Abstract
Serotonin is a bioamine regulating bone mass accrual differently depending on its site of synthesis. It decreases accrual when synthesized in the gut, and increases it when synthesized in the brain. The signal transduction events elicited by gut-derived serotonin once it binds to the Htr1b receptor present on osteoblasts have been identified and culminate in cAMP response element-binding protein (CREB) regulation of osteoblast proliferation. In contrast, we do not know how brain-derived serotonin favors bone mass accrual following its binding to the Htr2c receptor on neurons of the hypothalamic ventromedial nucleus (VMH). We show here—through gene expression analysis, serotonin treatment of wild-type and Htr2c−/− hypothalamic explants, and cell-specific gene deletion in the mouse—that, following its binding to the Htr2c receptor on VMH neurons, serotonin uses a calmodulin kinase (CaMK)-dependent signaling cascade involving CaMKKβ and CaMKIV to decrease the sympathetic tone and increase bone mass accrual. We further show that the transcriptional mediator of these events is CREB, whose phosphorylation on Ser 133 is increased by CaMKIV following serotonin treatment of hypothalamic explants. A microarray experiment identified two genes necessary for optimum sympathetic activity whose expression is regulated by CREB. These results provide a molecular understanding of how serotonin signals in hypothalamic neurons to regulate bone mass accrual and identify CREB as a critical determinant of this function, although through different mechanisms depending on the cell type, neuron, or osteoblast in which it is expressed.
Keywords: CAM kinases, CREB, bone mass accrual, brain-derived serotonin, hypothalamus
Serotonin is a bioamine synthesized in neurons of the brainstem and in enterochromaffin cells of the duodenum that does not cross the blood–brain barrier (Mann et al. 1992). In the brain, serotonin is a well-characterized neurotransmitter affecting cognitive functions (Heath and Hen 1995), whereas, in the periphery, it is thought to act locally in the gut and as a hormone whose spectrum of functions only begins to be defined (Walther et al. 2003, 2007; Gershon and Tack 2007; Yadav et al. 2009, 2010). An additional feature contributing to the wide variety of functions of serotonin is that it can bind to 14 different receptors that have overlapping but different patterns of expression. These receptors are classified in seven groups (Htr1–7), depending on their structure and the nature of the signaling events they trigger (Tohda et al. 2006).
Besides its well known role in influencing cognitive functions, brain-derived serotonin emerged recently as a regulator of three homeostatic functions: bone remodeling, appetite, and energy expenditure (Yadav et al. 2009). Briefly, following its binding to the Htr2c receptor in neurons of the ventromedial hypothalamic (VMH) nuclei, serotonin favors bone mass accrual by decreasing activity of the sympathetic nervous system, a negative regulator of this function (Takeda et al. 2002; Elefteriou et al. 2005), while, following its binding to the Htr1a and Htr2b receptors on neurons of the arcuate nuclei, serotonin favors appetite and decreases energy expenditure. To date, the nature of the molecular events elicited by serotonin in each of these hypothalamic nuclei is unknown, since all of these functions of brain-derived serotonin are inhibited by leptin signaling in the brainstem (Yadav et al. 2009). Elucidating the signal transduction events elicited by serotonin in hypothalamic neurons will ultimately increase our knowledge of leptin signaling in the brain.
The regulation of bone remodeling by serotonin is an excellent illustration of the fact that each pool of serotonin can have different functions. Indeed, and unlike its brain counterpart, gut-derived serotonin inhibits bone formation (Yadav et al. 2008). Gut-derived serotonin binds to the Htr1b receptor present on osteoblasts and, through a PKA-dependent pathway, inhibits cAMP response element-binding protein (CREB) phosphorylation and Creb expression, and, as a result, inhibits osteoblast proliferation (Yadav et al. 2008). This molecular knowledge surrounding the signaling of gut-derived serotonin in osteoblasts is a further incentive to improve our understanding of the signaling events—including the transcriptional cues—elicited by serotonin in neurons of the VMH nuclei.
CREB is a broadly expressed leucine zipper-containing transcription factor affecting differentiation and proliferation of multiple cell types (Peltier et al. 2007; Yadav et al. 2008). Although it acts in many different cellular contexts, CREB is a major regulator of multiple aspects of neurobiology, such as neuron survival, axon growth, and synaptic transmission (Ribar et al. 2000; Wayman et al. 2004, 2006; Mizuno et al. 2006). These neurobiological functions of CREB, together with the fact that it mediates the function of gut-derived serotonin, raise the testable hypothesis that CREB may also mediate some of the homeostatic functions of brain-derived serotonin.
We show here—through cell-based assays and cell-specific gene inactivation studies—that brain-derived serotonin uses a calmodulin kinase (CaMK) cascade involving CaMKKβ and CaMKIV to phosphorylate CREB following signaling through the Htr2c receptor in VMH neurons. We further show that, unlike what it is the case for signaling of gut-derived serotonin in osteoblasts, brain-derived serotonin facilitates expression and phosphorylation of CREB in VMH neurons. CREB, in turn, regulates the expression of genes necessary for optimal sympathetic activity. Moreover, by showing that CREB is implicated, albeit in a different manner, in the regulation of bone mass accrual through its expression in two cell types (VMH neurons and osteoblasts), this study underscores the growing importance of this transcription factor in the regulation of bone mass accrual.
Results
Expression of CaMKs in hypothalamic neurons
Htr2c is a G protein-coupled receptor using Ca2+ as a second messenger in a signal transduction pathway that requires the intervention of the calcium-binding protein CaM (Fink and Gothert 2007; Drago and Serretti 2009). To mediate its diverse functions, CaM can activate numerous enzymes, including several CaM kinases called CaMKI, CaMKII, or CaMKIV (Haribabu et al. 1995; Schulman et al. 1995; Tokumitsu and Soderling 1996). CaMKI and CaMKIV become fully active only when phosphorylated by other kinases called CaMK kinases (CaMKKs), of which two are known: CaMKKα and CaMKKβ (Bito et al. 1996; Edelman et al. 1996; Kitani et al. 1997; Corcoran and Means 2001; Soderling and Stull 2001; Tokumitsu et al. 2003). CaMKII, in contrast, is not phosphorylated by CaMKKβ (Hook and Means 2001). In the face of this complex and stepwise activation of CaM, we asked whether genes encoding CaMKKs and CaMKs were expressed in neurons of the VMH nuclei.
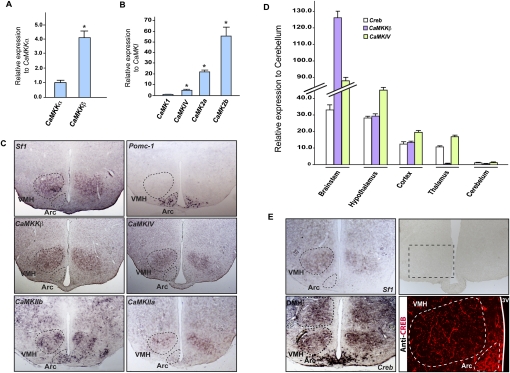
As shown in Figure 1A, when using real-time PCR as an assay, and even though both genes were expressed, CaMKKβ was by far the most highly expressed of the two CaMKKs in the hypothalamus. As for the CaMKs, CaMKIV was more highly expressed than CaMKI in the hypothalamus, and CaMKIIa and CaMKIIb were also highly expressed in the hypothalamus (Fig. 1B). We next used in situ hybridization to define the neuronal populations within the hypothalamus expressing these various genes. CaMKKβ, CaMKIV, and CaMKIIa were more highly expressed in Sf1-expressing neurons of the VMH nuclei than in Pomc1-expressing neurons of the arcuate nuclei, while CaMKIIb was equally expressed in VMH and arcuate neurons (Fig. 1C). Since calcium signaling often uses CREB as a transcriptional mediator, we studied whether Creb was also expressed in hypothalamic neurons. Real-time PCR verified that Creb is expressed in the hypothalamus (Fig. 1D), and Creb expression was observed in Sf1-expressing neurons of the VMH by in situ hybridization; CREB protein was detected by immunofluorescence (Fig. 1E). Taken together, these experiments demonstrate that the molecular components necessary for calcium-dependent signaling and gene activation exist in neurons of the VMH nuclei.
Figure 1.
