Summary
Recently, more than a thousand large intergenic non-coding RNAs (lincRNAs) have been reported. These RNAs are evolutionarily conserved in mammalian genomes and thus presumably function in diverse biological processes. Here, we report the identification of lincRNAs that are regulated by p53. One of these lincRNAs (lincRNA-p21) serves as a repressor in p53-dependent transcriptional responses. Inhibition of lincRNA-p21 affects the expression of hundreds of gene targets enriched for genes normally repressed by p53. The observed transcriptional repression by lincRNA-p21 is mediated through the physical association with hnRNP-K. This interaction is required for proper genomic localization of hnRNP-K at repressed genes and regulation of p53 mediated apoptosis. We propose a model whereby transcription factors activate lincRNAs that serve as key repressors by physically associating with repressive complexes and modulating their localization to sets of previously active genes.
Introduction
It has become increasingly clear that mammalian genomes encode numerous large non-coding RNAs (Mercer et al., 2009; Ponting et al., 2009; Mattick, 2009; Ponjavic et al., 2007). It has been recently reported the identification of more than a thousand large intergenic non-coding RNAs (lincRNAs) in the mouse genome (Carninci, 2008; Guttman et al., 2009). The approach to identify lincRNAs was by searching for a chromatin signature of actively transcribed genes, consisting of a histone 3–lysine 4 trimethylated (H3K4me3) promoter region and histone 3–lysine 36 trimethylation (H3K36me3) corresponding to the elongated transcript (Guttman et al., 2009). These lincRNAs show clear evolutionary conservation, implying that they are functional (Guttman et al., 2009; Ponjavic et al., 2007).
In an attempt to understand the potential biological roles of lincRNAs, a method to infer putative function based on correlation in expression between lincRNAs and protein-coding genes was developed. These studies led to preliminary hypotheses about the involvement of lincRNAs in diverse biological processes, from stem cell pluripotency to cell cycle regulation (Guttman et al., 2009). In particular, we observed a group of lincRNAs that are strongly associated with the p53 transcriptional pathway. p53 is an important tumor suppressor gene involved in maintaining genomic integrity (Vazquez et al., 2008). In response to DNA damage, p53 becomes stabilized and triggers a transcriptional response that causes either cell arrest or apoptosis (Riley et al., 2008).
The p53 transcriptional response involves both activation and repression of numerous genes. While p53 is known to transcriptionally activate numerous genes, the mechanisms by which p53 leads to gene repression have remained elusive. We recently reported evidence that many lincRNAs are physically associated with repressive chromatin modifying complexes and suggested that they may serve as repressors in transcriptional regulatory networks (Khalil et al., 2009). We therefore hypothesized that p53 may repress genes in part by directly activating lincRNAs, which in turn regulate downstream transcriptional repression.
Here we show that lincRNAs play a key regulatory role in the p53 transcriptional response. By exploiting multiple independent cell-based systems, we identify lincRNAs that are transcriptional targets of p53. Moreover, we find that one of these p53-activated lincRNAs - termed lincRNA-p21- serves as a transcriptional repressor in the p53 pathway and plays a role in triggering apoptosis. We further demonstrate that lincRNA-p21 binds to hnRNP-K. This interaction is required for proper localization of hnRNP-K and transcriptional repression of p53-regulated genes. Together, these results reveal insights into the p53 transcriptional response and lead us to propose that lincRNAs may serve as key regulatory hubs in transcriptional pathways.
Results
Numerous lincRNAs are activated in a p53-dependent manner
As a first attempt to dissect the functional mechanisms of lincRNAs, we focused on a strong association in the expression patterns of certain lincRNAs and genes in the p53 pathway (Guttman et al. 2009). In order to determine whether these lincRNAs are regulated by p53, we employed two independent experimental systems that allow us to monitor gene expression changes at different times following p53 induction (Ventura et al., 2007).
The first of these systems uses MEFs derived from mice where the endogenous p53 locus is inactivated by insertion of a transcriptional termination site flanked by loxP sites (LSL) in the first intron. This endogenous p53 locus (p53 LSL/LSL) is restorable by removal of the stop element by Cre-recombination (Ventura et al., 2007). The p53 LSL/LSL MEFs were treated with AdenoCre virus expressing the Cre recombinase to reconstitute the normal p53 allele or AdenoGFP control virus to maintain the inactive p53 LSL/LSL allele. Then we compared the transcriptional response between the p53-reconstituted and p53 LSL/LSL MEFs following 0, 3, 6 and 9 hours of DNA damage treatment with doxorubicin (we will refer to this system as ‘MEFs’) (Figure 1A). The second system uses a lung tumor cell line derived from mice expressing an oncogenic K-Ras mutation (K-RasG12D) and a restorable p53 knockout allele (p53 LSL/LSL), similar to that described above (D.F. and T.J. manuscript in preparation). We compared the transcriptional response at different times (0, 8, 16, 24, 40 and 48 hours) after restoration of p53 expression by Cre recombination (Experimental Procedures) (we will refer to this system as ‘KRAS’) (Figure 1A).
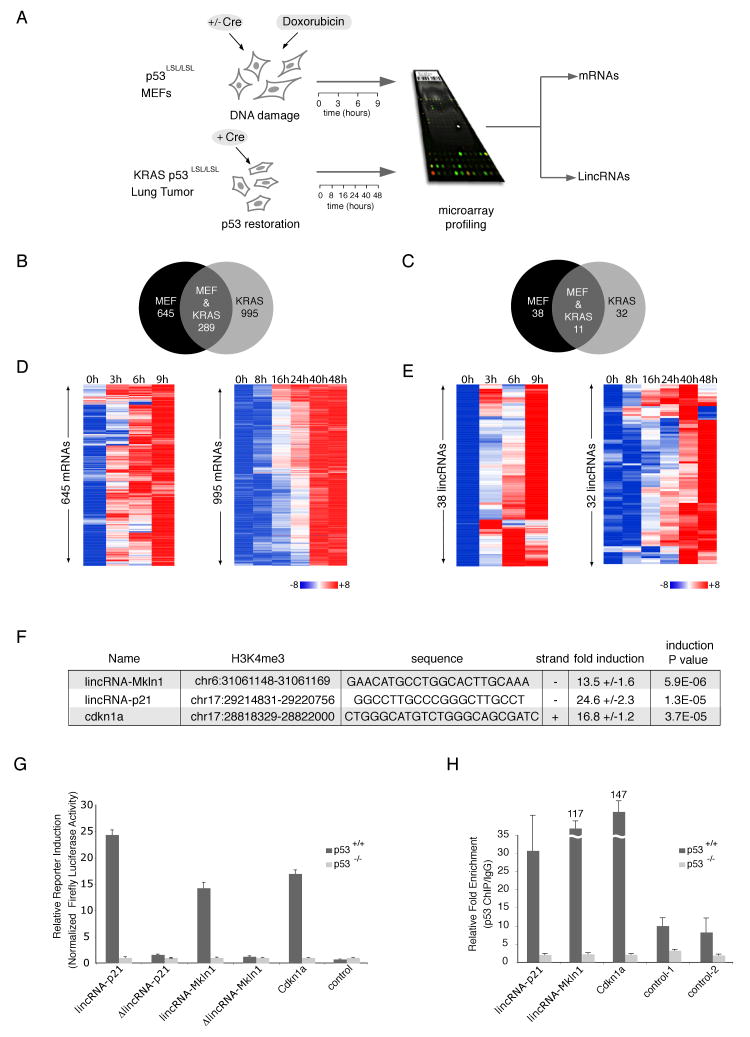
Figure 1. Several lincRNAs are p53 transcriptional targets.
(A). Experiment layout to monitor p53-dependent transcription. p53-restored (+Cre) or not restored (-Cre) p53LSL/LSL MEFs were treated with 500nM dox for 0, 3, 6 and 9 hours (top left). KRAS (p53LSL/LSL) tumor cells were treated with hydroxytamoxifen for p53 restoration for 0, 8, 16, 24, 40 or 48 hours (bottom left). RNA was subjected to microarray analysis of mRNAs and lincRNAs.
