Abstract
Replica molding with elastomeric polymers has been used routinely to replicate features less than 10 nm in size. Because the theoretical limit of this technique is set by polymer-surface interactions, atomic radii and accessible volumes, replication at sub-nm length scales should be possible. Using PDMS to create a mold and polyurethane to form the replica, we demonstrate replication of elementary steps 3-5 Å in height that define the minimum separation between molecular layers in the lattices of the ionic crystals potassium dihydrogen phosphate (KDP) and calcite (CaCO3). This work establishes the operation of replica molding at the molecular scale.
Replication of features above 10 nm by replica molding using elastomeric polymers has become routine.1, 2 Replication of sub-10 nm features is still a significant challenge because, at this length-scale, the feature size approaches that of the monomers used for replication. The theoretical limits to replica molding are set by the granularity of matter, and by the intermolecular interactions that determine the ability of molecular surfaces to come into conformal contact.3 In particular, whether a polymer can take on the curvature required to conform to molecular-scale variations in topography should depend on the interfacial energy of the polymer-surface contact. While there are some examples of replication below 10 nm,4-8 replication of molecular-scale features has not been achieved. Here we take advantage of the regular arrays of single molecular-height steps on the faces of ionic crystals to demonstrate that the very low interfacial free energy, flexibility and resistance to contamination of h-PDMS (polydimethylsiloxane) enables replica molding with sub-nanometer resolution.
Replica molding is the transfer of a topographical pattern from a “master” substrate into a polymer or other material to form the inverse mold, and the subsequent fabrication of a replica by solidifying a liquid precursor against the inverse mold.2 Due to its ease of application to a wide variety of materials, replica molding has been pursued as a general approach to repeatable production of nanostructured surfaces starting from a single, high-precision master.1, 2 Successful replication of features below 10 nm using elastomeric polymers, such as PDMS has been demonstrated for a number of materials.4-9 The lower limit on the size of features to which replica molding can be applied is, however, ultimately determined by atomic radii, molecular shapes, van der Waals interactions, and thermal and entropic effects,3 and remains largely unexplored. Because the step heights of ionic crystals are smaller than both the monomers and the radius of gyration of the elastomeric polymers used for replication, prior to this study, it was not obvious whether the molding process would be capable of replicating the atomic steps of the crystal surface. However, to aid in visualizing how large polymer molecules might replicate much smaller features, the polymers can be thought of as spherical or ellipsoidal particles bunched around a small step. As long as their spatial configuration is maintained after removal of the step, they will have replicated the underlying step structure, albeit with a distortion related to their specific size and interactions.
Thus, successful investigation in this lower limit requires surfaces with regular, well-defined features at the molecular scale. Ionic crystal surfaces present elementary steps with typical heights of ~ 0.3-0.8 nm, that provide a convenient and reproducible master with molecular-sized features that can be used to assess the performance of replica molding at this scale. The angstrom scale architectures achieved by the replication in this study can be significant to the nanotechnology industry seeking more stringent, scalable nano-fabrication methods because they show that new limits on the feature size accessible through stamp-based techniques can now be pursued. This work has implications for imprint lithography techniques, which are now on the semiconductor road map due to their ability to pattern high-resolution features at a significantly lower cost than next-generation photolithographic processes.
In this study, we implemented siloxane formulations that have been optimized previously for high resolution molding based on their viscosity, modulus, and surface hardness.10 The composite structure for replica molding comprising a thin (<100 μm) h-PDMS10 film formed in direct contact with single crystal surfaces, and backed by a significantly thicker (~3 mm) layer of “normal”-PDMS (n-PDMS). This design was chosen for three reasons. First, the large elastic modulus (~ 9 N mm-2) of h-PDMS10 allowed replication of shallow relief features with higher fidelity than is possible using n-PDMS; the high compressibility of n-PDMS often leads to deformation, buckling or collapse of sub-100 nm features formed on its surface.11 Second, the very low interfacial free energy of h-PDMS, and the flexibility of the composite structure, enables replication without any prior modification to the crystal or polymer surfaces.11 Third, h-PDMS is inert to ambient air, particles, and vapors; this lack of contamination reduces adhesion to both the crystal and the polyurethane (PU) replica formed from it, and thus eases release during replication.
