Abstract
In eukaryotic cells, proteins and RNA are transported between the nucleus and the cytoplasm by nuclear import and export receptors. Over the past decade, small molecules that inhibit the nuclear export receptor CRM1 have been identified, most notably leptomycin B. However, up to now no small molecule inhibitors of nuclear import have been described. Here we have used our automated Confocal Nanoscanning and bead picking method (CONA) for on-bead screening of a one bead/one compound library to identify the first such import inhibitor, karyostatin 1A. Karyostatin 1A binds importin β with high nanomolar affinity and specifically inhibits importin α/β mediated nuclear import at low micromolar concentrations in vitro and in living cells, without perturbing transportin mediated nuclear import or CRM1 mediated nuclear export. Surface plasmon resonance binding experiments suggest that karyostatin 1A acts by disrupting the interaction between importin β and the GTPase Ran. As a selective inhibitor of the importin α/β import pathway, karyostatin 1A will provide a valuable tool for future studies of nucleocytoplasmic trafficking.
Keywords: Confocal nanoscanning-bead picking, on-bead screening, one-bead one-compound libraries, High-throughput screening, importin beta, nuclear import inhibitors
Introduction
The nuclear envelope (NE) of eukaryotic cells is the membrane barrier that separates the nuclear events of DNA replication and mRNA biogenesis from cytoplasmic processes such as protein production and metabolism (1–2). Nucleocytoplasmic communication involves bidirectional, signal-dependent transport of RNAs and proteins across the NE through the nuclear pore complexes (NPCs), which are ~100 MDa protein assemblies comprising multiple copies of ~30 nucleoporins (Nups). Macromolecular trafficking across the NPC is mediated by transport receptors called karyopherins, or importins and exportins (3–4), which typically recognize linear stretches of amino acids called nuclear localization sequences (NLSs) and nuclear export sequences (NESs) on their protein cargoes. The β karyopherin protein family has over 20 members in vertebrates, including the exportin CRM1 and the importins importin β and transportin (5).
Importin α/β mediated nuclear import first involves recognition of NLSs by the adaptor importin α and binding of the latter to importin β through its N-terminal IBB domain (6). Subsequently the ternary cargo/receptor complex moves through the NPC via repeated transient interactions of importin β with phenylalanine-glycine (FG) repeat motifs present on certain Nups, culminating with its binding to Nup153 on the nucleoplasmic side of the NPC. Here, complex dissociation is thought to occur by the binding of RanGTP to importin β, which causes release of the IBB domain by an allosteric mechanism (7). The RanGTP:importin β complex then translocates into the cytoplasm and upon GTP hydrolysis, importin β is released for another round of nuclear import. The directionality of nucleocytoplasmic import is defined by nucleocytoplasmic compartmentalizion of the GEF (guanine exchange factor) and GAP (GTPase activating protein) that regulate the nucleotide state of Ran, with GTP hydrolysis providing the underlying energy that drives cargo concentration (2, 8).
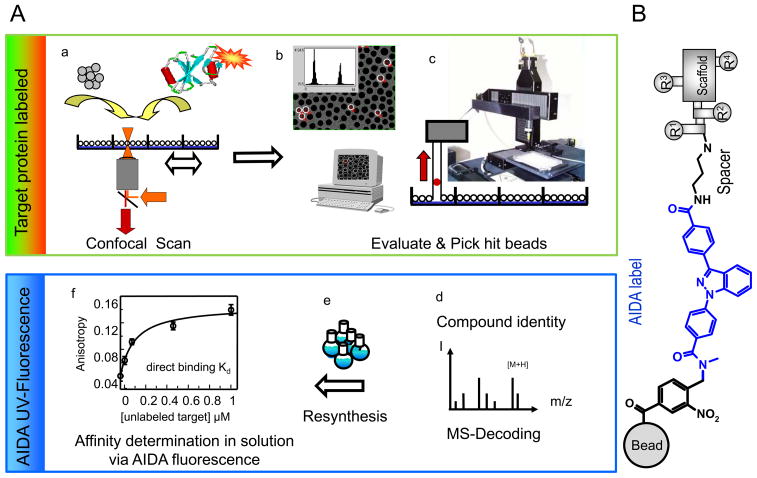
Confocal Nanoscanning (CONA) is an automated screening method developed for large one-bead one-compound libraries (Figure 1) (9). In this method, the binding of a fluorescently labeled protein to bead immobilized compounds is quantitatively detected with high optical resolution by large area confocal scanning of a bead monolayer in the wells of a microtiter plate. Beads that are positive for binding are robotically selected, and the compounds together with a tag comprising the UV-dye 4-{3-{4-[(3-Aminopropyl)-aminocarbonyl]-phenyl}-1H-indazol-1yl}-benzoic Acid (AIDA) are cleaved from the beads, identified by mass spectrometry, resynthesized, and used for binding studies via AIDA fluorescence. Fluorescence intensity profile analysis of beads offers high sensitivity detection of target protein-compound interactions on the bead surface and allows the exclusion of beads with strong autofluorescence. AIDA does not interfere with visible range fluorescent dyes and has a distinctive fragmentation pattern in MS that facilitates decoding of hit-compounds.
Figure 1. Affinity based screening of AIDA-tagged one-bead one-compound libraries by Confocal Nanoscanning (CONA) and fluorescence based secondary assays.
A) The screening process is performed in six steps (a–f): a) starting with distribution of 1 mg of resin from each AIDA-tagged sublibrary into the wells of a 96-well microtiter plate, followed by incubation with fluorescently tagged target protein. b) Automated confocal nanoscanning (CONA) identifies relevant hit-beads where the target protein has bound to compounds on the bead surface. c) Hit beads are isolated by the bead-picking device of the CONA screening instruments and the compounds are cleaved from the resin. d) MS-analysis of the hit-compounds allows structure assignment for each hit bead. e) The identified hits are re-synthesized in mg quantities with and without the AIDA tracer. f) The fluorescence from the UV-dye AIDA is used in a generic secondary assay to quantify the affinity of the hit-compounds for the target protein and for compound ranking. Note: This process falls into two phases: a phase where the green-red fluorescence on the target protein is first used to identify hits by on-bead screening (green box) and a second phase where the UV-signal from the tracer molecule is used quantify the obtained primary hits in a generic secondary assay (blue box).