(B) and (C). Venn diagrams showing the number of shared and distinct mRNAs (B) or lincRNAs (C) induced in a p53-dependent manner in the MEF or KRAS systems.
(D) and (E). mRNAs (D) and lincRNAs (E) activated by p53 induction (FDR < 0.05) in MEF or KRAS system. Colors represent transcripts above (red) or below (blue) the global median scaled to 8 fold activation or repression, respectively.
(F) Promoter region, conserved p53 binding motif, promoter orientation and p53-dependent fold induction in reporter assays of lincRNA promoters induced in a p53-dependent manner (values are average of at least three biological replicates (+/-STD); P values are determined by t-test)
(G) p53-dependent induction of lincRNA promoters requires the consensus p53 binding elements. Relative firefly luciferase expression driven by promoters with p53 consensus motif (lincRNA-p21, lincRNA-Mkln1) or with deleted motif (ΔlincRNA-p21 and ΔlincRNA-Mkln1) in p53-restored p53LSL/LSL (p53+/+) or p53LSL/LSL (p53-/-) cells. Values are relative to p53-/- and normalized by renilla levels (average of 3 replicates +/-STD).
(H) p53 specifically binds to p53 motifs in lincRNA promoters. p53 ChIP enrichment in p53+/+ and p53-/- MEFs on regions with p53 motifs (lincRNA-p21, lincRNA-Mkln1, Cdkn1a) or two irrelevant regions (controls). Enrichment values are relative to IgG and average of 3 replicates (+/-STD).
See also Figure S1 and Table S1.
To assess the transcriptional responses in each of these systems we isolated total RNA at each time point before and after p53 restoration and performed DNA microarray analysis to monitor protein-coding gene expression levels. In the MEF system and KRAS systems, we identified a total of 1067 (645 activated, 422 repressed) and 1960 (995 activated, 960 repressed) genes respectively, that were regulated in a p53-dependent manner (FDR < 0.05) (Figures 1B and 1D and Tables S1A and S1B). The sets of p53-induced genes identified in the two systems showed significant (P < 10-8) overlap, including such canonical p53 targets as Cdnk1a, Mdm2, PERP and FAS (Tables S1A and S1B). There are also a number of p53-induced genes unique to each system, likely reflecting specific properties of each cell-type (Levine et al., 2006) (Figure S1C, Tables S1A and S1B).
Functional analysis of the classes of genes that are enriched among the genes regulated by p53 in both the MEF and KRAS systems showed strong enrichment for known p53-regulated processes, such as cell-cycle and apoptosis (Figures S1A and S1B). Moreover, Gene Set Enrichment Analysis (GSEA) (Subramanian et al., 2005) of previously published microarray analyses revealed a significant overlap with the p53 regulated genes identified here (Tables S1H and S1I). Together these results demonstrate that these two systems are largely reflective of canonical p53 transcriptional responses.
We next examined lincRNAs regulated by p53 in these two systems across the same time course; by analogously using a custom tiling microarray representing 400 lincRNAs and analyzing the data with previously described statistical methods (Guttman et al. and Experimental Procedures). We found 38 and 32 lincRNAs induced by p53 in the MEF and KRAS systems, respectively (Figures 1C and 1E and Tables S1C to S1G). Interestingly, 11 lincRNAs were induced by p53 in both model systems (Figure 1C and Table S1C); many more than expected by chance (P < 10-6). These results confirm that, in a manner similar to canonical p53 protein coding gene targets, numerous lincRNAs are temporally regulated by p53.
lincRNAs are direct transcriptional targets of p53
We sought to identify lincRNAs that might be canonical p53 target genes. As a first approach, we examined the promoters of p53-induced lincRNA for enrichment of evolutionarily conserved p53-binding motifs (Garber et al., 2009) (Supplemental Experimental Procedures). The promoters of the p53-induced lincRNAs were highly enriched for conserved p53 motifs relative to the promoters of all lincRNAs (p<0.01). We selected two lincRNAs whose promoter regions contain highly conserved canonical p53-binding motifs (el-Deiry et al., 1992; Funk et al., 1992); we termed these lincRNA-p21 and lincRNA-Mkln1 (with the names referring to the neighboring gene). We next performed transcriptional reporter assays for these lincRNAs. Specifically, we cloned their promoters (as defined by the H3K4me3 peaks (Guttman et al., 2009)) into a luciferase reporter vector (Experimental Procedures) and transfected the constructs along with a vector to normalize transfection efficiency. Both promoters showed significant induction of firefly luciferase in p53-wild type but not in p53-null cells (p<0.01, Figure 1F and G).
To determine if the canonical p53-binding motif is required for the observed transactivation, we repeated these experiments in the absence of the p53-binding motif. Mutant promoters resulted in the abolition of the observed transactivation for both lincRNA-p21 and lincRNA-Mkln1 in p53+/+ cells (Fig 1F). Finally, we performed Chromatin Immunoprecipitation (ChIP) experiments to determine if p53 directly binds to the sites containing the consensus motifs in vivo. Indeed, p53 is bound to the site containing the consensus motif in the promoters of both lincRNA-p21 and lincRNA-Mkln1 in p53+/+ but not p53-/- MEFs treated with doxorubicin, and it is not enriched at negative control sites of irrelevant regions (Fig 1H, Supplemental Experimental Procedures). Together these results demonstrate that lincRNA-p21 and lincRNA-Mkln1 are bona fide p53 transcriptional targets.
lincRNA-p21 is induced by p53 in different cell systems
We were intrigued by the p53 transcriptional target lincRNA-p21, which resides ∼15Kb upstream of the gene encoding the critical cell cycle regulator Cdkn1a (also known as p21), a canonical transcriptional target of p53 (Riley et al., 2008) (Figure 2A-C, Figure S2A, Tables S1C, S1D, S1E). Given the proximity of lincRNA-p21 to the Cdkn1a gene, we sought to ensure that the lincRNA transcript is distinct from that of the Cdkn1a gene. To this end we cloned the full-length transcriptional unit of lincRNA-p21 using the 5′- and 3′- RACE method (Schaefer, 1995); the transcript contains two exons comprising 3.1Kb (Figure 2B). In support of lincRNA-p21 being an independent transcript, lincRNA-p21 is transcribed in the opposite orientation from the Cdkn1a gene. Furthermore, the analysis of chromatin structure in mouse embryonic stem (mES) cells (Mikkelsen et al., 2007) indicates that these are distinct genes with distinct promoters (Figure S2A).
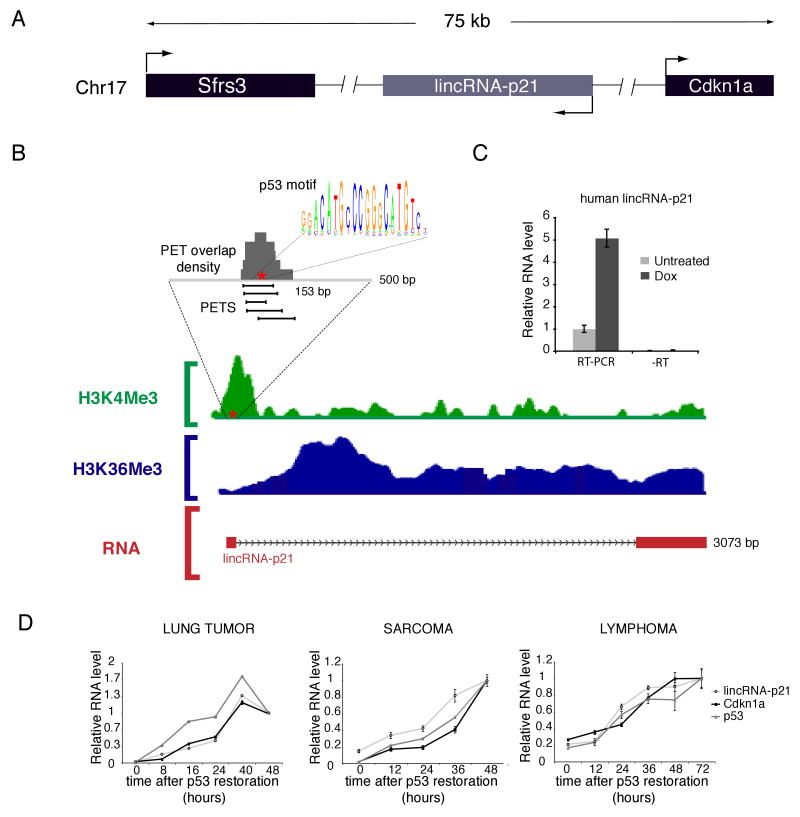
Figure 2. lincRNA-p21: a new p53 target gene induced in different tumor models.
