Abstract
Green photosynthetic bacteria harvest light and perform photosynthesis in low-light environments, and contain specialized antenna complexes to adapt to this condition. We performed small-angle neutron scattering (SANS) studies to obtain structural information about the photosynthetic apparatus, including the peripheral light-harvesting chlorosome complex, the integral membrane light-harvesting B808-866 complex, and the reaction center (RC) in the thermophilic green phototrophic bacterium Chloroflexus aurantiacus. Using contrast variation in SANS measurements, we found that the B808-866 complex is wrapped around the RC in Cfx. aurantiacus, and the overall size and conformation of the B808-866 complex of Cfx. aurantiacus is roughly comparable to the LH1 antenna complex of the purple bacteria. A similar size of the isolated B808-866 complex was suggested by dynamic light scattering measurements, and a smaller size of the RC of Cfx. aurantiacus compared to the RC of the purple bacteria was observed. Further, our SANS measurements indicate that the chlorosome is a lipid body with a rod-like shape, and that the self-assembly of bacteriochlorophylls, the major component of the chlorosome, is lipid-like. Finally, two populations of chlorosome particles are suggested in our SANS measurements.
Introduction
Photosynthesis is the ultimate source of all food and most energy resources on Earth, and photosynthetic organisms use solar energy to drive the synthesis of organic compounds such as biofuels and biomass. Photosynthetic organisms such as bacteria, algae, and plants use light-harvesting complexes (LHCs) to capture light energy and transfer the excitation energy to reaction centers (RCs), where electron transfer for photochemistry takes place. To perform these complex biological processes, various types of phototrophic bacteria employ distinct photosynthetic mechanisms that are simpler than those found in algae and higher plants. The photosynthetic apparatus includes LHCs and RCs, which together harvest solar energy and convert the excited light energy into chemical energy that is stored for use in cellular metabolism and function (1).
Several biophysical approaches, including x-ray crystallography, electron microscopy (EM), mass spectrometry, and many types of optical spectroscopy, have been used to investigate the structure of the photosynthetic machinery in various phototrophic organisms (2). In addition to these techniques, small-angle neutron scattering (SANS) has been demonstrated to be a reliable technique for investigating (bio)macromolecular assemblies in solution (3–5), with some advantages and disadvantages compared to small-angle x-ray scattering (SAXS) (6,7). However, to date there have been very few applications of SANS to photosynthetic complexes. Although both coherent and incoherent scattering lengths contribute to the neutron scattering, SANS is a coherent scattering method and thus considers only the coherent scattering length. One of the advantages of SANS compared to SAXS in structural studies is that SANS uses contrast variation. Deuterium has a much larger coherent scattering length (6.67 × 10−13 cm) than hydrogen (−3.74 × 10−13 cm), and, in contrast, hydrogen has much larger incoherent scattering length (25.18 × 10−13 cm) than deuterium (3.99 × 10−13 cm) (8). A few previous studies have examined photosynthetic pigment-protein complexes using SANS or neutron diffraction (9–13), and SANS studies have also been reported for various isoforms of isolated bacteriochlorophylls (BChls) (14–16). However, to our knowledge, no previous SANS reports have been concerned with the photosynthetic machinery of the green photosynthetic bacteria, including green sulfur bacteria (GSBs) and filamentous anoxygenic phototrophic (FAP) bacteria, which are known to efficiently harvest light in low-light environments (17).
In this study, we sought to obtain solution structural insights into the photosynthetic machinery of Chloroflexus (Cfx.) aurantiacus, one of the most investigated FAP bacteria (18). The proposed energy transfer in the photosynthetic machinery of Cfx. aurantiacus is shown in Fig. 1 A. In Cfx. aurantiacus, the light energy absorbed by the chlorosome is transferred to the chlorosome baseplate, the integral membrane light-harvesting B808-866 complex (19–22), and then to the RC. The absorption spectra of the intact membrane, chlorosome, B808-866 complex, and RC of Cfx. aurantiacus are shown in Fig. 1 B. The B808-866 complex of Cfx. aurantiacu has been proposed to function similarly to the B880 LHC I (LH1) of the purple photosynthetic bacteria (17). The crystal structure for the LH1-RC core complex of the purple bacterium Rhodopseudomonas (Rps.) palustris indicates that one RC is located within one LH1 ring (23,24), whereas a dimeric RC-LH1 complex was observed in Rhodobacter sphaeroides by atomic force microscopy (AFM) (25,26). The RC and B808-866 complex assembly is an essential element in the attempt to obtain more detailed structural information about the photosystem of Cfx. aurantiacus.
Figure 1.
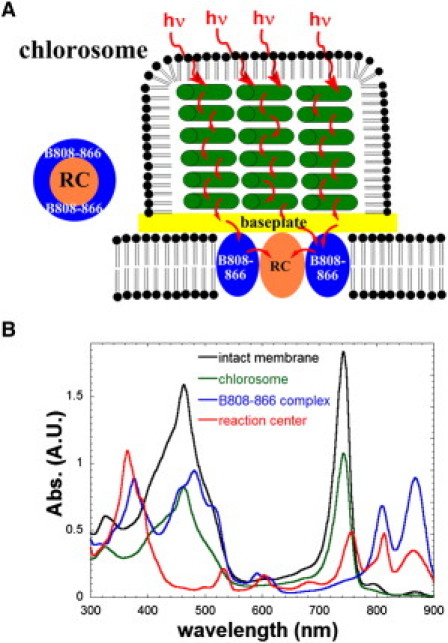
(A) The proposed photosystem. (B) Absorption spectra of the intact membrane, chlorosome, B808-866 complex, and RC of Cfx. aurantiacus.
The chlorosome, the peripheral light-harvesting antenna complex located on the cytoplasmic side of the inner membrane in green photosynthetic bacteria, has little protein associated and contains 135,000–300,000 self-assembled BChls (27). The BChl self-assemblies in the chlorosome can absorb light in the red to near-infrared region, and thus the green photosynthetic bacteria can transfer excitation energy efficiently under low-light intensity environments. No atomic-resolution structural information has been reported for the chlorosome of Cfx. aurantiacu. In this study, we investigated the size and overall conformation of the B808-866 complex using SANS and dynamic light scattering (DLS), and compared the solution conformation of the B808-866 complex and RC of Cfx. aurantiacus with the crystal structure for the LH1 and RC of Rps. palustris (23). Moreover, to verify SANS studies for Cfx. aurantiacus, we compared the solution conformation acquired by SANS with crystal structures for the RC and peripheral B800-850 LHC II (LH2) of the purple bacteria. Finally, the structural information of the chlorosome was probed by SANS.
Materials and Methods
Sample preparation
Chloroflexus aurantiacus strain J-10-fl was cultured anaerobically under low-light conditions as described previously (28). RC-lauryldimethylamine N-oxide (RC-LDAO), B808-866-octyl-β-D-glucopyranoside (βOG), and the chlorosome for Cfx. aurantiacus (19,29–32), and B800-850 LHC II (LH2)-LDAO mixtures (33) for Rhodobacter sphaeroides were prepared as reported previously. The critical micelle concentration (CMC) of LDAO (0.04% (w/v)) and βOG micelles (0.8%) (34,35) was included in the RC-LDAO and B808-866-βOG mixtures in 20 mM Tris-HCl buffer at pH 8.0, and the chlorosome were prepared in 20 mM Tris-HCl buffer at pH 8.0. Samples for contrast variation were prepared as follows: for Rb. sphaeroides: 100% D2O for RC-LDAO mixtures, and 0, 10, 20, 45, 80, and 100% D2O for LH2-LDAO mixtures; for Cfx. aurantiacus: 0, 25, 40, 60, 80, and 100% D2O for the chlorosome, and 0, 17, 40, 80, and 100% D2O for B808-866-βOG mixtures, and 5, 40, and 100% D2O for RC-LDAO mixtures.
