Abstract
RCK is a cytoplasmic regulatory domain of calcium-gated potassium channels. Binding of Ca2+ by RCK leads to channel activation through a series of yet unknown conformational changes. Structures of the K+ channel, MthK, and its cytoplasmic RCK domain revealed two binding sites for Ca2+ per dimer. We determined the crystal structure of RCK in complex with Cd2+ at 2.2 Å resolution. Cd2+ activates MthK more efficiently, and binds at the same binding sites for Ca2+ but with reduced coordination number. Two additional binding sites for Cd2+are found per dimer; one on the main Rossman-fold lobe, and the other on the small lobe of RCK. Using patch-clamp experiments, we demonstrate that Cd2+ binding to these novel sites enhances activation by Cd2+ and not by Ca2+. The structure reveals a large negatively charged surface patch in the proximity of the Ca2+/Cd2+ binding sites, charge neutralization of which appears to promote the channel open state.
Keywords: Potassium Channel, MthK, RCK, X-ray Structure
Introduction
Potassium channels control the flow of K+ across the cell membrane, and thus are central to a wide range of biological processes, including electrical signaling and electrolyte homeostasis. All K+ channels share the molecular architecture surrounding a central K+-selective pore (Doyle et al., 1998). Ligand-gated K+ channels respond to a variety of cellular homeostatic parameters by interacting with cytoplasmic ligands such as G-proteins, calmodulin, cyclic nucleotides or ions such as Mg2+ and Ca2+, each of which interacts with specific channel domains (Tang et al., 2004). Such ligand-binding domains are the RCK (regulator of conductance of K+) or the KTN (potassium transporter nucleotide-binding) domain, which are found in many prokaryotic K+ channels and transporters, as well as the eukaryotic BK channel (Jiang et al., 2001; Jiang et al., 2002; Roosild et al., 2004; Roosild et al., 2002). Ca2+-gated K+ channels such as the prokaryotic MthK, from Methanobacterium thermoautotrophicum, or the eukaryotic BK channel, contain C-terminal RCK domains that activate the channel upon binding of Ca2+ (Jiang et al., 2001; Jiang et al., 2002).
Cytoplasmic RCK is a soluble protein when expressed free of the transmembrane domain. It consists of a ∼230 aa α/β domain, made up of ∼115 aa N-terminal Rossman fold, connected to a smaller lobe of ∼75 aa via a helix-turn-helix motif. RCK proteins exist as highly stable dimers. At pH higher than 6.0 and depending on the Ca2+ concentrations, the canonical dimer can form tetramers, hexamers and octamers, but also dissociates into monomers at ∼ pH 9.0 (Dong et al., 2005; Kuo et al., 2007a; Ye et al., 2006). The reported crystal structure of MthK channel (Jiang et al., 2002) consists of two tail-to-tail interacting channels, each donating four RCK domains to form an octameric ring. Based on that, the authors suggested that the functional RCK exists as cytoplasmic octameric ring formed by interaction of four of the channel C-terminal RCK domains with four isolated cytoplasmic RCK proteins, translated from an alternative transcript of MthK starting from the codon for Met107. In that structure, two Ca2+ ions are bound by each dimer (in a total of 8 per channel). A similar octameric-ring structure has been revealed by the Ca2+-free structure of isolated RCK protein (without the transmembrane domain) (Ye et al., 2006). However, the above plausible model for channel assembly has not been confirmed yet structurally by solving the entire MthK channel in complex with isolated RCK proteins. Accordingly, two mechanisms for activation by Ca2+ have been proposed, the first assumes the invariable presence of an octameric ring that respond to Ca2+ binding by conformational changes, leading to channel opening (Jiang et al., 2002; Ye et al., 2006) and the second suggests that channel activation occurs as a result of oligomeric conversion of the RCK domains upon ligand binding (Kuo et al., 2007a; Roosild et al., 2002).
It was recently shown that MthK is activated by Cd2+ more effectively than by Ca2+ (Kuo et al., 2007a), however, it remained unknown whether Cd2+ utilizes the same binding sites on RCK as those for Ca2+. To study the mechanism of activation by Cd2+, we solved the crystal structure of the Cd2+-bound RCK. We found that Cd2+ is bound not only at the Ca2+ sites, but also at additional unique sites. To determine the involvement of these sites in channel activation by Cd2+, we knocked-out these sites by site directed mutagenesis, and followed activation by patch-clamping giant E. Coli spheroplasts expressing intact and the mutant channels.
