Abstract
STAT5 and IL-7 signaling are thought to control B-lymphopoiesis by regulating key transcription factor genes and activating VH gene segments at the Igh locus. Using conditional mutagenesis, we demonstrate that transgenic Bcl2 expression rescued the development of Stat5-deleted pro-B cells by compensating for the loss of Mcl-1. Ebf1 and Pax5 expression as well as VH gene recombination were normal in Bcl2-rescued pro-B cells lacking STAT5 or IL-7Rα. In agreement with this finding, STAT5-expressing pro-B cells contained little or no active chromatin at most VH genes. In contrast, Igk rearrangements were increased in STAT5-or IL-7Rα-deficient pro-B cells. Hence, STAT5 and IL-7 signaling control cell survival and the developmental ordering of immunoglobulin gene rearrangements by suppressing premature Igk recombination in pro-B cells.
Keywords: STAT5, IL-7 signaling, cell survival, Pax5, Ebf1, immunoglobulin gene rearrangements, chromatin activation
Introduction
Interleukin-7 (IL-7) is a nonredundant cytokine for T and B cell development1 and transmits its signal through the IL-7 receptor (IL-7R) consisting of the common γ chain (γc) chain and the IL-7Rα subunit (encoded by the Il7r gene). T-lymphopoiesis is strongly reduced in the thymus, and B-cell development is stringently arrested at the uncommitted pre-pro-B cell stage in the bone marrow of Il7r−/− mice2, 3. Transgenic expression of the anti-apoptotic Bcl-2 protein efficiently rescues T-lymphopoiesis4, 5, but not B-cell development in Il7r−/− mice5, 6, which indicates an important role of IL-7R signaling in controlling T-cell survival and early B-lymphopoiesis.
IL-7R signaling stimulates the Jak-STAT pathway leading to activation of the transcription factor STAT57. STAT5 consists of two highly related isoforms, STAT5A and STAT5B, which are encoded by separate genes and fulfill largely redundant roles within the lymphoid system7. Different strategies have been used for simultaneous inactivation of the two closely linked Stat5a and Stat5b genes. Teglund et al.8 disrupted the first protein-coding exons of Stat5a and Stat5b, which still results in expression of N-terminally truncated and partially functional STAT5 proteins (Stat5ΔN allele)9, 10. Consequently, T and B cell development is minimally affected in Stat5ΔN/ΔN mice11, 12. The subsequent generation of a conditional Stat5 allele (Stat5fl) containing Stat5a and Stat5b between loxP sites facilitated Cre-mediated deletion of both genes (Stat5− allele)13. Complete Stat5 inactivation causes perinatal lethality, severely reduced development of B and αβ T cells as well as the absence of γδ T-lymphocytes in Stat5−/− mice9, 10. Similar to Il7r−/− mice3, B-cell development is arrested at the uncommitted pre-pro-B cell stage in the bone marrow of Stat5−/− mice9, 10.
An instructive role of STAT5 in B-lymphopoiesis was suggested based on the partial rescue of B-cell development in Il7r−/− mice by a transgene expressing a constitutively active form of STAT5B14. STAT5 has furthermore been implicated in the regulation of the B-cell commitment gene Pax5 by binding to its promoter region15, 16. STAT5-mediated IL-7 signaling is also thought to activate expression of the B-cell specification factor EBF1 in common lymphoid progenitors (CLPs) and pre-pro-B cells3, 17, 18. Finally, IL-7 is sufficient to promote in vitro differentiation of wild-type CLPs to committed pro-B cells2. These data together with the inability of transgenic Bcl-2 expression to rescue B-lymphopoiesis in Il7r−/− mice5, 6 suggest that IL-7 signaling fulfills an instructive role in early B-cell development by STAT5-mediated activation of the B-cell regulatory genes Ebf1 and Pax5.
STAT5 and IL-7R signaling have also been implicated in controlling the chromatin accessibility of the immunoglobulin heavy-chain (Igh) gene in pro-B cells. The Igh locus consists of a 2.5 megabase (Mb) cluster of 195 variable (V) gene segments and a proximal 0.27 Mb region containing the diversity (D), joining (J) and constant (C) gene segments19. At the onset of B-cell development, the Igh locus is thought to undergo stepwise chromatin activation, whereby IL-7 signaling is reportedly responsible for activation of the large distal VHJ558 gene family20, 21. Consistent with this model, germline transcription and recombination of distal VH genes are strongly impaired, whereas proximal VH genes undergo normal germline transcription and rearrangements in B-cell progenitors of Il7r−/− and Stat5ΔN/ΔN mice22, 23. Hence, these data further support an instructive role for STAT5 and IL-7R signaling in early B-cell development.
Here we have investigated the cell intrinsic role of STAT5 in B-lymphopoiesis by conditional mutagenesis in adult mice. The B-cell developmental effect of STAT5 deficiency was previously studied in the fetal liver of Stat5−/− embryos or bone marrow of the rare growth-retarded Stat5−/− survivors9, 10, whose hematopoietic stem cells (HSCs) were severely impaired in their function9. As the Rag1 gene is not expressed in HSCs, but is subsequently activated in early lymphoid progenitors (ELPs)24, we used a Rag1cre knock-in allele25 for Cre-mediated deletion of Stat5 specifically at the onset of lymphopoiesis. Contrary to previous reports, our conditional mutagenesis data demonstrate that STAT5 and IL-7R signaling are not involved in the regulation of Ebf1 and Pax5 or in the chromatin activation and VH-DJH recombination of distal VHJ558 genes. Instead, the primary role of IL-7-mediated STAT5 activation during pro-B cell development is to maintain cell survival by activating the anti-apoptotic gene Mcl1 and to prevent premature Igk gene rearrangements by binding to the intronic iEκ enhancer.
RESULTS
STAT5 promotes pro-B cell development by controlling cell survival
To determine the activity of the Rag1cre allele in early hematopoiesis, we used a Cre reporter gene26 to show that Rag1-Cre-mediated deletion was initiated in 18% of multipotent progenitors (LSK cells) and was completed in all bone marrow pro-B cells and thymic pro-T (DN1/2) cells (Supplementary Fig. 1 online). As the Rag1cre allele deletes the floxed Stat5 allele not only in B-cells but also in all T-cells, Rag1cre/+ Stat5fl/fl mice became autoimmune within 2 weeks after birth and died at the age of 2-3 months due to autoimmune pathologies that are likely caused by the dramatic reduction of Treg cell numbers27 (Supplementary Fig. 2). Consequently, we first examined the B-cell phenotype caused by conditional Stat5 inactivation under non-autoimmune conditions. Fetal liver pro-B cells (c-Kit+CD19+IgM−) and pre-B cells (CD2+CD19+IgM−) were reduced 10-15 fold in Rag1cre/+ Stat5fl/fl compared to Stat5fl/fl embryos at day 18.5 (Supplementary Fig. 3a). Moreover, the bone marrow of T-cell-deficient Rag1cre/cre Stat5fl/fl mice contained similar amounts of CLPs, but 7-fold reduced numbers of pro-B cells compared to Stat5fl/fl mice, indicating that the loss of STAT5 interferes with the differentiation of CLPs to committed pro-B cells (Supplementary Fig. 3b).
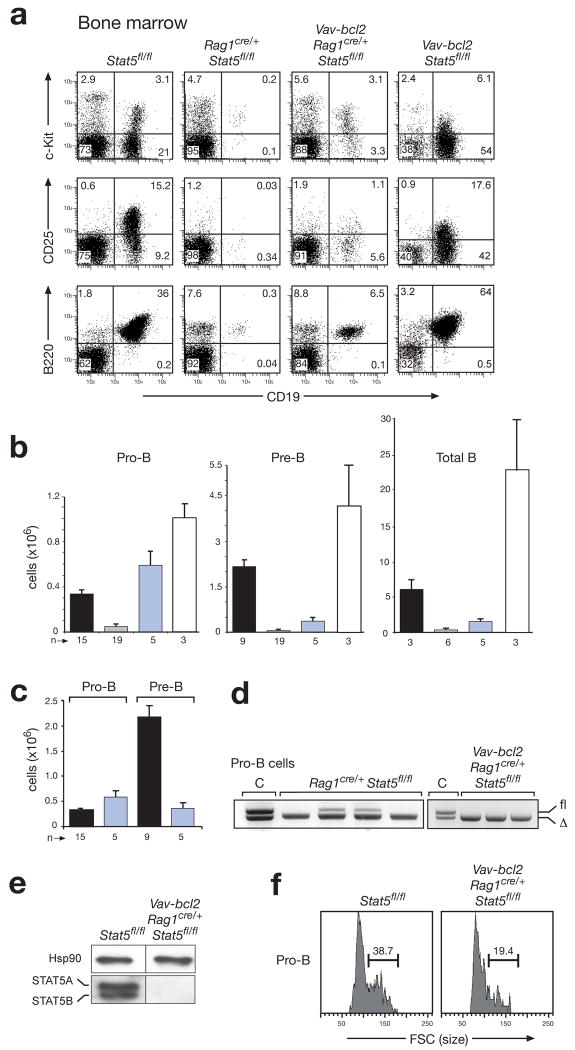
Pro-B, pre-B and total B cell numbers were similarly reduced in the bone marrow of Rag1cre/+ Stat5fl/fl mice compared to Stat5fl/fl mice (Fig. 1a-c). Deletion of the floxed Stat5 allele in the few residual pro-B cells of Rag1cre/+ Stat5fl/fl mice indicated that STAT5 is not absolutely required for pro-B cell development (Fig. 1d). Importantly, transgenic Bcl-2 expression efficiently rescued pro-B cell development (Fig. 1a,b) despite complete Stat5 deletion and STAT5 protein loss (Fig. 1d,e) in Vav-bcl2 Rag1cre/+ Stat5fl/fl mice. Hence, the pro-survival protein Bcl-2 can compensate for STAT5 deficiency in pro-B cell development. In contrast, pre-B cells and total B-cells were only minimally increased in Vav-bcl2 Rag1cre/+ Stat5fl/fl mice (Fig. 1a,b). Interestingly, no cell expansion was observed during the development of pro-B to pre-B cells in Vav-bcl2 Rag1cre/+ Stat5fl/fl mice (Fig. 1c) consistent with a decrease of larger, blasting pro-B cells (Fig. 1e). Hence, STAT5 is an essential mediator of the collaboration between IL-7 and pre-BCR signaling28 during this developmental transition.