General principle of contrast variation
The values of neutron scattering density for H2O and D2O are −5 × 109 cm−2 and 64 × 109 cm−2, respectively, and one can be achieve different solvent scattering densities by mixing different ratios of H2O and D2O in solution. Also, the average scattering density of various biological molecules is varied in different D2O/H2O ratios in solution. Contrast matching occurs when the average scattering density of a biological molecule matches the solvent scattering density in a D2O/H2O ratio. The matching point is 40–45% D2O for proteins, 65–70% D2O for nucleic acids, and 5–25% D2O for lipids.
SANS data collection and analysis
We performed SANS using the CG-3 Bio-SANS instruments of the Center for Structural Molecular Biology at the High Flux Isotope Reactor of Oak Ridge National Laboratory. The data were collected with a two-dimensional 0.4 m × 0.4 m position-sensitive He3 detector (Ordela, Oak Ridge, TN). The scattering data were collected at different sample-to-detector distances and covered the desired q range (q (momentum transfer vector) = 4πsinθ/λ, where 2θ is the scattering angle). Two sample-to-detector distances (1.1 m and 6.8 m), with neutron wavelength (λ) = 6.0 Å and wavelength spread (Δλ/λ) = 0.15, were applied to collect scattering data in 0.0064 < q < 0.064 Å−1 (6.8 m) and 0.026 < q < 0.35 Å−1 (1.1 m) for the samples reported here. Further, one additional instrument configuration, with neutron wavelength = 18 Å and sample-to-detector distance = 15.3 m, was used to collect the data in the low-q region (0.0009 < q < 0.01 Å−1) for the chlorosome. Both protein and buffer scattering profiles were collected by identical procedures and the buffer scattering profiles were subtracted for background correction. SANS data reduction further included normalization for flux, sample transmission, and sample thickness; correction for instrument-specific (dark) background; and azimuthal averaging, which provides scattering intensities per solid angle as a function of q. Detailed information on the data analysis and modeling is provided in the Supporting Material.
Results
SANS for the LH2 and RC of Rhodobacter sphaeroides
In nonsulfur purple bacteria, light harvesting is accomplished by two types of antenna complexes: the peripheral B800-850 LHC II (LH2) and core LHC I (LH1) in the RC-LH1 core complex. LH1 and LH2 have generally similar ring structures, although the LH2, composed of nine subunits of the αβ-heterodimer, has a smaller ring size than the LH1, which consists of 16 subunits of the αβ-heterodimer. In contrast to the photosynthetic machinery of Cfx. aurantiacus, the atomic-resolution structures for LH2 and RC of the purple bacteria are available. Therefore, we first performed SANS for the LH2 and RC of the purple bacterium Rb. sphaeroides and compared their solution conformation obtained by SANS with the crystal structures for the LH2 of Rps. acidophila (PDB ID: 1NKZ) (36) and the RC of Rb. sphaeroides (PDB ID: 1AIJ) (32). Fig. 2, A (SANS data) and B (Guinier fits), show SANS measurements for LH2-LDAO mixtures of Rb. sphaeroides in 0, 10, 20, 45, 80, and 100% D2O, and Fig. 2 E shows the SANS data and Guinier fit (inset) for RC-LDAO mixtures in 100% D2O. The weakest scattering for LH2-LDAO mixtures was acquired in 20% D2O, which can be treated as an average matching point between the matching points for the LH2 peptides (in 45% D2O) and bound LDAO micelles and cofactors, i.e., BChls and carotenoids (in 5–10% D2O). In addition, bound LDAO micelles and cofactors in the LH2-LDAO mixtures contributed to stronger scattering intensity in 45% D2O than in 10% D2O.
Figure 2.
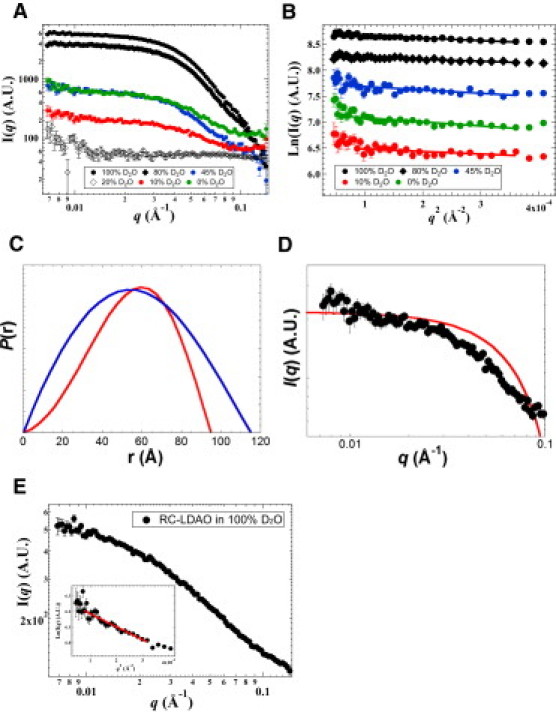
SANS for the LH2 and RC of Rb. sphaeroides. (A) SANS data for LH2-LDAO mixtures in 0, 10, 20, 45, 80, and 100% D2O. (B) The Guinier fit for LH2-LDAO mixtures in different concentrations of D2O. Data are shifted along the y axis for comparison. (C) The normalized P(r) profile for LH2-LDAO mixtures in 45% (blue curve) and 10% D2O (red curve). (D) The SANS data for LH2-LDAO mixtures in 10% D2O (•) and the predicted SANS pattern (red line) calculated from the crystal structure for LH2 of Rps. acidophila. (E) The SANS data and Guinier fit (inset) for RC-LDAO mixtures in 100% D2O.
Table 1 and Fig. 2 C show a larger radius of gyration, Rg, and longest distance within the particle, Dmax, for LH2-LDAO mixtures in 45% D2O than in 10% D2O, indicating that the LH2 is surrounded by LDAO micelles. The Dmax-values were obtained by several GNOM fits (37) with various Rmax-values input to acquire the most reasonable particle distance distribution function, P(r), because an inappropriate value of Dmax generates a rather unreasonable P(r) distribution. The GNOM program is an indirect Fourier transform (IFT) method, and the Rg- and Dmax-values determined by GNOM make use of the entire scattering curve and are less sensitive to intermolecular interactions (see the Supporting Material). Also, the larger Rg-value (36 ± 2 Å) estimated for RC-LDAO mixtures in 100% D2O compared to the value calculated from the crystal structure of Rb. sphaeroides (33.2 Å) with the program CRYSON (38) suggest that LDAO micelles occupy the periphery of the LH2. Assuming that the thickness of the LH2 particle is ∼47 Å (estimated from the crystal structure) (36), the ring diameter for LH2-LDAO mixtures in 45% D2O and 10% D2O is ∼105 Å (Dmax, 115 ± 5 Å) and ∼83 Å (Dmax, 95 ± 5 Å), respectively. Thus, an ∼11 Å detergent layer surrounding the LH2, with the increase of the total ring size by ∼22 Å, was suggested by SANS. The larger error for fitting the Rg-value for LH2-LDAO mixtures in 0% and 10% D2O is due to weaker scattering. Fig. 2 D illustrates that the predicted SANS curve calculated from the crystal structure of the LH2 of Rps. acidophila is comparable to the data collected for LH2-LDAO mixtures of Rb. sphaeroides in 10% D2O. Together with similar experimental and predicted Rg-values (Table 1) and no upturn curvature for the data of LH2-LDAO mixtures in 100% and 80% D2O (Fig. 2 A), our studies suggest a similar overall conformation for the LH2 of Rb. sphaeroides in solution and Rps. acidophila in the reported crystal structure (36) LH2 aggregates larger than the (αβ)9 oligomer reported previously (39,40) cannot be detected in our studies.
