Abstract
Neonatal sepsis is a major cause of neonatal mortality and morbidity. The current gold standard for diagnosis of sepsis, namely blood culture, suffers from low sensitivity and a reporting delay of approximately 48–72 h. Rapid detection of sepsis and institution of antimicrobial therapy may improve patient outcomes. Rapid and sensitive tests that can inform clinicians regarding the institution or optimization of antimicrobial therapy are urgently needed. The ideal diagnostic test should have adequate specificity and negative predictive value to reliably exclude sepsis and avoid unnecessary antibiotic therapy. We comprehensively searched for neonatal studies that evaluated molecular methods for diagnosis of sepsis. We identified 19 studies that were assessed with respect to assay methodology and diagnostic characteristics. In addition, we also reviewed newer molecular microbiological assays of relevance that have not been fully evaluated in neonates. Molecular methods offer distinct advantages over blood cultures, including increased sensitivity and rapid diagnosis. However, diagnostic accuracy and cost–effectiveness should be established before implementation in clinical practice.
Keywords: diagnosis, FISH, microarray, molecular, neonate, PCR, sepsis
Neonatal sepsis is a frequent life-threatening problem in neonatal intensive care units, particularly in very-low-birthweight infants (VLBW; infants with birthweight <1500 g) [1–3]. Early-onset sepsis (EOS; sepsis in infants <72 h old) occurs in 1.5–1.9% of VLBW infants, and late-onset sepsis (LOS; onset after 72 h of life) in approximately 20% [1]. Coagulase-negative staphylococci (CONS), Staphylococcus aureus and fungi are responsible for most neonatal infections [1]. Neonatal mortality in LOS is approximately 18% overall and 36% in Gram-negative sepsis. Sepsis also increases neonatal morbidities including patent ductus arteriosus, need for intravascular access, need for parenteral nutrition, bronchopulmonary dysplasia, necrotizing enterocolitis and length of hospital stay. In addition, sepsis significantly impairs long-term neurodevelopmental outcomes either by direct infection of the CNS or as a result of inflammatory injury [2]. In a large cohort of 6093 extremely low birthweight infants (ELBW; birthweight ≤1000 g), infected infants had a significantly higher incidence of adverse developmental outcomes at follow-up, including cerebral palsy, lower Bayley’s scores of infant development and vision impairment when compared with uninfected infants [3]. It is therefore critical that sepsis is diagnosed and treated early to improve neonatal outcomes. Delay in diagnosis and treatment can be life-threatening, but on the contrary overuse of antibiotics may foster antibiotic resistance.
Advances in molecular microbiology have fostered the development of newer molecular assays with the potential to diagnose sepsis rapidly and reliably. Molecular assays can be completed in less than 12 h and may be more sensitive than blood cultures. In addition, the significant increase in workload related to bloodstream infections for the clinical microbiological laboratory could potentially be offset by high-throughput molecular assays coupled with automation [4]. In this article, we will discuss the relevance and applicability of molecular assays in the diagnosis of neonatal sepsis, as well as published studies in the neonatal population.
Reliability of blood cultures in neonatal sepsis
Blood cultures are the gold standard in the diagnosis of neonatal sepsis, but suffer from the disadvantages of low sensitivity and reporting delay of 24–72 h. The diagnostic capabilities of blood culture systems have improved over the last decade with the advent of automated continuous blood culture monitoring systems. These systems use the release of CO2 (BACTEC™ FX/9000 series, Becton Dickinson, NJ, USA), colorimetry (BacT/ALERT series, bioMérieux, France) or pressure changes associated with the release and consumption of gases (VersaTREK, TREK Diagnostic Systems, OH, USA). In a direct comparison of sensitivities in the diagnosis of 179 episodes of bacteremia, BacT/ALERT and VersaTREK gave comparable results [5]. Although automated systems can save time, subcultures are required for specific biochemical or other assays, ultimately needed for pathogen identification.
Neonatal blood cultures present unique problems with regards to reliability. Fastidious organisms, maternal antibiotic treatment and small specimen volumes decrease the sensitivity of blood cultures. Furthermore, contamination of blood cultures by skin microbiota such as CONS may be problematic. Inadequate sample volume is a frequent problem in children and neonates, and the sensitivity of blood culture improves with increased blood volume [6–8]. In neonates, where low-grade bacteremia is common (<4 colony-forming units/ml), at least 1 ml is necessary for acceptable sensitivity and specificity of blood culture testing [9].
