Abstract
Cyclic AMP (cAMP) is a ubiquitous second messenger that regulates a variety of biological processes. The magnitude and duration of cAMP expression are regulated by both production and hydrolysis. Melanocyte-stimulating hormone (MSH) plays a crucial role in pigment cell differentiation via cAMP-regulated expression of the master transcription factor MITF. We report the identification of phosphodiesterase 4D3 as a direct target of the MSH/cAMP/MITF pathway. This creates a negative feedback loop that induces refractoriness to chronic stimulation of the cAMP pathway in melanocytes. This homeostatic pathway highlights a potent mechanism controlling melanocyte differentiation that may be amenable to pharmacologic manipulation for skin cancer prevention.
Keywords: cAMP, MITF, PDE4D, skin, pigmentation
Cyclic AMP (cAMP) is a second messenger that regulates key processes (such as cell growth, differentiation, and movement) and specialized actions unique to specific cell lineages (Houslay et al. 2007). cAMP is typically metabolized by phosphodiesterases (PDEs), which control the magnitude, duration, and subcellular localization of cAMP (Houslay et al. 2007). In melanocytes the cAMP/CREB signaling pathway is potently regulated by melanocyte-stimulating hormone (MSH) via the G protein-coupled receptor MC1R, which in turn transcriptionally activates expression of MITF (Bertolotto et al. 1998; Price et al. 1998), the master transcriptional regulator of melanocyte development (Levy et al. 2006). In vitro and in vivo, the up-regulation of cAMP in melanocytes leads to increased pigment production and protection of the skin against the deleterious effects of UV radiation (D'Orazio et al. 2006). Skin is the most common organ to be affected by cancer, and cutaneous malignancies most commonly occur in fair-skinned people with a limited capacity to tan (Fitzpatrick and Sober 1985; Fitzpatrick 1988; Sturm et al. 2003; Rijken et al. 2004). The role of pigmentation in modulating human skin cancer risk continues to be incompletely understood. However, darkly pigmented people or those who tan easily (Fitzpatrick phototypes 3–6) exhibit significantly diminished skin cancer risk relative to those with light skin who tan poorly (Fitzpatrick phototypes 1,2) (Lin and Fisher 2007).
Here we report that, in melanocytes, MITF directly regulates the transcription of the PDE4D3 gene, creating negative homeostatic control of the cAMP pathway and lineage differentiation. These findings provide a rationale for targeting PDE4D3 to modulate MITF expression and control skin pigmentation.
Results and Discussion
PDE4D expression is MITF-dependent in melanocytes
We used annotated microarray data (http://biogps.gnf.org/downloads) to search for PDEs that are expressed in the melanocyte lineage. Among the PDEs known to degrade cAMP (Conti and Beavo 2007; Omori and Kotera 2007), the following exhibited “present calls” in some or most of the melanoma lines: PDE1C, PDE3A, PDE3B, PDE4A, PDE4B, PDE4D, PDE6C, PDE7B, PDE8A, and PDE10A. When the expression of these PDEs was compared with that of MITF, we observed that two of them, PDE4D and PDE4B, exhibited fluctuations in expression that correlated with MITF expression, suggesting that they could be transcriptionally linked to MITF (Supplemental Fig. S1). In this analysis, we used tyrosinase, a known target of MITF, as a positive control (Bentley et al. 1994; Yasumoto et al. 1994). Interestingly, PDE4C did not correlate with MITF.
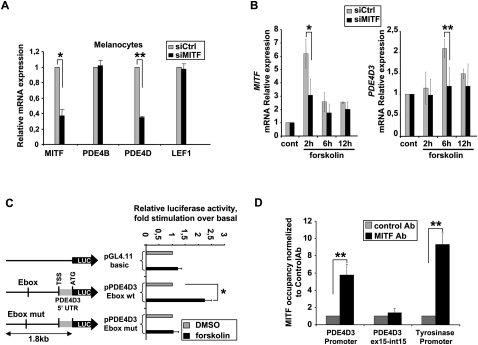
To address this possibility, we first investigated whether MITF was necessary for PDE4D and PDE4B expression (Fig. 1A). siRNA-mediated knockdown of MITF in three different primary human melanocyte cultures significantly suppressed expression of PDE4D, but not PDE4B. This experiment was carried out using primers able to recognize all isoforms of the PDE4D gene. Expression of LEF-1, an additional control, was also unaffected. The knockdown efficiency of MITF was also verified by Western blot (Supplemental Fig. S2).
Figure 1.
cAMP-induced up-regulation of PDE4D3 is dependent on MITF. (A) Primary human melanocytes were transfected with a nontargeting control siRNA (siCtrl) or a siRNA specific for MITF. Total mRNA was subjected to qPCR. The data are normalized to β-actin. (B) Primary human melanocytes were transfected with a nontargeting siRNA (siCtrl) or siRNAs targeting MITF and exposed to forskolin, as indicated. Total mRNA was subjected to qPCR. Results are expressed as fold stimulation and represent the mean ± SD of three independent experiments. (C) UACC62 cells were transiently transfected with reporter plasmids pPDE4D3 and pPDE4D3 Ebox mutated. After 24 h of forskolin stimulation, firefly luciferase was measured and normalized to the renilla luciferase control. Results are shown as fold stimulation and represent the mean ± SD of three experiments performed in triplicate. (TSS) Transcription start site. (D) ChIP from primary human melanocytes was performed using MITF or control antibody (see the Materials and Methods). RNA levels were analyzed by qPCR. Data are plotted as fold change versus control antibody, and each point is the mean ± SD of three independent experiments. P-values were obtained by a paired t-test. (*) P < 0.05; (**) P < 0.005.