Table 1.
Structural parameters acquired from SANS for the LH2 and RC of Rb. sphaeroides and estimated from the crystal structures for the RC and LH2 of the purple bacteria
| Samples |
Rg (Guinier) |
Rg (CRYSON) |
Rg (GNOM) |
Dmax (GNOM) |
|---|---|---|---|---|
| LH2-LDAO (Rb. sphaeroides) | ||||
| in 0% D2O | 38 ± 11 Å | 39 ± 3 Å | 110 ± 5 Å | |
| in 10% D2O | 30 ± 14 Å | 35 ± 5 Å | 95 ± 5 Å | |
| in 20% D2O | ND∗ | NDa | NDa | |
| in 45% D2O | 40 ± 4 Å | 38 ± 3 Å | 115 ± 5 Å | |
| in 80% D2O | 35 ± 2 Å | 37 ± 1 Å | 115 ± 2 Å | |
| in 100% D2O | 36 ± 2 Å | 37 ± 1 Å | 115 ± 3 Å | |
| LH2 (PDB ID: 1NKZ) Rps. acidophila | 33.2 Å | 34 ± 1 Å | 90 ± 2 Å | |
| RC-LDAO (Rb. sphaeroides) in 100% D2O | 38 ± 2 Å | 35± 1 Å | 95 ± 5 Å | |
| RC (Rb. Sphaeroides) (PDB ID: 1AIJ) | 29.4 Å | 30 ± 0.4 Å | 85 ± 2 Å | |
ND, a reasonable value cannot be obtained due to weak signal.
Further, the reduced frequency of both shorter and longer pair distances in the 10% D2O data as compared to the 45% D2O data (Fig. 2 C) suggests a hollow structure for LH2 (reduced frequency of the shorter pair distance) and LDAO micelles occupying the central cavity (increased frequency of the shorter pair distance) as well as surrounding the LH2 barrel (increased frequency of the larger pair distance).
Measurements of the B808-866 complex and RC of Cfx. aurantiacus
SANS with contrast variation was performed at 0, 17, 40, 80, and 100% D2O for B808-866-βOG mixtures (Fig. 3 A), and at 5, 40, and 100% D2O for RC-LDAO mixtures (Fig. S2 A). The overall size of the B808-866-βOG and RC-LDAO complex can be estimated from SANS in 100% D2O, in which the highest scattering contrast can be acquired. With the Guinier fit for the data in the low-q range (q < 0.02 Å−1), the Rg-value is 60 ± 1 Å (Rg × qmax = 0.8) for B808-866-βOG mixtures (Fig. 3 B) and 32 ± 1 Å (Rg × qmax = 0.7) for RC-LDAO mixtures (Fig. S2 B). Similar Rg-values for these biological complexes in 100% D2O were also acquired with the GNOM fits (Table 2).
Figure 3.
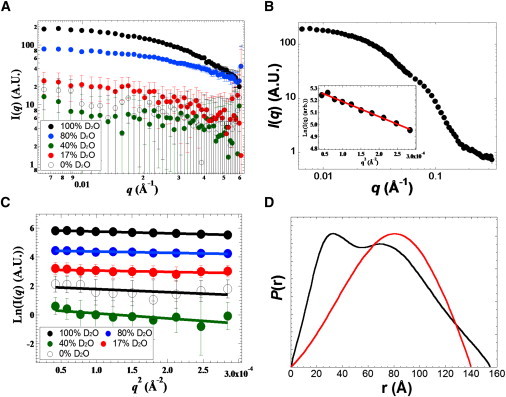
SANS for the B808-866 complex of Cfx. aurantiacus. (A) The SANS data for B808-866-βOG mixtures in 0, 17, 40, 80, and 100% D2O. (B) The SANS data and Guinier analysis (inset) for B808-866-βOG mixtures in 100% D2O. (C) The Guinier fit for B808-866-βOG mixtures in different concentrations of D2O. (D) The normalized P(r) profile for B808-866-βOG mixtures in 100% (black curve) and 17% D2O (red curve).
Table 2.
Structural parameters acquired from SANS for Cfx. aurantiacus and estimated from the crystal structure for the RC and LH1 of Rps. palustris
| Samples |
Rg (Guinier) |
Rc (modified Guinier) |
Rg (CRYSON) |
Rg (GNOM) |
Dmax (GNOM) |
|---|---|---|---|---|---|
| B808-866-βOG (Cfx. aurantiacu) | |||||
| in 100% D2O | 60 ± 1 Å | 56 ± 1 Å | 155 ± 5 Å | ||
| in 80% D2O | 54 ± 1 Å | 55 ± 1 Å | 155 ± 5 Å | ||
| in 40% D2O | 82 ± 30 Å | 65 ± 10 Å | 165 ± 10 Å | ||
| in 17% D2O | 52 ± 6 Å | 50 ± 2 Å | 140 ± 5 Å | ||
| in 0% D2O | 69 ± 20 Å | 60 ± 8 Å | 160 ± 10 Å | ||
| free βOG (in 100% D2O) | 23 ± 1 Å | 22 ± 1 Å | 70 ± 2.5 Å | ||
| LH1 (PDB ID: 1PYH) (Rps. palustris) | 47.2 Å | 47.4 Å | 124 ± 2 Å | ||
| RC-LDAO (Cfx.aurantiacus) | |||||
| in 100% D2O | 32 ± 1 Å | 28 ± 1 Å | 75 ± 5 Å | ||
| in 40% D2O | 28 ± 2 Å | 26 ± 1 Å | 70 ± 5 Å | ||
| in 5% D2O | 26 ± 10 Å | 23 ± 5 Å | 65 ± 5 Å | ||
| free LDAO (in 100% D2O) | 25 ± 3 Å | 26 ± 1 Å | 75 ± 3 Å | ||
| RC (PDB ID: 1PYH) (Rps. palustris) | 29.7 Å (or 27.1 Å w/o H-subunit) | 29.8 Å (or 27.2 Å w/o H-subunit) | 87 ± 2 Å (or 80 ± 2 Å w/o H-subunit) | ||
| Chlorosome (in 100% D2O) (Cfx. aurantiacus) | 196 ± 2 Å, 296 ± 6 Å | 242 ± 2 Å | 785 ± 5 Å | ||
To obtain more structural information about the B808-866 complex and RC of Cfx. aurantiacus, we analyzed SANS data for B808-866-βOG and RC-LDAO mixtures at the contrast point of βOG micelles (17% D2O) and LDAO micelles (5–10% D2O). The Rg-value for B808-866-βOG mixtures in 17% D2O is 52 ± 11 Å using the Guinier approximation (Fig. 3 C) and 50 ± 5 Å using the program GNOM (37) (Table 1). The Rg-value for RC-LDAO mixtures in 5% D2O is 26 ± 10 Å using the Guinier approximation and 23 ± 4 Å using the IFT approach (Table 2).
