Abstract
Centrioles are conserved microtubule-based organelles that lie at the core of the animal centrosome and play a crucial role in nucleating the formation of cilia and flagella in most eukaryotes. Centrioles have a complex ultrastructure with ninefold symmetry and a well-defined length. This structure is assembled from a host of proteins, including a variety of disease gene products. Over a century after the discovery of centrioles, the mechanisms underlying the assembly of these fascinating organelles, in particular the establishment of ninefold symmetry and the control of centriole length, are now starting to be uncovered.
Introduction
Centrioles are cylindrical structures composed of nine triplet microtubule ‘blades’ organized around a central cartwheel (Figure 1). In animal cells, centrioles can recruit microtubule-nucleating factors, called the pericentriolar material (PCM), to form a larger structure named the centrosome, which serves as the main microtubule-organizing center during both interphase and mitosis. Centrioles can also move to the cell surface and nucleate the formation of cilia and flagella: in this context, centrioles are called basal bodies (Figure 2). In recent years, genetic and functional genomic screens have identified genes essential for centriole assembly [1–11]. At the same time, proteomic analyses have identified a large list of centriole proteins, including several disease proteins, many of which remain to be further characterized [12–15]. Reflecting the fact that centrioles found in divergent eukaryotes are likely to derive from a common ancestral structure (Figure 3), and that the ultrastructural steps of centriole assembly appear to be largely conserved [16–20], many of the proteins identified in these studies are only conserved in species that assemble centrioles [13,21,22]. In addition to this conserved core of centriolar components, proteomic studies have identified a range of centriolar proteins that are unique to subsets of organisms. These differences likely reflect the different contexts in which centrioles are found, involving a variety of appendages, connecting fibers, or pericentriolar material, as well as differences in the regulation of centriole assembly (Figure 2). While great progress has been made during the past decade in understanding the molecular composition of centrioles, less is known about the assembly mechanisms that build a centriole from this large ‘parts list’. In this review, we will discuss recent work that has provided insight into the molecular mechanisms underlying the assembly of centrioles, focusing in particular on the establishment of ninefold symmetry, the control of centriole length, and the maturation of centrioles.
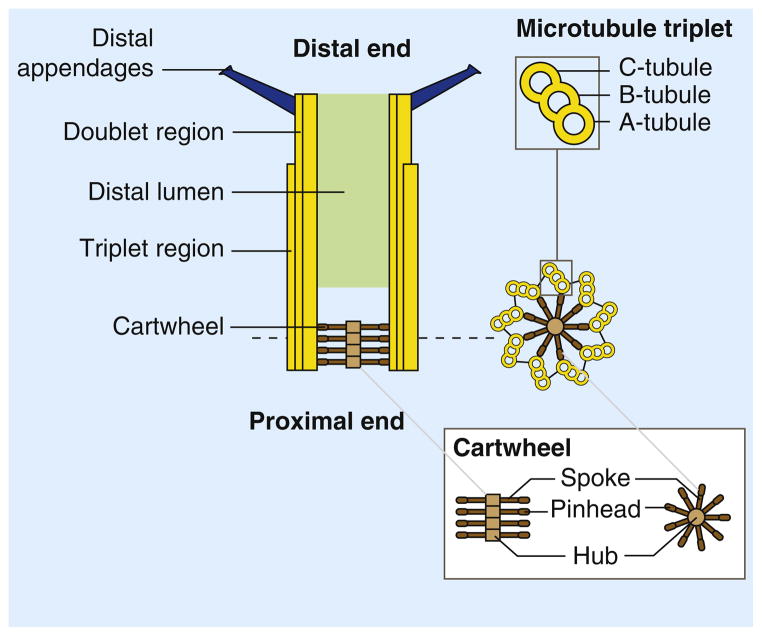
Figure 1. Centriole structure.
Centrioles are microtubule arrays composed of nine triplets of microtubules organized around a cartwheel structure. The triplets are connected to the cartwheel through the A-tubule, the first to assemble during centriole assembly and the only complete microtubule in a triplet. The B- and C-tubules are incomplete microtubules. In vertebrates and in Chlamydomonas, the C-tubule is shorter than the A- and B-tubules and the distal end of the centriole is thus formed by doublet microtubules [112,129]. The cartwheel is formed by a central hub from which emanate spokes terminated by a pinhead structure that binds the A-tubule of the microtubule triplet. The very distal end of the centriole is decorated by ninefold symmetric distal appendages (or transition fibers) required for anchoring the centrioles at the plasma membrane when they act as a basal bodies.
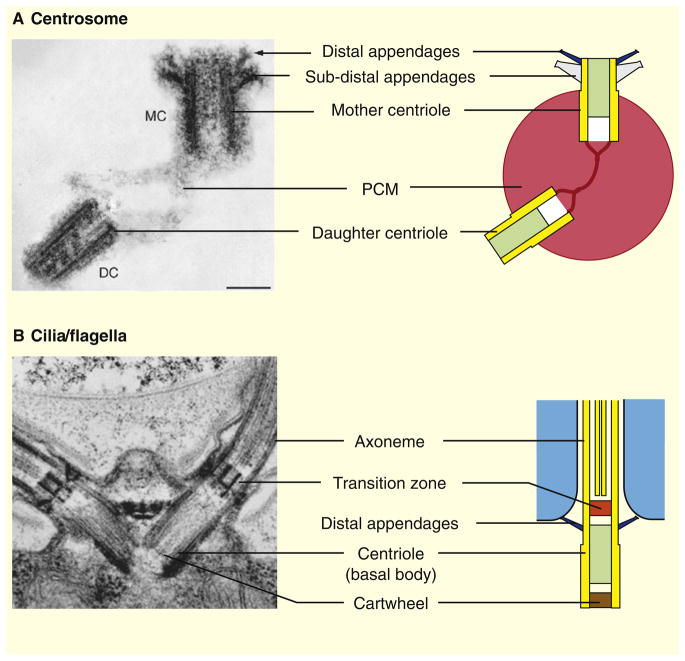
Figure 2. Centrioles in centrosomes and cilia/flagella.
(A) In animal cells, centrioles form the core structure of the centrosome, the main microtubule-organizing center. Quiescent cells (G0) or proliferating cells in the G1 phase of the cell cycle contain a single centrosome. The centrosome is formed by one mature centriole, the mother centriole (MC), and one non-mature centriole, the daughter centriole (DC), linked together and surrounded by a protein matrix called the pericentriolar material (PCM). In vertebrates, the mother centriole is decorated by two sets of ninefold symmetrical appendages: the distal and sub-distal appendages required for ciliogenesis and for the stable anchoring of microtubules at the centrosome, respectively. The distal appendages are conserved throughout eukaryotes, whereas the sub-distal appendages are found only in animal centrosomes. (B) In animals as well as in most other eukaryotes, centrioles are also required for the assembly of cilia/flagella. Centrioles, often referred to as basal bodies in this case, dock to the plasma membrane through their distal appendages and template the assembly of the nine outer microtubule doublets of the axoneme, the cytoskeletal core of cilia/flagella. A distinct structure called the transition zone separates the basal body from the axoneme. Shown are electron micrographs of (A) a human centrosome [130] and (B) the Chlamydomonas flagellar apparatus [24].
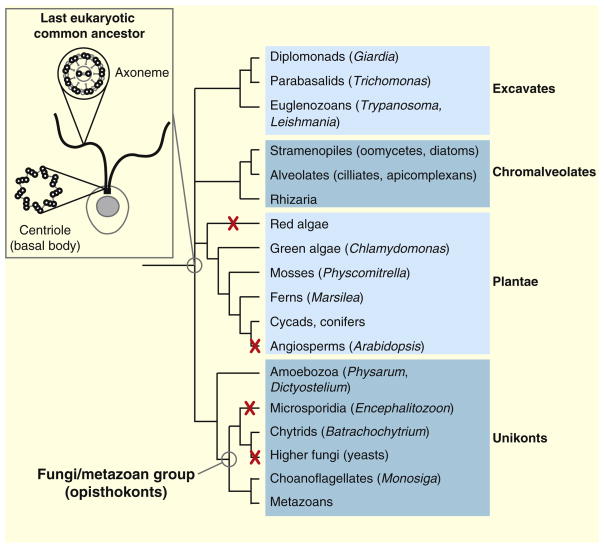
Figure 3. Conservation of the centriole and axoneme.
The centriole and the axoneme, i.e. the microtubule core of cilia and flagella, are conserved features of eukaryotes. These structures were probably present in the last eukaryotic common ancestor and are still found in most branches of the eukaryotic tree of life. Centrioles and axonemes were lost concomitantly during evolution of certain taxa, most notably angiosperms and higher fungi. Taxa in which all species have lost centrioles and axonemes are indicated by a red cross. Some taxa, such as amoebozoa, comprise species that form flagella (like Physarum) as well as species completely devoid of centrioles and axonemes (like Dictyostelium). The phylogenetic tree of life is adapted from the Tree of Life web project (http://tolweb.org/tree). The last eukaryotic common ancestor (inset) is schematically represented as a single-celled organism (the plasma membrane and the nucleus are in gray) bearing two motile flagella (in black). Schematic representations of cross-sections through the centriole and the axoneme are also shown.
Centriole Functions
It seems likely that centrioles have evolved for the primary purpose of growing cilia and flagella, which are important sensory and motile organelles found in almost all cells of the human body [23]. The ciliary microtubule doublets are continuous with the A- and B-tubules from centriole microtubule triplets (Figures 1 and 2) [24]. Proteins involved in the assembly of cilia are recruited to centrioles [25], and defects in several genes encoding centriolar proteins lead to ciliary disease phenotypes.