B) Library set-up: one-bead one-compound libraries were synthesized on 90 μm TentaGel beads, using a photo-cleavable linker as attachment site (black), followed by a chemically robust UV-tracer “AIDA” (blue), a 3-carbon atom spacer (black) and the actual screening compound. The screening compounds are built around a central scaffold, decorated with four combinatorial sites. According to the split-mix-and-divide synthesis protocol used, the last two combinatorial sites are identical for each compound in any one sublibrary.
The identification of CRM1 (exportin 1) as the protein target for the potent anti-tumor natural product leptomycin B (LMB) has spurred substantial interest in targeting the nuclear export and import machinery for drug discovery (10–15). LMB also has proved to be extremely useful for the analysis of nucleocytoplasmic transport of hundreds of endogenous, as well as viral proteins (16–17). Recent work has described additional small molecules that target CRM1 and inhibit nuclear export, specifically N-acetylacrylates (18). Also, a high affinity substrate-like peptide of 26 amino acids has been developed recently that binds to importin α and inhibits importin α/β mediated nuclear import and cell growth (19). However, to date no small molecule inhibitors of nuclear import have been described.
Here, we have identified a cohort of importin β-binding small molecules with a CONA screen. From this group, we have characterized one compound that selectively inhibits the importin α/β pathway in permeabilized and live cell nuclear import assays. We provide evidence that this compound, which we term karyostatin 1A, may function by inhibiting the binding of RanGTP to importin β. This compound should be useful for future structural and functional studies of nuclear import.
Results and Discussion
On-bead screening by Confocal Nanoscanning (CONA)
Importin β is a major β-karyopherin whose interactions with several components of the nuclear import machinery have been biochemically and structurally characterized. To explore the potential of inhibiting importin β function with small molecule high affinity binders, we performed a CONA on-bead screen (Figure 1) with Alexa-488 labeled importin β (9, 20–23) (Protein labelling and characterization are described in the supporting information text and in Figures S1–S4). We screened a 45,600 compound containing diversity-optimized subset from our much larger stock of one bead, one compound libraries (2.2 million compounds). The screened compound collection consisted of 96 sublibraries, each containing approximately 500 members, and each of which was based on heterocyclic scaffolds such as pyrroles, thiazoles, indoles, amino-prolines, etc. All compounds from one individual sublibrary contain the same building block in the last two combinatorial positions (R3 and R4; Figure 1, panel B, Figure S5, supporting information).
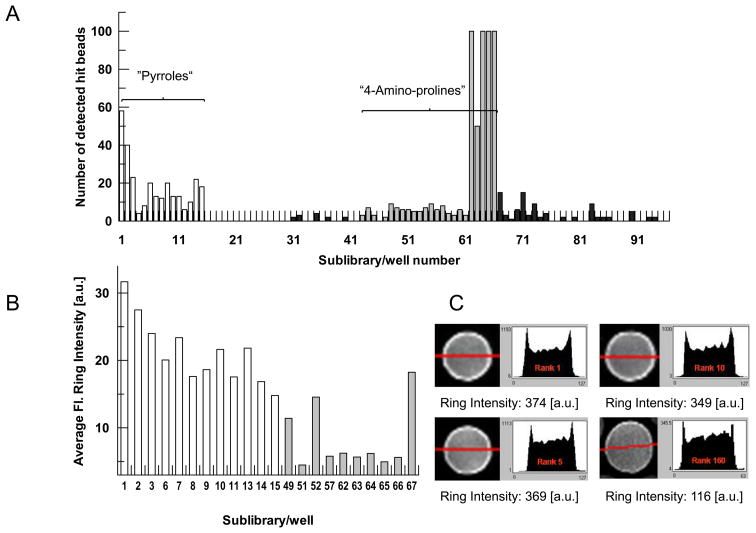
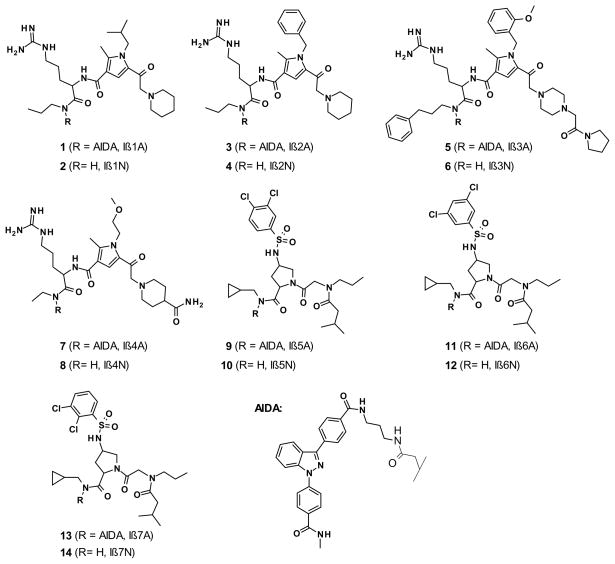
Using 60 nM Alexa-488 labeled importin β for the bead incubation, on-bead screening by CONA detected 928 hit beads, corresponding to a rather high hit-rate of 2% (Figure 2, panel A). The hits were clustered in sublibraries containing two scaffolds: pyrroles and 4-amino-prolines. All hit beads were ranked according to the intensity of fluorescently labeled target protein bound to the compounds on the bead surface (Figure 2, panels B and C). The 235 top ranked beads were then isolated using the bead picking feature of the PickoScreen instrument (Table S1 and Figure S6, supporting information). 153 of these contained pyrrole compounds and the remaining 82 hit beads came from 4-aminoproline sublibraries. Using HPLC-MS2 based structure elucidation, compound structures were successfully assigned to all isolated hit beads (Figures S7 and Figure S8, supporting information). Moreover, in the pyrrole subset, 17 compounds were identified twice (doublet), 7 compounds three times (triplet) and one compound four times (quadruplet), leaving 96 unique structures. The aminoproline subset of beads contained only 5 duplicates and 72 unique structures. Building block frequency analysis revealed a strong preference for either arginine or 4-aminoproline in the second combinatorial position (R2; Figure 1B) of the pyrrole hits and no clear preference in the first combinatorial position (R1; Figure 1B) (Figure S9, Supporting Information). The same building block frequency analysis for aminoprolines showed a preference for benzylamine in the first combinatorial position and a slight preference for chlorobenzylsulfonyl residues in the second combinatorial position (Figure S9, Supporting Information). Based on these decoding results, we resynthesized larger quantities of four pyrrole and three amino-proline AIDA/non-AIDA compound pairs, i.e., hit compounds with AIDA (compounds 1, 3, 5, 7, 9, 11, 13; designated Iβ1A to Iβ7A,) and without AIDA (compounds 2, 4, 6, 8, 10, 11, 14; designated Iβ1N to Iβ7N,) (Figure 3; Syntheses and characterization are described in the Supporting Information).