Expression of CaMKs and Creb in hypothalamic neurons. (A,B) Real-time PCR (qPCR) analysis of CaMKKα and CaMKKβ expression in wild-type (WT) hypothalamus, showing higher expression of CaMKKβ compared with CaMKKα (A), and CaMKI, CaMKIV, CaMKIIa, and CaMKIIb expression in hypothalamus, demonstrating a higher level of expression of CaMKIV, CaMKIIa, and CaMKIIb compared with CaMKI (B). We compared the level of expression of CaMKKα versus CaMKKβ, and CaMKIV, CaMKIIa, and CaMKIIb versus CaMKI after normalization with β-actin. Error bars represent SEM. Student's t-test (*) P < 0.05. (C) Coronal adjacent sections through the hypothalamus in 6-wk-old wild-type mice. Expression analysis of CaMKKβ, CaMKIV, CaMKIIa, and CaMKIIb in hypothalamus by in situ hybridization and compared with Sf1 and Pomc-1 probes, specific molecular markers of VMH and Arcuate (Arc) (outlined by dashed line), respectively. CaMKKβ and CaMKIV are more highly expressed in Sf1-expressing than Pomc-1-expressing neurons. (D) Real-time PCR (qPCR) analysis of Creb, CaMKKβ, and CaMKIV in brainstem, hypothalamus, cortex, thalamus, and cerebellum showing that Creb is expressed in most parts of the brain, including the hypothalamus. (E) Cross-sections through the hypothalamus in 6-wk-old wild-type mice. Expression analysis of Creb in hypothalamus by in situ hybridization and compared with Sf1 probes. Immunofluorescence analysis of CREB in hypothalamus.
Serotonin regulates CaMKs and CREB function and expression
As a first approach to determine whether serotonin transduces its signal through a CaM-dependent mechanism, we used a cell-based assay, taking advantage of the fact that P19 cells can differentiate into neurons following treatment with retinoic acid (McBurney 1993; Alam et al. 2009). Once they were differentiated into neurons, these cells were treated with either vehicle or serotonin (50 μM) for up to 45 min, and time-dependent phosphorylation of CaMKIV and CREB were analyzed by Western blot using specific antibodies.
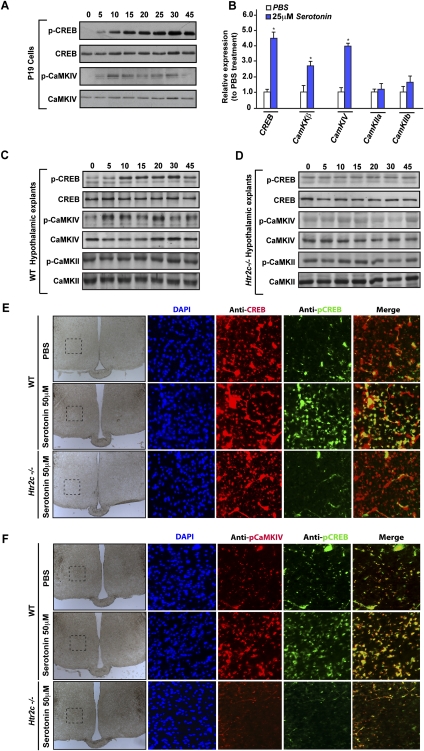
As shown in Figure 2, A–C, serotonin enhanced phosphorylation of CaMKIV (but not of CaMKII) and CREB in a time-dependent manner. Phosphorylation of CaMKIV was detected as early as 5 min after serotonin treatment, while phosphorylation of CREB on Ser 133, which was consistently weak after 5 min, became stronger 10 min after initiation of the serotonin treatment, and remained steady for 45 min (Fig. 2C). Serotonin not only affected phosphorylation of these proteins, but also enhanced expression of their mRNAs. Indeed, real-time PCR analysis showed a significant increase in the expression of CaMKKβ, CaMKIV, and Creb but not of CaMKIIa and CaMKIIb upon serotonin treatment (Fig. 2B). Remarkably, the up-regulation of Creb expression in neurons by brain-derived serotonin mirrors the inhibition of Creb expression in osteoblasts exerted by gut-derived serotonin (Yadav et al. 2008).
Figure 2.
Serotonin regulates CaMKs and Creb activity and expression. (A) P19 cells differentiated into neurons were treated with 50 μM serotonin for the indicated period of time. Western blot analysis of CREB and CaMKIV phosphorylation performed using p-CREB (s113) and p-CaMKIV (T196) antibodies. (B) Real-time PCR (qPCR) of Creb, CaMKKβ, CaMKIV, CaMKIIa, and CaMKIIb (relative to β-actin expression) in P19 cells after a 4-h treatment with 25 μM serotonin (n = 8 for each measurement). Error bars represent SEM. Student's t-test (*) P < 0.05. (C,D) Western blot analysis of CREB, CaMKIV, and CaMKII phosphorylation in wild-type (C) or in Htr2c−/− (D) hypothalamic explants following treatment with 50 μM serotonin for 5, 10, 15, 20, 30, and 45 min. (E,F) Analysis of the level of CREB phosphorylation by coimmunofluorescence using p-CREB (S133) and CREB antibodies (E) and CREB-CaMKIV phosphorylation using p-CREB (S133) and p-CaMKIV (T196) antibodies (F). Coimmunofluorescence was performed on coronal sections of wild-type (WT) or Htr2c−/− hypothalamic explants treated previously with 50 μM serotonin for 30 min. The first column represents large bright-field images of hypothalamic sections, and the black dashed line delimits the frame for the coimmunofluorescence analysis shown in the following rows. The last column shows the merge between the p-CREB and CREB or p-CREB and p-CaMKIV immunofluorescence images.
To establish the specificity of these results through another assay, we next generated hypothalamic explants obtained from wild-type mice or mice lacking the Htr2c receptor in VMH neurons (Htr2c−/− mice). These explants were then treated with either vehicle or serotonin (50 μM) for 5, 10, 15, 20, 30, and 45 min. As was the case in the cell culture assays, serotonin induced phosphorylation of CaMKIV—and, subsequently, of CREB on Ser 133—in wild-type hypothalamic explants (Fig. 2C). CaMKII was not phosphorylated following treatment of wild-type explants by serotonin (Fig. 2C). Coimmunofluorescence analysis also showed an increase of phospho-CREB in hypothalamic explants treated with serotonin, while total CREB level remained the same after treatment with serotonin or vehicle (Fig. 2E). As shown in Figure 2F, coimmunofluorescence showed colocalization of phospho-CREB and phospho-CaMKIV, and a similar increase in their levels after treatment of VMH neurons with serotonin. That serotonin did not affect phosphorylation of CaMKIV or CREB in Htr2c−/− hypothalamic explants (Fig. 2D–F) established the specificity of the phosphorylation events detected in wild-type hypothalamic explants.
CaMK signaling in VMH neurons favors bone mass accrual
The pattern of expression of CaMKKβ and CaMKIV and the ex vivo data presented above suggested that CaM signaling may, through CaMKIV, contribute to the serotonin-dependent regulation of bone mass accrual in VMH neurons. We focused on CaMKKβ and CaMKIV to address this question because they are phosphorylated following serotonin treatment of hypothalamic explants, while CaMKII is not (Fig. 2).
To determine to what extent this CaMK/CREB signaling module mediates brain-derived serotonin regulation of bone mass accrual, we generated and analyzed cell-specific gene inactivation models. Specifically, we crossed mice harboring a floxed allele of either CaMKKβ or CaMKIV with Sf1-Cre transgenic mice that express the Cre recombinase only in Sf1-expressing neurons of the VMH nuclei (Balthasar et al. 2004). Prior to analyzing their bone phenotypes, we verified through quantitative PCR (qPCR) and in situ hybridization that, in both CaMKKβSf1−/− and CaMKIVSf1−/− mice, we efficiently deleted these genes selectively in VMH neurons (Supplemental Fig. 1A–C,F,G).
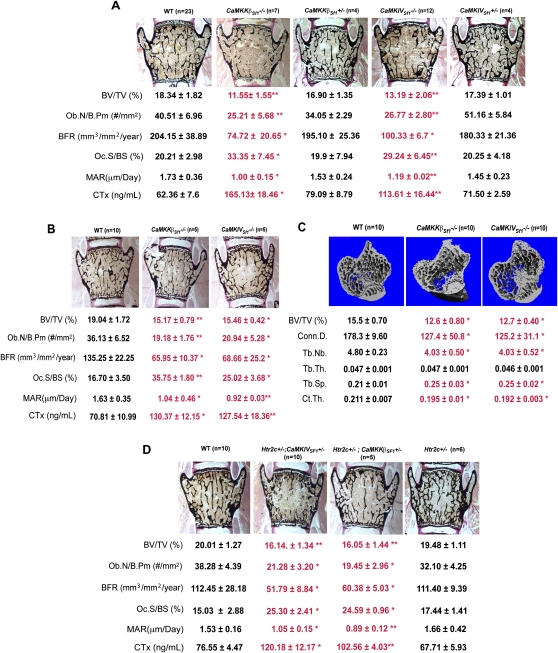
Whether they were analyzed by bone histology or microcomputed tomography (μCT), mice lacking either CaMKKβ or CaMKIV only in Sf1-expressing neurons of the VMH nuclei displayed, at both 8 and 12 wk of age, a severe low-bone-mass phenotype (Fig. 3A–C). This phenotype affected equally vertebrae and long bones, and trabecular and cortical bones, and was due to a concomitant decrease in bone formation parameters and an increase in osteoclast number (Fig. 3A–C). The increase in osteoclast number translated into an increase in bone resorption, as measured by the increase in serum levels of CTx, a biomarker of bone resorption in CaMKKβSf1−/− and CaMKIVSf1−/− (Guerrero et al. 1996). That these cellular anomalies are identical to the ones seen in mice lacking brain-derived serotonin or the Htr2c receptor (Yadav et al. 2009) provided strong correlative support to the hypothesis that serotonin uses a CaM-dependent mechanism to favor bone mass accrual following its signaling in VMH neurons.