Figure 1. Rescue of STAT5-deficient pro-B cells by transgenic Bcl-2 expression.
(a-c) Restoration of pro-B cell development by compensation of the STAT5 deficiency with transgenic Bcl-2 expression. Flow cytometric analysis (a) and cell counting were used to determine the absolute cell numbers (b,c) of pro-B (c-Kit+CD19+IgM−), pre-B (CD25+CD19+IgM−) and total B cells (B220+CD19+) in the bone marrow of Rag1cre/+ Stat5fl/fl (grey bar) and Vav-bcl2 Rag1cre/+ Stat5fl/fl mice (blue bar) compared to control Stat5fl/fl (black bar) and Vav-bcl2 Stat5fl/fl mice (white bar) at the age of 4-6 weeks. Bone marrow was isolated from the femur and tibia of the two hind legs. n, number of mice analyzed. (c) Reanalysis of the absolute numbers of pro-B and pre-B cells (b) revealed the absence of proliferative cell expansion during the pro-B to pre-B cell transition in Vav-bcl2 Rag1cre/+ Stat5fl/fl mice compared to control Stat5fl/fl mice. (d) PCR genotyping of the floxed (fl) and deleted (Δ) Stat5 alleles in sorted pro-B cells from the bone marrow of 4 Rag1cre/+ Stat5fl/fl and 3 Vav-bcl2 Rag1cre/+ Stat5fl/fl mice. Genomic Stat5fl/Δ DNA was used as control (C). (e) Immunoblot analysis of sorted CD19+ bone marrow B cells of the indicated genotypes with a mixture of STAT5A and STAT5B antibodies or a control Hsp90 antibody. (f) Size distribution of pro-B cells of the indicated genotypes, as determined by their forward scatter (FSC) profile.
The Bcl-2-mediated rescue of STAT5-deficient pro-B cells begged the question whether the STAT5 protein was already lost at the preceding pre-pro-B cell stage in Vav-bcl2 Rag1cre/+ Stat5fl/fl mice. Indeed, the floxed Stat5 allele was efficiently deleted, and the Stat5a mRNA was ~20-fold reduced in sorted pre-pro-B cells (Kit+B220+CD19−DX5−Ly6C−) of Vav-bcl2 Rag1cre/+ Stat5fl/fl compared to control mice (Supplementary Fig. 4). Moreover, intracellular staining of STAT5 protein revealed the same background fluorescence for pre-pro-B and pro-B cells of Vav-bcl2 Rag1cre/+ Stat5fl/fl mice, indicating that the pre-pro-B cells contained little or no STAT5 protein similar to the pro-B cells of these mice (Supplementary Fig. 5).
We next investigated whether the transcription factor EBF1 could rescue the STAT5 deficiency, as suggested by published data3, 17, 18. An Ikzf1Ebf1 transgene, that expresses Ebf1 under the control of the pan-hematopoietic Ikzf1 (Ikaros) locus (Supplementary Fig. 6a-d), rescued pro-B cell development in Ebf1−/− mice (Supplementary Fig. 6e), but could only partially restore the generation of Stat5-deficient pro-B cells in Rag1cre/+ Stat5fl/fl Ikzf1Ebf1 mice (Supplementary Fig. 7). Together, these data demonstrate that STAT5 fulfills a permissive rather than instructive role in pro-B cell development.
Normal development of mature B-cell types in the absence of STAT5
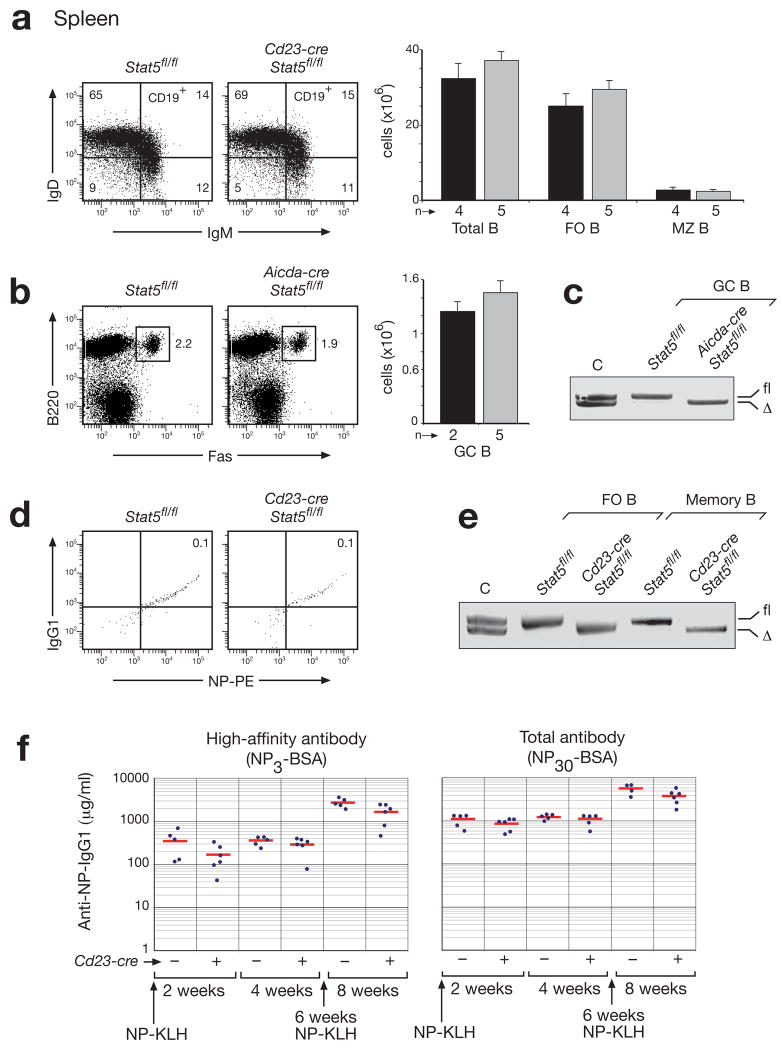
STAT5 was shown by gain-of-function experiments to control the differentiation and self-renewal potential of human memory B-cells29. To verify these findings by loss-of-function analysis in the mouse, we used the Cd23-cre and Aicda-cre lines26 to inactivate the floxed Stat5 allele in splenic B cells. Despite efficient Stat5 deletion, the mature (IgDhiIgMloCD19+), follicular (FO; CD21intCD23hi) and marginal zone (MZ; CD21hiCD23lo) B-cells were equally abundant in Cd23-cre Stat5fl/fl compared to control Stat5fl/fl mice, (Fig. 2a,e). Likewise, similar numbers of germinal center (GC) B-cells (B220+Fas+) were generated in the spleen of immunized Aicda-cre Stat5fl/fl and control Stat5fl/fl mice (Fig. 2b,c). NP-specific IgG1+ memory B-cells (Lin−CD38hiB220+CD19+IgG1+NP+) with complete Stat5 inactivation were also found in the spleen of Cd23-cre Stat5fl/fl mice eight weeks after immunization with NP-KLH (Fig. 2d,e and Supplementary Fig. 8). Finally, similar titers of anti-NP-IgG1 antibodies were detected in the sera of Cd23-cre Stat5fl/fl and control Stat5fl/fl mice 2 and 4 weeks after NP-KLH immunization as well as 2 weeks after NP-KLH boosting of the memory B-cell response (Fig. 2f). Together, these data indicate that STAT5 is dispensable for late B-lymphopoiesis, memory B-cell formation and plasma cell differentiation.
Figure 2. Normal development and immune responses of mature B cells in the absence of STAT5.
(a) Normal development of mature (IgDhiIgMloCD19+), follicular (FO; CD21intCD23hi) and marginal zone (MZ; CD21hiCD23lo) B cells in the spleen of nonimmunized Cd23-cre Stat5fl/fl mice (grey bar) compared to control Stat5fl/fl mice (black bar) at the age of 8-10 weeks. Note that the Cd23-cre line 5 initiates Cre-mediated gene deletion in immature B cells and results in complete deletion in MZ and FO B cells of the spleen26. Absolute numbers of the different B cell types and total B cells (CD19+B220+) are shown to the right together with the number (n) of mice analyzed. (b) Normal GC B cell formation in Aicda-cre Stat5fl/fl mice. The abundance of GC B cells (Fas+B220+) in control Stat5fl/fl (back bar) and Aicda-cre Stat5fl/fl (grey bar) mice was determined by flow cytometric analysis 15 days after immunization with sheep red blood cells. n, number of mice analyzed. (c) Efficient deletion of Stat5 in sorted GC B cells of Aicda-cre Stat5fl/fl mice 15 days after immunization. Genomic Stat5fl/Δ DNA was used as control (C). (d) Presence of memory B cells in the spleen of Cd23-cre Stat5fl/fl 8 weeks after the initial immunization with NP-KLH (100 μg in Alum) and 2 weeks after boosting with NP-KLH (10 μg in PBS). Memory B cells were identified as Lin−CD38hiB220+CD19+IgG1+NP+ cells as shown in Supplementary Fig. 8. (e) PCR genotyping of the floxed (fl) and deleted (Δ) Stat5 alleles in sorted FO and memory B cells from mice of the indicated genotypes. (f) NP-specific immune responses. Control Stat5fl/fl (−) and Cd23-cre Stat5fl/fl (+) mice were immunized with NP-KLH (100 μg in Alum) and boosted with NP-KLH (10 μg in PBS) 6 weeks later. After 2, 4 and 8 weeks, the serum titers of NP-specific antibodies were determined by ELISA using NP3-BSA- or NP30-BSA-coated plates for detecting high-affinity or total IgG1 antibodies, respectively. NP-specific IgG1 concentrations (μg/ml) were determined relative to a standard NP-binding IgG1 antibody.
