Abstract
Recent studies have shown that neuroglial progenitor/stem cells (NSCs) from different brain regions exhibit varying capacities for self-renewal and differentiation. In this study, we used neurofibromatosis-1 (NF1) as a model system to elucidate a novel molecular mechanism underlying brain region-specific NSC functional heterogeneity. We demonstrate that Nf1 loss leads to increased NSC proliferation and gliogenesis in the brainstem, but not in the cortex. Using Nf1 genetically engineered mice and derivative NSC neurosphere cultures, we show that this brain region-specific increase in NSC proliferation and gliogenesis results from selective Akt hyperactivation. The molecular basis for the increased brainstem-specific Akt activation in brainstem NSCs is the consequence of differential rictor expression, leading to region-specific mammalian target of rapamycin (mTOR)/rictor-mediated Akt phosphorylation and Akt-regulated p27 phosphorylation. Collectively, these findings establish mTOR/rictor-mediated Akt activation as a key driver of NSC proliferation and gliogenesis, and identify a unique mechanism for conferring brain region-specific responses to cancer-causing genetic changes.
Keywords: Neurofibromin, neural stem cell, regional heterogeneity, gliogenesis, Akt, mTOR
The capacity of neural stem/progenitor cells (NSCs) to self-renew, proliferate, and give rise to differentiated cell types is essential for normal brain development and maturation. In addition, NSCs have critical roles in numerous pathological states, including the response to nervous system injury, neurodegenerative disease, and brain tumor formation (Corbin et al. 2008; Waldau and Shetty 2008; Alcantara Llaguno et al. 2009; Yadirgi and Marino 2009). Recent studies have shown that NSCs from different regions of the CNS exhibit varying abilities to divide and generate lineage-restricted progeny (Klein et al. 2005; Armando et al. 2007). In these studies, NSCs from the forebrain proliferated faster than those from the hindbrain (Horiguchi et al. 2004; Kim et al. 2006) or spinal cord (Fu et al. 2005) and exhibited differing abilities to produce radial glia (Kulbatski and Tator 2009) and glial fibrillary acidic protein (GFAP)-positive cells (astrocytes) in vitro (Hitoshi et al. 2002). However, it is not known whether these intrinsic region-restricted NSC properties have relevance to human disease pathogenesis.
While brain tumors can arise in any location in the CNS in adults, glial cell tumors (astrocytomas or gliomas) are most frequently observed in the cerebellum, brainstem (BS), and optic pathway/hypothalamus in children (Louis et al. 2007). This anatomic predisposition is best illustrated by the neurofibromatosis-1 (NF1) inherited cancer syndrome, in which low-grade astrocytomas are located predominantly in the optic pathway/hypothalamus and BS of young children, with rare tumors developing in the cerebral cortex (Listernick et al. 1994; Pollack et al. 1996; Guillamo et al. 2003). In light of studies implicating cells with stem cell-like properties in the genesis of astrocytoma (for review, see Stiles and Rowitch 2008), it is conceivable that this regional distribution of tumors partly reflects the intrinsic heterogeneity of stem cells from different regions of the brain to expand in response to cancer-associated genetic changes. Based on the unique spatial distribution of gliomas in children with NF1, we chose to employ Nf1 genetically engineered mice and derivative NSCs to define the molecular basis for NSC heterogeneity in the brain.
NF1 is the most common cancer predisposition syndrome in which affected children develop gliomas. NF1-associated gliomas result from biallelic inactivation of the NF1 tumor suppressor gene, leading to total loss of NF1 protein (neurofibromin) function. Neurofibromin is a large cytoplasmic protein that negatively regulates RAS pathway signaling by inactivating the RAS proto-oncogene such that loss of NF1 gene expression is associated with increased RAS and RAS effector (Akt and MAPK) activity (Basu et al. 1992; DeClue et al. 1992). As predicted, examination of gliomas from individuals with NF1 confirmed loss of NF1 expression and increased RAS pathway activation (Gutmann et al. 2000; Lau et al. 2000). Similarly, Nf1-deficient primary mouse forebrain astrocytes exhibit high levels of RAS activation, resulting in increased Akt and mammalian target of rapamycin (mTOR) signaling (Dasgupta et al. 2005b; Sandsmark et al. 2007). Inhibition of Akt/mTOR activity in Nf1−/− astrocytes restores their proliferation to wild-type levels in vitro and blocks optic glioma growth in vivo (Hegedus et al. 2008).
Previous studies have shown that neurofibromin is an important regulator of NSC function. In this regard, Nf1 inactivation leads to an expansion in the number of proliferating spinal cord progenitors (Bennett et al. 2003), while Nf1−/− neural crest stem cells exhibit increased proliferation relative to their wild-type counterparts (Joseph et al. 2008). Similarly, Nf1−/− embryonic telencephalic NSCs exhibit increased proliferation in vitro (Dasgupta and Gutmann 2005), while Nf1 inactivation in brain lipid-binding protein (BLBP)-expressing neuroglial progenitors in vivo results in increased NSC proliferation and glial lineage differentiation (Hegedus et al. 2007). Moreover, the effect of neurofibromin loss on NSC glial differentiation was dependent on RAS and Akt signaling, and could be recapitulated by the expression of either activated K-RAS or Akt in BLBP+ cells in vivo.
In this study, we describe regional differences in brain NSC proliferation and glial differentiation following Nf1 gene inactivation in vitro and in vivo. Whereas Nf1−/− NSCs from the BS exhibit increased proliferation and glial cell differentiation in vitro and in vivo, we observed no effect of Nf1 gene inactivation on neocortex (CTX) NSC proliferation or gliogenesis. We further show that these cell-autonomous differences reflect the ability of BS NSCs to activate Akt in an mTOR/rictor-dependent fashion and culminate in Akt-mediated p27 phosphorylation. Collectively, these results demonstrate that intrinsic differences in NSC signaling cause brain region-specific abnormalities in glial differentiation, which may partially account for the propensity of glial cell tumors to arise in the BS of children with NF1.
Results
Nf1 inactivation results in increased BS NSC proliferation
Recently, we showed that Nf1 inactivation results in the increased proliferation of astrocytes from some brain regions (BS, optic nerve [ON], and cerebellum), whereas CTX astrocyte proliferation is unaffected by Nf1 loss (Yeh et al. 2009). Since gliomas have been hypothesized to originate from cells with stem cell-like properties, we generated neurosphere cultures from several regions of the postnatal brain. The ON and BS were chosen to represent regions where gliomas arise in children with NF1, while the CTX was included as a region where glioma formation is far less common. We used these different neurosphere populations to determine whether Nf1 loss had differential effects on NSC proliferation and gliogenesis as a function of brain region.
NSCs are defined as neurosphere cultures that express stem cell markers (e.g., BLBP and nestin) and retain the ability to undergo self-renewal and multi-lineage differentiation. For these experiments, we generated neurosphere cultures from postnatal day 1 (PN1) mouse CTX, BS, and ON. All of the resulting neurospheres from the CTX and BS were BLBP- and nestin-immunoreactive (Supplemental Fig. S1A). However, ON neurosphere cultures had limited capacity for proliferation and self-renewal (Supplemental Fig. S1B,C), and failed to generate Tuj-1+ neurons or O4+ oligodendrocytes (Supplemental Fig. S1D). In contrast, both CTX and BS neurospheres were capable of self-renewal, and generated neurons (Tuj-1+), astrocytes (GFAP+), and oligodendrocytes (O4+) (Supplemental Fig. S1E). Based on these results, ON neurospheres contained only glial progenitor cells and could not be classified as “true” NSCs. Therefore, we focused our analyses on NSCs from the BS and CTX as representative populations that derive from regions where gliomas commonly and rarely form, respectively, in children with NF1.