(A) Schematic representation of the chromosomal location of the lincRNA-p21 gene locus. Arrowheads indicate the orientation of transcription.
(B) Promoter and transcript structure of lincRNA-p21 gene locus. Chromatin structure at the lincRNA -p21 locus is shown as mES ChIP-Seq data (Mikkelsen et al., 2007); for each histone modification (H3K4me3, green; H3K36me3, blue), ChIP-seq results are plotted as number of DNA fragments obtained at each position relative to the genomic average. Red stars indicate the position of the p53-binding motif. The promoter region where p53 ChIP-PET fragments (black segments) map is enlarged (Wei et al., 2006). PET overlap density (grey) and p53 motif sequence are shown. The structure of the full-length lincRNA-p21 is represented with red boxes as exons and arrowed lines as the intronic sequence.
(C) Human lincRNA-p21 is induced by DNA damage. Relative RNA levels of human lincRNA-p21 determined by qRT-PCR (RT-PCR) or qPCR (-RT) from untreated human fibroblasts or 500nM DOX-treated for 14 hours. PCR primers map on the human region orthologus to the first exon of the mouse gene.
(D) lincRNA-p21 is induced by p53 in different tumor cell lines. lincRNA-p21, p53 and Cdkn1a relative RNA levels at different times after p53 restoration. Values are the median of 4 technical replicates.
Values in (C) and (D) are the median of 4 technical replicates (+/-STD).
See also Figure S2.
We next examined the transcriptional regulation of lincRNA-p21 in two additional cancer-derived cell lines. Specifically, we irradiated p53LSL/LSL mice to induce lymphomas and sarcomas, and then restored p53 expression in tumor derived-cells (Experimental Procedures) (Ventura et al., 2007). In cells derived in both tumor types, lincRNA-p21 was strongly induced following p53 restoration. Moreover, the induction of lincRNA-p21 followed similar kinetics as those of p53 and Cdnk1a, consistent with lincRNA-p21 being a primary transcriptional target of p53 (Figure 2D and S2B).
We further investigated the orthologous lincRNA-p21 locus in the human genome. We first mapped the promoter (H3K4me3 domain) of lincRNA-p21 to human genome. Interestingly, this region corresponds to one of four intergenic p53-binding sites identified from a study by Wei et al. (2006) (Figure 2B) performing p53 ChIP followed by sequencing. Next, we mapped the lincRNA-p21 exonic structures to the human genome to determine if this region is expressed and induced by DNA damage in human fibroblasts. Indeed, qRT-PCR showed that the orthologous 5′ exon region (adjacent to observed p53 ChIP binding site by Wei et al.) of human lincRNA-p21 is expressed and strongly induced in human fibroblasts upon DNA damage (Figure 2C and Supplemental Experimental Procedures).
Collectively, these results provide evidence that both the human and mouse lincRNA-p21 promoters are bound by p53 resulting in transcriptional activation in response to DNA damage. Moreover, lincRNA-p21 is induced by p53 in diverse biological contexts, including multiple different tumor types (Figures 2D and S2B), suggesting that lincRNA-p21 plays a role in the p53 pathway.
lincRNA-p21 as a repressor in the p53 pathway
We next investigated the consequence of the loss of lincRNA-p21 function in the context of the p53 response. We reasoned that, if lincRNA-p21 plays a role in carrying out the p53 transcriptional response, then inhibition of lincRNA-p21 would show effects that overlap with inhibition of p53 itself. To test this hypothesis, we performed RNAi-mediated knockdown of lincRNA-p21 and p53 separately, and monitored the resulting changes in mRNA levels by DNA microarray analysis.
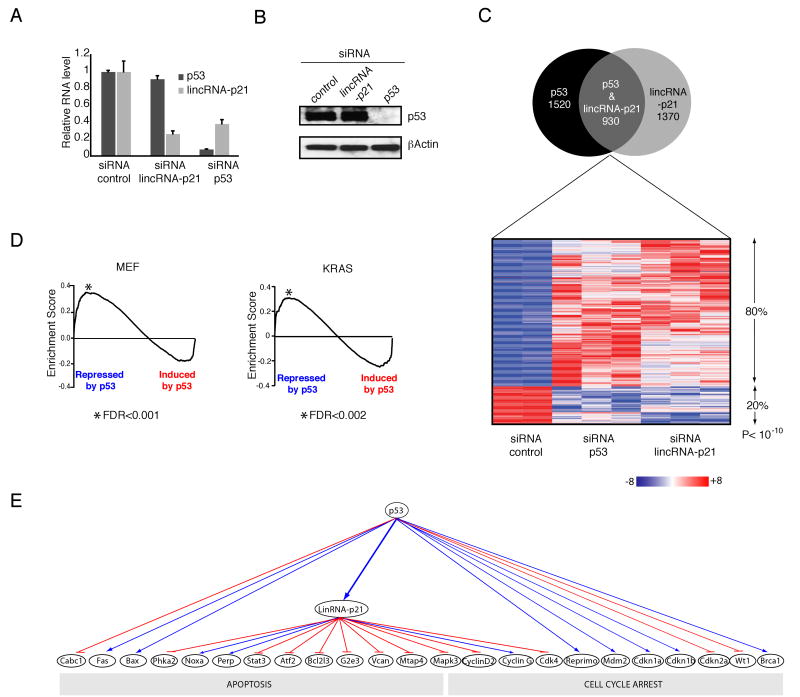
Toward this end, we first designed a pool of siRNA duplexes targeting lincRNA-p21, a pool targeting p53 or non-targeting control sequences. We validated that each pool was effective at knocking down its intended target genes in p53LSL/LSL restored MEFs (Figure 3A and 3B). We then used microarray analysis to examine the broader transcriptional consequences of knockdown of p53 and lincRNA-p21. We identified 1520 and 1370 genes that change upon knockdown of p53 and lincRNA-p21, respectively (relative to non-targeting control siRNA, FDR < 0.05). We observed a remarkable overlap of 930 genes in both the lincRNA-p21 and p53 knockdowns, vastly more than would be expected by chance (p<10-200) (Figures 3C and S3A and Table S2). Strikingly, 80% (745/930) of the common genes are derepressed in response to both p53 and lincRNA-p21 knockdown, much higher proportion than expected by chance (P < 10-10) (Figure 3C and Table S2) when compared to all genes affected by the p53 knockdown (Figure S3A). This observation suggests that lincRNA-p21 participates in downstream p53 dependent transcriptional repression.
Fig 3. lincRNA-p21 is a global repressor of genes in the p53 pathway.
A) RNAi-mediated knockdown of lincRNA-p21 and p53. Relative RNA levels determined by qRT-PCR in p53-reconstitued p53LSL/LSL MEFs transfected with the indicated siRNAs and treated with DOX (median of 4 technical replicates +/-STD).
(B) p53 protein levels after lincRNA-p21 and p53 knockdown from cells treated as in (A). βActin levels are shown as loading control.
(C) Many genes are corepressed by lincRNA-p21 and p53. Top: Venn diagram of differentially expressed genes (FDR< 0.05) upon p53 knockdown (left) or lincRNA-p21 knockdown (right); cells were treated as in (A) and subjected to microarray analysis. Bottom: expression level of genes in lincRNA-p21 and p53 siRNA-treated cells relative to control siRNA experiments. Expression values are displayed in shades of red or blue relative to the global median expression value across all experiments (linear scale).