Figure 2. Primary on-bead screening analysis.
A) Distribution of the number of hit-beads over one 96-well screening plate containing a diversity optimized subset of AIDA-tagged one-bead one-compound libraries. Each well number represents one sublibrary.
B) Quantitative analysis of relative fluorescence ring intensities of hit-beads from pyrrole and amino-proline containing wells. Fluorescence ring intensity, is the quantitative parameter, indicating the amount of fluorescently tagged target protein which has bound to the bead-immobilized compounds.
C) Four exemplary bead images with their corresponding fluorescence ring intensities and ranking.
Figure 3. Re-synthesized CONA derived hit compounds for importin β.
Based on the MS-analysis of the individual hit-compounds and the building block-frequency analysis seven hit-compounds were selected for re-synthesis with (Iβ1A to Iβ7A) and without the AIDA tag (Iβ1N to Iβ7N) for further investigations in follow-up assays.
Confirmation of identified hit compounds
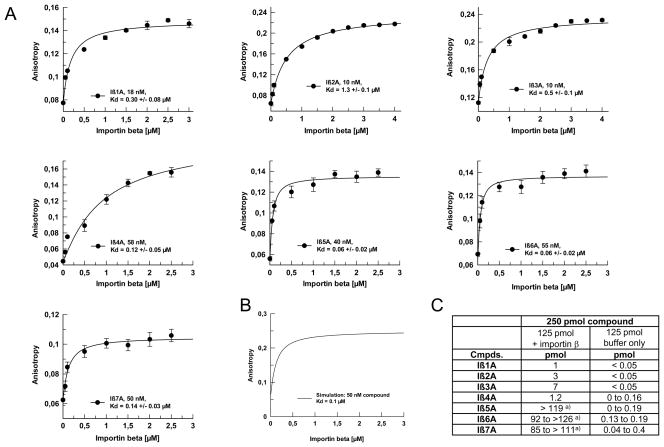
In a first confirmation step, we used the AIDA fluorescence of compounds Iβ1A to Iβ7A for measuring their affinities (Kd) for unlabeled importin β by fluorescence anisotropy (Figure 4). Using an average value of 0.73 ml/g for the specific protein and compound volume, the calculated start (free) and end (protein bound) anisotropy values for compounds Iβ1A to Iβ7A were 0.06 and 0.25, respectively (Figure 4, panel B, Supporting Information). The experimentally observed starting anisotropy values for compounds Iβ1A to Iβ7A were in good agreement with calculated values. Non-linear curve fitting based on a 1:1 binding stoichiometry of the the fluorescence anisotropy titration data resulted in Kd values ranging from 320 +/− 80 nM to 1.3 +/− 0.1 μM for the pyrrole compounds, Iβ1A to Iβ4A, (Figure 4, panel A). The affinities of the 4-aminoproline compounds, Iβ5A to Iβ7A, were generally higher and the determined Kds ranged from 60 +/− 20 nM to 140 +/− 30 nM. Two unrelated control compounds, containing AIDA and the spacer unit were also tested in the direct binding assay and did not produce any measurable affinity up to 4 μM of importin β (Figures S11, supporting information).
Figure 4. Determination of binding affinities (Kds) of hit compounds Iβ1A to Iβ7A for importin β.
A) Anisotropy measurements using the AIDA-derived fluorescence signal were carried out with increasing concentrations of importin β. The resulting titration data was fitted to a 1:1 interaction model.
B) Simulation of expected start- and end-anisotropy values for a ligand with a molecular weight of 1,000 Da and a globular shaped protein of 100 kDa, using the Perrin equation.
C) HPLC-quantified recoveries of AIDA-tagged hit compounds after size-exclusion chromatography experiments in presence and absence of a saturating amount of importin β.
In an attempt to measure the binding of the non-AIDA tagged compounds to importin β, we carried out competition titrations with the non-AIDA tagged compounds Iβ1N to Iβ7N. However, technical difficulties precluded robust conclusions from this method. When the binding of importin β to the AIDA tagged compounds Iβ1A to Iβ7A was monitored in presence of increasing amounts of the untagged compounds Iβ1N to Iβ7N, aggregation effects were observed at compound concentrations higher than 100 μM. In addition, high concentrations of non-AIDA compounds Iβ1N to Iβ7N yielded auto-fluorescence signals, further impairing a quantitative interpretation.
However, the activities of the AIDA-tagged compounds Iβ1A to Iβ7A were further confirmed by an adaptation of a previously described size-exclusion chromatography assay (24), in which compounds Iβ1A to Iβ7A were centrifuged through a size-exclusion matrix in presence or absence of importin β. The compound concentration in the filtrate after centrifugation was determined by HPLC (Figure 4, panel C). For all seven re-synthesized compounds, the recovery in the presence of importin β consistently exceeded the recovery without protein. The three 4-aminoproline compounds, however, showed higher recoveries (more than 68%), as compared to the pyrrole compounds (1–5%).
Iβ1A to Iβ4A specifically inhibit importin α/β mediated nuclear import in vitro
We next examined whether the importin β binding compounds Iβ1A to Iβ7A inhibited importin β mediated nuclear import in vitro. These compounds along, with the non AIDA compounds Iβ1N to Iβ7N, were tested in an assay involving permeabilized HeLa cells reconstituted with recombinant transport factors and FITC labeled NLS-conjugated BSA (FITC-BSA-NLS), an importin α/β dependent cargo (25). The AIDA-conjugated pyrrole compounds Iβ1A to Iβ4A, when tested at a concentration of 10 μM, inhibited importin β mediated nuclear import by 36% to 79% (Figure 5, panel A). However, neither the AIDA-tagged 4-aminoproline compounds Iβ5A to Iβ7A nor the non-AIDA compounds Iβ1N to Iβ7N showed significant inhibition of nuclear import (Figure 5, panel A). Thus, AIDA seems to be an integral part of the pharmacophore of the importin β inhibitors Iβ1A to Iβ4A, in contrast to similarly identified HuR binding compounds (21). The amino-proline compounds Iβ5A to Iβ7A, which do not inhibit import, might bind to importin β in a manner that does not perturb the transport functions measured in this assay. Alternatively, the higher hydrophobicity of the amino-proline compounds as compared to the pyrroles might cause sequestration of the former in the assay by binding to other cellular components.