Despite the presence of centrioles at the mitotic spindle poles, centrioles are in many cases dispensable for mitosis, even in species that normally contain them [26]. Cell-cycle progression and cytokinesis can be defective when centrioles are missing [27–29], but this could be due to indirect effects. For example, G1 arrest in mammalian cells following centriole ablation results from an increase in stress sensitivity rather than an absolute requirement for centrioles during cell-cycle progression [30].
While centrioles are dispensable for spindle assembly, they are more important for spindle positioning. When centrioles are experimentally ablated, spindles drift within the cell [28]. In vertebrates, centriole position appears to respond to planar cell polarity cues [31], consistent with the localization of some planar cell polarity proteins at centrioles [32]. Proper spindle positioning by centrioles is thought to be necessary for proper tissue development, because defects in spindle orientation caused by mutations in centriole-associated genes can lead to nephronophthisis, a cystic kidney disease associated with abnormally wide ducts [33].
An interesting possibility is that the centriole-positioning pathway may be specific for the mother centriole, which is the oldest centriole of the two in a G1 centrosome (Figure 1) and the only fully mature centriole (i.e. with the ability to act as a basal body) in a typical vertebrate cell. This is suggested by analysis of dividing stem cells in the Drosophila male germ line, in which the mitotic spindle is always oriented such that the older centriole is anchored on the side of the cell adjacent to the stem-cell niche [34]. Similar bias in mother centriole position is seen in radial glial progenitor cells in the mouse [35]. Remarkably, following depletion of ninein, a protein required for stable anchoring of microtubules at mother centrioles, this asymmetry in mother-centriole segregation is lost, eventually leading to premature depletion of the stem-cell pool [35,36]. In Chlamydomonas mutants defective in mother–daughter cohesion, mother centrioles move to their correct position while daughter centrioles do not, suggesting the mother centriole is uniquely responsive to the positioning pathway [37].
Initiation of Centriole Assembly
Regulation of Centriole Initiation
Initiation of centriole duplication is under tight regulation to ensure the control of centriole number (for more extensive coverage of this topic, see [38–40]). In mammalian cells, a single procentriole starts forming perpendicular to the wall of each parental centriole around the G1/S transition. Once the assembly of the two new procentrioles has been initiated, further centriole duplication is inhibited until the cells pass through mitosis [41]. The release of the tight association of procentrioles with the parental centrioles, termed disengagement, occurs in late mitosis in animal cells. Disengagement involves the protease separase and Polo-like kinase 1 (Plk1) and is a prerequisite for the next round of centriole duplication [42,43]. When centrioles are absent, new centrioles can form de novo, suggesting the role of pre-existing centrioles is not actually to template the procentrioles as long proposed, but rather to bias the spatial location where the procentrioles self-assemble [44–47]. When too many centrioles are present, cells can inhibit the synthesis of new centrioles, a mechanism that allows for correction of errors in centriole number [48].
The key regulator of centriole assembly is a kinase called Polo-like kinase 4 (Plk4) or SAK in Drosophila. Inhibiting Plk4 prevents centriole duplication in both human cells and flies [49,50]. Conversely, overexpression of Plk4 and SAK can trigger the assembly of supernumerary centrioles [46,47,49,50]. Interestingly, Plk4 orthologs are not found outside the Fungi/Metazoa group (Figure 3), which suggests that centriole duplication is triggered by different mechanisms in other eukaryotes, possibly involving other Polo-like kinases [22]. Also, no Plk4 ortholog is found in Caenorhabditis elegans, in which centriole duplication is instead triggered by a non-orthologous kinase called ZYG-1 [22,51]. ZYG-1 controls the recruitment of the centriole structural component SAS-6, which is a substrate for ZYG-1 [52–54]. The recruitment of ZYG-1 itself requires SPD-2, a component of the PCM essential for centriole duplication in C. elegans embryos [7,8,52,53]. Interestingly, SPD-2 family members are only found in the genomes of Unikonts, a branch of the eukaryotic tree comprising the Fungi/Metazoa group as well as Amoebozoa, such as the model organism Dictyostelium discoideum (Figure 3) [21,22,55]. Studies of SPD-2 orthologs in human and flies suggest that the primary function of SPD-2 and related proteins is to recruit PCM around the centrioles [56–58]. A SPD-2 ortholog is also found in the matrix associated with the Dictyostelium nuclear-associated body, a very distinctive structure that forms the core of the Dictyostelium centrosome [55]. In addition to its role in PCM recruitment, the human ortholog of SPD-2, called Cep192, is essential for centriole duplication, whereas its Drosophila ortholog appears dispensable for this process [57,58]. It is, however, possible that Cep192 affects centriole duplication indirectly through its ability to recruit PCM and microtubule-nucleating factors, as the PCM is known to play a role in centriole duplication [59]. In contrast, Drosophila Asterless (Asl) and related proteins are PCM components that appear to be more specifically required for centriole assembly [60,61]. In Drosophila, Asl localizes near the centriole wall in both proliferating cells and in testes and is required for centriole duplication in both cases [60,61]. Cep152, the vertebrate ortholog of Asl, is a component of the PCM in proliferating human cells [12]. Interestingly, zebrafish Cep152 was also found to be required for basal body assembly in multiciliated cells [61]. In these cells, up to several hundreds of basal bodies assemble at the same time around structures of unknown composition called deuterosomes as the cells undergo differentiation [62,63]. The defect observed in Cep152-depleted zebrafish supports the idea that the initiation of basal body assembly in multiciliated cells relies at least in part on the same mechanisms as centriole duplication in proliferating cells.
Establishment of the Ninefold Symmetry
Initiation of centriole assembly and establishment of the ninefold symmetry require a structure called the cartwheel (Figures 1 and 4). The cartwheel is located at the proximal end of basal bodies in a wide range of species. In vertebrate centrosomes, a cartwheel structure is present at the base of procentrioles but is no longer seen in daughter and mother centrioles [64,65]. The structure of the cartwheel has been best described in unicellular organisms. It is formed by a central hub from which emanate nine evenly spaced spokes, terminated by a pinhead structure to which microtubule triplets attach (Figure 1). In Chlamydomonas, the cartwheel is assembled prior to the addition of microtubules at the tip of each spoke [18]. Two components of the cartwheel have been described in this species. CrSAS-6/Bld12p, the homolog of C. elegans SAS-6, has been proposed to be part of the inner spokes or the hub of the cartwheel [66]. Bld10p, which also belongs to a conserved protein family, has been shown to form the outer spoke and the pinhead structure [67]. Recent studies of mutants defective for these genes have provided important clues on how the cartwheel assembles and sets centriole radial symmetry. When Chlamydomonas BLD12 is deleted, most cells lack a proper centriolar structure but approximately 20% of cells assemble defective centrioles that sometimes contain an abnormal number of triplets — 7, 8 or 10 — or have missing triplets. Strikingly, the hub is missing in bld12 mutant centrioles [66]. Similarly, depletion of SAS-6 by RNA interference (RNAi) in Paramecium results in the formation of centrioles with altered numbers of triplets that retain the cartwheel spokes but are lacking the central hub [68]. A Drosophila SAS-6 null mutant is also found to have a significant reduction in the number of centrioles and forms centrioles with structural defects, for example, missing triplets [69]. As in Chlamydomonas, the Paramecium and Drosophila SAS-6 orthologs localize to the central hub of the cartwheel [68,70]. Together these results support the hypothesis that proteins of the SAS-6 family are required to build the central hub, and that the hub plays a role in establishing the ninefold symmetry [39,66,69].
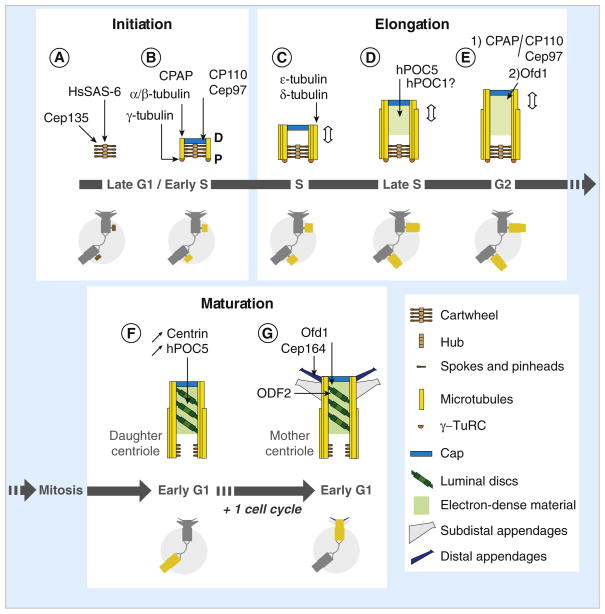
Figure 4. Model for centriole assembly within duplicating human centrosomes.
