Abstract
The generation of induced pluripotent stem cells (iPSCs) from somatic cells demonstrated that adult mammalian cells can be reprogrammed to a pluripotent state by the enforced expression of a few embryonic transcription factors. This discovery has raised fundamental questions about the mechanisms by which transcription factors influence the epigenetic conformation and differentiation potential of cells during reprogramming and normal development. In addition, iPSC technology has provided researchers with a unique tool to derive disease-specific stem cells for the study and possible treatment of degenerative disorders with autologous cells. In this review, we summarize the progress that has been made in the iPSC field over the last 4 years, with an emphasis on understanding the mechanisms of cellular reprogramming and its potential applications in cell therapy.
Keywords: ES cells, iPS cells, pluripotency, reprogramming
History of cellular reprogramming
The discovery of induced pluripotency represents the synthesis of scientific principles and technologies that have been developed over the last six decades. These are (1) the demonstration by somatic cell nuclear transfer (SCNT) that differentiated cells retain the same genetic information as early embryonic cells; (2) the development of techniques that allowed researchers to derive, culture, and study pluripotent cell lines; and (3) the observation that transcription factors are key determinants of cell fate whose enforced expression can switch one mature cell type into another. In this section, we briefly summarize these three areas of research and the influence they had on the generation of induced pluripotent stem cells (iPSCs).
Nuclear transfer and the cloning of animals
During mammalian development, cells gradually lose potential and become progressively differentiated to fulfill the specialized functions of somatic tissues. For example, only zygotes and blastomeres of early morulas retain the ability to give rise to all embryonic and extraembryonic tissues (Kelly 1977), and are therefore called “totipotent,” while cells of the inner cell mass (ICM) of the blastocyst can give rise to all embryonic but not all extraembryonic tissues, and are hence called “pluripotent.” Cells residing in adult tissues, such as adult stem cells, can only give rise to cell types within their lineage and are called either “multipotent” or “unipotent,” depending on the number of developmental options they have. Upon terminal differentiation, cells entirely lose their developmental potential.
During the 1950s, Briggs and King (Briggs and King 1952; King and Briggs 1955) established the technique of SCNT, or “cloning,” to probe the developmental potential of nuclei isolated from late-stage embryos and tadpoles by transplanting them into enucleated oocytes. This work, together with seminal experiments by Gurdon (Gurdon 1962; Gurdon et al. 1975), showed that differentiated amphibian cells indeed retain the genetic information necessary to support the generation of cloned frogs. The major conclusion from these and subsequent findings was that development imposes reversible epigenetic rather than irreversible genetic changes on the genome during cellular differentiation. The cloning of Dolly the sheep (Wilmut et al. 1997) and other mammals from adult cells, including terminally differentiated cells (Hochedlinger and Jaenisch 2002a; Eggan et al. 2004; Li et al. 2004; Inoue et al. 2005), showed that the genome of even fully specialized cells remains genetically totipotent; i.e., can support the development of an entire organism. However, most cloned animals exhibit subtle to severe phenotypic and gene expression abnormalities, suggesting that SCNT results in faulty epigenetic reprogramming (Wakayama and Yanagimachi 1999; Hochedlinger and Jaenisch 2002b; Humpherys et al. 2002; Ogonuki et al. 2002; Tamashiro et al. 2002; Gurdon et al. 2003).
Pluripotent cell lines and fusion hybrids
While SCNT is a powerful tool to probe the developmental potential of a cell, it is technically challenging and not well suited for genetic and biochemical studies. Thus, another major advance toward isolating iPSCs was the establishment of immortal pluripotent cell lines from teratocarcinomas, tumors of germ cell origin. These cell lines were called embryonal carcinoma cells (ECCs) (Stevens and Little 1954; Kleinsmith and Pierce 1964) and could be clonally expanded in culture while retaining pluripotency (Finch and Ephrussi 1967; Kahan and Ephrussi 1970). Importantly, when ECCs were fused with somatic cells, such as thymocytes, the resulting hybrid cells acquired biochemical and developmental properties of ECCs and extinguished features of the somatic fusion partner (Miller and Ruddle 1976, 1977). The dominance of the pluripotent state over the somatic state in hybrids suggested that soluble trans-acting factors must exist in ECCs that can confer a pluripotent state upon somatic cells, and that these factors should be identifiable.
Most ECC lines are aneuploid and contribute only poorly to adult somatic tissues (Brinster 1974) and rarely to the germline (Stewart and Mintz 1982; Bradley et al. 1984) upon injection into blastocysts and subsequent transfer into foster females, a key assay to probe the ability of nonhuman pluripotent cell lines to functionally integrate into tissues of the developing embryo (Table 1; Gardner 1968). The finding that ECCs can also be derived from teratocarcinomas experimentally induced by the transplantation of implantation-stage mouse embryos to extrauterine sites in histocompatible hosts motivated attempts to isolate pluripotent cells directly from such embryos. These efforts led to the derivation of embryonic stem cells (ESCs) from the ICM of mouse blastocysts (Evans and Kaufman 1981; Martin 1981) and, subsequently, also from human embryos (Thomson et al. 1998). In contrast to ECCs, ESCs are karyotypically normal and contribute efficiently to all adult tissues, including the germline. In fact, ESCs are capable of producing entirely ESC-derived animals after injection into tetraploid blastocysts (Nagy et al. 1990; Eggan et al. 2001), which by themselves develop into extraembryonic tissues only but fail to give rise to the embryo proper, thereby passing the most stringent developmental assay available in mice (Table 1). Notably, ESC lines can also be derived from cloned blastocysts generated by SCNT (Cibelli et al. 1998; Munsie et al. 2000; Wakayama et al. 2001), generating so-called NT-ESCs. In contrast to the abnormalities seen in directly cloned animals, NT-ESCs are transcriptionally and functionally indistinguishable from fertlization-derived ESCs, presumably because of a selection of faithfully reprogrammed cells in culture (Brambrink et al. 2006; Wakayama et al. 2006).
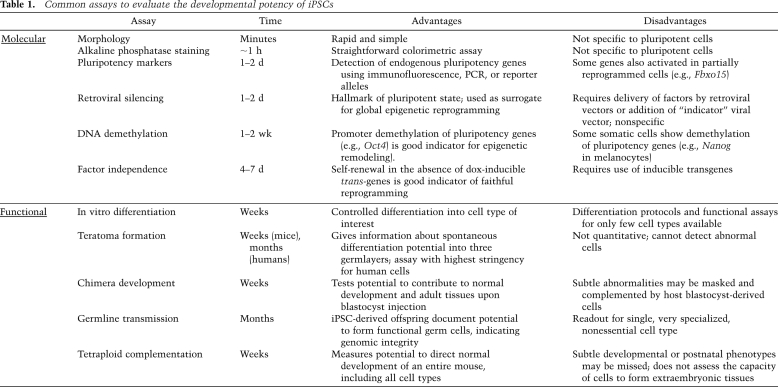
Table 1.
Common assays to evaluate the developmental potency of iPSCs
Pluripotent cell lines have also been derived from other embryonic and adult tissues upon explantation in culture. For example, epiblast-derived stem cells (EpiSCs) (Brons et al. 2007; Tesar et al. 2007) have been isolated from post-implantation embryos, embryonic germ cells (EGCs) (Matsui et al. 1992; Resnick et al. 1992) have been derived from primordial germ cells (PGCs) of the midgestation embryo, and multipotent germline stem cells (mGSCs) have been generated from explanted neonatal (Kanatsu-Shinohara et al. 2004) and adult (Guan et al. 2006; Seandel et al. 2007; Ko et al. 2009) mouse testicular cells. Common to all of these cell types is their origin from either early embryos or germ lineage cells, which appear to be the only cells that harbor an epigenetic conformation that is permissive for spontaneous conversion into a pluripotent state. A molecular commonality among these cell types is the expression of endogenous Oct4, which may thus serve as a valuable predictive marker for whether or not a cell can give rise to pluripotent cell lines. While ESCs, ECCs, mGSCs, and EGCs are pluripotent, only ESCs pass the most stringent developmental assay: tetraploid embryo complementation. This is because ESCs carry balanced parental imprints that are critical for normal development, whereas EGCs and mGSCs have erased imprints or paternal-only imprints, respectively, as a result of germline development (Hochedlinger and Jaenisch 2006). However, all of the pluripotent cell lines tested so far (ESCs, ECCs, and EGCs) have been shown to induce pluripotency in somatic cells after cellular fusion, demonstrating that they harbor dominant reprogramming activities (Tada et al. 1997, 2001; Cowan et al. 2005).
Transcription factors and lineage switching
The third principle that contributed to the discovery of induced pluripotency was the observation that lineage-associated transcription factors—which help to establish and maintain cellular identity during development by driving the expression of cell type-specific genes while suppressing lineage-inappropriate genes—can change cell fate when ectopically expressed in certain heterologous cells. This idea was first demonstrated by the formation of myofibers in fibroblast cell lines transduced with retroviral vectors expressing the skeletal muscle factor MyoD (Davis et al. 1987). Subsequently, Graf and colleagues (Xie et al. 2004; Laiosa et al. 2006) discovered that primary B and T cells could be converted efficiently into functional macrophages upon overexpression of the myeloid transcription factor C/EBPα. More recently, researchers have identified sets of transcription factors that induce the conversion of pancreatic acinar cells into insulin-producing β cells by overexpressing the pancreatic factors MafA, Pdx1, and Ngn3 (Zhou et al. 2008); the conversion of fibroblasts into neurons by the activation of the neural factors Ascl1, Brn2, and Myt1l (Vierbuchen et al. 2010); and the conversion of fibroblasts into cardiomyocytes by the cardiac factors Gata4, Mef2c, and Tbx5 (Ieda et al. 2010). Of note, these experiments proved that lineage conversions are not restricted to cell types within the same lineage or germ layer, since fibroblasts are mesodermal in origin, whereas neurons are derived from ectoderm. Some of the early transdifferentiation experiments provided the intellectual framework for a more systematic search for transcription factors that could induce the conversion of differentiated cells to a pluripotent state, which is discussed below.
iPSCs
To identify transcriptional regulators that can reprogram adult cells into pluripotent cells, Yamanaka and Takahashi (Tokuzawa et al. 2003) devised an elegant screen for factors within a pool of 24 pluripotency-associated candidate genes that could activate a dormant drug resistance allele integrated into the ESC-specific Fbxo15 locus. The combination of 24 factors, when coexpressed from retroviral vectors in mouse fibroblasts, indeed activated Fbxo15 and induced the formation of drug-resistant colonies with characteristic ESC morphology (Takahashi and Yamanaka 2006). Successive rounds of elimination of individual factors then led to the identification of the minimally required core set of four genes, comprising Klf4, Sox2, c-Myc, and Oct4. iPSCs generated by selection for Fbxo15 activation expressed markers of pluripotent stem cells such as SSEA-1 and Nanog, generated teratomas when injected subcutaneously into immunocompromised mice, and contributed to different tissues of developing embryos upon blastocyst injection (Takahashi and Yamanaka 2006), thereby fullfilling some criteria of pluripotency (Table 1). However, these iPSCs expressed lower levels of several key pluripotency genes compared with ESCs, showed incomplete promoter demethylation of ESC regulators such as Oct4, and failed to generate postnatal chimeras or contribute to the germline (Takahashi and Yamanaka 2006). These “first-generation” iPSCs therefore appeared to be only partially reprogrammed. Soon after this study, several laboratories, including Yamanaka's (Okita et al. 2007), were able to reproduce and improve upon these findings. For example, by selecting for the reactivation of the essential pluripotency genes Nanog or Oct4 instead of Fbxo15, iPSCs were generated that molecularly and functionally more closely resembled ESCs (Maherali et al. 2007; Okita et al. 2007; Wernig et al. 2007). More recently, rare iPSC lines have been identified that are even capable of generating “all-iPSC” mice upon injection into tetraploid blastocysts (Boland et al. 2009; Kang et al. 2009; Zhao et al. 2009; Stadtfeld et al. 2010b), suggesting that at least some iPSC clones have a developmental potency equivalent to ESCs.
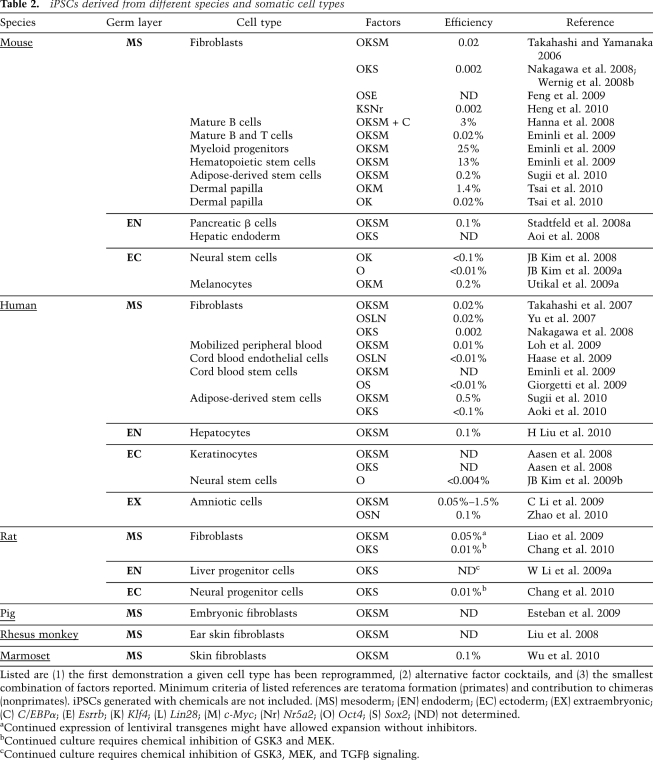
iPSCs have also been derived from a number of different species—including humans (Takahashi et al. 2007; Yu et al. 2007; Park et al. 2008b), rats (W Li et al. 2009a), and rhesus monkeys (Liu et al. 2008)—by expression of the four Yamanaka factors (Table 2), demonstrating that fundamental features of the transcriptional network governing pluripotency remain conserved during evolution. Similarly, iPSCs have been derived from other somatic cell populations, such as keratinocytes (Aasen et al. 2008; Maherali et al. 2008), neural cells (Eminli et al. 2008; JB Kim et al. 2008), stomach and liver cells (Aoi et al. 2008), and melanocytes (Utikal et al. 2009a), as well as from genetically labeled pancreatic β cells (Stadtfeld et al. 2008a) and terminally differentiated lymphocytes (see also Table 2; Hanna et al. 2008; Eminli et al. 2009), further underscoring the universality of induced pluripotency.
Table 2.
iPSCs derived from different species and somatic cell types
Listed are (1) the first demonstration a given cell type has been reprogrammed, (2) alternative factor cocktails, and (3) the smallest combination of factors reported. Minimum criteria of listed references are teratoma formation (primates) and contribution to chimeras (nonprimates). iPSCs generated with chemicals are not included. (MS) mesoderm; (EN) endoderm; (EC) ectoderm; (EX) extraembryonic; (C) C/EBPα; (E) Esrrb; (K) Klf4; (L) Lin28; (M) c-Myc; (Nr) Nr5a2; (O) Oct4; (S) Sox2; (ND) not determined.
aContinued expression of lentiviral transgenes might have allowed expansion without inhibitors.
bContinued culture requires chemical inhibition of GSK3 and MEK.
cContinued culture requires chemical inhibition of GSK3, MEK, and TGFβ signaling.
Technical advances in iPSC generation
iPSC derivation is ethically and legally less problematic and technically more feasible than SCNT. In order to use iPSCs as efficient research tools and ultimately translate this technology into clinical applications, suitable techniques of factor delivery and efficient identification of faithfully reprogrammed cells are crucial. Thus, recent advances in the area of iPSC generation and identification are discussed in the following section.