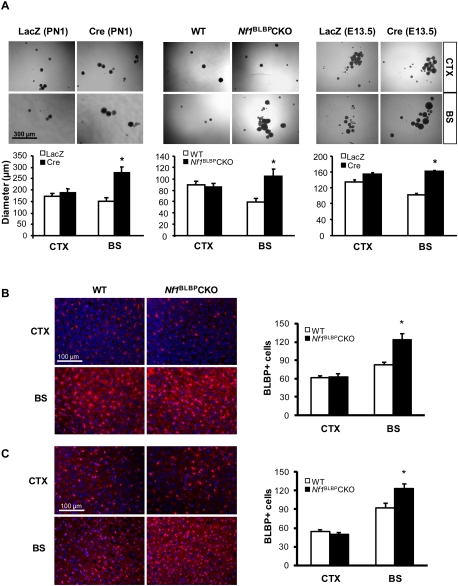
To determine whether Nf1 inactivation has different effects on CTX versus BS NSC proliferation, we first measured neurofibromin levels to exclude brain region-specific differences in overall Nf1 gene expression and found equivalent levels of neurofibromin expression (Supplemental Fig. S2A). Second, using Nf1−/− and wild-type NSC cultures from PN1 Nf1flox/flox mouse CTX and BS neurosphere cultures following Cre or LacZ gene adenovirus (Ad5) infection, respectively, Nf1−/− BS NSCs exhibited a 1.9-fold increased in proliferation compared with wild-type BS NSCs (Fig. 1A; Supplemental Fig. S2A). In contrast, there was no increase in CTX NSC proliferation after Nf1 inactivation. To demonstrate that these observations were not dependent on the developmental age or the method of Nf1 inactivation, we used Nf1−/− NSCs generated from PN1 Nf1flox/flox; BLBP-Cre (Nf1BLBPCKO) mice as well as embryonic day 13.5 (E13.5) Nf1−/− NSCs following Ad5-Cre infection. Compared with their matched wild-type counterparts, these Nf1−/− NSCs from the BS, but not the CTX, also exhibited increased proliferation (Fig. 1A). No change in self-renewal was observed in either BS or CTX NSCs following Nf1 inactivation (Supplemental Fig. S2B).
Figure 1.
Nf1 inactivation increases BS NSC proliferation. (A) Nf1 loss results in increased BS NSC neurosphere size, with no effect on CTX NSCs. Identical results were obtained with PN1 NSCs following acute Nf1 inactivation with Ad5-Cre (Cre), PN1 NSCs from Nf1BLBPCKO mice, or E13.5 NSCs after Nf1 inactivation. For controls, Nf1flox/flox NSCs were infected with Ad5-LacZ (LacZ). Phase contrast images show representative neurospheres, which are graphically illustrated below. Similarly, increased numbers of BLBP+ cells by immunofluorescence were observed in the BS of PN1 (B) and PN8 (C) mice, with no increase seen in the CTX. Sections were counterstained with DAPI. Graphs show the number of BLBP+ cells per surface area (0.1 mm2). Values denote the mean ± SEM. (*) P < 0.05. Bars: A, 300 μm; B,C, 100 μm.
Next, to assess the impact of Nf1 inactivation on NSCs in the subventricular zone (SVZ), an important germinal zone for neurogenesis in both adults and children, we isolated NSCs from the SVZ of PN1 mice. Whereas neurofibromin loss in BS NSCs resulted in a 1.8-fold increase in NSC proliferation, NSC proliferation was increased by 1.4-fold in SVZ NSCs in vitro (Supplemental Fig. S2C). Similarly, the number of proliferating (Ki67+) cells was increased by 2.8-fold in the BS of Nf1BLBPCKO mice compared with wild-type controls in vivo, while the number of Ki67+ cells in the SVZ was increased by only 1.3-fold (Supplemental Fig. S2D).
Finally, to demonstrate that this effect on NSC proliferation was also seen in vivo, the impact of Nf1 inactivation on the numbers of BLBP+ cells in PN1 and PN8 Nf1BLBPCKO mice was examined. Similar to the in vitro results, increased numbers of BLBP+ cells were observed in the BS of Nf1BLBPCKO mice at PN1 (1.5-fold) (Fig. 1B) and PN8 (1.3-fold) (Fig. 1C), whereas no significant change in BLBP+ cell number was observed in the CTX of Nf1BLBPCKO mice relative to wild-type mice. Cre-mediated Nf1 gene inactivation was confirmed by Cre immunohistochemistry and neurofibromin Western blotting (Supplemental Fig. S2E). Collectively, these data demonstrate that Nf1 inactivation increases NSC proliferation in a brain region-specific fashion.
Nf1 inactivation increases gliogenesis in the BS
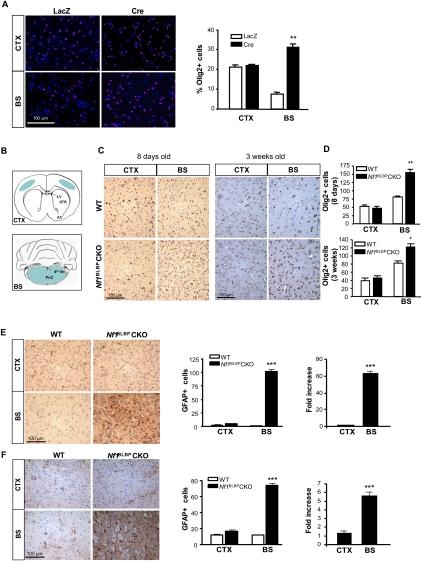
To determine whether Nf1 loss in NSCs results in increased glial lineage differentiation in a brain region-specific manner, we first quantified the number of Olig2+ glial progenitor cells in PN1 Nf1−/− and wild-type NSCs in vitro. The number of Olig2+ cells was increased by 2.4-fold to fourfold in dissociated Nf1−/− BS NSCs compared with wild-type BS NSCs (Fig. 2A; Supplemental Fig. S3A). Similar results were also observed using NSCs isolated from the BS and CTX of E16 pups (Supplemental Fig. S3B). We also noted a 1.2-fold increase in the number of Olig2+ cells following Nf1 loss in SVZ NSCs relative to wild-type SVZ NSCs (Supplemental Fig. S3A). In contrast, there was no significant change in Olig2+ cell number in NF1−/− CTX NSCs relative to wild-type CTX NSCs. Next, we examined the number of Olig2+ cells in Nf1BLBPCKO mice and control littermates in vivo. The brain regions used for direct cell counting are illustrated in Figure 2B. As observed in vitro, the number of Olig2+ cells was increased in the BS of PN8 (1.9-fold) and PN18 (1.5-fold) Nf1BLBPCKO mice compared with wild-type littermates in vivo (Fig. 2C,D).
Figure 2.
Nf1 inactivation increases the number of BS Olig2+ glial progenitors and GFAP+ astrocytes. (A) The number of Olig2+ cells (red) was increased in Nf1−/− BS NSCs (Cre) compared with wild-type NSCs (LacZ). No increases were observed in CTX NSCs. Cells were counterstained with DAPI (blue). (B) Schematic shows the brain regions used for direct cell counting (blue). (CC) Corpus callosum; (LV) lateral ventricle; (AC) anterior commissure; (Vn) ventricle; (PnC) pontine nucleus, caudal. (C) The number of Olig2+ glial progenitor cells was increased in the BS of Nf1BLBPCKO mice at both PN8 and PN18 compared with wild-type (WT) mice. There was no increase in the number of Olig2+ cells in the CTX of Nf1BLBPCKO mice. Graphs show the number of Olig2+ cells per surface area (0.1 mm2) (D). Increased numbers of GFAP+ cells were observed in the BS of Nf1BLBPCKO mice at both PN8 (E) and PN18 (F) compared with wild-type mice. There was no increase in the number of GFAP+ cells in the CTX of Nf1BLBPCKO mice. The number of GFAP+ cells per surface area (0.1 mm2; left) and fold change (Nf1BLBPCKO/WT; right) of GFAP+ cells are graphically represented. Values denote the mean ± SEM. (*) P < 0.005; (**) P < 0.001; (***) P < 0.0001. Bars, 100 μm.
To determine whether these Olig2+ cells represented glial progenitor cells or committed oligodendrocyte lineage cells, we performed double-immunofluorescence staining with Olig2 and established oligodendrocyte cell lineage markers, including CC1 (APC), PDGFR-α, and O4. First, we found that the number of APC (CC1)/Olig2 double-positive cells was not increased by Nf1 inactivation (Supplemental Fig. S3C). Second, the majority of the Olig2+ cells increased in the BS of Nf1BLBPCKO mice were not PDFR-α-immunoreactive (Supplemental Fig. S3D). Third, following Nf1-deficient NSC differentiation in vitro, the Olig2+ cells were not O4+ (Supplemental Fig. S3E). Fourth, while we were unable to perform NG2/Olig2 double-labeling experiments to determine whether the Olig2+ cells were also NG2+, we found that the total number of NG2+ cells was increased in both CTX and BS of Nf1BLBPCKO mice compared with wild-type controls (Supplemental Fig. S3F). Together, these data demonstrate that the Olig2+ cells increased following BS NSC Nf1 inactivation are glial progenitor cells, rather than committed oligodendrocyte progenitors.