(A) Centriole assembly begins during late G1 or early S phase with the assembly of the cartwheel, which depends on HsSAS-6 for the central hub and Cep135 (Bld10p ortholog) for the spokes and/or the pinheads. (B) CPAP (SAS-4 ortholog) triggers γ-tubulin-dependent nucleation of the A-tubules and their attachment to the pinheads of the cartwheel, possibly by participating in the recruitment of the γ-tubulin ring complex (γ-TuRC) at the proximal end of the cartwheel. Each A-tubule is nucleated by a γ-TuRC and grows unidirectionally from proximal (P) to distal (D). The A-tubule remains capped by the γ-TuRC throughout the assembly process but is lost from daughter and mother centrioles. A cap structure containing CP110 and Cep97 forms at the distal end of the procentriole. The cap is required to control procentriole microtubule growth and probably also to stabilize the nascent procentriole. (C) The B- and C-tubules form by a γ-TuRC-independent mechanism and grow bidirectionally until they reach the length of the A-tubule. The microtubule triplets are stabilized by ε- and δ-tubulin and centriole elongation begins. (D) During S phase, procentrioles elongate up to ~70% of their final length. This step is dependent on hPOC5, and possibly involves hPOC1 as well. (E) Procentriole elongation continues after the transition into G2. Two mechanisms control centriole length at this stage: 1) a balance between the activities of CPAP, which promotes α/β-tubulin incorporation at the distal end, and the cap structure containing CP110 and Cep97; 2) an Ofd1-dependent mechanism. (F) After mitosis, the procentrioles become daughter centrioles. Tilted discs surrounded by electron-dense material are observed from this stage onwards in the distal part of the centriole. Markers like centrin and hPOC5 accumulate within the distal lumen as cell cycle progresses, which could reflect the progressive maturation of the centriole. HsSAS-6 is no longer associated with the proximal part of the daughter centrioles, possibly correlating with the disassembly of the central hub of the cartwheel. In contrast, the spokes could be conserved to some extent as Cep135 remains associated with mother and daughter centrioles. (G) After the second mitosis, centriole maturation is completed when the distal and sub-distal appendages assemble. The assembly of both types of appendage depends on ODF2, a maturation-specific marker recruited at the distal end of the daughter centriole during the previous G2 phase. Assembly of the distal appendages is also dependent on Ofd1, and possibly as well on the distal appendage component Cep164. Schematic representations of the centrosome during the successive steps of centriole assembly and maturation are shown in the lower parts of panels A–G, in which the assembling centrioles are highlighted (brown when only the cartwheel is present, yellow for later stages).
HsSAS-6, the human homolog of SAS-6, is also essential for the initial steps of centriole assembly but, unlike its homologs in other species that remain associated with mature centrioles, is no longer found associated with daughter and mother centrioles [71,72]. Loss of HsSAS-6 from the procentrioles correlates with its degradation by the 26S proteasome at the end of mitosis, and possibly also with the loss of the cartwheel structure that occurs as procentrioles become daughter centrioles (Figure 4) [64,65,71]. In contrast, SAS-6 staining is retained at the proximal end of basal bodies from rat tracheal multiciliated cells, suggesting that the cartwheel may not disassemble in this case. Intriguingly, SAS-6 also localizes to the proximal region of ciliary axonemes in these cells, revealing a possible involvement in ciliary assembly or function [73].
In C. elegans, where SAS-6 was first identified, short centrioles composed of nine singlet rather than nine triplet microtubules are formed, and no recognizable cartwheel structure is observed. Microtubule singlets are instead seen to assemble around a structure that appears as a hollow cylinder by electron microscopy. The assembly of this structure, called the central tube, requires SAS-6 [53]. Though different at the ultrastructural level, the central tube and the cartwheel thus share at least one component and are likely to be similarly required for establishing the ninefold symmetry of centrioles.
Recruitment of SAS-6 within procentrioles in C. elegans requires SAS-5, a protein that physically interacts with SAS-6 and, like SAS-6, is essential for centriole duplication in this species [10,74]. Recently, a protein called Ana2 was shown to be the likely ortholog of SAS-5 in Drosophila [75]. Although poorly conserved at the amino acid level, Ana2 interacts with DSAS-6 and, like its C. elegans counterpart, is essential for centriole duplication [75–77]. Interestingly, the human ortholog of Ana2, called STIL or SIL, has been shown to be essential for proper mitotic spindle assembly and has been linked to microcephaly, reminiscent of the centriole duplication factor CPAP, the human ortholog of C. elegans SAS-4 (discussed further below) [75,78–80]. Analysis of the dynamic properties of SAS-5 in C. elegans, however, suggested that, rather than being a structural component of the centrioles, SAS-5 may be required for the recruitment of SAS-6 at procentriole assembly sites [10,74].
In addition to SAS-6-related proteins, the assembly of the cartwheel also depends on the conserved Bld10p/Cep135 family of proteins. Bld10p was originally identified as the product of a gene mutated in a Chlamydomonas strain that completely lacks basal bodies and was shown to be a component of the cartwheel spokes [9]. When the Chlamydomonas bld10 mutant is complemented by a truncated version of Bld10p, centrioles with eight triplets are often observed. These centrioles assemble around a cartwheel with shorter spokes, which seemingly leads to the formation of a centriole of smaller diameter than can only accommodate eight triplets. The cartwheel still forms nine spokes, one of which is not linked to a triplet [67]. Thus, if the central hub plays a critical role in attaining the correct ultrastructure by patterning centriole radial symmetry, the radial spokes, the length of which depends on Bld10p, do so by specifying centriole diameter.
Cep135, the human ortholog of Bld10p, also localizes to the cartwheel and is essential for centriole assembly [72,81]. In contrast, Bld10/Cep135 function is apparently dispensable for the initiation of centriole assembly in Drosophila, as centrioles duplicate normally in mutant flies lacking the Bld10/Cep135 homolog [82,83]. Depletion of Drosophila Bld10 by RNAi in S2 cells leads to a partial inhibition of centriole duplication, however, suggesting Bld10 could be required in some conditions [77]. Ultrastructural analysis reveals that sperm axonemes in bld10 mutant flies contain nine outer doublet microtubules, which shows that the centrioles from which they are assembled have a proper ninefold symmetry [83]. However, sperm centrioles are shorter in these mutants than in wild-type flies and most of the flagella lack the central pair of microtubules, leading to male sterility [22,82,83]. Whether the cartwheels assembled in the bld10 mutant are normal is not known.
Steps preceding cartwheel formation remain poorly understood. In Chlamydomonas and Paramecium, the cartwheel assembles on an amorphous, disk-like structure [18]. In addition to serving as a site of cartwheel assembly, the amorphous disk could play a role in establishing the ninefold symmetry by controlling the diameter of the centrioles, and thus the number of microtubule triplets that they can accommodate. In the Chlamydomonas bld12 mutant, 70% of the basal bodies that retain a circular assembly of triplet microtubules exhibit ninefold symmetry, despite lacking the central hub of the cartwheel, suggesting additional mechanisms influence the radial symmetry of the centrioles [66]. In Paramecium, Bld10 depletion by RNAi leads to formation of centrioles that lack cartwheel spokes but retain the central hub, suggesting the hub is connected to the microtubule cylinder by another structure, most likely the amorphous disk [68]. Similar disk-like structures have not been described in animal cells: instead, procentrioles appear to be linked to the wall of the parental centrioles by a connecting stalk [65].
Assembly of Centriole Microtubules
Assembly of centriole triplet microtubules seems to occur sequentially. Singlet microtubules, or A-tubules, first attach to the spokes of the cartwheel then doublets and triplets (incomplete B and C-tubules, respectively) assemble (Figures 1 and 4) [16,18,65]. Attachment of singlet microtubules is thought to require the conserved SAS-4 family of proteins, defined by C. elegans SAS-4 and required for centriole duplication in diverse organisms [5,72,84]. The likely homolog of SAS-4 in human cells is called CPAP. Though very divergent at the amino-acid level, CPAP is concentrated within the proximal lumen of the centrioles, where the cartwheel forms, and is required for centriole duplication, supporting the fact that it is the bona fide homolog of C. elegans SAS-4 [72,85]. In C. elegans embryos, the central tube forms during S phase and elongates during prophase, and microtubules start to assemble around it during prometaphase. In embryos depleted of SAS-4, the central tube forms and elongates, but microtubules fail to attach [53]. Centriolar SAS-4 increases as S phase progresses, suggesting that SAS-4 interacts with the central tube and that the amount of centriolar SAS-4 increases as the central tube elongates. Interestingly, centriolar SAS-4 remains in dynamic exchange with the cytoplasmic pool until prophase/prometaphase, where it becomes stably associated with the centrioles [86]. This suggests that centriolar SAS-4 may be stabilized by the assembly of centriolar microtubules. This model is further supported by the observation that stabilization of centriolar SAS-4 requires γ-tubulin, which is believed to nucleate centriole microtubules, as well as β-tubulin. Moreover, γ-tubulin is required for the accumulation of SAS-4 in the pericentriolar matrix, suggesting a possible interaction between these two proteins [86]. In human cells, CPAP has been shown to co-immunoprecipitate with γ-tubulin [87]. γ-tubulin function in centriole assembly also appears to be conserved, as γ-tubulin is required for centriole duplication in a wide range of eukaryotes [72,88–90]. γ-tubulin function appears to be critical in the control of centriole assembly, because the introduction of specific mutations in γ-tubulin is sufficient to induce the assembly of extra centrioles in Tetrahymena [91].
A recent study utilizing cryo-electron microscopy provides important new insights into the mechanism of procentriole microtubule assembly in human centrosomes [65]. In nascent procentrioles, the proximal or minus end of the A-tubule is capped by a conical structure resembling the γ-tubulin ring complex (γ-TuRC), a structure known to nucleate microtubules in animal cells (Figure 4). This suggests that each A-tubule is nucleated by a γ-TuRC and then grows unidirectionally from the proximal to the distal end. Supporting this hypothesis, the distal or plus end of the A-tubules from assembling procentrioles show outwardly curved extensions characteristic of growing microtubule extremities. In contrast, the incomplete B- and C-tubules are never capped at their proximal end, suggesting that their assembly is initiated by a different mechanism. The B- and C-tubules appear to start assembling at variable positions along the A- or B-tubules, respectively, and undergo bidirectional growth as suggested by the presence of curved extensions at both the proximal and distal ends of B- and C-tubules before they reach their final length. The proximal ends of the B- and C-tubules become blunt as they reach the proximal extremity of the A-tubule, suggesting that a mechanism of stabilization occurs at that time [65].