PDE4D is a complex gene encoding multiple isoforms (Supplemental Fig. S3) with distinct regulation and tissue distribution (D'Sa et al. 2002; Richter et al. 2005). To identify the specific isoforms expressed in primary melanocytes, isoform-specific primers were used and revealed expression of seven of the nine PDE4Ds known to be expressed in humans (all except PDE4D4 and PDE4D8) (Supplemental Fig. S4A). Since MITF is regulated by signaling pathways using cAMP (Bertolotto et al. 1998; Price et al. 1998), we examined the effect of forskolin on the expression of the PDE4D isoforms in melanocytes. Only PDE4D3 mRNA was consistently up-regulated upon forskolin stimulation (Supplemental Fig. S4B). We observed induction of PDE4D3 mRNA levels with a slight delay (first seen at 6 h), consistent with the possibility of a mechanistic intermediate between forskolin/cAMP and PDE4D3 transcription. Slight suppression of PDE4D3 levels at 2 h was also observed in fibroblasts (Supplemental Fig. S5A), suggesting a mechanism unrelated to M-MITF. Treatment of normal human fibroblasts with forskolin failed to induce the expression of PDE4D (using common region primers) or PDE4D3 (Supplemental Fig. S5A), although it did induce phosphorylation of CREB, confirming the functionality of forskolin in fibroblasts (Supplemental Fig. S5B).
In rat cortical neurons, it has been observed previously that cAMP signaling can directly target the expression of Pde4b2 and Pde4d1/2 but not Pde4d3 (D'Sa et al. 2002). Although we observed PDE4B2 to be expressed in melanocytes, its expression did not change in response to cAMP signaling (data not shown), suggesting that tissue-restricted mechanisms may regulate the precise PDE species involved in feedback loops of different cell lineages.
MITF regulates the transcription of PDE4D3
To assess whether MITF is involved in the regulation of PDE4D3 expression, melanocytes were stimulated with forskolin in the presence of siRNA targeting MITF (or control siRNA). Whereas PDE4D3 was up-regulated by forskolin in control siRNA-treated cells, it was not up-regulated in melanocytes transfected with either of two siRNAs targeting MITF (Fig. 1B; Supplemental Fig. S6A), indicating that up-regulation of PDE4D3 is MITF-dependant in melanocytes. The MITF knockdown efficiency was verified by Western blot (Supplemental Fig. S6B). Although MITF-dependent PDE4D3 up-regulation by forskolin was a consistent finding, we observed some variability in the kinetics of its induction (between 6 and 12 h) that was linked to the donor source of the primary human melanocytes. It is currently unknown if these varied kinetics correlate with differing pigmentation features.
To further assess the regulation of PDE4D3 by MITF, UACC62 melanoma cells were transfected with a luciferase reporter driven by 1.8 kb upstream of the translation start site of PDE4D3. We observed stimulation of luciferase activity by forskolin, and this up-regulation was lost when the consensus MITF-binding site (Ebox) was mutated (CACATG to GAGATG) (Fig. 1C). The ability of MITF to bind to the Ebox of the 1.8-kb promoter region of PDE4D3 was verified by an electropheretic mobility shift assay (Supplemental Fig. S7). The supershift observed by adding an MITF antibody in the binding reaction was successfully chased by a nonbiotinylated Ebox wild-type probe and failed when using the Ebox mutated probe.
To investigate if MITF directly occupies the promoter region of the PDE4D3 gene, we performed chromatin immunoprecipitation (ChIP) using genomic DNA from primary human melanocytes. When the relative occupancy of MITF on the PDE4D3 promoter was quantified, a significant enrichment was seen that was comparable with MITF occupancy measured at the tyrosinase promoter (Fig. 1D). As a negative control, detection of the exon-14/intron-14 boundary of PDE4D3 failed to show enrichment of MITF binding. The transcriptional regulation of PDE4D3 by MITF is particularly notable because MITF is a central regulator of the pigmentation cascade via targeting of numerous enzymes and other factors important for melanocyte-specific synthesis, maturation, and transport of melanin. Interestingly, examination of a gene expression data set from MITF-overexpressing melanoma cells showed PDE4D up-regulation by MITF (Hoek et al. 2008). Also, PDE4D was found to be regulated by the MC1R pathway in mouse melanocytes (Le Pape et al. 2008).