To determine whether the increase in Olig2+ cells reflected an overall increase in gliogenesis, we quantified the number of GFAP+ cells (astrocytes) and found increased numbers of GFAP+ cells in the BS of Nf1BLBPCKO mice at PN8 (63.5-fold) (Fig. 2E) and PN18 (5.6-fold) (Fig. 2F) compared with wild-type mice. In contrast, there was no difference in the number of GFAP+ cells in the CTX or SVZ (Supplemental Fig. 3G) of Nf1BLBPCKO mice relative to wild-type controls. We confirmed these results using additional astrocytic markers (clusterin [ApoJ] [Fagan et al. 1999; Bachoo et al. 2004] and WT-1 [Schittenhelm et al. 2008]): Increased numbers of WT-1+ and clusterin+ cells were observed in the BS, but not in the CTX, of Nf1BLBPCKO mice (Supplemental Fig. S4A,B). Moreover, the numbers of NeuN+ (neurons) and APC+ (oligodendrocytes) cells were not changed in either the CTX or the BS of PN18 Nf1BLBPCKO mice relative to wild-type mice (Supplemental Fig. S4C,D). These data demonstrate that Nf1 inactivation increases gliogenesis specifically in the BS.
Ras activation increases NSC proliferation and gliogenesis in the BS
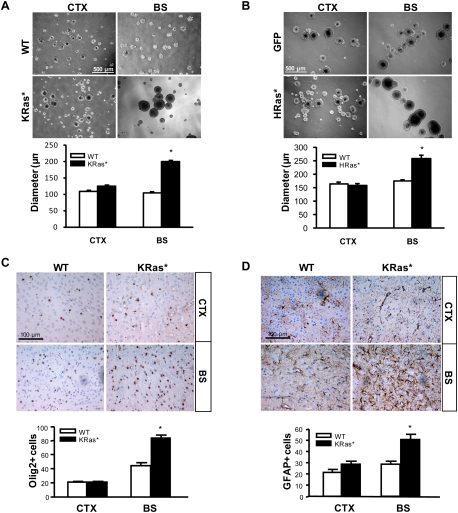
Because Ras activity is increased in Nf1−/− embryonic NSCs (Dasgupta and Gutmann 2005), we examined the activation state of the three different Ras isoforms (K-Ras, H-Ras, and N-Ras) expressed in E11.5 NSCs following Nf1 loss, and found increased activation of all three Ras isoforms in Nf1−/− NSCs relative to wild-type NSCs (Supplemental Fig. S5A). To recapitulate Ras activation by Nf1 loss in vitro, we infected PN1 NSCs from the CTX and BS with either constitutively active K-Ras (KRas*) or H-Ras (HRas*). The expression and activation of these transduced Ras molecules were confirmed by Western blotting and Ras activity assays (Supplemental Fig. S5B–D). Similar to Nf1−/− NSCs, only BS NSCs exhibited increased proliferation following either activated K-Ras (1.9-fold) (Fig. 3A) or H-Ras (1.5-fold) (Fig. 3B) transduction. No increased proliferation was observed in CTX NSCs following either K-Ras or H-Ras expression. To provide an in vivo correlate for these in vitro observations, we also examined the effect of K-Ras activation on gliogenesis in PN18 Lox-stop-lox (LSL)-K-RasG12D; BLBP-Cre (KRas*) mice. Similar to Nf1BLBPCKO mice (Fig. 2), KRas* mice exhibited increased numbers of Olig2+ glial progenitor cells (1.9-fold) (Fig. 3C) and GFAP+ astrocytes (1.8-fold) (Fig. 3D) in the BS, but not the CTX, compared with wild-type mice. These data show that Ras activation in NSCs recapitulates the brain region-specific effects of Nf1 loss on NSC proliferation and gliogenesis.
Figure 3.
Ras expression results in increased NSC proliferation and gliogenesis only in the BS. Neurosphere size (NSC proliferation) was increased in KRas*expressing (A) and HRas*expressing (B) BS NSCs. There was no increase in the diameters of KRas* or HRas* CTX NSCs. (C) The number of Olig2+ cells was increased in the BS of PN18 KRas* mice. There was no change in the number of Olig2+ cells in the CTX. (D) The number of GFAP+ astrocytes was increased in the BS of KRas* mice. Olig2+ and GFAP+ cell numbers per surface area (0.1 mm2) are graphically represented. Values denote the mean ± SEM. (*) P < 0.01. Bars: A,B, 500 μm; C,D, 100 μm.
The brain region-specific effects of Nf1 loss on NSC proliferation and gliogenesis are Akt-dependent
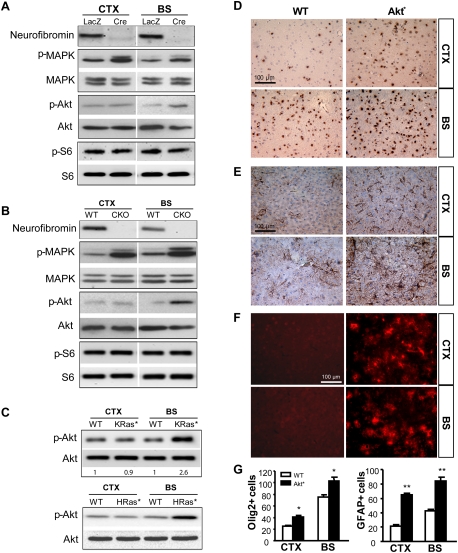
Because MAPK and Akt are the two major Ras downstream targets relevant to neurofibromin growth regulation, we measured their activation status in PN1 Nf1−/− and wild-type NSCs using phospho-specific antibodies. Whereas MAPK activation was increased in both CTX and BS Nf1-deficient NSCs relative to wild-type NSCs (Fig. 4A,B), Akt activation was increased only in Nf1−/− NSCs from the BS. In contrast, S6 activation (phospho-S6 [p-S6]) was not increased in either BS or CTX NSCs following Nf1 inactivation (Fig. 4A,B). Similar to Nf1 loss, increased Akt activation was observed in BS, but not CTX, NSCs expressing either constitutively active K-Ras (KRas*) or H-Ras (HRas*) compared with wild-type NSCs (Fig. 4C).
Figure 4.
The region-specific effects of Nf1 loss on NSC proliferation and gliogenesis are Akt-dependent. (A) MAPK activation was increased in both CTX and BS NSCs (PN1 Nf1flox/flox) after Nf1 inactivation (Cre). Infection with Ad5-LacZ (LacZ) was used as a wild-type (WT) control. The level of activated Akt (Ser 473) was increased only in BS Nf1−/− (Cre) NSCs relative to wild-type (LacZ) BS NSCs. No change in p-S6 was observed. (B) MAPK activation was increased in both CTX and BS NSC cultures from Nf1BLBPCKO (CKO) mice. Increased Akt activation was observed only in BS NSCs. No change in p-S6 was observed. (C) Akt activation was similarly increased only in BS NSCs expressing KRas* (2.6-fold increase, P = 0.0034) and HRas*. The number of Olig2+ cells (D) as well as GFAP+ cells (E) was increased in both the CTX and the BS of PN18 Akt*mice. (F) GFP immunohistochemistry shows that the Akt transgene is highly expressed in the Akt* mouse brain. (G) The numbers of Olig2+ and GFAP+ cells per surface area (0.1 mm2) are quantitated. Values represent mean ± SEM. (*) P < 0.05; (**) P < 0.01. Bars, 100 μm.
To determine whether induced expression of an activated Akt in brain NSCs would result in increased gliogenesis in both the BS and the CTX, we generated mice with activated Akt (myr-Akt) expression in BLBP+ cells (BLBP-Cre × LSL-myr-Akt mice [Akt* mice]) (Hegedus et al. 2007). In contrast to Nf1BLBPCKO and KRas* mice, Akt* mice exhibited increased numbers of Olig2+ cells in both the CTX (1.7-fold) and the BS (1.4-fold) compared with wild-type mice (Fig. 4D,G). Similarly, the number of GFAP+ cells was also increased in both the CTX (threefold) and the BS (twofold) (Fig. 4E,G). Transgene expression was confirmed by GFP immunohistochemistry (Fig. 4F). Together, these data demonstrate that Akt activation is the major determinant underlying the region-specific effects of Nf1 inactivation on brain NSC proliferation and gliogenesis.
Differential regulation of Akt by mTOR underlies the brain region-specific effects of Nf1 loss on NSC proliferation and gliogenesis
To determine the mechanism of differential Akt regulation in BS versus CTX NSCs, we first measured the levels of numerous upstream regulators of Ras and Akt by Western blotting (Fig. 5A) and quantitative RT–PCR (qRT–PCR) (Supplemental Fig. S6A). We found no significant differences in p120 Ras-GAP, PTEN, or phospho-PDK1 expression in NSCs from the CTX versus the BS (Fig. 5A), as well as no differences in Ppp2r2b (protein phosphatase 2), Prkce (PKCɛ), Prkcq (PKCθ), Tenc1 (C1-TEN), Rasal2 (Ras protein activator-like 2), or Rasgrp1 (Ras-GRP) mRNA expression in NSCs from the CTX versus the BS (Supplemental Fig. S6A).
Figure 5.