Figure 5. Effect of compounds Iβ1A to Iβ4A on in vitro nuclear import using permeabilized cells.
Importin α/β mediated in vitro nuclear import using recombinant transport factors. Transport factors were added to permeabilized HeLa suspension cells together with cargo, energy and compounds. DMSO concentration was kept at 1% across the samples. Following a 30-minute reaction, nuclear fluorescence was analyzed by flow cytometry. The data points represent the average of 3 to 7 independent experiments. A. Compounds Iβ1A to Iβ7A and Iβ1N to Iβ7N were added at 10 μM final concentration together with FITC-BSA-NLS cargo and recombinantly expressed transport factors. B. Compounds Iβ1A to Iβ3A were added at 10 μM, 3.3 μM and 1 μM concentrations with FITC-BSA-NLS cargo and recombinantly expressed transport factors. C. Compounds Iβ1A to Iβ4A were added at 10 μM final concentration together with FITC-BSA-NLS cargo and cytosol as a source of nuclear transport factors. WGA: wheat germ agglutinin. D. Nuclear import assay with FITC-BSA-NLS cargo and recombinantly expressed transport factors. Compounds Iβ1A to Iβ4A were added at 10 μM final concentration only after the nuclear import reactions were terminated by hexokinase/glucose, followed by 30 minutes of incubation at 30°C to test for the loss of intranuclear FITC-BSA-NLS. E. Transportin mediated nuclear import using recombinant transport factors. Compounds Iβ1A to Iβ4A were added at 10 μM final concentration to permeabilized HeLa suspension cells together with FITC-M9-nucleoplasmin cargo.
We examined the concentration dependence for inhibition of importin α/β mediated nuclear import by the three most active compounds Iβ1A to Iβ3A, and found IC50 values ranging from 5 μM to 9 μM (Figure 5, panel B). We also examined transport inhibition in permeabilized cells reconstituted with cytosol instead of recombinant transport factors. In this case, significant inhibition of importin α/β mediated nuclear import by 10 μM compound was observed for Iβ1A to Iβ3A (27–46% inhibition) but was not seen for Iβ4A. The weaker inhibition in this assay might be due to additional promiscuous interactions of these compounds with other cytosolic components (Figure 5, panel C).
We also determined that inhibition of cargo accumulation with Iβ1A to Iβ4A does not result from nonspecific effects on the permeability barrier of the NE, since addition of the compounds following a nuclear import reaction did not lead to a reduction in the level of imported cargo (Figure 5, panel D). The effect of Iβ1A to Iβ4A on transportin mediated nuclear import was also examined. Transportin, although structurally similar to importin β, recognizes different cargoes, such as the M9 sequence comprising the NLS of hnRNP A1. Despite the homology of the nuclear import receptors transportin and importin β, none of the four compounds Iβ1A to Iβ4A inhibited transportin-mediated nuclear import at a compound concentration of 10 μM in the permeabilized cell nuclear import assay using recombinant transport factors (Figure 5, panel E). This suggests that pyrrole compounds Iβ1A–Iβ4A target importin β specifically rather than the Ran system, which underlies all karyopherin-mediated nucleocytoplasmic transport.
Structure-activity relationships
The frequency with which a specific building block occurred in a combinatorial position within the primary hit structures gave a first indication of structure-activity relationships for the identified hit compounds. Based on these results, we designed and tested a set of 10 additional pyrrole compounds, Iβ8A to Iβ18A, containing a variety of residues in the four combinatorial positions (Figure S10, supporting information). Five of these were strong inhibitors (> 50% inhibition) in the in vitro nuclear import assay and confirmed the dominant role of arginine in the second combinatorial position already seen from set of primary hits. An N-ethylcyclohexylamine in the fourth combinatorial position proved to be a second crucial element. (For more detailed interpretation of the SAR results, see Supporting Information).
Iβ1A inhibits importin α/β mediated nuclear import in living cells
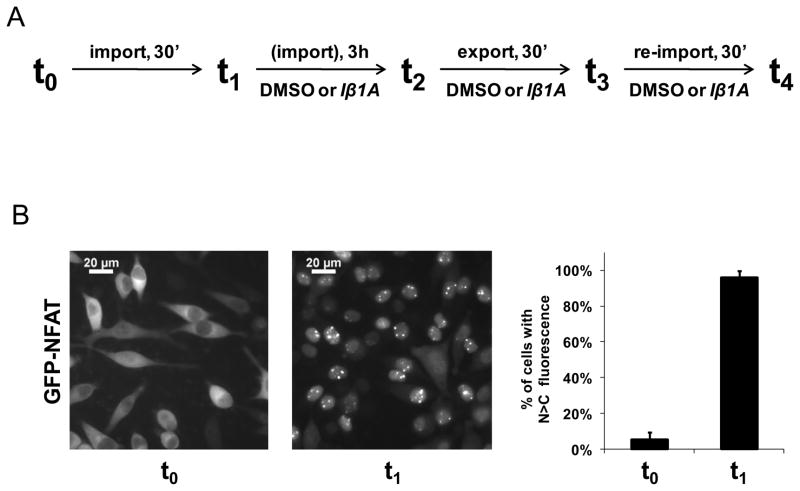
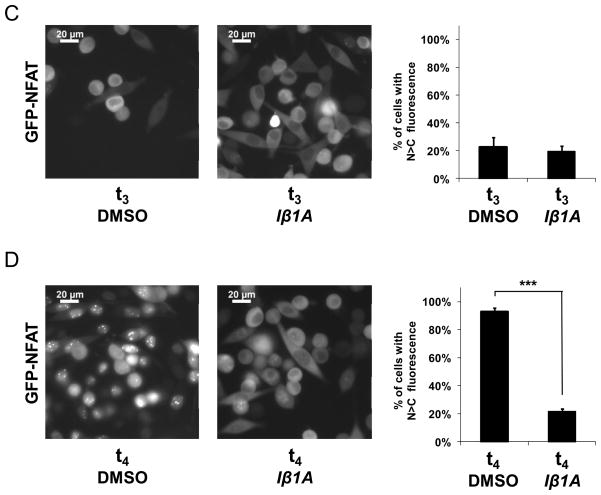
Iβ1A, the pyrrole compound exhibiting the best combination of solubility and in vitro inhibitory characteristics, was further tested in living cells for its effects on the nuclear import and export of a well-characterized karyopherin cargo, green fluorescent protein fused to nuclear factor of activated T-cells (GFP-NFAT) (26–27) (Figure 6, panel A). 24 hours after the induction of its expression, GFP-NFAT had accumulated to substantial levels in the cytoplasm of HeLa cells (Figure 6, panel B). Nuclear import of GFP-NFAT was induced by the addition of ionomycin. After 30 min, GFP-NFAT localized to the nucleus in close to 100% of the cells (Figure 6, panel B). DMSO or 25 μM Iβ1A were then incubated with cells for 3 h, and subsequently, nuclear export was induced by incubating cells with medium lacking ionomycin but containing either 25 μM Iβ1A or DMSO for 30 min. GFP-NFAT was exported to the cytoplasm in the presence of Iβ1A as completely as with DMSO (Figure 6, panel C). Ionomycin was then re-administered to the same cells. In the presence of 25 μM Iβ1A, there was inhibition of the re-import of GFP-NFAT in about 80% of the cells but not in the control (Figure 6, panel D). Together, these data show that 25 μM Iβ1A significantly inhibits importin α/β mediated nuclear import but not CRM1 mediated nuclear export of GFP-NFAT in living cells. We cannot, however, exclude the possibility that Iβ1A interferes with one or more of the other members of the β-karyopherin family in vivo.