Crystal surfaces used in this study included the {100} face of KDP (KH2PO4) and the {104} face of calcite (CaCO3), for which the elementary step heights are 0.37 and 0.31 nm respectively.12, 13 We first used {100} KDP vicinal faces (Figure 1), because they present a variety of step sizes from elementary steps of 0.37-nm height, to macrosteps comprising closely bunched elementary steps and ranging in height from several nanometers to microns.13 We also used the {104} faces of calcite crystals because they consist of regular, continuous arrays of 0.31 nm high elementary steps.12 Figure 1B and C are models of {100} KDP and {104} calcite and Figure 1D and E are AFM images of steps on these two crystal faces.
Figure 1.
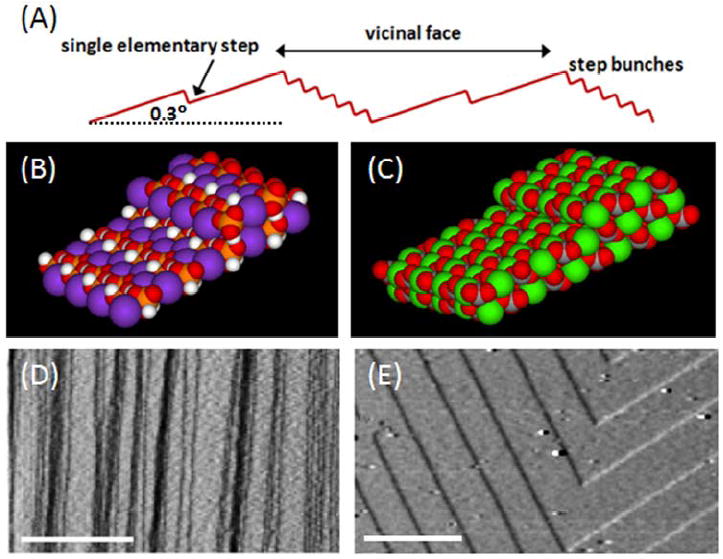
(A) Schematic of a vicinal crystal surface used as a master for replication. It consists of macrosteps, which are bunched elementary steps, separated by atomically flat terraces and elementary steps. On both KDP and calcite surfaces, the macrosteps can vary from a few to hundreds of elementary steps. The slope of the vicinal surface relative to the crystallographically defined terraces depends on growth conditions and the nature of the step source. In this study, crystals had nominal vicinal slopes of 0.1° and 0.3° for calcite and KDP, respectively. (B) Space filling model of the (100) surface of KDP with a single elementary step. The height of a single elementary step is 0.37 nm. The purple spheres are potassium; white spheres are hydrogen; red spheres are oxygen; and the orange spheres are phosphate. (C) Space filling model of the (104) surface of calcite with a single elementary step. The height of a single elementary step is 0.31 nm. The green spheres are calcium; red spheres are oxygen; and the buried grey spheres are carbon. Note that the models of KDP and calcite assume the steps are a simple truncation of the surface layers, which is consistent with atomically resolved images of both crystals. (D) and (E) AFM deflection images of steps showing (D) the arrays of macrosteps and elementary steps on KDP {100} and (E) the regular array of elementary steps on calcite {104}. Scale bars are (D) 2 μm and (E) 500 nm. The step heights on both crystals surfaces are one-half the unit cell heights. (For analysis of macrostep and elementary step heights on KDP {100} see Fig. S1 in the Supplementary Information (SI).)
Figure 2A illustrates the procedure used to replicate crystal surfaces. Figure 2B is a schematic showing the KDP growth habit; Figure 2C and D are photographs of the original KDP crystal and the PU replica generated from it. The surfaces of the crystals were sufficiently mechanically robust that the polymer replicas were easily separated from the crystal without loss of replica fidelity. This ease of separation arises from the low surface energy of h-PDMS (γ = 22-24 × 10-3 J/m2 14, 15) (Compare this, for example, to KDP{100} for which γ = 470 × 10-3 J/m2 16).
Figure 2.