Factor delivery into target cells
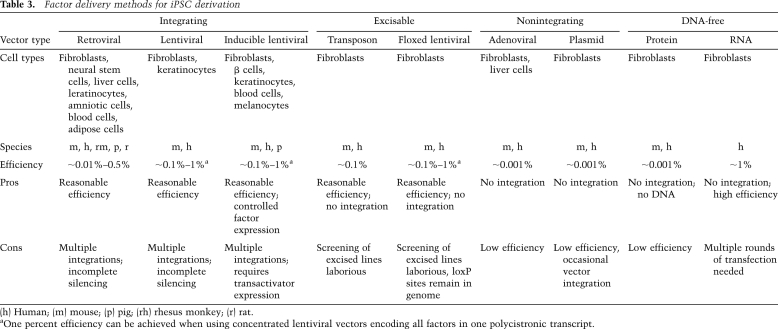
A number of different approaches have been devised to shuttle reprogramming factors into somatic cells (Table 3), which can affect the efficiency of reprogramming and the quality of resultant iPSCs. For example, the first studies on iPSCs used constitutively active retroviral vectors that stably integrated into the host cell genome to introduce c-Myc, Klf4, Oct4, and Sox2 (Takahashi and Yamanaka 2006; Maherali et al. 2007; Okita et al. 2007; Wernig et al. 2007). While retroviral transgenes are usually silenced toward the end of reprogramming (Stadtfeld et al. 2008b), due to the activation of both DNA (Lei et al. 1996) and histone (Matsui et al. 2010) methyltransferases, this process is often incomplete, resulting in partially reprogrammed cell lines that continue to depend on exogenous factor expression and fail to activate the corresponding endogenous genes (Takahashi and Yamanaka 2006; Mikkelsen et al. 2008; Sridharan et al. 2009). In addition, residual activity or reactivation of viral transgenes in iPSC-derived somatic cells can interfere with their developmental potential (Takahashi and Yamanaka 2006) and frequently leads to the formation of tumors in chimeric animals (Okita et al. 2007). This issue becomes exacerbated when constitutively active lentiviral vectors are used to produce iPSCs, which are even less efficiently silenced in pluripotent cells than retroviral vectors and can thus cause a differentiation block (Brambrink et al. 2008; Sommer et al. 2010). The use of inducible lentiviral vectors, whose expression can be controlled by the inert drug doxycycline, diminishes the risk of continued transgene expression and allows for the selection of fully reprogrammed iPSCs, since cells that depend on exogenous factor expression readily stop proliferating upon doxycycline withdrawal (Brambrink et al. 2008; Stadtfeld et al. 2008b). Lentiviral vectors are also more efficient than retroviral vectors at infecting different somatic cell types and can be used to express polycistronic cassettes encoding all four reprogramming factors, thus increasing reprogramming efficiency (Carey et al. 2009; Sommer et al. 2009).
Table 3.
Factor delivery methods for iPSC derivation
(h) Human; (m) mouse; (p) pig; (rh) rhesus monkey; (r) rat.
aOne percent efficiency can be achieved when using concentrated lentiviral vectors encoding all factors in one polycistronic transcript.
Inducible vector systems have been employed to generate so-called “secondary” reprogramming systems, which do not rely on direct factor delivery into target cells. These systems entail differentiating “primary” iPSC clones, generated with doxycyline-inducible lentiviral vectors or transposons, into genetically homogeneous somatic cells using either in vitro differentiation (for human cells) (Hockemeyer et al. 2008; Maherali et al. 2008) or blastocyst injection (for mice) (Wernig et al. 2008a; Woltjen et al. 2009). These somatic cells are then cultured in doxycycline-containing media, thus triggering the formation of “secondary” iPSCs at efficiencies that depend on the specific cell type used but are generally several orders of magnitude higher than the efficiencies obtained after primary infection. Secondary systems therefore (1) allow for the reprogramming of large quantities of genetically homogeneous cells for biochemical studies and cells that are difficult to culture or transduce, and (2) facilitate the comparison of genetically matched iPSCs derived from different somatic cell types.
In a modification of the conventional secondary system, mouse strains lacking individual reprogramming transgenes have been generated as a screening platform for the identification of small molecules that can substitute for a given reprogramming factor (Markoulaki et al. 2009). Because lentiviral transgenes, however, often exhibit heterogeneous expression patterns in secondary cells, several primary iPSC clones need to be screened to identify the ones that efficiently reactivate the factors, a process that can be quite cumbersome. The recent development of “reprogrammable” mouse strains, which contain a single inducible polycistronic transgene in a defined genomic position, has solved this issue, and also enables the breeding of animals into desired mutant backgrounds for mechanistic studies (Carey et al. 2010; Stadtfeld et al. 2010a).
Integration-free iPSCs
Approaches to derive iPSCs free of transgenic sequences are aimed at circumventing the potentially harmful effects of leaky transgene expression and insertional mutagenesis. This is particularly important when considering iPSC technology in a therapeutic setting. Techniques to generate integration-free iPSCs can be subdivided into three categories: (1) those that use vectors that do not integrate into the host cell genome, (2) those that use integrating vectors that can be subsequently removed from the genome, and (3) those that do not use nucleic acid-based vectors at all (Table 3).
The first integration-free iPSCs were generated from adult mouse hepatocytes using nonintegrating adenoviral vectors (Stadtfeld et al. 2008c), and from mouse embryonic fibroblasts (MEFs) transfected with plasmids (Okita et al. 2008). These experiments provided the proof of principle that transient expression of the four classical reprogramming factors is indeed sufficient to induce pluripotency in somatic cells. The absence of common integration sites in iPSCs produced with retroviruses (Varas et al. 2009) or lentiviruses (Winkler et al. 2010) corroborated this conclusion, and showed that insertional mutagenesis at best plays a supportive role during iPSC formation. Human fibroblasts have also been reprogrammed into iPSCs with adenoviral vectors (Zhou and Freed 2009) and Sendai virus (Fusaki et al. 2009), as well as with polycistronic minicircle vectors (Jia et al. 2010) and self-replicating selectable episomes (Yu et al. 2009), albeit the latter system required the simultaneous overexpression of additional factors, including another potent oncogene (Oct4, Sox2, c-Myc, and Klf4, together with Nanog, Lin28, and SV40LT).
Reprogramming efficiencies with current nonintegrating methods are several orders of magnitude lower (∼0.001%) than those achieved with integrating vectors (0.1%–1%) (Table 3), most likely because factor expression is not maintained for a sufficient length of time to allow complete epigenetic remodeling. To avoid this issue, several laboratories have developed integration-dependent gene delivery vectors with incorporated loxP sites that can be subsequently excised from the host genome by transient expression of Cre recombinase (Kaji et al. 2009; Soldner et al. 2009). This approach enables the efficient generation of iPSCs from different cell types, especially if polycistronic vectors are used (Chang et al. 2009; Sommer et al. 2010). It remains to be seen, however, whether short vector sequences, which inevitably remain in the host cell DNA after excision, affect cellular function. Transgene-free iPSCs can also be generated with piggyBac transposons, mobile genetic elements that can be introduced into and removed from the host genome by transient expression of transposase (Woltjen et al. 2009; Yusa et al. 2009). The low error rate of this process allows for a seamless excision, but requires characterization of integration sites in iPSCs before and after transposon removal. It also remains unclear if transposase expression can induce nonspecific genomic alterations in iPSCs (Stadtfeld and Hochedlinger 2009).
Successful reprogramming has been achieved recently without the use of viral or plasmid vectors at all. Specifically, iPSCs have been derived from both mouse and human fibroblasts by delivering the reprogramming factors as purified recombinant proteins (Zhou et al. 2009) or as whole-cell extracts isolated from either ESCs (Cho et al. 2010) or genetically engineered HEK293 cells (D Kim et al. 2009). While the use of purified proteins represents an attractive approach for the generation of transgene-free iPSCs, its efficiency is extremely low and, in the recombinant protein approach, required the addition of the histone deacetylase (HDAC) inhibitor valproic acid (VPA) to the culture media. A more efficient and safer way of producing integration-free iPSCs may be the introduction of modified RNA molecules encoding for the reprogramming factors into somatic cells, which has been validated recently (Warren et al. 2010).
To improve the overall low efficiencies of generating iPSCs with most nonintegrating approaches, screens for chemical compounds that promote reprogramming have been performed. This led to the identification of a number of molecules that significantly increase reprogramming efficiencies in the context of Oct4, Klf4, Sox2, and c-Myc overexpression (for review, see Desponts and Ding 2010; Li and Ding 2010). Notably, some of these molecules can also replace individual reprogramming factors, raising the possiblility of deriving iPSCs solely with chemicals (Desponts and Ding 2010; Li and Ding 2010). However, it should be noted that chemical substitution of a reprogramming factor is, in most cases, associated with a significant decrease in the number of iPSC clones generated, indicating that no single chemical compound is able to entirely replace the function of a transcription factor. Another potential caveat of chemical reprogramming approaches is the introduction of genetic or epigenetic abnormalities into resultant iPSCs, especially since many of the reported compounds are potent modulators of DNA and chromatin modifications.
Identification of iPSC colonies
The inherently low efficiency of iPSC derivation benefits from selection approaches that distinguish successfully reprogrammed clones from partially reprogrammed or simply transformed colonies. The reactivation of endogenous pluripotency-associated genes—such as Fbxo15 (Takahashi and Yamanaka 2006), Nanog or Oct4 (Maherali et al. 2007; Okita et al. 2007; Wernig et al. 2007), and Utf1 (Pfannkuche et al. 2010)—linked to drug selection cassettes has been successfully employed for this purpose. As mentioned above, Fbxo15 selection generates partially reprogrammed cells (Takahashi and Yamanaka 2006), likely because activation of this gene occurs early in the reprogramming process when the majority of cells are not yet faithfully reprogrammed (Stadtfeld et al. 2008b). A general limitation of any drug selection approach is that it requires genetic engineering of cells or mice. To circumvent this problem, lentiviral vector systems have been developed that carry promoter fragments of pluripotency genes whose activity can be selected for, and that, in principle, can be applied to a wide range of murine and human cell types (Hotta et al. 2009).
For human iPSCs, expression of surface markers such as TRA-1-81 has been shown to enrich for reprogrammed cells (Lowry et al. 2008). A more stringent approach to identify faithfully reprogrammed human iPSCs without the use of drug selection combines the detection of surface markers with that of “indicator retroviruses” expressing fluorescent proteins, which become silenced upon acquisition of pluripotency (Chan et al. 2009).
Importantly, high-quality iPSCs can be derived from unmodified somatic cells without drug selection or fluorescent reporters at all by simply using morphological criteria (Blelloch et al. 2007; Maherali et al. 2007; Meissner et al. 2007), although this approach requires careful characterization of the resultant cell lines. This “no selection” approach is therefore most powerful when combined with doxycycline-inducible vectors, as cells that have entered a self-sustaining pluripotent state can be easily selected for by removal of doxycycline (Brambrink et al. 2008; Stadtfeld et al. 2008b), even though, in rare cases, doxycycline-independent partially reprogrammed cells have been reported (Mikkelsen et al. 2008).
For mouse iPSCs, the expression status of the imprinted Gtl2 gene has been described recently as a refined marker that allows for the prospective identification of clones that support the development of tetraploid embryo complementation mice and therefore appear developmentally indistinguishable from ESCs (L Liu et al. 2010; Stadtfeld et al. 2010b). Whether the human homolog MEG-3 or any other gene has similar predictive value in human iPSCs remains to be tested.
Mechanisms underlying iPSC formation
In the following section, we introduce models that have been developed to explain the low efficiency of reprogramming at a cellular level. We then discuss key molecular events that may act as barriers during the reprogramming process, and speculate on the role of the individual reprogramming factors as well as on supporting and antagonizing factors during epigenetic remodeling. This is followed by a discussion of different pluripotent states that have been identified recently and that can be interconverted by some of the same transcription factors. Finally, we address the question of whether iPSCs are molecularly and functionally equivalent to fertilization-derived ESCs.
Elite vs. stochastic models
The derivation of iPSCs from most studied somatic cells is extremely inefficient (0.01%–0.1%) and occurs at a slow speed (∼2 wk) (Brambrink et al. 2008; Stadtfeld et al. 2008b). Even in the context of secondary systems, in which somatic cells homogenously express the factors, the efficiency of fibroblast reprogramming generally does not exceed 1%–5% (Maherali et al. 2008; Wernig et al. 2008a; Stadtfeld et al. 2010a), although one report documented an efficiency of up to 20% (Woltjen et al. 2009). Two opposing, but mutually nonexclusive, models have been put forward to explain these observations (Yamanaka 2009).
The so-called “elite” or “deterministic” model proposes that the efficiency of iPSC derivation is low because only a few cells in a somatic cell culture are susceptible to reprogramming. Somatic stem or progenitor cells, present in most adult tissues and possibly also in explanted cell populations, are the most obvious candidate cells, as they are rare and developmentally closer to pluripotent cells than differentiated cells. In contrast, the “stochastic” model poses that all somatic cells are equally amenable to factor-mediated reprogramming, but have to go through a series of stochastic epigenetic events to acquire pluripotency. Only a few cells may pass all of these roadblocks, resulting in the overall low efficiency.
The elite model by itself is difficult to sustain, since iPSC can be derived from several defined somatic cell types, including fully differentiated B and T lymphocytes (Hanna et al. 2008; Eminli et al. 2009) as well as pancreatic β cells (Stadtfeld et al. 2008a). Moreover, when following clonal populations of early B cells and monocytes expressing the reprogramming factors, almost all cell clones ultimately give rise to daughter cells that form iPSCs, even though this process requires several weeks to months (Hanna et al. 2009b). The latter observation suggests that continuous cell proliferation allows rare cells in a homogenous cell population to acquire stochastic changes that facilitate their conversion into a pluripotent state (Hanna et al. 2009b). In an independent set of experiments, however, clonally plated hematopoietic stem and progenitor cells were shown to give rise to iPSCs with significantly higher efficiencies and within a shorter time frame than mature lymphocytes and myeloid cells (10%–40% vs. 0.01%–1%), regardless of the proliferative state of the cells at the time of factor expression (Eminli et al. 2009), suggesting that the differentiation state itself may also influence the susceptibility of cells to form iPSCs. These seemingly contradictory results may be reconciled if one assumes that adult progenitor and stem cells require fewer stochastic events to induce pluripotency than terminally differentiated cells (Fig. 1A). Hence, reprogramming of progenitor or stem cells may occur in a shorter period of time, which is consistent with the observed fast up-regulation of pluripotency markers upon factor expression (Eminli et al. 2009). It would be important to perform similar long-term clonal assays as described above for monocytes and B cells with defined immature cell types to test this hypothesis. It should also be informative to follow the reprogramming of defined mature and immature cell populations at single-cell resolution by using, for example, time-lapse microscopy, as has been done recently for fibroblasts (Araki et al. 2010; Smith et al. 2010).
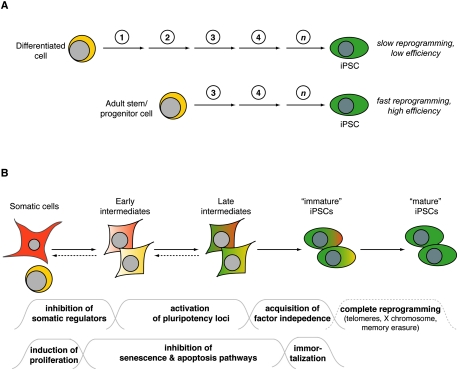
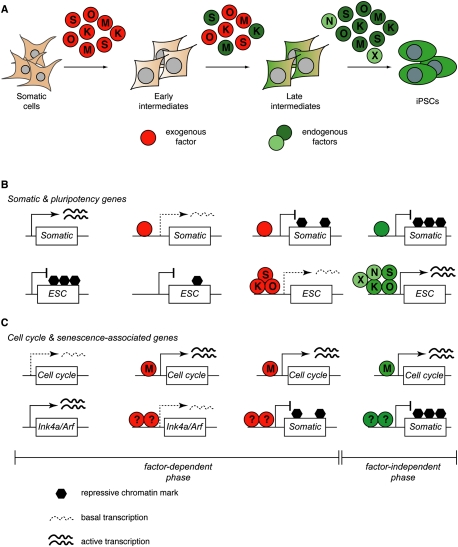
Figure 1.
Models of cellular reprogramming. (A) Mature cells, such as lymphocytes, reprogram into iPSCs at lower efficiencies than immature cells, such as hematopoietic stem cells. This may be due to a lower number of stochastic epigenetic events (represented by circled numbers and arrows) that are required in immature cells to acquire pluripotency. The precise number and nature of such changes is unclear (represented by “n”). (B) Scheme summarizing major changes that characterize the transition of somatic cells into iPSCs. The early steps are reversible, as indicated by the dashed reverse arrows. “Immature iPSCs” are defined as cells that have already acquired pluripotency but still retain an epigenetic memory of their cell type of origin, while “mature iPSCs” have lost this memory. The wavelines below indicate assumed reprogramming roadblocks that cells are facing at different stages. Failure to pass any of these roadblocks may result in cells that arrest at that stage or, alternatively, undergo senescence or apoptosis.
In conclusion, a modified stochastic model that integrates an elite component might be most accurate to explain the low efficiency of reprogramming. Consistent with this interpretation is the recent observation that subpopulations of fibroblasts give rise to iPSCs much sooner than the bulk population when following individual cells with live cell imaging (Smith et al. 2010). Similarly, a low expression status of the tumor suppressor gene Arf in fibroblast populations marks cells that are more amenable to reprogramming than cells with high Arf expression (Utikal et al. 2009b), suggesting that “elite” cells may also exist in cell populations with no proven differentiation hierarchy. Whether these elite populations represent fibroblasts that have undergone fewer numbers of cell division, contaminating nonfibroblast cell types, or immature mesenchymal cells remains to be tested.
Reprogramming roadblocks
The low efficiency and slow kinetics of iPSC derivation is in contrast to somatic cell lineage switching triggered by transcription factor overexpression, such as the conversion of B cells into macrophages induced by C/EBPα, which occurs at efficiencies of up to 100% and within 48 h (Xie et al. 2004; Bussmann et al. 2009). This suggests that the induction of pluripotency by defined factors faces more barriers than lineage conversion, possibly because of a higher degree of transcriptional and epigenetic similarity among mature cell types than between mature cells and pluripotent cells. Thus, what are the major molecular changes a somatic cell has to undergo during reprogramming into an iPSC?