Development of committed pro-B cells in the absence of IL-7 signaling
As STAT5 is a downstream mediator of IL-7 signaling7, we next determined the role of the IL-7R in controlling pro-B cell development. Consistent with published reports2, 3, the pro-B, pre-B and total B cells were drastically reduced in the bone marrow of adult Il7r−/− mice (Supplementary Fig. 9a). Importantly, the bone marrow from the two hind legs (femur and tibia) of 4-6-week-old Il7r−/− mice contained on average 5,000 committed c-Kit+CD19+IgM− pro-B cells, indicating that IL-7R signaling is not strictly required for the formation of committed pro-B cells (Supplementary Fig. 9a). Transgenic Bcl-2 expression resulted in a 5-10-fold increase of pro-B, pre-B and total B cells in Vav-bcl2 Il7r−/− mice (Supplementary Fig. 9a). Moreover, the majority of the Bcl-2-rescued Il7r−/− pro-B cells were small in size and thus failed to proliferate (Supplementary Fig. 9b). Hence, the development of committed pro-B cells was only partially rescued by transgenic Bcl-2 expression in Vav-bcl2 Il7r−/− mice in contrast to its efficient rescue in Vav-bcl2 Rag1cre/+ Stat5fl/fl mice.
STAT5- and IL-7R-dependent gene regulation in pro-B cells
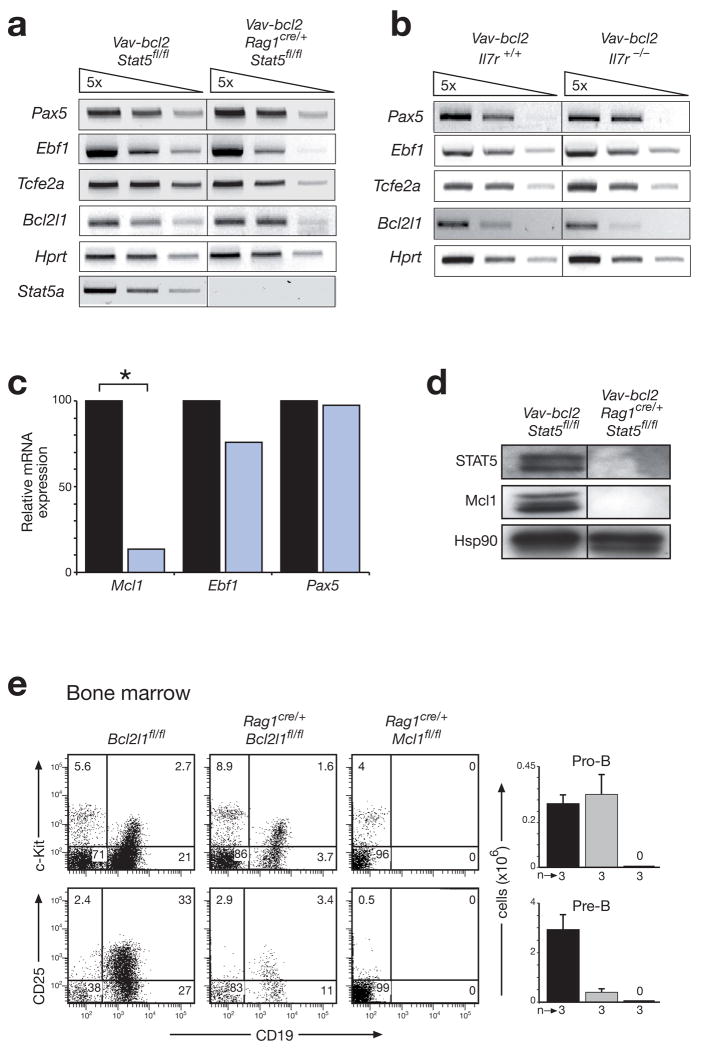
The Bcl-2-mediated rescue of STAT5- and IL-7Rα-deficient pro-B cells provided the unique opportunity to FACS-sort sufficient pro-B cells (Supplementary Fig. 10) for investigating the transcriptional role of STAT5 and IL-7R signaling in early B-cell development. Semiquantitative RT-PCR analysis demonstrated that the transcription factor genes Pax5, Ebf1 and Tcfe2a (E2A) were similarly expressed in sorted bone marrow pro-B cells of Vav-bcl2 Rag1cre/+ Stat5fl/fl and control Vav-bcl2 Stat5fl/fl mice (Fig. 3a,c). Similar results were obtained with bone marrow pro-B cells of Vav-bcl2 Il7r−/− and Vav-bcl2 Il7r+/+ mice (Fig. 3b) as well as with the rare pro-B cells isolated from the fetal liver of Stat5−/−embryos at day 18.5 (Supplementary Fig. 11). As all three transcription factors are essential for early B-lymphopoiesis, their normal expression in Stat5 and Il7r mutant pro-B cells further supports a permissive rather than instructive role for STAT5 and IL-7R signaling in pro-B cell development.
Figure 3. STAT5 controls the survival of pro-B cells by activating the Mcl1 gene.
(a,b) Normal expression of B cell transcription factor genes in the absence of STAT5 (a) or IL-7R signaling (b). Expression of the indicated genes was determined by semiquantitative RT-PCR analysis using 5-fold serial dilutions of cDNA prepared from FACS-sorted pro-B cells (Supplementary Fig. 10) of 4-6-week-old mice of the indicated genotypes. Tcfe2a (E2A); Bcl2l1 (Bcl-x). (c) Reduced Mcl1 expression in STAT5-deficient pro-B cells. Transcripts of the indicated genes were analyzed by real-time RT-PCR of cDNA prepared from sorted pro-B cells of control Vav-bcl2 Stat5fl/fl (black bar) and Vav-bcl2 Rag1cre/+ Stat5fl/fl (blue bar) mice. The transcript levels of control pro-B cells were set to 100%. Statistical significance was determined by the student’s t test (*, p < 0.05). (d) Loss of Mcl-1 expression in the absence of STAT5. Whole cell extracts of sorted CD19+ bone marrow cells from mice of the indicated genotypes were analyzed by immunoblot analysis with STAT5, Mcl-1 and Hsp90 antibodies. (e) Distinct functions of Mcl1 and Bcl2l1 in early B cell development. Pro-B and pre-B cells were analyzed by flow cytometric analysis in the bone marrow of control Bcl2l1fl/fl (black bar), Rag1cre/+ Bcl2l1fl/fl (grey bar) and Rag1cre/+ Mcl1fl/fl mice (denoted by 0). Absolute cell numbers are shown to the right together with the number (n) of mice analyzed. A more comprehensive FACS analysis is shown in Supplementary Fig. 12.
Expression of the pro-survival gene Bcl2l1 (encoding Bcl-xL) was unaffected by the loss of STAT5 or IL-7Rα (Fig. 3a,b). In contrast, transcripts of the Mcl1 gene were 8-fold reduced and the Mcl-1 protein was undetectable in pro-B cells of Vav-bcl2 Rag1cre/+ Stat5fl/fl mice compared to Vav-bcl2 Stat5fl/fl mice (Fig. 3c,d), suggesting that STAT5 mediates the survival of pro-B cells by activating the Mcl1 gene. As pro-B cells are still generated in the bone marrow of Cd19-cre Mcl1fl/fl mouse30, we examine the role of Mcl1 in Rag1cre/+ Mcl1fl/fl mice. These mice entirely failed to produce pro-B, pre-B and B cells in their bone marrow (Fig. 3e and Supplementary Fig. 12). In contrast, pro-B cells were normally generated in Rag1cre/+ Bcl2l1fl/fl mice despite efficient Bcl2l1 deletion, whereas pre-B cells were strongly reduced, thus indicating a critical function for Bcl-xL in pre-B cells and later stages of B-cell development (Fig. 3e and Supplementary Fig. 12). Together these data demonstrate that STAT5 controls the survival of pro-B cells by regulating the Mcl1 but not the Bcl2l1 gene.