Materials and Methods
Expression and Purification
The free RCK domain of MthK was expressed as previously described (Dong et al., 2005). Briefly, we used the gene for MthK cloned into the pQE70 vector (Qiagen) with a thrombine-cleavable C-terminal hexa-histidine tag. The channel was overexpressed in E. coli, BL21 DE3 strain (Invitrogen), by induction at A600 = ∼ 1.0 with 0.4 mM isopropyl-β-D-thiogalactopyranoside (IPTG) for 5 hours at 25 °C. Cells were lysed with a Microfluidizer (Microfluidics) in 20 mM Tris pH8.0, 500 mM KCl, 20 mM imidazole, 1 mM PMSF and 1mM binzamidine. The excessive soluble RCK domain was bound to a HiTrap IMAC FF column (GE Healthcare) loaded with Co2+, washed with 20 mM imidazole and 500 mM NaCl, and eluted with a gradient to 500 mM imidazole. The eluted sample was desalted back to the washing solution and the 6xHis tag was cleaved off by treatment with thrombin overnight at 4 °C. The cleaved sample was further purified by size exclusion chromatography (SEC) on a 26/60 Superdex-200 column (GE Healthcare) at pH 9.5 with 200 mM NaCl in the running buffer. For crystallization, the sample was concentrated to ∼5 mg/mL and the buffer was exchanged to 20 mM Tris pH 7.4, 100 mM NaCl, 1 mM EDTA
Crystallization and Structure Determination
Crystals were grown by the hanging-drop vapor diffusion method after mixing the protein stock solution with equal volume of the reservoir solution (1 M NH4 formate, 100 mM sodium acetate at pH 4.6 and 10% PEG 4000). For the co-crystallization, 10 mM of Ca2+ or 5 mM Cd2+ were added to the reservoir solution. X-ray data for native, Ca2+- and Cd2+-cocrystals of RCK were collected on beamline 9-1 at the SSRL synchrotron (Table 1).
Table 1. X-ray Data Collection Statistics on RCK Crystals.
| Data | 5 mM Cd | 10 mM Ca | Native |
| Space group | p21 | p21 | p21 |
| Unit cell dimensions (a, b, c in Å, angles in °) | 58.02, 38.29, 96.63, 90.00, 95.10, 90.00 | 58.136, 38.31, 96, 88, 90.00, 95.28, 90.00 | 58.44, 38.33, 96.92, 90.00, 94.88, 90.00 |
| Resolution in Å | 57.8 - 2.2 (2.32 - 2.2) | 57-9 - 2.3 (2.42 - 2.3) | 58.22 - 2.0 (2.11 - 2.00) |
| Wilson B-factor | 36.3 | 33.9 | 25.9 |
| reflections observed | 80, 630 (11, 993) | 71, 400 (10, 564) | 111, 034 (16, 475) |
| unique reflections | 21, 805 (3, 163) | 19, 216 (2, 789) | 29, 003 (4, 198) |
| % Completeness | 99.4 (99.1) | 99.5 (99.8) | 98.6 (98.5) |
| Multiplicity | 3.7 (3.8) | 3.7 (3.8) | 3.8 (3.9) |
| Mean((I)/σ(I)) | 10.8 (4.7) | 10.4 (6.6) | 12.0 (6.7) |
| Rsym # | 0.039 (0.159) | 0.044 (0.104) | 0.035 (0.107) |
*Parenthesis refer to highest resolution shell
#ΣhklΣi∣Ii(hkl) - <I(hkl)> ∣ / ΣhΣiIi(hkl), where Ii(hkl) is the ith measurement and <I(hkl)> is the mean of all measurements of I(hkl) for Miller indices hkl.
The structure was solved by the isomorphous Fourier difference method initially by rigid-body refinement in REFMAC5.2 (Murshudov et al., 1997) using the protein coordinates of the dimeric apo-RCK structure (PDB ID: 2AEJ), allowing movement only between the two subunits of the asymmetric unit. This yielded an R-factor of 0.3 and free-R value of 0.31 at 2.4 Å resolution. Additional restrained refinement at 2.2 Å resolution, using the protein atoms only, yielded an R-factor of 0.23 and free-R value of 0.3. At this point, Fourier difference maps were calculated to locate the positions of the Cd2+ using the phases of this initially refined model and the following coefficients: F(Cd)-F(proteincalc), F(Cd)-F(Ca), F(Ca)-F(native) as well as the anomalous coefficients F(Cd)+-F(Cd)-. These coefficients were used after scaling all of the corresponding datasets against each other using the programs SCALEIT (Howell and Smith, 1992) and FHSCAL (Kraut et al., 1962) of the CCP4 (CCP4, 1994). Four cadmium sites, corresponding to the highest peaks of the difference maps (all > 4.0 σ), were located (Figure 1). Addition of these Cd2+ sites and of 36 structural water molecules improved the free-R values by 1% in a restrained refinement. By defining TLS groups for each monomer of the ASU using the TLSMD server (Painter and Merritt, 2006) (Residues 116-232, 233-257, 258-339 for molecule A and residues 116-232, 233-259, 260-334 for molecule B) and refining the structure in a TLS& Restrained mode using REFMA5.2 (Murshudov et al., 1997) the free-R value dropped to 0.265. After several cycles of manual model editing and addition of structural water molecules using the programs O (Jones et al., 1991) and TLS & Restrained refinement with REFMAC, the R-factor and the free-R values converged to 0.18 and 0.25 (Table 2). The refined structure has been deposited in the PDB with the ID 3KXD.