Studies in fibroblasts suggest that reprogramming follows an organized sequence of events, which begins with the down-regulation of somatic markers (Stadtfeld et al. 2008b) and morphological changes reminiscent of a mesenchymal-to-epithelial transition (MET). In accordance, interference with genes involved in MET, such as E-cadherin and BMP receptor signaling, abrogate reprogramming (Li et al. 2010; Samavarchi-Tehrani et al. 2010). These events are followed by the activation of the early pluripotency markers SSEA-1, alkaline phosphatase, and Fbxo15 before bona fide pluripotency genes such as Nanog or Oct4 become expressed and cells gain independence from exogenous factor expression (Brambrink et al. 2008; Stadtfeld et al. 2008b). Notably, the acquisition of pluripotency may not be complete upon independence of exogenous factor expression and the activation of endogenous pluripotency genes but may continue for several rounds of cell divisions, as suggested by the finding that early- and late-passage iPSCs exhibit discernible differences in telomere length (Marion et al. 2009b) as well as global transcriptional and DNA methylation patterns (Chin et al. 2009; Polo et al. 2010). The transition to a pluripotent state is also accompanied by genome-wide remodeling of chromatin modifications, such as DNA and histone tail methylation from a somatic to an ESC-like state (Maherali et al. 2007; Mikkelsen et al. 2008), although the temporal sequence of these changes remains elusive.
Only a subset of fibroblasts expressing reprogramming factors down-regulates somatic markers and activates pluripotency genes (Wernig et al. 2008a; Stadtfeld et al. 2010a), indicating that many cells are refractory to reprogramming or become so as a consequence of factor expression. Consistent with this observation, reprogramming intermediates isolated based on combinations of the aforementioned markers have an increased probability of forming iPSC colonies (Stadtfeld et al. 2008b), suggesting that these cells have overcome several transcriptional and epigenetic barriers that normally prevent the induction of pluripotency. Of note, refractory fibroblast populations do not give rise to iPSCs even after prolonged culture, despite homogeneous factor expression (M Stadtfeld and K Hochedlinger, unpubl.), which is in contrast to observations made in pro-B cells (Hanna et al. 2009b) and may reflect cell type-specific responses to reprogramming factors. Collectively, these results suggest that the extinction of the somatic program and the subsequent activation of endogenous pluripotency genes may be roadblocks during iPSC formation (Fig. 1B).
A case in point for the importance of endogenous factor activation might be the (stochastic) epigenetic derepression of the endogenous Nanog locus by the reprogramming factors. Nanog and related pluripotency genes are activated late during reprogramming and may thus be limiting for efficient conversion of somatic cells into iPSCs. In agreement with this hypothesis, simultaneous overexpression of Nanog (Hanna et al. 2009b; Silva et al. 2009; Heng et al. 2010) as well as other pluripotency-associated transcriptional regulators—including Tbx3 (Han et al. 2010) and Sall4 (Tsubooka et al. 2009)—with Oct4, Sox2, Klf4, and c-Myc, has been shown to enhance and/or accelerate reprogramming. It is interesting to note that Nanog expression also appears to be a limiting factor during fusion-mediated reprogramming (Silva et al. 2006), suggesting common mechanisms.
The promoters of pluripotency genes such as nanog and Oct4 are stably silenced by DNA methylation in somatic cells (Gidekel and Bergman 2002), which likely interferes with transcription factor binding and gene activation during reprogramming. It is unclear how this roadblock is overcome during iPSC derivation, but mechanisms based on either passive or active DNA demethylation have been postulated (Hochedlinger and Plath 2009). In one scenario, an unknown reprogramming factor (or one of its targets) might antagonize the activity of the DNA methyltransferase Dnmt1, which maintains somatic DNA methylation patterns. This may lead to the progressive loss of DNA methylation with cell division. Since genome-wide demethylation is toxic for somatic cells (Li et al. 1992), this process would have to happen in a promoter-specific fashion, perhaps by the rapid binding of reprogramming factors to target sites on the newly synthesized and hence temporarily unmethylated DNA strand (Hochedlinger and Plath 2009). Otherwise, cell death pathways such as the p53 pathway would have to be inactivated to ensure survival of cells (Jackson-Grusby et al. 2001). In support of this notion, fibroblasts undergoing reprogramming have been shown to inactivate the ink4–arf pathway (H Li et al. 2009; Utikal et al. 2009b).
In a second scenario, DNA is actively demethylated at key pluripotency promoters by an as-yet-uncharacterized enzymatic activity. Evidence for a role of active DNA demethylation during development comes from studies of germ cells (Hajkova et al. 2008, 2010) and zygotes (Mayer et al. 2000; Oswald et al. 2000) as well as some adult somatic cells, including neurons and T lymphocytes (Bruniquel and Schwartz 2003; Miller and Sweatt 2007). In the context of reprogramming, active DNA demethylation has been observed upon SCNT, although the molecules involved remain elusive (Simonsson and Gurdon 2004). The deaminase Aid has been proposed recently to play a role in the demethylation of the NANOG and OCT4 promoters after cell fusion of mouse ESCs and human fibroblasts (Bhutani et al. 2010; Popp et al. 2010). Consistent with this finding, Aid, together with Gadd45 and Mbd4, has been shown to elicit DNA demethylation in zebrafish (Rai et al. 2008). Yet another class of enzymes potentially involved in active DNA demethylation is the Tet proteins, which catalyze the conversion of 5-methylcytosine to 5-hydroxymethylcytosine (Tahiliani et al. 2009; Ito et al. 2010). This modification could be recognized by glycosylases, resulting in demethylation of DNA. However, a role for Tet proteins in iPSC formation has yet to be established.
The slow kinetics of somatic pluripotency gene activation after enforced factor expression in fibroblasts (∼8–12 d) (Brambrink et al. 2008; Stadtfeld et al. 2008b) is in contrast to the rapid activation of these genes after SCNT (Egli et al. 2009) or cell fusion (∼24–48 h) (Han et al. 2008). This observation might argue for a passive, replication-dependent mode of DNA demethylation during iPSC derivation. Alternatively, DNA demethylation might be involved, but key components of its machinery might be limiting until late during reprogramming.
In contrast to ESCs, which are immortal, fibroblasts and most other somatic cell types have a restricted proliferative potential and undergo apoptosis, growth arrest, or stress-induced senescence in culture (Collado et al. 2007). The acquisition of cellular immortality therefore appears to be yet another roadblock during iPSC cell generation. Increased proliferation and up-regulation of genes driving cell cycle progression are indeed early events seen during fibroblast reprogramming (Mikkelsen et al. 2008). However, the observed dependence of cells at intermediate stages of reprogramming on exogenous factor expression for continuous growth (Brambrink et al. 2008; Stadtfeld et al. 2008b) suggests that stable immortalization occurs late during the process (Fig. 1B). In agreement with the idea that the onset of cellular senescence is a barrier during fibroblast reprogramming is the finding that expression of the reprogramming factors in p53 or ink4a/arf-deficient immortalized fibroblasts leads to dramatic increases in reprogramming efficiency and speed (Banito et al. 2009; Hong et al. 2009; Kawamura et al. 2009; H Li et al. 2009; Utikal et al. 2009b). In addition to its role in conferring immortality on fibroblasts, p53 loss may also contribute to increased reprogramming by inhibiting DNA damage-induced apoptosis (Marion et al. 2009a). It is interesting to note, however, that loss of p53 can elicit different responses in cells expressing the four factors. In fibroblasts, the main effect of p53 loss appears to be inhibition of senescence and cell death, while, in blood cells expressing reprogramming factors, p53 loss mainly contributes to reprogramming by accelerating cell cycle progression (Hanna et al. 2009b).
The analysis of partially reprogrammed cells has been informative in identifying additional roadblocks of reprogramming. For example, Mikkelsen et al. (2008) showed that the activity of lineage-associated transcription factors and DNA hypermethylation can be barriers for the transition of partially reprogrammed cells into a fully pluripotent state. It is unclear whether observations made in partially reprogrammed cells, which are immortal cell lines, are representative of events that take place in regular reprogramming intermediates, which are unstable and transient cell populations. Nevertheless, support for an antagonizing role of somatic transcription factors during reprogramming comes from the observation that knockdown of the B-cell factor Pax5, which has been shown previously to elicit dedifferentiation of B cells into hematopoietic progenitors (Mikkola et al. 2002), significantly improves the reprogramming efficiency of mature B lymphocytes (Hanna et al. 2008).
A question that remains to be addressed in this context is whether reprogramming into iPSCs reverses normal development and thus entails dedifferentiation into a progenitor-like state before acquiring pluripotency. The appearance of partially reprogrammed cells, which exhibit similar phenotypes regardless of starting cell type (Mikkelsen et al. 2008), argues against dedifferentiation into lineage-specific progenitors. However, a definitive answer to this question will require the analysis of well-defined somatic cell populations, such as lymphocytes, and growth of cells in media that allow for the detection of potentially rare and transient progenitor cell populations. It has also been speculated that the enforced expression of transcription factors may generate artificial cell states that do not resemble any progenitor cell found in vivo (Nagy and Nagy 2010).
Role of reprogramming factors during epigenetic remodeling
How might individual reprogramming factors overcome the described roadblocks during iPSC derivation? To date, no studies have been performed that provide a comprehensive map of transcription factor binding, chromatin modifications, and gene expression in transient reprogramming intermediates to address this question. Nevertheless, extensive information is available on transcription factor binding in established human and mouse ESCs (Chen et al. 2008; J Kim et al. 2008; Kunarso et al. 2010) and iPSCs (Mikkelsen et al. 2008; Sridharan et al. 2009) that should be informative to build models on how they may act during reprogramming. These data suggest, for example, that Oct4, Nanog, Sox2, and Klf4 are part of a core pluripotency network that serves two main purposes: the repression of genes associated with differentiation, and the activation of ESC-specific targets (Boyer et al. 2005; Loh et al. 2006). Gene suppression by pluripotency factors in ESCs is associated with the recruitment of repressive chromatin remodeling complexes, such as NuRD (Kaji et al. 2006) and Polycomb (Boyer et al. 2006; Lee et al. 2006), to target promoters, leading to histone deacetylation and H3K27 trimethylation. A functional role for components of the Polycomb complex during epigenetic remodeling has been suggested by a recent report investigating fusion-mediated reprogramming between ESCs and lymphocytes (Pereira et al. 2010). ESCs lacking either PRC1 or PRC2 activity failed to remodel the genome of their somatic fusion partners, suggesting that these molecules might also be important during iPSC derivation.
Intriguingly, the mechanism by which pluripotency factors mediate transcriptional repression in pluripotent cells involves binding of single factors to target gene promoters (Fig. 2; J Kim et al. 2008; Sridharan et al. 2009). In contrast, occupancy by multiple factors (J Kim et al. 2008) leads to recruitment of the basal transcription machinery and, consequently, strong gene activation. This dual control of target gene regulation might explain why somatic genes are usually silenced before pluripotency genes become activated: While repressive complexes can form immediately upon binding of individual exogenously expressed factors to cognate DNA sequences, key components of the more elaborate activating complexes, such as Nanog or Dax1 (Wang et al. 2006), may be limiting or absent at early stages of reprogramming and may become available only late, when their respective endogenous genomic loci have become activated (Fig. 2). The binding of pluripotency factors to their target genes might be facilitated by nucleosome remodelers such as Chd1 (Gaspar-Maia et al. 2009) and BAF (Singhal et al. 2010), both of which increase reprogramming efficiencies and kinetics when overexpressed. Once the majority of core pluripotency factors are expressed, they presumably engage in positive feedback and feed-forward loops of their own and other pluripotency promoters (Chen et al. 2008; J Kim et al. 2008) in order to sustain pluripotency in the absence of exogenous factor expression (Fig. 2). This model is supported by the observation that endogenous pluripotency genes, telomerase, and the silenced X chromosome in female cells become reactivated, while retroviruses are silenced, toward the end of the reprogramming process (Stadtfeld et al. 2008b), although the precise order of these events has yet to be established.
Figure 2.
Putative role of reprogramming factors during iPSC formation. (A) Scheme depicting the expression of exogenous (red circles) and endogenous (dark-green circles) pluripotency factors at the protein level during different stages of reprogramming. The reprogramming process is initiated predominantly by the exogenous factors, which are gradually replaced by endogenous proteins as well as their targets, such as Nanog (N) or as-yet-unidentified factors (X) (light-green circles). The endogenous loci of some reprogramming factors (such as c-Myc, Klf4, and Sox2) are expressed in some somatic cell types, and the corresponding endogenous proteins might thus become available before activation of the Oct4 locus. (B) Scheme illustrating how the reprogramming factors may exert the rapid repression of somatic genes and the gradual activation of pluripotency (ESC) genes, two processes assumed to be mediated largely by Klf4, Sox2, and Oct4. Somatic gene silencing is associated by single-factor binding to promoter regions, while ESC gene activation involves the establishment of multiprotein complexes. The initial loss of repressive marks (such as DNA methylation and H3K27 histone trimethylation) at ESC promoters might be a passive process driven by multiple rounds of cell division. (C) Scheme showing activation of genes promoting cell division (such as cyclins) by c-Myc and repression of the Ink4a/Arf tumor suppressor locus conferring immortality by an as-yet-undefined combination of reprogramming factors.
Studies on bulk populations undergoing reprogramming as well as on partially reprogrammed cells suggest that incorrect binding of transcription factors to targets might be another reason for the inability of many cells to acquire pluripotency (Sridharan et al. 2009). This report confirms the previously noted cooperativity of Oct4, Sox2, and Klf4 in pluripotent cells. It further suggests that c-Myc plays an independent role early during reprogramming by inducing cellular proliferation and driving a concomitant switch toward an energy metabolism typical of cancer cells (Fig. 2C; Mikkelsen et al. 2008; Sridharan et al. 2009). Accordingly, for Myc to exert its enhancing effects, it has to be expressed only during the first few days of reprogramming (Sridharan et al. 2009). Consistent with this finding, premature expression of c-Myc and Klf4 in fibroblasts prior to activation of all four factors increases reprogramming efficiencies and speed, while early expression of Sox2 and Oct4 has no effect (Markoulaki et al. 2009). Thus, c-Myc might not be involved directly in inducing pluripotency, but rather in priming somatic cells for subsequent pluripotency factor expression. This could occur by facilitating the binding of Oct4 and Sox2 to target genes; for example, by establishing or maintaining activating histone methylation (CH Lin et al. 2009) and acetylation (Knoepfler 2008) marks. In addition, c-Myc might play a role later during reprogramming by solidifying pluripotency gene expression, as is suggested by its presence on promoters of many genes and microRNAs (miRNAs) highly expressed in ESCs (Chen et al. 2008; J Kim et al. 2008; Judson et al. 2009).
Consistent with c-Myc's role as a facilitator of early stages of reprogramming, exogenous c-Myc can be omitted entirely from the reprogramming cocktail (Nakagawa et al. 2008; Wernig et al. 2008b), albeit at the cost of reduced reprogramming efficiency and speed. Another reason why exogenous c-Myc expression is dispensable might be the observation that endogenous c-Myc is already expressed at low levels in many somatic cells, and may hence become readily activated by the remaining factors. Consistent with this notion, somatic cells that express other reprogramming factors endogenously do not require administration of the corresponding exogenous factor(s), as has been demonstrated for neural stem cells that can be converted into iPSCs by Oct4 alone (Table 2; JB Kim et al. 2009a).
In this context, it is important to mention that Oct4, Sox2, Klf4, and c-Myc are not the only factor combination that can generate iPSCs. For example, human iPSCs have been derived by enforced expression of Oct4, Sox2, Nanog, and Lin28 (Yu et al. 2007). This suggests that different routes may lead to a common pluripotent ground state, or, alternatively, that different transcription factors activate the same program by reinforcing each other's synthesis. Indeed, lin28 represses let-7 miRNAs (Viswanathan et al. 2008), which are negative regulators of c-Myc translation (Akao et al. 2006; HH Kim et al. 2009), thus establishing a link between the two reprogramming cocktails (Oct4, Klf4, Sox2, and c-Myc, and Oct4, Sox2, Nanog, and Lin28). Similarly, Sox2 and Klf4 can be replaced by a number of closely related proteins, such as Sox1 and Klf2 (Nakagawa et al. 2008), respectively, suggesting that they recognize similar DNA-binding motifs. However, some of the classical reprogramming factors can be replaced by seemingly unrelated molecules—such as Klf4 by Esrrb (Feng et al. 2009) and Oct4 by the orphan nuclear receptor Nr5a2 (Heng et al. 2010)—during mouse fibroblast reprogramming. The mechanisms by which these alternative factors function during reprogramming remain elusive.