Normal Igh recombination in pro-B cells lacking STAT5 or IL-7Rα
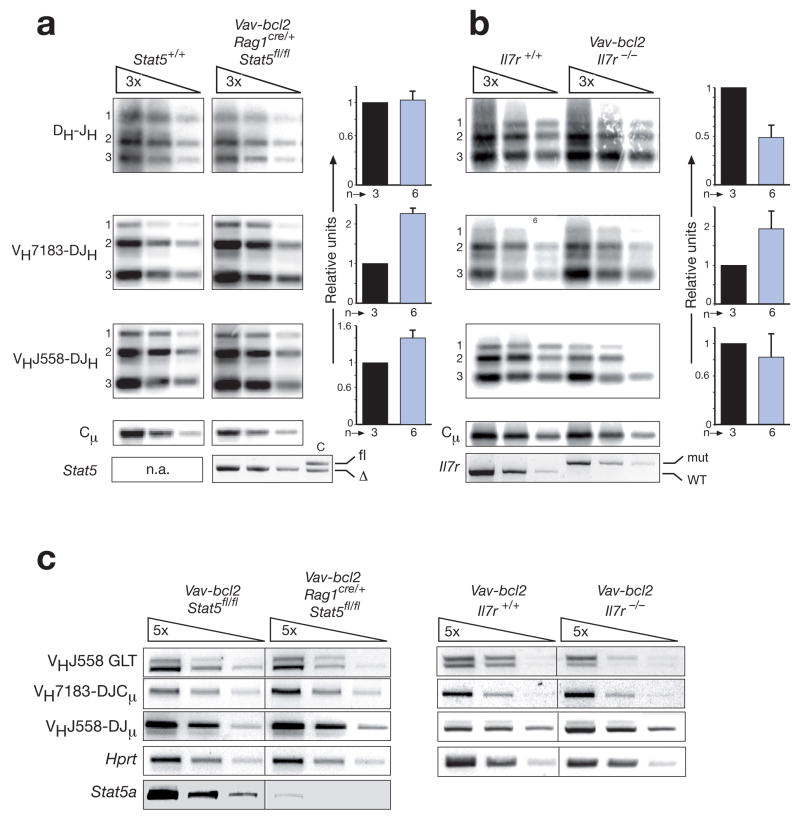
We next investigated the role of STAT5 and IL-7R signaling in controlling distal VH-DJH recombination of the Igh locus by semiquantitative PCR detection and quantification of the different Igh rearrangements in FACS-sorted pro-B cells (c-Kit+CD19+IgM−) from Vav-bcl2 Rag1cre/+ Stat5fl/fl, Vav-bcl2 Il7r−/−, control wild-type and Vav-bcl2 mice (Fig. 4a,b and Supplementary Fig. 13). As expected20, 22, 23, we observed no or only a minor (~2-fold) difference in the frequency of DH-JH and proximal VH7183-DJH rearrangements in Vav-bcl2 Rag1cre/+ Stat5fl/fl and Vav-bcl2 Il7r−/− pro-B cells compared to wild-type and Vav-bcl2 pro-B cells (Fig. 4a,b and Supplementary Fig. 13). Surprisingly however, we could not detect a significant difference in the frequency of distal VHJ558-DJH rearrangements between STAT5-deficient, Il7r mutant and control pro-B cells (Fig. 4a,b and Supplementary Fig. 13). RT-PCR analysis confirmed this result as rearranged VH7183-DJCμ and VHJ558-DJμ transcripts were expressed at similar levels in Vav-bcl2 Rag1cre/+ Stat5fl/fl and Vav-bcl2 Il7r−/− pro-B cells compared to control Vav-bcl2 pro-B cells (Fig. 4c). Moreover, germline transcripts (GLT) of the distal VHJ558 genes were also expressed in pro-B cells of the different genotypes (Fig. 4c). In contrast, Bertolino et al.23 reported a 5-fold reduction of distal VHJ558-DJH rearrangements in B220+IgM− bone marrow B-lymphocytes (pro-B and pre-B cells) of Stat5ΔN/ΔN mice8 and a 10-fold reduction of VHJ558 germline transcripts in cultured Stat5ΔN/ΔN pro-B cells compared to wild-type control cells. However, our own analysis of ex vivo sorted pro-B cells from the bone marrow of Stat5ΔN/ΔN mice failed to demonstrate any effect of the N-terminally truncated (ΔN) STAT5A and STAT5 proteins on VHJ558 germline transcription, VHJ558-DJH recombination and expression of rearranged VHJ558-DJμ transcripts (Supplementary Fig. 14; see legend for detailed explanation of the discrepancy). We therefore conclude that neither STAT5 nor IL-7R signaling play a critically important role in controlling VH-DJH recombination at the Igh locus.
Figure 4. Normal Igh gene recombination in pro-B cells lacking STAT5 or IL-7Rα.
(a) PCR analysis of DH-JH, VH7183-DJH and VHJ558-DJH rearrangements in c-Kit+CD19+IgM− pro-B cells, which were FACS-sorted from the bone marrow of control Stat5+/+ (black bar) and Vav-bcl2 Rag1cre/+ Stat5fl/fl mice (blue bar) at the age of 4-6 weeks as described in Supplementary Fig. 10. Input DNA was normalized by PCR amplification of an Igh Cμ fragment. Threefold serial DNA dilutions were analyzed by semiquantitative PCR and Southern blotting. The radioactive signals of each rearrangement type were quantified for each DNA dilution by phosphorimager analysis and normalized relative to the corresponding value of the Cμ PCR fragment. For each experiment, the average of the normalized signals obtained with the three DNA dilutions was determined, and the average value of all experiments is displayed as a bar graph for each rearrangement type to the right. The average values obtained with wild-type (Stat5+/+) pro-B cells were set to 1, and the number (n) of independent experiments is shown. Numbers along the left margin indicate rearrangements to the JH1, JH2 and JH3 gene segments. n.a., not applicable. (b) PCR analysis of Igh rearrangements in sorted pro-B cells from 4-6-week-old Il7r+/+ (black bar) and Vav-bcl2 Il7r−/− mice (blue bar) was performed and evaluated as described in (a). (c) RT-PCR analysis. Expression of the VHJ558 germline transcripts (GLT) as well as the rearranged VH7183-DJCμ and VHJ558-DJμ transcripts was analyzed by semiquantitative RT-PCR analysis using 5-fold serial dilutions of cDNA prepared from FACS-sorted pro-B cells of the indicated genotypes.
Absence of detectable active chromatin at most VH genes in Rag2−/− pro-B cells
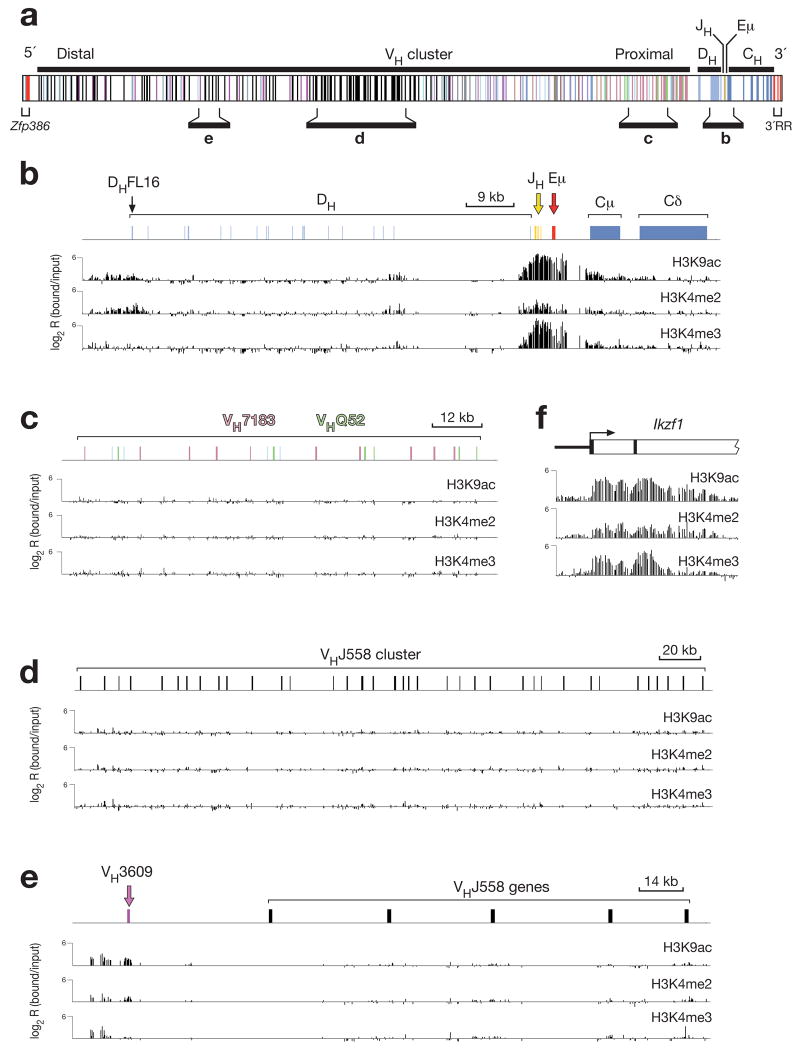
STAT5 and IL-7 signaling have previously been implicated in controlling the accessibility of distal VH genes to the recombination machinery by promoting histone acetylation20, 21, 23. We therefore studied this aspect of STAT5 and IL-7 signaling by determining whether the distal VH genes contain active chromatin in IL-7-treated pro-B cells. As chromatin activation of the distal VH genes is thought to precede V(D)J recombination20, 21, we used Rag2−/− pro-B cells for high-resolution ChIP-chip mapping of active histone modifications along the Igh locus. To this end, bone marrow pro-B cells from Rag2−/− mice were expanded for only 5 days in the presence of IL-7 and OP9 stromal cells prior to chromatin immunoprecipitation (ChIP) with antibodies that recognized acetylated lysine 9 (H3K9ac), dimethylated lysine 4 (H3K4me2) or trimethylated lysine 4 (H3K4me3) on histone H3. The precipitated DNA was subjected to either T7-based linear amplification31 or whole genome amplification (WGA)32 prior to fluorescent labeling and hybridization together with the input DNA probe onto a high-density 50-mer oligonucleotide array containing the non-repetitive sequences of the Igh locus (Fig. 5a). Probes prepared with the two different protocols gave rise to similar results as shown in Fig. 5 (linear amplification) and Supplementary Fig. 15 (WGA amplification). The three active histone marks H3K9ac, H3K4me2 and H3K4me3 were strongly enriched in a region centered on the intronic Eμ enhancer and extending from the DQ52 element to the Cμ gene (Fig. 5b and Supplementary Fig. 15b). Active histone modifications were absent throughout the central DH region and at all proximal VH7183 and VHQ52 genes in Rag2−/− pro-B cells (Fig. 5b,c and Supplementary Fig. 15b,c), as published20, 21, 33. Surprisingly however, we detected no or only very low enrichment of active histone marks at distal VHJ558 genes despite the fact that the Rag2−/− pro-B cells had been stimulated with IL-7 for 5 days (Fig. 5d and Supplementary Fig. 15d). In contrast, the distal VH3609 genes, which are interspersed with the VHJ558 genes, carried relatively high levels of active H3K9ac and H3K4me2 marks (Fig. 5e and Supplementary Fig. 15e), although the H3K4me3 modification was barely detectable in contrast to its abundance at promoters of other expressed B-cell-specific genes31 such as the Ikzf1 gene (Fig. 5f and Supplementary Fig. 15f).