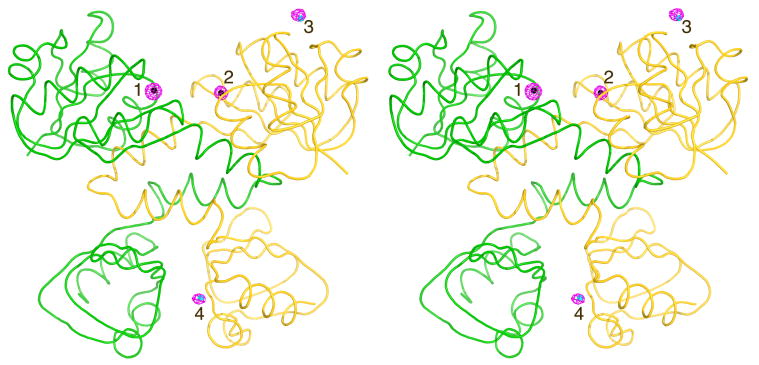
Figure 1.
Stereo view of the Cadmium Sites in the Dimeric Structure of RCK. RCK dimer is shown with molecule A in yellow and molecule B in green. Difference electron density maps contoured at 4.5 σ (pink mesh), calculated using the coefficients [F(Cd)-F(Ca)] and phases from the initial protein model after rigid body refinement, show the position of four Cd2+ sites. Two of them occupy the previously found Ca2+ sites (Sites 1 in molecule B and Site 2 in molecule A, both colored black) and two sites are unique to Cd2+ (cyan, 3 and 4).
Table 2. Refinement Statistics for RCK-Cd Complex.
| Resolution (Å) | 57.8 - 2.2 |
| Mean B-value | 27.8 |
| # Reflections used / 5% Free set | 20697 / 1108 |
| Rwork / Rfree | 0.18 / 0.25 |
| # Non H atoms | 3501 |
| # Water molecules | 112 |
| RMS deviation in bond length | 0.021 |
| RMS deviation in angles | 1.877 |
| Ramachandran Plot Analysis (%) | |
| Core, allowed, generously allowed, disallowed | 94.3, 5.7, 0.0, 0.0 |
Molecular illustrations
All structural images were produced with PyMol (DeLano, 2002). Electrostatic calculations were performed with APBS (Baker et al., 2001) and displayed in PyMol.
Preparation of E. coli giant spheroplasts
The generation of giant E. coli spheroplasts was previously described (Kuo et al., 2007b). Briefly, a freshly grown colony of E. coli FRAG1 ΔKch (Kuo et al., 2003) was inoculated in modified LB (LB5) medium (10 g/L tryptone, 5 g/L yeast extract, 5 g/L NaCl) and grown to OD of 0.3-0.4. Then, 0.5 mL was diluted by 4.5 mL of pre-warmed LB5 supplemented with 60 μg/mL cephalexin to block cell fission. After 2-3 h of incubation, 0.5 mM IPTG was added to the culture to induce the expression of MthK for 1.5-2 h. Unseptated filaments (“snakes”) were harvested by spinning down 2 mL of the culture, at 3000×g for 1 min. The pellet was resuspended in 0.5 mL of 0.8 M sucrose. Then, the following were added sequentially: 30 μL of 1 M Tris-HCl pH 8.0; 24 μL of 0.5 mg/mL lysozyme; 6 μL of 5 mg/mL DNase I (NEB); and 6 μL of 125 mM EDTA-NaOH pH 8.0. After 8 min of incubation at room temperature, 100 μL of stop solution (10 mM Tris-HCl pH 8.0, 0.7 M sucrose, 20 mM MgCl2) were added to terminate digestion. Spheroplasts are stored at -80 °C.
Patch-Clamp recording
The electrophysiological recording method was previously described (Kuo et al., 2007b). The patch-clamp pipette was filled with a solution containing KCl 150 mM, 10 mM Tris-Hepes pH 7.5, 20mM MgCl2, 5 mM EGTA and 500 mM sucrose. The same solution is also added to the chamber for seal formation. Typically a seal of 2 Giga Ohm is achieved, and the seal is then tested by gentle suctions to confirm the presence of mechanosensitive channels. Then the membrane patch is excised by tapping the manipulator, achieving an inside-out configuration (the intracellular side of the membrane is exposed to the perfusion system). Perfusion solutions used contain the basic components: 500 mM sucrose, 150 mM KCl and 10 mM buffer (Tris-Mes for pH 6.0 and pH 6.5 or Tris-Hepes for pH 7.5 and for pH 8.5) and various concentrations of CaCl2 or CdCl2 as needed for activation. All the recordings were done at a holding membrane potential of -50 mV. At this holding potential, activation of channels by calcium induces the flow of potassium ions from the pipette (“extracellular”) to the bath (“intracellular”), in the direction corresponding to inward rectification. The effective concentrations resulting 50% of the maximal activity (EC50) values were calculated by fitting the data to the Hill equation using Origin8.0.
Results
Overall Structure of RCK
Crystals of native RCK with or without Ca2+ or Cd2+ grew in a few days at 20°C, however, seeding was found to improve both the reproducibility of growth and the crystal quality. Both the native and co-crystals grew in P21 space group with similar unit cell dimensions and a dimer in the asymmetric unit (Table 1).