Antagonists and cooperative factors
The finding that iPSC formation is slow in comparison with SCNT and cell fusion suggests that additional cooperative factors of Oct4, Sox2, c-Myc, and Klf4 must exist and thus should be identifiable. Indeed, a number of modulators of the reprogramming process have been identified that can be divided into transcription factors, miRNAs, chromatin regulators, and growth factors, as well as small compounds targeting any of these molecules. An understanding of how these modulators interact with the reprogramming process has provided additional insight into the mechanisms of iPSC formation.
One group of cooperating factors appears to enhance reprogramming by impairing the activity or expression of components of the p53 pathway, further emphasizing the importance of overcoming cellular senescence and/or cell death pathways during iPSC formation. For instance, overexpression of SV40 large T antigen (Mali et al. 2008; Park et al. 2008c) or Rem2 GTPase (Edel et al. 2010) has been shown to enhance reprogramming by inhibiting p53. The culturing of reprogramming cells in hypoxic conditions (Utikal et al. 2009b; Yoshida et al. 2009) or in the presence of ascorbic acid (Esteban et al. 2010) also seems to target the p53 pathway and thus may provide safer ways to boost iPSC generation. Interestingly, these culture conditions also bypass the requirement for c-Myc overexpression, thus indicating a possible functional redundancy between c-Myc overexpression and loss of p53.
Another class of modulators comprises components of signaling pathways. For example, addition of recombinant TGFβ to fibroblast cultures almost completely abrogates iPSC formation, whereas inhibition of TGFβ signaling by chemically antagonizing its receptor significantly increases reprogramming efficiency and kinetics. This was seen in both mouse (Ichida et al. 2009; Maherali and Hochedlinger 2009) and human fibroblasts (T Lin et al. 2009), with human cells requiring simultaneous inhibition of MAPK signaling. In mice, TGFβ inhibition also abolishes the requirement for exogenous c-Myc and, to a lesser extent, Sox2 expression. While the effect of Tgfβ signal inhibition might be partly explained by facilitating the activation of the Nanog locus (Ichida et al. 2009), another interpretation is that blocking TGFβ signaling might be critical to initiate a MET that occurs early during reprogramming (Li et al. 2010; Samavarchi-Tehrani et al. 2010).
Similarly to Tgfβ inhibition, activation of Wnt signaling increases the efficiency of fibroblast reprogramming in the absence of exogenous c-Myc (Marson et al. 2008). This is consistent with c-Myc being a downstream effector of Wnt signaling (He et al. 1998). Furthermore, chemical inhibition of GSK3, which is an inhibitor of the Wnt signaling pathway, allows for the reprogramming of human fibroblasts without exogenous Sox2 (W Li et al. 2009b) and, when combined with inhibition of MAPK signaling, enhances reprogramming of neural stem cells and triggers the conversion of partially reprogrammed cells into iPSCs (Silva et al. 2008). Of note, the combined inhibition of Gsk3 and MAPK signaling has been shown previously by the Smith laboratory (Ying et al. 2008) to be sufficient to maintain murine ESCs in a self-renewing, pluripotent state by suppressing their differentiation. Consistently, inhibition of MAPK/Erk signaling antagonizes differentiation of the ICM in the mouse blastocyst (Chazaud et al. 2006; Nichols et al. 2009b). The finding that inhibition of differentiation pathways enhances reprogramming is in further agreement with the observations that adult progenitor cells give rise to iPSCs more efficiently than their differentiated progeny (Eminli et al. 2009), and that inhibition of lineage-specific factors (Hanna et al. 2008; Mikkelsen et al. 2008) can facilitate the reprogramming process.
iPSC formation requires the extinction of a somatic epigenetic state and the establishment of a pluripotent epigenetic state, suggesting that the manipulation of epigenetic regulators or modifications should influence reprogramming as well. Indeed, a number of chromatin-modifying agents have been identified that enhance the overall efficiency of reprogramming and also substitute for individual reprogramming factors or combinations of factors. For instance, inhibition of HDACs by VPA or butyrate allows for the efficient derivation of mouse iPSCs from fibroblasts in the absence of either c-Myc (Huangfu et al. 2008a) or Klf4 (Mali et al. 2010), and human iPSCs in the absence of both SOX2 and C-MYC (Huangfu et al. 2008b). Whether HDAC treatment enhances reprogramming by increasing the levels of endogenous c-Myc and Klf4 expression remains unclear. Likewise, inhibition of the histone methyltransferase G9a can replace either c-Myc or Sox2 during the reprogramming of mouse neural progenitor cells (NPCs) and fibroblasts, and it can substitute for Oct4 when administered to NPCs transduced with retroviruses expressing c-Myc, Klf4, and Sox2 (Shi et al. 2008). This effect is consistent with G9a's role in the post-implantation embryo, where it is involved in the stable epigenetic silencing of Oct4 (Feldman et al. 2006).
Lastly, a number of miRNAs have been identified that influence the reprogramming process. These include the previously discussed let-7 family, which blocks reprogramming by interfering with activation of the pluripotency factors c-Myc, lin28, and Sall4, while miR-294 activates these genes and thereby has a promoting effect on reprogramming (Melton et al. 2010). Both let-7 inhibition (Melton et al. 2010) and enforced miR-294 expression (Judson et al. 2009) have also been shown to functionally replace exogenous c-Myc expression during reprogramming. Recently, several miRNAs suppressing mesenchymal gene expression have been reported to enhance the reprogramming of fibroblasts, possibly by promoting a MET (Samavarchi-Tehrani et al. 2010).
Reprogramming factors and alternative pluripotent states
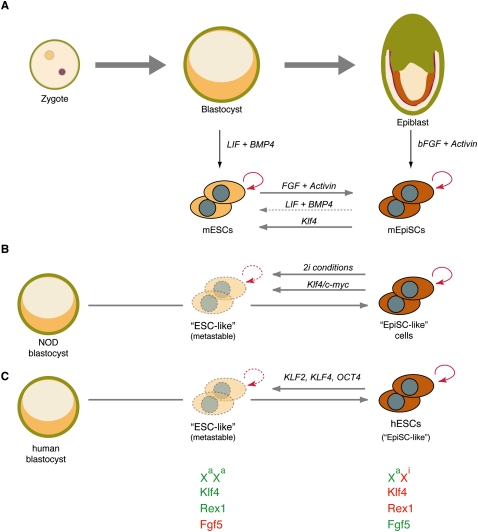
Pluripotent cell lines exist in two distinct states that are characterized by different growth factor requirements and developmental properties (Fig. 3). Murine ESCs established from the ICM of preimplantation blastocysts in the presence of LIF and BMP exist in a more primitive or “naïve” pluripotent state and fulfill all criteria of pluripotency (Table 1; Nichols and Smith 2009). In contrast, EpiSCs derived from post-implantation embryos in the presence of bFGF and activin represent a more advanced or “primed” pluripotent state that exhibits some pluripotency criteria, such as teratoma formation, but fails to contribute efficiently to tissues in mice (Brons et al. 2007; Tesar et al. 2007). ESCs exposed to bFGF readily give rise to EpiSCs (Guo et al. 2009), probably reflecting their normal differentiation path, whereas EpiSCs cultured in LIF and BMP reprogram into ESC-like cells at low frequency (Bao et al. 2009). Together, these results indicate that environmental changes are sufficient to interconvert these closely related cell types. Forced expression of Klf4 can promote the conversion of EpiSCs into ESC-like cells in the presence of LIF and BMP (Guo et al. 2009), presumably by reinforcing the LIF/Stat3 pathway (Hall et al. 2009).
Figure 3.
Transitions between alternative pluripotent states. (A) Model showing early developmental stages of the mouse embryo, from zygote to blastocyst and, subsequently, to post-implantation epiblast. ESCs are derived from the ICM (orange crescent) of the blastocyst and require LIF and BMP4 for indefinite self-renewal in vitro (indicated by the curved red arrow). EpiSCs are derived from epiblast stage embryos and require bFGF and activin for their propagation. ESCs readily differentiate into EpiSCs upon the switch to appropriate culture conditions, while the reverse transition is rare but can be enhanced significantly by enforced expression of Klf4. (B) Explant cultures of blastocysts from nonpermissive mouse strains (such as NOD) do not give rise to stable ESC lines in LIF and BMP4. However, a metastable ESC-like state can be attained by forced expression of Klf4 and c-Myc, or by repression of both MAPK and GSK3 signaling (“2i condition”). (C) Human ESCs resemble mouse EpiSCs in their epigenetic configuration (one inactive X chromosome [Xi], and one active X chromosome [Xa]) and marker gene expression (such as Fgf5). A metastable murine ESC-like state can be induced in these cells by overexpression of KLF2, KLF4, and OCT4. Active genes are shown in green, and inactive genes are shown in red.
Enforced expression of combinations of the repromming factors have also been shown to facilitate the derivation of ESC-like cells from mouse strains that have thus far been considered refractory for ESC derivation, such as the nonobese diabetic (NOD) strain. While explanted blastocysts from NOD animals give rise to cells that are unstable in conventional ESC culture conditions, overexpression of either Klf4 or c-Myc endows these cells with a murine ESC-like state (Hanna et al. 2009a). However, this state is “metastable,” as it depends on the continuous expression of factors. Of note, the effect of Klf4/c-Myc overexpression can be mimicked by treatment of NOD stem cells with either drugs that induce Klf4 expression (Hanna et al. 2009a) or, alternatively, compounds that inhibit MAPK and GSK3 (Nichols et al. 2009a).
Human ESCs and iPSCs also resemble EpiSCs more closely than ESCs, suggesting that previously used culture conditions capture a primed rather than a naïve pluripotent state. Recent data from the Jaenisch laboratory (Hanna et al. 2010) indicate that overexpression of Oct4, Klf4, and Klf2, together with LIF and inhibitors of MAPK and GSK3 signaling, induces a metastable naïve murine ESC-like state in human ESCs. In contrast to human ESCs grown in regular ESC conditions, treated cells showed responsiveness to LIF as well as reactivation of the silenced X chromosome, indicating the acquisition of murine ESC-like signaling and epigenetic patterns. Human ESCs with two active X chromosomes are also observed when cells are derived in 5% oxygen (Lengner et al. 2010), suggesting that physiological oxygen levels help to maintain human ESCs in a developmentally more immature state. A metastable state that shares some morphological and molecular features with murine ESCs can also be directly induced in human somatic cells by overexpression of Oct4, Sox2, Klf4, c-Myc, and Nanog, and culture in the presence of LIF (Buecker et al. 2010).
Equivalency of iPSCs and ESCs?
The artificial nature of induced pluripotency raises the question of whether iPSCs and blastocyst-derived ESCs are molecularly and functionally equivalent. Analyses of genome-wide expression patterns and global histone modifications have shown a high degree of similarity between ESCs and iPSCs (Maherali et al. 2007; Okita et al. 2007; Wernig et al. 2007; Mikkelsen et al. 2008). However, substantial differences between the two cell types have been reported as well. For example, a reduced and more variable neuronal potential has been described for a number of human iPSC lines, regardless of whether they carried reprogramming transgenes in their genome or not (Hu et al. 2010). Likewise, an increased propensity of iPSC-derived neural cells to form tumors after transplantation into the brains of immunocompromised mice has been observed (Miura et al. 2009). In addition, human iPSC-derived early blood progenitor cells appear to undergo premature senescence (Feng et al. 2010). At the molecular level, gene-specific and global differences in DNA methylation (Deng et al. 2009; Doi et al. 2009; Pick et al. 2009) and in the expression of mRNAs and miRNAs (Chin et al. 2009; Wilson et al. 2009) have been reported between both mouse and human ESCs and iPSCs. Importantly, a contribution of genetic background (Brambrink et al. 2006) and residual transgene expression (Takahashi and Yamanaka 2006; Soldner et al. 2009), which can influence the properties of pluripotent cells, to the observed phenotypes could not be excluded in these studies.
A recent comparison of global mRNA and miRNA expression in genetically matched mouse ESCs and iPSCs revealed no consistent gene expression differences, with the exception of transcripts within the imprinted Dlk1–Dio3 gene cluster (Stadtfeld et al. 2010b). This suggests that many of the previously observed transcriptional differences were due to experimental variables. Specifically, maternally expressed genes in this cluster were aberrantly silenced in the majority of iPSCs derived from fibroblasts, blood cells, and keratinocytes. In fact, the expression status of the Dlk1–Dio3 locus served as a useful indicator for the quality of iPSC clones: While all iPSCs formed teratomas and contributed to low-grade chimeras, only cells with an ESC-equivalent expression pattern of Dlk1–Dio3 had the capacity to contribute to high-grade chimeras and form viable “all-iPSC” mice after injection into tetraploid blastocysts (L Liu et al. 2010; Stadtfeld et al. 2010b). It is noteworthy that iPSCs with normal Dlk1–Dio3 expression have so far been derived only from fibroblasts, which may be linked to high expression levels of this cluster in these cells. Importantly, the Dlk1–Dio3 cluster is normally expressed in NT-ESCs derived from different cell types (Stadtfeld et al. 2010b), indicating that SCNT more frequently generates faithfully reprogrammed cells than factor-mediated reprogramming. Although the molecular reasons for the susceptibility of the Dlk1–Dio3 cluster to acquire epigenetic abnormalities during iPSC formation are unclear, binding of reprogramming factors to specific sequences in this locus followed by the recruitment of repressive chromatin modifiers is likely involved. It remains an open question whether other epigenetic and/or genetic abnormalities, which are not detectable in the undifferentiated state or with current technology, are accrued during iPSC formation, especially in human cells. A recent study comparing human ESCs and iPSCs found no consistent differences in global gene expression and histone modification patterns (Guenther et al. 2010). However, these findings do not rule out that subtle differences exist, but they may have been masked by the use of stem cells that were not genetically matched.
A related issue is the question of whether iPSCs retain an “epigenetic memory” of their cell type of origin, which is defined here as remaining epigenetic marks originating from the starting cell that influence transcription in resultant iPSCs. Previous experiments using SCNT suggested that cloned embryos exhibit gene expression patterns reflective of their cell type of origin (Ng and Gurdon 2005, 2008). A recent study found gene expression differences indicative of a transcriptional memory in human iPSCs derived from fibroblasts, adipose tissue, and keratinocytes (Marchetto et al. 2009). While the analyzed cell lines were derived in independent laboratories and with different technologies, which can confound gene expression analyses (Newman and Cooper 2010), another study confirmed and extended this finding by comparing genetically matched iPSCs derived from granulocytes, muscle progenitors, fibroblasts, and lymphocytes (Polo et al. 2010). iPSCs derived from these cell types exhibited discernible gene expression and DNA methylation patterns as well as differentiation biases into hematopoietic cells in vitro, some of which could be attributed to their cell type of origin. A parallel study corroborated these conclusions and further discovered that analysis of DNA methylation patterns in a given iPSC clone could predict the somatic cell from which it was derived (Kim et al. 2010). Notably, continuous passaging of iPSCs (Polo et al. 2010) or the treatment of cells with chromatin-modifying drugs (Kim et al. 2010) attenuated these differences. Together, these results suggest that low-passage iPSCs retain a transient epigenetic memory of their cell type of origin that can influence their differentiation potential.
To date, no comparison of genetically matched human ESCs and iPSCs has been described. Nevertheless, recent data suggest that female human iPSCs fail to reactivate the epigenetically silenced X chromosome (Tchieu et al. 2010). Female fibroblast populations are mosaic in that 50% of cells carry an active maternal X chromosome and an inactive paternal X chromosome, whereas 50% of cells carry an active paternal X and an inactive maternal X. iPSCs derived from female mouse fibroblasts faithfully reactivate the silenced X chromosome and undergo random X inactivation upon differentiation (Maherali et al. 2007). In contrast, human iPSCs derived from female dermal fibroblasts fail to reactivate the silenced X chromosome and keep the same X chromosome inactivated in differentiated cells (Tchieu et al. 2010). These results indicate differences in the reprogramming process between mouse and human cells. Moreover, the data show that female human iPSCs can also retain an epigenetic memory of their cell type of origin. In contrast to mouse iPSCs, however, this memory does not become attenuated with passage. This observation could be useful in disease modeling and potential cell therapy of X-linked disorders such as RETT syndrome or Duchenne's muscular dystrophy, as it allows for the derivation of phenotypically wild-type and isogenic mutant cell lines from carriers harboring these mutations. At the same time, the possible clinical use of iPSCs derived from female carriers with unidentified X-linked mutations might be complicated by this finding.
Therapeutic potential of iPSCs
In addition to being an exciting research tool to probe mammalian development and epigenetic reprogramming, iPSCs have therapeutic potential for both custom-tailored cell therapy and so-called “disease modeling.” These two concepts are illustrated in Figure 4 and are discussed in the last section.
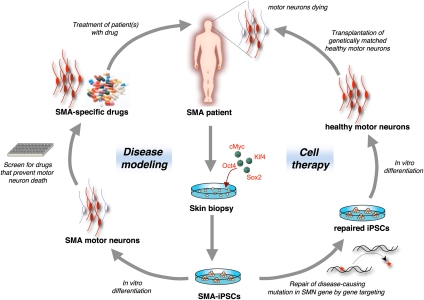
Figure 4.