Inhibition of PDE4 sustains CREB phosphorylation and MITF expression in melanocytes
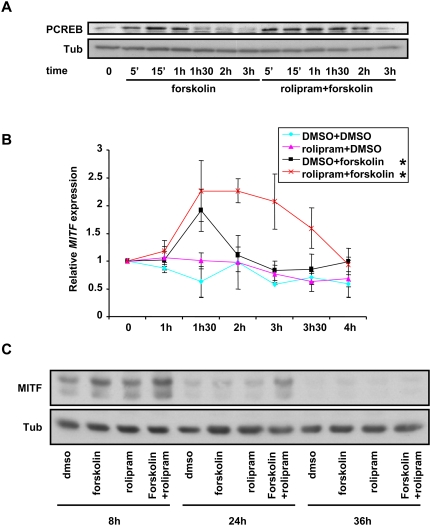
To explore the physiological role of PDE4D3 in melanocytes, we investigated the impact of its inhibition by rolipram, a known PDE4 inhibitor that has been studied previously as an anti-inflammatory and anti-depressant agent (Bobon et al. 1988; Griswold et al. 1993). We tested the toxicity of this inhibitor and its effect on the cells' morphology. We observed that it does not significantly affect human primary melanocyte growth or alter cellular morphology (Supplemental Fig. S8A,B). We observed the same results when the cells were exposed to forskolin or the combination of forskolin and rolipram, although the combination of forskolin and rolipram modestly increased pigmentation by 48 h (data not shown). At the cAMP signaling level, we observed that pretreatment of melanocytes with rolipram prolonged CREB phosphorylation after forskolin stimulation (Fig. 2A). Several PDE4D isoforms are activated via PKA phosphorylation, creating a short-term feedback loop (Sette and Conti 1996; MacKenzie et al. 2002; Richter et al. 2005). This phenomenon could explain why rolipram extends the duration of forskolin-induced CREB phosphorylation. PDE4D3 has also been shown to be inhibited by phosphorylation by ERK2, although this inhibition is ablated by subsequent PKA phosphorylation (Hoffmann et al. 1999), an event that is likely to occur in melanocytes (Busca et al. 2000).
Figure 2.
Inhibition of PDE4 potentiates forskolin-induced CREB phosphorylation in primary human melanocytes. (A) Extracts from primary human melanocytes placed in minimal media for 14 h before being treated as indicated were immunoblotted using antibodies against phospho-CREB and α-tubulin. (B) Primary human melanocytes were exposed to the indicated conditions for 30 min before being rinsed and placed in minimal media for the indicated times. The results are normalized to β-actin and are shown as fold induction of MITF. Each data point is the mean ± SD of three experiments. (*) P < 0.05. (C) Extracts from primary human melanocytes were placed in minimal media for 14 h before being exposed for 30 min to rolipram or DMSO prior to being treated with forskolin for 30 min. Subsequently, the cells were rinsed and placed in minimal media for the indicated times. The protein extracts were submitted to immunoblotting using MITF and α-tubulin antibodies.
Since MITF expression is directly regulated by CREB, we investigated the combined effect of rolipram and forskolin on MITF mRNA expression. Primary melanocytes were pretreated with DMSO or rolipram for 30 min, and then exposed to forskolin or DMSO for up to 4 h before mRNA was collected and analyzed by quantitative PCR (qPCR) (Fig. 2B). Whereas the combination of rolipram plus forskolin only minimally affected peak MITF levels, the duration of MITF induction was significantly prolonged. We also verified the effect of PDE4 inhibition of MITF protein expression, and we observed that rolipram did not significantly affect the expression level of MITF after 8 h of stimulation, but prolonged its expression. Indeed, at 24 h, MITF expression remained higher than control only when the cells were exposed to the combination of rolipram and forskolin (Fig. 2C). These results suggest that PDE4 activity plays a measurably important role in modulating MITF, and suggest a testable strategy to modulate skin pigmentation.
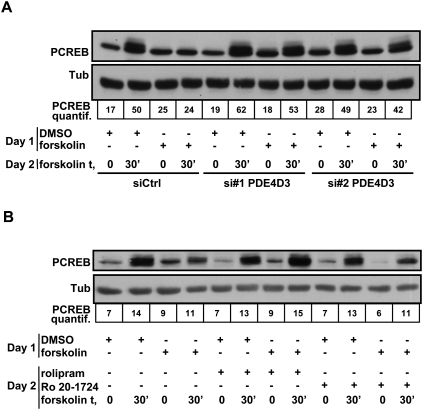
To further examine whether PDE4D3 regulation by MITF is a physiologically relevant negative feedback loop, we asked whether prior treatment with forskolin may render melanocytes resistant to repeated forskolin treatment due to negative feedback mediated by PDE4D3. Primary melanocytes were treated on day 1 for 4 h with forskolin (or DMSO control) and then returned to minimal media until repeat forskolin challenge on day 2 in the presence or absence of pretreatment with either siRNA directed against PDE4D3 or control siRNA. Melanocytes pretreated with DMSO on day 1 were sensitive to forskolin on day 2, as measured by phosphorylation of CREB. Melanocytes treated with forskolin on day 1 were insensitive to forskolin the next day, as assessed by lack of CREB phosphorylation. However, sensitivity to repeated forskolin challenge was restored by siRNA-mediated knockdown of PDE4D3 (Fig. 3A). The efficiency of PDE4D3 knockdown was assessed by qPCR (Supplemental Fig. S9). The same phenomenon was observed when looking at the expression of a target of MITF: PMEL17. When primary human melanocytes were exposed to DMSO on day 1, we observed an up-regulation of PMEL17 mRNA expression on day 2 after 8 h of forskolin treatment. However, if the cells were exposed previously to forskolin, we did not observe an up-regulation of PMEL17 after forskolin treatment. The sensitivity to forskolin was restored when the cells were transfected with an siRNA specific for PDE4D3 but not when they were transfected with a siRNA directed against PDE4D5. This experiment suggests that PDE4D3 is the main isoform of PDE4D preventing chronic activation of the cAMP pathway in human primary melanocytes (Supplemental Fig. S10). The same experiment was also performed by adding the PDE4 inhibitors rolipram or Ro 20-1724 prior to forskolin stimulation on day 2. Here, the drug treatments similarly restored CREB phosphorylation by forskolin on day 2 (Fig. 3B). We conclude that up-regulation of PDE4D3 by the MITF autoregulatory loop normally limits repetitive stimulation of the melanogenic pathway.