An ideal diagnostic test for neonatal sepsis should be rapid, sensitive and specific, while providing detection of all organisms relevant in neonatal sepsis and limiting the effects of maternal exposure to antibiotics. Inflammatory markers, for example, acute-phase reactants, cytokines and chemokines (e.g., C-reactive protein, procalcitonin, IL-1, IL-6, IL-8, IL-10, granulocyte colony-stimulating factor [G-CSF], TNF-α and soluble ICAM-1 [sICAM-1]), have been evaluated in the diagnosis of sepsis [4–19]. Individually or in combination these markers may be useful in the diagnosis of sepsis, but may be falsely elevated in inflammatory states such as surgery or trauma, and do not provide antibiotic susceptibility data to guide therapy [8].
Search strategy
We searched PubMed using the following keywords: neonate, the root word neonat*, newborn, sepsis, infection, pathogens, low birthweight infants, molecular methods, polymerase chain reaction, hybridization, FISH, microarrays and spectroscopy. All articles concerning neonatal sepsis were retrieved. We also searched the reference lists of the retrieved articles and personal files. We identified 19 articles relevant to neonatal sepsis, of which two were published only as abstracts and one article was in Chinese (Table 1).
Table 1.
Neonatal studies using molecular methods for the diagnosis of sepsis.
| Study (year) | Population | Samples | Type of molecular assay | SEN (%) | SPEC (%) | PPV (%) | NPV (%) | Detection time (h) | Limitations | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|
| Laforgia et al. (1997) | Neonates | Blood (n = 33) | Broad-range conventional PCR | 100 | 93.1 | 66.6 | 100 | Rapid detection | Small sample size | [35] |
| Jordan and Durso (2000) | Neonates | Blood (n = 548) | Broad-range conventional PCR and DNA dot blot analysis after 5 h preamplification culture | 96 | 99.4 | 88.9 | 99.8 | Rapid detection | [33] | |
| Shang et al. (2001) | Neonates | Blood (22) and CSF (4) 30 healthy children were controls | Broad-range PCR followed by reverse hybridization with Gram-specific probes | ≤6 | Test indices not reported | [59] | ||||
| Villaneuva-Uy et al. (2003) | Neonates with LOS | Blood (n = 61) | Broad-range 16S rRNA conventional PCR | 78 | 100 | 100 | 83 | 9 | Only abstract | [40] |
| Tong et al. (2004) | Neonates | Blood (n = 285) | 16S rRNA-based PCR followed by hybridization to chips with 18 probes | 100 | 96.8 | 47.1 | 100 | [60] | ||
| Jordan and Durso (2005) | Neonates | Blood (n = 86) | Real-time 16S rRNA PCR | 96 | 100 | 100 | 94.2 | ~4 | Did not detect Haemophilus influenzae or enterococci | [80] |
| Yadav et al. (2005) | Neonates | Blood (n = 100) | Broad-range 16S rRNA PCR | 100 | 95 | 69 | 100 | Rapid detection | [36] | |
| Makhoul et al. (2005) | Neonates with LOS | Blood (n = 215) | Staphylococcal 16S rRNA PCR (both Staphylococcus aureus and CONS) | 69.2 | 100 | 100 | 98 | <4 | Low sensitivity | [48] |
| Makhoul et al. (2006) | Neonates with LOS | Blood (n = 148) | Staphylococcal 16S rRNA PCR (both S. aureus and CONS) | 57.1 | 94.7 | 53.3 | 95.4 | <4 | Low sensitivity | [50] |
| Jordan et al. (2006) | Near term infants (>34 weeks) | Blood (n = 1233) | Conventional PCR based on 16S rRNA assay followed by pyrosequencing | 41.1 | 97.5 | 18.9 | 99.2 | Rapid detection | Low sensitivity | [34] |