(D) Genes derepressed by lincRNA-p21 and p53 knockdown overlap with the genes repressed by p53 restoration in the MEF and KRAS systems. The black line represents the observed enrichment score profile of genes in the lincRNA-p21/p53 derepressed gene set to the MEF or KRAS gene sets respectively.
(E) Genes corregulated by lincRNA-p21 and p53 are part of the p53 biological response. Examples of genes affected by lincRNA-p21 and/or p53 siRNA-knockdown (FDR< 0.05). Downregulated and upregulated genes are indicated with blue arrows and red lines respectively. See also Figure S3 and Table S2.
To demonstrate that the observed derepression upon lincRNA-p21 knockdown is indeed p53-dependent and not due to off target effects of the RNAi-mediated knockdown, we performed several additional experiments and analyses. First we repeated the knockdown experiments with four individual siRNAs targeting lincRNA-p21, transfected separately rather than in a pool and confirmed the derepression effect on select target genes (Figure S3F and Table S2). Second we confirmed that the same genes that were derepressed in the lincRNA-p21 and p53 knockdown experiments correspond to genes that are normally repressed upon p53 induction in both the KRAS and MEF systems, in the absence of RNAi treatment (GSEA FDR < 0.002) (Figure 3D). Third we demonstrated that enforced expression of lincRNA-p21 (Experimental Procedures) also perturbed the expression of genes that are normally regulated by p53 in both the KRAS and MEF systems (GSEA FDR < 0.01) (Figure S3H). Finally, we repeated the siRNA experiments in the absence of p53 (dox/-AdCre) and demonstrated that derepression of these genes did not occur upon siRNA-mediated knockdown in the absence of p53 (Figure S3I). Collectively, these results indicate that lincRNA-p21 acts to repress many genes in p53-dependent transcriptional response.
LincRNA-p21 regulates apoptosis
The activation of the p53 pathway has two major phenotypic outcomes: growth arrest and apoptosis (Levine et al., 2006). Consistent with this, our microarray analysis demonstrates that p53 and lincRNA-p21 both regulate a number of apoptosis and cell-cycle regulator genes (Figures 3E, S3G and Tables S2A and S2B). Thus, we aimed to determine the physiological role of lincRNA-p21 in these processes.
Toward this end, we used RNAi-mediated knockdown of lincRNA-p21 in dox-treated or untreated primary MEFs. We similarly performed RNAi-mediated knockdown of p53 (as a positive control) or used the non-targeting siRNA pool (as a negative control) under the same conditions. We observed a significant increase in viability following DNA damage of cells treated with siRNAs targeting either lincRNA-p21 or p53 compared to those treated with the control siRNA pool (Figure 4A and 4B). The increase in viability was greater for knockdown of p53, but was still highly significant for knockdown of lincRNA-p21 (P< 0.01). We observed similar results using three individual siRNA duplexes targeting lincRNA-p21, as well as two different control siRNA pools (Figures 4B, S4A-C). These results suggest that lincRNA-p21 plays a physiological role in regulating cell viability upon DNA damage in this system, although they do not discriminate whether the effect is due to misregulation of the cell cycle or apoptosis.
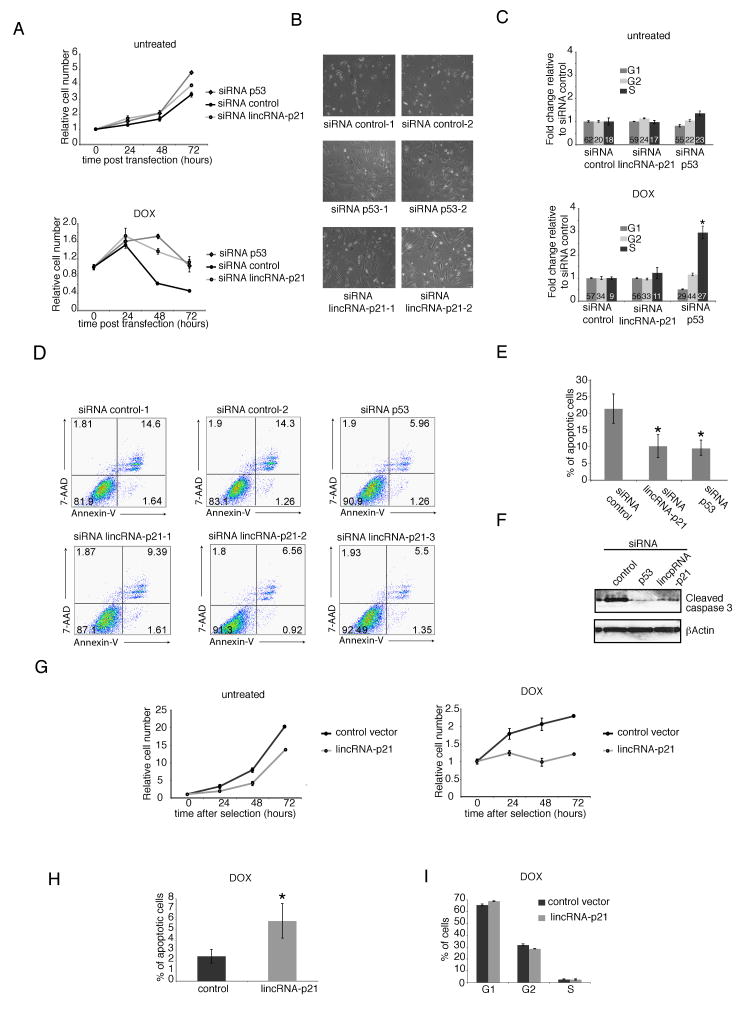
Fig 4. lincRNA-p21 is required for proper apoptotic induction.
(A) Increased cell viability of lincRNA-p21 depleted cells. Relative number of siRNA-transfected MEFs treated with 400nM DOX from 24h after transfection (right) or untreated (left) determined by MTT assay.
(B) Knockdown of lincRNA-p21 with individual siRNAs increases cell viability. Images of MEFs treated with different individual siRNAs after 48h of DOX treatment (72h post transfection).
(C) LincRNA-p21 knockdown doesn't affect cell cycle regulation. Relative cell numbers in each cell cycle phase determined by FACS of BrdU incorporation and PI staining of MEFs treated as in (A). Numbers inside bars represent percentages of cells in each phase.
(D) LincRNA-p21 knockdown causes a decrease in cellular apoptosis. p53-reconstituted p53LSL/LSL MEFs transfected with three individual siRNAs targeting lincRNA-p21 (bottom), two independent control siRNAs (top left and middle) or a siRNA pool targeting p53 (top right). 24h after transfection cells were treated with 400nM doxorubicin and 14h later harvested and subjected to FACS analysis. X-axis represents Annexin-V and y-axis 7-AAD staining. The percentage of cells in each quadrant is indicated.
(E) Decreased apoptosis caused by lincRNA-p21 knockdown. Percentage of Annexin-V positive cells (FACS) at 38h post transfection (14h of 400nM DOX treatment) in MEFs treated as in (A).
(F) LincRNA-p21 knockdown in p53-reconstituted p53LSL/LSL MEFs causes decrease in Caspase 3 cleavage. Levels of cleaved Caspase 3 or control βActin in p53 reconstituted-p53LSL/LSL MEFs treated with the indicated siRNA pools and 500nM DOX for 14h.
(G) Decreased cell viability caused by lincRNA-p21 overexpression. Relative numbers of LKR cells overexpressing lincRNA-p21 or control plasmid determined by MTT assay.
(H) Overexpression of lincRNA-p21 causes cellular apoptosis under DNA damage induction. Percentage of Annexin-V-positive LKR cells overexpressing lincRNA-p21 or control vector treated with 500nM DOX.