Studies in Chlamydomonas and Paramecium revealed a role for tubulin family members δ- and ε-tubulin in the formation or stabilization of the B- and C-tubules [1,3,92,93]. In the Chlamydomonas bld2-1 mutant, which expresses a truncated form of ε-tubulin, doublet and triplet microtubules are missing [3]. In Chlamydomonas and Paramecium cells defective for δ-tubulin, the C-tubule is often missing and most centrioles are composed of doublet microtubules [1,92]. The requirement for δ-tubulin in C-tubule formation can be bypassed by suppressor mutations in α-tubulin, suggesting that δ-tubulin may be required for triplet stabilization rather than for C-tubule assembly [94]. Furthermore, genes encoding ε- and δ-tubulins are absent from the Drosophila genome, despite the fact that centrioles containing triplet microtubules are formed in this species [95]. To date, there is still no mechanistic clue as to how the formation of the incomplete B- and C-tubules is achieved.
Centriole Elongation and Length Control
Temporal Regulation of Centriole Elongation
At the stage at which B- and C-tubules start to assemble, procentrioles are short, with their length slightly exceeding the length of the cartwheel (about 70–100 nm; Figure 4) [16,18,59]. In subsequent stages of assembly, procentrioles undergo elongation to eventually reach the full length (400–500 nm in most species). Centriole length appears to be under active control, based not only on the limited variation of length observed in any given cell type but also on the fact that length undergoes dynamic changes at well-defined stages in the assembly process. In Chlamydomonas, for instance, procentrioles are assembled during mitosis and elongate during the following G2 phase [11]. In mammalian proliferating cells, procentrioles assembled at the G1/S transition or during early S phase start to elongate during S phase, and elongation proceeds further during G2 and mitosis [19,20,96]. Centriole elongation seems to be coupled to cell-cycle progression. In mammalian cells arrested in S phase by drugs that inhibit DNA replication, procentrioles elongate to reach approximately 70% of the full length, which corresponds to the length of late S-phase procentrioles in untreated cells, but do not elongate further, suggesting that completion of centriole elongation requires transition into G2 [59,97,98]. In addition, studies of a conserved centriolar component called hPOC5 suggest that procentriole elongation and cell-cycle progression are also coupled earlier during S phase [13,98]. In human cells depleted of hPOC5, centriole elongation is inhibited, the procentrioles remaining about the length of early S-phase procentrioles. In addition, cell-cycle progression is impaired and the cells accumulate in S phase. It is not known whether these two events are linked or whether hPOC5 regulates centriole elongation and cell-cycle progression independently [98].
Centriole elongation can also be restricted to certain cell types or certain developmental stages, like in Drosophila and in Apicomplexans [95,99]. In these species, centriole elongation mostly occurs prior to the formation of cilia and flagella. In the Drosophila uncoordinated mutant (unc), centrioles fail to elongate in ciliated sensory neurons and in sperm, which leads to inhibition of ciliogenesis and, as a result, to impaired sensory functions and male sterility. Interestingly, the product of the unc gene, which localizes specifically to the basal bodies in these two cell types, does not have orthologs outside dipteran insects [100], suggesting that centriole elongation in these organisms may rely on somewhat different molecular mechanisms.
Centriole Length Control
How do microtubule triplets elongate and what molecular mechanism determines their final size? In animal cells, incorporation of tubulin dimers into centriolar microtubules appears to occur beneath a distal cap containing the CP110 protein [72]. CP110 is conserved among animals and is essential for centriole duplication [22,72,101]. CP110, together with another centriolar protein called Cep97, has also been implicated in controlling the formation of primary cilia in mammalian cells. Depletion of these proteins in the ciliated RPE1 cell line increases the proportion of cells that form a primary cilium. Conversely, overexpression of CP110 prevents primary cilium assembly in conditions that normally induce ciliogenesis [102]. Interestingly, in non-ciliated cell lines like U2OS or HeLa, inhibition of either CP110 or Cep97 induces the assembly of elongated structures that were originally proposed to be primary cilia as well, but were recently found to correspond to abnormally elongated centrioles [102–104]. Three recent reports showed that similar structures are assembled following overexpression of CPAP, the human homolog of SAS-4 [85,103,104]. These centriole-like structures, which can be up to several microns in length, are formed by excessive elongation of centriole microtubules from both parental centrioles and procentrioles. Ultrastructural analyses revealed that, in spite of their abnormal sizes, elongated centrioles induced by CPAP overexpression or CP110 depletion often resemble genuine centrioles. This is also supported by immunofluorescence data showing that markers of specific subparts of the centriolar structure are properly localized in elongated centrioles. Taken together, these data led the authors of these studies to propose a model in which CPAP and CP110 are required for controlling the length of the centrioles. CPAP would promote elongation, possibly by favoring tubulin incorporation at the plus end of centriole microtubules, whereas CP110 capping activity would limit microtubule growth [85,103,104]. Consistent with this hypothesis, CPAP/SAS-4 family members contain a domain that can bind tubulin dimers and induce microtubule depolymerization, which is essential for CPAP to promote centriole elongation [104–106]. Interestingly, CPAP overexpression induces high rates of centriole elongation in G2 phase and mitosis but not in S-phase-arrested cells [85,104], which could reflect differences in the mechanisms underlying centriole elongation in S and G2 or M phases.
More recently, another centriolar component called Ofd1 was found to play a role in controlling the length of mammalian centrioles [107]. Ofd1 is a conserved centriolar protein known to be mutated in different types of ciliopathies [13,108,109]. Studies of knockout mice lacking the Ofd1 gene show that Ofd1 is essential for primary cilium formation in the embryonic node and in kidneys [110]. This phenotype is recapitulated in a mouse embryonic stem cell line deficient for Ofd1 [107]. In addition, about one third of the cells exhibit long centrioles comparable to those observed following depletion of either CP110 or Cep97, or following overexpression of CPAP. In most cases, only the mother centriole is elongated in Ofd1-deficient cells even though in control cells Ofd1 localizes to the distal ends of mother and daughter centrioles, as well as procentrioles. Interestingly, CP110 and Cep97 localization at the distal end of centrioles is unaffected by Ofd1 depletion, suggesting that these proteins act in a separate pathway for centriole length control. However, abnormal elongation of the mother centriole occurs during G2 but not S phase in Ofd1 cells, just as in the context of CP110 depletion and CPAP overexpression [107].
Finally, centriole elongation could require another conserved family of proteins called POC1 proteins [15,82]. When human POC1 is overexpressed in U2OS cells arrested in S phase, which is when centriole elongation is normally initiated in these cells, hPOC1-containing filaments reminiscent of those induced by CPAP overexpression are observed [15]. These filaments contain centrin and γ-tubulin, suggesting that they may correspond to abnormally elongated centrioles as well, although ultrastructural analyses are still needed to determine how similar they are to bona fide centrioles. A role for POC1 proteins in centriole elongation is also supported by the study of two mutant lines defective for the Drosophila POC1 homolog. In these mutants, spermatid centrioles are shorter than in wild type, suggesting a partial impairment of centriole elongation [82]. In contrast, centriole length is unaffected by POC1 depletion in Tetrahymena [111]: in a Tetrahymena POC1 deletion strain, basal bodies exhibit breaks in microtubule blades and these defects are further enhanced by growing the mutant cells at higher temperature, leading to loss of most of the basal bodies. In addition, basal bodies in poc1Δ cells are more sensitive to nocodazole, suggesting that Tetrahymena POC1 may play a role in centriole stabilization rather than in length control per se [111].
Building the Distal End
Besides elongation of the microtubule triplets, centriole elongation involves the assembly of intra-luminal structures in the distal end of centrioles. Little is known about the function and molecular composition of these structures, and they exhibit a remarkable degree of ultrastructural diversity among species. In mammalian cells, the distal lumen of centrioles is filled with a periodic stack of tilted discs (Figure 4) [112,113]. In Paramecium, the lumen contains a helical structure [16], whereas in Tetrahymena, another ciliate, a cylindrical electron-dense structure is observed [111]. The lumen of Chlamydomonas basal bodies appears to be filled with fibers connecting the microtubule triplets to each other [114]. Despite this variability in their architecture, intraluminal structures in diverse eukaryotes seem to have common properties. In particular, the distal lumen of centrioles in mammalian cells, in ciliates and in Chlamydomonas all contain centrin proteins [115–118]. Centrin proteins are calcium-binding proteins related to calmodulin that are found associated with centrioles in most species and are present in the genomes of all species that assemble motile cilia [88,114,115,119–121]. Their precise role remains elusive, however, possibly because they have several distinct binding partners within centrioles. Human centrin proteins, for instance, bind directly to hPOC5 and to a related centriolar component called hSFI1, and co-immunoprecipitate CP110 [98,101,122].
Centriole Maturation
A centriole is mature when it is able to nucleate a cilium or a flagellum. Vertebrate centrosomes contain only one mature centriole that bears two sets of ninefold symmetrical appendages (Figures 2 and 4). Only the mother centriole can attach to the plasma membrane and nucleate a primary cilium. In mouse cells defective for the centriolar protein ODF2, these appendages are not assembled, mother centrioles fail to anchor to the plasma membrane, and ciliogenesis is inhibited [123]. The most distal set of appendages is likely to be involved in centriole anchoring. Similar structures, often referred to as transition fibers, decorate the distal end of mature centrioles in most eukaryotic species that assemble cilia (Figure 2). A protein called Cep164 that specifically localizes to the distal appendages has been identified in human cells. Although it is not known whether Cep164 is essential for the assembly of the distal appendages, ciliogenesis is inhibited in RPE1 cells depleted of Cep164 [124]. Cep164 is found in the genome of most ciliated species, which suggests that distal appendages and transition fibers are probably related at the molecular level [21]. Assembly of the distal appendages was recently shown to depend on Ofd1, as Ofd1-deficient cells lack the distal appendages and show no Cep164 staining. Remarkably, by studying the effects of five disease-associated Ofd1 mutations in Ofd1-deficient cells, Singla and coworkers [107] identify mutations that affect centriole length but not the assembly of the distal appendages, which suggests that these two processes are controlled independently by Ofd1.