Figure 3.
Forskolin pretreatment induces forskolin resistance that is reversible by PDE4D3 knockdown or rolipram treatment. (A) Primary human melanocytes were transfected with a nontargeting siRNA (siCtrl) or siRNAs targeting PDE4D3. Twenty-four hours after transfection, the cells were placed in minimal media (MM) for 14 h, and were subsequently treated with forskolin for 4 h before being returned to minimal media. The following day, the cells were exposed to forskolin for the indicated times. Cell lysates were subjected to Western blotting using antibodies specific for phospho-CREB and α-tubulin. The intensity of the phospho-CREB Western blot was measured and normalized to α-tubulin. (B) Primary human melanocytes were incubated in minimal media for 14 h, treated with forskolin or DMSO for 4 h, and returned to minimal media. The following day, the cells were exposed to DMSO or rolipram or Ro 20-1724 for 4 h and then rechallenged with forskolin for the indicated times. Cell lysates were subjected to Western blotting using antibodies specific for phospho-CREB and α-tubulin. The intensity of the phospho-CREB Western blot was measured and normalized to α-tubulin.
PDE4 inhibition synergizes with forskolin to induce skin pigmentation
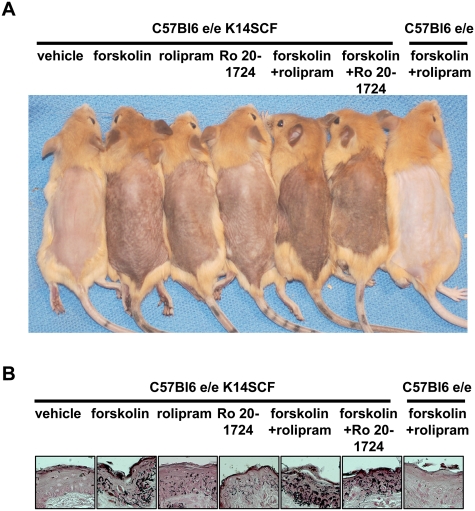
To study the relevance of the regulation of PDE4D3 by MITF in vivo, we asked if topical PDE4 inhibitors may enhance skin pigmentation in C57bl6e/e K14 SCF mice (Kunisada et al. 1998; D'Orazio et al. 2006). This mouse strain carries a frameshift mutation in MC1R, causing the red hair phenotype, as well as a transgene (K14-SCF) that produces “humanized” skin containing epidermal melanocytes (in contrast to wild-type mice that lack melanocytes in the interfollicular epidermis) (Kunisada et al. 1998). After 5 d, a modest degree of melanization was observed in the mice exposed to either forskolin or two different PDE4 inhibitors: rolipram and Ro 20-1724. However, the combination of forskolin with either of the PDE4 inhibitors induced robust darkening (Fig. 4A). C57bl6e/e mice harboring no melanocytes in the epidermis served as a negative control. Fontana-Masson staining of skin showed that skin darkness correlated with increased melanin pigment (Fig. 4B). Darkening of the skin was quantified using reflective colorimetry (Supplemental Table 1; Park et al. 1999), and showed that the combination of forskolin and rolipram or forskolin and Ro 20-1724, applied once daily for five consecutive days, induced pigmentation comparable with the effect observed previously after 21 d of forskolin treatment alone (D'Orazio et al. 2006).
Figure 4.
PDE4 inhibition synergizes with forskolin to induce sunless tanning in redhead/fair-skinned mice. (A) C57bl6e/e;K14-SCF and C57bl6e/e mice were treated for five consecutive days with forskolin alone, rolipram alone, Ro 20-1724 alone, or the combinations of forskolin/rolipram or forskolin/Ro 20-1724, once per day. A representative group of mice out of three independent groups receiving the same treatment was photographed at day 5. (B) Fontana-Masson-stained skin sections (for melanin) (black deposits, 63× magnification).