(I) LincRNA overexpression doesn't affect cell cycle regulation. Cell cycle analysis of DOX-treated LKR cells overexpressing lincRNA-p21 or control plasmid.
All values are the average of 3 biological replicates (+/-STD). *=P< 0.001 relative to controls.
Also see Figure S4.
To distinguish between these two possibilities, we first examined whether cell cycle regulation in response to DNA damage is affected by knockdown of p53 and lincRNAp-21. Specifically, we assayed 5-bromo-2-deoxyuridine (BrdU) incorporation and propidium iodide staining of the cells by FACS analysis. Consistent with the ability of p53 to inhibit cell-cycle progression, knockdown of p53 caused a significant increase in BrdU incorporation in response to DNA damage (P<0.01). In contrast, knockdown of lincRNA-p21 neither showed significant changes in BrdU levels nor in the percentages of cells in any of the cell cycle phases (S, G1 or G2) with or without dox treatment (Figure 4C). These results suggest that lincRNA-p21 does not substantially contribute to cell cycle arrest upon DNA damage.
We then examined the impact of lincRNA-p21 and p53 knockdowns on apoptosis. To this end, we assayed the proportion of the cell population undergoing apoptosis by measuring Annexin-V by FACS analysis. We observed a significant decrease in the number of apoptotic cells following DNA damage in both the lincRNA-p21 and p53 depleted cells relative to the siRNA control (P<0.01) (Figures 4D and 4E). We also observed a decrease in Caspase 3 cleavage following knockdown of both p53 or lincRNA-p21, relative to controls (Figure 4F). We next sought to determine whether, conversely to lincRNA-p21 knockdown, the enforced expression of lincRNA-p21 would result in an increased apoptosis. Indeed, lincRNA-p21 overexpression in a lung cancer cell line harboring a KRAS mutation (referred to as LKR) and in NIH/3T3 MEFs caused a significant decrease in cell viability (Experimental Procedures and Figures 4G and S4E). This decrease in viability was due to increased apoptosis in response to DNA damage (P<0.01) and not to an effect in cell cycle regulation (Figures 4H, 4I and S4G). Together these results demonstrate a reproducible and similar reduction of apoptotic cells in response to DNA damage in both lincRNA-p21 and p53 knockdown experiments.
Although MEFs typically respond to DNA damage by undergoing cell-cycle arrest rather than apoptosis (Kuerbitz et al., 1992), several additional lines of evidence are consistent with the observed apoptosis phenotype in response to knockdown on p53 and lincRNA-p21. First, certain critical cell-cycle regulators, such as Cdkn1a/p21, Cdkn2a and Reprimo are regulated by p53 but not lincRNA-p21. For example, knockdown of lincRNA-p21 perturbs neither the transcript levels of Cdkn1a/p21 nor the protein stability (Figure S3E); this may explain why lincRNA-p21 knockdown is insufficient to cause a cell-cycle phenotype, yet the p53 knockdown is. Second, we observed that both lincRNA-p21 and p53 knockdowns resulted in the repression of apoptosis genes (Noxa and Perp) and derepression of cell survival genes (Bcl2l3, Stat3 and Atf2 among others) (Figure 3E and Table S2). Moreover, the decrease of apoptotic cells in response to knockdown of lincRNA-p21 was comparable to that caused by knockdown of p53 (Figure 4D-E and S4D and S4E). Third, the apoptosis phenotype is dependent on the dosage of dox-induced DNA damage (Figure S4D). Thus, the apoptosis response is both p53-dependent and lincRNA-p21-dependent, with this dependence confirmed in multiple cell-types and conditions (Figures 4B, 4D, 4F, 4H and S4A-C). Collectively, these observations demonstrate that lincRNA-p21 plays an important role in the p53-dependent induction of cell death.
lincRNA-p21 functions through interaction with hnRNP-K
We next wanted to investigate the mechanism by which lincRNA-p21 mediates transcriptional repression. We have recently reported that many lincRNAs regulate gene expression through their interaction with several chromatin regulatory complexes (Khalil et al., 2009). We thus hypothesized that lincRNA-p21 could affect gene expression in a similar manner.
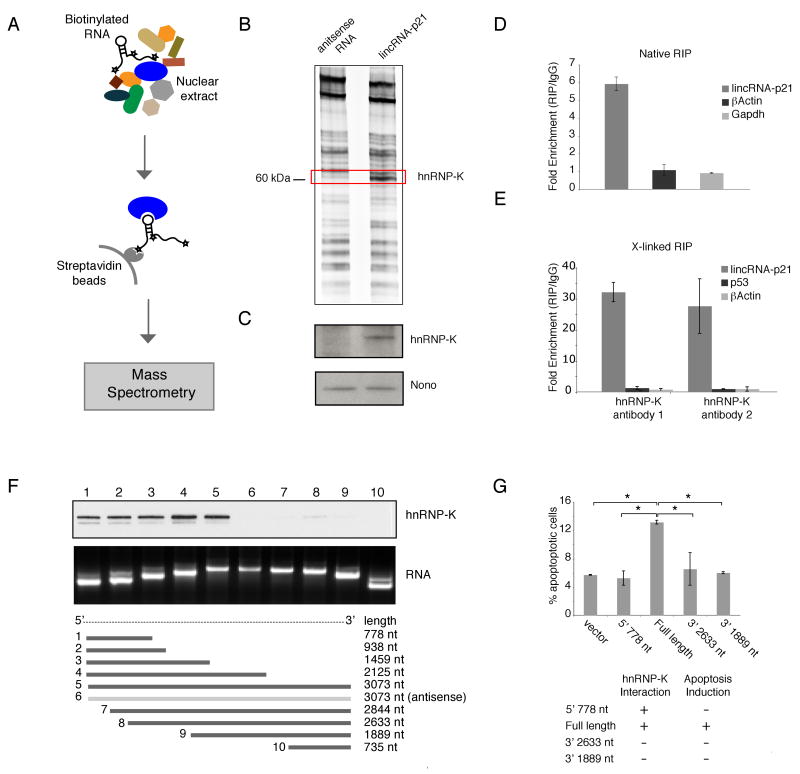
To test this, we first performed nuclear fractionation experiments and confirmed that lincRNA-p21 is enriched in the nucleus (Figure S5A). We next sought to identify proteins that are associated with lincRNA-p21 by an RNA-pulldown experiment. Specifically we incubated in vitro synthesized biotinylated lincRNA-p21 and antisense lincRNA-p21 transcripts (negative control) with nuclear cell extracts and isolated co-precipitated proteins with streptavidin beads (Experimental Procedures). We resolved the RNA-associated proteins on a SDS-PAGE gel, cut out the bands specific to lincRNA-p21 and subjected them to mass spectrometry (Figure 5A, 5B). In all six biological replicates, mass spectrometry analysis identified heterogeneous nuclear ribonucleoprotein K (hnRNP-K) as specifically associated with the sense (but not antisense) strand of lincRNA-p21. We independently verified this interaction by western blot analysis (Figure 5C). hnRNP-K has been shown to play various roles in the p53 pathway (Kim et al., 2008; Moumen et al., 2005). Interestingly, among these roles, Kim et al. (2008) demonstrated that hnRNP-K is a component of a repressor complex that acts in the p53 pathway; consistent with our evidence that lincRNA-p21 plays a role in global repression in this pathway.
Figure 5. lincRNA-p21 physically interacts with hnRNP-K.
(A) Schematic representation of RNA pull-down experiments to identify associated proteins. Biotinylated lincRNA-p21 or antisense RNA were incubated with nuclear extracts, targeted with streptavidin beads, washed, and associated proteins resolved in a gel. Specific bands were cutout and identified by Mass spectrometry.
(B) lincRNA-p21 and hnRNP-K specifically interact in vitro. SDS-PAGE gel of proteins bound to lincRNA-p21 (right lane) or antisense RNA (left lane). The highlighted region was submitted for Mass spectrometry identifying hnRNP-K as the band unique to lincRNA-p21.
(C) Western blot analysis of the specific association of hnRNP-K with lincRNA-p21. A nonspecific protein (NONO) is shown as control.