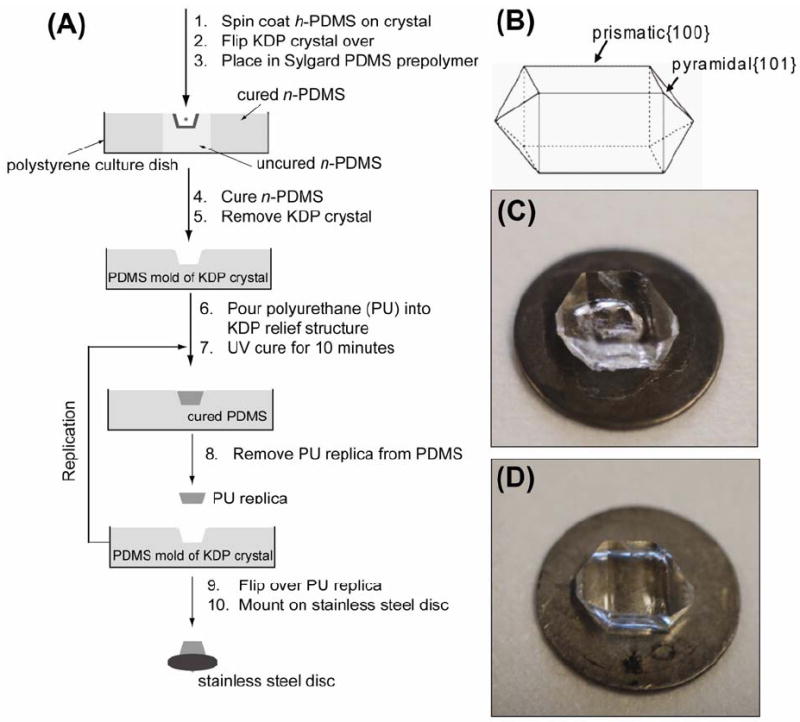
(A) Schematic of the replication procedure of a crystal surface on the {100} and {101} oriented surfaces of KDP using a PDMS mold and PU replica. (B) Schematic of macroscopic KDP crystal showing location of the {100} and {101} faces. The elementary and macro-steps replicated on the KDP surface are located on the {100} face. (C) Photograph of the original KDP crystal supported on a stainless steel disk (15-mm diameter). The hole seen in the image is on the underside of the crystal and represents the original location of the seed crystal used during crystal growth. (D) A PU replica of the original crystal in (C) supported on a stainless steel disk. The hole of the seed crystal is now missing from the PU replica because the {100} and {101} surfaces are replicated in part (A). (Only the {100} surface was subsequently imaged due to geometric constraints.) The same procedure was used to replicate the surface of calcite{104}.
After being used to mold PDMS, AFM imaging of the crystal surfaces demonstrated no apparent damage, and the replicated steps could be easily re-imaged. Figure 3A and B are comparisons of AFM images of the original KDP {100} and calcite {104} crystal surfaces with their corresponding PU replicas, along with AFM height profiles line-scans and roughness measurements for each type of surface. Single elementary steps on both KDP{100} and calcite {104} surfaces were well replicated as indicated in the height profile plots. AFM images in Figure 3A and B of the elastomeric replica show three elementary steps located on a ~2-μm long terrace between two macrosteps on KDP, and two elementary steps on calcite over ~3-μm. For KDP, vertical dimensions and line-scans of the replicated macrosteps and of the inverse h-PDMS replica used as a mold ranged from ~ 2-40 nm (~ 5-110 elementary steps) (see SI Fig. S2). For calcite {104}, under the conditions used in this study, surfaces grew on rhombohedral-shaped dislocation hillocks composed of a continuous array of elementary steps that formed a “spiral staircase” generated by a screw dislocation (Figure 4).12 These hillocks have two distinct step types that grow at different speeds, and result in two corresponding terrace widths17 (Figure 4). Our AFM measurements gave a step height on the crystal in fluid of 0.33 ± 0.05 nm (n=10), which, within experimental variation, is equal to the accepted value of 0.31 nm.12 Figure 4B and C show that, just as with KDP, these growth hillocks can be successfully replicated down to the level of elementary steps. The measured heights of replicated steps were 0.37 ± 0.12 nm (n=14), which is also equal to the true step height to within experimental variation.
Figure 3.