Potential applications of iPSCs. Shown are the potential applications of iPSC technology for cell therapy and disease modeling using SMA as an example. In SMA patients, motor neurons are afflicted and die, causing the devastating symptoms of the disease. SMA-specific iPSCs could be coaxed into motor neurons in vitro in order to establish a culture model of the disease that may lead to the identification of novel drugs that prevent the abnormal death of motor neurons in patients. Alternatively, if known, the disease-causing mutation could be repaired (in this case the SMA gene) in iPSCs by gene targeting prior to their differentiation into healthy motor neurons, followed by transplantation into the patient's brain.
iPSCs and cell therapy
Organ transplantation among nonrelated individuals is complicated by the limited availability of matched tissues and the requirement for life-long treatment with immunosuppresive drugs that can have serious side effects. iPSCs might potentially circumvent these problems, as they could be coaxed into the desired cell types that would already be genetically matched with the patient. Another key advantage of iPSCs over current transplantation approaches is the possibility of repairing disease-causing mutations by homologous recombination, a technology that has been used with limited success in adult stem cells because of notorious difficulties in growing them outside the body.
Promising experiments in mice indeed suggest that the treatment of genetic disorders with iPSCs is feasible. Specifically, Jaenisch and colleagues (Hanna et al. 2007) showed that iPSCs can be used to rescue the defects seen in an animal model of sickle cell anemia. Sickle cell anemia is the result of a single point mutation in the hemoglobin gene, rendering red blood cells nonfunctional. In this proof-of-concept study, skin cells from the mouse model, which recapitulates the human condition, were first reprogrammed into iPSCs. The disease-causing mutation was subsequently fixed in iPSCs by gene targeting, and the repaired cells were then coaxed into blood-forming progenitors. These now healthy progenitors were transplanted back into anemic mice, where they produced normal red blood cells and cured the disease (Hanna et al. 2007). In principle, this approach could be applied to any disease in humans for which the underlying mutation is known, and that can be treated by cell transplantation. This conclusion is further supported by the phenotypic correction of hemophilia A in mice transplanted with heterologous, iPSC-derived endothelial progenitor cells (Xu et al. 2009).
Drug development with iPSCs
The study and treatment of many degenerative diseases—such as type I diabetes, Alzheimer's disease, and Parkinson's disease—is limited by the accessibility of the affected tissues, as well as the inability to grow the relevant cell types in culture for extended periods of time. The idea behind so-called “disease modeling” is to derive iPSCs from patients' skin cells and then differentiate them in vitro into the affected cell types, thereby recapitulating the disease in a Petri dish (Fig. 4). The advantage of this approach over currently used strategies is that the very cell type that is compromised can be recreated in culture to be studied, even when the cell type is long gone from the patient. Moreover, because iPSCs grow indefinitely in culture, they provide an unlimited source for any desired specialized cells. Ultimately, the goal of this approach is to use these “Petri dish” models of disease to identify novel drugs to treat the disease; for example, drugs that prevent the pathological death of motor neurons in patients suffering from amyotrophic lateral sklerosis (ALS) and spinal muscular atrophy (SMA), or the abnormal loss of insulin-producing β cells in diabetes patients.
Another problem associated with studying degenerative diseases that iPSCs may overcome is the fact that researchers are usually confined to cell material from patients whose disease has already progressed to advanced stages at the time of diagnosis. Since iPSCs likely need to pass through the same differentiation steps in vitro as patients' cells went through as they became sick, this technology may allow researchers to recapitulate and capture the very early stages of the disease. This may inform researchers when and why things went awry in the patient in the first place.
How far is research from identifying new drugs with iPSCs? In fact, several laboratories have already derived iPSCs from patients suffering from Huntington's and Parkinson's disease, ALS, juvenile diabetes, muscular dystrophy, Fanconi anemia, Down syndrome, and others (Dimos et al. 2008; Park et al. 2008a; Raya et al. 2009; Soldner et al. 2009), which will facilitate these studies. Moreover, three promising reports showed that iPSCs derived from patients suffering from the devastating disorders SMA (Ebert et al. 2009), familial dysautonomia (FD) (Lee et al. 2009), and LEOPARD syndrome (Carvajal-Vergara et al. 2010) recapitulated the cell abnormalities in a Petri dish as they are seen in patients. Remarkably, when the cultured cells were exposed to experimental drugs for these diseases, the “symptoms” were partially alleviated in culture. This principle can now be applied to many other diseases and cell types for which we currently do not have treatments, and may result in the development of drugs from which not just one individual, as in cell therapy, but many patients may benefit.
Challenges for disease modeling and cell therapy
Despite successes in animal models, iPSC technology is not yet ready for transplanting cells into patients. The main issue is safety concerns; iPSCs, like ESCs, tend to form teratomas, and current differentiation protocols cannot efficiently eliminate residual undifferentiated cells (Wernig et al. 2008c). However, this problem should be surmountable by devising positive or negative selection strategies using FACS or drug selection approaches, as has been done before with cultured cells (Schuldiner et al. 2003). Most patient-specific iPSCs have been generated with integrating vectors, which may not get silenced efficiently or could disrupt endogenous genes, which also pose potential impediments for the use of human iPSCs in cell therapy. It will thus be critical to further improve the transgene-free approaches discussed previously to derive new patient-specific iPSC lines. For diseases that require gene targeting to repair mutant alleles, more efficient targeting strategies need to be developed. Regular targeting approaches in human ESCs are quite inefficient, and may result in karyotypic abnormalities due to extensive culturing (Draper et al. 2004). The use of zinc finger nucleases to target endogenous genes in human ESCs led to a significant increase in efficiency, and may hence become the method of choice for manipulating human ESCs (Hockemeyer et al. 2009; Zou et al. 2009). Another advance has been the recognition that human ESCs can be transiently converted into a murine ESC-like state that is more amenable to homologous recombination and single-cell subcloning by exposing cells to drugs or overexpressing certain reprogramming factors (Buecker et al. 2010; Hanna et al. 2010). Last, the observation that many mouse iPSCs harbor epigenetic abnormalities (Stadtfeld et al. 2010b) and retain a transient epigenetic memory of their donor cells (Kim et al. 2010; Polo et al. 2010) will require a careful molecular and functional evaluation of human iPSCs for similar potential aberrations, as well as for possible rare, nonrecurrent mutations.
Disease modeling faces some of the same challenges as cell therapy, but it also raises additional questions. While promising preliminary data have been published on the modeling of monogenic diseases (see above), it remains unclear if multigenic diseases such as diabetes or Alzheimer's disease are equally amenable to in vitro modeling. Another question that needs to be addressed is whether late-onset diseases such as Alzheimer's and Parkinson's disease can be recapitulated in vitro within a few weeks or whether one needs to accelerate the phenotype by exposing cells to different types of environmental or genetic stress to unveil a phenotype. This challenge is underscored by the observation that neurons derived from “Parkinson iPSCs” did not exhibit obvious abnormalities compared with wild-type neurons (Soldner et al. 2009). Many diseases develop in a non-cell-autonomous manner and involve the interaction of multiple different cell types. Even though it should be possible, in principle, to derive all of the relevant cell types involved in disease from iPSCs, current differentiation strategies into functional cell types are inefficient and limited to a few tissues. Nevertheless, the idea of probing the effect of different cell types on the development of a disease has been demonstrated by different laboratories studying mouse and human models of ALS, another condition characterized by the loss of motor neurons (Di Giorgio et al. 2007, 2008; Marchetto et al. 2008). Specifically, Di Giorgio et al. (2007, 2008) and Marchetto et al. (2008) engineered ESCs to carry the ALS-specific SOD1 mutation and coaxed them in vitro into both motor neurons and astrocytes, which have been speculated to synergize during disease development in vivo. Coculture of both cell types indeed resulted in significantly more cell death of motor neurons, thus indicating that astrocytes, in addition to motor neurons themselves, contribute to the pathophysiology of ALS.
Concluding thoughts
The generation of iPSCs 5 years ago provided researchers with a unique platform to dissect the mechanisms of cellular reprogramming, which largely remained elusive for the past six decades. While many questions remain, interesting insights have been gained into the process of reprogramming, such as the finding that cells undergo defined sequential molecular events in an apparently stochastic manner, and these events are influenced by the choice and number of transcription factors as well as the starting cell type. The ease with which iPSCs can be generated with improved methodology has facilitated the development of chemical and siRNA screens as well as biochemical studies that should further unravel the mechanisms of this process.
The discovery of iPSCs has also influenced our view of normal development, as it demonstrated that activation of only a few transcription factors can potently change cell fate and, hence, that mammalian cells must have developed epigenetic mechanisms to efficiently lock in a cell once it has differentiated. These mechanisms are often broken in cancer cells, which exhibit features of stem cells and signs of dedifferentiation. Notably, many signaling pathways mutated in cancer cells have been shown recently to affect the formation of iPSCs, indicating remarkable similarities between tumorigenesis and cellular reprogramming.
The isolation of iPSCs has also sparked new interest in interconverting mature cell types directly into each other, which has already led to a number of remarkable examples for pancreatic, muscle, and neural cell types. It is likely that many other direct cell switches will be achieved in the near future. It remains to be tested, however, whether transdifferentiation works in the human system as well, and whether lineage-converted cells are functionally equivalent to their in vivo counterparts.
Despite numerous technical advances in the derivation of human iPSCs, relatively little is known about their molecular and functional equivalence to ESCs, which could affect their potential therapeutic utility. Addressing this question will require a careful analysis of the genomic and epigenomic integrity of human iPSCs, as well as the development of optimized differentiation protocols and reliable assays to evaluate the functionality of iPSC-derived specialized cells.
Acknowledgments
We thank Kathrin Plath, George Daley, Gustavo Mostoslavsky, Effie Apostolou, Nimet Maherali, and Jose Polo for critical reading of the manuscript and providing helpful comments. We apologize to those colleagues whose work we could not cite.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1963910.
References
- Aasen T, Raya A, Barrero MJ, Garreta E, Consiglio A, Gonzalez F, Vassena R, Bilic J, Pekarik V, Tiscornia G, et al. 2008. Efficient and rapid generation of induced pluripotent stem cells from human keratinocytes. Nat Biotechnol 26: 1276–1284 [DOI] [PubMed] [Google Scholar]
- Akao Y, Nakagawa Y, Naoe T 2006. let-7 microRNA functions as a potential growth suppressor in human colon cancer cells. Biol Pharm Bull 29: 903–906 [DOI] [PubMed] [Google Scholar]
- Aoi T, Yae K, Nakagawa M, Ichisaka T, Okita K, Takahashi K, Chiba T, Yamanaka S 2008. Generation of pluripotent stem cells from adult mouse liver and stomach cells. Science 321: 699–702 [DOI] [PubMed] [Google Scholar]
- Aoki T, Ohnishi H, Oda Y, Tadokoro M, Sasao M, Kato H, Hattori K, Ohgushi H 2010. Generation of induced pluripotent stem cells from human adipose-derived stem cells without c-MYC. Tissue Eng Part A 16: 2197–2206 [DOI] [PubMed] [Google Scholar]
- Araki R, Jincho Y, Hoki Y, Nakamura M, Tamura C, Ando S, Kasama Y, Abe M 2010. Conversion of ancestral fibroblasts to induced pluripotent stem cells. Stem Cells 28: 213–220 [DOI] [PubMed] [Google Scholar]
- Banito A, Rashid ST, Acosta JC, Li S, Pereira CF, Geti I, Pinho S, Silva JC, Azuara V, Walsh M, et al. 2009. Senescence impairs successful reprogramming to pluripotent stem cells. Genes Dev 23: 2134–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao S, Tang F, Li X, Hayashi K, Gillich A, Lao K, Surani MA 2009. Epigenetic reversion of post-implantation epiblast to pluripotent embryonic stem cells. Nature 461: 1292–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhutani N, Brady JJ, Damian M, Sacco A, Corbel SY, Blau HM 2010. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature 463: 1042–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blelloch R, Venere M, Yen J, Ramalho-Santos M 2007. Generation of induced pluripotent stem cells in the absence of drug selection. Cell Stem Cell 1: 245–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland MJ, Hazen JL, Nazor KL, Rodriguez AR, Gifford W, Martin G, Kupriyanov S, Baldwin KK 2009. Adult mice generated from induced pluripotent stem cells. Nature. 461: 91–94 [DOI] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. 2005. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122: 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. 2006. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441: 349–353 [DOI] [PubMed] [Google Scholar]
- Bradley A, Evans M, Kaufman MH, Robertson E 1984. Formation of germ-line chimaeras from embryo-derived teratocarcinoma cell lines. Nature 309: 255–256 [DOI] [PubMed] [Google Scholar]
- Brambrink T, Hochedlinger K, Bell G, Jaenisch R 2006. ES cells derived from cloned and fertilized blastocysts are transcriptionally and functionally indistinguishable. Proc Natl Acad Sci 103: 933–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brambrink T, Foreman R, Welstead GG, Lengner CJ, Wernig M, Suh H, Jaenisch R 2008. Sequential expression of pluripotency markers during direct reprogramming of mouse somatic cells. Cell Stem Cell 2: 151–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs R, King TJ 1952. Transplantation of living nuclei from blastula cells into enucleated frogs' eggs. Proc Natl Acad Sci 38: 455–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinster RL 1974. The effect of cells transferred into the mouse blastocyst on subsequent development. J Exp Med 140: 1049–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brons IG, Smithers LE, Trotter MW, Rugg-Gunn P, Sun B, Chuva de Sousa Lopes SM, Howlett SK, Clarkson A, Ahrlund-Richter L, Pedersen RA, et al. 2007. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature 448: 191–195 [DOI] [PubMed] [Google Scholar]
- Bruniquel D, Schwartz RH 2003. Selective, stable demethylation of the interleukin-2 gene enhances transcription by an active process. Nat Immunol 4: 235–240 [DOI] [PubMed] [Google Scholar]
- Buecker C, Chen HH, Polo JM, Daheron L, Bu L, Barakat TS, Okwieka P, Porter A, Gribnau J, Hochedlinger K, et al. 2010. A murine ESC-like state facilitates transgenesis and homologous recombination in human pluripotent stem cells. Cell Stem Cell 6: 535–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussmann LH, Schubert A, Vu Manh TP, De Andres L, Desbordes SC, Parra M, Zimmermann T, Rapino F, Rodriguez-Ubreva J, Ballestar E, et al. 2009. A robust and highly efficient immune cell reprogramming system. Cell Stem Cell 5: 554–566 [DOI] [PubMed] [Google Scholar]