(D) Association between endogenous lincRNA-p21 and hnRNP-K in the nucleus of DNA damaged MEFs in native conditions. RNA Immunoprecipitation (RIP) enrichment is determined as RNA associated to hnRNP-K IP relative to IgG control.
(E) Physical association between lincRNA-p21 and hnRNP-K after chemical crosslinking of life cells. hnRNP-K was immunoprecipitated from nuclear extracts of formaldehyde-crosslinked DNA-damaged MEFs, and associated RNAs detected by RT-qPCR. The relative enrichment is calculated as in (D) and is the median of 3 technical replicates of a representative experiment (+/- STD).
(F) lincRNA-p21 binds hnRNP-K through its 5′ terminal region. RNAs corresponding to different fragments of lincRNA-p21 or its antisense sequence (middle and bottom panels) were treated as in (A) and associated hnRNP-K was detected by western blot (top panel).
Also see Figure S5.
(G) Percentage of Annexin-V positive LKR cells overexpressing the indicated lincRNA-p21 fragments or empty vector as control (average of 3 replicates (+/-STD)). *= P< 0.001.
To further validate the interaction between lincRNA-p21 and hnRNP-K in our cell-based systems we performed RNA immunoprecipitation (RIP) with an antibody against hnRNP-K from nuclear extracts of MEFs subjected to DNA damage. We observed an enrichment of lincRNA-p21 (but not other unrelated RNAs) with hnRNP-K antibody as compared with the non-specific antibody (IgG control) (Fig 5D). We further performed analogous RIP experiments, with formaldehyde cross-linked cells followed by stringent washing conditions (Ule et al., 2005) to rule out potential non-specific interactions. Consistent with a bona fide interaction, we observed a greater and very significant enrichment of lincRNA-p21 in the hnRNP-K RIP relative to the IgG control RIP with two hnRNP-K different antibodies (Figure 5E).
We further performed deletion-mapping experiments to determine whether hnRNP-K interacts within a specific region of lincRNA-p21. To this end we carried out RNA pull-down experiments with truncated versions of lincRNA-p21 followed by western blot detection of bound hnRNP-K. These analyses identified a 780nt region at the 5′-end of lincRNA-p21 required for the interaction with hnRNP-K (Figure 5F). Interestingly, RNA folding analyses of this region based on sequence conservation and compensatory changes across 14 mammalian species (Hofacker, 2003) predict a highly stable 280nt structure of lincRNA-p21 with deep evolutionary conservation (Figure S5B and C). Together, the RNA pull-down, native RIP, cross-linked RIP and deletion mapping results demonstrate a specific association between hnRNP-K and lincRNA-p21.
We next sought to determine the functional relevance of the interaction between lincRNA-p21 and hnRNP-K. To do so, we monitored the ability of different truncated versions of lincRNA-p21 to induce cellular apoptosis when overexpressed in LKR cells (Experimental Procedures). This revealed that the deletion of the 5′ end of lincRNA-p21, which mediates the hnRNP-K interaction, abolishes the ability of lincRNA-p21 to induce apoptosis (Figure 5G). Interestingly, the 780nt fragment at the 5′-end of lincRNA-p21 alone does not induce apoptosis, indicating that this fragment is required but not sufficient for lincRNA-p21 mediated cellular apoptosis.
We hypothesized that hnRNP-K is required for proper transcriptional repression of target genes shared between p53 and lincRNA-p21. If so, knockdown of hnRNP-K should result in derepression of these shared targets. We tested this hypothesis by performing siRNA-mediated knockdown of hnRNP-K, lincRNA-p21 and p53 in p53-restored p53LSL/LSL MEFs, treating the cells with dox and profiling the changes in gene expression by microarray analysis.
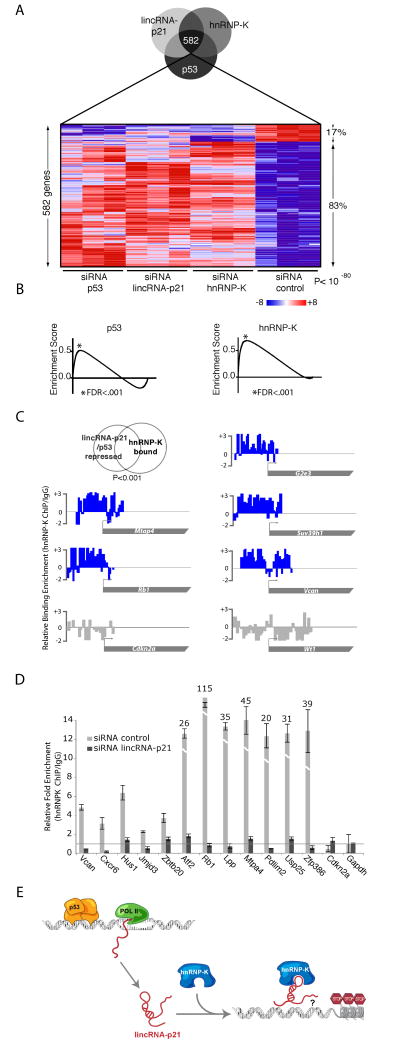
Consistent with our previous data, we observed a strong overlap of 582 genes affected in the hnRNP-K, lincRNA-p21 and p53 knockdowns (FDR<0.05 Figures 6 and S6). Remarkably, 83% of these common genes were derepressed in all three knockdown experiments (Figures 6A and S6D). The genes previously identified as co-regulated by lincRNA-p21 and p53 also were strongly enriched (GSEA, FDR <10-4) among those regulated by hnRNP-K (Figure 6B, Table S3). Thus, lincRNA-p21 and hnRNP-K play roles in repressing a significant common set of genes in the p53-dependent response to DNA damage.
Figure 6. LincRNA-p21 and hnRNP-K corepress genes in the p53 transcriptional response.
(A) Many genes are coregulated by p53, LincRNA-p21 and hnRNP-K. Genes affected by knockdown of lincRNA-p21, p53 or hnRNP-K in p53-restored-DNA-damaged p53LSL/LSL MEFs determined by microarray analysis (FDR<0.05). Shades of red or blue represent expression values relative to global median across experiments. Percentages of up- and down-regulated genes are indicated.
(B) Genes repressed by lincRNA-p21 are significantly enriched in genes repressed by p53 and hnRNPK. GSEA comparing the genes upregulated on knockdown of LincRNA-p21 and those upregulated upon knockdown of p53 (upper panel) or hnRNP-K (lower panel). The black line represents the observed enrichment score profile of genes in the lincRNA-p21 gene set to the p53 or hnRNP-K gene sets respectively.
(C) hnRNP-K associates to promoters of genes corepressed by lincRNA-p21 and p53. Examples of promoters of genes repressed by p53 and lincRNA-p21 (G2e3, Mtap4, Suv39h1 and Vcan) or repressed by lincRNA-p21 but not p53 (Rb1) bound by hnRNP-K (blue) determined by ChIP-chip of hnRNP-K in dox-trated p53-reconstituted p53LSL/LSL MEFs (FDR<0.05). Cdkn2a and Wt1 are negative controls (grey).
(D) hnRNP-K binding to lincRNA-p21 and p53 corepressed genes is dependent on lincRNA-p21. Relative enrichment of hnRNP-K (ChIP-qPCR) in the indicated promoter regions in p53-reconstituted p53LSL/LSL MEFs transfected with siRNA lincRNA-p21 or siRNA control and dox-treated determined by ChIP-qPCR (representative of two biological replicates shown +/- STD).
(E) Proposed models for the function of licRNA-p21 in the p53 transcriptional response. Induction of p53 activates the transcription of lincRNA-p21 by binding to its promoter (top left). LincRNA-p21 binds to hnRNP-K, and this interaction imparts specificity to genes repressed by p53 induction (top right). See also Figure S6 and Table S3.