Human epidemiological data and mouse models have both demonstrated correlations between pigmentation and skin cancer risk (D'Orazio et al. 2006; Lin and Fisher 2007). Whereas skin pigmentation may thus be protective against UV carcinogenesis, the standard method of enhancing skin pigmentation (via UV exposure) carries simultaneous carcinogenic risk. Parenteral peptide analogs of MSH exhibit strong skin-darkening effects (Lerner and McGuire 1961), although they require systemic delivery. Prior topical application of forskolin was seen to darken the skin of redhead/light-skinned mice and confer significant protection against subsequent UV carcinogenic challenge (D'Orazio et al. 2006). However, forskolin exhibits very poor human skin penetration and thus has limited efficacy (data not shown). The development of drugs that function as antagonists of inhibitors (such as PDE4D inhibitors) may represent easier drug discovery than identification of enzyme stimulators (such as forskolin, which activates adenylate cyclase). Aside from their potential roles in protection against skin cancer and photo-aging, the use of topical agents affecting pigmentation may provide opportunities to better understand signaling and trafficking components of the pigmentation response through the ability to pharmacologically and synchronously activate melanogenesis. The homeostatic pathway reported here (Fig. 5) uses a tissue-restricted transcription factor (MITF) to target expression of a factor that down-regulates a major stimulatory pathway. MITF is an amplified oncogene in certain melanomas (Garraway et al. 2005), and this PDE4D3 pathway may thus be significantly altered, carrying therapeutic implications worthy of study. PDEs modulate numerous signaling pathways in distinct lineage contexts. It is thus plausible that comparable lineage-specific transcriptional mechanisms may exist in other cell types that provide feedback control of cellular signaling
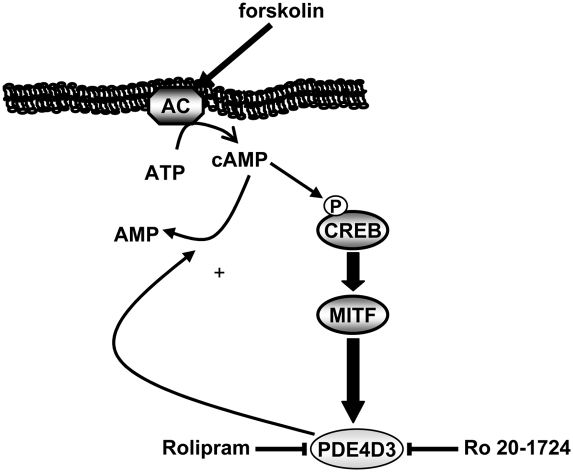
Figure 5.
Graphical representation of the cAMP–MITF–PDE4D3 feedback loop.
Materials and methods
Cell culture
Primary human melanocytes from neonatal foreskins were established in Ham's F10 (Invitrogen) medium containing 7% fetal bovine serum, penicillin/streptomycin/L-glutamine (Invitrogen), 1 × 10−4 M 3-isobutyl-1-methylxanthine (Sigma), 50 ng mL−1 12-O-tetradecanoylphorbol-13-acetate (Sigma), 1 μM Na3VO4, and 1 × 10−3 M N6,2′-O-dibutyryladenosine 3:5-cyclic monophosphate (Sigma). Minimal media was made with Ham's F10 medium supplemented with 3% fetal bovine serum and penicillin/streptomycin/L-glutamine. UACC62 human melanoma cells were obtained from NCI and grown in RPMI medium supplemented with 10% fetal bovine serum and penicillin/streptomycin/L-glutamine.
ChIP
ChIP was performed as described previously (Du et al. 2004). The immunoprecipitations were performed with MITF polyclonal antibody and Placental Protein 4 (control antibody) purchased from Assay Designs. qPCR was carried out using primers specific for the promoter region of human PDE4D3 (5′-GAGAACAGCCAGCCTTATTATGGG-3′ and 5′-CTGCTCTGCAGGACAAGATTACCA-3′) or spanning the junction of exon 14 and intron 14 of human PDE4D3 (5′-ACGTGGCATGGAGATAAGCCC-3′ and 5′- AACCAAATGCTAAAGCGGTAGCTC-3′). The human tyrosinase promoter primers used were 5′-GTGGGATACGAGCCAATTCGAAAG-3′ and 5′-TCCCACCTCCAGCATCAAACACTT-3′.
RT–PCR
cDNA were obtained using total mRNA for primary human melanocytes and sk-n-sh using SuperScript III Reverse Transcriptase (Invitrogen) according to the manufacturer's recommendations. Human heart quick-clone cDNA was purchased from Clontech. For primer sequences, refer to the primer table in the Supplemental Material.
qPCR
Primary human melanocytes were starved for 14 h before being treated as specified in each experiment. RNA was isolated using RNeasy plus minikits from Qiagen, and was subjected to one-step RT–PCR using QuantiTect Probe RT–PCR kits from Qiagen and iQ Sybr-Green Supermix (Bio-Rad). For each reaction, 20 ng of RNA was subjected to the following steps: reverse transcription for 30 min at 48°C, hot-start Taq activation for 8.5 min at 95°C, and 40 cycles of PCR reaction for 15 sec at 95°C and for 30 sec at 62°C. The results are the average of three independent experiments. For primer sequences, refer to the primer table in the Supplemental Material (Supplemental Table 2).