The structure has a bilobe architecture with a Rossman fold for the N-terminal main lobe, connected to a smaller C-terminal lobe via a helix-turn-helix which forms the dimer interface common to all RCK structures of MthK reported to date (Figure 1). The two subunits of the dimer in the ASU are very similar with root mean square deviation (RMSD) of 0.57 Å, calculated using the 3dSS server (Sumathi et al., 2006). The structure does not form a tetramer or an octamer via crystallographic symmetry, and is most similar to the low-pH dimeric Ca2+-free (PDB ID: 2AEJ) and Ca2+-bound (PDB ID: 2AEF) structures (Dong et al., 2005). In all of these dimeric structures, the crystallographic packing between dimers does not seem to be functionally relevant. Structural alignment with the dimeric Ca2+-free structure results in RMSD values of 0.62 Å for the A subunits, 0.37 Å for subunits B. Similar values were obtained for the Ca2+ bound dimeric structure, 2AEF. In contrast, we found the ligand-free octameric-ring structure of RCK (PDB ID 2FY8) significantly different; with RMSD values ranging from 2.0 to 2.3 Å for chains A through G, while surprisingly chain H of that structure is much more similar with an RMSD value of 0.68 Å. Four binding sites for Cd2+ were located (Figure 1) using F(Cdobs)-F(proteincalc) difference maps but were also confirmed by the Cd2+ anomalous difference maps and additional difference maps calculated using the Ca2+ and the native (apo) data sets (See Materials and Methods).
Cadmium Coordination
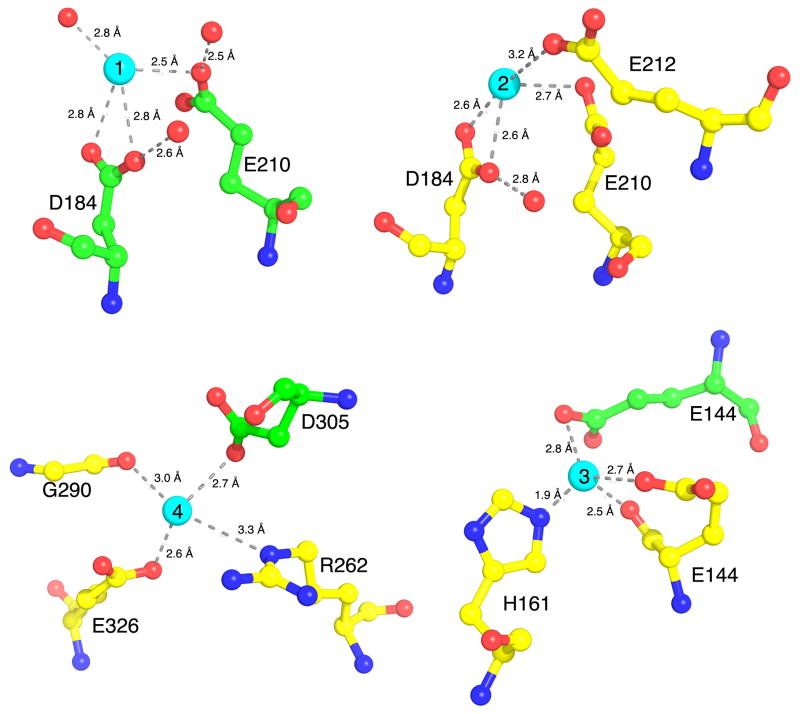
The 2Fo-Fc electron density peaks for Cd2+ in Sites 1 (16σ) and 2 (10.5 σ), are significantly higher than those seen in Sites 3 (9.5 σ) and 4 (6.0 σ). This may indicate that the affinity for Cd2+ is higher in Sites 1 and 2, the known sites for binding Ca2+ (Jiang et al., 2002). Figure 2 shows that the binding geometry in Sites 1 and 2 is similar; both employing the carboxyls of E210 and the bidentate ligand, D184, except that Site 1 has an additional coordination via a water molecule while Site 2 has an additional longer coordination via E212. In Site 3, the Cd2+ is coordinated through a short (1.9 Å) contact with the imidazole-nitrogen of H161 and through E144 as a bidentate ligand from subunit A (via its carboxyl and carbonyl groups). Additional lattice contact is made via E144 as a monodentate ligand from subunit B (of a crystallographically symmetrical dimer). Interestingly, in the current structure Site 3 exists only on subunit A since E144 from subunit B is involved in crystal contact that converges at this point. As a result, E144 from subunit B has an extended conformation compared to that of E144 from subunit A (Figure 2). This, however, is likely a crystallization artifact and at higher concentration of Cd2+ or in different solution parameters, two sites may form (one on each subunit), each engaging a single E144 as a bindetate ligand in a compact conformation. Consistent with this interpretation we noticed that the concentration of Cd2+ is critical to the crystal formation and crystals significantly lose diffraction quality at 10 mM or completely deform at 20 mM Cd2+, whereas Ca2+, which does not seem to bind at this site, is completely tolerable at 20 mM.
Figure 2.
Interactions of Cadmium with RCK. The crystal structure of RCK in complex with Cd2+ reveals four unique binding sites per RCK dimer. Sites 1 and 2 are in similar location as the Ca2+ binding sites, located on subunits B (green) and A (yellow), respectively. In contrast, Sites 3 and Site 4 are unique to Cd2+. Interacting amino acids residues are shown as ball-and-stick format, with yellow carbons for chain A and green carbons for chain B, red balls for oxygens or water molecules and blue for nitrogen atoms. Cd2+ are shown as cyan spheres. Interaction distances are shown in Å.