We further reasoned that if hnRNP-K is involved in the repression of genes corepressed by p53 and lincRNA-p21, then hnRNP-K might also be physically bound to the promoters of these genes. To test this we performed ChIP experiments with antibodies against hnRNP-K, followed by hybridization to DNA tiling microarrays covering 30,000 gene promoters. We identified 1621 promoter regions with significant occupancy by hnRNP-K (FDR< 0.05) (Figure 6, Table S3). Notably, these promoter regions exhibit a significant overlap with genes that were differentially expressed upon hnRNP-K knockdown (GSEA FDR < 0.001, Figure S6E). Moreover, hnRNP-K localizes to a significant fraction (FDR < 0.001) of the genes corepressed by lincRNA-p21 and p53 (Figure 6C); suggesting these are primary sites of hnRNP-K regulation.
We next wanted to determine if lincRNA-p21 plays a role in hnRNP-K localization at promoters of p53-repressed genes. To this end we determined whether siRNA-mediated knockdown of lincRNA-p21 affected the localization of hnRNP-K after induction of p53. Specifically, we performed hnRNP-K ChIP in dox-treated p53-restored p53LSL/LSL MEFs either transfected with siRNAs targeting lincRNA-p21 or non-targeting control siRNAs. These experiments revealed that the depletion of lincRNA-p21 causes a significant reduction in the association of hnRNP-K at the promoter regions of genes that are normally repressed in a lincRNA-p21 and p53-dependent fashion, as determined by ChIP-qPCR (Figure 6D). Specifically, 12 of the 15 tested promoter regions exhibited loss of hnRNP-K enrichment, in two biological replicate experiments, upon depletion of lincRNA-p21.
Thus, hnRNP-K is bound to the promoters of genes that are normally repressed in a p53- and lincRNA-p21-dependent manner, and this localization requires lincRNA-p21.
Collectively, our results indicate that lincRNA-p21 is a direct p53 transcriptional target in response to DNA damage, acts to repress genes that are down-regulated as part of the canonical p53 transcriptional response, is necessary for p53 dependent apoptotic responses to DNA damage in our cell-based systems, and functions at least in part through interaction with hnRNP-K by modulating hnRNP-K localization.
Discussion
It is clear that mammalian genomes encode numerous large non-coding RNAs (Carninci, 2008; Guttman et al., 2009; Mattick, 2009; Ponjavic et al., 2007;). Here, we demonstrate that numerous lincRNAs are key constituents in the p53-dependent transcriptional pathway. Moreover, we observed that some of these lincRNAs are bound by p53 in their promoter regions and sufficient to drive p53-dependent reporter activity that requires the consensus p53-binding motif, suggesting that these lincRNAs are bona fide p53 transcriptional targets.
Having discovered multiple lincRNAs in the p53 pathway, we decided to focus on one such lincRNA in particular: lincRNA-p21. Intrigued by its properties (genomic location upstream of p21; p53-dependent activation requiring the consensus p53 motif, which is bound by p53; and conserved p53-dependent activation of this gene in both human and mouse cell-based systems), we explored the functional roles of lincRNA-p21. Our studies revealed a role for lincRNA-p21 in a p53-dependent apoptotic response following DNA damage.
We further observed that siRNA-mediated inhibition of lincRNA-p21 affects the expression of hundreds of gene targets that are enriched for genes normally repressed by p53 in both the MEF and RAS cell-based systems. Strikingly, the vast majority of these common target genes are derepressed upon inhibition of either p53 or lincRNA-p21 – suggesting that lincRNA-p21 functions as a downstream repressor in the p53 transcriptional response.
We gained mechanistic clues into how lincRNA-p21 functions to repress such a large subset of the p53 transcriptional response by biochemical experiments that identified a specific interaction between lincRNA-p21 and hnRNP-K. This interaction is supported by RNA-pulldown, native RIP, cross-linked RIP and deletion mapping experiments. Moreover, we identified a 780nt 5′ region of lincRNA-p21 that is required for hnRNP-K binding and subsequent induction of apoptosis. Interestingly, this region is much more highly conserved than the remainder of the transcript. This suggests that patches of conservations, previously determined to be unique to lincRNAs, (Guttman et al., 2009), may point to functional elements for binding interactions within lincRNAs (as was also recently determined for Xist binding to PRC2) (Zhao et al., 2008).
hnRNP-K is known to interact with other repressive complexes such as the Histone H1.2 or members or the Polycomb-group (PcG) (Kim et al., 2008; Denisenko and Bomsztyk, 1997). The physical interaction between lincRNA-p21 and hnRNP-K is likely required for lincRNA-p21-mediated gene repression, as loss of hnRNP-K function results in the derepression of the same genes that are repressed by both p53 and lincRNA-p21. Importantly, genome-wide ChIP-chip analysis revealed hnRNP-K binding at the promoters of these corepressed gene loci, suggestive of direct regulation by hnRNP-K and lincRNA-p21. Moreover, we observed a lincRNA-p21 dependent binding of hnRNP-K at several of these corepressed promoter regions. While hnRNP-K has been previously shown to activate one gene in the p53 pathway (Moumen et al., 2005), our analyses suggest it plays a much more widespread role in repression. Together, these results implicate lincRNA-p21 as an important repressor in the p53 pathway, by interacting with and modulating the localization of hnRNP-K.
Our results raise the possibility that many transcriptional programs (beyond the p53-pathway) may involve inducing protein factors that activate specific sets of downstream genes and lincRNAs that repress previously active sets of genes. The notion of a non-coding RNA being involved in silencing specific gene loci is consistent with our recent observation that many lincRNAs (including lincRNA-p21) bind to chromatin complexes (such as PRC2) and are required to mediate repression at key gene loci (Khalil et al., 2009). Moreover, there are several examples of lincRNAs involved in repression of known target genes – including HOTAIR dependent repression of HOXD genes (Rinn et al., 2007) and XIST, AIR and KcNQto1, involved in genomic imprinting and silencing of several genes in cis (Nagano et al., 2008; Pandey et al., 2008; Zhao et al., 2008).
The precise mechanism by which lincRNA-p21 contributes to repression at specific loci remains to be defined. Various possibilities include that: (i) lincRNA-p21 might direct a protein complex to specific loci by Crick-Watson base pairing; (ii) lincRNAs might act by forming DNA-DNA-RNA triple helical structures, which do not require Crick-Watson base-pairing, such as reported for a large non-coding RNA that forms a triple-helix upstream of the Dihydrofolate Reductase (DHFR) promoter resulting in repression of DHFR (Martianov et al., 2007); or (iii) lincRNAs might alter the binding specificity of DNA-binding proteins (such as hnRNP-K), to influence their target preference (Figure 6D). Further experiments are needed to distinguish between these and other possibilities.
Aside from the general interest in gene regulation, we note that lincRNA-p21 and several other lincRNAs function in an important pathway for cancer. It is tempting to speculate that other lincRNAs may also play key roles in numerous other tumor-suppressor and oncogenic pathways, representing a hitherto unknown paradigm in cellular transformation and metastasis. It will be important for future studies to determine if lincRNAs genes can serve as tumor suppressor genes or oncogenes.
In summary, lincRNAs may point to new mechanisms of gene regulation, components in disease pathways and potential targets for the development of therapies.
Experimental Procedures
Cell lines and in vivo models
KRAS Lung tumor-derived cell lines were isolated from individual tumors (D.F. and T.J. manuscript in preparation). Isolation of matched p53+/+ and p53-/- MEFs, p53LSL/LSL MEFs, Lymphomas and Sarcomas and p53 restoration were done as described (Ventura et al., 2007). Primary wt MEFs and NIH/3T3 MEF cells were purchased from ATCC. Transfection, infection and treatment conditions are described in Supplemental Experimental Procedures.