Data analysis of B808-866-βOG mixtures
The maximal diameter of the LH1 of R. rubrum has been reported to be 116 Å by EM (41), and Table 2 shows that the predicted Dmax for the LH1 of Rps. palustris from the crystal structure is 124 ± 2 Å. As the thickness of the LH1 particle in the reported crystal structure (23) is ∼47 Å, the diameter for the LH1 ring is estimated to be ∼115 Å, which is similar to the data reported by EM. The Dmax estimated from SANS for B808-866-βOG mixtures in 17% D2O, where only B808-866 polypeptides contribute to the scattering profile, is 140 ± 5 Å, and the diameter for the B808-866 complex is estimated to be 132 ± 5 Å. Similar to the P(r) profile obtained for the LH2 of Rb. sphaeroides (Fig. 2 C), a hollow structure is also suggested for the B808-866 complex as reduced frequency of the shorter pair distance in the P(r) profile (Fig. 3 D, red curve). The Guinier fit for the scattering data (q < 0.017 Å−1 for B808-866-βOG mixtures in 0, 17, 40, 80, and 100% D2O) is shown in Fig. 3 C. The Rg-value for the B808-866 complex determined from SANS is somewhat larger than the LH1 estimated from the crystal structure of the LH1-RC cocomplex: 47.2 Å (Table 2). These results suggest that the ring size of the B808-866 complex is a bit larger than that of the LH1. Further, the Dmax of B808-866-βOG mixtures in 100% D2O is estimated to be 155 ± 5 Å (Fig. 3 D), and the diameter for the B808-866 complex with bound micelles is ∼148 Å. These results indicate that the overall size of B808-866-βOG mixtures is increased with the micelle bound, and the detergent layer increases ∼15 Å of the overall size of the B808-866-βOG mixtures, in similarity to the detergent layer reported for the spinach LHC II-βOG complex (10). Thus it is possible to identify the thickness of the micelle layer using contrast variation, although the size of bound micelles cannot be accurately calculated due to the weak signal/noise ratio of the scattering signal in 40% D2O. Also, a peak at the shorter pair distance (∼30 Å) in the P(r) profile of B808-866-βOG mixtures in 100% D2O (Fig. 3 D, black curve) may be attributed to the presence of free βOG micelles in B808-866-βOG mixtures, although the detergent included in the samples reported here was prepared around its CMC. Additionally, assuming free βOG micelles as a polydisperse system of cylinders, the Rg (22–23 Å) and maximal length of free βOG micelles (0.8%) are estimated to be smaller than those of the bound micelles (Table S1).
To further investigate the particle size of B808-866-βOG mixtures, we employed DLS in addition to SANS. SANS, via elastic coherent neutron scattering, and DLS, via quasi-elastic light scattering, have been used to investigate polymer-surfactant interactions (42,43). Although both SANS and DLS can probe the size of the particles, different approaches are employed to acquire the information. The particle size estimated from SANS is generated from neutrons interacting with nuclei in the particle, whereas the hydrodynamic diameter (d(H)) estimated by DLS refers to how a particle diffuses within the medium, i.e., the translational diffusion coefficient. The translational diffusion coefficient depends not only on the size of the particle core, but also on any surface structure, as well as the concentration and type of ions in the medium. Similar temperatures (24–25°C) were used for both SANS and DLS. Whereas 1–2 mg/mL B808-866-βOG mixtures were used in SANS (Fig. 3), a lower concentration of samples (0.2 mg/L) was used in DLS. Fig. S1 shows the average d(H) of free βOG micelles and B808-866-βOG mixtures measured by DLS. The d(H) of B808-866-βOG mixtures was estimated to be 140 ± 10 Å by DLS, and Dmax of the mixtures was 155 ± 5 Å as estimated by SANS. Also, the d(H) of free βOG micelles (0.8%) is ∼50 Å by both DLS and SANS (Fig. S1 and Table S1), and the d(H) from our measurements is in agreement with the reported size of free βOG micelles (14,34).
Data analyses of RC-LDAO mixtures
Fig. S2 B shows the Guinier fit for the scattering data (q < 0.02 Å−1 for RC-LDAO mixtures in 5, 40, and 100% D2O) and a larger error for estimating the Rg-value for RC-LDAO mixture in 5% D2O (Table 2) that is due to weak scattering. Fig. S2 C shows that the shape in 100% D2O data (black curve) has a maximum in the P(r) profile toward smaller distances and a somewhat narrow tail, as expected for a slightly elongated and cylindrical particle as shown in the crystal structure for RC of the purple bacteria (23,32). The P(r) profile in the 40% D2O data (blue curve) has a bell-shaped curve, as expected for an isometric (or spherical) particle, and has a maximum at larger pair distances than the P(r) profile in 100% D2O data. It is possible that LDAO micelles do not conform to the cylindrical shape of the RC. Also, the Dmax-value is larger for RC-LDAO mixtures in 40% D2O (matching protein scattering) than in 5% D2O (matching LDAO micelles scattering; Table 2), indicating that LDAO micelles are wrapped around the RC in RC-LDAO mixtures. Using the atomic coordinates for the LH1-RC core complex of Rps. palustris (PDB ID: 1PYH) (23) and the RC of Rb. sphaeroides (PDB ID: 1AIJ) (32), the Rg-values for the RC of Rps. palustris and Rb. sphaeroides are estimated to be 29.7 Å (Table 1) and 29.4 Å (Table 2), respectively. Our studies indicate a smaller RC of Cfx. aurantiacus than the purple bacteria (Table 2 and Fig. S2 D), consistent with the biochemical characterization that only the L and M subunits, not the H subunit, are identified in the RC of Cfx. aurantiacus (30). Moreover, the Rg- and Dmax-values for the RC of Cfx. aurantiacus determined from our SANS measurements are smaller than the values calculated from the crystal structure for the RC of Rps. palustris without the H subunit (Table 2).
The chlorosome is a specialized lipid body with a rod-like shape
Most of the available structural information about the chlorosome has been provided by cryo-EM, AFM, FT-IR, fluorescence, solid-state NMR, SAXS, and molecular modeling (45–48). It has been generally accepted that the majority of the mass of the chlorosome is BChls, and other components, such as proteins in the outer envelope (a monolayer lipid membrane) of the chlorosome, are proportionally minor. Here, we report SANS for the chlorosome with contrast points in 0, 25, 40, 60, 80, and 100% D2O. Note that previous SANS studies for isolated BChls were performed in (perdeuterated) organic solvents or D2O only (14–16), whereas our studies for the intact chlorosome were measured in the biological buffer (20 mM Tris-HCl buffer at pH 8.0). Fig. 4 A shows that scattering curves in the buffer with different D2O concentrations are similar, suggesting that the chlorosome is rather uniform in composition. Further, the cross-sectional radius (Rc) calculated from scattering data in different D2O buffer is also comparable (Fig. S3).