In addition to distal appendages, the mother centriole in vertebrate cells is decorated by ninefold symmetrical sub-distal appendages. Sub-distal appendages are thought to play a role in microtubule anchoring [125] and, as with distal appendages, their assembly requires the ODF2 protein [123].
Centriole maturation is coupled to cell-cycle progression. In vertebrate cells, the full maturation of centrioles takes 1.5 cell cycles (Figure 4). A centriole formed during the previous cell cycle remains immature until mitosis, when the two sets of appendages are assembled at its distal end [20]. In cells arrested in S phase for prolonged periods, the daughter centrioles never acquire the appendages, suggesting that the transition through G2 or mitosis is essential for centriole maturation [126]. The maturation of centrioles in vertebrate cells may be regulated so that each cell has only one mature centriole at a time, a condition that seems crucial for the proper development of certain organs [35]. In contrast, biflagellate cells, like Chlamydomonas, possess two mature, appendage-bearing centrioles per cell. Intriguingly, the study of δ-tubulin-deficient cells suggests that the two mature centrioles are not strictly equivalent. In these cells, the maturation of centrioles is slower, and many cells carry only one flagellum, which is nucleated by the older centriole [1]. In some algae, centrioles undergo a series of functional transitions at each generation, going through as many as three different states such that daughter, mother, and grandmother centrioles nucleate flagella with different structures and motile activities [127]. These studies indicate that centrioles can count up to at least three generations, but the mechanisms by which this occurs are completely unknown. In vertebrate cells, some proteins, including centrin and hPOC5, are seen to associate with daughter centrioles in increasing amounts as the cell cycle progresses. In addition, the centriolar pools of centrin and hPOC5 are highly phosphorylated [98,128]. Such modifications in centriole composition may be part of the maturation process.
Conclusions
During the past decade, there has been great progress in our understanding of the biology of centrioles, yet we have only scratched the surface. The identification of major molecular players, such as SAS-6, SAS-4/CPAP, Bld10p/Cep135 and Plk4/SAK, the master regulator of centriole duplication [38,49], has allowed us to gain a much clearer picture of the initial steps of centriole assembly and how these steps are regulated, as well as the molecular bases of the ninefold symmetry. With this understanding of the upstream cues for assembly, as well the growing laundry list of centriole component proteins, we are now poised to begin to understand how centrioles are actually built, at a mechanistic level.
Acknowledgments
The authors would like to thank P. Gönczy (ISREC/EPFL, Switzerland), E. Kannegaard (UCSF) and K. Wemmer (UCSF) for critical comments on the manuscript. Our work is supported by NIH grant R01GM077004.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dutcher SK, Trabuco EC. The UNI3 gene is required for assembly of basal bodies of Chlamydomonas and encodes delta-tubulin, a new member of the tubulin superfamily. Mol Biol Cell. 1998;9:1293–1308. doi: 10.1091/mbc.9.6.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gönczy P, Echeverri C, Oegema K, Coulson A, Jones SJ, Copley RR, Duperon J, Oegema J, Brehm M, Cassin E, et al. Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature. 2000;408:331–336. doi: 10.1038/35042526. [DOI] [PubMed] [Google Scholar]
- 3.Dutcher SK, Morrissette NS, Preble AM, Rackley C, Stanga J. Epsilon-tubulin is an essential component of the centriole. Mol Biol Cell. 2002;13:3859–3869. doi: 10.1091/mbc.E02-04-0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kirkham M, Muller-Reichert T, Oegema K, Grill S, Hyman A. SAS-4 is a C. elegans centriolar protein that controls centrosome size. Cell. 2003;112:575–587. doi: 10.1016/s0092-8674(03)00117-x. [DOI] [PubMed] [Google Scholar]
- 5.Leidel S, Gönczy P. SAS-4 is essential for centrosome duplication in C. elegans and is recruited to daughter centrioles once per cell cycle. Dev Cell. 2003;4:431–439. doi: 10.1016/s1534-5807(03)00062-5. [DOI] [PubMed] [Google Scholar]
- 6.Dammermann A, Muller-Reichert T, Pelletier L, Habermann B, Desai A, Oegema K. Centriole assembly requires both centriolar and pericentriolar material proteins. Dev Cell. 2004;7:815–829. doi: 10.1016/j.devcel.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 7.Kemp CA, Kopish KR, Zipperlen P, Ahringer J, O’Connell KF. Centrosome maturation and duplication in C. elegans require the coiled-coil protein SPD-2. Dev Cell. 2004;6:511–523. doi: 10.1016/s1534-5807(04)00066-8. [DOI] [PubMed] [Google Scholar]
- 8.Pelletier L, Ozlu N, Hannak E, Cowan C, Habermann B, Ruer M, Muller-Reichert T, Hyman AA. The Caenorhabditis elegans centrosomal protein SPD-2 is required for both pericentriolar material recruitment and centriole duplication. Curr Biol. 2004;14:863–873. doi: 10.1016/j.cub.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 9.Matsuura K, Lefebvre PA, Kamiya R, Hirono M. Bld10p, a novel protein essential for basal body assembly in Chlamydomonas: localization to the cartwheel, the first ninefold symmetrical structure appearing during assembly. J Cell Biol. 2004;165:663–671. doi: 10.1083/jcb.200402022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leidel S, Delattre M, Cerutti L, Baumer K, Gönczy P. SAS-6 defines a protein family required for centrosome duplication in C. elegans and in human cells. Nat Cell Biol. 2005;7:115–125. doi: 10.1038/ncb1220. [DOI] [PubMed] [Google Scholar]
- 11.Piasecki BP, Lavoie M, Tam LW, Lefebvre PA, Silflow CD. The Uni2 phosphoprotein is a cell cycle regulated component of the basal body Mmaturation pathway in Chlamydomonas reinhardtii. Mol Biol Cell. 2008;19:262–273. doi: 10.1091/mbc.E07-08-0798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andersen JS, Wilkinson CJ, Mayor T, Mortensen P, Nigg EA, Mann M. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 2003;426:570–574. doi: 10.1038/nature02166. [DOI] [PubMed] [Google Scholar]
- 13.Keller LC, Romijn EP, Zamora I, Yates JR, 3rd, Marshall WF. Proteomic analysis of isolated chlamydomonas centrioles reveals orthologs of ciliary-disease genes. Curr Biol. 2005;15:1090–1098. doi: 10.1016/j.cub.2005.05.024. [DOI] [PubMed] [Google Scholar]
- 14.Kilburn CL, Pearson CG, Romijn EP, Meehl JB, Giddings TH, Jr, Culver BP, Yates JR, 3rd, Winey M. New Tetrahymena basal body protein components identify basal body domain structure. J Cell Biol. 2007;178:905–912. doi: 10.1083/jcb.200703109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keller LC, Geimer S, Romijn E, Yates J, 3rd, Zamora I, Marshall WF. Molecular architecture of the centriole proteome: the conserved WD40 domain protein POC1 is required for centriole duplication and length control. Mol Biol Cell. 2009;20:1150–1166. doi: 10.1091/mbc.E08-06-0619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dippell RV. The development of basal bodies in paramecium. Proc Natl Acad Sci USA. 1968;61:461–468. doi: 10.1073/pnas.61.2.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robbins E, Jentzsch G, Micali A. The centriole cycle in synchronized HeLa cells. J Cell Biol. 1968;36:329–339. doi: 10.1083/jcb.36.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cavalier-Smith T. Basal body and flagellar development during the vegetative cell cycle and the sexual cycle of Chlamydomonas reinhardii. J Cell Sci. 1974;16:529–556. doi: 10.1242/jcs.16.3.529. [DOI] [PubMed] [Google Scholar]
- 19.Kuriyama R, Borisy GG. Centriole cycle in Chinese hamster ovary cells as determined by whole-mount electron microscopy. J Cell Biol. 1981;91:814–821. doi: 10.1083/jcb.91.3.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vorobjev IA, Chentsov Yu S. Centrioles in the cell cycle. I. Epithelial cells. J Cell Biol. 1982;93:938–949. doi: 10.1083/jcb.93.3.938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hodges ME, Scheumann N, Wickstead B, Langdale JA, Gull K. Reconstructing the evolutionary history of the centriole from protein components. J Cell Sci. 2010;123:1407–1413. doi: 10.1242/jcs.064873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carvalho-Santos Z, Machado P, Branco P, Tavares-Cadete F, Rodrigues-Martins A, Pereira-Leal JB, Bettencourt-Dias M. Stepwise evolution of the centriole-assembly pathway. J Cell Sci. 2010;123:1414–1426. doi: 10.1242/jcs.064931. [DOI] [PubMed] [Google Scholar]