Figure 5. Mapping of active histone modifications along the Igh locus in Rag2−/− pro-B cells.
(a) Structure of the Igh locus indicating the location of variable (VH), diversity (DH), joining (JH) and constant (CH) gene segments together with the intronic (Eμ) enhancer, the 5’ DNase I hypersensitive sites (red) and 3’ regulatory region (3’RR; red). Different colors indicate the members of distinct VH gene families, of which the VHJ558 (black), VH3609 (pink), VHQ52 (green) and VH7183 (rosa) genes are relevant for this study. The diagram of the Igh locus is based on the mm8 genome sequence and the published VH gene assembly19 of the C57BL/6 mouse. (b-f) Mapping of active chromatin marks in the DH-Cμ domain (b) and representative proximal (c) and distal (d,e) VH gene regions of the Igh locus as well as at the 5’ end of the Ikzf1 (Ikaros) gene (f) in Rag2−/− pro-B cells. Antibodies recognizing acetylated lysine 9 (H3K9ac), dimethylated lysine 4 (H3K4me2) or trimethylated lysine 4 (H3K4me3) on histone H3 were used for ChIP-chip analysis31 of Rag2−/− bone marrow pro-B cells, which were expanded in vitro for 5 days in the presence of IL-7 and OP9 stromal cells. The ChIP-precipitated DNA was amplified with the T7-based linear amplification protocol31 prior to fluorescent labeling and co-hybridization with the input DNA probe onto a high-density 50-mer oligonucleotide array containing the mouse Igh locus at 100-bp resolution (produced by NimbleGen Systems). The logarithmic ratio (log2) of the hybridization intensities between antibody-precipitated and input DNA (bound/input) is shown as a bar for each oligonucleotide on the microarray. The results shown are representative of two ChIP-chip experiments, which were performed with independently prepared DNA probes from Rag2−/− pro-B cells. A scale bar is shown in kb. Supplementary Fig. 15 shows the result of a similar ChIP-chip analysis, which was performed with DNA probes amplified with the WGA method32. The mm8 sequence coordinates for the displayed regions of the Igh locus on chromosome 12 are 116,520,000-113,586,000 (a), 113,938,000-113,852,500 (b), 114,234,000-114,086,000 (c), 115,441,500-115,087,000 (d) and 115,859,000-115,780,000 (e).
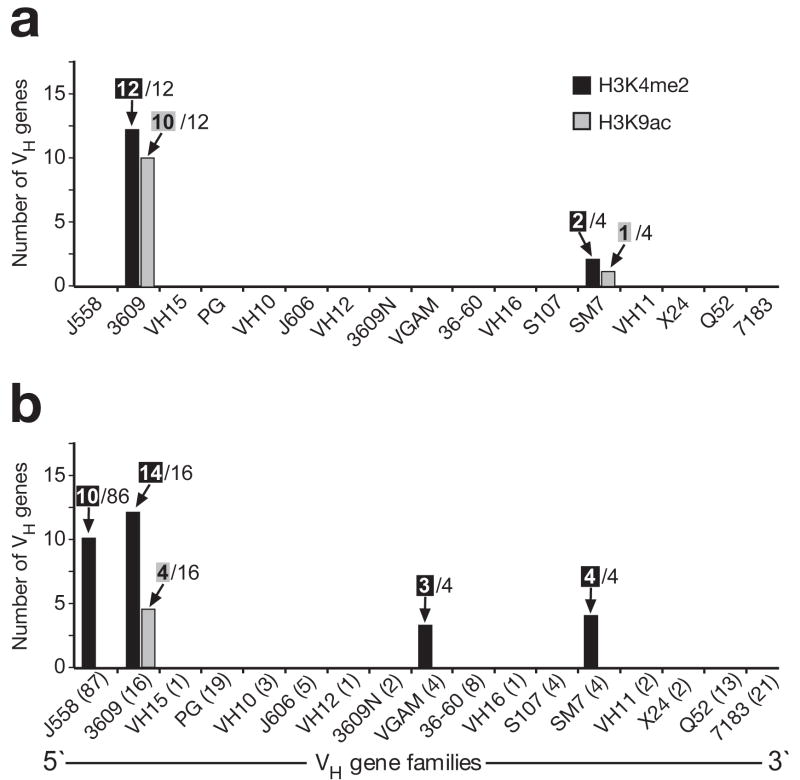
Systematic peak-finder analysis demonstrated that Rag2−/− pro-B cells carried active H3K9ac marks at 10 VH3609 genes and H3K4me2 modifications at all 12 VH3609 genes present on the microarray hybridized with linearly amplified probes (Fig. 6a). Except for 2 VHSM7 genes, no significant peaks of active histone modifications could be detected at all other VH genes including the large VHJ558 gene family (Fig. 6a). A similar picture was obtained with the microarray hybridized with WGA-amplified probes. The H3K4me2 marks were present at 14 of 16 VH3609 genes, all 4 VHSM7 genes, 3 VHGAM genes and only at 10 of 86 VHJ558 genes, whereas significant peaks of H3K9ac could be detected only at 4 VH3609 genes (Fig. 6b). In summary, we conclude that IL-7-stimulated Rag2−/− pro-B cells lack active histone modifications at most VH genes including the large VHJ558 gene family with the notable exception of the VH3609 and VHSM7 genes. Importantly, ChIP-chip analysis also revealed a similar chromatin profile of the Igh locus in wild-type pro-B cells undergoing Igh rearrangements (Supplementary Fig. 16). Hence, the VH gene cluster of the rearranging Igh locus is characterized by a distinct chromatin structure that differs from that of other expressed B-cell-specific genes.
Figure 6. Distribution of active histone marks along the VH gene cluster in Rag2−/− pro-B cells.
Peak-finder analysis was used to systematically evaluate the ChIP-chip data, which were obtained with linearly amplified (a) or WGA-amplified (b) probes as shown in Fig. 5 and Supplementary Fig. 15, respectively. The number of VH genes, which contained a peak of H3K4me2 (black bar) or H3K9ac (gray bar) above the threshold of the peak-finder analysis, are shown together with the number of all genes of each VH gene family that were present on the microarray. The VH gene families are arranged according to their position in the Igh locus. The number of all annotated genes is shown in brackets (b) for each VH gene family of the C57BL/6 mouse19.
Increased Igk recombination in pro-B cells lacking STAT5 or IL-7Rα
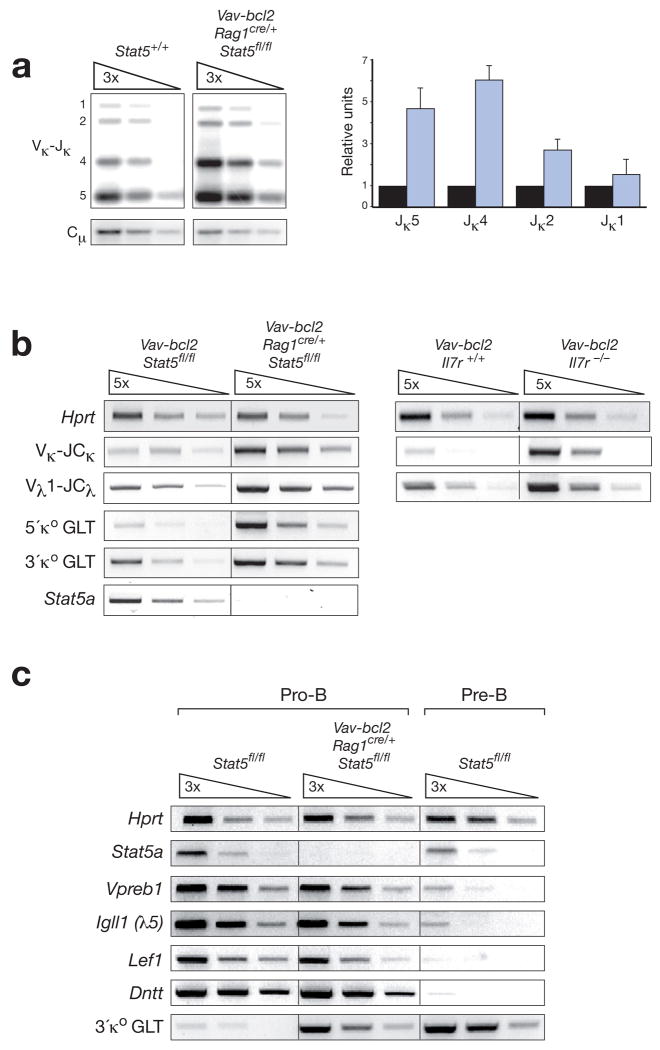
As pro-B cells undergo differentiation and Igk recombination upon IL-7 withdrawal34, 35, we tested the hypothesis that STAT5 and IL-7R signaling may repress Igk gene rearrangements at the pro-B cell stage. PCR analysis and quantification of Igk rearrangements revealed that Vκ-Jκ recombination involving the downstream Jκ4 and Jκ5 elements was 5-6-fold increased in FACS-sorted pro-B cells from Vav-bcl2 Rag1cre/+ Stat5fl/fl mice compared to wild-type mice (Fig. 7a). Increased Igk recombination likely resulted in secondary rearrangements, which could explain the preferential usage of downstream Jκ elements in Bcl-2-rescued STAT5-deficient pro-B cells (Fig. 7a). Consistent with enhanced Igk recombination, RT-PCR analysis revealed a ~10-fold increase of rearranged Vκ-JCκ transcripts in sorted pro-B cells of Vav-bcl2 Rag1cre/+ Stat5fl/fl mice compared to control Vav-bcl2 pro-B cells (Fig. 7b). The proximal and distal κo germline transcripts (GLT) were also 5- to 20-fold induced in STAT5-deficient pro-B cells (Fig. 7b) and reached the expression level of wild-type pre-B cells (Fig. 7c). In addition to the Vκ-JCκ transcripts, rearranged Vλ1-JCλ mRNA were strongly increased in pro-B cells of Vav-bcl2 Rag1cre/+ Stat5fl/fl as well as Vav-bcl2 Il7r−/− mice (Fig. 7b), indicating that Igk and Igl rearrangements or transcription were similarly activated in Bcl-2-rescued pro-B cells lacking STAT5 or IL-7Rα. Importantly, the pro-B cells of Vav-bcl2 Rag1cre/+ Stat5fl/fl mice expressed the ‘pro-B cell-specific’ genes Vpreb1, Igll1 (λ5), Dntt (TdT) and Lef1 (Fig. 7c) and exhibited the same cell surface phenotype (c-Kit+CD19+CD25−CD2−MHCII−) as wild-type pro-B cells (Supplementary Fig. 17). Hence, the STAT5-deficient pro-B cells did not acquire pre-B cell characteristics other than increased IgL gene recombination and expression, indicating that STAT5 deficiency alone does not promote the progression from pro-B to pre-B cells. Moreover, ChIP analysis of IL-7-stimulated wild-type pro-B cells demonstrated that STAT5 specifically interacted with the 3’ region of the intronic iEκ enhancer among all Igk regulatory elements tested (Supplementary Fig. 18) in agreement with recently published data obtained with Irf4−/− Irf8−/− pre-B cells36. In summary, our loss-of-function data identified a critical role for STAT5 and IL-7R signaling in suppressing premature Igk and Igl recombination in pro-B cells.