Figure 3.
CaMK signaling cascade in VMH neurons favors bone mass accrual. (A,B) Histomorphometric analysis of vertebrae of wild-type (WT), CaMKKβSf1−/−, CaMKIVSf1−/−, CaMKKβSf1+/−, and CaMKIVSf1+/− mice at 12 wk (A) and 8 wk (B) of age. Mineralized bone matrix is stained in black by von Kossa reagent. Histomorphometric parameters—bone volume over trabecular volume (BV/TV%), number of osteoblasts per bone perimeter (Ob.N/B.Pm [number of square millimeters]), and bone formation rate (BFR)—are decreased in CaMKKβSf1−/− and CaMKIVSf1−/− in comparison with wild-type littermate mice, whereas osteoclast surface per bone surface (Oc.S/BS [percent]), mineral apposition rate (MAR [micrometers per day]), and CTx (nanograms per milliliter), a biomarker of bone resorption, are increased. (C) μCT analysis of long bones of 12-wk-old CaMKKβSf1−/− and CaMKIVSf1−/− mice shows a lower bone density (BV/TV%), a lower trabecular number (Tb.Nb.), a higher trabecular separation (Tb.Sp.), and a significant decrease in connectivity density (Conn.D.) and cortical thickness (Ct.Th.), but no changes in the trabecular thickness (Tb.Th.). (D) Histomorphometric analysis of vertebrae of Htr2c+/−; CaMKKβSf1+/−, Htr2c+/−; CaMKIVSf1+/−, and Htr2c+/− in 12-wk-old mice present the same changes observed in CaMKKβSf1−/− and CaMKIVSf1−/− described above to the wild-type (WT) littermate mice. Error bars represent SEM. Student's t-test (*) P < 0.05; (**) P < 0.001.
That serotonin cannot affect CaMK phosphorylation in Htr2c−/− hypothalami explants strongly suggested the existence of an interaction occurring in VMH neurons—where Htr2c is most expressed—between this receptor signaling and CaM kinase signaling. To demonstrate that this was the case, we relied on genetic interaction experiments, and generated mutant mice lacking one allele of Htr2c and one allele of either CaMKKβ or CaMKIV (Htr2c+/−; CaMKKβSf1+/− and Htr2c+/−; CaMKIVSf1+/− mice) in Sf1-expressing neurons only (Fig. 3D). In both cases, the cell specificity in these mutant mouse strains was conferred by the VMH-specific deletion of CaMKKβ and CaMKIV, although a cell-specific deletion of Htr2c is needed to demonstrate formally that this is the case. We note, however, that Htr2c is much more expressed in VMH neurons than in any other neurons of the hypothalamus (Yadav et al. 2009).
Both mutant mouse strains displayed, at 3 mo of age, the same low-bone-mass phenotype that was observed in either Tph2−/− (Yadav et al. 2009), Htr2c−/− (Yadav et al. 2009), CaMKKβSf1−/−, or CaMKIVSf1−/− mice (Fig. 3A). These experiments provide genetic evidence that CaMKKβ and CaMKIV act downstream from serotonin signaling through Htr2c in Sf1-expressing neurons of the VMH nuclei to favor bone mass accrual. Importantly, and consistent with the previously described functions of serotonin signaling in Sf1-expressing neurons of the VMH nuclei, neither CaMKKβSf1−/− nor CaMKIVSf1−/− mice had appetite or energy expenditure abnormalities when fed a normal chow diet at the time they developed their low-bone-mass phenotype (Supplemental Fig. 2A–E).
CREB mediates serotonin regulation of bone mass accrual in VMH neurons
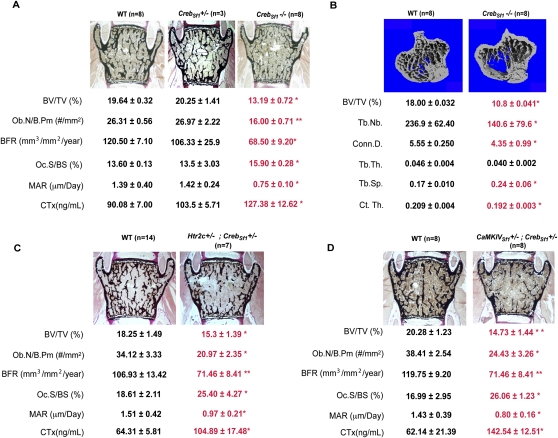
The cell-based assays presented in Figure 2 showing that serotonin favors CREB phosphorylation on Ser 133 suggested that CREB is a transcriptional mediator of serotonin regulation of bone mass in VMH neurons. To establish formally that this is the case in vivo, we first deleted Creb in VMH neurons (CrebSf1−/− mice). We verified through qPCR and in situ hybridization that, in CrebSf1−/− mice, we efficiently deleted this gene selectively in VMH neurons (Supplemental Fig. 1D,E).
Twelve-week-old CrebSf1−/− mice demonstrated a marked decrease in bone mass, affecting vertebrae and long bones, and trabecular and cortical bones. Histomorphometry revealed that this was due to a decrease in bone formation parameters and an increase in bone resorption parameters, as is the case in mice lacking serotonin signaling or CaMKIV in VMH neurons (Fig. 4A,B). The similarities in the phenotypes of these various mouse mutant strains, together with the evidence presented above, strongly suggested that Creb lies downstream from the CaMK-dependent serotonin signaling in VMH neurons. To establish that this is the case, we generated compound heterozygous mice lacking, on the one hand, one allele of Creb in Sf1-expressing neurons of the VMH nuclei, and, on the other hand, either one allele of Htr2c or one allele of CaMKIV in the same neurons. Histomorphometry analyses of vertebrae showed that Htr2c+/− ; CrebSf1+/− and CaMKIVSf1+/− ; CrebSf1+/− mice had a low-bone-mass phenotype similar to the one seen in Htr2c−/− and CaMKIVSf1−/− mice (Fig. 4C,D). Again, this phenotype was due to a decrease in bone formation parameters, and to an increase in bone resorption parameters (Fig. 4C,D). Taken together, these data demonstrate that brain-derived serotonin mediates its regulation of bone mass accrual through the activation of the transcription factor CREB in VMH neurons of the hypothalamus.
Figure 4.
CREB mediates serotonin regulation of bone mass accrual in VMH neurons. (A,B) Histomorphometric analysis of vertebrae (A) and μCT analysis of long bones (B) of wild-type and CrebSf1−/− mice at 12 wk old. Mineralized bone matrix is stained in black by von Kossa reagent. (A) Bone volume over trabecular volume (BV/TV%), number of osteoblasts per bone perimeter (Ob.N/B.Pm [number of square millimeters]), and bone formation rate (BFR) are all decreased in CrebSf1−/− compared with wild-type and CrebSf1+/− mice, whereas osteoclast surface per bone surface (Oc.S/BS [percent]), mineral apposition rate (MAR [micrometers per day]), and CTx (nanograms per milliliter), a biomarker of bone resorption, are increased. (B) μCT analysis shows a lower bone density in long bones (BV/TV%) of 12-wk-old CrebSf1−/− mice, along with lower trabecular number (Tb.Nb.) and cortical thickness (Ct.Th.), higher trabecular separation (Tb.Sp.), and decreased connectivity density (Conn.D.). (C,D) Histomorphometric analyses of vertebrae of 12-wk-old Htr2c+/−; CrebSf1+/− (C) and CaMKIVSf1+/− ; CrebSf1+/− (D) mice present similar changes as described above. Error bars represent SEM. Student's t-test (*) P < 0.05; (**) P < 0.001.
CREB regulates expression of genes determining sympathetic activity in VMH neurons
Having established that CREB, through its expression in VMH neurons, is the transcriptional mediator of brain serotonin regulation of bone mass accrual, we then asked which genes and pathways controlled by Creb expression in VMH neurons could account for this function of serotonin. To that end, we isolated hypothalami of wild-type and CrebSf1−/− mice and performed a microarray experiment.