Gel electrophoresis and immunoblotting
Primary human melanocytes were cultured in six-well dishes and exposed to different agents for the times indicated in each figure legend. Subsequently, the cells were lysed in buffer containing 50 mM Tris (pH 7.4), 150 mM NaCl, 1% Triton X-100, 10 μM leupeptin, 1 mM AEBSF, 100 U/mL aprotinin, 10 mM NaF, and 1 mM Na3VO4. Samples (30 μg) were resolved by 10% SDS-PAGE, transferred to nitrocellulose membranes, and then exposed to the appropriate antibodies: anti-phospho-CREB (Cell Signaling Technologies), anti-tubulin (Sigma Aldrich), and anti-MITF (C5). Proteins were visualized with the ECL system from Perkin Elmer Life Sciences using horseradish peroxidase-conjugated anti-rabbit or anti-mouse secondary antibody. Western blot assays shown are representative of at least three experiments.
Construction of pPDE4D3 reporter and mutagenesis
A 1.8-kb fragment upstream of the translation start site of PDE4D3 was amplified from BAC clone RP11-A7, purchased from Children's Hospital Oakland Research Institute using primers 5′-CCCAAGCTTGGTCATCTGCAGCTAAATGGTTAC-3′ and 5′-CATGCCATGGTCGCAGATCTTCTGTCATTAATA-3′. The fragment was digested with HindIII and NcoI and inserted into pGL4.11 (Promega). Site-directed mutagenesis was performed using the QuickChange method from Stratagene according to the supplier's recommendations. The primers used were 5′-TCCAAGGCAAAATATGAAAAGCTCCGAGATGGTTTTGATAATAACAAAATAAAG-3′ and 5′- CTTTATTTTGTTATTATCAAAACCATCTCGGAGCTTTTCATATTTTGCCTTGGA-3′.
Transfection and dual luciferase reporter assay
UACC62 melanoma cells were seeded onto 24-well dishes and transfected the following day in 1.5 μL of Lipofectamine 2000 (Invitrogen) with 0.5 μg each of pGL4.11, pPDE4D3, and pPDE4D3mut. Test plasmids were transfected with pGL2cmv renilla luciferase reporter to control for the variability in transfection efficiency. After 24 h of transfection, the cells were exposed to 20 μM fsk for 24 h. Cell lysates were prepared 48 h later, and the activity of firefly and renilla luciferase was measured using the Dual Luciferase kit (Promega) according to the manufacturer's recommendations.
siRNA transfection
Primary human melanocytes were seeded in six-well dishes transfected three times with HiPerFect (Qiagen) according to the manufacturer's protocols and 100 pmol of double-stranded siRNA per well (0.5 × 106 cells) at 24-h intervals. Twelve hours after the last transfection, the cells were placed in minimal media for 14 h and treated as indicated in the figures. Nontargeting siRNA (Silencer Negative Control; Ambion) used were si#1, MITF, 5′-GGCUUUCUAGAAAGAAUAA-3′ (Ambion); and si#2, MITF, 5′-GGUGAAUCGGAUCAUCAAG-3′ (Carreira et al. 2005) (Ambion). si PDE4D3 pool 1 was a mix of 5′-CACGAUAGCUGCUCAAACA-3′ (Qiagen) and 5′-UAACGUAGGAGACAAGAAA-3′ (Qiagen), and si PDE4D3 pool 2 was a mix of 5′-UGAUGCACGUGAAUAAUUU-3′ (Qiagen) and 5′-GAGUUGGAAUUCAUCUGUA-3′ (Qiagen) (Lynch et al. 2005).
Mice
C57BL6e/e K14SCF and C57BL6e/e mice were described previously (D'Orazio et al. 2006) . Animals between 5 and 10 wk of age were depilated in the dorsal area using bee's wax. Crude extracts of ground-up Plectranthus barbatus (forskolin) (ATZ natural) and rolipram and Ro20-1724 (Sigma-Aldrich) were dissolved in 70% ethanol and 30% propylene glycol at a final concentration of 98 mM for forskolin and 18 mM for rolipram and Ro20-1724. Eighty microliters of the different drugs or combinations was applied to the skin once a day for 5 d. Skin reflective colorimetry measurements were assessed with a CR-400 Colorimeter (Minolta Corporation). The instrument was calibrated against the white standard background provided by the manufacturer before use. Degree of melanization (darkness) is described as the colorimetric measurement on the *L axis (white–black axis) of the Center Internationale d'Eclairage (CIE) L*a*b* color system (Park et al. 1999). The measurement is based on the ability of colors to reflect light. The instrument emits flashes and quantitatively measures reflected light. The results range from 0 to 100, where 0 represents no reflected light (black) and 100 represents maximal reflected light (white).