Site 4 provides one carboxylate-oxygen of E326, the carbonyl-oxygen of G290, the Nε atom of R262 for the Cd2+ coordination. An additional coordination is made with D305 from subunit B of the dimer, therefore Site 4 is located at the dimer interface.
Activation by Cd2+is Enhanced by Binding at Unique Sites
To test for the involvement of the structurally identified sites for Cd2+ in channel gating, we performed patch-clamp experiments on E. Coli giant spheroplasts expressing MthK. This technique has been used extensively to study the function of bacterial mechanosensitive channels (Blount et al., 1999; Martinac et al., 1987) and has recently been applied very successfully in studying gating of potassium channels including MthK (Kuo et al., 2007a; Kuo et al., 2007b; Kuo et al., 2008). MthK undergoes desensitization via its N-terminus portion corresponding to positions 2-17 of its sequence, deletion of which completely removes desensitization (Kuo et al., 2008). This sequence does not influence the channel conductance and in fact complicates the analysis of the activation by Ca2+ or Cd2+. To avoid that, we used MthK lacking its 2-17 residues (Δ2-17 MthK, referred to as the wt channel) for the K+ currents recordings. Additional site-specific mutations of the Δ2-17 MthK construct were made to perturb the structurally observed sites for Cd2+.
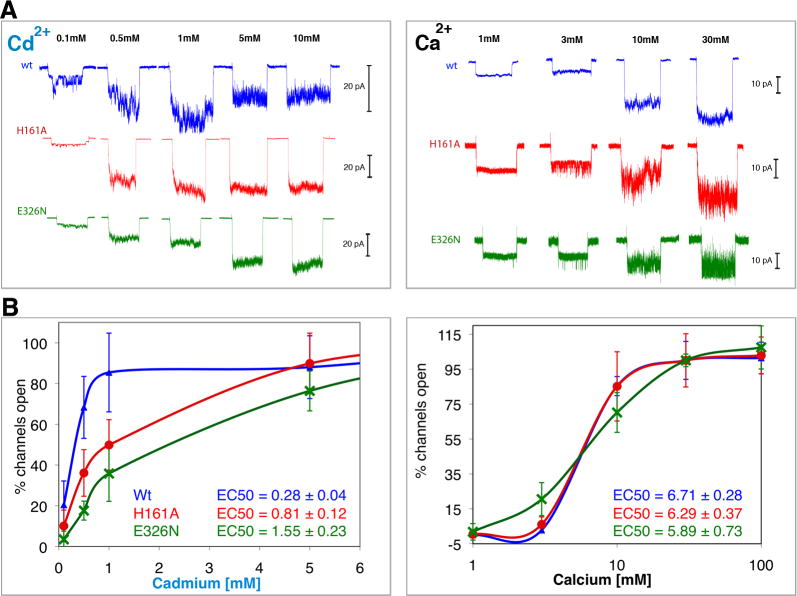
Figure 3 shows K+ currents measured from intact and the mutant channels H161A and E326N which should perturb the binding of Cd2+ in Sites 3 and 4, respectively. It can bee seen that although H161 is not required for activation of MthK by Cd2+, it promotes activation since the mutation H161A leads to a ∼3 fold increase in the EC50 value (0.801 ± 0.12 vs. 0.275 ± 0.04 mM for wt). In contrast, there is no significant difference in the Ca2+ EC50 values measured between H161A and wt channels (6.71 ± 0.28 vs. 6.29 ± 0.37 mM, respectively). This suggests that H161A does not perturb the structure significantly and validates the uniqueness of Site 3 for Cd2+. As mentioned above, Cd2+ is partly coordinated by E144 from a symmetry related dimer in Site 3. Since the crystal packing between RCK dimers in this structure does not seem to be physiologically relevant, in solution, a structural water molecule, such as seen in Site 1, is likely to substitute that coordination for Cd2+ in Site 3.
Figure 3.
Involvement of the unique Cd2+ sites in activation of MthK. Potassium currents were measured via patch-clamp experiments on giant E. coli spheroplasts expressing MthK channels activated by Cd2+ (left) and Ca2+ (right). To avoid N-terminal desensitization, Δ2-17Mthk channels were used (A) Typical macroscopic currents observed upon ligand activation of excised patches of wt (blue) and of H161A (red) and E326N (green), the putative knock-outs of the unique Cd2+-bindig Sites 3 and 4, respectively. Each trace corresponds to a single patch activated for 10 seconds, followed by a resting time of at least another 10 seconds before changing the ligand concentration. The maximum number of open channels was reached at ∼1 mM Cd2+, above which inhibition is observed. However, to achieve that with Ca2+ more than 30 mM was required. To better appreciate the effect of the treatments it is necessary to account for the various numbers of channels that exist in different patches. This number was derived dividing the peak amplitude of a patch by the unitary conductance (of a single channel) measured at each ligand concentration as described (Kuo et al., 2007b) (B) The percentages of open channels, averaged from 5 to 10 patches at each concentration (SD are shown as error bars), are plotted against concentrations of Cd2+ and Ca2+ (in logarithmic scale for clarity). These plots show that mutations at H161 and E326 significantly reduce efficiency of activation by Cd2+ but not by Ca2+.