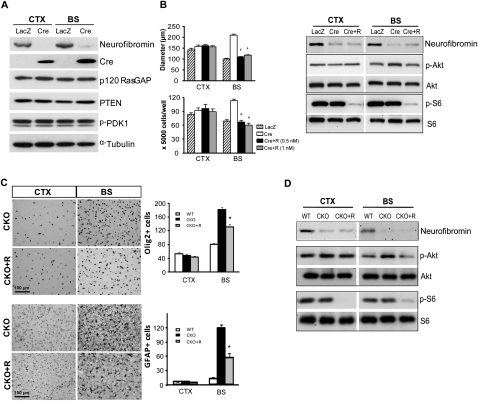
Region-specific effects of rapamycin on Nf1−/− NSC proliferation and gliogenesis. (A) The levels of p120 Ras-GAP, PTEN, and p-PDK1 are equivalently expressed in Nf1−/− NSCs (Cre) compared with wild-type (LacZ). Western blotting of neurofibromin and Cre recombinase shows loss of neurofibromin expression in Ad5-Cre-infected Nf1flox/flox NSCs. α-Tubulin was used as an internal loading control. (B) Decreased Nf1−/− BS NSC proliferation was observed following treatment with rapamycin. There was no effect of rapamycin treatment (Cre + R) on CTX NSCs. Akt activation (Ser 473; p-Akt) was reduced to control levels in rapamycin-treated (1 nM) Nf1−/− BS NSCs. In both BS and CTX NSCs, S6 activation (p-S6) was inhibited by rapamycin treatment. (C) Decreased numbers of Olig2+ glial progenitors and GFAP+ astrocytes were observed in the BS of PN8 Nf1BLBPCKO mice treated with rapamycin (CKO + R) in vivo. (D) Akt activation was decreased following rapamycin treatment in the BS of Nf1BLBPCKO mice in vivo. p-S6 was inhibited by rapamycin treatment in both the BS and CTX in vivo. Values represent mean ± SEM. (*) P < 0.0001. Bar, 100 μm.
Next, we explored the possibility that mTOR complexes might differentially regulate Akt activity through raptor or rictor function (Guertin and Sabatini 2007). Using rapamycin to inhibit mTOR function in vitro, Nf1−/− BS NSC proliferation was reduced by twofold in response to rapamycin treatment (0.5–1 nM), whereas CTX NSC proliferation was not affected (Fig. 5B). In these experiments, rapamycin treatment (1 nM) reduced Akt hyperphosphorylation exclusively in BS NSCs, whereas S6 activation was inhibited in Nf1−/− NSCs from both the BS and CTX (Fig. 5B). To provide an in vivo correlate for these in vitro findings, pregnant females and their resulting pups were treated with rapamycin, and the numbers of Olig2+ and GFAP+ cells were quantified at PN8. Consistent with the hypothesis that mTOR regulates Akt-mediated gliogenesis, we found that rapamycin treatment reduced the number of Olig2+ (1.4-fold) and GFAP+ (2.1-fold) cells in the BS of Nf1BLBPCKO mice (Fig. 5C). Similar to Nf1−/− NSCs, we found that Akt hyperactivation in the BS of Nf1BLBPCKO mice was reduced following rapamycin treatment in vivo, while S6 activation was inhibited in both the BS and CTX in vivo (Fig. 5D).
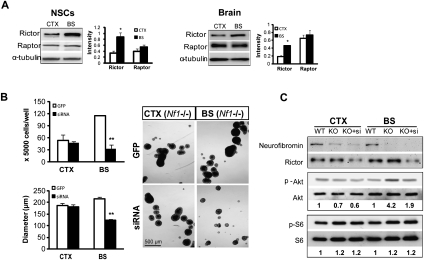
Based on reports demonstrating that rictor-containing mTOR complexes positively regulate Akt through Ser 473 phosphorylation (Hresko and Mueckler 2005), we measured raptor and rictor expression in vitro and in vivo. Surprisingly, rictor expression was higher in BS compared wtih CTX in both NSC cultures (threefold) and brain tissue (2.6-fold), whereas no differences in raptor expression were observed (Fig. 6A). To determine whether increased rictor expression in BS NSCs reflected differential rictor/raptor transcription, we examined the mRNA expression levels of rictor and raptor in CTX and BS NSCs using qRT–PCR. No differences were observed in the mRNA expression of rictor (P = 0.3962) or raptor (P = 0.7442) between CTX and BS NSCs.
Figure 6.
Differences in mTOR regulation of Akt underlie the brain region-specific differences in Nf1−/− NSC proliferation. (A) Rictor expression was higher in the BS compared with the CTX in vitro (NSCs; threefold) and in vivo (brain; 2.6-fold). There was no difference in raptor expression. Results were quantitated by scanning densitometry using α-tubulin as an internal loading control. (B) Rictor silencing by lentiviral rictor siRNA (siRNA) decreased neurosphere size in Nf1−/− BS NSC cultures. No change was observed in Nf1−/− CTX NSCs following rictor silencing. Lentivirus-GFP (GFP) served as a control for viral infection. (C) The fold change in p-Akt and p-S6 (after normalization to total Akt and S6, respectively) relative to wild-type (WT) controls is indicated below the blots. Error bars denote the mean ± SEM. (*) P < 0.05; (**) P < 0.01. Bar, 500 μm.
To determine whether mTOR/rictor function mediates the increased proliferation and gliogenesis observed in Nf1−/− NSCs, we performed shRNAi knockdown (KD) experiments. Rictor KD decreased the proliferation (as assessed by direct counting and neurosphere diameter measurements) and Akt activation of NF1−/− BS NSCs to wild-type levels, with no effect on Nf1−/− CTX NSCs (Fig. 6B,C). Consistent with its role in TORC2 signaling, rictor KD had no effect on S6 phosphorylation (Fig. 6C). In contrast, raptor KD had no effect on Nf1−/− BS NSC proliferation (neurosphere diameter and cell number) or gliogenesis (Olig2+ cell number) and did not change Akt activation (data not shown). Together, these results demonstrate that the ability of BS NSCs to proliferate following Nf1 loss results from mTOR/rictor-mediated Akt activation.
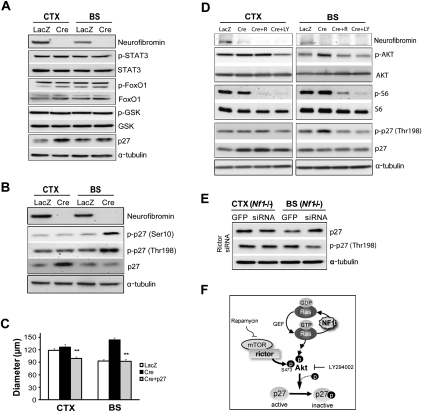
Since we observed no differential changes in the activation status of mTOR downstream effectors (PKCα, S6 kinase, S6, and 4EBP1) after Nf1 inactivation in the BS versus the CTX (Supplemental Fig. S6B), we reasoned that downstream targets of Akt, other than mTOR, must be responsible for the increased proliferation of BS NSCs following Nf1 loss. We first measured the activation status of STAT3, FOXO1, and GSK3β in PN1 Nf1−/− and wild-type NSCs from CTX and BS using phospho-specific antibodies, but found no differential activation of these signaling molecules. However, p27 expression was increased in Nf1−/− CTX NSCs relative to wild-type CTX NSCs, whereas no such increase was observed in Nf1−/− BS NSCs (Fig. 7A).
Figure 7.
Differential phosphorylation of p27 results from mTOR/rictor/Akt-mediated activation in Nf1−/− BS NSCs. (A) p27 expression was increased exclusively in Nf1−/− BS NSCs compared with wild type, whereas no regional difference in the level of phospho-STAT3, FoxO1, or GSK was observed following Nf1 loss. Ad5-LacZ (LacZ) served as a control for viral infection. (B) Nf1 loss resulted in increased p27 phosphorylation (p-p27; Ser 10 and Thr 198) in Nf1−/− BS NSCs. (C) Ectopic p27 expression in Nf1−/− BS NSCs restored proliferation to wild-type levels. (D) p27 phosphorylation (p-p27; Thr 198) in Nf1−/− BS NSCs was reduced by rapamycin (1 nM) and LY294002 (20 μM) treatment. (E) p27 expression in Nf1−/− BS NSCs was increased following rictor siRNA silencing. In contrast, p27 phosphorylation (Thr 198) in Nf1−/− BS NSCs was reduced following rictor siRNA treatment. No change in p27 expression or phosphorylation was observed following rictor KD in Nf1−/− CTX NSCs. α-Tubulin was included as an internal loading control for Western blotting. (F) Rictor-mediated Akt activation regulates p27 phosphorylation and function in Nf1−/− BS NSCs. Error bars denote the mean ± SEM. (**) P < 0.01.