| Wu et al. (2008) | Neonates | Blood (n = 600) | Real-time PCR with Gram-specific probes followed by sequencing | 100 | 97.1 | 68 | 100 | ~3 | [81] | |
| Enomoto et al. (2009) | Newborn (23–41 weeks GA) | Blood, CSF, urine, BAL, skin, ascites, pharyngeal mucus (n = 130) | Multiplex PCR targeting eight pathogens | 50 | 93 | 38 | 96 | 3.5–4.5 | Low sensitivity | [51] |
| Reier-Nielsen et al. (2009) | Newborn (Bwt >1000 g and ≤7 days) | Blood (n = 48) | Broad-range 16S rRNA PCR followed by sequencing of PCR products | 66.6 | 85.7 | 40 | 94.7 | Rapid detection | Low sensitivity and PPV | [30] |
| Chan et al. (2009) | Preterm infants (29–37 weeks GA and >72 h old) | Blood (n = 218), peritoneal fluid (n = 1) and CSF (n = 1) | Real-time PCR with universal primers and Gram-specific probes | 78.6 | 97.2 | 86.8 | 95 | 5–29 | Low sensitivity | [53] |
| Paolucci et al. (2009) | Neonates with LOS | Blood (n = 34) | Commercial LightCycler® SeptiFast system | 75 | 86.7 | 42.9 | 96.3 | 8 | [41] | |
| Dutta et al. (2009) | Neonates (mean GA 32.1 weeks, mean Bwt 1529 g) | Blood (n = 242) | Broad-range conventional PCR after 5 h preamplification culture | 96.2 | 96.3 | 87.7 | 98.8 | 8 | PCR positivity after antibiotics also evaluated | [38] |
| Chen et al. (2009) | Neonates | Blood (n = 190) and CSF (n = 5) | Broad-range 16S rRNA based real-time fluorescent PCR (FQ-PCR) | 100 | 94.4 | 60 | 100 | 3–4 | [39] | |
| Fungal PCR | ||||||||||
| Tirodker et al. (2003) | Neonates and children | Blood (n = 70) NICU-46, PICU-17 | Fungal conventional PCR targeting 18S rRNA of fungi | 76.1 | 77.2 | 43.5 | 93.6 | 5 | Malassezia not detected | [82] |
| Briones et al. (2003) | Neonates with LOS | Blood (n = 61) | Fungal conventional PCR targeting ITS3 and ITS4 regions of the 5S rRNA | 95 | 95 | 90 | 97 | 9 | Only abstract | [42] |
BAL: Bronchoalveolar lavage; Bwt: Birthweight; CONS: Coagulase-negative staphylococci; CSF: Cerebrospinal fluid; FQ: Fluorescence quantitative; GA: Gestational age; ITS: Internal transcribed spacer regions; LOS: Late-onset sepsis; NICU: Neonatal intensive-care unit; NPV: Negative predictive value; PICU: Pediatric intensive care unit; PPV: Positive predictive value; SEN: Sensitivity; SPEC: Specificity.
Molecular assays for the diagnosis of neonatal sepsis
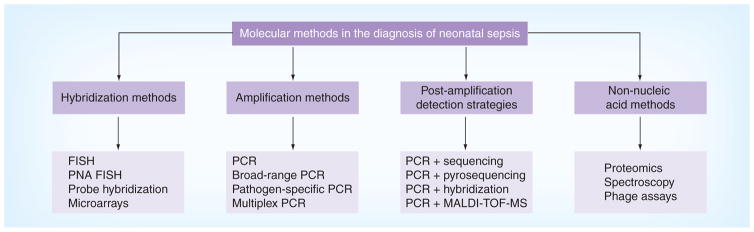
Molecular pathogen detection methods are based on hybridization (e.g., FISH or microarrays) or amplification (e.g., PCR) (Figure 1). These methods also differ by whether pathogens can be directly identified from patient samples or after initial growth in blood cultures. Sensitivity of any molecular assay is dependent on the yield of bacterial DNA from the extraction process and the presence of inhibitors. The quality of the assay is also affected by contamination (bacterial, fungal or human DNA), laboratory cross-contamination and low levels of pathogen DNA in low-grade bacteremia [10]. Diagnosis of fungal infections pose a challenge as isolation of fungal DNA is difficult owing to thicker fungal cell walls, and fungemia is also usually low grade [11].
Figure 1. Molecular methods in the diagnosis of sepsis.
MALDI-TOF MS: Matrix-assisted laser desorption ionization time-of-flight mass spectrometry; PNA: Peptide nucleic acid.