Figure 4.
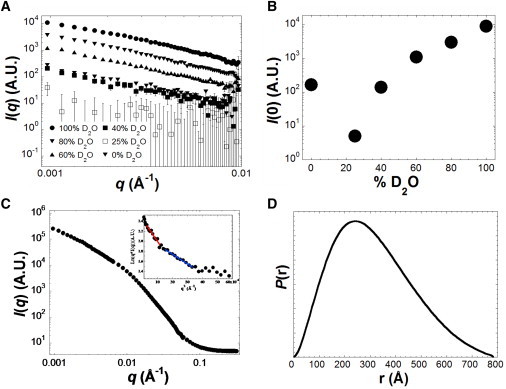
SANS for the chlorosome of Cfx. aurantiacus. (A) The SANS data for the chlorosome in 0, 25, 40, 60, 80, and 100% D2O at 20 mM Tris buffer at pH 8.0. (B) The total scattering intensity (I(0)) plot for the chlorosome in different concentrations of D2O. (C) The scattering data and Guinier analysis of the chlorosome in 100% D2O with two fits shown (inset). (D) The P(r) profile for the chlorosome in 100% D2O.
The lowest scattering intensity of the chlorosome was obtained in 25% D2O, which is close to the contrast point of lipids (Fig. 4 B). The results suggest that the chlorosome is a lipid body, and that the self-assembly of BChls (the major components of the chlorosome) is lipid-like. The lipid property of BChls can be attributed to the C173-hydrophobic long-chain ester. The data analysis shown in Fig. 4, C and D, was performed for the samples in 100% D2O, in which the maximum scattering signal was obtained. Our studies suggest that the scattering signals in the low-q region (0.0009 < q < 0.01 Å−1) can best be interpreted using a modified Guinier analysis for rod-like particles, presumably composed of the self-assembly of BChls, with the Rc estimated from the modified Guinier fits (Fig. 4 C, inset): 196 ± 2 Å with Rc × qmax ∼ 1.1 and 296 ± 6 Å with Rc × qmax ∼ 1.0. The Dmax is estimated to be 785 ± 5 Å (Fig. 4 D). In contrast, the scattering data cannot be reasonably fitted by the Guinier fit for a compact particle or for a platelet-like particle (Fig. S4). Further, an upturned curvature was detected in the very low-q range (∼0.001–0.002 Å−1) of the modified Guinier plot (Fig. 4 C, inset), suggesting the presence of very large particles in solution whose size cannot be accurately estimated. Together, the two different modified Guinier fits for the chlorosome may be interpreted as indicating the presence of two major populations of the chlorosome in solution: one in a monomeric form with Rc ∼ 200 Å, and the other likely in a dimeric form with Rc ∼ 300 Å.
Discussion
Two general types of LHCs in photosynthetic organisms are the protein-pigment complexes, which include various LHCs in higher plants, algae, and bacteria, and the pigment-pigment complex, i.e., the chlorosome. Both types of LHCs—the protein-pigment complex (the B808-866 complex) and the pigment-pigment complex (the chlorosome)—have been identified in Cfx. aurantiacus. In this study, we performed SANS to elucidate the photosystem of Cfx. aurantiacus and the purple bacteria, and investigated a variety of LHCs, including the LH2 of the purple bacteria, the chlorosome and the B808-866 complex of Cfx. aurantiacus, and the RC from the purple bacteria and Cfx. aurantiacus. The LHC and RC of the purple bacteria have been investigated extensively, and LH1 is reported to be a large (αβ)16 cyclic structure that surrounds the RC in several purple bacteria, including Rps. palustris (23), Rps. viridis (49), and Rhodospirillum rubrum (41,50,51). Earlier studies indicated that the B808-866 complex, containing a 5.6 kDa α-subunit and a 4.8 kDa β-subunit, has spectral features similar to those of LH2 (shown in Fig. S5), but higher sequence similarity to LH1 than to LH2 (52,53). Further, a ring-like structure for the B808-866-βOG complex with a diameter of 22 nm (220 Å) was estimated previously by EM, and a hydrodynamic radius of 12 nm was estimated by DLS (19).
Our SANS and DLS data indicate somewhat different results. In 100% D2O, in which the bound micelle molecules were taken into consideration for size estimation, SANS estimated that Dmax for B808-866-βOG mixtures is 155 ± 5 Å (Table 2). Further, with contrast variation, Rg is 51 ± 9 Å and Dmax is 140 ± 5 Å for B808-866-βOG mixtures in 17% D2O, in which only peptide chains are visible. Assuming a similar thickness of the LH1 and B808-866 particle (∼47 Å based on the crystal structure of LH1) (23), the ring diameter is 132 ± 5 Å for the B808-866 complex and 115 ± 2 Å for LH1 by calculating from the values of Dmax. Further, a similar particle size of B808-866-βOG mixtures was suggested by DLS (Table S1). The B808-866 complex is estimated to be 15–20 Å larger than the LH1 of Rps. palustris, but is significantly smaller (by 90 Å) than the previously reported value (220 Å). Although more structural insights are required before we can understand the assembly of the B808-866 complex, the somewhat larger ring size compared to LH1 may be partially related to the two types of BChls (B808 and B866) in the B808-866 complex versus one group of BChls (B880) in LH1. It is possible that the second group of BChls and/or the protein scaffold lead to the larger ring size for the B808-866 complex compared to LH1.
Because of certain differences in the intrinsic properties between H2O and D2O, it is possible that the smaller size of the B808-866 complex shown in our studies compared to the previously reported value is due to the presence of D2O in SANS. We compared scattering parameters obtained from our studies for protein-micelle mixtures in different ratios of D2O, and Table 1 shows similar values of Rg and Dmax for LH2-LDAO mixtures in D2O versus in H2O. Although the scattering signal collected for B808-866-βOG mixtures in H2O was not strong enough to obtain reliable structural parameters, to our knowledge no SANS or other structural studies of biomolecules and protein-micelle complexes have reported a twofold size decrease in D2O versus H2O. Alternatively, it has been reported that D2O stabilizes hydrophobic interactions and favors aggregation for some proteins (44). Thus, we consider that our experimental results indicating a similar particle size for the B808-866 complex of Cfx. aurantiacus and the LH1 of Rps. palustris are unlikely due to the presence of D2O in our measurements.