Previous studies have shown that p27 expression can be regulated at both the transcriptional and post-translational levels (Lin et al. 2009; Prasad et al. 2009). Whereas there were no differences in p27 mRNA expression between Nf1-deficient BS and CTX NSCs by qRT–PCR in vitro or in vivo (P = 0.46), we found that p27 phosphorylation on both Ser 10 and Thr 198 residues was increased following Nf1 loss exclusively in BS NSCs (Fig. 7B). Since increased p27 phosphorylation on Thr 198 and Ser 10 leads to its nuclear export and subsequent degradation (Fujita et al. 2002; Ishida et al. 2002), we ectopically expressed p27 in Nf1−/− BS NSCs by viral transduction to mimic the increased p27 levels seen in Nf1−/− CTX NSCs, and found that p27 expression reduced Nf1−/− BS NSC proliferation to wild-type levels (Fig. 7C).
To determine whether p27 phosphorylation was regulated by mTOR and Akt in Nf1−/− BS NSCs, we examined Thr 198 p27 phosphorylation following mTOR (rapamycin) and PI3-kinase (LY294002) inhibition. We found that p27 phosphorylation was inhibited by rapamycin and LY294002 in Nf1−/− BS NSCs (Fig. 7D). Similarly, p27 phosphorylation at Ser 10 was also inhibited by rapamycin and LY294002 treatment in Nf1−/− BS NSCs (data not shown). Finally, using the identical lysates from the experiment shown in Figure 6, B and C, we found that, following rictor KD (but not raptor KD) (data not shown), p27 expression was increased and p27 phosphorylation (Thr 198) was decreased in Nf1−/− BS NSCs (Fig. 7E). These results demonstrate that mTOR/rictor/Akt regulates p27 function through phosphorylation in Nf1−/− NSCs (Fig. 7F), and collectively establish mTOR/rictor-mediated positive regulation of Akt activity as the major determinant responsible for mediating the brain region-specific effects of neurofibromin loss on NSC proliferation and gliogenesis.
Discussion
Neural stem cell functional diversity in the CNS is dictated by the developmental age of the animal as well as by spatially restricted signals present in specific brain regions. In addition, NSCs from different regions of the brain may be intrinsically distinct by virtue of cell-autonomous properties innate to these unique NSC populations. These brain region-specific cell-autonomous differences are well illustrated by the NF1 tumor predisposition syndrome, in which gliomas most commonly arise in the ON and BS, but rarely occur in the cortex. In this study, we demonstrate that NSCs from the BS proliferate faster and differentiate into glial lineage cells following Nf1 gene inactivation, whereas Nf1 loss in CTX NSCs does not result in increased proliferation or gliogenesis. These differences do not reflect the mouse developmental age, as similar region-specific effects of Nf1 loss were observed in both embryonic and early postnatal NSCs. Moreover, these differences were also found in young mice in vivo following Nf1 inactivation in BLBP+ NSCs at E9.5. It is also unlikely that these differences result from unique microenvironmental influences present in the BS or CTX, since these brain region-specific differences were observed in isolated NSC populations in vitro.
Recently, we showed that astrocytes from the BS exhibit increased proliferation following Nf1 inactivation, whereas no such increase in proliferation was observed in Nf1-deficient neocortical astrocytes (Yeh et al. 2009). To account for these brain region-specific responses to neurofibromin loss, we found that astrocytes from the CTX expressed significantly less neurofibromin relative to astrocytes from the BS and ON. Furthermore, Nf1 loss in GFAP+ cells had no effect on neocortical astrocyte proliferation in vitro or astrocyte numbers in vivo. In contrast to astrocytes, we found no differences in Nf1 gene or neurofibromin protein expression between NSCs from the CTX and the BS. This finding implicates other mechanisms in dictating the response of NSCs from different brain regions to Nf1 inactivation.
To identify the mechanism underlying the brain region-specific effect of Nf1 loss on NSC function, we sought to determine whether constitutive RAS activation in NSCs in vitro and in vivo could mimic the effects of neurofibromin loss. The finding that RAS activation in NSCs similarly increases NSC proliferation and gliogenesis in an identical brain region-specific manner supports the notion that RAS hyperactivation is the primary driver of NSC proliferation and glial differentiation following Nf1 gene inactivation. In contrast to Nf1−/− astrocytes in which only K-RAS hyperactivation was observed (Dasgupta et al. 2005a), neurofibromin loss in NSCs results in increased activity of all three RAS isoforms (H-RAS, K-RAS, and N-RAS). While we found that both activated K-RAS and H-RAS expression in NSCs led to increased BS (but not CTX) NSC proliferation, it is not clear whether the three RAS isoforms perform overlapping redundant functions with respect to NSC function. Current studies are focused on exploring the consequences of N-RAS activation on NSC function in the BS and CTX in vitro and in vivo.
Next, we determined whether differential Akt activation was responsible for the observed differences in NSC behavior. Akt is a potent regulator of NSC proliferation in many brain regions (Otaegi et al. 2006; Peltier et al. 2007), such that hyperactivation of PI3-kinase and Akt increases the proliferation of Pten-deficient NSCs (Groszer et al. 2001). We showed previously that Akt activation was increased in Nf1-deficient embryonic NSCs (Dasgupta and Gutmann 2005), and that activated Akt expression in BLBP+ cells resulted in increased gliogenesis in vivo (Hegedus et al. 2007). In the present study, we found differential Akt activation in BS NSCs relative to NSCs from the CTX. This brain region-specific Akt activation was also observed following K-RASG12D and H-RASG12V expression in NSCs in vitro or K-RASG12D expression in BLBP+ cells in vivo (data not shown), suggesting that deregulated neurofibromin/RAS signaling in NSCs is not responsible for the differences in brain region-specific NSC Akt activation. In contrast, expression of activated Akt in BLBP+ cells in vivo was sufficient to increase BLBP+ NSC numbers and gliogenesis, arguing that Akt activation alone accounts for the brain region-restricted differences in Nf1−/− NSC behavior. The observation that Akt is a central driver of gliogenesis is consistent with prior reports demonstrating that inhibition of PI3-kinase/Akt activation with LY294002 decreased the number of GFAP+ cells in the olfactory bulb (Otaegi et al. 2006) as well as changed the subcellular localization of Olig2, which is critical for the induction of astrocyte differentiation (Setoguchi and Kondo 2004).
In this study, we determined that BS-specific NSC Akt activation results from differences in mTOR regulation. mTOR forms complexes with raptor and rictor to negatively and positively regulate Akt activity, respectively (Guertin and Sabatini 2007). The mTOR complex containing rictor (TORC2) positively regulates Akt activity through phosphorylation on Ser 473 (Hresko and Mueckler 2005; Sarbassov et al. 2005), which can be inhibited by rapamycin treatment (Sarbassov et al. 2006). Our finding that raptor and rictor are expressed at different levels in BS versus CTX NSCs is further underscored by an examination of publically available Unigene EST expression data sets: In both mouse and human tissues, rictor levels were higher than raptor levels in several tissues, whereas raptor expression predominated in other tissues (DY Lee and DH Gutmann, unpubl.; http://www.ncbi.nlm.nih.gov/UniGene/ESTProfileViewer). The consequence of this differential rictor expression is preferential Akt activation in BS NSCs, such that pharmacologic (rapamycin inhibition) or genetic silencing with rictor KD restores Nf1−/− BS NSC proliferation and gliogenesis to wild-type levels.
Previously, we found increased mTOR activation in Nf1−/− astrocytes and Nf1-deficient mouse optic gliomas (Dasgupta et al. 2005b; Sandsmark et al. 2007). In astrocytes, mTOR regulates cell growth by activating Rac1, rather than by activating Akt, as seen in NSCs. This observation argues that different cell types in the CNS employ mTOR pathway signaling in unique fashions to regulate cell growth. These critical distinctions must be considered when designing therapies to target brain tumors. In this regard, rictor is overexpressed in gliomas, resulting in Akt hyperactivation (Masri et al. 2007), and PI3-kinase/Akt, but not mTOR, signaling regulates stem cell-like phenotypes in gliomas (Bleau et al. 2009).