Hybridization-based FISH techniques
FISH involves identifying the organism after hybridization to fluorescence-labeled DNA probes and then visualizing the samples under the fluorescent or confocal microscope. Preliminary steps include incubation of the sample in blood culture media and then hybridization to probes after permeabilizing the cells. Probes usually target rRNA gene sequences and detect pathogens within 1–2 h. Pathogen-specific probes are available commercially or custom designed for the target microorganism. Gescher and colleagues designed a panel of FISH probes that were genus-specific and species-specific for Gram-positive cocci (staphylococci, streptococci, enterococci and others) to identify organisms from blood cultures. This FISH assay had an overall sensitivity of 98.7%, specificity of 99% and achieved rapid and reliable detection of Gram-positive cocci from clinical blood culture specimens [12]. Kudo and colleagues used FISH to identify microorganisms in blood cultures of 60 patients with suspected sepsis [13]. FISH assays identified more organisms than blood cultures (FISH 41.7% vs blood cultures 11.7%), especially in those patients who had been treated with antibiotics (FISH 61.9% vs blood cultures 4.7%). Thus far, no studies have been reported using FISH assays in neonatal specimens.
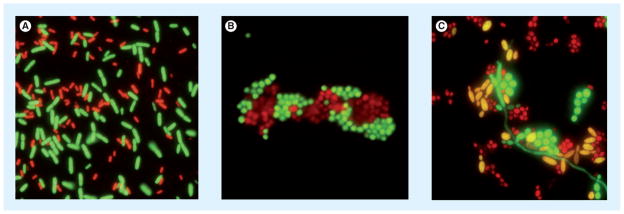
Peptide nucleic acid (PNA) probes are similar to FISH probes but are synthetic oligomers, where the negatively charged sugar-phosphate backbone of DNA is replaced with a noncharged polyamide or ‘peptide’ backbone. This structural change confers electrical neutrality, improves hybridization to the target and is less sensitive to impurities than FISH (AdvanDx, Boston, MA, USA) (Figure 2) [14]. Forrest et al. evaluated the usefulness of PNA FISH for the identification of Enterococcus fecalis and other enterococci commonly reported as Gram-positive cocci in pairs and chains. PNA FISH technique allowed rapid organism identification and institution of antimicrobial therapy, and decreased 30-day mortality [15]. PNA FISH has also been used to differentiate Candida albicans (most strains sensitive to fluconazole) and Candida glabrata (higher incidence of resistance to fluconazole), and was found to be faster than conventional methods, allowed for the optimization of anti-fungal therapy and reduced therapeutic costs [16,17]. Differentiation between CONS and Staphylococcus aureus in cultures positive for Gram-positive cocci by the PNA FISH technique resulted in reduced mortality, antibiotic usage and hospital stay [18,19].
Figure 2. Peptide nucleic acid FISH.
(A) A mixed culture of Escherichia coli and Pseudomonas aeruginosa was fixed to the slide and hybridized with the E. coli/P. aeruginosa probes. E. coli appears green and P. aeruginosa red. (B) A mixed culture of Staphylococcus aureus and Staphylococcus epidermidis was fixed to the slide and hybridized with specific PNA FISH probes. S. aureus appears green and S. epidermidis red. (C) A mixed culture of Candida albicans, Candida tropicalis and Candida glabrata was fixed to the slide and Yeast Traffic Light probe was applied. C. albicans appears green, C. tropicalis yellow and C. glabrata red.
PNA FISH images were supplied by AdvanDx, MA, USA.
Amplification methods
PCR amplifies specific target regions in the bacterial genome. Foremost among these regions is the 16S rRNA gene, an ubiquitous gene that is preserved in all bacteria and comprises both conserved and variable regions [20]. The conserved regions are targeted by universal primers for identifying bacterial infection, and the variable regions can be utilized in genus- or species-specific assays [6,21]. Amplified target regions are then subjected to downstream applications such as sequencing or microarray/probe hybridization.
DNA extraction methods
Pathogen DNA extraction is the critical step for the success of PCR and other downstream applications as it is imperative to produce high-quality DNA devoid of inhibitors or contaminants [22]. PCR can be applied directly to the patient’s blood sample, but it is more often performed after preliminary growth in blood culture media. In the latter method, DNA is extracted using one of several available protocols or with a commercially available DNA extraction kit [23–25]. Hogg et al. compared four different methods of DNA extraction in 29 methicillin-resistant S. aureus (MRSA)/methicillin-susceptible S. aureus (MSSA)-positive blood cultures and noted that the optimal method was the benzyl alcohol-based extraction [26]. DNA is extracted in less than 2 h and PCR inhibitors (e.g., hemoglobin, anticoagulants, SDA and leukocyte DNA) routinely present in blood culture media were removed. The resulting high-quality DNA showed high specificity (99.2%) and sensitivity (100%) for MRSA when used in downstream PCR assays. Millar et al. compared ten different DNA extraction methods from blood culture that included commercial kits and found that a simple wash/alkali/heat lysis was the most effective method in removing sodium polyanetholesulfonate, a known PCR inhibitor present in blood culture bottles [23,25,26]. Compared with manual DNA extraction methods, automated systems allow standardization, reduce hands-on time, allow high-throughput assays and potentially limit technician-dependent errors [27].