- Carey BW, Markoulaki S, Hanna J, Saha K, Gao Q, Mitalipova M, Jaenisch R 2009. Reprogramming of murine and human somatic cells using a single polycistronic vector. Proc Natl Acad Sci 106: 157–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey BW, Markoulaki S, Beard C, Hanna J, Jaenisch R 2010. Single-gene transgenic mouse strains for reprogramming adult somatic cells. Nat Methods 7: 56–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvajal-Vergara X, Sevilla A, D'Souza SL, Ang YS, Schaniel C, Lee DF, Yang L, Kaplan AD, Adler ED, Rozov R, et al. 2010. Patient-specific induced pluripotent stem-cell-derived models of LEOPARD syndrome. Nature 465: 808–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan EM, Ratanasirintrawoot S, Park IH, Manos PD, Loh YH, Huo H, Miller JD, Hartung O, Rho J, Ince TA, et al. 2009. Live cell imaging distinguishes bona fide human iPS cells from partially reprogrammed cells. Nat Biotechnol 27: 1033–1037 [DOI] [PubMed] [Google Scholar]
- Chang CW, Lai YS, Pawlik KM, Liu K, Sun CW, Li C, Schoeb TR, Townes TM 2009. Polycistronic lentiviral vector for ‘hit and run’ reprogramming of adult skin fibroblasts to induced pluripotent stem cells. Stem Cells 27: 1042–1049 [DOI] [PubMed] [Google Scholar]
- Chang MY, Kim D, Kim CH, Kang HC, Yang E, Moon JI, Ko S, Park J, Park KS, Lee KA, et al. 2010. Direct reprogramming of rat neural precursors cells and fibroblasts into pluripotent stem cells. PLoS ONE 5: e9838 doi: 10.1371/journal.pone.0009838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chazaud C, Yamanaka Y, Pawson T, Rossant J 2006. Early lineage segregation between epiblast and primitive endoderm in mouse blastocysts through the Grb2–MAPK pathway. Dev Cell 10: 615–624 [DOI] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. 2008. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133: 1106–1117 [DOI] [PubMed] [Google Scholar]
- Chin MH, Mason MJ, Xie W, Volinia S, Singer M, Peterson C, Ambartsumyan G, Aimiuwu O, Richter L, Zhang J, et al. 2009. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell 5: 111–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HJ, Lee CS, Kwon YW, Paek JS, Lee SH, Hur J, Lee EJ, Roh TY, Chu IS, Leem SH, et al. 2010. Induction of pluripotent stem cells from adult somatic cells by protein-based reprogramming without genetic manipulation. Blood 116: 386–395 [DOI] [PubMed] [Google Scholar]
- Cibelli JB, Stice SL, Golueke PJ, Kane JJ, Jerry J, Blackwell C, Ponce de Leon FA, Robl JM 1998. Transgenic bovine chimeric offspring produced from somatic cell-derived stem-like cells. Nat Biotechnol 16: 642–646 [DOI] [PubMed] [Google Scholar]
- Collado M, Blasco MA, Serrano M 2007. Cellular senescence in cancer and aging. Cell 130: 223–233 [DOI] [PubMed] [Google Scholar]
- Cowan CA, Atienza J, Melton DA, Eggan K 2005. Nuclear reprogramming of somatic cells after fusion with human embryonic stem cells. Science 309: 1369–1373 [DOI] [PubMed] [Google Scholar]
- Davis RL, Weintraub H, Lassar AB 1987. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 51: 987–1000 [DOI] [PubMed] [Google Scholar]
- Deng J, Shoemaker R, Xie B, Gore A, LeProust EM, Antosiewicz-Bourget J, Egli D, Maherali N, Park IH, Yu J, et al. 2009. Targeted bisulfite sequencing reveals changes in DNA methylation associated with nuclear reprogramming. Nat Biotechnol 27: 353–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desponts C, Ding S 2010. Using small molecules to improve generation of induced pluripotent stem cells from somatic cells. Methods Mol Biol 636: 207–218 [DOI] [PubMed] [Google Scholar]
- Di Giorgio FP, Carrasco MA, Siao MC, Maniatis T, Eggan K 2007. Non-cell autonomous effect of glia on motor neurons in an embryonic stem cell-based ALS model. Nat Neurosci 10: 608–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giorgio FP, Boulting GL, Bobrowicz S, Eggan KC 2008. Human embryonic stem cell-derived motor neurons are sensitive to the toxic effect of glial cells carrying an ALS-causing mutation. Cell Stem Cell 3: 637–648 [DOI] [PubMed] [Google Scholar]
- Dimos JT, Rodolfa KT, Niakan KK, Weisenthal LM, Mitsumoto H, Chung W, Croft GF, Saphier G, Leibel R, Goland R, Wichterle H, Henderson CE, Eggan K 2008. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science 321: 1218–1221 [DOI] [PubMed] [Google Scholar]
- Doi A, Park IH, Wen B, Murakami P, Aryee MJ, Irizarry R, Herb B, Ladd-Acosta C, Rho J, Loewer S, et al. 2009. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat Genet 41: 1350–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper JS, Smith K, Gokhale P, Moore HD, Maltby E, Johnson J, Meisner L, Zwaka TP, Thomson JA, Andrews PW 2004. Recurrent gain of chromosomes 17q and 12 in cultured human embryonic stem cells. Nat Biotechnol 22: 53–54 [DOI] [PubMed] [Google Scholar]
- Ebert AD, Yu J, Rose FF Jr, Mattis VB, Lorson CL, Thomson JA, Svendsen CN 2009. Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature 457: 277–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edel MJ, Menchon C, Menendez S, Consiglio A, Raya A, Izpisua Belmonte JC 2010. Rem2 GTPase maintains survival of human embryonic stem cells as well as enhancing reprogramming by regulating p53 and cyclin D1. Genes Dev 24: 561–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggan K, Akutsu H, Loring J, Jackson-Grusby L, Klemm M, Rideout WM III, Yanagimachi R, Jaenisch R 2001. Hybrid vigor, fetal overgrowth, and viability of mice derived by nuclear cloning and tetraploid embryo complementation. Proc Natl Acad Sci 98: 6209–6214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggan K, Baldwin K, Tackett M, Osborne J, Gogos J, Chess A, Axel R, Jaenisch R 2004. Mice cloned from olfactory sensory neurons. Nature 428: 44–49 [DOI] [PubMed] [Google Scholar]
- Egli D, Sandler VM, Shinohara ML, Cantor H, Eggan K 2009. Reprogramming after chromosome transfer into mouse blastomeres. Curr Biol 19: 1403–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eminli S, Utikal J, Arnold K, Jaenisch R, Hochedlinger K 2008. Reprogramming of neural progenitor cells into induced pluripotent stem cells in the absence of exogenous Sox2 expression. Stem Cells 26: 2467–2474 [DOI] [PubMed] [Google Scholar]
- Eminli S, Foudi A, Stadtfeld M, Maherali N, Ahfeldt T, Mostoslavsky G, Hock H, Hochedlinger K 2009. Differentiation stage determines potential of hematopoietic cells for reprogramming into induced pluripotent stem cells. Nat Genet 41: 968–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteban MA, Xu J, Yang J, Peng M, Qin D, Li W, Jiang Z, Chen J, Deng K, Zhong M, et al. 2009. Generation of induced pluripotent stem cell lines from Tibetan miniature pig. J Biol Chem 284: 17634–17640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteban MA, Wang T, Qin B, Yang J, Qin D, Cai J, Li W, Weng Z, Chen J, Ni S, et al. 2010. Vitamin C enhances the generation of mouse and human induced pluripotent stem cells. Cell Stem Cell 6: 71–79 [DOI] [PubMed] [Google Scholar]
- Evans MJ, Kaufman MH 1981. Establishment in culture of pluripotential cells from mouse embryos. Nature 292: 154–156 [DOI] [PubMed] [Google Scholar]
- Feldman N, Gerson A, Fang J, Li E, Zhang Y, Shinkai Y, Cedar H, Bergman Y 2006. G9a-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nat Cell Biol 8: 188–194 [DOI] [PubMed] [Google Scholar]
- Feng B, Jiang J, Kraus P, Ng JH, Heng JC, Chan YS, Yaw LP, Zhang W, Loh YH, Han J, et al. 2009. Reprogramming of fibroblasts into induced pluripotent stem cells with orphan nuclear receptor Esrrb. Nat Cell Biol 11: 197–203 [DOI] [PubMed] [Google Scholar]
- Feng Q, Lu SJ, Klimanskaya I, Gomes I, Kim D, Chung Y, Honig GR, Kim KS, Lanza R 2010. Hemangioblastic derivatives from human induced pluripotent stem cells exhibit limited expansion and early senescence. Stem Cells 28: 704–712 [DOI] [PubMed] [Google Scholar]
- Finch BW, Ephrussi B 1967. Retention of multiple developmental potentialities by cells of a mouse testicular teratocarcinoma during prolonged culture in vitro and their extinction upon hybridization with cells of permanent lines. Proc Natl Acad Sci 57: 615–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusaki N, Ban H, Nishiyama A, Saeki K, Hasegawa M 2009. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc Jpn Acad 85: 348–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner RL 1968. Mouse chimeras obtained by the injection of cells into the blastocyst. Nature 220: 596–597 [DOI] [PubMed] [Google Scholar]
- Gaspar-Maia A, Alajem A, Polesso F, Sridharan R, Mason MJ, Heidersbach A, Ramalho-Santos J, McManus MT, Plath K, Meshorer E, et al. 2009. Chd1 regulates open chromatin and pluripotency of embryonic stem cells. Nature 460: 863–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidekel S, Bergman Y 2002. A unique developmental pattern of Oct-3/4 DNA methylation is controlled by a cis-demodification element. J Biol Chem 277: 34521–34530 [DOI] [PubMed] [Google Scholar]
- Giorgetti A, Montserrat N, Aasen T, Gonzalez F, Rodríguez-Pizà I, Vassena R, Raya A, Boué S, Barrero MJ, Corbella BA, et al. 2009. Generation of induced pluripotent stem cells from human cord blood using OCT4 and SOX2. Cell Stem Cell 5: 353–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan K, Nayernia K, Maier LS, Wagner S, Dressel R, Lee JH, Nolte J, Wolf F, Li M, Engel W, et al. 2006. Pluripotency of spermatogonial stem cells from adult mouse testis. Nature 440: 1199–1203 [DOI] [PubMed] [Google Scholar]
- Guenther MG, Frampton GM, Soldner F, Hockemeyer D, Mitalipova M, Jaenisch R, Young RA 2010. Chromatin structure and gene expression programs of human embryonic and induced pluripotent stem cells. Cell Stem Cell 7: 249–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G, Yang J, Nichols J, Hall JS, Eyres I, Mansfield W, Smith A 2009. Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development 136: 1063–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurdon JB 1962. The developmental capacity of nuclei taken from intestinal epithelium cells of feeding tadpoles. J Embryol Exp Morphol 10: 622–640 [PubMed] [Google Scholar]
- Gurdon JB, Laskey RA, Reeves OR 1975. The developmental capacity of nuclei transplanted from keratinized skin cells of adult frogs. J Embryol Exp Morphol 34: 93–112 [PubMed] [Google Scholar]
- Gurdon JB, Byrne JA, Simonsson S 2003. Nuclear reprogramming and stem cell creation. Proc Natl Acad Sci 100: 11819–11822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase A, Olmer R, Schwanke K, Wunderlich S, Merkert S, Hess C, Zweigerdt R, Gruh I, Meyer J, Wagner S, et al. 2009. Generation of induced pluripotent stem cells from human cord blood. Cell Stem Cell 5: 434–441 [DOI] [PubMed] [Google Scholar]
- Hajkova P, Ancelin K, Waldmann T, Lacoste N, Lange UC, Cesari F, Lee C, Almouzni G, Schneider R, Surani MA 2008. Chromatin dynamics during epigenetic reprogramming in the mouse germ line. Nature 452: 877–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajkova P, Jeffries SJ, Lee C, Miller N, Jackson SP, Surani MA 2010. Genome-wide reprogramming in the mouse germ line entails the base excision repair pathway. Science 329: 78–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J, Guo G, Wray J, Eyres I, Nichols J, Grotewold L, Morfopoulou S, Humphreys P, Mansfield W, Walker R, et al. 2009. Oct4 and LIF/Stat3 additively induce Kruppel factors to sustain embryonic stem cell self-renewal. Cell Stem Cell 5: 597–609 [DOI] [PubMed] [Google Scholar]
- Han DW, Do JT, Gentile L, Stehling M, Lee HT, Schöler HR 2008. Pluripotential reprogramming of the somatic genome in hybrid cells occurs with the first cell cycle. Stem Cells 26: 445–454 [DOI] [PubMed] [Google Scholar]
- Han J, Yuan P, Yang H, Zhang J, Soh BS, Li P, Lim SL, Cao S, Tay J, Orlov YL, et al. 2010. Tbx3 improves the germ-line competency of induced pluripotent stem cells. Nature 463: 1096–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J, Wernig M, Markoulaki S, Sun CW, Meissner A, Cassady JP, Beard C, Brambrink T, Wu LC, Townes TM, Jaenisch R 2007. Treatment of sickle cell anemia mouse model with iPS cells generated from autologous skin. Science 318: 1920–1923 [DOI] [PubMed] [Google Scholar]
- Hanna J, Markoulaki S, Schorderet P, Carey BW, Beard C, Wernig M, Creyghton MP, Steine EJ, Cassady JP, Foreman R, et al. 2008. Direct reprogramming of terminally differentiated mature B lymphocytes to pluripotency. Cell 133: 250–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J, Markoulaki S, Mitalipova M, Cheng AW, Cassady JP, Staerk J, Carey BW, Lengner CJ, Foreman R, Love J, et al. 2009a. Metastable pluripotent states in NOD-mouse-derived ESCs. Cell Stem Cell 4: 513–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J, Saha K, Pando B, van Zon J, Lengner CJ, Creyghton MP, van Oudenaarden A, Jaenisch R 2009b. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature 462: 595–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J, Cheng AW, Saha K, Kim J, Lengner CJ, Soldner F, Cassady JP, Muffat J, Carey BW, Jaenisch R 2010. Human embryonic stem cells with biological and epigenetic characteristics similar to those of mouse ESCs. Proc Natl Acad Sci 107: 9222–9227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He TC, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT, Morin PJ, Vogelstein B, Kinzler KW 1998. Identification of c-MYC as a target of the APC pathway. Science 281: 1509–1512 [DOI] [PubMed] [Google Scholar]
- Heng JC, Feng B, Han J, Jiang J, Kraus P, Ng JH, Orlov YL, Huss M, Yang L, Lufkin T, et al. 2010. The nuclear receptor Nr5a2 can replace Oct4 in the reprogramming of murine somatic cells to pluripotent cells. Cell Stem Cell 6: 167–174 [DOI] [PubMed] [Google Scholar]
- Hochedlinger K, Jaenisch R 2002a. Monoclonal mice generated by nuclear transfer from mature B and T donor cells. Nature 415: 1035–1038 [DOI] [PubMed] [Google Scholar]
- Hochedlinger K, Jaenisch R 2002b. Nuclear transplantation: Lessons from frogs and mice. Curr Opin Cell Biol 14: 741–748 [DOI] [PubMed] [Google Scholar]
- Hochedlinger K, Jaenisch R 2006. Nuclear reprogramming and pluripotency. Nature 441: 1061–1067 [DOI] [PubMed] [Google Scholar]
- Hochedlinger K, Plath K 2009. Epigenetic reprogramming and induced pluripotency. Development 136: 509–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D, Soldner F, Cook EG, Gao Q, Mitalipova M, Jaenisch R 2008. A drug-inducible system for direct reprogramming of human somatic cells to pluripotency. Cell Stem Cell 3: 346–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D, Soldner F, Beard C, Gao Q, Mitalipova M, DeKelver RC, Katibah GE, Amora R, Boydston EA, Zeitler B, et al. 2009. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat Biotechnol 27: 851–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong H, Takahashi K, Ichisaka T, Aoi T, Kanagawa O, Nakagawa M, Okita K, Yamanaka S 2009. Suppression of induced pluripotent stem cell generation by the p53–p21 pathway. Nature 460: 1132–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotta A, Cheung AY, Farra N, Vijayaragavan K, Seguin CA, Draper JS, Pasceri P, Maksakova IA, Mager DL, Rossant J, et al. 2009. Isolation of human iPS cells using EOS lentiviral vectors to select for pluripotency. Nat Methods 6: 370–376 [DOI] [PubMed] [Google Scholar]
- Hu BY, Weick JP, Yu J, Ma LX, Zhang XQ, Thomson JA, Zhang SC 2010. Neural differentiation of human induced pluripotent stem cells follows developmental principles but with variable potency. Proc Natl Acad Sci 107: 4335–4340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D, Maehr R, Guo W, Eijkelenboom A, Snitow M, Chen AE, Melton DA 2008a. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat Biotechnol 26: 795–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D, Osafune K, Maehr R, Guo W, Eijkelenboom A, Chen S, Muhlestein W, Melton DA 2008b. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat Biotechnol 26: 1269–1275 [DOI] [PubMed] [Google Scholar]
- Humpherys D, Eggan K, Akutsu H, Friedman A, Hochedlinger K, Yanagimachi R, Lander ES, Golub TR, Jaenisch R 2002. Abnormal gene expression in cloned mice derived from embryonic stem cell and cumulus cell nuclei. Proc Natl Acad Sci 99: 12889–12894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichida JK, Blanchard J, Lam K, Son EY, Chung JE, Egli D, Loh KM, Carter AC, Di Giorgio FP, Koszka K, et al. 2009. A small-molecule inhibitor of tgf-βsignaling replaces sox2 in reprogramming by inducing nanog. Cell Stem Cell 5: 491–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, Srivastava D 2010. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 142: 375–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Wakao H, Ogonuki N, Miki H, Seino K, Nambu-Wakao R, Noda S, Miyoshi H, Koseki H, Taniguchi M, et al. 2005. Generation of cloned mice by direct nuclear transfer from natural killer T cells. Curr Biol 15: 1114–1118 [DOI] [PubMed] [Google Scholar]
- Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y 2010. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 466: 1129–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson-Grusby L, Beard C, Possemato R, Tudor M, Fambrough D, Csankovszki G, Dausman J, Lee P, Wilson C, Lander E, et al. 2001. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat Genet 27: 31–39 [DOI] [PubMed] [Google Scholar]
- Jia F, Wilson KD, Sun N, Gupta DM, Huang M, Li Z, Panetta NJ, Chen ZY, Robbins RC, Kay MA, et al. 2010. A nonviral minicircle vector for deriving human iPS cells. Nat Methods 7: 197–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judson RL, Babiarz JE, Venere M, Blelloch R 2009. Embryonic stem cell-specific microRNAs promote induced pluripotency. Nat Biotechnol 27: 459–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahan BW, Ephrussi B 1970. Developmental potentialities of clonal in vitro cultures of mouse testicular teratoma. J Natl Cancer Inst 44: 1015–1036 [PubMed] [Google Scholar]
- Kaji K, Caballero IM, MacLeod R, Nichols J, Wilson VA, Hendrich B 2006. The NuRD component Mbd3 is required for pluripotency of embryonic stem cells. Nat Cell Biol 8: 285–292 [DOI] [PubMed] [Google Scholar]
- Kaji K, Norrby K, Paca A, Mileikovsky M, Mohseni P, Woltjen K 2009. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature 458: 771–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M, Inoue K, Lee J, Yoshimoto M, Ogonuki N, Miki H, Baba S, Kato T, Kazuki Y, Toyokuni S, et al. 2004. Generation of pluripotent stem cells from neonatal mouse testis. Cell 119: 1001–1012 [DOI] [PubMed] [Google Scholar]
- Kang L, Wang J, Zhang Y, Kou Z, Gao S 2009. iPS cells can support full-term development of tetraploid blastocyst-complemented embryos. Cell Stem Cell 5: 135–138 [DOI] [PubMed] [Google Scholar]
- Kawamura T, Suzuki J, Wang YV, Menendez S, Morera LB, Raya A, Wahl GM, Belmonte JC 2009. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature 460: 1140–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly SJ 1977. Studies of the developmental potential of four- and eight-cell stage mouse blastomeres. J Exp Zool 200: 365–376 [DOI] [PubMed] [Google Scholar]
- Kim J, Chu J, Shen X, Wang J, Orkin SH 2008. An extended transcriptional network for pluripotency of embryonic stem cells. Cell 132: 1049–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JB, Zaehres H, Wu G, Gentile L, Ko K, Sebastiano V, Arauzo-Bravo MJ, Ruau D, Han DW, Zenke M, et al. 2008. Pluripotent stem cells induced from adult neural stem cells by reprogramming with two factors. Nature 454: 646–650 [DOI] [PubMed] [Google Scholar]
- Kim D, Kim CH, Moon JI, Chung YG, Chang MY, Han BS, Ko S, Yang E, Cha KY, Lanza R, et al. 2009. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell 4: 472–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HH, Kuwano Y, Srikantan S, Lee EK, Martindale JL, Gorospe M 2009. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev 23: 1743–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JB, Sebastiano V, Wu G, Araúzo-Bravo MJ, Sasse P, Gentile L, Ko K, Ruau D, Ehrich M, van den Boom D, et al. 2009a. Oct4-induced pluripotency in adult neural stem cells. Cell 136: 411–419 [DOI] [PubMed] [Google Scholar]
- Kim JB, Greber B, Araúzo-Bravo MJ, Meyer J, Park KI, Zaehres H, Schöler HR 2009b. Direct reprogramming of human neural stem cells by OCT4. Nature 461: 649–643 [DOI] [PubMed] [Google Scholar]
- Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, Kim J, Aryee MJ, Ji H, Ehrlich LI, et al. 2010. Epigenetic memory in induced pluripotent stem cells. Nature doi: 10.1038/nature09342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King TJ, Briggs R 1955. Changes in the nuclei of differentiating gastrula cells, as demonstrated by nuclear transplantation. Proc Natl Acad Sci 41: 321–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinsmith LJ, Pierce GB Jr 1964. Multipotentiality of single embryonal carcinoma cells. Cancer Res 24: 1544–1551 [PubMed] [Google Scholar]
- Knoepfler PS 2008. Why myc? An unexpected ingredient in the stem cell cocktail. Cell Stem Cell 2: 18–21 [DOI] [PubMed] [Google Scholar]
- Ko K, Tapia N, Wu G, Kim JB, Bravo MJ, Sasse P, Glaser T, Ruau D, Han DW, Greber B, et al. 2009. Induction of pluripotency in adult unipotent germline stem cells. Cell Stem Cell 5: 87–96 [DOI] [PubMed] [Google Scholar]
- Kunarso G, Chia NY, Jeyakani J, Hwang C, Lu X, Chan YS, Ng HH, Bourque G 2010. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat Genet 42: 631–634 [DOI] [PubMed] [Google Scholar]
- Laiosa CV, Stadtfeld M, Xie H, de Andres-Aguayo L, Graf T 2006. Reprogramming of committed T cell progenitors to macrophages and dendritic cells by C/EBPα and PU.1 transcription factors. Immunity 25: 731–744 [DOI] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. 2006. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125: 301–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G, Papapetrou EP, Kim H, Chambers SM, Tomishima MJ, Fasano CA, Ganat YM, Menon J, Shimizu F, Viale A, et al. 2009. Modelling pathogenesis and treatment of familial dysautonomia using patient-specific iPSCs. Nature 461: 402–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei H, Oh SP, Okano M, Juttermann R, Goss KA, Jaenisch R, Li E 1996. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development 122: 3195–3205 [DOI] [PubMed] [Google Scholar]
- Lengner CJ, Gimelbrant AA, Erwin JA, Cheng AW, Guenther MG, Welstead GG, Alagappan R, Frampton GM, Xu P, Muffat J, et al. 2010. Derivation of pre-X inactivation human embryonic stem cells under physiological oxygen concentrations. Cell 141: 872–883 [DOI] [PubMed] [Google Scholar]
- Li W, Ding S 2010. Small molecules that modulate embryonic stem cell fate and somatic cell reprogramming. Trends Pharmacol Sci 31: 36–45 [DOI] [PubMed] [Google Scholar]
- Li E, Bestor TH, Jaenisch R 1992. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69: 915–926 [DOI] [PubMed] [Google Scholar]
- Li J, Ishii T, Feinstein P, Mombaerts P 2004. Odorant receptor gene choice is reset by nuclear transfer from mouse olfactory sensory neurons. Nature 428: 393–399 [DOI] [PubMed] [Google Scholar]
- Li C, Zhou J, Shi G, Ma Y, Yang Y, Gu J, Yu H, Jin S, Wei Z, Chen F, et al. 2009. Pluripotency can be rapidly and efficiently induced in human amniotic fluid-derived cells. Hum Mol Genet 18: 4340–4349 [DOI] [PubMed] [Google Scholar]
- Li H, Collado M, Villasante A, Strati K, Ortega S, Canamero M, Blasco MA, Serrano M 2009. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature 460: 1136–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Wei W, Zhu S, Zhu J, Shi Y, Lin T, Hao E, Hayek A, Deng H, Ding S 2009a. Generation of rat and human induced pluripotent stem cells by combining genetic reprogramming and chemical inhibitors. Cell Stem Cell 4: 16–19 [DOI] [PubMed] [Google Scholar]
- Li W, Zhou H, Abujarour R, Zhu S, Young Joo J, Lin T, Hao E, Scholer HR, Hayek A, Ding S 2009b. Generation of human-induced pluripotent stem cells in the absence of exogenous Sox2. Stem Cells 27: 2992–3000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Liang J, Ni S, Zhou T, Qing X, Li H, He W, Chen J, Li F, Zhuang Q, et al. 2010. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell 7: 51–63 [DOI] [PubMed] [Google Scholar]
- Liao J, Cui C, Chen S, Ren J, Chen J, Gao Y, Li H, Jia N, Cheng L, Xiao H, et al. 2009. Generation of induced pluripotent stem cell lines from adult rat cells. Cell Stem Cell 4: 11–15 [DOI] [PubMed] [Google Scholar]
- Lin CH, Lin C, Tanaka H, Fero ML, Eisenman RN 2009. Gene regulation and epigenetic remodeling in murine embryonic stem cells by c-Myc. PLoS ONE 4: e7839 doi: 10.1371/journal.pone.0007839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T, Ambasudhan R, Yuan X, Li W, Hilcove S, Abujarour R, Lin X, Hahm HS, Hao E, Hayek A, et al. 2009. A chemical platform for improved induction of human iPSCs. Nat Methods 6: 805–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Zhu F, Yong J, Zhang P, Hou P, Li H, Jiang W, Cai J, Liu M, Cui K, et al. 2008. Generation of induced pluripotent stem cells from adult rhesus monkey fibroblasts. Cell Stem Cell 3: 587–590 [DOI] [PubMed] [Google Scholar]
- Liu H, Ye Z, Kim Y, Sharkis S, Jang YY 2010. Generation of endoderm-derived human induced pluripotent stem cells from primary hepatocytes. Hepatology 51: 1810–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Luo GZ, Yang W, Zhao X, Zheng Q, Lv Z, Li W, Wu HJ, Wang L, Wang XJ, Zhou Q 2010. Activation of the imprinted Dlk1–Dio3 region correlates with pluripotency levels of mouse stem cells. J Biol Chem. 285: 19483–19490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, et al. 2006. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet 38: 431–440 [DOI] [PubMed] [Google Scholar]
- Loh YH, Agarwal S, Park IH, Urbach A, Huo H, Heffner GC, Kim K, Miller JD, Ng K, Daley GQ 2009. Generation of induced pluripotent stem cells from human blood. Blood 113: 5476–5479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry WE, Richter L, Yachechko R, Pyle AD, Tchieu J, Sridharan R, Clark AT, Plath K 2008. Generation of human induced pluripotent stem cells from dermal fibroblasts. Proc Natl Acad Sci 105: 2883–2888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maherali N, Hochedlinger K 2009. Tgfβ signal inhibition cooperates in the induction of iPSCs and replaces Sox2 and cMyc. Curr Biol 19: 1718–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold K, Stadtfeld M, Yachechko R, Tchieu J, Jaenisch R, et al. 2007. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell 1: 55–70 [DOI] [PubMed] [Google Scholar]
- Maherali N, Ahfeldt T, Rigamonti A, Utikal J, Cowan C, Hochedlinger K 2008. A high-efficiency system for the generation and study of human induced pluripotent stem cells. Cell Stem Cell 3: 340–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Ye Z, Hommond HH, Yu X, Lin J, Chen G, Zou J, Cheng L 2008. Improved efficiency and pace of generating induced pluripotent stem cells from human adult and fetal fibroblasts. Stem Cells 26: 1998–2005 [DOI] [PubMed] [Google Scholar]
- Mali P, Chou BK, Yen J, Ye Z, Zou J, Dowey S, Brodsky RA, Ohm JE, Yu W, Baylin SB, et al. 2010. Butyrate greatly enhances derivation of human induced pluripotent stem cells by promoting epigenetic remodeling and the expression of pluripotency-associated genes. Stem Cells 28: 713–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetto MC, Muotri AR, Mu Y, Smith AM, Cezar GG, Gage FH 2008. Non-cell-autonomous effect of human SOD1 G37R astrocytes on motor neurons derived from human embryonic stem cells. Cell Stem Cell 3: 649–657 [DOI] [PubMed] [Google Scholar]
- Marchetto MC, Yeo GW, Kainohana O, Marsala M, Gage FH, Muotri AR 2009. Transcriptional signature and memory retention of human-induced pluripotent stem cells. PLoS ONE 4: e7076 doi: 10.1371/journal.pone.0007076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion RM, Strati K, Li H, Murga M, Blanco R, Ortega S, Fernandez-Capetillo O, Serrano M, Blasco MA 2009a. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature 460: 1149–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion RM, Strati K, Li H, Tejera A, Schoeftner S, Ortega S, Serrano M, Blasco MA 2009b. Telomeres acquire embryonic stem cell characteristics in induced pluripotent stem cells. Cell Stem Cell 4: 141–154 [DOI] [PubMed] [Google Scholar]
- Markoulaki S, Hanna J, Beard C, Carey BW, Cheng AW, Lengner CJ, Dausman JA, Fu D, Gao Q, Wu S, et al. 2009. Transgenic mice with defined combinations of drug-inducible reprogramming factors. Nat Biotechnol 27: 169–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marson A, Foreman R, Chevalier B, Bilodeau S, Kahn M, Young RA, Jaenisch R 2008. Wnt signaling promotes reprogramming of somatic cells to pluripotency. Cell Stem Cell 3: 132–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GR 1981. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci 78: 7634–7638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui Y, Zsebo K, Hogan BL 1992. Derivation of pluripotential embryonic stem cells from murine primordial germ cells in culture. Cell 70: 841–847 [DOI] [PubMed] [Google Scholar]
- Matsui T, Leung D, Miyashita H, Maksakova IA, Miyachi H, Kimura H, Tachibana M, Lorincz MC, Shinkai Y 2010. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature 464: 927–931 [DOI] [PubMed] [Google Scholar]
- Mayer W, Niveleau A, Walter J, Fundele R, Haaf T 2000. Demethylation of the zygotic paternal genome. Nature 403: 501–502 [DOI] [PubMed] [Google Scholar]
- Meissner A, Wernig M, Jaenisch R 2007. Direct reprogramming of genetically unmodified fibroblasts into pluripotent stem cells. Nat Biotechnol 25: 1177–1181 [DOI] [PubMed] [Google Scholar]
- Melton C, Judson RL, Blelloch R 2010. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature 463: 621–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A 2008. Dissecting direct reprogramming through integrative genomic analysis. Nature 454: 49–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkola I, Heavey B, Horcher M, Busslinger M 2002. Reversion of B cell commitment upon loss of Pax5 expression. Science 297: 110–113 [DOI] [PubMed] [Google Scholar]
- Miller RA, Ruddle FH 1976. Pluripotent teratocarcinoma-thymus somatic cell hybrids. Cell 9: 45–55 [DOI] [PubMed] [Google Scholar]
- Miller RA, Ruddle FH 1977. Teratocarcinoma X friend erythroleukemia cell hybrids resemble their pluripotent embryonal carcinoma parent. Dev Biol 56: 157–173 [DOI] [PubMed] [Google Scholar]
- Miller CA, Sweatt JD 2007. Covalent modification of DNA regulates memory formation. Neuron 53: 857–869 [DOI] [PubMed] [Google Scholar]
- Miura K, Okada Y, Aoi T, Okada A, Takahashi K, Okita K, Nakagawa M, Koyanagi M, Tanabe K, Ohnuki M, et al. 2009. Variation in the safety of induced pluripotent stem cell lines. Nat Biotechnol 27: 743–745 [DOI] [PubMed] [Google Scholar]
- Munsie MJ, Michalska AE, O'Brien CM, Trounson AO, Pera MF, Mountford PS 2000. Isolation of pluripotent embryonic stem cells from reprogrammed adult mouse somatic cell nuclei. Curr Biol 10: 989–992 [DOI] [PubMed] [Google Scholar]
- Nagy A, Nagy K 2010. The mysteries of induced pluripotency: Where will they lead? Nat Methods 7: 22–24 [DOI] [PubMed] [Google Scholar]
- Nagy A, Gocza E, Diaz EM, Prideaux VR, Ivanyi E, Markkula M, Rossant J 1990. Embryonic stem cells alone are able to support fetal development in the mouse. Development 110: 815–821 [DOI] [PubMed] [Google Scholar]
- Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T, Okita K, Mochiduki Y, Takizawa N, Yamanaka S 2008. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol 26: 101–106 [DOI] [PubMed] [Google Scholar]
- Newman AM, Cooper JB 2010. Lab-specific gene expression signatures in pluripotent stem cells. Cell Stem Cell 7: 258–262 [DOI] [PubMed] [Google Scholar]
- Ng RK, Gurdon JB 2005. Epigenetic memory of active gene transcription is inherited through somatic cell nuclear transfer. Proc Natl Acad Sci 102: 1957–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng RK, Gurdon JB 2008. Epigenetic memory of an active gene state depends on histone H3.3 incorporation into chromatin in the absence of transcription. Nat Cell Biol 10: 102–109 [DOI] [PubMed] [Google Scholar]
- Nichols J, Smith A 2009. Naive and primed pluripotent states. Cell Stem Cell 4: 487–492 [DOI] [PubMed] [Google Scholar]
- Nichols J, Jones K, Phillips JM, Newland SA, Roode M, Mansfield W, Smith A, Cooke A 2009a. Validated germline-competent embryonic stem cell lines from nonobese diabetic mice. Nat Med 15: 814–818 [DOI] [PubMed] [Google Scholar]
- Nichols J, Silva J, Roode M, Smith A 2009b. Suppression of Erk signalling promotes ground state pluripotency in the mouse embryo. Development 136: 3215–3222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogonuki N, Inoue K, Yamamoto Y, Noguchi Y, Tanemura K, Suzuki O, Nakayama H, Doi K, Ohtomo Y, Satoh M, et al. 2002. Early death of mice cloned from somatic cells. Nat Genet 30: 253–254 [DOI] [PubMed] [Google Scholar]
- Okita K, Ichisaka T, Yamanaka S 2007. Generation of germline-competent induced pluripotent stem cells. Nature 448: 313–317 [DOI] [PubMed] [Google Scholar]
- Okita K, Nakagawa M, Hyenjong H, Ichisaka T, Yamanaka S 2008. Generation of mouse induced pluripotent stem cells without viral vectors. Science 322: 949–953 [DOI] [PubMed] [Google Scholar]
- Oswald J, Engemann S, Lane N, Mayer W, Olek A, Fundele R, Dean W, Reik W, Walter J 2000. Active demethylation of the paternal genome in the mouse zygote. Curr Biol 10: 475–478 [DOI] [PubMed] [Google Scholar]