Figure 7. STAT5 and IL-7 signaling repress Igk and Igl gene rearrangements in pro-B cells.
(a) Increased Igk gene rearrangements in pro-B cells sorted from Vav-bcl2 Rag1cre/+ Stat5fl/fl mice (blue bar) compared to wild-type mice (black bar). Vκ gene rearrangements to the Jκ1 to Jκ5 gene segments (indicated by numbers along the left margin) were analyzed by semiquantitative PCR of 3-fold serially diluted DNA followed by Southern blot detection. The radioactive signals corresponding to these rearrangements were quantified by phosphorimager analysis, normalized to the value of the control Cμ PCR fragment and displayed in the bar graph to the right. The normalized value obtained for each Vκ-Jκ rearrangement in wild-type pro-B cells was set to 1. The quantification of 3 independent experiments is shown. (b) RT-PCR analysis of Igk transcripts. The expression of rearranged Vκ-JCκ and Vλ1-JCλ transcripts as well as the two κo germline transcripts (GLT) were analyzed by semiquantitative RT-PCR using 5-fold serial dilutions of cDNA prepared from FACS-sorted pro-B cells of the indicated genotypes. The 5’ and 3’ κo GLTs are transcribed from distinct promoters (see Supplementary Fig. 18a) and give rise to 1.1-kb and 0.8-kb spliced transcripts, respectively. (c) Expression of pro-B cell-specific genes in Bcl-2-rescued STAT5-deficient pro-B cells. FACS-sorted pro-B cells (c-Kit+CD19+CD25−IgM−) and pre-B cells (CD19+CD25+c-Kit−IgM−) of control Stat5+/+ or Vav-bcl2 Rag1cre/+ Stat5fl/fl mice were analyzed by semiquantitative RT-PCR for expression of the ‘pro-B cell-specific’ genes Vpreb1, Igll1 (λ5), Lef1 and Dntt (TdT)48 and the ‘pre-B cell-specific’ 3’ κo GLT.
DISCUSSION
STAT5 and IL-7 signaling have been considered to control early B-cell development in an instructive manner by regulating the B-cell-specific transcription factor genes Ebf1 and Pax5 (ref. 3, 16, 17) and by controlling distal VH-DJH recombination at the Igh locus20-23. Here we have reached a different conclusion, as conditional mutagenesis and transgenic Bcl-2 rescue experiments revealed a critical role for STAT5 and IL-7 signaling in controlling cell survival and repressing premature Igk recombination during pro-B cell development.
Cell survival control is known to be a primary function of IL-7R signaling in T-cell development4, 5. In contrast, the development of pro-B cells (defined as CD43+HSA+B220+IgM− cells) is apparently not rescued by transgenic Bcl-2 expression in Il7r−/− mice6, contributing to the notion of instructive IL-7R signaling in pro-B cell development. We observed, however, a partial rescue of committed pro-B cells (defined as c-Kit+CD19+IgM− cells) in Vav-bcl2 Il7r−/− mice, indicating that Bcl-2 expression can substitute for the survival, but not the proliferation function of IL-7R signaling in early B-lymphopoiesis. Moreover, transgenic Bcl-2 expression efficiently rescued pro-B cell development in Vav-bcl2 Rag1cre/+ Stat5fl/fl mice, demonstrating that STAT5 mediates the survival function of IL-7 signaling at the onset of B-lymphopoiesis.
The pro-B to pre-B cell transition requires pre-BCR signaling, which also modulates IL-7 responsiveness via an ERK/MAP kinase-dependent pathway to facilitate selective cell expansion under low IL-7 conditions28, 37. The absence of this cell expansion in Vav-bcl2 Rag1cre/+ Stat5fl/fl mice suggests that IL-7R signaling critically depends on STAT5 for its collaboration with the pre-BCR. STAT5 may thereby synergize with pre-BCR signaling by promoting the proliferation and/or survival of pre-BCR+ cells. STAT5 has also been implicated in late B-cell development, as retroviral expression of a constitutive active form of STAT5B induces the proliferation and differentiation of human tonsillar B-cells to memory B-cells29. Contrary to these in vitro gain-of-function studies, our conditional Stat5 inactivation experiments demonstrate that STAT5 is not essential for the in vivo generation of functional memory B-cells or other mature B cell types.
STAT5 directly regulates the pro-survival gene Bcl2l1 (Bcl-xL) in erythroid cells in response to erythropoietin signaling38 and in myeloid cells upon GM-CSF stimulation39. Bcl2l1 is, however, not an activated STAT5 target gene in early B-cell development, as it is normally expressed in Bcl-2-rescued STAT5- and IL-7Rα-deficient pro-B cells. Notably, the Bcl-xL protein is more highly expressed in pre-B cells than in pro-B cells40. Accordingly, pre-B cells but not pro-B cells were lost upon conditional inactivation of Bcl2l1, which demonstrates a critical survival function for Bcl-xL in pre-B cells as suggested by previous analysis of Bcl2l1−/− mice41. In contrast, the pro-survival protein Mcl-1 was no longer expressed in Bcl-2-rescued STAT5-deficient pro-B cells, and its conditional loss entirely abrogates pro-B cell development, indicating that STAT5 mediates its survival function by activating the Mcl1 gene in pro-B cells.
STAT5 and IL-7 signaling have also been implicated in the regulation of the B-cell-specific transcription factor genes Ebf1 and Pax5. Ebf1 can be induced in Il7−/− pre-pro-B cells by IL-7 stimulation3 and in BaF/3 progenitor cells by expression of constitutive active STAT5A18, suggesting that IL-7 signaling induces B-cell specification in pre-pro-B cells by activating the Ebf1 gene. However, retroviral expression of Ebf1 in Il7r−/− HSCs leads to a minimal rescue of B-cell development3, and STAT5 regulates only indirectly the activity of the Ebf1 promoter18. Our Bcl-2 rescue experiments demonstrate, however, that neither IL-7R signaling nor STAT5 are essential for normal Ebf1 transcription in bone marrow pro-B cells. STAT5 is also thought to regulate Pax5 by binding to sequences upstream of its distal promoter15, 16. However like Ebf1, Pax5 was also normally expressed in Stat5−/− fetal liver pro-B cells as well as in Bcl-2-rescued bone marrow pro-B cells lacking STAT5 or IL-7Rα, consistent with the fact that we could not detect STAT5 binding to any regulatory element of the Pax5 locus42.