Broad-range PCR assays
PCR amplification strategies targeting conserved regions, such as pan-bacterial (16S rRNA) or pan-fungal (internal transcribed spacer [ITS] regions) assays, are useful when followed by sequencing or hybridization [28–30]. Broad-range or universal PCR primers are used to detect the presence or absence of bacterial or fungal DNA in patient samples. A pan-fungal PCR assay based on the rRNA sequences for two major fungal organisms (Candida and Aspergillus) followed by hybridization to a specific probe has been described [31,32]. In the diagnosis of neonatal sepsis, broad-range PCR assays targeting the 16S rRNA gene have been widely utilized (Table 1) [30,33–37].
In the earliest study of neonatal sepsis using PCR, Laforgia et al. evaluated conventional PCR, targeting a universal region of 16S rRNA, and compared it with blood cultures [35]. Venous blood samples from 33 newborns suspected of early-onset sepsis were analyzed along with spiked blood samples from healthy adult volunteers as positive controls. PCR amplicons were detected in all infected adult blood samples as well as culture-positive neonatal blood samples. Two cases that produced negative blood cultures were PCR-positive, and the authors attributed the negative blood culture results to low inocula. Yadav et al. also compared another broad-range PCR targeting 16S rRNA with blood cultures in 100 neonates with suspected sepsis. This PCR assay had 100% sensitivity and 95.6% specificity [36]. In a large study evaluating PCR methods in the diagnosis of suspected neonatal sepsis, Jordan et al. compared broad-range PCR assays with conventional blood cultures in 548 paired blood samples. PCR showed a high sensitivity (96%), specificity (99.4%), positive predictive value (PPV; 88.9%) and negative predictive value (NPV; 99.8%) [33]. In another large study of 1233 near-term infants (>34 weeks gestational age), PCR using 16S rDNA had similar specificity (97.5%) and NPV (99.2%), but failed to detect a significant number of culture-positive cases (ten out of 17) [34]. The low sensitivity was attributed to small sample volume, and variation in sample collection and sample preparation. Reier-Nilsen and colleagues reported sensitivity, specificity, PPV and NPV of 66.7, 87.5, 95.4 and 75%, respectively, using 16S rDNA-targeted PCR compared with conventional BACTEC Peds Plus/F blood cultures [30]. The study population consisted of 48 neonates with suspected sepsis in the first week of life. Two patients in their study who had negative or inconclusive PCR results produced positive cultures and were clinically diagnosed with sepsis. The low sensitivity was attributed to the low levels of bacteremia in neonatal sepsis. Wu et al. evaluated 830 blood samples from neonates with suspected sepsis using a broad-range 16S rRNA gene-based real-time PCR assay [37]. The real-time PCR assay was positive in all 20 positive blood culture samples, while 30 noninfectious blood samples were negative by both PCR and blood culture. The sensitivity of the real-time PCR assay was 100%, specificity was 97.2% and the index of accurate diagnosis was 0.97 when compared with blood culture.
Dutta et al. evaluated 242 neonates with clinical sepsis using a 16S rRNA gene-based PCR before antibiotic therapy and 12, 24 and 48 h after antibiotic therapy [38]. PCR assay in samples before antibiotic treatment had sensitivity, specificity, PPV and NPV of 96.2, 96.3, 87.7 and 98.8%, respectively. Contrary to previous theories that PCR results are not influenced by antibiotic therapy, only 12% of samples that were PCR positive prior to the start of antibiotic therapy were positive after 12 h of therapy, and none were positive after 24 or 48 h following initiation of antibiotic therapy. Chen et al. used a real-time fluorescent PCR assay to evaluate blood (n = 190) and CSF (n = 5) samples from neonates suspected to have sepsis and meningitis [39]. The sensitivity and specificity of this assay were 100 and 94.4%, respectively, and PCR yielded more positive results than cultures. Villanueva-Uy et al. evaluated 61 newborn infants with suspected late-onset sepsis by conventional PCR using broad-range 16S rRNA primers, and specificity of PCR was computed at 100%, sensitivity at 78%, PPV at 100% and NPV at 83% [40]. Of note, six PCR-negative samples grew Pseudomonas species. Paolucci et al. used a commercially available LightCycler® SeptiFast assay (Roche Molecular Diagnostics Inc., Branchburg, NJ, USA) for diagnosing sepsis in 34 neonates older than 3 days [41]. This assay had a rapid turnaround time of approximately 8 h compared with 48–72 h. The sensitivity, specificity, PPV and NPV were 75, 86.7, 42.9 and 96.3%, respectively.