Fig. 5 A shows that the volume of the reconstructed model for the B808-866 complex of Cfx. aurantiacus derived from the SANS data collected for B808-866-βOG mixtures in 17% D2O is reasonably superimposed on the atomic-resolution structure for the LH1 of Rps. palustris, and Fig. 5 B shows the predicted scattering pattern calculated from the reconstructed model in Fig. 5 A and the crystal structure for the LH1 of Rps. palustris. The experimental data for B808-866-βOG mixtures in 17% D2O is better fitted by the predicted SANS profile for the reconstructed model of the B808-866 complex (red curve) than for the crystal structure of LH1 (blue curve), and the predicted SANS profiles also suggest that the B808-866 complex is slightly larger than LH1. Comparisons of SANS for the RC of Cfx. aurantiacus and the crystal structure for the RC of Rps. palustris are shown in Fig. 5 C. The relatively weak features found in the reconstructed model shown in Fig. 5 C can be attributed to the weak signal/noise ratio of the scattering signal for the RC of Cfx. aurantiacus in 5% D2O (Fig. S2 A), whereas the volume of the reconstructed model for the RC of Cfx. aurantiacus generated from data collected for RC-LDAO mixtures in 5% D2O is roughly comparable to the crystal structure for the RC of Rps. palustris. Fig. 5 D shows that the data collected for RC-LDAO mixtures in 5% D2O can be better fitted by the predicted SANS profile for the reconstructed model (red curve) than the crystal structure for the RC of Rps. palustris with (blue curve) or without (green curve) the H-subunit, and suggests that the RC of Cfx. aurantiacus is smaller than the crystal structure for the RC of Rps. palustris (with or without the H-subunit attached). The results indicate that a more compact complex for the RC of Cfx. aurantiacus is formed via the assembly of the L- and M-subunits. Overall, our results suggest that the B808-866 complex of Cfx. aurantiacus is comparable in size to the LH1 of Rps. palustris (the B808-866 complex is ∼10% larger), whereas the RC of Cfx. aurantiacus is ∼20% smaller than the RC of Rps. palustris without the H-subunit (Table 2).
Figure 5.
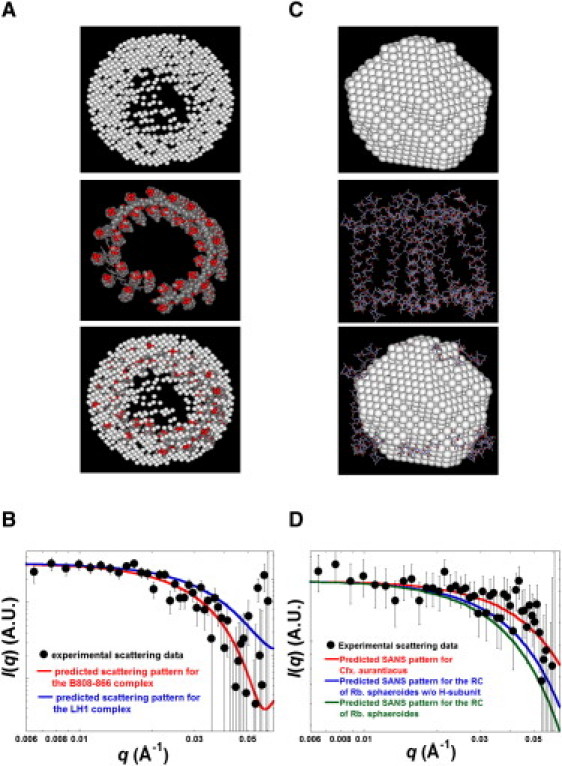
Comparisons of the SANS studies for B808-866 complex and RC of Cfx. aurantiacus and the crystal structure for LH1 and RC of Rps. palustris (PDB ID: 1PYH). (A) The reconstructed model for the B808-866 complex and the crystal structure for the LH1 of Rps. palustris. (C) The reconstructed model for the RC of Cfx. aurantiacus and the crystal structure for the RC of Rps. palustris without the H-subunit. (B) The predicted SANS patterns calculated from the reconstructed model for the B808-866 complex and the crystal structure for the LH1 fit the data for B808-866-βOG mixtures in 17% D2O. (D) The predicted SANS patterns calculated from the reconstructed model for the RC of Cfx. aurantiacus and the crystal structure for the RC of Rps. palustris with or without the H-subunit fit the data for RC-LDAO mixtures in 5% D2O.
Comparing our results with previous SANS studies, Worcester et al. (15,16) showed that isolated BChls in organic solvents form cylindrical micelles with Rc = 50–100 Å, and Wang et al. (14) demonstrated that isolated BChl c and BChl a form dimers with Rg = 17.0 ± 0.5 Å and 16.5 ± 0.5 Å, respectively. Additionally, these authors reported the presence of large aggregates in isolated BChls (14–16), consistent with our observations of much larger chlorosome particles in solution (Fig. 4 C). Further, the Rc of the chlorosome estimated by SANS is similar to the size reported by cryo-EM and AFM (54).
The structural information can be used to gain further insights into the RC-B808-866 cocomplex of Cfx. aurantiacus and detailed structural information about the chlorosome, including pigment arrangements. Considering the uniqueness of the chlorosome among antenna LHCs, the more-detailed structural information provided by SANS and other biophysical approaches will be of great value. For example, a lamellar shape in the BChls packing in the chlorosome of Cfx. aurantiacus and some green bacteria was recently suggested by SAXS (45,55), and it is essential to acquire detailed structural information about the chlorosome using multiple biophysical and biochemical approaches.
Conclusions
In this study, we employed SANS to obtain structural insights into the chlorosome and the building block for the B808-866 complex and RC of Cfx. aurantiacus, as well as the LH2 and RC of the purple bacterium Rb. sphaeroides. The latter was used to compare the structural information obtained by SANS measurements and reported crystal structures. Our studies indicate that the chlorosome is a rod-shaped lipid body, and suggest that two populations of the chlorosomes are present in solution. Further, SANS and DLS indicate that the size and conformation of the B808-866 complex of Cfx. aurantiacus and the LH1 of the purple bacterium Rps. palustris are comparable, with the B808-866 complex being ∼10% larger than the LH1. Consistent with the H-subunit's absence in the RC of Cfx. aurantiacus, our studies indicate that the RC of Cfx. aurantiacus is ∼20% smaller than the RC from the purple bacteria without the H-subunit attached. As B808-866/RC and LH1/RC are roughly comparable in size, the overall arrangement for the RC-B808-866 cocomplex of Cfx. aurantiacus and the RC-LH1 core complex of Rps. palustris is probably arranged in a similar way. To our knowledge, this is the first SANS report regarding the overall photosynthetic machinery of Cfx. aurantiacus.
Acknowledgments
The authors thank Dr. Pratim Biswas from the Department of Energy, Environment and Chemical Engineering, Washington University in St. Louis, for the use of the DLS instrument.
This study is based on work supported as part of the Photosynthetic Antenna Research Center, an Energy Frontier Research Center funded by the Office of Science, Office of Basic Energy Sciences, U.S. Department of Energy, under award No. DE-SC 0001035. The SANS studies at Oak Ridge National Laboratory's Center for Structural Molecular Biology were supported by the Office of Biological and Environmental Research, using facilities supported by the U.S. Department of Energy, managed by UT-Battelle, LLC, under contract No. DE-AC05-00OR22725.
Footnotes
Yueyong Xin's present address is College of Life Sciences, HangZhou Normal University, HangZhou, China.