STAT5 and IL-7 signaling have also been associated with V(D)J recombination of the Tcrg and Igh loci. Il7r−/− mice and Stat5−/− embryos are unable to generate γδ T cells due to a severe defect in Vγ-Jγ recombination9, 10, as STAT5 induces chromatin accessibility and germline transcription at the Tcrg locus by direct binding to the Jγ promoters43. A similar role has been suggested for STAT5 and IL-7R signaling in controlling the chromatin accessibility and rearrangements of distal VH genes at the Igh locus20, 22, 23. In contrast to these published data, we could not detect any significant difference in distal VHJ558-DJH rearrangements in Bcl-2-rescued Il7r−/− and Stat5Δ/Δ pro-B cells compared to control cells. In an attempt to explain these discrepancies, we note that Corcoran et al.22 detected similar numbers of pro-B cells in the bone marrow of wild-type and Il7r−/− mice in marked contrast to our study and other reports2, 3. The CD43+B220+ ‘pro-B cells’ analyzed by Corcoran et al.22 were likely a mixture of pro-B cells, recirculating CD43+ B1 cells, NK cell progenitors and plasmacytoid dendritic cells44, 45, which could explain the apparent decrease of distal VHJ558-DJH recombination in this impure cell population isolated from Il7r−/− mice. As Bertolino et al.23 also observed decreased germline transcripts and rearrangements of distal VHJ558 genes in B220+IgM− B-lymphocytes (pro-B and pre-B cells) of Stat5ΔN/ΔN mice, we repeated these experiments to realize that VHJ558 germline transcription and recombination is normal in Stat5ΔN/ΔN pro-B cells similar to Bcl-2-rescued STAT5-deficient pro-B cells. Finally, Chowdhury and Sen20 described a role for IL-7 signaling in controlling the chromatin accessibility of distal VH genes in Rag2−/− pro-B cells. Our extensive ChIP-chip analyses revealed, however, no or only very low levels of the active histone modifications H3K4me2 and H3K9ac at most VH genes in IL-7-stimulated Rag2−/− pro-B cells. Active chromatin was, however, consistently present at VH3609 and VHSM7 genes. As wild-type pro-B cells exhibited a similar H3K4me2 and H3K9ac modification pattern throughout the VH gene cluster as Rag2−/− pro-B cells, we conclude that germline transcription and VH-DJH recombination can proceed in the absence of overt chromatin activation of VH genes. As active chromatin at VH genes can currently only be analyzed at the cell population level, our data are also compatible with the remote possibility that each individual pro-B cell is able to induce active chromatin only at a small random subset of VH genes, which would escape detection in the entire pro-B cell population. In contrast to the VH genes, the three active marks H3K9ac, H3K4me2 and H3K4me3 accumulate to high levels at the intronic Eμ enhancer and JH elements, which suggests the following sequence of epigenetic events during Igh recombination. The V(D)J recombinase consisting of RAG1 and RAG2 is likely tethered to the JH elements through specific binding of H3K4me3 by the C-terminal PHD finger of RAG2 (ref. 46) and may act from this proximal location to undergo synapse formation with DH and VH elements. Following DH-JH recombination in B-lymphoid progenitors, the rearranged DH element is incorporated into the active chromatin domain at the JH-Eμ region, which facilitates subsequent VH-DJH recombination upon juxtaposition of VH genes by Pax5-dependent contraction of the Igh locus in committed pro-B cells47. This model could explain why active chromatin is asymmetrically distributed at the proximal JH region but not throughout the VH gene cluster in pro-B cells.
Under physiological conditions, the expression of IL-7Rα is down-regulated in response to pre-BCR signaling, which renders small pre-B cells unresponsive to IL-7 signals, leading to the loss of STAT5 activation37, 48. Moreover, pro-B cells upon IL-7 withdrawal initiate pre-B cell differentiation and Igk rearrangements34, 35. Here we have shown that the loss of STAT5 and IL-7 signaling is sufficient to increase Igk germline transcription and rearrangements in Bcl-2-rescued pro-B cells in the absence of further differentiation. As Igk rearrangements can be detected in only 15% of wild-type pro-B cells49, the observed 5-6 fold increase in recombination frequency predicts that almost all Bcl-2-rescued pro-B cells undergo Igk recombination in the absence of STAT5. Hence, an important function of STAT5 and IL-7 signaling is the suppression of premature Igk rearrangements in pro-B cells. STAT5 directly binds to the intronic iEκ enhancer in wild-type pro-B cells (this study) and Irf4−/− Irf8−/− pre-B cells36, where it likely represses the recombination-activating function of the iEκ enhancer50. Importantly, our genetic in vivo analysis demonstrates a key role for STAT5 and IL-7 signaling in suppressing Igk germline transcription as well as recombination in pro-B cells and thus provides strong complementary evidence to a recently published in vitro study36, which describes an inverse correlation between STAT5 activation and Igk germline transcription during the developmental transition of Irf4−/− Irf8−/− large pre-B cells to small pre-B cells.
Constitutive activation of STAT5 is associated with human leukemias and myeloproliferative disorders7. In particular, the presence of activated STAT5 is a characteristic feature of B-cell acute lymphophastic leukemia (B-ALL) carrying the BCR-ABL1 chromosomal translocation7. A critical role of STAT5 in the generation of this B-ALL subset is indicated by the fact that retroviral BCR-ABL (p185) expression in Stat5−/− hematopoietic progenitors fails to induce leukemia in the mouse10. Our study has now provided a molecular explanation suggesting that the oncogenic function of STAT5 in B-ALL is likely to control the survival of leukemic B-lymphocytes by activating the Mcl1 gene.
METHODS
Methods and any associated references are available in the online version of the paper at http://www.nature.com/natureimmunology/.
Supplementary Material
Acknowledgments
We are thank L. Hennighausen for providing the Stat5fl/fl and Bcl2l1fl/fl mice, T. Rabbitts for the Rag1cre/cre mouse, J. Adams for the Vav-bcl2 mouse and R. Grosschedl for the Ebf1−/− mouse. We also thank L. Hennighausen, R. Moriggl, T. Decker and A. Rolink for insightful discussions, M. Fuxa for advice on Ig recombination analysis, A. Ebert for help with ChIP analyses, I. Tamir for bioinformatical analysis of ChIP-chip data and G. Stengl for FACS sorting. This research was supported by Boehringer Ingelheim, the Austrian Science Fund (grant P16701-BO9), the Austrian GEN-AU initiative (financed by the Bundesminsterium für Bildung und Wissenschaft) and the EU FP6 funding for the Epigenome Network of Excellence. Stephen Malin was the recipient of a Marie-Curie Fellowship.
Footnotes
AUTHOR CONTRIBUTIONS S. Malin performed most experiments; S. McManus contributed the ChIP-chip analyses; C.C. generated and characterized the Ikzf1Ebf1 transgenic mouse; A.D. carried out the histological analysis; M.N. did the bioinformatical analysis of the Igh locus; P.B. and A.S. provided the Mcl1fl/fl mouse; and M.B. and S. Malin planned the project, designed the experiments and wrote the manuscript.
COMPETING INTERESTS STATEMENT The authors declare that they have no competing financial interests.
Note: Supplementary Information is available on the Nature Immunology website.
Accession code. The microarray data discussed in this paper are available at the GEO repository at NCBI under the accession number GSE18278.
References
- 1.von Freeden-Jeffry U, et al. Lymphopenia in interleukin (IL)-7 gene-deleted mice identifies IL-7 as a nonredundant cytokine. J Exp Med. 1995;181:1519–1526. doi: 10.1084/jem.181.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miller JP, et al. The earliest step in B lineage differentiation from common lymphoid progenitors is critically dependent upon interleukin 7. J Exp Med. 2002;196:705–711. doi: 10.1084/jem.20020784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kikuchi K, Lai AY, Hsu C-L, Kondo M. IL-7 receptor signaling is necessary for stage transition in adult B cell development through up-regulation of EBF. J Exp Med. 2005;201:1197–1203. doi: 10.1084/jem.20050158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maraskovsky E, et al. Bcl-2 can rescue T lymphocyte development in interleukin-7 receptor-deficient mice but not in mutant rag-1−/− mice. Cell. 1997;89:1011–1019. doi: 10.1016/s0092-8674(00)80289-5. [DOI] [PubMed] [Google Scholar]
- 5.Akashi K, Kondo M, von Freeden-Jeffry U, Murray R, Weissman IL. Bcl-2 rescues T lymphopoiesis in interleukin-7 receptor-deficient mice. Cell. 1997;89:1033–1041. doi: 10.1016/s0092-8674(00)80291-3. [DOI] [PubMed] [Google Scholar]
- 6.Maraskovsky E, Peschon JJ, McKenna H, Teepe M, Strasser A. Overexpression of Bcl-2 does not rescue impaired B lymphopoiesis in IL-7 receptor-deficient mice but can enhance survival of mature B cells. Int Immunol. 1998;10:1367–1375. doi: 10.1093/intimm/10.9.1367. [DOI] [PubMed] [Google Scholar]
- 7.Hennighausen L, Robinson GW. Interpretation of cytokine signaling through the transcription factors STAT5A and STAT5B. Genes Dev. 2008;22:711–721. doi: 10.1101/gad.1643908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Teglund S, et al. Stat5a and Stat5b proteins have essential and nonessential, or redundant, roles in cytokine responses. Cell. 1998;93:841–850. doi: 10.1016/s0092-8674(00)81444-0. [DOI] [PubMed] [Google Scholar]