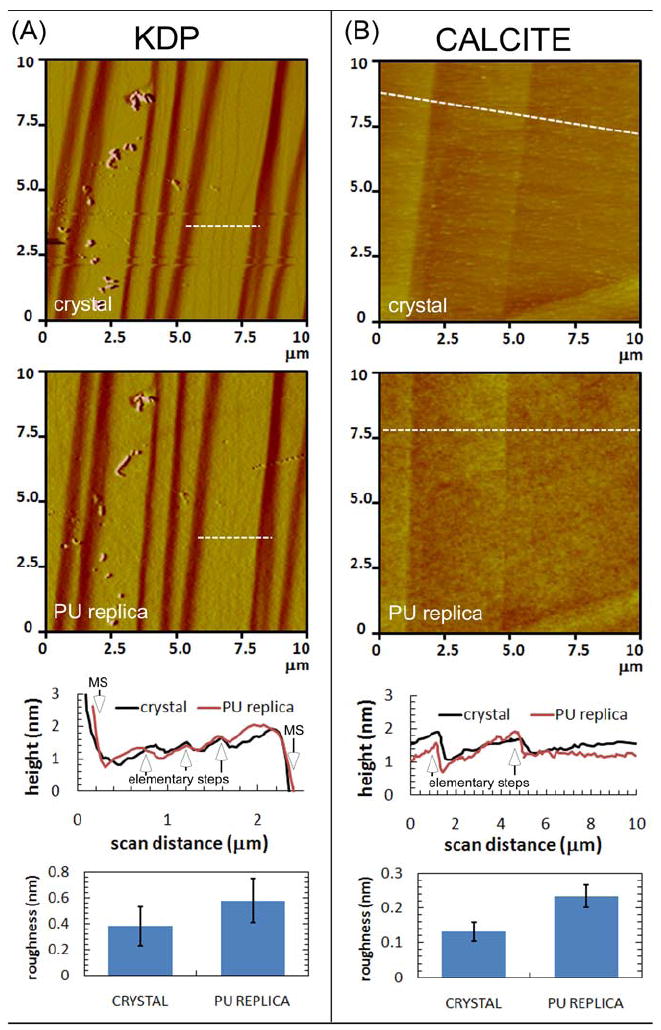
(A) AFM images, height profiles, and root-mean-squared (rms) surface roughness measured by AFM for KDP and (B) calcite crystal and PU replica surfaces. In AFM images, the white dashed lines indicate the location of the line-scans reported in the corresponding height profile plots where the elementary steps and macrosteps (MS) are indicated. RMS roughness values were obtained from 5 μm AFM scans with error bars indicating ± one standard deviation (n=10).
Figure 4.
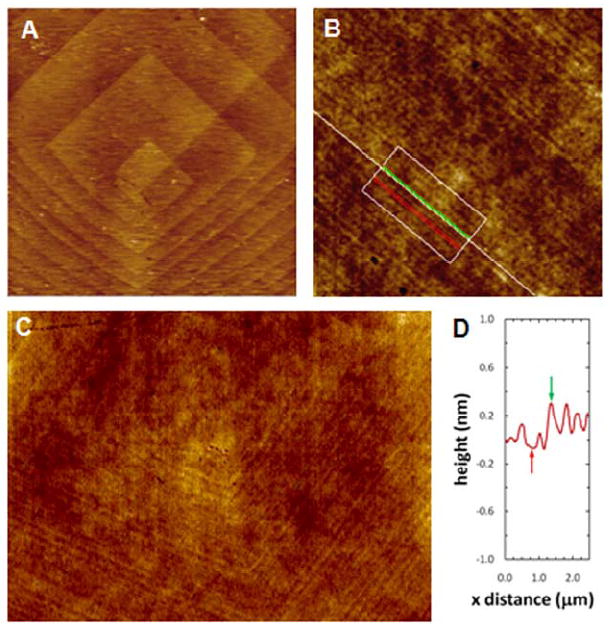
AFM images and height profile of a calcite crystal and replica. AFM height images (A-C) and height profile (D) of a dislocation hillock on (A) calcite {104} face and (B-C) PU replica of the same crystal face. Height profile in (D) was taken within the box and perpendicular to the lines in (B). The average measured height of steps on the PU replica was 0.37±0.12 nm, which is equal to the elementary step height on calcite{104} (½ the unit cell or 0.31 nm) to within experimental error. Image sizes are (A) 10 × 10 μm, (B) 12 × 12 μm, (C) 20 × 14 μm.
The measured step heights on the crystal and replica were indistinguishable to within the experimental variation. Using the known dimensions of elementary steps KDP {100}13 as a vertical calibration standard, we measured the height of steps on the crystal and PU replica to be 0.37±0.09 nm (n=10) and 0.38 ± 0.07nm (n=13), respectively, indicating no significant shrinkage of the polymer. (Polymer shrinkage upon curing varies among polymers and depends on a number of experimental parameters, however PDMS generally undergoes very mild shrinkage of ~1-2 %15 and the PU adhesive also has a low shrinkage of 1.5%18.)