Supporting Material
References
- 1.Blankenship R.E. Blackwell Science Ltd.; Oxford: 2002. Molecular Mechanisms of Photosynthesis. [Google Scholar]
- 2.Messinger J., Alia A., Govindjee Special educational issue on ‘Basics and application of biophysical techniques in photosynthesis and related processes’. Photosynth. Res. 2009;101:89–92. doi: 10.1007/s11120-009-9471-x. [DOI] [PubMed] [Google Scholar]
- 3.Thiyagarajan P., Tiede D.M. Detergent micelle structure and micelle-micelle interactions determined by small-angle neutron scattering under solution conditions used for membrane protein crystallization. J. Phys. Chem. 1994;98:10343–10351. [Google Scholar]
- 4.Owen T., Pynn R., Butler A. Metal-dependent self-assembly of a microbial surfactant. Langmuir. 2007;23:9393–9400. doi: 10.1021/la700671p. [DOI] [PubMed] [Google Scholar]
- 5.Mutch K.J., Duijneveldt J.S., Heenan R.K. Small-angle neutron scattering study of microemulsion-polymer mixtures in the protein limit. Langmuir. 2008;24:3053–3060. doi: 10.1021/la702913y. [DOI] [PubMed] [Google Scholar]
- 6.Koch M.H., Vachette P., Svergun D.I. Small-angle scattering: a view on the properties, structures and structural changes of biological macromolecules in solution. Q. Rev. Biophys. 2003;36:147–227. doi: 10.1017/s0033583503003871. [DOI] [PubMed] [Google Scholar]
- 7.Svergun D.I., Koch M.H.J. Small-angle scattering studies of biological macromolecules in solution. Rep. Prog. Phys. 2003;66:1735–1782. [Google Scholar]
- 8.Pieper J., Renger G. Protein dynamics investigated by neutron scattering. Photosynth. Res. 2009;102:281–293. doi: 10.1007/s11120-009-9480-9. [DOI] [PubMed] [Google Scholar]
- 9.Wang Z.Y., Muraoka Y., Nozawa T. Determination of the B820 subunit size of a bacterial core light-harvesting complex by small-angle neutron scattering. Biochemistry. 2003;42:11555–11560. doi: 10.1021/bi034436d. [DOI] [PubMed] [Google Scholar]
- 10.Cardoso M.B., Smolensky D., O'Neill H. Insight into the structure of light-harvesting complex II and its stabilization in detergent solution. J. Phys. Chem. B. 2009;113:16377–16383. doi: 10.1021/jp905050b. [DOI] [PubMed] [Google Scholar]
- 11.Prince S.M., Howard T.D., Isaacs N.W. Detergent structure in crystals of the integral membrane light-harvesting complex LH2 from Rhodopseudomonas acidophila strain 10050. J. Mol. Biol. 2003;326:307–315. doi: 10.1016/s0022-2836(02)01361-x. [DOI] [PubMed] [Google Scholar]
- 12.Tiede D.M., Littrell K., Thiyagarajan P. Solution structure of a biological bimolecular electron transfer complex: characterization of the photosynthetic reaction center-cytochrome c2 protein complex by small angle neutron scattering. J. Appl. Cryst. 2000;33:560–564. [Google Scholar]
- 13.Tiede D.M., Thiyagarajan P. Characterization of photosynthetic supramolecular assemblies using small angle neutron scattering. In: Amez J., Hoff A., editors. Biophysical Techniques in Photosynthesis. Vol. 3. Kluwer Academic Publishers; Dordrecht, The Netherlands: 1996. pp. 375–390. [Google Scholar]
- 14.Wang Z.Y., Umetsu M., Nozawa T. A small-angle neutron scattering study on the small aggregates of bacteriochlorophylls in solutions. Biochim. Biophys. Acta. 1997;1320:73–82. [Google Scholar]
- 15.Worcester D.L., Michalski T.J., Katz J.J. Small-angle neutron scattering studies of chlorophyll micelles: models for bacterial antenna chlorophyll. Proc. Natl. Acad. Sci. USA. 1986;83:3791–3795. doi: 10.1073/pnas.83.11.3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Worcester D.L., Michalski T.J., Katz J.J. Structure, red-shifted absorption and electron-transport properties of specific aggregates of chlorophylls. Physica B. 1989;156:502–504. [Google Scholar]
- 17.Blankenship R.E., Matsuura K. Antenna complexes from green photosynthetic bacteria. In: Green B.R., Parson W.W., editors. Anoxygenic Photosynthetic Bacteria. Kluwer Academic Publishers; Dordrecht, The Netherlands: 2003. pp. 195–217. [Google Scholar]
- 18.Hanada S., Pierson B.K. The family Chloroflexaceae. Prokaryotes. 2006;7:815–842. [Google Scholar]
- 19.Xin Y., Lin S., Blankenship R.E. Purification and characterization of the B808-866 light-harvesting complex from green filamentous bacterium Chloroflexus aurantiacus. Photosynth. Res. 2005;86:155–163. doi: 10.1007/s11120-005-5103-2. [DOI] [PubMed] [Google Scholar]
- 20.Wechsler T.D., Brunisholz R.A., Zuber H. Isolation and protein chemical characterization of the B806-866 antenna complex of the green thermophilic bacterium Chloroflexus aurantiacus. J. Photochem. Photobiol. B. 1991;8:189–197. doi: 10.1016/1011-1344(91)80058-p. [DOI] [PubMed] [Google Scholar]
- 21.Feick R.G., Fuller R.C. Topography of the photosynthetic apparatus of Chloroflexus aurantiacus. Biochemistry. 1984;23:3693–3700. [Google Scholar]
- 22.Montano G.A., Xin Y.Y., Blankenship R.E. Carotenoid and bacteriochlorophyll energy transfer in the b808–866 complex from Chloroflexus aurantiacus. J. Phys. Chem. B. 2004;108:10607–10611. [Google Scholar]
- 23.Roszak A.W., Howard T.D., Cogdell R.J. Crystal structure of the RC-LH1 core complex from Rhodopseudomonas palustris. Science. 2003;302:1969–1972. doi: 10.1126/science.1088892. [DOI] [PubMed] [Google Scholar]
- 24.Richter M.F., Baier J., Köhler J. Refinement of the x-ray structure of the RC LH1 core complex from Rhodopseudomonas palustris by single-molecule spectroscopy. Proc. Natl. Acad. Sci. USA. 2007;104:20280–20284. doi: 10.1073/pnas.0704599105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bahatyrova S., Frese R.N., Hunter C.N. The native architecture of a photosynthetic membrane. Nature. 2004;430:1058–1062. doi: 10.1038/nature02823. [DOI] [PubMed] [Google Scholar]
- 26.Sener M., Hsin J., Schulten K. Structural model and excitonic properties of the dimeric RC-LH1-PufX complex from Rhodobacter sphaeroides. Chem. Phys. 2009;357:188–197. doi: 10.1016/j.chemphys.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Montaño G.A., Bowen B.P., Blankenship R.E. Characterization of Chlorobium tepidum chlorosomes: a calculation of bacteriochlorophyll c per chlorosome and oligomer modeling. Biophys. J. 2003;85:2560–2565. doi: 10.1016/s0006-3495(03)74678-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pierson B.K., Castenholz R.W. A phototrophic gliding filamentous bacterium of hot springs, Chloroflexus aurantiacus, gen. and sp. nov. Arch. Microbiol. 1974;100:5–24. doi: 10.1007/BF00446302. [DOI] [PubMed] [Google Scholar]