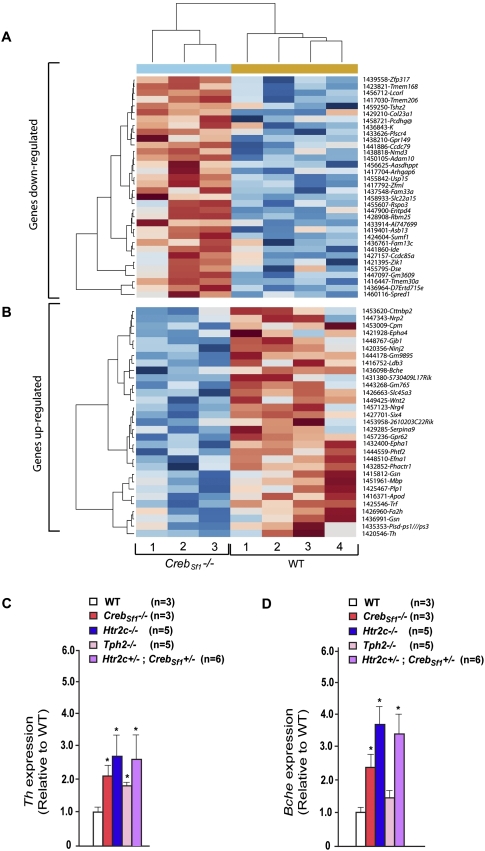
As shown in Figure 5, A and B, there is a large number of genes whose expression was altered by the absence of Creb in Sf1-expressing neurons. Among them, two were of particular interest for the present study. Indeed, in keeping with the fact that brain-derived serotonin favors bone mass accrual by decreasing the sympathetic tone, we were excited to discover that the expression of tyrosine hydroxylase (Th), a rate-limiting enzyme of catecholamine synthesis, and butyrylcholinesterase (Bche), an enzyme inactivating acetylcholine, the main neurotransmitter of the parasympathetic nervous system (van Koppen and Kaiser 2003; Nathanson 2008), were increased in the hypothalami of mice lacking Creb in VMH neurons. We verified this by real-time PCR of mRNA isolated from VMH neurons showing that expression of the two genes was increased in CrebSf1−/− hypothalami (Fig. 5C,D). Moreover, and consistent with the notion that serotonin signaling through Htr2c in VMH neurons regulates bone mass accrual in a Creb-dependent manner, we also observed an increase of Th and Bche expression in hypothalami of Tph2−/−, Htr2c−/−, and Htr2c+/− ; CrebSf1+/− mice (Fig. 5C,D). Binding sites for CREB have already been described in the Th promoter, and three consensus binding sites are present in the mouse Bche promoter (Cambi et al. 1989; Lonze and Ginty 2002). Importantly, two of these putative CREB-binding sites are also present in the human Bche promoter (Supplemental Fig. 3). Further work will determine how CREB regulates expression of these genes in vivo. Nevertheless, taken together, these data establish that CREB transcriptional activity in VMH neurons is regulated by serotonin signaling, and identify two genes whose expression in VMH neurons is regulated by CREB that determine the activity of the sympathetic tone: the mediator of serotonin regulation of bone mass accrual (Yadav et al. 2009).
Figure 5.
Microarray experiment comparing expression of genes in hypothalami of wild-type mice and of mice lacking Creb only in VMH neurons. (A,B) Heat map of the microarray analysis of four wild-type hypothalami versus hypothalami of theee mice lacking Creb only in VMH neurons (CrebSf1−/− mice). Genes significantly (P < 0.05) up-regulated (A) or down-regulated (B) in hypothalamus of CrebSf1−/− mice compared with that of wild-type littermates. These genes are ordered by fold change from maximum to the threshold of 1.20. The list of the genes up-regulated or down-regulated are mentioned in the last column. (C,D) Real-time PCR (qPCR) analysis of Th (C) and Bche (D) expression in hypothalami of wild-type (WT), CrebSf1−/−, Htr2c−/−, Tph2−/−, and Htr2c+/−; CrebSf1+/− mice at 12 wk of age.
The sympathetic nervous system acts downstream from the CaMK/CREB signaling pathway to regulate bone mass
The only known peripheral mediator of the regulation of bone mass by brain-derived serotonin is the sympathetic tone acting on osteoblasts through the β2 adrenergic receptor (Adrβ2) (Yadav et al. 2009). In osteoblasts, the sympathetic tone has two functions: It inhibits osteoblast proliferation by recruiting for that purpose members of the molecular clock, while it favors bone resorption by increasing the expression of RankL (Takeda et al. 2002; Elefteriou et al. 2005; Fu et al. 2005). That the microarray experiment identified Th as a CREB target gene in VMH neurons was an incentive to study whether the sympathetic tone was mediating the regulation of bone mass accrual by CaM signaling and CREB in Sf1-expressing neurons of the VMH nuclei.
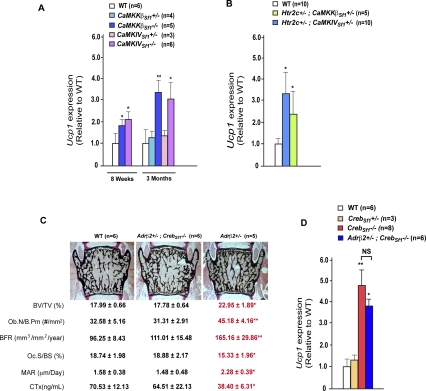
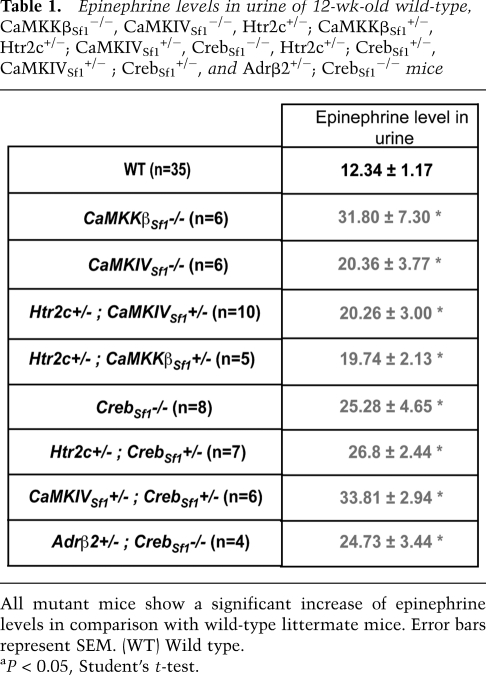
A first line of evidence, albeit indirect, supporting this hypothesis came from the cellular nature of the bone phenotype observed in mice lacking CaMK signaling in VMH neurons. Indeed, the low-bone-mass phenotype of CaMKKßSf1−/− and CaMKIVSf1−/− was caused by a concomitant decrease in bone formation parameters and increase in bone resorption parameters (Fig. 3). These cellular abnormalities are identical to those seen in mice lacking serotonin signaling in the brain whose bone phenotype is secondary to an increased sympathetic activity (Yadav et al. 2009). The hypothesis that the sympathetic tone may be increased in mice lacking CaMK signaling in VMH neurons was supported by the fact that Ucp1 expression in brown fat and urinary elimination of epinephrine sympathetic activity was high in mice lacking CaMKKβ or CaMKIV in Sf1-expressing neurons only (Fig. 6A; Table 1). The same was true for mutant mice lacking, in Sf1-expressing neurons, one allele of Htr2c and one allele of either CaMKKβ or CaMKIV (Htr2c+/−; CaMKKβSf1+/− and Htr2c+/−; CaMKIVSf1+/− mice (Fig. 6B; Table1).
Figure 6.
CaMK/CREB signaling cascade regulates bone remodeling by inhibiting the sympathetic tone. (A,B,D) Real-time PCR (qPCR) analysis of Ucp1 expression in brown adipose tissue in wild-type (WT), CaMKKβSf1−/−, and CaMKIVSf1−/− mice at 8 wk, and wild-type (WT), CaMKKβSf1−/−, CaMKKβSf1+/−, CaMKIVSf1−/−, and CaMKIVSf1+/− mice 12 wk of age (A), and in 12-wk-old Htr2c+/−; CaMKKβSf1+/− and Htr2c+/−; CaMKIVSf1+/−,− (B) and CrebSf1−/−, CrebSf1+/−, Adrβ2+/−; CrebSf1−/−, and Adrβ2+/ (D) mice. Error bars represent SEM. Student's t-test (*) P < 0.05, (**) P < 0.001. (C) Histomorphometric analysis of Adrβ2+/−; CrebSf1−/− do not show any significant difference in comparison with wild-type mice for bone volume over trabecular volume (BV/TV%), number of osteoblasts per bone perimeter (Ob.N/B.Pm [number of square millimeters]), bone formation rate (BFR), osteoclast surface per bone surface (Oc.S/BS [percent]), mineral apposition rate (MAR [micrometers per day]), and CTx (nanograms per milliliter), a biomarker of bone resorption.