Figure 6. Effect of Iβ1A on nucleocytoplasmic export and import of GFP-NFAT in living cells.
A. Schematic representation of the experimental timeline to test the in vivo effect of Iβ1A on nuclear export and import of GFP-NFAT in HeLa cells. “Import” or “re-import” denotes conditions where nuclear import of GFP-NFAT is triggered with the addition of 1 μM ionomycin to the cells. “Export” denotes conditions where the ionomycin is washed out with cell culture medium. B. Cytoplasmic localization of GFP-NFAT from untreated, stably transfected HeLa cells and its nuclear translocation 30 minutes after inducing import. C. Localization of GFP-NFAT in the presence of either 0.25% DMSO or 25 μM Iβ1A 30 minutes after nuclear export was induced. A three-hour incubation period with DMSO or Iβ1A in the presence of ionomycin preceded the initiation of export. D. Localization of GFP-NFAT in the presence of either 0.25% DMSO or 25 μM Iβ1A 30 minutes after nuclear re-import of GFP-NFAT was induced with ionomycin on the same cells that underwent export in C.
Images in B., C. and D. were collected of HeLa cells expressing GFP-NFAT by fluorescence microscopy. The graphs depict the percentage of cells with predominantly nuclear GFP-NFAT, as determined by visual inspection of at least 200 cells for each condition.
Concentrations higher than 25 μM of Iβ1A (50–100 μM) triggered Iβ1A aggregation and precipitation in the cell culture medium. Lower concentrations of Iβ1A yielded weaker inhibition of nuclear import. Inhibition was seen in about 22% of the cells at 12.5 μM compound, but was not seen at 6.2 μM and 3.1 μM Iβ1A (data not shown). We also analyzed compound Iβ3A in this assay but due to its lower solubility, no inhibitory effect could be detected at any tested concentration (data not shown).
Iβ1A disrupts the binding of importin β to RanGTP
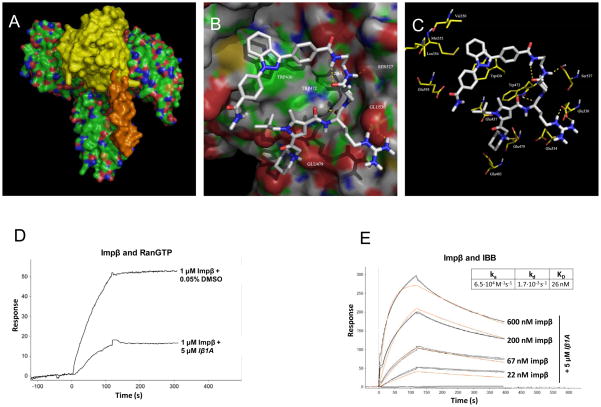
To rationalize the mode of action of our compounds and the observed SAR we performed molecular modeling and docking studies with the best compound Iβ1A. First, possible binding sites were identified using the program STP (http://opus.bch.ed.ac.uk/stp) and a computational protocol was designed to probe possible binding orientations of Iβ1A using FRED (http://www.eyesopen.com/), followed by fully flexible docking with RosettaLigand (http://www.rosettacommons.org/). The only suitable small molecule binding site, identified by STP is located in the interface between importin alpha and RanGTP (Figure 7A). According to the docking results, Iβ1A makes crucial pi-stacking interactions with Trp430 as well as forming a series of hydrogen bonds, most notably the arginine in Iβ1A interacts with Glu530 in importin β, explaining the preference observed for this specific residue in the second combinatorial position (Figure 7B, C).
Figure 7. Molecular modeling and surface plasmon resonance measurements of the Iβ1A-importin beta interaction.
A. Interaction between importin β (green, coloured by atom type), importin α (orange) and Ran-GTP (yellow).
B. Docked structure of Iβ1A within its identified binding site Importin β as obtained by fully flexible protein-ligand docking using the program RosettaLigand (http://www.rosettacommons.org/).
C. Pharmacophore model derived from fully flexible molecular docking studies.
D. Binding of importin β at 1 μM concentration to GST-RanQ69LGTP non-covalently immobilized to a GST antibody chip in the presence of 5 μM Iβ1A or 0.05% DMSO. Binding of importin β at various concentrations in the presence of 5 μM Iβ1A to GST-IBB (E.) non-covalently immobilized to a GST antibody chip. Black lines represent actual data collected in duplicates and red lines are theoretical simulations derived from global fit on the dataset.
Inspired by the docking studies we turned to surface plasmon resonance (SPR) for measuring the effect of Iβ1A on the interaction of importin β with three of its binding partners (GST-tagged): a Ran mutant incapable of GTP hydrolysis (RanQ69LGTP), the C-terminal region of Nup153 (amino acids 895–1,475) and the IBB (Importin Beta Binding) domain of importin α (Figure 7 panels D, E and Figure S12 and S13, supporting information). GST-RanQ69LGTP, GST-Nup153, and GST-IBB were non-covalently immobilized on a Biacore sensor chip containing surface bound GST antibody. In the absence of Iβ1A, importin β bound to all three of its binding partners. The apparent Kds measured in the SPR experiments were about one order of magnitude higher than previously published values (28–30). We attribute this to the requirement for omitting strong reducing agents from the SPR assay, and the oxidation sensitive nature of importin β (31–32). The activity of importin β with our experimental conditions was nonetheless suitable for evaluating the effects of Iβ1A on binding to the various ligands. While Iβ1A at a concentration of 5μM had no effect on the binding of importin β to either GST-IBB or GST-Nup153-C (Figure 7, panel E and Figure S13 panel D), Iβ1A essentially abolished binding of importin β to GST-RanQ69LGTP, since with 1 μM importin β, an ~ 70% drop in response units was observed with 5 μM Iβ1A as compared to the DMSO control (Figure 7, panel D).