The polymer physics that governs the replication process at sub 5-nm resolution is poorly understood.19 The ultimate resolution of the replica is controlled by local granularity of the “master” at the atomic scale, impurities in the master or polymer, reconstruction of the polymer surface due to attractive/repulsive interactions with the surface to be replicated, differences in adhesion across the surface of the master due, for example, to wetting/dewetting phenomena, and changes in polymer dynamics, both in the polymerization and melt stages, due to proximity to the master surface. All of these factors depend critically on properties of the polymers such as the cross-link density of both h-PDMS and n-PDMS.7
The work presented here shows that polymers with monomers having an average radius of ~ 1 nm, average bond lengths of 0.2 nm, and average distance between cross-links of ~ 1 nm7 are able to replicate vertical features on a solid substrate with dimensions significantly smaller than the average monomer size. However, this length scale is also below the lateral resolution of AFM imaging and, therefore, the convolution of the shape of the tip with the topography of the surface becomes a key factor in evaluating the fidelity of this process and the interpretation of the results. In particular, the large difference between the lateral and vertical resolution of AFM renders features such as atomic steps, which are only of order 0.1 nm in height but many 10’s or 100’s of nanometers in length, clearly visible even when variations along the step due to the atomic-scale corrugation of the surface are on par with the step height itself.20 The tips used in this study have a nominal radius of 2 nm. Consequently, provided lateral variations in surface height have a characteristic dimension below this value, extended features are easily imaged as continuous structures.
The importance of long-range continuity in feature replication is further demonstrated by considering the surface roughness of the replicas. The average rms roughness of the h-PDMS replica of KDP and calcite as measured by AFM imaging was 0.6 and 0.2 nm, respectively, over a 5 μm scale on terraces between single atomic steps, while the rms roughness of the crystal over the same area was about one-half of these values (see Figure 3A and B). Given that the rms roughness of the replica is comparable to the step height of 0.37 nm, clearly only features that extend over significantly greater lengths can be recognized in the replica. The roughness, however, should depend on many of the same factors that determine the lower limit of replication, particularly the polymer-surface interactions, which affect both the interfacial energy and the degree of polymer adhesion. Indeed, previous work on h-PDMS demonstrated that the average rms roughness of ~ 0.35 nm for replication of features on test-grade, polished silicon wafers was dependent upon polymer formulation.7, 19 Moreover, the average roughness for a composite PDMS replica of a flat Si/SiO2 wafer having an rms roughness of 0.13 ± 0.03 nm was only 0.23 ± 0.05 nm,8 which is an increase of ~100% but remains smaller than the lattice spacing of most crystals. Consequently, while results presented here show that atomic scale features on crystals can be replicated, the comparable dimensions of rms replica roughness and large AFM probe radius limit replication of such features to those that extend laterally over length scales significantly longer than either the probe size or the characteristic lateral dimension of the roughness.
Results presented here demonstrate that elastomer-based replica molding is capable of providing information about molecular-scale features on crystal surfaces. A comparison of the feature height to both the tip radius and the surface roughness of the replicas indicated, however, that the extended character of these surface features is a key element in the fidelity of the replication process. A reduction in surface roughness, either through variations in polymer formulation or by choice of a crystal surface that provides both low interfacial energy and weak polymer adhesion would be required before replication of features with lateral dimensions at the molecular level can be addressed. Consequently, whether the structure of crystal surfaces or features with lateral dimensions of atomic scale can be replicated at true atomic scale remains an open question.
Supplementary Material
Acknowledgments
We thank Dr. Raymond Friddle for his assistance in preparing calcite crystals for this study. This research was supported by NIH (GM065364), and by DARPA (subaward to GMW from the Center for Optofluidic Integration at the California Institute of Technology). The research used MRSEC and NSEC facilities supported by NSF (DMR-0213805 and PHY-0117795) and at the Center for Nanoscale Systems (CNS: NSE ECS-0335765). R. M. R. acknowledges NIH for a postdoctoral fellowship (1 F32 NS060356). Crystal fabrication and AFM analysis were supported by Office of Science, Office of Basic Energy Sciences of the U.S. Department of Energy by Lawrence Livermore National Laboratory under Contract DE-AC52-07NA27344 and by the Molecular Foundry, Lawrence Berkeley National Laboratory under Contract No. DE-AC02-05CH11231.