- Park IH, Arora N, Huo H, Maherali N, Ahfeldt T, Shimamura A, Lensch MW, Cowan C, Hochedlinger K, Daley GQ 2008a. Disease-specific induced pluripotent stem cells. Cell 134: 877–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park IH, Lerou PH, Zhao R, Huo H, Daley GQ 2008b. Generation of human-induced pluripotent stem cells. Nat Protoc 3: 1180–1186 [DOI] [PubMed] [Google Scholar]
- Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, Lerou PH, Lensch MW, Daley GQ 2008c. Reprogramming of human somatic cells to pluripotency with defined factors. Nature 451: 141–146 [DOI] [PubMed] [Google Scholar]
- Pereira CF, Piccolo FM, Tsubouchi T, Sauer S, Ryan NK, Bruno L, Landeira D, Santos J, Banito A, Gil J, et al. 2010. ESCs require PRC2 to direct the successful reprogramming of differentiated cells toward pluripotency. Cell Stem Cell 6: 547–556 [DOI] [PubMed] [Google Scholar]
- Pfannkuche K, Fatima A, Gupta MK, Dieterich R, Hescheler J 2010. Initial colony morphology-based selection for iPS cells derived from adult fibroblasts is substantially improved by temporary UTF1-based selection. PLoS ONE 5: e9580 doi: 10.1371/journal.pone.0009580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pick M, Stelzer Y, Bar-Nur O, Mayshar Y, Eden A, Benvenisty N 2009. Clone- and gene-specific aberrations of parental imprinting in human induced pluripotent stem cells. Stem Cells 27: 2686–2690 [DOI] [PubMed] [Google Scholar]
- Polo JM, Liu S, Figueroa ME, Kulalert W, Eminli S, Tan KY, Apostolou E, Stadtfeld M, Li Y, Shioda T, et al. 2010. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat Biotechnol 28: 848–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp C, Dean W, Feng S, Cokus SJ, Andrews S, Pellegrini M, Jacobsen SE, Reik W 2010. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature 463: 1101–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai K, Huggins IJ, James SR, Karpf AR, Jones DA, Cairns BR 2008. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell 135: 1201–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raya A, Rodriguez-Piza I, Guenechea G, Vassena R, Navarro S, Barrero MJ, Consiglio A, Castella M, Rio P, Sleep E, et al. 2009. Disease-corrected haematopoietic progenitors from Fanconi anaemia induced pluripotent stem cells. Nature 460: 53–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick JL, Bixler LS, Cheng L, Donovan PJ 1992. Long-term proliferation of mouse primordial germ cells in culture. Nature 359: 550–551 [DOI] [PubMed] [Google Scholar]
- Samavarchi-Tehrani P, Golipour A, David L, Sung HK, Beyer TA, Datti A, Woltjen K, Nagy A, Wrana JL 2010. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell 7: 64–77 [DOI] [PubMed] [Google Scholar]
- Schuldiner M, Itskovitz-Eldor J, Benvenisty N 2003. Selective ablation of human embryonic stem cells expressing a ‘suicide’ gene. Stem Cells 21: 257–265 [DOI] [PubMed] [Google Scholar]
- Seandel M, James D, Shmelkov SV, Falciatori I, Kim J, Chavala S, Scherr DS, Zhang F, Torres R, Gale NW, et al. 2007. Generation of functional multipotent adult stem cells from GPR125+ germline progenitors. Nature 449: 346–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Do JT, Desponts C, Hahm HS, Scholer HR, Ding S 2008. A combined chemical and genetic approach for the generation of induced pluripotent stem cells. Cell Stem Cell 2: 525–528 [DOI] [PubMed] [Google Scholar]
- Silva J, Chambers I, Pollard S, Smith A 2006. Nanog promotes transfer of pluripotency after cell fusion. Nature 441: 997–1001 [DOI] [PubMed] [Google Scholar]
- Silva J, Barrandon O, Nichols J, Kawaguchi J, Theunissen TW, Smith A 2008. Promotion of reprogramming to ground state pluripotency by signal inhibition. PLoS Biol 6: e253 doi: 10.1371/journal.pbio.0060253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J, Nichols J, Theunissen TW, Guo G, van Oosten AL, Barrandon O, Wray J, Yamanaka S, Chambers I, Smith A 2009. Nanog is the gateway to the pluripotent ground state. Cell 138: 722–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhal N, Graumann J, Wu G, Araúzo-Bravo MJ, Han DW, Greber B, Gentile L, Mann M, Schöler HR 2010. Chromatin-remodeling components of the BAF complex facilitate reprogramming. Cell 141: 943–955 [DOI] [PubMed] [Google Scholar]
- Simonsson S, Gurdon J 2004. DNA demethylation is necessary for the epigenetic reprogramming of somatic cell nuclei. Nat Cell Biol 6: 984–990 [DOI] [PubMed] [Google Scholar]
- Smith ZD, Nachman I, Regev A, Meissner A 2010. Dynamic single-cell imaging of direct reprogramming reveals an early specifying event. Nat Biotechnol 28: 521–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldner F, Hockemeyer D, Beard C, Gao Q, Bell GW, Cook EG, Hargus G, Blak A, Cooper O, Mitalipova M, et al. 2009. Parkinson's disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell 136: 964–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer CA, Stadtfeld M, Murphy GJ, Hochedlinger K, Kotton DN, Mostoslavsky G 2009. Induced pluripotent stem cell generation using a single lentiviral stem cell cassette. Stem Cells 27: 543–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer CA, Sommer AG, Longmire TA, Christodoulou C, Thomas DD, Gostissa M, Alt FW, Murphy GJ, Kotton DN, Mostoslavsky G 2010. Excision of reprogramming transgenes improves the differentiation potential of iPS cells generated with a single excisable vector. Stem Cells 28: 64–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridharan R, Tchieu J, Mason MJ, Yachechko R, Kuoy E, Horvath S, Zhou Q, Plath K 2009. Role of the murine reprogramming factors in the induction of pluripotency. Cell 136: 364–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Hochedlinger K 2009. Without a trace? PiggyBac-ing toward pluripotency. Nat Methods 6: 329–330 [DOI] [PubMed] [Google Scholar]
- Stadtfeld M, Brennand K, Hochedlinger K 2008a. Reprogramming of pancreatic β cells into induced pluripotent stem cells. Curr Biol 18: 890–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Maherali N, Breault DT, Hochedlinger K 2008b. Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell 2: 230–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Nagaya M, Utikal J, Weir G, Hochedlinger K 2008c. Induced pluripotent stem cells generated without viral integration. Science 322: 945–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Maherali N, Borkent M, Hochedlinger K 2010. A reprogrammable mouse strain from gene-targeted embryonic stem cells. Nat Methods 7: 53–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Apostolou E, Akutsu H, Fukuda A, Follett P, Natesan S, Kono T, Shioda T, Hochedlinger K 2010. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature 465: 175–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens LC, Little CC 1954. Spontaneous testicular teratomas in an inbred strain of mice. Proc Natl Acad Sci 40: 1080–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart TA, Mintz B 1982. Recurrent germ-line transmission of the teratocarcinoma genome from the METT-1 culture line to progeny in vivo. J Exp Zool 224: 465–469 [DOI] [PubMed] [Google Scholar]
- Sugii S, Kida Y, Kawamura T, Suzuki J, Vassena R, Yin YQ, Lutz MK, Berggren WT, Izpisúa Belmonte JC, Evans RM 2010. Human and mouse adipose-derived cells support feeder-independent induction of pluripotent stem cells. Proc Natl Acad Sci 107: 3558–3563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada M, Tada T, Lefebvre L, Barton SC, Surani MA 1997. Embryonic germ cells induce epigenetic reprogramming of somatic nucleus in hybrid cells. EMBO J 16: 6510–6520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada M, Takahama Y, Abe K, Nakatsuji N, Tada T 2001. Nuclear reprogramming of somatic cells by in vitro hybridization with ES cells. Curr Biol 11: 1553–1558 [DOI] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A 2009. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324: 930–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126: 663–676 [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S 2007. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131: 861–872 [DOI] [PubMed] [Google Scholar]
- Tamashiro KL, Wakayama T, Akutsu H, Yamazaki Y, Lachey JL, Wortman MD, Seeley RJ, D'Alessio DA, Woods SC, Yanagimachi R, et al. 2002. Cloned mice have an obese phenotype not transmitted to their offspring. Nat Med 8: 262–267 [DOI] [PubMed] [Google Scholar]
- Tchieu J, Kuoy E, Chin MH, Trinh H, Patterson M, Sherman SP, Aimiuwu O, Lindgren A, Zack JA, Clark AT, et al. 2010. Female human iPS cells retain an inactive X-chromosome. Cell Stem Cell 19: 329–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, McKay RD 2007. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature 448: 196–199 [DOI] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM 1998. Embryonic stem cell lines derived from human blastocysts. Science 282: 1145–1147 [DOI] [PubMed] [Google Scholar]
- Tokuzawa Y, Kaiho E, Maruyama M, Takahashi K, Mitsui K, Maeda M, Niwa H, Yamanaka S 2003. Fbx15 is a novel target of Oct3/4 but is dispensable for embryonic stem cell self-renewal and mouse development. Mol Cell Biol 23: 2699–2708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SY, Clavel C, Kim S, Ang YS, Grisanti L, Lee DF, Kelley K, Rendl M 2010. Oct4 and klf4 reprogram dermal papilla cells into induced pluripotent stem cells. Stem Cells 28: 221–228 [DOI] [PubMed] [Google Scholar]
- Tsubooka N, Ichisaka T, Okita K, Takahashi K, Nakagawa M, Yamanaka S 2009. Roles of Sall4 in the generation of pluripotent stem cells from blastocysts and fibroblasts. Genes Cells 14: 683–694 [DOI] [PubMed] [Google Scholar]
- Utikal J, Maherali N, Kulalert W, Hochedlinger K 2009a. Sox2 is dispensable for the reprogramming of melanocytes and melanoma cells into induced pluripotent stem cells. J Cell Sci 122: 3502–3510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utikal J, Polo JM, Stadtfeld M, Maherali N, Kulalert W, Walsh RM, Khalil A, Rheinwald JG, Hochedlinger K 2009b. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature 460: 1145–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varas F, Stadtfeld M, de Andres-Aguayo L, Maherali N, di Tullio A, Pantano L, Notredame C, Hochedlinger K, Graf T 2009. Fibroblast-derived induced pluripotent stem cells show no common retroviral vector insertions. Stem Cells 27: 300–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M 2010. Direct conversion of fibroblasts to functional neurons by defined factors. Nature 463: 1035–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan SR, Daley GQ, Gregory RI 2008. Selective blockade of microRNA processing by Lin28. Science 320: 97–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakayama T, Yanagimachi R 1999. Cloning of male mice from adult tail-tip cells. Nat Genet 22: 127–128 [DOI] [PubMed] [Google Scholar]
- Wakayama T, Tabar V, Rodriguez I, Perry AC, Studer L, Mombaerts P 2001. Differentiation of embryonic stem cell lines generated from adult somatic cells by nuclear transfer. Science 292: 740–743 [DOI] [PubMed] [Google Scholar]
- Wakayama S, Jakt ML, Suzuki M, Araki R, Hikichi T, Kishigami S, Ohta H, Van Thuan N, Mizutani E, Sakaide Y, et al. 2006. Equivalency of nuclear transfer-derived embryonic stem cells to those derived from fertilized mouse blastocysts. Stem Cells 24: 2023–2033 [DOI] [PubMed] [Google Scholar]
- Wang J, Rao S, Chu J, Shen X, Levasseur DN, Theunissen TW, Orkin SH 2006. A protein interaction network for pluripotency of embryonic stem cells. Nature 444: 364–368 [DOI] [PubMed] [Google Scholar]
- Warren L, Manos PD, Ahfeldt T, Loh Y, Li H, Lau F, Ebina W, Smith ZD, Meissner A, Daley GQ, et al. 2010. Highly efficient reprogramming to pluripotency and directed differentiation of human cells using synthetic modified mRNA. Cell Stem Cell (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein BE, Jaenisch R 2007. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature 448: 318–324 [DOI] [PubMed] [Google Scholar]
- Wernig M, Lengner CJ, Hanna J, Lodato MA, Steine E, Foreman R, Staerk J, Markoulaki S, Jaenisch R 2008a. A drug-inducible transgenic system for direct reprogramming of multiple somatic cell types. Nat Biotechnol 26: 916–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernig M, Meissner A, Cassady JP, Jaenisch R 2008b. c-Myc is dispensable for direct reprogramming of mouse fibroblasts. Cell Stem Cell 2: 10–12 [DOI] [PubMed] [Google Scholar]
- Wernig M, Zhao JP, Pruszak J, Hedlund E, Fu D, Soldner F, Broccoli V, Constantine-Paton M, Isacson O, Jaenisch R 2008c. Neurons derived from reprogrammed fibroblasts functionally integrate into the fetal brain and improve symptoms of rats with Parkinson's disease. Proc Natl Acad Sci 105: 5856–5861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmut I, Schnieke AE, McWhir J, Kind AJ, Campbell KH 1997. Viable offspring derived from fetal and adult mammalian cells. Nature 385: 810–813 [DOI] [PubMed] [Google Scholar]
- Wilson KD, Venkatasubrahmanyam S, Jia F, Sun N, Butte AJ, Wu JC 2009. MicroRNA profiling of human-induced pluripotent stem cells. Stem Cells Dev 18: 749–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler T, Cantilena A, Metais JY, Xu X, Nguyen AD, Borate B, Antosiewicz-Bourget JE, Wolfsberg TG, Thomson JA, Dunbar CE 2010. No evidence for clonal selection due to lentiviral integration sites in human induced pluripotent stem cells. Stem Cells 28: 687–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woltjen K, Michael IP, Mohseni P, Desai R, Mileikovsky M, Hamalainen R, Cowling R, Wang W, Liu P, Gertsenstein M, et al. 2009. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature 458: 766–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Zhang Y, Mishra A, Tardif SD, Hornsby PJ 2010. Generation of induced pluripotent stem cells from newborn marmoset skin fibroblasts. Stem Cell Res 4: 180–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H, Ye M, Feng R, Graf T 2004. Stepwise reprogramming of B cells into macrophages. Cell 117: 663–676 [DOI] [PubMed] [Google Scholar]
- Xu D, Alipio Z, Fink LM, Adcock DM, Yang J, Ward DC, Ma Y 2009. Phenotypic correction of murine hemophilia A using an iPS cell-based therapy. Proc Natl Acad Sci 106: 808–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka S 2009. Elite and stochastic models for induced pluripotent stem cell generation. Nature 460: 49–52 [DOI] [PubMed] [Google Scholar]
- Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A 2008. The ground state of embryonic stem cell self-renewal. Nature 453: 519–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida Y, Takahashi K, Okita K, Ichisaka T, Yamanaka S 2009. Hypoxia enhances the generation of induced pluripotent stem cells. Cell Stem Cell 5: 237–241 [DOI] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. 2007. Induced pluripotent stem cell lines derived from human somatic cells. Science 318: 1917–1920 [DOI] [PubMed] [Google Scholar]
- Yu J, Hu K, Smuga-Otto K, Tian S, Stewart R, Slukvin II, Thomson JA 2009. Human induced pluripotent stem cells free of vector and transgene sequences. Science 324: 797–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusa K, Rad R, Takeda J, Bradley A 2009. Generation of transgene-free induced pluripotent mouse stem cells by the piggyBac transposon. Nat Methods 6: 363–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao XY, Li W, Lv Z, Liu L, Tong M, Hai T, Hao J, Guo CL, Ma QW, Wang L, et al. 2009. iPS cells produce viable mice through tetraploid complementation. Nature 461: 86–90 [DOI] [PubMed] [Google Scholar]
- Zhao HX, Li Y, Jin HF, Xie L, Liu C, Jiang F, Luo YN, Yin GW, Li Y, Wang J, et al. 2010. Rapid and efficient reprogramming of human amnion-derived cells into pluripotency by three factors OCT4/SOX2/NANOG. Differentiation doi: 10.1016/j.diff.2010.03.002 [DOI] [PubMed] [Google Scholar]
- Zhou W, Freed CR 2009. Adenoviral gene delivery can reprogram human fibroblasts to induced pluripotent stem cells. Stem Cells 27: 2667–2674 [DOI] [PubMed] [Google Scholar]
- Zhou Q, Brown J, Kanarek A, Rajagopal J, Melton DA 2008. In vivo reprogramming of adult pancreatic exocrine cells to β-cells. Nature 455: 627–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Wu S, Joo JY, Zhu S, Han DW, Lin T, Trauger S, Bien G, Yao S, Zhu Y, et al. 2009. Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell 4: 381–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Maeder ML, Mali P, Pruett-Miller SM, Thibodeau-Beganny S, Chou BK, Chen G, Ye Z, Park IH, Daley GQ, et al. 2009. Gene targeting of a disease-related gene in human induced pluripotent stem and embryonic stem cells. Cell Stem Cell 5: 97–110 [DOI] [PMC free article] [PubMed] [Google Scholar]