Fungal PCR
Briones et al. evaluated 61 newborn infants with late-onset sepsis using primers for the ITS regions ITS3 and ITS4 of the 5.8S rRNA gene (240–430 bp long) by conventional PCR, and this technique had a turnaround time of 9 h instead of greater than 48 h for yeast cultures. The sensitivity, specificity, PPV and NPV of this PCR assay for detection of candidemia were 95, 95, 90 and 97%, respectively [42].
Pathogen-specific PCR assays
The diversity of organisms causing neonatal sepsis limits the utility of pathogen-specific single target PCR assays. Specific PCR-based assays target species-specific or genus-specific sequences, selected using information from documented complete genome sequences. Pathogen-specific PCR assays are typically used when infections with rare organisms are suspected and reliable routine diagnostic tests are not available (e.g., rickettsiosis, brucellosis and Q fever). Tissue samples for such pathogens may include heart valves and tissue biofilms [43]. Genus-specific assays are useful in conditions where species determination may delay the final identification and where genus-level identification may be sufficient for therapeutic considerations (e.g., invasive Aspergillosis and Candidiasis) [44,45].
Pathogen-specific assays may be useful in perinatal settings where specific pathogens are suspected (e.g., Group B streptococcus [GBS] and Escherichia coli in early-onset sepsis and CONS in late-onset sepsis). Bergseng et al. developed a PCR assay targeting the sip gene, a locus present in all strains of GBS, to detect GBS colonization in pregnant women in Norway [46]. Cultures and PCR were compared in vaginal and rectal swabs from 251 pregnant women and real-time PCR was performed with sensitivity and specificity of 97 and 99%, respectively. Two women who tested positive by PCR but not by cultures were later found to have GBS colonization. This pathogen-specific PCR assay underscores the potential of PCR in the rapid diagnosis of intrapartum GBS colonization that could prompt initiation of earlier prophylaxis for the prevention of neonatal GBS infections.
Another pathogen-specific assay targeting S. aureus has demonstrated promise in pediatric and adult infections. Thomas et al. evaluated a duplex real-time TaqMan® PCR targeting the species-specific nuc gene and the mecA gene encoding methicillin-resistance in 120 blood cultures (reconstituted OmniMix™ HS lyophilized beads Taqman PCR [TakaRa Bio Inc., Otsu, Japan] in Smart Cycler II [Cepheid, Sunnyvale, CA, USA]). This group reported sensitivity and specificity of 98 and 100%, respectively, for S. aureus (nuc gene) and sensitivity and specificity of 97 and 100%, respectively, for MRSA (mecA gene) [47]. Makhoul et al. studied Staphylococcus-specific PCR in the diagnosis of both early- and late-onset neonatal sepsis [48–50]. In a study of 215 clinical samples of late-onset neonatal sepsis, including 13 cases of staphylococcal bacteremia, sensitivity, specificity, PPV and NPV were 69.2, 100, 100 and 98%, respectively [48]. In a subsequent report that evaluated 148 clinical samples in 111 neonates with sepsis after 3 days of age, Staphylococcus-specific PCR showed a sensitivity, specificity, PPV and NPV of 57.1, 94.7, 53.3 and 95.4%, respectively, compared with Staphylococcus-positive blood cultures [50]. However, Staphylococcus-specific PCR may not be useful in early-onset sepsis due to the paucity of staphylococcal infections [49].