- 23.Marshall WF, Nonaka S. Cilia: tuning in to the cell’s antenna. Curr Biol. 2006;16:R604–R614. doi: 10.1016/j.cub.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 24.Ringo DL. Flagellar motion and fine structure of the flagellar apparatus in Chlamydomonas. J Cell Biol. 1967;33:543–571. doi: 10.1083/jcb.33.3.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deane JA, Cole DG, Seeley ES, Diener DR, Rosenbaum JL. Localization of intraflagellar transport protein IFT52 identifies basal body transitional fibers as the docking site for IFT particles. Curr Biol. 2001;11:1586–1590. doi: 10.1016/s0960-9822(01)00484-5. [DOI] [PubMed] [Google Scholar]
- 26.Debec A, Sullivan W, Bettencourt-Dias M. Centrioles: active players or passengers during mitosis? Cell Mol Life Sci. 2010;67:2173–2194. doi: 10.1007/s00018-010-0323-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Piel M, Nordberg J, Euteneuer U, Bornens M. Centrosome-dependent exit of cytokinesis in animal cells. Science. 2001;291:1550–1553. doi: 10.1126/science.1057330. [DOI] [PubMed] [Google Scholar]
- 28.Hinchcliffe EH, Miller FJ, Cham M, Khodjakov A, Sluder G. Requirement of a centrosomal activity for cell cycle progression through G1 into S phase. Science. 2001;291:1547–1550. doi: 10.1126/science.1056866. [DOI] [PubMed] [Google Scholar]
- 29.Mikule K, Delaval B, Kaldis P, Jurcyzk A, Hergert P, Doxsey S. Loss of centrosome integrity induces p38-p53-p21-dependent G1-S arrest. Nat Cell Biol. 2007;9:160–170. doi: 10.1038/ncb1529. [DOI] [PubMed] [Google Scholar]
- 30.Uetake Y, Loncarek J, Nordberg JJ, English CN, La Terra S, Khodjakov A, Sluder G. Cell cycle progression and de novo centriole assembly after centrosomal removal in untransformed human cells. J Cell Biol. 2007;176:173–182. doi: 10.1083/jcb.200607073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Montcouquiol M, Kelley MW. Planar and vertical signals control cellular differentiation and patterning in the mammalian cochlea. J Neurosci. 2003;23:9469–9478. doi: 10.1523/JNEUROSCI.23-28-09469.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park TJ, Haigo SL, Wallingford JB. Ciliogenesis defects in embryos lacking inturned or fuzzy function are associated with failure of planar cell polarity and Hedgehog signaling. Nat Genet. 2006;38:303–311. doi: 10.1038/ng1753. [DOI] [PubMed] [Google Scholar]
- 33.Simons M, Walz G. Polycystic kidney disease: cell division without a c(l)ue? Kidney Int. 2006;70:854–864. doi: 10.1038/sj.ki.5001534. [DOI] [PubMed] [Google Scholar]
- 34.Yamashita YM, Mahowald AP, Perlin JR, Fuller MT. Asymmetric inheritance of mother versus daughter centrosome in stem cell division. Science. 2007;315:518–521. doi: 10.1126/science.1134910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang X, Tsai JW, Imai JH, Lian WN, Vallee RB, Shi SH. Asymmetric centrosome inheritance maintains neural progenitors in the neocortex. Nature. 2009;461:947–955. doi: 10.1038/nature08435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mogensen MM, Malik A, Piel M, Bouckson-Castaing V, Bornens M. Microtubule minus-end anchorage at centrosomal and non-centrosomal sites: the role of ninein. J Cell Sci. 2000;113:3013–3023. doi: 10.1242/jcs.113.17.3013. [DOI] [PubMed] [Google Scholar]
- 37.Feldman JL, Geimer S, Marshall WF. The mother centriole plays an instructive role in defining cell geometry. PLoS Biol. 2007;5:e149. doi: 10.1371/journal.pbio.0050149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bettencourt-Dias M, Glover DM. Centrosome biogenesis and function: centrosomics brings new understanding. Nat Rev Mol Cell Biol. 2007;8:451–463. doi: 10.1038/nrm2180. [DOI] [PubMed] [Google Scholar]
- 39.Strnad P, Gönczy P. Mechanisms of procentriole formation. Trends Cell Biol. 2008;18:389–396. doi: 10.1016/j.tcb.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 40.Nigg EA, Raff JW. Centrioles, centrosomes, and cilia in health and disease. Cell. 2009;139:663–678. doi: 10.1016/j.cell.2009.10.036. [DOI] [PubMed] [Google Scholar]
- 41.Wong C, Stearns T. Centrosome number is controlled by a centrosome-intrinsic block to reduplication. Nat Cell Biol. 2003;5:539–544. doi: 10.1038/ncb993. [DOI] [PubMed] [Google Scholar]
- 42.Tsou MF, Stearns T. Mechanism limiting centrosome duplication to once per cell cycle. Nature. 2006;442:947–951. doi: 10.1038/nature04985. [DOI] [PubMed] [Google Scholar]
- 43.Tsou MF, Wang WJ, George KA, Uryu K, Stearns T, Jallepalli PV. Polo kinase and separase regulate the mitotic licensing of centriole duplication in human cells. Dev Cell. 2009;17:344–354. doi: 10.1016/j.devcel.2009.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marshall WF, Vucica Y, Rosenbaum JL. Kinetics and regulation of de novo centriole assembly. Implications for the mechanism of centriole duplication. Curr Biol. 2001;11:308–317. doi: 10.1016/s0960-9822(01)00094-x. [DOI] [PubMed] [Google Scholar]
- 45.La Terra S, English CN, Hergert P, McEwen BF, Sluder G, Khodjakov A. The de novo centriole assembly pathway in HeLa cells: cell cycle progression and centriole assembly/maturation. J Cell Biol. 2005;168:713–722. doi: 10.1083/jcb.200411126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rodrigues-Martins A, Riparbelli M, Callaini G, Glover DM, Bettencourt-Dias M. Revisiting the role of the mother centriole in centriole biogenesis. Science. 2007;316:1046–1050. doi: 10.1126/science.1142950. [DOI] [PubMed] [Google Scholar]
- 47.Peel N, Stevens NR, Basto R, Raff JW. Overexpressing centriole-replication proteins in vivo induces centriole overduplication and de novo formation. Curr Biol. 2007;17:834–843. doi: 10.1016/j.cub.2007.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marshall WF. Stability and robustness of an organelle number control system: modeling and measuring homeostatic regulation of centriole abundance. Biophys J. 2007;93:1818–1833. doi: 10.1529/biophysj.107.107052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Habedanck R, Stierhof YD, Wilkinson CJ, Nigg EA. The Polo kinase Plk4 functions in centriole duplication. Nat Cell Biol. 2005;7:1140–1146. doi: 10.1038/ncb1320. [DOI] [PubMed] [Google Scholar]
- 50.Bettencourt-Dias M, Rodrigues-Martins A, Carpenter L, Riparbelli M, Lehmann L, Gatt MK, Carmo N, Balloux F, Callaini G, Glover DM. SAK/PLK4 is required for centriole duplication and flagella development. Curr Biol. 2005;15:2199–2207. doi: 10.1016/j.cub.2005.11.042. [DOI] [PubMed] [Google Scholar]
- 51.O’Connell K, Caron C, Kopish K, Hurd D, Kemphues K, Li Y, White J. The C. elegans zyg-1 gene encodes a regulator of centrosome duplication with distinct maternal and paternal roles in the embryo. Cell Motil Cytoskeleton. 2001;105:547–558. doi: 10.1016/s0092-8674(01)00338-5. [DOI] [PubMed] [Google Scholar]
- 52.Delattre M, Canard C, Gönczy P. Sequential protein recruitment in C. elegans centriole formation. Curr Biol. 2006;16:1844–1849. doi: 10.1016/j.cub.2006.07.059. [DOI] [PubMed] [Google Scholar]
- 53.Pelletier L, O’Toole E, Schwager A, Hyman AA, Muller-Reichert T. Centriole assembly in Caenorhabditis elegans. Nature. 2006;444:619–623. doi: 10.1038/nature05318. [DOI] [PubMed] [Google Scholar]
- 54.Kitagawa D, Busso C, Fluckiger I, Gönczy P. Phosphorylation of SAS-6 by ZYG-1 is critical for centriole formation in C. elegans embryos. Dev Cell. 2009;17:900–907. doi: 10.1016/j.devcel.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 55.Schulz I, Erle A, Graf R, Kruger A, Lohmeier H, Putzler S, Samereier M, Weidenthaler S. Identification and cell cycle-dependent localization of nine novel, genuine centrosomal components in Dictyostelium discoideum. Cell Motil Cytoskeleton. 2009;66:915–928. doi: 10.1002/cm.20384. [DOI] [PubMed] [Google Scholar]
- 56.Gomez-Ferreria MA, Rath U, Buster DW, Chanda SK, Caldwell JS, Rines DR, Sharp DJ. Human Cep192 is required for mitotic centrosome and spindle assembly. Curr Biol. 2007;17:1960–1966. doi: 10.1016/j.cub.2007.10.019. [DOI] [PubMed] [Google Scholar]
- 57.Dix CI, Raff JW. Drosophila Spd-2 recruits PCM to the sperm centriole, but is dispensable for centriole duplication. Curr Biol. 2007;17:1759–1764. doi: 10.1016/j.cub.2007.08.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu F, Lawo S, Bird A, Pinchev D, Ralph A, Richter C, Muller-Reichert T, Kittler R, Hyman AA, Pelletier L. The mammalian SPD-2 ortholog Cep192 regulates centrosome biogenesis. Curr Biol. 2008;18:136–141. doi: 10.1016/j.cub.2007.12.055. [DOI] [PubMed] [Google Scholar]
- 59.Loncarek J, Hergert P, Magidson V, Khodjakov A. Control of daughter centriole formation by the pericentriolar material. Nat Cell Biol. 2008;10:322–328. doi: 10.1038/ncb1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Varmark H, Llamazares S, Rebollo E, Lange B, Reina J, Schwarz H, Gonzalez C. Asterless is a centriolar protein required for centrosome function and embryo development in Drosophila. Curr Biol. 2007;17:1735–1745. doi: 10.1016/j.cub.2007.09.031. [DOI] [PubMed] [Google Scholar]