Table 1.
Epinephrine levels in urine of 12-wk-old wild-type, CaMKKβSf1−/−, CaMKIVSf1−/−, Htr2c+/−; CaMKKβSf1+/−, Htr2c+/−; CaMKIVSf1+/−, CrebSf1−/−, Htr2c+/−; CrebSf1+/−, CaMKIVSf1+/− ; CrebSf1+/−, and Adrβ2+/−; CrebSf1−/− mice
All mutant mice show a significant increase of epinephrine levels in comparison with wild-type littermate mice. Error bars represent SEM. (WT) Wild type.
aP < 0.05, Student's t-test.
To provide a formal proof that the sympathetic tone is downstream from CREB in Sf1-expressing neurons of the VMH nuclei, we generated CrebSf1−/− mice lacking one allele of Adrβ2, the adrenergic receptor used by the sympathetic nervous system to regulate bone mass (Takeda et al. 2002). Analysis of Adrβ2+/− ; CrebSf1−/− mice at 12 wk of age showed bone mass, bone formation, and bone resorption parameters indistinguishable from those of wild-type littermates (Fig. 6C). Ucp1 expression in brown fat and urinary levels of epinephrine were increased in Adrβ2+/− ; CrebSf1−/− mice as well as in CrebSf1−/− mice (Fig. 6D; Table 1). Taken together, these results demonstrated that it is, at least in part, by regulating the activity of sympathetic neurons that Creb, through its expression in VMH neurons, favors bone mass accrual.
Discussion
In this study, we present expression analysis, cell-based, and cell-specific genetic evidence that all support the notion that brain-derived serotonin favors bone mass accrual by using a CaMK cascade with CREB as its transcriptional effector in neurons of the VMH nuclei.
The initial study revealing the critical role of brain-derived serotonin during bone remodeling stopped short of determining the nature of the signaling events elicited by serotonin within VMH neurons (Yadav et al. 2009). The fact that Htr2c belongs to a subfamily of serotonergic receptors using calcium as a second messenger (Fink and Gothert 2007; Drago and Serretti 2009), along with the importance of CREB in neurobiology (Dash et al. 1990; Bourtchuladze et al. 1994; Yin et al. 1994, 1995; Bartsch et al. 1995; Petrij et al. 1995; Nestler et al. 2002; Peters et al. 2003; Josselyn and Nguyen 2005), led us to explore the possibility that serotonin could use a CaM signaling pathway in VMH neurons to phosphorylate and activate CREB. This would ultimately result in a decrease of the activity of the sympathetic nervous system, and an increase in bone mass accrual.
Given the complexity of the problem studied, we relied on cell-specific gene deletion in order to perform a rigorous analysis. Using gene expression analysis, serotonin treatment of wild-type and Htr2c−/− hypothalamus, cell-specific gene deletion, and genetic epistasis experiments, we show here that, following its binding to Htr2c on VMH neurons, serotonin triggers the activation of CaMKKβ, which phosphorylates and activates CaMKIV. This results in phosphorylation of CREB on Ser 133 and a decrease in the activity of the sympathetic nervous system peripherally (Fig. 7). We cannot exclude, at the present time, that other signaling pathways may be involved in the regulation of bone mass accrual. For instance, CaMKIIb is also expressed in VMH neurons. However, CaMKIIb is not phosphorylated by CaMKKβ (Hook and Means 2001), nor is it phosphorylated following serotonin treatment of hypothalamic explants. Moreover, there is no available floxed CaMKIIb allele—a reagent absolutely necessary for rigorous analysis, since this gene is expressed elsewhere in the hypothalamus.
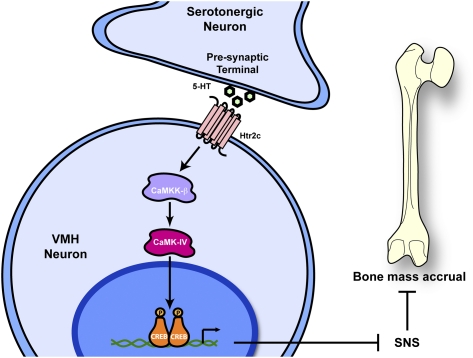
Figure 7.
Model. Brain-derived serotonin (5-HT) regulates bone mass accrual through a CaM/CREB signaling cascade in VMH neurons. Serotonin is released from the presynaptic terminal of serotonergic neurons of the brainstem. Serotonin, following its binding to the Htr2c receptor expressed in VMH neurons, activates the CaM signaling pathway and leads to the activation of CREB by its phosphorylation. CREB favors bone mass accrual by inhibiting the activity of sympathetic neurons.
Unexpectedly, over the course of the last 2 years, serotonin has emerged as a major regulator of bone mass accrual by exerting complex and antagonistic influences on this physiological function. Indeed, while gut-derived serotonin inhibits bone formation following its binding to the Htr1b receptor on osteoblasts, brain-derived serotonin instead favors bone mass accrual by acting on VMH neurons following its binding to the Htr2c receptor (Yadav et al. 2008, 2009, 2010). To the best of our knowledge, serotonin may be the only molecule exerting, to such an extent, opposite influences on the same physiological function, depending on its site of synthesis. In a broader context, since leptin inhibits bone mass accrual by inhibiting serotonin synthesis in and release from brainstem neurons, this study further improves our molecular knowledge of the mechanisms of action used by leptin in the brain to fulfill one of its function (Karsenty 2006).
Another unexpected feature of the serotonin regulation of bone mass accrual is that CREB phosphorylation and function are regulated by serotonin in opposite manners, depending its site of synthesis. Indeed, gut-derived serotonin inhibits expression and phosphorylation of CREB in osteoblasts, while brain-derived serotonin does exactly the opposite in VMH neurons. We further show that, in VMH neurons, CREB regulates the expression of genes needed for the synthesis of catecholamines (the mediators of the sympathetic tone), and chief among them is Th. This observation—along with the increase of Th and Bche expression in Tph2−/−, Htr2c−/−, and Htr2c+/− ; Creb Sf1+/− hypothalami—reveals for the first time that serotonin regulates, in a Creb-dependent manner, catecholamine synthesis in VMH neurons. It also indicates that, at least in VMH neurons, CREB inhibits Th and Bche expression.
In broader terms, this study and a previous one (Yadav et al. 2008) identified CREB as the first transcription factor regulating bone mass accrual through its expression in two different cell types: one present in bone (the osteoblast), and one present in the brain (the VMH neurons). By analogy to what is seen during development and cell differentiation, a general expectation in the field of molecular physiology has been that the transcriptional control of the main functions of specialized cell types would be executed by cell-specific transcription factors. The validity of this hypothesis has been verified in many instances. Indeed, if we look at bone biology, cell-specific transcription factors like Runx2 or ATF4 do affect bone formation by osteoblasts, osteoclast differentiation, or bone extracellular matrix mineralization (Ducy et al. 1997, 1999; Yang et al. 2004; Elefteriou et al. 2005, 2006). However, growing evidence points toward a more complex picture involving both cell-specific and broadly expressed transcription factors in the control of specialized cells functions. Regarding bone physiology in this context is the role of the members of the AP-1 family of proteins (Ducy et al. 1997, 2000; Sabatakos et al. 2000; Jochum et al. 2001; Fu et al. 2005; Wagner and Eferl 2005) in osteoblasts and osteoclasts, Schnurri-3 as a regulator of Runx2 postnatally (Jones et al. 2006), Foxo1 in osteoblasts in the regulation of oxidative stress (Rached et al. 2010a,b), and CREB in regulating bone mass accrual through either its osteoblast or neuronal expression (Yadav et al. 2008). It is likely that this complex pattern involving few cell-specific and numerous non-cell-specific transcription factors will not be limited to skeleton physiology, but may become the rule rather than the exception in most cell lineages when it comes to regulating cell functions.
Materials and methods
Mice generation
Genetic backgrounds of mice were as follows: Crebsf1−/−, Crebsf1+/−, CaMKKβsf1−/−, CaMKKβsf1+/−, CaMKIVsf1−/−, CaMKIVsf1+/−, CaMKKβsf1−/−, Htr2c+/− (C57BL/6J:75%; 129/Sv:25%), Htr2c+/−; CaMKKβSf1+/−, Htr2c+/−; CaMKIVSf1+/−, CaMKIVSf1+/−; CrebSf1+/−, Adrβ2+/−; CrebSf1−/−, and Htr2c+/−; CrebSf1+/− (C57BL/6J:87.5%; 129/Sv:12.5%). Control littermates were used in all experiments. Mice genotypes were determined by PCR; primer sequences are available on request.
Molecular studies
RNA isolation, cDNA preparation, and qPCR analysis were carried out following standard protocols using oligos from SA Biosciences Qiagen Company. Genotypes of all of the mice were determined by PCR. All primer sequences for genotyping and DNA probes for Southern hybridization are available on request.