The strong inhibition of the interaction of importin β with RanGTP but not its interaction with the importin α IBB domain or with Nup153 by Iβ1A suggest that the most likely mechanism for inhibition of nuclear import by Iβ1A involves compromised dissociation of importin α/β/cargo complexes at the nuclear side of the NPC and a failure in cargo unloading into the nucleus, since these critically depend on binding of RanGTP to importin β (33). However, we cannot rule out other effects involving the binding of RanGTP to importin β, such as importin β recycling to the cytoplasm.
In summary, using Confocal Nanoscanning we have identified several structurally related novel small-molecule inhibitors for importin β mediated nuclear import that will provide valuable tools for further investigations. Our in vitro and in vivo results with Iβ1A are of particular importance as, along with peptidomimetic inhibitors recently identified in our laboratory (Ambrus G, Whitby LR, Singer EL et al., unpublished data), they provide the first bona fide small molecule inhibitor of importin α/β mediated import, and establish the chemical druggability of the importin β pathway for inducing biological effects. We suggest naming these nuclear transport inhibitors karyostatins and have therefore have designated Iβ1A, the prototype of importin β inhibitors, as karyostatin 1A. Future work will be directed towards optimization of these compounds. The currently available set of compounds, although containing valuable SAR information, shows a suboptimal behaviour in cellular assays due to the rather hydrophobic nature of AIDA as well as some of the building block combinations. Truncation of the compounds at the less sensitive positions and replacement of AIDA by less extended hydrophobic heterocycles may lead to further optimized compounds with better efficacy in cellular assays
Materials and Methods
On-bead screening by Confocal Nanoscanning and bead picking
A detailed description of the PS02 instrument and its application for on-bead screening by automated confocal nanoscanning has recently been published (9). For library screening, 1 mg of beads from the respective sublibraries were placed in 1.5 ml tubes (Eppendorf) and swollen in 200μl of screening buffer, containing 20 mM Hepes pH 7.4, 110 mM potassium acetate, 2 mM magnesium acetate, 0.2 mM DTT, 0.1 mM CHAPS, 2% DMSO 2% (v/v), 2% Acetonitrile 2% (v/v), 0.1% Pluronic (w/v) (Molecular Probes), 0.2% BSA (w/v) (Sigma). The samples were then vortexed, remaining bead clusters broken up by short sonication and the beads transferred to the wells of 96-well microtiter plate. The samples were incubated for 1 h with 200 μl of screening buffer containing 60 nM Alexa488 labeled importin β under constant agitation. After that the beads were left to settle for a 1 min to form a monolayer at the well bottom of the microtiter plate was placed on the sample holder of the PS02 instrument. Confocal nanoscanning was performed at 5 μm resolution, the confocal scan height was set to 25 μm above the well-bottom and the scanning was performed using the appropriate instrumental settings and filter sets for Alexa488 (488 nm Argon ion laser excitation, dichroic mirror 565DRLP, and emission filter 535RDF50).
The acquired scan images were quantitatively analyzed using the instrument’s BeadEval software (Perkin Elmer) and the beads were ranked according to their fluorescence ring intensities according to previously published procedures (9). Finally, the beads with the highest fluorescent ring intensities were isolated using the PS02’s bead picking device and placed into autosampler glass vials (8002-SC-H/i3μ, Glastechnik Gräfenroda).
Cleavage of isolated hit compounds by photolysis
For photolysis, 40 μl of 1% TFA/methanol (Merck) was added to each vial and the closed vials were placed in a Stratalinker 1800 UV illumination cabinet (Stratagene), and exposed for 120 minutes to 365 nm UV illumination at a power of 1070 μJ/min. After photolysis, the solvents were removed under vacuum and the compounds re-dissolved in 5 μl of 30% acetonitrile in water, containing 0.1 % TFA for μHPLC/MS analysis.
MS-analysis and decoding of hit-structures
The samples were analyzed on a μHPLC/MS instrument, consisting of a HP1100 HPLC system (Hewlett-Packard), equipped with a 30 nL flow cell UV detector (LC Packings) set to 214 nm and a LCQ ion trap mass spectrometer (Finnigan Corp) operated in MS mode for mass determination and in CID dependent scan mode for fragmentation of selected ions. 2.5 μl of each sample were injected and analyzed on a Hypersil C18 (5μm, 150×0.8 mm) column (Lc Packings) at a flow rate of 100μl/min and with a linear gradient from 10% to 95% acetonitrile in water containing 0.1 % TFA over 15 min. The analytes were passed into the electrospray source of the LCQ via a fused silica capillary (340 μm o.d.×50 μm i.d.). The source was operated at 4.5 kV with the heated capillary set at 220°C and sheath nitrogen gas flow rate at 80. In the MS mode the ion time was set at 500 ms and the target number of ions at 5×107; in the CID mode the ion time was at 500 ms and the target number of ions at 2×107. In both modes 3 microscans/spectrum were performed. The electron multiplier was set at −1000 V and all spectra were collected in the positive-ion mode.
Mass spectra analysis and interpretation of the individual LC separated peaks was performed using the Xcalibur software (Thermo Finnigan, Version 1.3). After identification of the monoisotopic mass of the apparent end product, structures were assigned using a lookup table containing the expected molecular masses as well as masses for the expected MS-fragment’s for each library compound.
