, McQueen M, Schneiter K, Celedon JC, Weiss ST, Silverman EK, Lange C. Testing and estimating gene-environment interactions in family-based association studies. Biometrics. 2008;64:458–467. doi: 10.1111/j.1541-0420.2007.00925.x. URL http://dx.doi.org/10.1111/j.1541-0420. 2007.00925.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material