Histology
Sections containing VMH from bregma −1.06 to −2.06 mm according to the Franklin and Paxinos (1997) mouse brain atlas. In situ hybridization was performed as described in Oury et al. (2006). Briefly, mice were anesthetized and perfused transcardially with ice-cold saline followed by 4% PFA. Brains were dissected, fixed in 4% PFA overnight at 4°C, cryoprotected by overnight immersion in a 20% sucrose solution, embedded in Shandon Cryomatrix (Thermo Electron Corporation), and frozen at −80°C. Twenty-micrometer cryostat sections were cut in the coronal plane.
Hypothalamic explants
Animals were anesthetized with 1.2% Avertin (2,2,2-tribromoethanol) in saline solution (0.9% NaCl) used at a concentration of 0.2 mL per 10 g body weight. The depth of anesthesia was determined by the animal's respiratory pattern and by pinching the animal's foot to test reflex response. Mice were killed by cervical dislocation. Brains were dissected in cold ACSF solution and left in the same solution for 30 min before being sectioned at 500 μm using a chopper at the level of the VMH nuclei of the hypothalamus (from bregma −1.22 to −1.70 mm). The resulting slices were incubated in ACSF for 1 h, and were treated with either vehicle or serotonin (50 μM) for up to 45 min, followed by precise dissection of the hypothalamus. Phosphorylation of CaMKIV and CREB was detected by Western blot after 5, 10, 15, 30, and 45 min using P-CREB (S133) rabbit antibody (Cell Signaling [87G3]), CREB rabbit antibody (Cell Signaling [48H2]), or P-CaMKIV (T196),(Abcam [ab59424]) or CaMKIV rabbit antibody (Cell Signaling).
Immunofluorescence
Immunofluorescence analysis of CREB phosphorylation in hypothalamus was performed after 30 min of treatment with PBS or serotonin on 500-μm brain slices of wild-type or Htr2c−/− mice (as described previously). After treatment, the brain slices were fixed in 4% PFA overnight at 4°C, cryoprotected by overnight immersion in a 20% sucrose solution, embedded in Shandon Cryomatrix (Thermo Electron Corporation), and frozen at −80°C. Twenty-micrometer cryostat sections were cut in the coronal plane. For analysis of the CREB/P-CREB level, sections were dried for 20 min at room temperature, blocked in donkey serum, and then incubated in P-CREB (S133) rabbit antibody (Cell Signaling) and CREB mouse antibody (Cell Signaling) for 24 h at 4°C. Sections were rinsed and incubated with a donkey anti-rabbit-Cy2 antibody (1:1000; Cy2; Jackson ImmunoResearch) and a donkey anti-mouse Cy3 antibody (1:1000; Cy3; Jackson ImmunoResearch). For the analysis of P-CaMKIV/P-CREB, the sections were dried for 20 min at room temperature, blocked in donkey serum, and then incubated in P-CREB (S133) rabbit antibody (Cell Signaling [87G3]) for 24 h at 4°C, rinsed in PBS, and incubated with a donkey anti-rabbit Cy2 antibody (1:1000; Cy2; Jackson ImmunoResearch). Following P-CREB staining, sections were rinsed overnight at 4°C and processed for P-CaMKIV (Abcam [T196]) in the conditions described above for P-CREB using a secondary antibody donkey anti-rabbit- Cy3 antibody (1:1000; Cy3; Jackson ImmunoResearch). All slides were mounted with fluoro-gel II with DAPI (Electron Microscopy Sciences).
Microarray analysis
We microdissected wild-type and CrebSf1−/− mice hypothalami (from bregma −1.22 to −1.70 mm). Hypothalami RNA were extracted by Trizol and analyzed using Genechip Mouse Genome 430 2.0 Array. Data analyses were done by Precision Biomarker using the Affymetrix Expression Console software, and were normalized by robust multiarray average expression measure. The criteria for increased or decreased were P-value <0.05. The heat map in Figure 5, A and B, is a false-color image derived from the selected gene expression values with a dendrogram added to the left side and the top. Dendrograms are based on hierarchical clustering. Vertical lines indicate the distance between samples (columns) or genes (rows). The color red indicates higher relative expression as compared with blue. This heat map was generated using R.
Galactosidase staining
Galactosidase staining was performed on the tissues obtained from the Sf1-Cre/rosa26 mice following standard procedures. Briefly, tissue samples were dissected after intracardial perfusion with ice-cold 4% paraformaldehyde in PBS, and fixed for 1–2 h The samples were then washed three times with washing buffer (0.2% nonidet P-40, 0.1% sodium deoxycholate, 100 mM phosphate buffer at pH 7.4, 2 mM magnesium chloride) for 15–30 min each and then stained overnight (12–16 h) at 37°C in freshly prepared LacZ staining solution containing 0.2% nonidet P-40, 0.1% sodium deoxycholate, 100 mM phosphate buffer (pH 7.4), 2 mM magnesium chloride, 3 mM potassium ferricyanide, 3 mM potassium ferrocyanide, and 0.5 mg/mL X-gal (5-bromo-4-chloro-3-indolyl-D-galactopyranoside) protected from light. After staining overnight, tissues were photographed before being processed for paraffin embedding. Paraffin blocks were sectioned at 5- to 7-μm thickness, deparaffinized, counterstained with eosin, cleared in xylene, and mounted in DPX.
Cell culture and cell differentiation
P19 cells were cultured as described (McBurney 1993; Alam et al. 2009). To induce neuronal differentiation, P19 cells were allowed to aggregate in bacteriological dishes at 1 × 105 cells per milliliter in the presence of 1 μM all-trans-retinoic acid (Sigma-Aldrich) in α-MEM/10% FBS. After 4 d of aggregation, cells were dissociated into single cells by 0.25% trypsin and 1 μM EDTA solution, and were replated in a tissue culture dish at a density of 5 × 105 cells per milliliter. Cells were allowed to adhere and were cultured in the absence of 1 μM retinoic acid. Media were replaced every 48 h. After differentiation, cells were starved for at least 12 h with serum-free medium, then treated with 50 μM serotonin for up to 45 min. Phosphorylation of CaMKIV and CREB was analyzed by Western blot analysis every 5 min using P-CREB (S133) rabbit antibody (Cell Signaling [87G3]), CREB rabbit antibody (Cell Signaling [48H2]), and P-CaMKIV (T196) (Abcam [ab59424]) and CaMKIV rabbit antibody (Cell Signaling).
Bone histomorphometry analysis
Bone histomorphometry was performed as described previously (Yadav et al. 2009). Briefly, lumbar vertebrae were dissected, fixed for 24 h in 10% formalin, dehydrated in graded ethanol, and embedded in methyl methacrylate resin according to standard protocols. Von Kossa/Von Gieson staining was performed using 7-μm sections for bone volume over tissue volume (BV/TV) measurement. Bone formation rate (BFR) was analyzed by the calcein double-labeling method. Calcein (Sigma Chemical Co.) was dissolved in calcein buffer (0.15 M NaCl, 2% NaHCO3) and injected twice at 0.125 mg/g body weight on days 1 and 4, and mice were then killed on day 6. Four-micrometer sections were cleared in xylene and used for BFR measurements. For the analysis of osteoblast and osteoclast parameters, 4-μm sections were stained with toluidine blue and tartrate-resistant acid phosphatase (TRAP), respectively. Histomorphometric analyses were performed using the Osteomeasure Analysis System (Osteometrics).
μCT analysis
μCT analysis was performed on distal tibia using a μCT) system (VivaCT 40, SCANCO Medical AG). Tibial bone specimens were stabilized with gauze in a 2-mL centrifuge tube filled with 70% ethanol and fastened in the specimen holder of the μCT scanner. One-hundred μCT slices, corresponding to a 1.05-mm region distal from the growth plate, were acquired at an anisotropic spatial resolution of 10.5 μm. A global threshold technique was applied to binarize grayscale μCT images, where the minimum between the bone and bone marrow peaks in the voxel gray value histogram was chosen as the threshold value. The trabecular bone compartment was segmented by a semiautomatic contouring method, and was subjected to a model-independent morphological analysis (Hildebrand et al. 1999) by the standard software provided by the manufacturer of the μCT scanner. Three-dimensional morphological parameters—including bone volume fraction (BV/TV), trabecular thickness (Tb.Th.), and connectivity density (Conn.D)—were evaluated. The connectivity density is a quantitative description of the trabecular connection (Feldkamp et al. 1989; Gundersen et al. 1993).
Physiological measurements
For food intake studies, mice were housed individually in metabolic cages (Nalgene) and fed ad libitum. After a 30-h acclimation to the apparatus, data were collected for 24 h and analyzed as recommended by the manufacturer of the energy expenditure apparatus (Columbus Instruments). Food consumption was determined by weighing the powdered chow before and after the 2-h period. Oxygen consumption (VO2) and respiratory exchange ratio (RER) were measured by indirect calorimetry method using a six-chamber Oxymax system (Columbus Instruments).