Affinity measurements of AIDA-tagged hit compounds in solution
For affinity measurements in homogenous solution, the AIDA tagged hit compounds were dissolved in DMSO and subsequently diluted into assay buffer (20 mM Hepes pH 7.4, 110 mM potassium acetate, 2 mM magnesium acetate, 0.2 mM DTT, 0.1 mM CHAPS), keeping the final DMSO concentration at 5%. The titration of AIDA compounds were performed on a SPEX Fluorolog τ3 spectro- fluorometer in quartz cuvettes with 2 mm thickness and 10 mm path length (Hellma, 119.004F-QS) at 25° C. The polarization measurements (titrations) of the AIDA-tagged compounds were performed at excitation and emission wavelengths set to 336 nm and 397 nm, respectively using magic angle settings and an excitation bandwidth of 5 nm. The voltage of the emission channel PMTs was set to 950 V. A WG345 long-pass filter was used in the emission channel to reduce straylight influences. Measurements were performed in T-format. For each titration point, a cycle of 3 measurements was performed. Each cycle consisted of 9 repeated measurements at vertically and horizontally oriented polarizers with an integration time of 5 seconds. The iteration limit for the calculated anisotropy was set to 0.1%. The samples were titrated by adding increasing amounts of importin β into a solution containing ~ 50 nM of AIDA tagged compound. Equilibrium dissociation constants (Kd values) were obtained by performing a nonlinear least square regression fit of the anisotropy data sets, with the software package GraFit 5.0 and assuming a 1:1 binding model
Nuclear import assay using permeabilized cells
Nuclear import assays using permeabilized cells were carried out essentially as previously described (34). See Supporting Information for more details.
Nuclear import assay in living cells using stably transfected HeLa cells
The in vivo nuclear transport assay with GFP-NFAT stably transfected into HeLa cells have been carried out essentially as described before (26–27). See Supporting Information for more detail.
Protein expression and purification
Expression and purification of importin β, importin α, Ran, NTF2, transportin, M9-nucloplasmin (35), GST-IBB, GST-RanQ69L and GST-Nup153 (34) has been described previously (25, 36). GTP loading of GSTRanQ69L was carried out as described earlier. FITC-BSA-NLS (36) and FITC-M9-nucloplasmin (35) cargo coupling and labeling was done following previously published methods.
Surface plasmon resonance measurements
In the surface plasmon resonance experiments were carried out essentially as described previously (30). See Supporting Information for more detail.
Compound Characterization
For compound characterization see Supporting Information.
Supplementary Material
Acknowledgments
The authors would like to thank Gino Cingolani for his advice on surface plasmon resonance measurements. This work was supported by NIH grants GM041955 and NS059460 to LG, and CHRP postdoctoral fellowship F07-SRI-214 to GA.
References
- 1.Terry LJ, Shows EB, Wente SR. Crossing the nuclear envelope: hierarchical regulation of nucleocytoplasmic transport. Science. 2007;318:1412–1416. doi: 10.1126/science.1142204. [DOI] [PubMed] [Google Scholar]
- 2.Stewart M. Molecular mechanism of the nuclear protein import cycle. Nat Rev Mol Cell Biol. 2007;8:195–208. doi: 10.1038/nrm2114. [DOI] [PubMed] [Google Scholar]
- 3.Weis K. Importins and exportins: how to get in and out of the nucleus. Trends Biochem Sci. 1998;23:235. doi: 10.1016/s0968-0004(98)01204-3. [DOI] [PubMed] [Google Scholar]
- 4.Cook A, Bono F, Jinek M, Conti E. Structural biology of nucleocytoplasmic transport. Annu Rev Biochem. 2007;76:647–671. doi: 10.1146/annurev.biochem.76.052705.161529. [DOI] [PubMed] [Google Scholar]
- 5.Bednenko J, Cingolani G, Gerace L. Nucleocytoplasmic transport: navigating the channel. Traffic. 2003;4:127–135. doi: 10.1034/j.1600-0854.2003.00109.x. [DOI] [PubMed] [Google Scholar]
- 6.Gorlich D, Henklein P, Laskey RA, Hartmann E. A 41 amino acid motif in importin-alpha confers binding to importin-beta and hence transit into the nucleus. EMBO J. 1996;15:1810–1817. [PMC free article] [PubMed] [Google Scholar]
- 7.Lee SJ, Matsuura Y, Liu SM, Stewart M. Structural basis for nuclear import complex dissociation by RanGTP. Nature. 2005;435:693–696. doi: 10.1038/nature03578. [DOI] [PubMed] [Google Scholar]
- 8.Nachury MV, Weis K. The direction of transport through the nuclear pore can be inverted. Proc Natl Acad Sci U S A. 1999;96:9622–9627. doi: 10.1073/pnas.96.17.9622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hintersteiner M, Buehler C, Uhl V, Schmied M, Muller J, Kottig K, Auer M. Confocal Nanoscanning, Bead Picking (CONA): PickoScreen Microscopes for Automated and Quantitative Screening of One-Bead One-Compound Libraries. J Comb Chem. 2009;11:886–894. doi: 10.1021/cc900059q. [DOI] [PubMed] [Google Scholar]
- 10.Kudo N, Wolff B, Sekimoto T, Schreiner EP, Yoneda Y, Yanagida M, Horinouchi S, Yoshida M. Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Experimental Cell Research. 1998;242:540–547. doi: 10.1006/excr.1998.4136. [DOI] [PubMed] [Google Scholar]
- 11.Nishi K, Yoshida M, Fujiwara D, Nishikawa M, Horinouchi S, Beppu T. Leptomycin B targets a regulatory cascade of crm1, a fission yeast nuclear protein, involved in control of higher order chromosome structure and gene expression. J Biol Chem. 1994;269:6320–6324. [PubMed] [Google Scholar]
- 12.Kwon YJ, Genovesio A, Kim NY, Kim HC, Jung S, David-Watine B, Nehrbass U, Emans N. High-content classification of nucleocytoplasmic import or export inhibitors. J Biomol Screening. 2007;12:621–627. doi: 10.1177/1087057107301319. [DOI] [PubMed] [Google Scholar]
- 13.Wolff B, Sanglier JJ, Wang Y. Leptomycin B is an inhibitor of nuclear export: inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem Biol. 1997;4:139–147. doi: 10.1016/s1074-5521(97)90257-x. [DOI] [PubMed] [Google Scholar]