Based on the observation that mTOR acts solely as a regulator of Akt function in NSCs, we examined Akt downstream targets that are regulated by Akt. No changes in GSK3β, FOXO1, S6, S6K, 4EBP1, PKCα, and STAT3 phosphorylation were found in Nf1−/− BS NSCs relative to those from the CTX. In contrast, we found increased expression of the p27 cyclin-dependent kinase inhibitor following Nf1 inactivation in CTX NSCs, but not in BS NSCs. p27 function is regulated by Akt-mediated phosphorylation at residues Thr 198 and Ser 10, resulting in cytoplasmic localization and inhibition of its activity (Fujita et al. 2002; Motti et al. 2004; Crean et al. 2006). Akt also regulates Skp2 (S-phase kinase-associated protein-2), a SCF F-box protein, which binds to p27 and targets it for ubiquitination and degradation (Lin et al. 2009). Consistent with a model in which mTOR/rictor-mediated Akt activation in Nf1−/− BS NSCs results in down-regulation of p27 expression, we identified an Akt-dependent increase in p27 phosphorylation (Ser 10 and Thr 198) and a reduction in nuclear (active) p27 in Nf1−/− BS, but not CTX, NSCs (DY Lee and DH Gutmann, unpubl.). Second, p27 re-expression in Nf1−/− BS NSCs restored proliferation to levels observed in wild-type BS NSCs. Third, p27 induction (as seen in Nf1−/− CTX NSCs) was observed only in Nf1−/− BS NSCs following rictor shRNAi silencing. These findings establish mTOR/rictor/Akt regulation of p27 function as a key determinant of Nf1-deficient brain region-specific NSC proliferation and differentiation.
The relevance of p27 to gliomagenesis is further underscored by several observations: First, p27 loss in O2A glial progenitors results in a 2.6-fold increase in the number of GFAP+ astrocytes generated following in vitro differentiation and a 2.8-fold increase in p27-null mice in vivo (Casaccia-Bonnefil et al. 1997). Second, p27 loss in mice increases the number and proliferation of transient amplifying (type C) neuroglial progenitors in the SVZ in vivo (Doetsch et al. 2002). Third, mutant epidermal growth factor receptor (EGFR) signaling, as observed in some high-grade gliomas, has been shown to regulate glioma cell growth by suppressing p27 expression through activation of the PI3-kinase/Akt pathway (Narita et al. 2002). While these findings provide compelling support for p27 as a critical downstream Akt effector that controls NSC proliferation and perhaps gliomagenesis, it is also possible that other molecules are involved in specifying glial differentiation. In this regard, Akt is known to regulate gliogenesis by modulating the phosphorylation/expression of specific transcription factors important for glial differentiation, including N-CoR and Olig2 (Hermanson et al. 2002; Setoguchi and Kondo 2004). Future studies will be required to address the role of these Akt-regulated transcription factors in gliogenesis and glioma formation.
Our finding that Nf1 loss results in increased numbers of Olig2+ cells is important in light of several recent reports implicating Olig2 in gliomagenesis. First, Olig2 is expressed universally in diffuse gliomas (Ligon et al. 2004), including low-grade pilocytic astrocytomas, such as those arising in children with NF1 (Takei et al. 2008). Second, the vast majority of the proliferating cells in diffuse gliomas are Olig2+ cells (Rhee et al. 2009). Third, Olig2 function is required for neuroglial progenitor proliferation as well as astrocytoma formation in genetically engineered mouse glioma models in vivo (Ligon et al. 2007). Collectively, these observations, coupled with our findings, suggest a critical role for this population of neuroglial progenitors in glioma development.
Over the last several years, it has become widely recognized that neural stem cells residing in the SVZ of adults may be the cell of origin for gliomas (Jackson et al. 2006; Alcantara Llaguno et al. 2009). In these studies, targeting NSCs in the SVZ niche with glioma-associated oncogenic mutation or tumor suppressor gene inactivation is sufficient for gliomagenesis. It was therefore unanticipated that Nf1 loss in SVZ NSCs in vitro and in vivo resulted in more modest increases in proliferation and Olig2+ progenitor cell generation. While further experimentation will be required, it is possible that different stem cell niches are uniquely affected by both the specific cancer-associated genetic change (Nf1 loss vs. combined Nf1, p53, and Pten inactivation) and the developmental age of the animal (young vs. adult mice). These determinants may be particularly relevant to the distinct spatial and temporal pattern of gliomagenesis in patients with NF1 (optic pathway and BS gliomas arising in children <10 yr of age) (Listernick et al. 2007).
In summary, we provide a mechanistic explanation for the differential response of spatially distinct populations of brain NSCs to neurofibromin loss. These findings are particularly germane to glioma formation in children with NF1, whose brain tumors usually develop in the optic pathway and BS, but rarely in the cortex. We showed recently that stromal signals expressed by microglia (meningioma-expressed antigen-5 [MGEA5]) or multiple other cellular elements (CXCL12) in a spatially restricted fashion along the optic pathway uniquely promote the proliferation and survival of Nf1-deficient, but not wild-type, astrocytes (Daginakatte and Gutmann 2007; Warrington et al. 2007). In this regard, we propose that tumor formation requires a permissive microenvironment composed of cell types and molecular signals that facilitate cell growth in receptive preneoplastic cell populations. Our demonstration of a receptive preneoplastic cell type defined by differential mTOR-mediated Akt activation broadens our understanding of the mechanisms underlying NSC heterogeneity, and offers new insights relevant to the design of future therapeutics that target specific populations of cancer cells.
Materials and methods
Mice
BLBP-Cre transgenic mice were intercrossed with Nf1flox/flox mice to generate BLBP-Cre; Nf1flox/flox (Nf1BLBPCKO) mice. To generate transgenic mice that express KRas* or Akt*, BLBP-Cre transgenic mice were intercrossed with LSL-K-RasG12D mice and LSL-myr-Akt mice, respectively. All strains were maintained on a C57BL/6 background and used in accordance with approved animal studies protocols at Washington University.
Culture of NSCs and generation of Nf1−/− neurospheres
CTX and BS regions were dissected from PN1 mouse pups as well as E13.5 mouse embryos of each genotype, and NSCs were generated as reported previously (Dasgupta and Gutmann 2005). Briefly, CTX tissues were derived from the somatosensory cortex, excluding the white matter tracts and the corpus callosum, while BS tissues included the midbrain, pons, and medulla after careful removal of the cerebellar peduncles (Fig. 2B). NSCs from Nf1flox/flox mouse brains were treated with Ad5-Cre or Ad5-LacZ diluted in NSC medium. After loss of neurofibromin, expression was verified by Western blotting.
Measurement of NSC proliferation
At least 10 neurospheres from each genotype were trypsinized and plated into individual wells of ultralow-binding 24-well plates with complete NSC medium containing EGF and FGF. After 10–12 d, the size (diameter) of the resulting secondary neurospheres was determined using Metamorph 7.1 (Molecular Devices).
Generation of NSCs expressing constitutively active K-Ras and H-Ras
K-Ras-expressing NSCs were generated by infecting PN1 LSL-K-RasG12D neurospheres with Ad5-Cre. As a control, NSCs isolated from wild-type mouse brains were infected with Ad5-Cre. H-Ras-expressing NSCs were generated following infection with MSCV containing H-RasG12V (MSCV-H-RasG12V-GFP). MSCV-GFP was used as a control for these experiments.
Immunohistochemistry and immunocytochemistry
Brain tissues were obtained following intracardiac perfusion with PBS and 4% PFA solution, and paraffin sections prepared as described previously (Hegedus et al. 2007). Sections were treated with antigen retrieval solution and stained with appropriate antibodies (Supplemental Table S1). Immunocytochemistry was performed on trypsinized NSCs plated onto 50 μg/mL poly-D-lysine-coated and 10 μg/mL fibronectin-coated 24-well plates in defined NSC medium containing N2 and B27 without growth factors. After 5 d, cells were fixed and stained with appropriate primary antibodies (Supplemental Table S1) overnight at 4°C. For fluorescence detection, suitable Alexa Fluor-tagged secondary antibodies (Molecular Probes) were used, and cells were counterstained with DAPI.
Western blotting
Western blots were performed as described previously (Dasgupta and Gutmann 2005; Hegedus et al. 2007) using the primary antibodies listed in Supplemental Table S2. Appropriate HRP-conjugated secondary antibodies (Cell Signaling) were used for detection by enhanced chemiluminescence (New England Biolabs). Each experiment was performed with samples from at least three independent groups.
Pharmacologic inhibition studies
Neurospheres were dissociated into single cells by trypsinization and plated onto ultralow-binding 24-well plates at a density of 1 × 104 cells per well. Cells were treated with rapamycin (0.5–1 nM) and LY294002 (20 μM) for 5–6 d in vitro.
For the in vivo experiments, rapamycin (1 mg/kg per day) was injected intraperitoneally into pregnant females from E16 to E18. Postnatally, 0.5 mg/kg rapamycin was injected subcutaneously at PN1 and PN3, and the pups were perfused at PN8.
Retrovirus infection
Rictor siRNA (NM030168.2; TRCN0000123397) or GFP-containing pLKO.1 plasmid was cotransfected with VSV-G and Δ8.9 helper DNA into HEK293T cells using the FuGENEHD transfection reagent (Roche). p27-MSCV was cotransfected with T-helper DNA into HEK293T cells. Nf1−/− neurospheres (Ad5-Cre-infected Nf1flox/flox cells) were dissociated into single cells by trypsin and transduced with viral supernatants from HEK293T cells. Transfection efficiency was evaluated by Western blotting. Each experiment was performed at least two times with identical results.