Multiplex PCR assays
Advances in PCR-based diagnosis of sepsis include multiplex PCR approaches that utilize multiple primer pairs for multiple targets in a single PCR reaction. Only one neonatal study by Enomoto et al. has applied this technique targeting eight major pathogens implicated in neonatal sepsis (GBS, E. coli, Pseudomonas aeruginosa, MRSA, Ureaplasma urealyticum, herpes simplex virus, cytomegalovirus and C. albicans) [51]. A total of 130 clinical samples from neonates undergoing evaluation for sepsis were tested by the multiplex PCR assay, which had a rapid detection time of less than 5 h. This multiplex PCR assay also had a high specificity (93%), NPV (96%) and a 90% concordance rate with blood cultures.
PCR assays that rapidly differentiate between Gram-positive and Gram-negative organisms may have clinical value in optimizing antibiotic therapy [52,53]. Chan et al. evaluated a Gram-specific probe-based amplification strategy in 218 suspected episodes of septicemia in preterm infants [53]. This PCR assay had a sensitivity and specificity of 86.4 and 99.0% for Gram-negative infections and sensitivity and specificity of 73.7 and 98.5% for Gram-positive infections, respectively. In addition, five intra-abdominal infections that were negative by cultures were found to be positive by PCR in less than 5 h.
A combined approach consisting of an initial multiplexed broad-range PCR assay followed by a pathogen-specific assay has been applied to the diagnosis of fungal infections. The ITS regions (ITS 1–4) of the rDNA sequences in fungi are interspersed between the highly conserved regions and may have sufficient heterogeneity for species-specific or genus-specific identification [54,55]. In 24 cases of keratomycoses, PCR amplification of the ITS region followed by sequencing significantly reduced time to diagnosis (24 h vs 5–10 days), with 71% sensitivity [55]. Multiplexing with two probes labeled with two different fluorescent dyes or combining one pan-bacterial probe with one species-specific probe have been useful in the diagnosis of polymicrobial infections [56].
Commercially available multiplex systems have recently been tested in the diagnosis of adult sepsis. The LightCycler SeptiFast and SeptiTest multiplex PCR system were developed to detect 25 different pathogens. Westh and colleagues conducted a multi-center study to compare the performance of SeptiFast with blood cultures on 558 paired blood samples from 359 patients with suspected sepsis. The SeptiFast assay was concordant with blood cultures in 78% of positive and in 83% of negative results [57]. Dierkes et al. also compared the SeptiFast system with blood cultures from 101 blood samples from 77 patients [58]. In this study, concordance was reported to be 62% for negative and 13% for positive results. In addition, 9% were found to be positive only by blood cultures and 13% were found to be positive by SeptiFast only. Advantages of the SeptiFast system include reduced time to positive results (18–21 h) compared with blood cultures (2 days) and lower contamination rates, but cost–effectiveness needs to be assessed [57,58]. The detection capabilities of the assay are limited by the primers selected (limiting the number of microorganisms targeted), and the system is further limited by the inability to perform antimicrobial susceptibility testing. However, multiplex PCR assays may be an invaluable adjunct to blood cultures, as they provide rapid information for the early recognition and management of sepsis.
Post-amplification detection strategies
PCR amplification methods combined with sequencing or hybridization have been evaluated in two neonatal studies. Shang et al. used a universal 16S rRNA gene-based PCR followed by reverse hybridization with Gram-positive and Gram-negative specific probes [59]. Neonatal blood samples that were infected were identified by this method, while controls tested negative. Tong et al. performed 16S rRNA gene amplification and hybridized the resulting amplicons to probes on glass slides [60]. When blood specimens from neonates with suspected sepsis were compared with blood cultures, PCR had a sensitivity of 100% and specificity of 96.8%. PCR also identified microorganisms that correlated with blood culture results. One neonatal study by Jordan et al. utilized DNA pyrosequencing to differentiate between bacterial pathogens (643 bacterial isolates and 15 whole blood samples) and found it to be an accurate method for differentiating pathogens causing neonatal sepsis [61].
Ibis biosensor T5000 (Ibis Biosciences, Abbott Laboratories, IL, USA) combines nucleic acid amplification (usually broad-range PCR primers targeting rDNA or housekeeping genes) followed by high-performance electrospray ionization mass spectrometry that determines base composition of the amplicons [62,63]. The advantages of this system include the ability to identify all known bacteria and major families of fungi and viruses that are pathogenic to humans in a rapid (4–6 h) and high-throughput format.