- 61.Blachon S, Gopalakrishnan J, Omori Y, Polyanovsky A, Church A, Nicastro D, Malicki J, Avidor-Reiss T. Drosophila asterless and vertebrate Cep152 Are orthologs essential for centriole duplication. Genetics. 2008;180:2081–2094. doi: 10.1534/genetics.108.095141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lemullois M, Boisvieux-Ulrich E, Laine MC, Chailley B, Sandoz D. Development and functions of the cytoskeleton during ciliogenesis in metazoa. Biol Cell. 1988;63:195–208. doi: 10.1016/0248-4900(88)90058-5. [DOI] [PubMed] [Google Scholar]
- 63.Dawe HR, Farr H, Gull K. Centriole/basal body morphogenesis and migration during ciliogenesis in animal cells. J Cell Sci. 2007;120:7–15. doi: 10.1242/jcs.03305. [DOI] [PubMed] [Google Scholar]
- 64.Alvey PL. Do adult centrioles contain cartwheels and lie at right angles to each other? Cell Biol Int Rep. 1986;10:589–598. doi: 10.1016/0309-1651(86)90136-0. [DOI] [PubMed] [Google Scholar]
- 65.Guichard P, Chretien D, Marco S, Tassin AM. Procentriole assembly revealed by cryo-electron tomography. EMBO J. 2010;29:1565–1572. doi: 10.1038/emboj.2010.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nakazawa Y, Hiraki M, Kamiya R, Hirono M. SAS-6 is a cartwheel protein that establishes the 9-fold symmetry of the centriole. Curr Biol. 2007;17:2169–2174. doi: 10.1016/j.cub.2007.11.046. [DOI] [PubMed] [Google Scholar]
- 67.Hiraki M, Nakazawa Y, Kamiya R, Hirono M. Bld10p constitutes the cartwheel-spoke tip and stabilizes the 9-fold symmetry of the centriole. Curr Biol. 2007;17:1778–1783. doi: 10.1016/j.cub.2007.09.021. [DOI] [PubMed] [Google Scholar]
- 68.Jerka-Dziadosz M, Gogendeau D, Klotz C, Cohen J, Beisson J, Koll F. Basal body duplication in Paramecium: the key role of Bld10 in assembly and stability of the cartwheel. Cytoskeleton. 2010;67:161–171. doi: 10.1002/cm.20433. [DOI] [PubMed] [Google Scholar]
- 69.Rodrigues-Martins A, Bettencourt-Dias M, Riparbelli M, Ferreira C, Ferreira I, Callaini G, Glover DM. DSAS-6 organizes a tube-like centriole precursor, and its absence suggests modularity in centriole assembly. Curr Biol. 2007;17:1465–1472. doi: 10.1016/j.cub.2007.07.034. [DOI] [PubMed] [Google Scholar]
- 70.Gopalakrishnan J, Guichard P, Smith AH, Schwarz H, Agard DA, Marco S, Avidor-Reiss T. Self-assembling SAS-6 multimer is a core centriole building block. J Biol Chem. doi: 10.1074/jbc.M109.092627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Strnad P, Leidel S, Vinogradova T, Euteneuer U, Khodjakov A, Gönczy P. Regulated HsSAS-6 levels ensure formation of a single procentriole per centriole during the centrosome duplication cycle. Dev Cell. 2007;13:203–213. doi: 10.1016/j.devcel.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kleylein-Sohn J, Westendorf J, Le Clech M, Habedanck R, Stierhof YD, Nigg EA. Plk4-induced centriole biogenesis in human cells. Dev Cell. 2007;13:190–202. doi: 10.1016/j.devcel.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 73.Vladar EK, Stearns T. Molecular characterization of centriole assembly in ciliated epithelial cells. J Cell Biol. 2007;178:31–42. doi: 10.1083/jcb.200703064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Delattre M, Leidel S, Wani K, Baumer K, Bamat J, Schnabel H, Feichtinger R, Schnabel R, Gönczy P. Centriolar SAS-5 is required for centrosome duplication in C. elegans. Nat Cell Biol. 2004;6:656–664. doi: 10.1038/ncb1146. [DOI] [PubMed] [Google Scholar]
- 75.Stevens NR, Dobbelaere J, Brunk K, Franz A, Raff JW. Drosophila Ana2 is a conserved centriole duplication factor. J Cell Biol. 2010;188:313–323. doi: 10.1083/jcb.200910016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Goshima G, Wollman R, Goodwin SS, Zhang N, Scholey JM, Vale RD, Stuurman N. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science. 2007;316:417–421. doi: 10.1126/science.1141314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dobbelaere J, Josue F, Suijkerbuijk S, Baum B, Tapon N, Raff J. A genome-wide RNAi screen to dissect centriole duplication and centrosome maturation in Drosophila. PLoS Biol. 2008;6:e224. doi: 10.1371/journal.pbio.0060224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pfaff KL, Straub CT, Chiang K, Bear DM, Zhou Y, Zon LI. The zebra fish cassiopeia mutant reveals that SIL is required for mitotic spindle organization. Mol Cell Biol. 2007;27:5887–5897. doi: 10.1128/MCB.00175-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bond J, Roberts E, Springell K, Lizarraga SB, Scott S, Higgins J, Hampshire DJ, Morrison EE, Leal GF, Silva EO, et al. A centrosomal mechanism involving CDK5RAP2 and CENPJ controls brain size. Nat Genet. 2005;37:353–355. doi: 10.1038/ng1539. [DOI] [PubMed] [Google Scholar]
- 80.Kumar A, Girimaji SC, Duvvari MR, Blanton SH. Mutations in STIL, encoding a pericentriolar and centrosomal protein, cause primary microcephaly. Am J Hum Genet. 2009;84:286–290. doi: 10.1016/j.ajhg.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ohta T, Essner R, Ryu JH, Palazzo RE, Uetake Y, Kuriyama R. Characterization of Cep135, a novel coiled-coil centrosomal protein involved in microtubule organization in mammalian cells. J Cell Biol. 2002;156:87–99. doi: 10.1083/jcb.200108088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Blachon S, Cai X, Roberts KA, Yang K, Polyanovsky A, Church A, Avidor-Reiss T. A proximal centriole-like structure is present in Drosophila spermatids and can serve as a model to study centriole duplication. Genetics. 2009;182:133–144. doi: 10.1534/genetics.109.101709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mottier-Pavie V, Megraw TL. Drosophila bld10 is a centriolar protein that regulates centriole, basal body, and motile cilium assembly. Mol Biol Cell. 2009;20:2605–2614. doi: 10.1091/mbc.E08-11-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Basto R, Lau J, Vinogradova T, Gardiol A, Woods CG, Khodjakov A, Raff JW. Flies without centrioles. Cell. 2006;125:1375–1386. doi: 10.1016/j.cell.2006.05.025. [DOI] [PubMed] [Google Scholar]
- 85.Kohlmaier G, Loncarek J, Meng X, McEwen BF, Mogensen MM, Spektor A, Dynlacht BD, Khodjakov A, Gönczy P. Overly long centrioles and defective cell division upon excess of the SAS-4-related protein CPAP. Curr Biol. 2009;19:1012–1018. doi: 10.1016/j.cub.2009.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dammermann A, Maddox PS, Desai A, Oegema K. SAS-4 is recruited to a dynamic structure in newly forming centrioles that is stabilized by the gamma-tubulin-mediated addition of centriolar microtubules. J Cell Biol. 2008;180:771–785. doi: 10.1083/jcb.200709102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hung LY, Tang CJ, Tang TK. Protein 4.1 R-135 interacts with a novel centrosomal protein (CPAP) which is associated with the gamma-tubulin complex. Mol Cell Biol. 2000;20:7813–7825. doi: 10.1128/mcb.20.20.7813-7825.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ruiz F, Beisson J, Rossier J, Dupuis-Williams P. Basal body duplication in Paramecium requires gamma-tubulin. Curr Biol. 1999;9:43–46. doi: 10.1016/s0960-9822(99)80045-1. [DOI] [PubMed] [Google Scholar]
- 89.Shang Y, Li B, Gorovsky MA. Tetrahymena thermophila contains a conventional gamma-tubulin that is differentially required for the maintenance of different microtubule-organizing centers. J Cell Biol. 2002;158:1195–1206. doi: 10.1083/jcb.200205101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Haren L, Remy MH, Bazin I, Callebaut I, Wright M, Merdes A. NEDD1-dependent recruitment of the gamma-tubulin ring complex to the centrosome is necessary for centriole duplication and spindle assembly. J Cell Biol. 2006;172:505–515. doi: 10.1083/jcb.200510028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shang Y, Tsao CC, Gorovsky MA. Mutational analyses reveal a novel function of the nucleotide-binding domain of gamma-tubulin in the regulation of basal body biogenesis. J Cell Biol. 2005;171:1035–1044. doi: 10.1083/jcb.200508184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Garreau de Loubresse N, Ruiz F, Beisson J, Klotz C. Role of delta-tubulin and the C-tubule in assembly of Paramecium basal bodies. BMC Cell Biol. 2001;2:4. doi: 10.1186/1471-2121-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dupuis-Williams P, Fleury-Aubusson A, de Loubresse N, Geoffroy H, Vayssie L, Galvani A, Espigat A, Rossier J. Functional role of epsilon-tubulin in the assembly of the centriolar microtubule scaffold. J Cell Biol. 2002;158:1183–1193. doi: 10.1083/jcb.200205028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fromherz S, Giddings TH, Jr, Gomez-Ospina N, Dutcher SK. Mutations in alpha-tubulin promote basal body maturation and flagellar assembly in the absence of delta-tubulin. J Cell Sci. 2004;117:303–314. doi: 10.1242/jcs.00859. [DOI] [PubMed] [Google Scholar]