The mutation E326N (on the small lobe of RCK) seems to mildly destabilize the channel as indicated by the change in the activation curves by Ca2+ and Cd2+ and by the significant reduction in the number of active patches found. This is consistent with the location of Site 4 at the dimer interface, which is important for the stability of all RCK domains. As shown in Figure 3, the mutation E326N also increases the Cd2+ EC50 values by ∼5.6 fold (1.55 ± 0.23 mM vs. 0.275 ± 0.04 mM for wt), but not significantly the Ca2+ EC50 values (5.89 ± 0.73 mM vs. 6.29 ± 0.37 mM for the wt).
Taken together, the data show that Cd2+ is an order of magnitude more effective than Ca2+ in activating MthK. The unique utilization of Sites 3 and 4 (involving H161 and E326, respectively) by Cd2+ to enhance activation of MthK largely explains the difference in the efficiency of activation between Cd2+ and Ca2+, i.e. EC50 values of 0.275 ± 0.04 and 6.29 ± 0.37 mM, respectively.
Charge Neutralization Promotes Activation of MthK
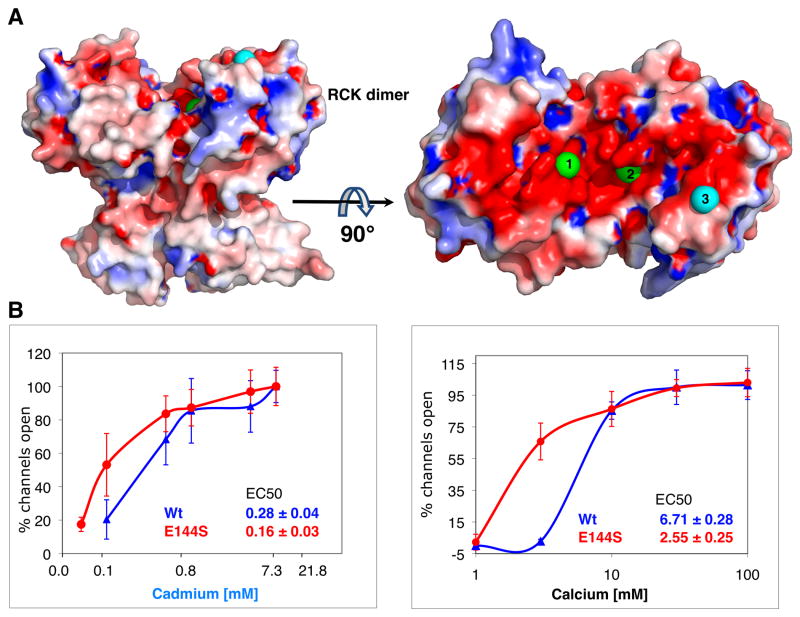
Mapping the electrostatic properties of RCK on the surface of its dimeric structure reveals a striking patch of negatively charge surface at the Cd2+/Ca2+ binding cleft and its proximity (Figure 4a). This negatively charged surface of RCK may be unique to Ca2+-gated potassium channels. It is not present in other RCK-like dimeric KTN structure of KtrA (Roosild et al., 2002) (PDB ID 1LSU) or of KefC (Roosild et al., 2009) (PDB ID 3EYW). The corresponding regions of the KtrA and KefC structures bind the ligand NADH+, but have a rather neutral or positively charged binding surface (Supplementary Figure 1).
Figure 4.
Involvement of a negatively charged surface of RCK in activation. (A) The electrostatic surface of the dimeric RCK (contoured at ± 5 kT/e with negative charges in red and positive charges in blue) is rendered in a similar view as in Figure 1. Rotation of 90° about a horizontal axis shows that the dimer face, at which the cations bind at Sites 1, 2 and 3, is remarkably negatively charged. The two Cd2+ metal ions bound at the calcium-binding sites (1 and 2) are shown as green spheres, while Cd2+ at Site 3 is shown as cyan sphere. (B) Activation curves of wt (blue) and E144S (red) by Cd2+ (left) and Ca2+ (right). Potassium currents were measured via patch-clamp experiments on giant E. coli spheroplasts expressing MthK. The percentages of open channels, averaged from 5 to 10 patches at each concentration (SD are shown as error bars) are plotted against the cation concentrations in logarithmic scale. EC50 values for both Cd2+ and Ca2+ decrease significantly by the E144S mutation (reduction of two charge values per dimer) indicating that the negative charge on this patch promotes the closed state channel.