Bioassays
Quantitative determination of CTx serum levels, a marker of osteoclast activity, was analyzed by enzyme immunoassay (Guerrero et al. 1996) (RatLaps EIA, Immunodiagnostic Systems). Urinary epinephrine contents were measured in acidified spot urine samples by EIA (Bi-CAT kit, Alpco), and Creatinine (Metra Creatinine kit, Quidel Corp.) was used to standardize theurine samples.
Statistical analyses
Results are given as mean ± standard error of the mean, except in human studies, in which standard deviations were used. Statistical analyses were performed using unpaired, two-tailed Student's t-test. For all experiments (*) P ≤ 0.05, (**) P ≤ 0.01, and (***) P ≤ 0.001.
Acknowledgments
We thank G. Ren for mouse genotyping and Dr. Patricia Ducy for critical reading of the manuscript. This work was supported by grants from the NIH (G.K., V.K.Y., and A.R.M.), a Rodan fellowship from IBMS (V.K.Y.), HFSP (F.O.), foundation Bettencourt Schueller (F.O.), and the Philippe foundation (F.O.).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1977210.
Supplemental material is available at http://www.genesdev.org.
References
- Alam AH, Suzuki H, Tsukahara T 2009. Expression analysis of Fgf8a &Fgf8b in early stage of P19 cells during neural differentiation. Cell Biol Int 33: 1032–1037 [DOI] [PubMed] [Google Scholar]
- Balthasar N, Coppari R, McMinn J, Liu SM, Lee CE, Tang V, Kenny CD, McGovern RA, Chua SC Jr, Elmquist JK, et al. 2004. Leptin receptor signaling in POMC neurons is required for normal body weight homeostasis. Neuron 42: 983–991 [DOI] [PubMed] [Google Scholar]
- Bartsch D, Ghirardi M, Skehel PA, Karl KA, Herder SP, Chen M, Bailey CH, Kandel ER 1995. Aplysia CREB2 represses long-term facilitation: Relief of repression converts transient facilitation into long-term functional and structural change. Cell 83: 979–992 [DOI] [PubMed] [Google Scholar]
- Bito H, Deisseroth K, Tsien RW 1996. CREB phosphorylation and dephosphorylation: A Ca2+- and stimulus duration-dependent switch for hippocampal gene expression. Cell 87: 1203–1214 [DOI] [PubMed] [Google Scholar]
- Bourtchuladze R, Frenguelli B, Blendy J, Cioffi D, Schutz G, Silva AJ 1994. Deficient long-term memory in mice with a targeted mutation of the cAMP-responsive element-binding protein. Cell 79: 59–68 [DOI] [PubMed] [Google Scholar]
- Cambi F, Fung B, Chikaraishi D 1989. 5′ flanking DNA sequences direct cell-specific expression of rat tyrosine hydroxylase. J Neurochem 53: 1656–1659 [DOI] [PubMed] [Google Scholar]
- Corcoran EE, Means AR 2001. Defining Ca2+/calmodulin-dependent protein kinase cascades in transcriptional regulation. J Biol Chem 276: 2975–2978 [DOI] [PubMed] [Google Scholar]
- Dash PK, Hochner B, Kandel ER 1990. Injection of the cAMP-responsive element into the nucleus of Aplysia sensory neurons blocks long-term facilitation. Nature 345: 718–721 [DOI] [PubMed] [Google Scholar]
- Drago A, Serretti A 2009. Focus on HTR2C: A possible suggestion for genetic studies of complex disorders. Am J Med Genet B Neuropsychiatr Genet 150B: 601–637 [DOI] [PubMed] [Google Scholar]
- Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G 1997. Osf2/Cbfa1: A transcriptional activator of osteoblast differentiation. Cell 89: 747–754 [DOI] [PubMed] [Google Scholar]
- Ducy P, Starbuck M, Priemel M, Shen J, Pinero G, Geoffroy V, Amling M, Karsenty G 1999. A Cbfa1-dependent genetic pathway controls bone formation beyond embryonic development. GenesDev 13: 1025–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducy P, Amling M, Takeda S, Priemel M, Schilling AF, Beil FT, Shen J, Vinson C, Rueger JM, Karsenty G 2000. Leptin inhibits bone formation through a hypothalamic relay: A central control of bone mass. Cell 100: 197–207 [DOI] [PubMed] [Google Scholar]
- Edelman AM, Mitchelhill KI, Selbert MA, Anderson KA, Hook SS, Stapleton D, Goldstein EG, Means AR, Kemp BE 1996. Multiple Ca2+–calmodulin-dependent protein kinase kinases from rat brain. Purification, regulation by Ca2+–calmodulin, and partial amino acid sequence. J Biol Chem 271: 10806–10810 [DOI] [PubMed] [Google Scholar]
- Elefteriou F, Ahn JD, Takeda S, Starbuck M, Yang X, Liu X, Kondo H, Richards WG, Bannon TW, Noda M, et al. 2005. Leptin regulation of bone resorption by the sympathetic nervous system and CART. Nature 434: 514–520 [DOI] [PubMed] [Google Scholar]
- Elefteriou F, Benson MD, Sowa H, Starbuck M, Liu X, Ron D, Parada LF, Karsenty G 2006. ATF4 mediation of NF1 functions in osteoblast reveals a nutritional basis for congenital skeletal dysplasiae. Cell Metab 4: 441–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldkamp LA, Goldstein SA, Parfitt AM, Jesion G, Kleerekoper M 1989. The direct examination of three-dimensional bone architecture in vitro by computed tomography. J Bone Miner Res 4: 3–11 [DOI] [PubMed] [Google Scholar]
- Fink KB, Gothert M 2007. 5-HT receptor regulation of neurotransmitter release. Pharmacol Rev 59: 360–417 [DOI] [PubMed] [Google Scholar]
- Franklin KBJ, Paxinos G 1997. The mouse brain in stereotaxic coordinates. Academic Press, San Diego, CA [Google Scholar]
- Fu L, Patel MS, Bradley A, Wagner EF, Karsenty G 2005. The molecular clock mediates leptin-regulated bone formation. Cell 122: 803–815 [DOI] [PubMed] [Google Scholar]
- Gershon MD, Tack J 2007. The serotonin signaling system: From basic understanding to drug development for functional GI disorders. Gastroenterology 132: 397–414 [DOI] [PubMed] [Google Scholar]
- Guerrero R, Diaz Martin MA, Diaz Diego EM, Disla T, Rapado A, de la Piedra C 1996. New biochemical markers of bone resorption derived from collagen breakdown in the study of postmenopausal osteoporosis. Osteoporos Int 6: 297–302 [DOI] [PubMed] [Google Scholar]
- Gundersen HJ, Boyce RW, Nyengaard JR, Odgaard A 1993. The Conneulor: Unbiased estimation of connectivity using physical disectors under projection. Bone 14: 217–222 [DOI] [PubMed] [Google Scholar]
- Haribabu B, Hook SS, Selbert MA, Goldstein EG, Tomhave ED, Edelman AM, Snyderman R, Means AR 1995. Human calcium–calmodulin dependent protein kinase I: cDNA cloning, domain structure and activation by phosphorylation at threonine-177 by calcium–calmodulin dependent protein kinase I kinase. EMBO J 14: 3679–3686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath MJ, Hen R 1995. Serotonin receptors. Genetic insights into serotonin function. Curr Biol 5: 997–999 [DOI] [PubMed] [Google Scholar]
- Hildebrand T, Laib A, Müller R, Dequeker J, Rüegsegger P 1999. Direct three-dimensional morphometric analysis of human cancellous bone: Microstructural data from spine, femur, iliac crest, and calcaneus. J Bone Miner Res 14: 1167–1174 [DOI] [PubMed] [Google Scholar]
- Hook SS, Means AR 2001. Ca2+/CaM-dependent kinases: From activation to function. Annu Rev Pharmacol Toxicol 41: 471–505 [DOI] [PubMed] [Google Scholar]
- Jochum W, Passegue E, Wagner EF 2001. AP-1 in mouse development and tumorigenesis. Oncogene 20: 2401–2412 [DOI] [PubMed] [Google Scholar]
- Jones DC, Wein MN, Oukka M, Hofstaetter JG, Glimcher MJ, Glimcher LH 2006. Regulation of adult bone mass by the zinc finger adapter protein Schnurri-3. Science 312: 1223–1227 [DOI] [PubMed] [Google Scholar]
- Josselyn SA, Nguyen PV 2005. CREB, synapses and memory disorders: Past progress and future challenges. Curr Drug Targets CNS Neurol Disord 4: 481–497 [DOI] [PubMed] [Google Scholar]