Statistical analysis
Each experiment was performed with samples from at least three independent groups. Statistical significance (P < 0.05) was determined using the Student's t-test and GraphPad Prism 4.0 software (GraphPad, Inc.).
Acknowledgments
We thank Scott Gianino for excellent technical assistance. These studies were supported by a grant from the National Institutes of Health (R21 NS058433 and R01 NS065547 to D.H.G.). D.Y.L. was partly supported by a Children's Tumor Foundation Young Investigators Award.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1957110.
Supplemental material is available at http://www.genesdev.org.
References
- Alcantara Llaguno S, Chen J, Kwon CH, Jackson EL, Li Y, Burns DK, Alvarez-Buylla A, Parada LF 2009. Malignant astrocytomas originate from neural stem/progenitor cells in a somatic tumor suppressor mouse model. Cancer Cell 15: 45–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armando S, Lebrun A, Hugnot JP, Ripoll C, Saunier M, Simonneau L 2007. Neurosphere-derived neural cells show region-specific behaviour in vitro. Neuroreport 18: 1539–1542 [DOI] [PubMed] [Google Scholar]
- Bachoo RM, Kim RS, Ligon KL, Maher EA, Brennan C, Billings N, Chan S, Li C, Rowitch DH, Wong WH, et al. 2004. Molecular diversity of astrocytes with implications for neurological disorders. Proc Natl Acad Sci 101: 8384–8389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu TN, Gutmann DH, Fletcher JA, Glover TW, Collins FS, Downward J 1992. Aberrant regulation of ras proteins in malignant tumour cells from type 1 neurofibromatosis patients. Nature 356: 713–715 [DOI] [PubMed] [Google Scholar]
- Bennett MR, Rizvi TA, Karyala S, McKinnon RD, Ratner N 2003. Aberrant growth and differentiation of oligodendrocyte progenitors in neurofibromatosis type 1 mutants. J Neurosci 23: 7207–7217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleau AM, Hambardzumyan D, Ozawa T, Fomchenko EI, Huse JT, Brennan CW, Holland EC 2009. PTEN/PI3K/Akt pathway regulates the side population phenotype and ABCG2 activity in glioma tumor stem-like cells. Cell Stem Cell 4: 226–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casaccia-Bonnefil P, Tikoo R, Kiyokawa H, Friedrich V Jr, Chao MV, Koff A 1997. Oligodendrocyte precursor differentiation is perturbed in the absence of the cyclin-dependent kinase inhibitor p27Kip1. Genes Dev 11: 2335–2346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbin JG, Gaiano N, Juliano SL, Poluch S, Stancik E, Haydar TF 2008. Regulation of neural progenitor cell development in the nervous system. J Neurochem 106: 2272–2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crean JK, Furlong F, Mitchell D, McArdle E, Godson C, Martin F 2006. Connective tissue growth factor/CCN2 stimulates actin disassembly through Akt/protein kinase B-mediated phosphorylation and cytoplasmic translocation of p27Kip-1. FASEB J 20: 1712–1714 [DOI] [PubMed] [Google Scholar]
- Daginakatte GC, Gutmann DH 2007. Neurofibromatosis-1 (Nf1) heterozygous brain microglia elaborate paracrine factors that promote Nf1-deficient astrocyte and glioma growth. Hum Mol Genet 16: 1098–1112 [DOI] [PubMed] [Google Scholar]
- Dasgupta B, Gutmann DH 2005. Neurofibromin regulates neural stem cell proliferation, survival, and astroglial differentiation in vitro and in vivo. J Neurosci 25: 5584–5594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta B, Li W, Perry A, Gutmann DH 2005a. Glioma formation in neurofibromatosis 1 reflects preferential activation of K-RAS in astrocytes. Cancer Res 65: 236–245 [PubMed] [Google Scholar]
- Dasgupta B, Yi Y, Chen DY, Weber JD, Gutmann DH 2005b. Proteomic analysis reveals hyperactivation of the mammalian target of rapamycin pathway in neurofibromatosis 1-associated human and mouse brain tumors. Cancer Res 65: 2755–2760 [DOI] [PubMed] [Google Scholar]
- DeClue JE, Papageorge AG, Fletcher JA, Diehl SR, Ratner N, Vass WC, Lowy DR 1992. Abnormal regulation of mammalian p21ras contributes to malignant tumor growth in von Recklinghausen (type 1) neurofibromatosis. Cell 69: 265–273 [DOI] [PubMed] [Google Scholar]
- Doetsch F, Verdugo JM, Caille I, Alvarez-Buylla A, Chao MV, Casaccia-Bonnefil P 2002. Lack of the cell-cycle inhibitor p27Kip1 results in selective increase of transit-amplifying cells for adult neurogenesis. J Neurosci 22: 2255–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagan AM, Holtzman DM, Munson G, Mathur T, Schneider D, Chang LK, Getz GS, Reardon CA, Lukens J, Shah JA, et al. 1999. Unique lipoproteins secreted by primary astrocytes from wild type, apoE−/−, and human apoE transgenic mice. J Biol Chem 274: 30001–30007 [DOI] [PubMed] [Google Scholar]
- Fu SL, Ma ZW, Yin L, Iannotti C, Lu PH, Xu XM 2005. Region-specific growth properties and trophic requirements of brain- and spinal cord-derived rat embryonic neural precursor cells. Neuroscience 135: 851–862 [DOI] [PubMed] [Google Scholar]
- Fujita N, Sato S, Katayama K, Tsuruo TJ 2002. Akt-dependent phosphorylation of p27Kip1 promotes binding to 14–3–3 and cytoplasmic localization. Biol Chem 277: 28706–28713 [DOI] [PubMed] [Google Scholar]
- Groszer M, Erickson R, Scripture-Adams DD, Lesche R, Trumpp A, Zack JA, Kornblum HI, Liu X, Wu H 2001. Negative regulation of neural stem/progenitor cell proliferation by the Pten tumor suppressor gene in vivo. Science 294: 2186–2189 [DOI] [PubMed] [Google Scholar]
- Guertin DA, Sabatini DM 2007. Defining the role of mTOR in cancer. Cancer Cell 12: 9–22 [DOI] [PubMed] [Google Scholar]
- Guillamo JS, Creange A, Kalifa C, Grill J, Rodriguez D, Doz F, Barbarot S, Zerah M, Sanson M, Bastuji-Garin S, et al. 2003. Prognostic factors of CNS tumours in Neurofibromatosis 1 (NF1): A retrospective study of 104 patients. Brain 126: 152–160 [DOI] [PubMed] [Google Scholar]
- Gutmann DH, Donahoe J, Brown T, James CD, Perry A 2000. Loss of neurofibromatosis 1 (NF1) gene expression in NF1-associated pilocytic astrocytomas. Neuropathol Appl Neurobiol 26: 361–367 [DOI] [PubMed] [Google Scholar]
- Hegedus B, Dasgupta B, Shin JE, Emnett RJ, Hart-Mahon EK, Elghazi L, Bernal-Mizrachi E, Gutmann DH 2007. Neurofibromatosis-1 regulates neuronal and glial cell differentiation from neuroglial progenitors in vivo by both cAMP- and Ras-dependent mechanisms. Cell Stem Cell 1: 443–457 [DOI] [PubMed] [Google Scholar]
- Hegedus B, Banerjee D, Yeh TH, Rothermich S, Perry A, Rubin JB, Garbow JR, Gutmann DH 2008. Preclinical cancer therapy in a mouse model of neurofibromatosis-1 optic glioma. Cancer Res 68: 1520–1528 [DOI] [PubMed] [Google Scholar]
- Hermanson O, Jepsen K, Rosenfeld MG 2002. N-CoR controls differentiation of neural stem cells into astrocytes. Nature 419: 934–939 [DOI] [PubMed] [Google Scholar]
- Hitoshi S, Tropepe V, Ekker M, van der Kooy D 2002. Neural stem cell lineages are regionally specified, but not committed, within distinct compartments of the developing brain. Development 129: 233–244 [DOI] [PubMed] [Google Scholar]
- Horiguchi S, Takahashi J, Kishi Y, Morizane A, Okamoto Y, Koyanagi M, Tsuji M, Tashiro K, Honjo T, Fujii S, et al. 2004. Neural precursor cells derived from human embryonic brain retain regional specificity. J Neurosci Res 75: 817–824 [DOI] [PubMed] [Google Scholar]
- Hresko RC, Mueckler M 2005. mTOR.RICTOR is the Ser473 kinase for Akt/protein kinase B in 3T3-L1 adipocytes. J Biol Chem 280: 40406–40416 [DOI] [PubMed] [Google Scholar]
- Ishida N, Hara T, Kamura T, Yoshida M, Nakayama K, Nakayama KI 2002. Phosphorylation of p27Kip1 on serine 10 is required for its binding to CRM1 and nuclear export. J Biol Chem 277: 14355–14358 [DOI] [PubMed] [Google Scholar]