- 95.Callaini G, Whitfield WG, Riparbelli MG. Centriole and centrosome dynamics during the embryonic cell cycles that follow the formation of the cellular blastoderm in Drosophila. Exp Cell Res. 1997;234:183–190. doi: 10.1006/excr.1997.3618. [DOI] [PubMed] [Google Scholar]
- 96.Chretien D, Buendia B, Fuller SD, Karsenti E. Reconstruction of the centrosome cycle from cryoelectron micrographs. J Struct Biol. 1997;120:117–133. doi: 10.1006/jsbi.1997.3928. [DOI] [PubMed] [Google Scholar]
- 97.Rattner JB, Phillips SG. Independence of centriole formation and DNA synthesis. J Cell Biol. 1973;57:359–372. doi: 10.1083/jcb.57.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Azimzadeh J, Hergert P, Delouvee A, Euteneuer U, Formstecher E, Khodjakov A, Bornens M. hPOC5 is a centrin-binding protein required for assembly of full-length centrioles. J Cell Biol. 2009;185:101–114. doi: 10.1083/jcb.200808082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Morrissette NS, Sibley LD. Cytoskeleton of apicomplexan parasites. Microbiol Mol Biol Rev. 2002;66:21–38. doi: 10.1128/MMBR.66.1.21-38.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Baker JD, Adhikarakunnathu S, Kernan MJ. Mechanosensory-defective, male-sterile unc mutants identify a novel basal body protein required for ciliogenesis in Drosophila. Development. 2004;131:3411–3422. doi: 10.1242/dev.01229. [DOI] [PubMed] [Google Scholar]
- 101.Chen Z, Indjeian VB, McManus M, Wang L, Dynlacht BD. CP110, a cell cycle-dependent CDK substrate, regulates centrosome duplication in human cells. Dev Cell. 2002;3:339–350. doi: 10.1016/s1534-5807(02)00258-7. [DOI] [PubMed] [Google Scholar]
- 102.Spektor A, Tsang WY, Khoo D, Dynlacht BD. Cep97 and CP110 suppress a cilia assembly program. Cell. 2007;130:678–690. doi: 10.1016/j.cell.2007.06.027. [DOI] [PubMed] [Google Scholar]
- 103.Schmidt TI, Kleylein-Sohn J, Westendorf J, Le Clech M, Lavoie SB, Stierhof YD, Nigg EA. Control of centriole length by CPAP and CP110. Curr Biol. 2009;19:1005–1011. doi: 10.1016/j.cub.2009.05.016. [DOI] [PubMed] [Google Scholar]
- 104.Tang CJ, Fu RH, Wu KS, Hsu WB, Tang TK. CPAP is a cell-cycle regulated protein that controls centriole length. Nat Cell Biol. 2009;11:825–831. doi: 10.1038/ncb1889. [DOI] [PubMed] [Google Scholar]
- 105.Huang B, Mengersen A, Lee VD. Molecular cloning of cDNA for caltractin, a basal body-associated Ca2+-binding protein: homology in its protein sequence with calmodulin and the yeast CDC31 gene product. J Cell Biol. 1988;107:133–140. doi: 10.1083/jcb.107.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cormier A, Clement MJ, Knossow M, Lachkar S, Savarin P, Toma F, Sobel A, Gigant B, Curmi PA. The PN2-3 domain of centrosomal P4.1-associated protein implements a novel mechanism for tubulin sequestration. J Biol Chem. 2009;284:6909–6917. doi: 10.1074/jbc.M808249200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Singla V, Romaguera-Ros M, Garcia-Verdugo JM, Reiter JF. Ofd1, a human disease gene, regulates the length and distal structure of centrioles. Dev Cell. 2010;18:410–424. doi: 10.1016/j.devcel.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ferrante MI, Giorgio G, Feather SA, Bulfone A, Wright V, Ghiani M, Selicorni A, Gammaro L, Scolari F, Woolf AS, et al. Identification of the gene for oral-facial-digital type I syndrome. Am J Hum Genet. 2001;68:569–576. doi: 10.1086/318802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Coene KL, Roepman R, Doherty D, Afroze B, Kroes HY, Letteboer SJ, Ngu LH, Budny B, van Wijk E, Gorden NT, et al. OFD1 is mutated in X-linked Joubert syndrome and interacts with LCA5-encoded lebercilin. Am J Hum Genet. 2009;85:465–481. doi: 10.1016/j.ajhg.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ferrante MI, Zullo A, Barra A, Bimonte S, Messaddeq N, Studer M, Dolle P, Franco B. Oral-facial-digital type I protein is required for primary cilia formation and left-right axis specification. Nat Genet. 2006;38:112–117. doi: 10.1038/ng1684. [DOI] [PubMed] [Google Scholar]
- 111.Pearson CG, Osborn DP, Giddings TH, Jr, Beales PL, Winey M. Basal body stability and ciliogenesis requires the conserved component Poc1. J Cell Biol. 2009;187:905–920. doi: 10.1083/jcb.200908019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Paintrand M, Moudjou M, Delacroix H, Bornens M. Centrosome organization and centriole architecture: their sensitivity to divalent cations. J Struct Biol. 1992;108:107–128. doi: 10.1016/1047-8477(92)90011-x. [DOI] [PubMed] [Google Scholar]
- 113.Ibrahim R, Messaoudi C, Chichon FJ, Celati C, Marco S. Electron tomography study of isolated human centrioles. Microsc Res Tech. 2009;72:42–48. doi: 10.1002/jemt.20637. [DOI] [PubMed] [Google Scholar]
- 114.Geimer S, Melkonian M. The ultrastructure of the Chlamydomonas reinhardtii basal apparatus: identification of an early marker of radial asymmetry inherent in the basal body. J Cell Sci. 2004;117:2663–2674. doi: 10.1242/jcs.01120. [DOI] [PubMed] [Google Scholar]
- 115.Paoletti A, Moudjou M, Paintrand M, Salisbury JL, Bornens M. Most of centrin in animal cells is not centrosome-associated and centrosomal centrin is confined to the distal lumen of centrioles. J Cell Sci. 1996;109:3089–3102. doi: 10.1242/jcs.109.13.3089. [DOI] [PubMed] [Google Scholar]
- 116.Geimer S, Melkonian M. Centrin scaffold in Chlamydomonas reinhardtii revealed by immunoelectron microscopy. Eukaryot Cell. 2005;4:1253–1263. doi: 10.1128/EC.4.7.1253-1263.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Stemm-Wolf AJ, Morgan G, Giddings TH, Jr, White EA, Marchione R, McDonald HB, Winey M. Basal body duplication and maintenance require one member of the Tetrahymena thermophila centrin gene family. Mol Biol Cell. 2005;16:3606–3619. doi: 10.1091/mbc.E04-10-0919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ruiz F, Garreau de Loubresse N, Klotz C, Beisson J, Koll F. Centrin deficiency in Paramecium affects the geometry of basal-body duplication. Curr Biol. 2005;15:2097–2106. doi: 10.1016/j.cub.2005.11.038. [DOI] [PubMed] [Google Scholar]
- 119.Salisbury JL, Suino KM, Busby R, Springett M. Centrin-2 is required for centriole duplication in mammalian cells. Curr Biol. 2002;12:1287–1292. doi: 10.1016/s0960-9822(02)01019-9. [DOI] [PubMed] [Google Scholar]
- 120.Steinkötter J. PhD thesis. Mathematisch-Naturwissenschaftliche Fakultät; Cologne: 1997. Centrin und centrinbindende Proteine in grünalgen. [Google Scholar]
- 121.Azimzadeh J, Bornens M. The centrosome in evolution. In: Nigg EA, editor. Centrosomes in Development and Disease. Weinheim: Wiley-VCH; 2004. pp. 93–122. [Google Scholar]
- 122.Kilmartin JV. Sfi1p has conserved centrin-binding sites and an essential function in budding yeast spindle pole body duplication. J Cell Biol. 2003;162:1211–1221. doi: 10.1083/jcb.200307064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ishikawa H, Kubo A, Tsukita S. Odf2-deficient mother centrioles lack distal/subdistal appendages and the ability to generate primary cilia. Nat Cell Biol. 2005;7:517–524. doi: 10.1038/ncb1251. [DOI] [PubMed] [Google Scholar]
- 124.Graser S, Stierhof YD, Lavoie SB, Gassner OS, Lamla S, Le Clech M, Nigg EA. Cep164, a novel centriole appendage protein required for primary cilium formation. J Cell Biol. 2007;179:321–330. doi: 10.1083/jcb.200707181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Piel M, Meyer P, Khodjakov A, Rieder CL, Bornens M. The respective contributions of the mother and daughter centrioles to centrosome activity and behavior in vertebrate cells. J Cell Biol. 2000;149:317–330. doi: 10.1083/jcb.149.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Guarguaglini G, Duncan PI, Stierhof YD, Holmstrom T, Duensing S, Nigg EA. The forkhead-associated domain protein Cep170 interacts with Polo-like kinase 1 and serves as a marker for mature centrioles. Mol Biol Cell. 2005;16:1095–1107. doi: 10.1091/mbc.E04-10-0939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Beech P, Heimann K, Melkonian M. Development of the flagellar apparatus during the cell cycle in unicellular algae. Protoplasma. 1991;164:23–37. [Google Scholar]
- 128.Paoletti A, Bordes N, Haddad R, Schwartz C, Chang F, Bornens M. Fission yeast cdc31p is a component of the half-bridge and controls SPB duplication. Mol Biol Cell. 2003;14:2793–2808. doi: 10.1091/mbc.E02-10-0661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.O’Toole ET, Giddings TH, McIntosh JR, Dutcher SK. Three-dimensional organization of basal bodies from wild-type and delta-tubulin deletion strains of Chlamydomonas reinhardtii. Mol Biol Cell. 2003;14:2999–3012. doi: 10.1091/mbc.E02-11-0755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Bornens M. Centrosome composition and microtubule anchoring mechanisms. Curr Opin Cell Biol. 2002;14:25–34. doi: 10.1016/s0955-0674(01)00290-3. [DOI] [PubMed] [Google Scholar]