Intriguingly, Site 3 (involving H161) is part of this negatively charge patch, (purple sphere in Figure 4a), at which Cd2+ is partly coordinated by E144. To determine the involvement of this negatively-charged patch in activation, we reduced its effective charge by knocking-out Site 3 (i.e. E144S). Comparison of K+ currents from intact with mutant E144S MthK is shown in Figure 4b. The Cd2+ EC50 values for E144S had large errors possibly emanating from the increased open probability, yet the enhanced activation by Cd2+ is noticeable from the shift to the left of the activation curve for E144S compared to the intact channel. Remarkably, although Ca2+ does not bind at Site 3, activation by Ca2+ is significantly enhanced by E144S compared to the intact MthK, with Ca2+ EC50 of 2.55 ± 0.25 for E144S vs. 6.71 ± 0.28 mM for the intact MthK. Given the functional channel activity, the tertiary structure has not been significantly changed, and thus the data indicate that the reduction in the negative charge of the patch by E144S increases the open probability of the channel independent of Ca2+. This may suggests that the large negatively-charged surface of the RCK dimer facing Sites 1, 2 and 3 is utilized similarly by Ca2+ and Cd2+ to promote the closed state of the channel. Therefore a plausible model for activation can be imagined through which binding of a divalent cation (two Ca2+ or three Cd2+) at this negatively-charged patch reduces the overall charge of that RCK dimer face, which in turn leads to channel opening. This model explains the increased efficiency of Cd2+, which appears to bind an additional unique site at this face of the dimer, thereby decreasing its charge more effectively than Ca2+.
Discussion
Although Ca2+ activates the MthK channel in a concentration-dependent manner, the effective concentration is in the milimolar range. The bacterial cytosolic concentrations range from 0.1 to 1 μM (Norris et al., 1996), thus it is still questionable whether Ca2+ is the physiological ligand for this channel. It was recently shown that MthK is activated by Cd2+ more effectively than by Ca2+ (Kuo et al., 2007a), however, the cytosolic concentration of Cd2+ are presumably even lower than those of Ca2+. Moreover, unlike calcium which serves as a ligand and cofactor in numerous biological processes, cadmium is toxic with unknown metabolism (Kazantzis, 2004), with exceptions such as that identified in binding the marine diatom carbonic anhydrase (Lane and Morel, 2000). Although these are the only activating ligands discovered so far, the physiological relevance of Ca2+ and Cd2+ in activating MthK remains to be understood.
Two Ca2+-binding sites in RCK reside at the bottom of a cleft formed between the two Rossman-fold lobes of the dimer (Sites 1 and 2, Figure 1). Although these Ca2+-binding sites do not possess the EF-hand structural motif, the pentagonal bipyramid geometry of 7 oxygens (bidentate D184, E210, E212 and 3 water molecules) coordinating the cation in the center is maintained. Surprisingly the structure we report here shows that although Cd2+ is more affective than Ca2+ in activating MthK it is bound by longer contacts in Sites 1 and 2. Moreover, Cd2+ has reduced coordination number (only 4) while Site 1 completely lacks the contact with E212. Our data suggest that employment of two other binding sites (Sites 3, 4) by Cd2+ accounts for its enhanced ability to activate MthK despite its apparent reduced affinity for Sites 1 and 2.
Biochemical and structural studies on isolated RCK or the full-length MthK channel suggest that RCK domains assemble as an octameric-ring that gates the channel (Jiang et al., 2002; Ye et al., 2006). Other biochemical data is inconsistent with the gating-ring model of activation, showing that isolated RCK exists as hexamer at pH ∼8.0 and dissociates to monomers at pH ∼9.0, at which MthK can still be effectively activated by Ca2+ (Kuo et al., 2007a; Kuo et al., 2008). This inconsistency may stem from the oligomerization data limited to isolated (“free”) RCK in solution, rather than the full-length channel in the membrane.
Regardless of the oligomeric state of RCK in the active channel, binding of Ca2+ by RCK obviously induces conformational changes ultimately required for channel opening. The various structures of RCK provide us with a range of conformational states RCK assembly can adopt, some of which are likely to reflect the conformational changes for channel opening. However, significant conformational changes are observed between subunits of the same crystal structure independent of the ligand-bound state, such as the ‘open dimer’ seen in the ligand-free octameric structure (Ye et al., 2006), therefore limiting the extent at which we can correlate conformational changes with ligand binding.
Herein we identified a negatively charged patch on the surface of the dimeric RCK, which appears to play a role in channel activation. Based on our structural and electrophysiological data, we propose an alternative model for activation of MthK via cation-charge-neutralization that conforms to both Ca2+- and Cd2+-mediated activation. Ca2+ possesses two binding sites located on that patch. The additional binding-site uniquely for Cd2+ (Site 3) found on this patch makes Cd2+ more efficient in reducing this negative charge upon binding, and thus explains its enhanced capacity compared to Ca2+ to activate the channel. The data presented here would serve as a valuable framework for future studies to substantiate this model and identify further mechanistic details leading to channel opening.