- Jackson EL, Garcia-Verdugo JM, Gil-Perotin S, Roy M, Quinones-Hinojosa A, VandenBerg S, Alvarez-Buylla A 2006. PDGFRα-positive B cells are neural stem cells in the adult SVZ that form glioma-like growths in response to increased PDGF signaling. Neuron 51: 187–199 [DOI] [PubMed] [Google Scholar]
- Joseph NM, Mosher JT, Buchstaller J, Snider P, McKeever PE, Lim M, Conway SJ, Parada LF, Zhu Y, Morrison SJ 2008. The loss of Nf1 transiently promotes self-renewal but not tumorigenesis by neural crest stem cells. Cancer Cell 13: 129–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HT, Kim IS, Lee IS, Lee JP, Snyder EY, Park KI 2006. Human neurospheres derived from the fetal central nervous system are regionally and temporally specified but are not committed. Exp Neurol 199: 222–235 [DOI] [PubMed] [Google Scholar]
- Klein C, Butt SJ, Machold RP, Johnson JE, Fishell G 2005. Cerebellum- and forebrain-derived stem cells possess intrinsic regional character. Development 132: 4497–4508 [DOI] [PubMed] [Google Scholar]
- Kulbatski I, Tator CH 2009. Region specific differentiation potential of adult rat spinal cord neural stem/precursors and their plasticity in response to in vitro manipulation. J Histochem Cytochem 57: 405–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau N, Feldkamp MM, Roncari L, Loehr AH, Shannon P, Gutmann DH, Guha A 2000. Loss of neurofibromin is associated with activation of RAS/MAPK and PI3-K/AKT signaling in a neurofibromatosis 1 astrocytoma. J Neuropathol Exp Neurol 59: 759–767 [DOI] [PubMed] [Google Scholar]
- Ligon KL, Alberta JA, Kho AT, Weiss J, Kwaan MR, Nutt CL, Louis DN, Stiles CD, Rowitch DH 2004. The oligodendroglial lineage marker OLIG2 is universally expressed in diffuse gliomas. J Neuropathol Exp Neurol 63: 499–509 [DOI] [PubMed] [Google Scholar]
- Ligon KL, Huillard E, Mehta S, Kesari S, Liu H, Alberta JA, Bachoo RM, Kane M, Louis DN, Depinho RA, et al. 2007. Olig2-regulated lineage-restricted pathway controls replication competence in neural stem cells and malignant glioma. Neuron 53: 503–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HK, Wang G, Chen Z, Teruya-Feldstein J, Liu Y, Chan CH, Yang WL, Erdjument-Bromage H, Nakayama KI, Nimer S, et al. 2009. Phosphorylation-dependent regulation of cytosolic localization and oncogenic function of Skp2 by Akt/PKB. Nat Cell Biol 11: 420–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Listernick R, Charrow J, Greenwald M, Mets M 1994. Natural history of optic pathway tumors in children with neurofibromatosis type 1: A longitudinal study. J Pediatr 125: 63–66 [DOI] [PubMed] [Google Scholar]
- Listernick R, Ferner RE, Liu GT, Gutmann DH 2007. Optic pathway gliomas in neurofibromatosis-1: Controversies and recommendations. Ann Neurol 61: 189–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis DN, Ohgaki H, Wiestler OD, Cavenee WK 2007. WHO classification of tumors of the central nervous system. International Agency for Research on Cancer, Lyon [Google Scholar]
- Masri J, Bernath A, Martin J, Jo OD, Vartanian R, Funk A, Gera J 2007. mTORC2 activity is elevated in gliomas and promotes growth and cell motility via overexpression of rictor. Cancer Res 67: 11712–11720 [DOI] [PubMed] [Google Scholar]
- Motti ML, De Marco C, Califano D, Fusco A, Viglietto G 2004. Akt-dependent T198 phosphorylation of cyclin-dependent kinase inhibitor p27kip1 in breast cancer. Cell Cycle 3: 1074–1080 [PubMed] [Google Scholar]
- Narita Y, Nagane M, Mishima K, Huang HJ, Furnari FB, Cavenee WK 2002. Mutant epidermal growth factor receptor signaling down-regulates p27 through activation of the phosphatidylinositol 3-kinase/Akt pathway in glioblastomas. Cancer Res 62: 6764–6769 [PubMed] [Google Scholar]
- Otaegi G, Yusta-Boyo MJ, Vergano-Vera E, Mendez-Gomez HR, Carrera AC, Abad JL, Gonzalez M, de la Rosa EJ, Vicario-Abejon C, de Pablo F 2006. Modulation of the PI 3-kinase-Akt signalling pathway by IGF-I and PTEN regulates the differentiation of neural stem/precursor cells. J Cell Sci 119: 2739–2748 [DOI] [PubMed] [Google Scholar]
- Peltier J, O'Neill A, Schaffer DV 2007. PI3K/Akt and CREB regulate adult neural hippocampal progenitor proliferation and differentiation. Dev Neurobiol 67: 1348–1361 [DOI] [PubMed] [Google Scholar]
- Pollack IF, Shultz B, Mulvihill JJ 1996. The management of brainstem gliomas in patients with neurofibromatosis 1. Neurology 46: 1652–1660 [DOI] [PubMed] [Google Scholar]
- Prasad RC, Wang XL, Law BK, Davis B, Green G, Boone B, Sims L, Law M 2009. Identification of genes, including the gene encoding p27Kip1, regulated by serine 276 phosphorylation of the p65 subunit of NF-κB. Cancer Lett 275: 139–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee W, Ray S, Yokoo H, Hoane ME, Lee CC, Mikheev AM, Horner PJ, Rostomily RC 2009. Quantitative analysis of mitotic Olig2 cells in adult human brain and gliomas: Implications for glioma histogenesis and biology. Glia 57: 510–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandsmark DK, Zhang H, Hegedus B, Pelletier CL, Weber JD, Gutmann DH 2007. Nucleophosmin mediates mammalian target of rapamycin-dependent actin cytoskeleton dynamics and proliferation in neurofibromin-deficient astrocytes. Cancer Res 67: 4790–4799 [DOI] [PubMed] [Google Scholar]
- Sarbassov DD, Guertin DA, Ali SM, Sabatini DM 2005. Phosphorylation and regulation of Akt/PKB by the rictor–mTOR complex. Science 307: 1098–1101 [DOI] [PubMed] [Google Scholar]
- Sarbassov DD, Ali SM, Sengupta S, Sheen JH, Hsu PP, Bagley AF, Markhard AL, Sabatini DM 2006. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell 22: 159–168 [DOI] [PubMed] [Google Scholar]
- Schittenhelm J, Beschorner R, Simon P, Tabatabai G, Herrmann C, Schlaszus H, Capper D, Weller M, Meyermann R, Mittelbronn M 2008. WT1 expression distinguishes astrocytic tumor cells from normal and reactive astrocytes. Brain Pathol 18: 344–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setoguchi T, Kondo T 2004. Nuclear export of OLIG2 in neural stem cells is essential for ciliary neurotrophic factor-induced astrocyte differentiation. J Cell Biol 166: 963–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiles CD, Rowitch DH 2008. Glioma stem cells: A midterm exam. Neuron 58: 832–846 [DOI] [PubMed] [Google Scholar]
- Takei H, Yogeswaren ST, Wong KK, Mehta V, Chintagumpala M, Dauser RC, Lau CC, Adesina AM 2008. Expression of oligodendroglial differentiation markers in pilocytic astrocytomas identifies two clinical subsets and shows a significant correlation with proliferation index and progression free survival. J Neurooncol 86: 183–190 [DOI] [PubMed] [Google Scholar]
- Waldau B, Shetty AK 2008. Behavior of neural stem cells in the Alzheimer brain. Cell Mol Life Sci 65: 2372–2384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrington NM, Woerner BM, Daginakatte GC, Dasgupta B, Perry A, Gutmann DH, Rubin JB 2007. Spatiotemporal differences in CXCL12 expression and cyclic AMP underlie the unique pattern of optic glioma growth in neurofibromatosis type 1. Cancer Res 67: 8588–8595 [DOI] [PubMed] [Google Scholar]
- Yadirgi G, Marino S 2009. Adult neural stem cells and their role in brain pathology. J Pathol 217: 242–253 [DOI] [PubMed] [Google Scholar]
- Yeh TH, Lee DY, Gianino SM, Gutmann DH 2009. Microarray analyses reveal regional astrocyte heterogeneity with implications for neurofibromatosis type 1 (NF1)-regulated glial proliferation. Glia 57: 1239–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]