- 14.Yashiroda Y, Yoshida M. Nucleo-cytoplasmic transport of proteins as a target for therapeutic drugs. Curr Med Chem. 2003;10:741–748. doi: 10.2174/0929867033457791. [DOI] [PubMed] [Google Scholar]
- 15.Kau TR, Silver PA. Nuclear transport as a target for cell growth. Drug Discov Today. 2003;8:78–85. doi: 10.1016/s1359-6446(02)02562-x. [DOI] [PubMed] [Google Scholar]
- 16.Kobayashi T, Kamitani W, Zhang G, Watanabe M, Tomonaga K, Ikuta K. Borna disease virus nucleoprotein requires both nuclear localization and export activities for viral nucleocytoplasmic shuttling. Journal of Virology. 2001;75:3404–3412. doi: 10.1128/JVI.75.7.3404-3412.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar S, Saradhi M, Chaturvedi NK, Tyagi RK. Intracellular localization and nucleocytoplasmic trafficking of steroid receptors: an overview. Mol Cell Endocrinol. 2006;246:147–156. doi: 10.1016/j.mce.2005.11.028. [DOI] [PubMed] [Google Scholar]
- 18.Van Neck T, Pannecouque C, Vanstreels E, Stevens M, Dehaen W, Daelemans D. Inhibition of the CRM1-mediated nucleocytoplasmic transport by N-azolylacrylates: Structure-activity relationship and mechanism of action. Bioorg Med Chem. 2008;16:9487–9497. doi: 10.1016/j.bmc.2008.09.051. [DOI] [PubMed] [Google Scholar]
- 19.Kosugi S, Hasebe M, Entani T, Takayama S, Tomita M, Yanagawa H. Design of Peptide Inhibitors for the Importin alpha/beta Nuclear Import Pathway by Activity-Based Profiling. Chemistry & Biology (Cambridge, MA, United States) 2008;15:940–949. doi: 10.1016/j.chembiol.2008.07.019. [DOI] [PubMed] [Google Scholar]
- 20.Meisner NC, Hintersteiner M, Uhl V, Weidemann T, Schmied M, Gstach H, Auer M. The chemical hunt for the identification of drugable targets. Curr Opin Chem Biol. 2004/8;8:424–431. doi: 10.1016/j.cbpa.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 21.Meisner NC, Hintersteiner M, Seifert JM, Bauer R, Benoit Roger M, Widmer A, Schindler T, Uhl V, Lang M, Gstach H, Auer M. Terminal Adenosyl Transferase Activity of Posttranscriptional Regulator HuR Revealed by Confocal On-Bead Screening. J Mol Biol. 2009;386:435–450. doi: 10.1016/j.jmb.2008.12.020. [DOI] [PubMed] [Google Scholar]
- 22.Hintersteiner M, Auer M. Single-bead, single-molecule, single-cell fluorescence technologies for drug screening and target validation. Annals of the New York Academy of Sciences. 2008;1130:1–11. doi: 10.1196/annals.1430.055. [DOI] [PubMed] [Google Scholar]
- 23.Hintersteiner M, Kimmerlin T, Kalthoff F, Stoeckli M, Garavel G, Seifert JM, Meisner NC, Uhl V, Buehler C, Weidemann T, Auer M. Single Bead Labeling Method for Combining Confocal Fluorescence On-Bead Screening and Solution Validation of Tagged One-Bead One-Compound Libraries. Chemistry & Biology (Cambridge, MA, United States) 2009;16:724–735. doi: 10.1016/j.chembiol.2009.06.011. [DOI] [PubMed] [Google Scholar]
- 24.Muckenschnabel I, Falchetto R, Mayr LM, Filipuzzi I. SpeedScreen: label-free liquid chromatography-mass spectrometry-based high-throughput screening for the discovery of orphan protein ligands. Anal Biochem. 2004 Jan 15;324:241–249. doi: 10.1016/j.ab.2003.09.040. [DOI] [PubMed] [Google Scholar]
- 25.Melchior F, Sweet DJ, Gerace L. Analysis of Ran/TC4 function in nuclear protein import. Methods Enzymol. 1995;257:279–291. doi: 10.1016/s0076-6879(95)57032-2. [DOI] [PubMed] [Google Scholar]
- 26.Kehlenbach RH, Dickmanns A, Kehlenbach A, Guan T, Gerace L. A role for RanBP1 in the release of CRM1 from the nuclear pore complex in a terminal step of nuclear export. J Cell Biol. 1999;145:645–657. doi: 10.1083/jcb.145.4.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kehlenbach RH, Dickmanns A, Gerace L. Nucleocytoplasmic shuttling factors including Ran and CRM1 mediate nuclear export of NFAT In vitro. J Cell Biol. 1998;141:863–874. doi: 10.1083/jcb.141.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bischoff FR, Gorlich D. RanBP1 is crucial for the release of RanGTP from importin beta-related nuclear transport factors. FEBS Lett. 1997;419:249–254. doi: 10.1016/s0014-5793(97)01467-1. [DOI] [PubMed] [Google Scholar]
- 29.Ben-Efraim I, Gerace L. Gradient of increasing affinity of importin beta for nucleoporins along the pathway of nuclear import. J Cell Biol. 2001;152:411–417. doi: 10.1083/jcb.152.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mitrousis G, Olia AS, Walker-Kopp N, Cingolani G. Molecular basis for the recognition of snurportin 1 by importin beta. J Biol Chem. 2008;283:7877–7884. doi: 10.1074/jbc.M709093200. [DOI] [PubMed] [Google Scholar]
- 31.Adam SA, Gerace L. Cytosolic proteins that specifically bind nuclear location signals are receptors for nuclear import. Cell. 1991;66:837–847. doi: 10.1016/0092-8674(91)90431-w. [DOI] [PubMed] [Google Scholar]
- 32.Adam EJ, Adam SA. Identification of cytosolic factors required for nuclear location sequence-mediated binding to the nuclear envelope. J Cell Biol. 1994;125:547–555. doi: 10.1083/jcb.125.3.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kutay U, Izaurralde E, Bischoff FR, Mattaj IW, Gorlich D. Dominant-negative mutants of importin-beta block multiple pathways of import and export through the nuclear pore complex. EMBO J. 1997;16:1153–1163. doi: 10.1093/emboj/16.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bednenko J, Cingolani G, Gerace L. Importin beta contains a COOH-terminal nucleoporin binding region important for nuclear transport. J Cell Biol. 2003;162:391–401. doi: 10.1083/jcb.200303085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Melchior F, Paschal B, Evans J, Gerace L. Inhibition of nuclear protein import by nonhydrolyzable analogues of GTP and identification of the small GTPase Ran/TC4 as an essential transport factor. J Cell Biol. 1993;123:1649–1659. doi: 10.1083/jcb.123.6.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cassany A, Gerace L. Reconstitution of nuclear import in permeabilized cells. Methods Mol Biol. 2009;464:181–205. doi: 10.1007/978-1-60327-461-6_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.