- 29.Tang K.H., Wen J., Blankenship R.E. Role of the AcsF protein in Chloroflexus aurantiacus. J. Bacteriol. 2009;191:3580–3587. doi: 10.1128/JB.00110-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xin Y., Lin S., Blankenship R.E. Femtosecond spectroscopy of the primary charge separation in reaction centers of Chloroflexus aurantiacus with selective excitation in the QY and Soret bands. J. Phys. Chem. A. 2007;111:9367–9373. doi: 10.1021/jp073900b. [DOI] [PubMed] [Google Scholar]
- 31.Feick R.G., Fitzpatrick M., Fuller R.C. Isolation and characterization of cytoplasmic membranes and chlorosomes from the green bacterium Chloroflexus aurantiacus. J. Bacteriol. 1982;150:905–915. doi: 10.1128/jb.150.2.905-915.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stowell M.H., McPhillips T.M., Feher G. Light-induced structural changes in photosynthetic reaction center: implications for mechanism of electron-proton transfer. Science. 1997;276:812–816. doi: 10.1126/science.276.5313.812. [DOI] [PubMed] [Google Scholar]
- 33.Cogdell R.J., Crofts A.R. Analysis of the pigment content of an antenna pigment-protein complex from three strains of Rhodopseudomonas sphaeroides. Biochim. Biophys. Acta. 1978;502:409–416. doi: 10.1016/0005-2728(78)90074-9. [DOI] [PubMed] [Google Scholar]
- 34.Lorber B., Bishop J.B., DeLucas L.J. Purification of octyl β-D-glucopyranoside and re-estimation of its micellar size. Biochim. Biophys. Acta. 1990;1023:254–265. doi: 10.1016/0005-2736(90)90421-j. [DOI] [PubMed] [Google Scholar]
- 35.Herrmann K.W. Non-ionic-cationic micellar properties of dimethyldodecylamide oxide. J. Phys. Chem. 1962;66:295–300. [Google Scholar]
- 36.Papiz M.Z., Prince S.M., Isaacs N.W. The structure and thermal motion of the B800-850 LH2 complex from Rps.acidophila at 2.0A resolution and 100K: new structural features and functionally relevant motions. J. Mol. Biol. 2003;326:1523–1538. doi: 10.1016/s0022-2836(03)00024-x. [DOI] [PubMed] [Google Scholar]
- 37.Semenyuk A.V., Svergun D.I. GNOM—a program package for small-angle scattering data processing. J. Appl. Cryst. 1991;24:537–540. [Google Scholar]
- 38.Svergun D.I., Richard S., Zaccai G. Protein hydration in solution: experimental observation by x-ray and neutron scattering. Proc. Natl. Acad. Sci. USA. 1998;95:2267–2272. doi: 10.1073/pnas.95.5.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Savage H., Cyrklaff M., Sinning I. Two-dimensional structure of light harvesting complex II (LHII) from the purple bacterium Rhodovulum sulfidophilum and comparison with LHII from Rhodopseudomonas acidophila. Structure. 1996;4:243–252. doi: 10.1016/s0969-2126(96)00029-9. [DOI] [PubMed] [Google Scholar]
- 40.Koepke J., Hu X., Michel H. The crystal structure of the light-harvesting complex II (B800-850) from Rhodospirillum molischianum. Structure. 1996;4:581–597. doi: 10.1016/s0969-2126(96)00063-9. [DOI] [PubMed] [Google Scholar]
- 41.Karrasch S., Bullough P.A., Ghosh R. The 8.5 A projection map of the light-harvesting complex I from Rhodospirillum rubrum reveals a ring composed of 16 subunits. EMBO J. 1995;14:631–638. doi: 10.1002/j.1460-2075.1995.tb07041.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Inomoto N., Osaka N., Shibayama M. Interaction of nanogel with cyclodextrin or protein: study by dynamic light scattering and small-angle neutron scattering. Polymer (Guildf.). 2009;50:541–546. [Google Scholar]
- 43.Mears S.J., Cosgrove T., Howell I. Dynamic light scattering and small-angle neutron scattering studies on the poly(ethylene oxide)/sodium dodecyl sulfate/polystyrene latex system. Langmuir. 1998;14:4997–5003. [Google Scholar]
- 44.Bonneté F., Madern D., Zaccaï G. Stability against denaturation mechanisms in halophilic malate dehydrogenase “adapt” to solvent conditions. J. Mol. Biol. 1994;244:436–447. doi: 10.1006/jmbi.1994.1741. [DOI] [PubMed] [Google Scholar]
- 45.Psencík J., Collins A.M., Butcher S.J. Structure of chlorosomes from the green filamentous bacterium Chloroflexus aurantiacus. J. Bacteriol. 2009;191:6701–6708. doi: 10.1128/JB.00690-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ganapathy S., Oostergetel G.T., de Groot H.J. Alternating syn-anti bacteriochlorophylls form concentric helical nanotubes in chlorosomes. Proc. Natl. Acad. Sci. USA. 2009;106:8525–8530. doi: 10.1073/pnas.0903534106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jochum T., Reddy C.M., Balaban T.S. The supramolecular organization of self-assembling chlorosomal bacteriochlorophyll c, d, or e mimics. Proc. Natl. Acad. Sci. USA. 2008;105:12736–12741. doi: 10.1073/pnas.0802719105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Egawa A., Fujiwara T., Akutsu H. Structure of the light-harvesting bacteriochlorophyll c assembly in chlorosomes from Chlorobium limicola determined by solid-state NMR. Proc. Natl. Acad. Sci. USA. 2007;104:790–795. doi: 10.1073/pnas.0605911104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scheuring S., Seguin J., Rigaud J.L. Nanodissection and high-resolution imaging of the Rhodopseudomonas viridis photosynthetic core complex in native membranes by AFM. Atomic force microscopy. Proc. Natl. Acad. Sci. USA. 2003;100:1690–1693. doi: 10.1073/pnas.0437992100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jamieson S.J., Wang P., Bullough P.A. Projection structure of the photosynthetic reaction centre-antenna complex of Rhodospirillum rubrum at 8.5 A resolution. EMBO J. 2002;21:3927–3935. doi: 10.1093/emboj/cdf410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Walz T., Ghosh R. Two-dimensional crystallization of the light-harvesting I-reaction centre photounit from Rhodospirillum rubrum. J. Mol. Biol. 1997;265:107–111. doi: 10.1006/jmbi.1996.0714. [DOI] [PubMed] [Google Scholar]
- 52.Wechsler T.D., Brunisholz R.A., Zuber H. The complete amino acid sequence of the antenna polypeptide B806-866-β from the cytoplasmic membrane of the green bacterium Chloroflexus aurantiacus. FEBS Lett. 1987;210:189–194. [Google Scholar]
- 53.Zuber H., Cogdell R.J. Structure and organization of purple bacteria antenna complexes. In: Blankenship R.E., Madigan M.T., Bauer C.E., editors. Anoxygenic Photosynthetic Bacteria. Kluwer Academic Publishers; Dordrecht, The Netherlands: 1995. pp. 315–348. [Google Scholar]
- 54.Martinez-Planells A., Arellano J.B., Garcia-Gil J. Determination of the topography and biometry of chlorosomes by atomic force microscopy. Photosynth. Res. 2002;71:83–90. doi: 10.1023/A:1014955614757. [DOI] [PubMed] [Google Scholar]
- 55.Psencík J., Ikonen T.P., Tuma R. Lamellar organization of pigments in chlorosomes, the light harvesting complexes of green photosynthetic bacteria. Biophys. J. 2004;87:1165–1172. doi: 10.1529/biophysj.104.040956. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