Promoter reporter assays
LincRNA promoters were cloned into pGL3-basic vector (Promega) and motif deletions were performed by mutagenesis. p53-reconstituted or control p53LSL/LSL MEFs were transfected with 800ng of pGL3 and 30ng of TK-Renilla plasmid per 24 well. 24 hours later cells were treated with 500nM dox for 13 hours and cell extracts were assayed for firefly and renilla luciferase activities (Promega E1910).
lincRNA and gene-expression profiling and informatic analyses
RNA isolation, lincRNA expression profiling and ChIP-chip analyses (Nimblegen arrays) as well as Affymetrix gene-expression profiling and analyses were performed as described (Guttman et al. 2009) and Supplemental Experimental Procedures). Structure predictions were performed using the Vienna RNA package (Hofacker, 2003).
Antibodies
Anti-p53:Novocastra (NCL-p53-CM5p) (western blot) and Vector Labs (CM-5) (ChIP). Anti-hnRNP-K: Santa Cruz Biotechnology (sc-25373) (western blot) and Abcam (Ab70492 and Ab39975) (ChIP and RIP). Control rabbit IgG Abcam (Ab37415-5) (RIP and ChIP-chip).
Viability and apoptosis assays and cell cycle analysis
MTT assays were performed using Cell Proliferation Kit I from Roche (11465007001). For apoptosis quantification, the Apoptosis Detection Kit I from BD Biosciences (cat#559763) and FACS (van Engeland et al., 1996) were used. Cell cycle analysis was performed as described (Brugarolas et al., 1995).
Cloning, RNA pull-down, deletion mapping, RIP, ChIP
5′ and 3′ RACE cloning of lincRNA-p21 was performed from total RNA of dox-treated MEFs using RLM-RACE Kit (Ambion). RNA pull-down and deletion mapping were performed as described (Rinn et al., 2007) with 1mg of mES nuclear extract and 50 pmol of biotinylated RNA. Mass Spectrometry was performed as described (Shevchenko et al., 1996). Native RIP was carried out as described (Rinn et al., 2007). For cross-linked RIP, cells were cross-linked with 1% formaldehyde, antibody incubated overnight, recovered with protein G Dynabeads and washed with RIPA buffer. After reverse-crosslink, RNA was analyzed by qRT-PCR. p53 ChIP and hnRNP-K ChIP experiments were performed as previously described ((Rinn et al., 2007) and Supplemental Experimental Procedures).
RNA interference and lincRNA-p21 overexpression
siRNA transfections were done with 75nM of siRNA and Lipofectamine 2000 (Invitrogen). For overexpression, lincRNA-p21 or truncated forms were cloned into the pBABE vector. After transfection cells were selected with 2μg/ml puromycin.
Full-length sequence of LincRNA-p21 has been deposited in GenBank (Accession # HM210889 (bankit1350506). All primary data are available at the Gene Expression Omnibus (GSE21761).
Supplementary Material
Acknowledgments
We would like to thank Loyal A. Goff (MIT) for bioinformatic support, Nadya Dimitrova (MIT) for input on the manuscript, David Garcia (MIT) for experimental assistance and Sigrid Hart (Broad Institute) for illustration support. J.L. Rinn is a Damon Runyon-Rachleff, Searle and Smith Family Foundation Scholar. J.L. Rinn and A.R. are Richard Merkin Foundation Scholars. This work was supported by the NIH Directors New Innovator Award, Smith Family Foundation, Damon Runyon Cancer Foundation, Searle Scholar Program and NIH 1R01CA119176-01.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Brugarolas J, Chandrasekaran C, Gordon JI, Beach D, Jacks T, Hannon GJ. Radiation-induced cell cycle arrest compromised by p21 deficiency. Nature. 1995;377:552–557. doi: 10.1038/377552a0. [DOI] [PubMed] [Google Scholar]
- Carninci P. Non-coding RNA transcription: turning on neighbours. Nat Cell Biol. 2008;10:1023–1024. doi: 10.1038/ncb0908-1023. [DOI] [PubMed] [Google Scholar]
- Denisenko ON, Bomsztyk K. The product of the murine homolog of the Drosophila extra sex combs gene displays transcriptional repressor activity. Mol Cell Biol. 1997;17:4707–4717. doi: 10.1128/mcb.17.8.4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- el-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. Definition of a consensus binding site for p53. Nat Genet. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- Funk WD, Pak DT, Karas RH, Wright WE, Shay JW. A transcriptionally active DNA-binding site for human p53 protein complexes. Mol Cell Biol. 1992;12:2866–2871. doi: 10.1128/mcb.12.6.2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber M, Guttman M, Clamp M, Zody MC, Friedman N, Xie X. Identifying novel constrained elements by exploiting biased substitution patterns. Bioinformatics. 2009;25:i54–62. doi: 10.1093/bioinformatics/btp190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker IL. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31:3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Choi J, Heo K, Kim H, Levens D, Kohno K, Johnson EM, Brock HW, An W. Isolation and characterization of a novel H1.2 complex that acts as a repressor of p53-mediated transcription. J Biol Chem. 2008;283:9113–9126. doi: 10.1074/jbc.M708205200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuerbitz SJ, Plunkett BS, Walsh WV, Kastan MB. Wild-type p53 is a cell cycle checkpoint determinant following irradiation. Proc Natl Acad Sci U S A. 1992;89:7491–7495. doi: 10.1073/pnas.89.16.7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine AJ, Hu W, Feng Z. The P53 pathway: what questions remain to be explored? Cell Death Differ. 2006;13:1027–1036. doi: 10.1038/sj.cdd.4401910. [DOI] [PubMed] [Google Scholar]
- Martianov I, Ramadass A, Serra Barros A, Chow N, Akoulitchev A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 2007;445:666–670. doi: 10.1038/nature05519. [DOI] [PubMed] [Google Scholar]
- Mattick JS. The genetic signatures of noncoding RNAs. PLoS Genet. 2009;5:e1000459. doi: 10.1371/journal.pgen.1000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moumen A, Masterson P, O'Connor MJ, Jackson SP. hnRNP K: an HDM2 target and transcriptional coactivator of p53 in response to DNA damage. Cell. 2005;123:1065–1078. doi: 10.1016/j.cell.2005.09.032. [DOI] [PubMed] [Google Scholar]
- Nagano T, Mitchell JA, Sanz LA, Pauler FM, Ferguson-Smith AC, Feil R, Fraser P. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322:1717–1720. doi: 10.1126/science.1163802. [DOI] [PubMed] [Google Scholar]
- Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, Nagano T, Mancini-Dinardo D, Kanduri C. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32:232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- Ponjavic J, Ponting CP, Lunter G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res. 2007;17:556–565. doi: 10.1101/gr.6036807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer BC. Revolutions in rapid amplification of cDNA ends: new strategies for polymerase chain reaction cloning of full-length cDNA ends. Anal Biochem. 1995;227:255–273. doi: 10.1006/abio.1995.1279. [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Jensen ON, Podtelejnikov AV, Neubauer G, Mortensen P, Mann M. A strategy for identifying gel-separated proteins in sequence databases by MS alone. Biochem Soc Trans. 1996;24:893–896. doi: 10.1042/bst0240893. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule J, Jensen K, Mele A, Darnell RB. CLIP: a method for identifying protein-RNA interaction sites in living cells. Methods. 2005;37:376–386. doi: 10.1016/j.ymeth.2005.07.018. [DOI] [PubMed] [Google Scholar]
- van Engeland M, Ramaekers FC, Schutte B, Reutelingsperger CP. A novel assay to measure loss of plasma membrane asymmetry during apoptosis of adherent cells in culture. Cytometry. 1996;24:131–139. doi: 10.1002/(SICI)1097-0320(19960601)24:2<131::AID-CYTO5>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Vazquez A, Bond EE, Levine AJ, Bond GL. The genetics of the p53 pathway, apoptosis and cancer therapy. Nat Rev Drug Discov. 2008;7:979–987. doi: 10.1038/nrd2656. [DOI] [PubMed] [Google Scholar]
- Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, Newman J, Reczek EE, Weissleder R, Jacks T. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445:661–665. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- Wei CL, Wu Q, Vega VB, Chiu KP, Ng P, Zhang T, Shahab A, Yong HC, Fu Y, Weng Z, et al. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.