Histology
Animals were euthanized by CO2 inhalation prior to skin sampling. Dorsal skin biopsies were kept in 10% buffered formalin until paraffin embedding and sectioning. Fontana-Masson staining was performed according to routine procedures.
Statistics
P-values were calculated with a paired, two-sided t-test. P-values were noted in each statistical analysis when P was <0.05.
Acknowledgments
We thank Andre Rosowsky, Kathleen C. Robinson, and Su-Jean Seo for critical comments on the manuscript, and Scott Granter, Hans Widlund, Akinori Kawakami, Rosa Veguilla, and Satoru Yokoyama for discussions and help with technical aspects of the study. We also thank Riham Carden and Abraham Cooper, who participated in early portions of the project. We thank Takahiro Kunisada for providing the C57BL6 K14SCF mouse. M.K. acknowledges the Philippe Foundation for their support. D.E.F. gratefully acknowledges support for this research from the National Institutes of Health (NIAMS AR043369-15 and 1AR058469), the Melanoma Research Alliance, the US/Israel Binational Science Foundation, The Adelson Medical Research Foundation, and the Doris Duke Charitable Foundation.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1937710.
Supplemental material is available at http://www.genesdev.org.
References
- Bentley NJ, Eisen T, Goding CR 1994. Melanocyte-specific expression of the human tyrosinase promoter: Activation by the microphthalmia gene product and role of the initiator. Mol Cell Biol 14: 7996–8006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolotto C, Abbe P, Hemesath TJ, Bille K, Fisher DE, Ortonne JP, Ballotti R 1998. Microphthalmia gene product as a signal transducer in cAMP-induced differentiation of melanocytes. J Cell Biol 142: 827–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobon D, Breulet M, Gerard-Vandenhove MA, Guiot-Goffioul F, Plomteux G, Sastre-y-Hernandez M, Schratzer M, Troisfontaines B, von Frenckell R, Wachtel H 1988. Is phosphodiesterase inhibition a new mechanism of antidepressant action? A double blind double-dummy study between rolipram and desipramine in hospitalized major and/or endogenous depressives. Eur Arch Psychiatry Neurol Sci 238: 2–6 [DOI] [PubMed] [Google Scholar]
- Busca R, Abbe P, Mantoux F, Aberdam E, Peyssonnaux C, Eychene A, Ortonne JP, Ballotti R 2000. Ras mediates the cAMP-dependent activation of extracellular signal-regulated kinases (ERKs) in melanocytes. EMBO J 19: 2900–2910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreira S, Goodall J, Aksan I, La Rocca SA, Galibert MD, Denat L, Larue L, Goding CR 2005. Mitf cooperates with Rb1 and activates p21Cip1 expression to regulate cell cycle progression. Nature 433: 764–769 [DOI] [PubMed] [Google Scholar]
- Conti M, Beavo J 2007. Biochemistry and physiology of cyclic nucleotide phosphodiesterases: Essential components in cyclic nucleotide signaling. Annu Rev Biochem 76: 481–511 [DOI] [PubMed] [Google Scholar]
- D'Orazio JA, Nobuhisa T, Cui R, Arya M, Spry M, Wakamatsu K, Igras V, Kunisada T, Granter SR, Nishimura EK, et al. 2006. Topical drug rescue strategy and skin protection based on the role of Mc1r in UV-induced tanning. Nature 443: 340–344 [DOI] [PubMed] [Google Scholar]
- D'Sa C, Tolbert LM, Conti M, Duman RS 2002. Regulation of cAMP-specific phosphodiesterases type 4B and 4D (PDE4) splice variants by cAMP signaling in primary cortical neurons. J Neurochem 81: 745–757 [DOI] [PubMed] [Google Scholar]
- Du J, Widlund HR, Horstmann MA, Ramaswamy S, Ross K, Huber WE, Nishimura EK, Golub TR, Fisher DE 2004. Critical role of CDK2 for melanoma growth linked to its melanocyte-specific transcriptional regulation by MITF. Cancer Cell 6: 565–576 [DOI] [PubMed] [Google Scholar]
- Fitzpatrick TB 1988. The validity and practicality of sun-reactive skin types I through VI. Arch Dermatol 124: 869–871 [DOI] [PubMed] [Google Scholar]
- Fitzpatrick TB, Sober AJ 1985. Sunlight and skin cancer. N Engl J Med 313: 818–820 [DOI] [PubMed] [Google Scholar]
- Garraway LA, Widlund HR, Rubin MA, Getz G, Berger AJ, Ramaswamy S, Beroukhim R, Milner DA, Granter SR, Du J, et al. 2005. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature 436: 117–122 [DOI] [PubMed] [Google Scholar]
- Griswold DE, Webb EF, Breton J, White JR, Marshall PJ, Torphy TJ 1993. Effect of selective phosphodiesterase type IV inhibitor, rolipram, on fluid and cellular phases of inflammatory response. Inflammation 17: 333–344 [DOI] [PubMed] [Google Scholar]