Footnotes
Supporting Information Available. Replication Methods, AFM images of crystal surfaces and replica with surface roughness measurements. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Gates BD, Xu QB, Love JC, Wolfe DB, Whitesides GM. Annu Rev Mater Res. 2004;34:339–372. [Google Scholar]
- 2.Gates BD, Xu QB, Stewart M, Ryan D, Willson CG, Whitesides GM. Chem Rev. 2005;105(4):1171–1196. doi: 10.1021/cr030076o. [DOI] [PubMed] [Google Scholar]
- 3.The ultimate practical limit to replica molding has not been established. It should be below that accessible by photolithography, and is thus relevant to future nanomanufacturing. It may also be relevant to a number of areas of molecular chemistry.
- 4.Deng ZX, Mao CD. Angew Chem Int Ed. 2004;43(31):4068–4070. doi: 10.1002/anie.200460257. [DOI] [PubMed] [Google Scholar]
- 5.Gabai R, Ismach A, Joselevich E. Adv Mat. 2007;19(10):1325–1330. [Google Scholar]
- 6.Gates BD, Whitesides GM. J Am Chem Soc. 2003;125(49):14986–14987. doi: 10.1021/ja0367647. [DOI] [PubMed] [Google Scholar]
- 7.Hua F, Sun YG, Gaur A, Meitl MA, Bilhaut L, Rotkina L, Wang JF, Geil P, Shim M, Rogers JA, Shim A. Nano Lett. 2004;4(12):2467–2471. [Google Scholar]
- 8.Xu QB, Mayers BT, Lahav M, Vezenov DV, Whitesides GM. J Am Chem Soc. 2005;127(3):854–855. doi: 10.1021/ja043436d. [DOI] [PubMed] [Google Scholar]
- 9.Lin RS, Rogers JA. Nano Lett. 2007;7(6):1613–1621. doi: 10.1021/nl070559y. [DOI] [PubMed] [Google Scholar]
- 10.Schmid H, Michel B. Macromolecules. 2000;33(8):3042–3049. [Google Scholar]
- 11.Odom TW, Love JC, Wolfe DB, Paul KE, Whitesides GM. Langmuir. 2002;18(13):5314–5320. [Google Scholar]
- 12.Teng HH, Dove PM, Orme CA, De Yoreo JJ. Science. 1998;282(5389):724–727. doi: 10.1126/science.282.5389.724. [DOI] [PubMed] [Google Scholar]
- 13.Thomas TN, Land TA, Martin T, Casey WH, DeYoreo JJ. J Cryst Growth. 2004;260(3-4):566–579. [Google Scholar]
- 14.Chaudhury MK, Whitesides GM. Langmuir. 1991;7(5):1013–1025. [Google Scholar]
- 15.Choi KM, Rogers JA. J Am Chem Soc. 2003;125(14):4060–4061. doi: 10.1021/ja029973k. [DOI] [PubMed] [Google Scholar]
- 16.Stack AG, Rustad JR, DeYoreo JJ, Land TA, Casey WH. J Phys Chem B. 2004;108(47):18284–18290. [Google Scholar]
- 17.Teng HH, Dove PM, DeYoreo JJ. Geochim Cosmo Acta. 1999;63(17):2507–2512. [Google Scholar]
- 18.http://www.norlandprod.com/
- 19.Hua F, Gaur A, Sun YG, Word M, Jin N, Adesida I, Shim M, Shim A, Rogers JA. IEEE Trans Nan. 2006;5(3):301–308. [Google Scholar]
- 20.Friddle RW, Weaver ML, Qiu SR, Wierzbicki A, Casey WH, De Yoreo JJ. Proc Natl Acad Sci USA. 2010;107(1):11–15. doi: 10.1073/pnas.0908205107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zaitseva NP, DeYoreo JJ, Dehaven MR, Vital RL, Montgomery KE, Richardson M, Atherton LJ. J Cryst Growth. 1997;180(2):255–262. [Google Scholar]
- 22.The imaging of the KDP crystal and h-PDMS replica surfaces by AFM were conducted using as little force on the substrate as possible. To achieve this, we: 1) used a soft cantilever, 2) minimized setpoint deflection (i.e., force) enough to maintain imaging contact with the surface, and 3) scanned slow enough that the piezo-PID controller could track the surface and correct the position of the tip quick enough to prevent the tip from plowing into the surface. We performed the imaging in repulsive contact mode (meaning the tip and substrate were pushing against each other). The force exerted by the AFM tip on the surface is ~0.05 nN.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