- 9.Yao Z, et al. Stat5a/b are essential for normal lymphoid development and differentiation. Proc Natl Acad Sci USA. 2006;103:1000–1005. doi: 10.1073/pnas.0507350103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoelbl A, et al. Clarifying the role of Stat5 in lymphoid development and Abelson-induced transformation. Blood. 2006;107:4898–4906. doi: 10.1182/blood-2005-09-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moriggl R, et al. Stat5 is required for IL-2-induced cell cycle progression of peripheral T cells. Immunity. 1999;10:249–259. doi: 10.1016/s1074-7613(00)80025-4. [DOI] [PubMed] [Google Scholar]
- 12.Sexl V, et al. Stat5a/b contribute to interleukin 7-induced B-cell precursor expansion, but abl- and bcr/abl-induced transformation are independent of Stat5. Blood. 2000;96:2277–2283. [PubMed] [Google Scholar]
- 13.Cui Y, et al. Inactivation of Stat5 in mouse mammary epithelium during pregnancy reveals distinct functions in cell proliferation, survival, and differentiation. Mol Cell Biol. 2004;24:8037–8047. doi: 10.1128/MCB.24.18.8037-8047.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goetz CA, Harmon IR, O’Neil JJ, Burchill MA, Farrar MA. STAT5 activation underlies IL7 receptor-dependent B cell development. J Immunol. 2004;172:4770–4778. doi: 10.4049/jimmunol.172.8.4770. [DOI] [PubMed] [Google Scholar]
- 15.Hirokawa S, Sato H, Kato I, Kudo A. EBF-regulating Pax5 transcription is enhanced by STAT5 in the early stage of B cells. Eur J Immunol. 2003;33:1824–1829. doi: 10.1002/eji.200323974. [DOI] [PubMed] [Google Scholar]
- 16.Goetz CA, et al. Restricted STAT5 activation dictates appropriate thymic B versus T cell lineage commitment. J Immunol. 2005;174:7753–7763. doi: 10.4049/jimmunol.174.12.7753. [DOI] [PubMed] [Google Scholar]
- 17.Dias S, Silva H, Jr, Cumano A, Vieira P. Interleukin-7 is necessary to maintain the B cell potential in common lymphoid progenitors. J Exp Med. 2005;201:971–979. doi: 10.1084/jem.20042393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roessler S, et al. Distinct promoters mediate the regulation of Ebf1 gene expression by IL-7 and Pax5. Mol Cell Biol. 2007;27:579–594. doi: 10.1128/MCB.01192-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnston CM, Wood AL, Bolland DJ, Corcoran AE. Complete sequence assembly and characterization of the C57BL/6 mouse Ig heavy chain V region. J Immunol. 2006;176:4221–4234. doi: 10.4049/jimmunol.176.7.4221. [DOI] [PubMed] [Google Scholar]
- 20.Chowdhury D, Sen R. Stepwise activation of the immunoglobulin μ heavy chain gene locus. EMBO J. 2001;20:6394–6403. doi: 10.1093/emboj/20.22.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson K, Angelin-Duclos C, Park S, Calame KL. Changes in histone acetylation are associated with differences in accessibility of VH gene segments to V-DJ recombination during B-cell ontogeny and development. Mol Cell Biol. 2003;23:2438–2450. doi: 10.1128/MCB.23.7.2438-2450.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Corcoran AE, Riddell A, Krooshoop D, Venkitaraman AR. Impaired immunoglobulin gene rearrangement in mice lacking the IL-7 receptor. Nature. 1998;391:904–907. doi: 10.1038/36122. [DOI] [PubMed] [Google Scholar]
- 23.Bertolino E, et al. Regulation of interleukin 7-dependent immunoglobulin heavy-chain variable gene rearrangements by transcription factor STAT5. Nat Immunol. 2005;6:836–843. doi: 10.1038/ni1226. [DOI] [PubMed] [Google Scholar]
- 24.Igarashi H, Gregory SC, Yokota T, Sakaguchi N, Kincade PW. Transcription from the RAG1 locus marks the earliest lymphocyte progenitors in bone marrow. Immunity. 2002;17:117–130. doi: 10.1016/s1074-7613(02)00366-7. [DOI] [PubMed] [Google Scholar]
- 25.McCormack MP, Forster A, Drynan L, Pannell R, Rabbitts TH. The LMO2 T-cell oncogene is activated via chromosomal translocations or retroviral insertion during gene therapy but has no mandatory role in normal T-cell development. Mol Cell Biol. 2003;23:9003–9013. doi: 10.1128/MCB.23.24.9003-9013.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kwon K, et al. Instructive role of the transcription factor E2A in early B lymphopoiesis and germinal center B cell development. Immunity. 2008;28:751–762. doi: 10.1016/j.immuni.2008.04.014. [DOI] [PubMed] [Google Scholar]
- 27.Yao Z, et al. Nonredundant roles for Stat5a/b in directly regulating Foxp3. Blood. 2007;109:4368–4375. doi: 10.1182/blood-2006-11-055756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fleming HE, Paige CJ. Pre-B cell receptor signaling mediates selective response to IL-7 at the pro-B to pre-B cell transition via an ERK/MAP kinase-dependent pathway. Immunity. 2001;15:521–531. doi: 10.1016/s1074-7613(01)00216-3. [DOI] [PubMed] [Google Scholar]
- 29.Scheeren FA, et al. STAT5 regulates the self-renewal capacity and differentiation of human memory B cells and controls Bcl-6 expression. Nat Immunol. 2005;6:303–313. doi: 10.1038/ni1172. [DOI] [PubMed] [Google Scholar]
- 30.Opferman JT, et al. Development and maintenance of B and T lymphocytes requires antiapoptotic MCL-1. Nature. 2003;426:671–676. doi: 10.1038/nature02067. [DOI] [PubMed] [Google Scholar]
- 31.Schebesta A, et al. Transcription factor Pax5 activates the chromatin of key genes involved in B cell signaling, adhesion, migration and immune function. Immunity. 2007;27:49–63. doi: 10.1016/j.immuni.2007.05.019. [DOI] [PubMed] [Google Scholar]
- 32.O’Geen H, Nicolet CM, Blahnik K, Green R, Farnham PJ. Comparison of sample preparation methods for ChIP-chip assays. BioTechniques. 2006;41:577–580. doi: 10.2144/000112268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chakraborty T, et al. Repeat organization and epigenetic regulation of the DH-Cμ domain of the immunoglobulin heavy-chain gene locus. Mol Cell. 2007;27:842–850. doi: 10.1016/j.molcel.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 34.Grawunder U, Haasner D, Melchers F, Rolink A. Rearrangement and expression of κ light chain genes can occur without μ heavy chain expression during differentiation of pre-B cells. Int Immunol. 1993;5:1609–1618. doi: 10.1093/intimm/5.12.1609. [DOI] [PubMed] [Google Scholar]
- 35.Johnson K, et al. Regulation of immunoglobulin light-chain recombination by the transcription factor IRF-4 and the attenuation of interleukin-7 signaling. Immunity. 2008;28:335–345. doi: 10.1016/j.immuni.2007.12.019. [DOI] [PubMed] [Google Scholar]
- 36.Mandal M, et al. Ras orchestrates exit from the cell cycle and light-chain recombination during early B cell development. Nat Immunol. 2009;10:1110–1117. doi: 10.1038/ni.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marshall AJ, Fleming HF, Wu GE, Paige CJ. Modulation of the IL-7 dose-response threshold during pro-B cell differentiation is dependent on pre-B cell receptor expression. J Immunol. 1998;161:6038–6045. [PubMed] [Google Scholar]
- 38.Socolovsky M, Fallon AE, Wang S, Brugnara C, Lodish HF. Fetal anemia and apoptosis of red cell progenitors in Stat5a−/−5b−/− mice: a direct role for Stat5 in Bcl-XL induction. Cell. 1999;98:181–191. doi: 10.1016/s0092-8674(00)81013-2. [DOI] [PubMed] [Google Scholar]
- 39.Kieslinger M, et al. Antiapoptotic activity of Stat5 required during terminal stages of myeloid differentiation. Genes Dev. 2000;14:232–244. [PMC free article] [PubMed] [Google Scholar]
- 40.Grillot DAM, et al. bcl-x exhibits regulated expression during B cell development and activation and modulates lymphocyte survival in transgenic mice. J Exp Med. 1996;183:381–391. doi: 10.1084/jem.183.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Motoyama N, et al. Massive cell death of immature hematopoietic cells and neurons in bcl-x-deficient mice. Science. 1995;267:1506–1510. doi: 10.1126/science.7878471. [DOI] [PubMed] [Google Scholar]
- 42.Decker T, et al. Stepwise activation of enhancer and promoter regions of the B cell commitment gene Pax5 in early lymphopoiesis. Immunity. 2009;30:508–520. doi: 10.1016/j.immuni.2009.01.012. [DOI] [PubMed] [Google Scholar]
- 43.Ye S-K, et al. The IL-7 receptor controls the accessibility of the TCRγ locus by Stat5 and histone acetylation. Immunity. 2001;15:813–823. doi: 10.1016/s1074-7613(01)00230-8. [DOI] [PubMed] [Google Scholar]
- 44.Rolink A, et al. A subpopulation of B220+ cells in murine bone marrow does not express CD19 and contains natural killer cell progenitors. J Exp Med. 1996;183:187–194. doi: 10.1084/jem.183.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tudor KS, Payne KJ, Yamashita Y, Kincade PW. Functional assessment of precursors from murine bone marrow suggests a sequence of early B lineage differentiation events. Immunity. 2000;12:335–345. doi: 10.1016/s1074-7613(00)80186-7. [DOI] [PubMed] [Google Scholar]
- 46.Matthews AG, et al. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature. 2007;450:1106–1110. doi: 10.1038/nature06431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fuxa M, et al. Pax5 induces V-to-DJ rearrangements and locus contraction of the immunoglobulin heavy-chain gene. Genes Dev. 2004;18:411–422. doi: 10.1101/gad.291504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schebesta M, Pfeffer PL, Busslinger M. Control of pre-BCR signaling by Pax5-dependent activation of the BLNK gene. Immunity. 2002;17:473–485. doi: 10.1016/s1074-7613(02)00418-1. [DOI] [PubMed] [Google Scholar]
- 49.Novobrantseva TI, et al. Rearrangement and expression of immunoglobulin light chain genes can precede heavy chain expression during normal B cell development in mice. J Exp Med. 1999;189:75–87. doi: 10.1084/jem.189.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu Y, Davidson L, Alt FW, Baltimore D. Deletion of the Igκ light chain intronic enhancer/matrix attachment region impairs but does not abolish VκJκ rearrangement. Immunity. 1996;4:377–385. doi: 10.1016/s1074-7613(00)80251-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.