Supplementary Material
Acknowledgments
This work is supported by the NIH GM56653. Elvira Valera was the recipient of a fellowship from the Ministerio de Ciencia e Innovación, Instituto de Salud Carlos III (Spain).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blount P, Sukharev SI, Moe PC, Martinac B, Kung C. Mechanosensitive channels of bacteria. Methods Enzymol. 1999;294:458–82. doi: 10.1016/s0076-6879(99)94027-2. [DOI] [PubMed] [Google Scholar]
- CCP4. Collaborative Computational Project, Number 4. The CCP4 Suite: Programs for Protein Crystallography. Acta Cryst D Biol Crystallogr. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos, CA, USA: 2002. [Google Scholar]
- Dong J, Shi N, Berke I, Chen L, Jiang Y. Structures of the MthK RCK domain and the effect of Ca2+ on gating ring stability. J Biol Chem. 2005;280:41716–24. doi: 10.1074/jbc.M508144200. [DOI] [PubMed] [Google Scholar]
- Doyle DA, Morais Cabral J, Pfuetzner RA, Kuo A, Gulbis JM, Cohen SL, Chait BT, MacKinnon R. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- Howell L, Smith D. Normal Probability Analysis. J Appl Cryst. 1992;25:81–86. [Google Scholar]
- Jiang Y, Pico A, Cadene M, Chait BT, MacKinnon R. Structure of the RCK domain from the E. coli K+ channel and demonstration of its presence in the human BK channel. Neuron. 2001;29:593–601. doi: 10.1016/s0896-6273(01)00236-7. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Lee A, Chen J, Cadene M, Chait BT, MacKinnon R. Crystal structure and mechanism of a calcium-gated potassium channel. Nature. 2002;417:515–22. doi: 10.1038/417515a. [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Cryst A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kazantzis G. Cadmium, osteoporosis and calcium metabolism. Biometals. 2004;17:493–498. doi: 10.1023/b:biom.0000045727.76054.f3. [DOI] [PubMed] [Google Scholar]
- Kraut J, Sieker LC, High DF, Freer ST. Chymotrypsinogen: A three-dimensioanl Fourier synthesis at 5 Å resolution. Proc Natl Acad Sci U S A. 1962;48:1417–1424. doi: 10.1073/pnas.48.8.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo MM, Saimi Y, Kung C. Gain-of-function mutations indicate that Escherichia coli Kch forms a functional K+ conduit in vivo. EMBO J. 2003;22:4049–4058. doi: 10.1093/emboj/cdg409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo MM, Baker KA, Wong L, Choe S. Dynamic oligomeric conversions of the cytoplasmic RCK domains mediate MthK potassium channel activity. Proc Natl Acad Sci U S A. 2007a;104:2151–2156. doi: 10.1073/pnas.0609085104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo MM, Saimi Y, Kung C, Choe S. Patch clamp and phenotypic analyses of a prokaryotic cyclic nucleotide-gated K+ channel using Escherichia coli as a host. J Biol Chem. 2007b;282:24294–24301. doi: 10.1074/jbc.M703618200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo MM, Maslennikov I, Molden B, Choe S. The desensitization gating of the MthK K+ channel is governed by its cytoplasmic amino terminus. PLoS Biol. 2008;6:2286–2296. doi: 10.1371/journal.pbio.0060223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane TW, Morel FM. A biological function for cadmium in marine diatoms. Proc Natl Acad Sci U S A. 2000;97:4627–4631. doi: 10.1073/pnas.090091397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinac B, Buechner M, Delcour AH, Adler J, Kung C. Pressure-sensitive ion channel in Escherichia coli. Proc Natl Acad Sci U S A. 1987;84:2297–301. doi: 10.1073/pnas.84.8.2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Cryst D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Norris V, Grant S, Freestone P, Canvin J, Sheikh FN, Toth I, Trinei M, Modha K, Norman RI. Calcium signalling in bacteria. J Bacteriol. 1996;178:3677–3682. doi: 10.1128/jb.178.13.3677-3682.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Cryst D Biol Crystallogr. 2006;D62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- Roosild TP, Le KT, Choe S. Cytoplasmic gatekeepers of K+-channel flux: a structural perspective. Trends Biochem Sci. 2004;29:39–45. doi: 10.1016/j.tibs.2003.11.008. [DOI] [PubMed] [Google Scholar]
- Roosild TP, Miller S, Booth IR, Choe S. A mechanism of regulating transmembrane potassium flux through a ligand-mediated conformational switch. Cell. 2002;109:781–791. doi: 10.1016/s0092-8674(02)00768-7. [DOI] [PubMed] [Google Scholar]
- Roosild TP, Castronovo S, Miller S, Li C, Rasmussen T, Bartlett W, Gunasekera B, Choe S, Booth IR. KTN (RCK) domains regulate K+ channels and transporters by controlling the dimer-hinge conformation. Structure. 2009;17:893–903. doi: 10.1016/j.str.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumathi K, Ananthalakshmi P, Roshan MN, Sekar K. 3dSS: 3D structural superposition. Nucleic Acids Res. 2006;34:W128–132. doi: 10.1093/nar/gkl036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang XD, Santarelli LC, Heinemann SH, Hoshi T. Metabolic regulation of potassium channels. Annu Rev Physiol. 2004;66:131–159. doi: 10.1146/annurev.physiol.66.041002.142720. [DOI] [PubMed] [Google Scholar]
- Ye S, Li Y, Chen L, Jiang Y. Crystal structures of a ligand-free MthK gating ring: insights into the ligand gating mechanism of K+ channels. Cell. 2006;126:1161–1173. doi: 10.1016/j.cell.2006.08.029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.