- Karsenty G 2006. Convergence between bone and energy homeostases: Leptin regulation of bone mass. Cell Metab 4: 341–348 [DOI] [PubMed] [Google Scholar]
- Kitani T, Okuno S, Fujisawa H 1997. Molecular cloning of Ca2+/calmodulin-dependent protein kinase kinase β. J Biochem 122: 243–250 [DOI] [PubMed] [Google Scholar]
- Lonze BE, Ginty DD 2002. Function and regulation of CREB family transcription factors in the nervous system. Neuron 35: 605–623 [DOI] [PubMed] [Google Scholar]
- Mann JJ, McBride PA, Brown RP, Linnoila M, Leon AC, DeMeo M, Mieczkowski T, Myers JE, Stanley M 1992. Relationship between central and peripheral serotonin indexes in depressed and suicidal psychiatric inpatients. Arch Gen Psychiatry 49: 442–446 [DOI] [PubMed] [Google Scholar]
- McBurney MW 1993. P19 embryonal carcinoma cells. Int J Dev Biol 37: 135–140 [PubMed] [Google Scholar]
- Mizuno K, Ris L, Sanchez-Capelo A, Godaux E, Giese KP 2006. Ca2+/calmodulin kinase kinase α is dispensable for brain development but is required for distinct memories in male, though not in female, mice. Mol Cell Biol 26: 9094–9104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathanson NM 2008. Synthesis, trafficking, and localization of muscarinic acetylcholine receptors. Pharmacol Ther 119: 33–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestler EJ, Barrot M, DiLeone RJ, Eisch AJ, Gold SJ, Monteggia LM 2002. Neurobiology of depression. Neuron 34: 13–25 [DOI] [PubMed] [Google Scholar]
- Oury F, Murakami Y, Renaud JS, Pasqualetti M, Charnay P, Ren SY, Rijli FM 2006. Hoxa2- and rhombomere-dependent development of the mouse facial somatosensory map. Science 313: 1408–1413 [DOI] [PubMed] [Google Scholar]
- Peltier J, O'Neill A, Schaffer DV 2007. PI3K/Akt and CREB regulate adult neural hippocampal progenitor proliferation and differentiation. Dev Neurobiol 67: 1348–1361 [DOI] [PubMed] [Google Scholar]
- Peters M, Mizuno K, Ris L, Angelo M, Godaux E, Giese KP 2003. Loss of Ca2+/calmodulin kinase kinase β affects the formation of some, but not all, types of hippocampus-dependent long-term memory. J Neurosci 23: 9752–9760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrij F, Giles RH, Dauwerse HG, Saris JJ, Hennekam RC, Masuno M, Tommerup N, van Ommen GJ, Goodman RH, Peters DJ, et al. 1995. Rubinstein-Taybi syndrome caused by mutations in the transcriptional co-activator CBP. Nature 376: 348–351 [DOI] [PubMed] [Google Scholar]
- Rached MT, Kode A, Silva BC, Jung DY, Gray S, Ong H, Paik JH, DePinho RA, Kim JK, Karsenty G, et al. 2010a. FoxO1 expression in osteoblasts regulates glucose homeostasis through regulation of osteocalcin in mice. J Clin Invest 120: 357–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rached MT, Kode A, Xu L, Yoshikawa Y, Paik JH, Depinho RA, Kousteni S 2010b. FoxO1 is a positive regulator of bone formation by favoring protein synthesis and resistance to oxidative stress in osteoblasts. Cell Metab 11: 147–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribar TJ, Rodriguiz RM, Khiroug L, Wetsel WC, Augustine GJ, Means AR 2000. Cerebellar defects in Ca2+/calmodulin kinase IV-deficient mice. J Neurosci 20: RC107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatakos G, Sims NA, Chen J, Aoki K, Kelz MB, Amling M, Bouali Y, Mukhopadhyay K, Ford K, Nestler EJ, et al. 2000. Overexpression of ΔFosB transcription factor(s) increases bone formation and inhibits adipogenesis. Nat Med 6: 985–990 [DOI] [PubMed] [Google Scholar]
- Schulman H, Heist K, Srinivasan M 1995. Decoding Ca2+ signals to the nucleus by multifunctional CaM kinase. Prog Brain Res 105: 95–104 [DOI] [PubMed] [Google Scholar]
- Soderling TR, Stull JT 2001. Structure and regulation of calcium/calmodulin-dependent protein kinases. Chem Rev 101: 2341–2352 [DOI] [PubMed] [Google Scholar]
- Takeda S, Elefteriou F, Levasseur R, Liu X, Zhao L, Parker KL, Armstrong D, Ducy P, Karsenty G 2002. Leptin regulates bone formation via the sympathetic nervous system. Cell 111: 305–317 [DOI] [PubMed] [Google Scholar]
- Tohda M, Nomura M, Nomura Y 2006. Molecular pathopharmacology of 5-HT2C receptors and the RNA editing in the brain. J Pharmacol Sci 100: 427–432 [DOI] [PubMed] [Google Scholar]
- Tokumitsu H, Soderling TR 1996. Requirements for calcium and calmodulin in the calmodulin kinase activation cascade. J Biol Chem 271: 5617–5622 [DOI] [PubMed] [Google Scholar]
- Tokumitsu H, Inuzuka H, Ishikawa Y, Kobayashi R 2003. A single amino acid difference between α and β Ca2+/calmodulin-dependent protein kinase kinase dictates sensitivity to the specific inhibitor, STO-609. J Biol Chem 278: 10908–10913 [DOI] [PubMed] [Google Scholar]
- van Koppen CJ, Kaiser B 2003. Regulation of muscarinic acetylcholine receptor signaling. Pharmacol Ther 98: 197–220 [DOI] [PubMed] [Google Scholar]
- Wagner EF, Eferl R 2005. Fos/AP-1 proteins in bone and the immune system. Immunol Rev 208: 126–140 [DOI] [PubMed] [Google Scholar]
- Walther DJ, Peter JU, Bashammakh S, Hortnagl H, Voits M, Fink H, Bader M 2003. Synthesis of serotonin by a second tryptophan hydroxylase isoform. Science 299: 76. [DOI] [PubMed] [Google Scholar]
- Walther A, Petri E, Peter C, Czabanka M, Martin E 2007. Selective serotonin-receptor antagonism and microcirculatory alterations during experimental endotoxemia. J Surg Res 143: 216–223 [DOI] [PubMed] [Google Scholar]
- Wayman GA, Kaech S, Grant WF, Davare M, Impey S, Tokumitsu H, Nozaki N, Banker G, Soderling TR 2004. Regulation of axonal extension and growth cone motility by calmodulin-dependent protein kinase I. J Neurosci 24: 3786–3794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayman GA, Impey S, Marks D, Saneyoshi T, Grant WF, Derkach V, Soderling TR 2006. Activity-dependent dendritic arborization mediated by CaM-kinase I activation and enhanced CREB-dependent transcription of Wnt-2. Neuron 50: 897–909 [DOI] [PubMed] [Google Scholar]
- Yadav VK, Ryu JH, Suda N, Tanaka KF, Gingrich JA, Schutz G, Glorieux FH, Chiang CY, Zajac JD, Insogna KL, et al. 2008. Lrp5 controls bone formation by inhibiting serotonin synthesis in the duodenum. Cell 135: 825–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav VK, Oury F, Suda N, Liu ZW, Gao XB, Confavreux C, Klemenhagen KC, Tanaka KF, Gingrich JA, Guo XE, et al. 2009. A serotonin-dependent mechanism explains the leptin regulation of bone mass, appetite, and energy expenditure. Cell 138: 976–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav VK, Balaji S, Suresh PS, Liu XS, Lu X, Li Z, Guo XE, Mann JJ, Balapure AK, Gershon MD, et al. 2010. Pharmacological inhibition of gut-derived serotonin synthesis is a potential bone anabolic treatment for osteoporosis. Nat Med 16: 308–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Matsuda K, Bialek P, Jacquot S, Masuoka HC, Schinke T, Li L, Brancorsini S, Sassone-Corsi P, Townes TM, et al. 2004. ATF4 is a substrate of RSK2 and an essential regulator of osteoblast biology; implication for Coffin-Lowry syndrome. Cell 117: 387–398 [DOI] [PubMed] [Google Scholar]
- Yin JC, Wallach JS, Del Vecchio M, Wilder EL, Zhou H, Quinn WG, Tully T 1994. Induction of a dominant negative CREB transgene specifically blocks long-term memory in Drosophila. Cell 79: 49–58 [DOI] [PubMed] [Google Scholar]
- Yin JC, Del Vecchio M, Zhou H, Tully T 1995. CREB as a memory modulator: Induced expression of a dCREB2 activator isoform enhances long-term memory in Drosophila. Cell 81: 107–115 [DOI] [PubMed] [Google Scholar]