- Hoek KS, Schlegel NC, Eichhoff OM, Widmer DS, Praetorius C, Einarsson SO, Valgeirsdottir S, Bergsteinsdottir K, Schepsky A, Dummer R, et al. 2008. Novel MITF targets identified using a two-step DNA microarray strategy. Pigment Cell Melanoma Res 21: 665–676 [DOI] [PubMed] [Google Scholar]
- Hoffmann R, Baillie GS, MacKenzie SJ, Yarwood SJ, Houslay MD 1999. The MAP kinase ERK2 inhibits the cyclic AMP-specific phosphodiesterase HSPDE4D3 by phosphorylating it at Ser579. EMBO J 18: 893–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houslay MD, Baillie GS, Maurice DH 2007. cAMP-specific phosphodiesterase-4 enzymes in the cardiovascular system: A molecular toolbox for generating compartmentalized cAMP signaling. Circ Res 100: 950–966 [DOI] [PubMed] [Google Scholar]
- Kunisada T, Lu SZ, Yoshida H, Nishikawa S, Mizoguchi M, Hayashi S, Tyrrell L, Williams DA, Wang X, Longley BJ 1998. Murine cutaneous mastocytosis and epidermal melanocytosis induced by keratinocyte expression of transgenic stem cell factor. J Exp Med 187: 1565–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Pape E, Wakamatsu K, Ito S, Wolber R, Hearing VJ 2008. Regulation of eumelanin/pheomelanin synthesis and visible pigmentation in melanocytes by ligands of the melanocortin 1 receptor. Pigment Cell Melanoma Res 21: 477–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner AB, McGuire JS 1961. Effect of α- and β-melanocyte stimulating hormones on the skin colour of man. Nature 189: 176–179 [DOI] [PubMed] [Google Scholar]
- Levy C, Khaled M, Fisher DE 2006. MITF: Master regulator of melanocyte development and melanoma oncogene. Trends Mol Med 12: 406–414 [DOI] [PubMed] [Google Scholar]
- Lin JY, Fisher DE 2007. Melanocyte biology and skin pigmentation. Nature 445: 843–850 [DOI] [PubMed] [Google Scholar]
- Lynch MJ, Baillie GS, Mohamed A, Li X, Maisonneuve C, Klussmann E, van Heeke G, Houslay MD 2005. RNA silencing identifies PDE4D5 as the functionally relevant cAMP phosphodiesterase interacting with βarrestin to control the protein kinase A/AKAP79-mediated switching of the β2-adrenergic receptor to activation of ERK in HEK293B2 cells. J Biol Chem 280: 33178–33189 [DOI] [PubMed] [Google Scholar]
- MacKenzie SJ, Baillie GS, McPhee I, MacKenzie C, Seamons R, McSorley T, Millen J, Beard MB, van Heeke G, Houslay MD 2002. Long PDE4 cAMP specific phosphodiesterases are activated by protein kinase A-mediated phosphorylation of a single serine residue in Upstream Conserved Region 1 (UCR1). Br J Pharmacol 136: 421–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omori K, Kotera J 2007. Overview of PDEs and their regulation. Circ Res 100: 309–327 [DOI] [PubMed] [Google Scholar]
- Park SB, Suh DH, Youn JI 1999. A long-term time course of colorimetric evaluation of ultraviolet light-induced skin reactions. Clin Exp Dermatol 24: 315–320 [DOI] [PubMed] [Google Scholar]
- Price ER, Horstmann MA, Wells AG, Weilbaecher KN, Takemoto CM, Landis MW, Fisher DE 1998. α-Melanocyte-stimulating hormone signaling regulates expression of microphthalmia, a gene deficient in Waardenburg syndrome. J Biol Chem 273: 33042–33047 [DOI] [PubMed] [Google Scholar]
- Richter W, Jin SL, Conti M 2005. Splice variants of the cyclic nucleotide phosphodiesterase PDE4D are differentially expressed and regulated in rat tissue. Biochem J 388: 803–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijken F, Bruijnzeel PL, van Weelden H, Kiekens RC 2004. Responses of black and white skin to solar-simulating radiation: Differences in DNA photodamage, infiltrating neutrophils, proteolytic enzymes induced, keratinocyte activation, and IL-10 expression. J Invest Dermatol 122: 1448–1455 [DOI] [PubMed] [Google Scholar]
- Sette C, Conti M 1996. Phosphorylation and activation of a cAMP-specific phosphodiesterase by the cAMP-dependent protein kinase. Involvement of serine 54 in the enzyme activation. J Biol Chem 271: 16526–16534 [DOI] [PubMed] [Google Scholar]
- Sturm RA, Duffy DL, Box NF, Chen W, Smit DJ, Brown DL, Stow JL, Leonard JH, Martin NG 2003. The role of melanocortin-1 receptor polymorphism in skin cancer risk phenotypes. Pigment Cell Res 16: 266–272 [DOI] [PubMed] [Google Scholar]
- Yasumoto K, Yokoyama K, Shibata K, Tomita Y, Shibahara S 1994. Microphthalmia-associated transcription factor as a regulator for melanocyte-specific transcription of the human tyrosinase gene. Mol Cell Biol 14: 8058–8070 [DOI] [PMC free article] [PubMed] [Google Scholar]