Chakravorty et al. developed a novel technique that overcomes limitations in multiplex PCR assays, namely the narrow range of probe regions needed for multiplexing and the inability of the PCR instrument to detect more than six fluorophores simultaneously [64]. They developed a class of mismatch tolerant molecular beacons that had an extended hybridization range for bacterial 16S rRNA. By analyzing the melting temperatures (Tm) of multiple molecular beacon probes, a Tm signature of common bacterial species was produced. This method was evaluated in 270 clinical cultures, including 106 patient blood cultures, and was 95–97% concordant with blood culture results with no misidentifications and 100% specificity. Luna et al. performed amplification of the 16S rRNA gene by targeting the conserved regions flanking two variable regions, V1 and V6, and subsequent DNA pyrosequencing provided variable region sequences suitable for genus and species-level identification [65]. A total of 414 isolates from 312 pediatric patients that were difficult to identify by conventional biochemical and morphometric methods were identified by DNA pyrosequencing in approximately 90% of the samples.
DNA microarray platform
Microarrays are matrices of probes for specific genomic targets of organisms, and have been used for direct identification of organisms from patient samples [66–68]. The ability to combine multiple assays on a single chip is a distinct advantage. Microarrays in addition to pathogen identification can evaluate antibiotic resistance genes and virulence markers, for example, S. aureus methicillin resistance.
One neonatal study by Shang et al. evaluated 16S rRNA-based PCR amplification followed by DNA microarray hybridization in neonatal blood samples derived from 172 infants with clinically suspected sepsis [68]. Compared with blood cultures, this method had 100% sensitivity and 97.9% specificity. In the 17 samples that were positive by PCR, microarray hybridization identified the organisms responsible for neonatal sepsis. This method may hold significant promise for the rapid identification of pathogens responsible for neonatal sepsis.
Other studies evaluating this molecular method in adults with sepsis may also be of relevance to sepsis diagnosis in neonates. Cleven and coworkers designed a DNA microarray consisting of 120 species-specific probes for the evaluation of three important pathogens causing bacterial sepsis, namely S. aureus, E. coli and P. aeruginosa [66]. On evaluation of 42 clinical isolates, three reference strains and 13 positive blood cultures, this microarray was highly specific for the organisms targeted, and testing for antibiotic resistance genes correlated with phenotypic antibiotic resistance assays. Low sensitivity of microarrays may be due to the low amount of pathogen DNA in patient samples, which could potentially be improved by PCR amplification before microarray hybridization. Palka-Santini et al. evaluated a large-scale multiplex PCR (LSplex PCR) using primers that amplify DNA genome segments of nine pathogenic species [69]. The amplicons were then hybridized to corresponding probes on a microarray chip. This technique improves the sensitivity of detection by 100- to 1000-fold. In another study, Palka-Santini et al. improved postprocessing of positive blood cultures using PCR amplification of gene segments of S. aureus followed by microarray hybridization. This method was able to identify S. aureus from Gram-negative bacteria and CONS including antibiotic susceptibility on the same day as positive blood cultures [67].
Tissari and colleagues evaluated a DNA-based microarray platform for pathogen identification from positive blood cultures (Prove-it™ Sepsis assay, Mobidiag, Helsinki, Finland) [70]. This assay amplified gyrB, parE and mecA genes of 50 bacterial species. On evaluation of 2107 positive blood cultures from two centers (UK and Finland), this assay was 18 h faster than the conventional method and had a sensitivity and specificity of 94.7 and 98.8%, respectively. For S. aureus bacteremia, the sensitivity and specificity were 100%. Inacio and colleagues used a novel method for the rapid diagnosis of invasive fungal infections. This technique is characterized by loop-mediated isothermal DNA amplification using nonspecific primers for the 26S rRNA gene followed by hybridization to species-specific probes on a nylon membrane [71]. This new method was reliable in the rapid identification of invasive fungal infections.
Marlowe and colleagues used a rRNA probe matrix consisting of DNA probes targeting rRNA sequences of bacteria and fungi [72]. This novel rapid molecular approach to the identification of bacteria and yeast in blood cultures was highly sensitive (100%) and specific (96%), and discordant with blood cultures by the BacT/Alert® 3D system (bioMérieux Clinical Diagnostics, Marcy l’Etoile, France) in only 1.75% of samples.
Non-nucleic acid methods
New phage-based pathogen detection assays are available, such as MicroPhage’s MRSA/MSSA blood culture test (MicroPhage, Longmont, CO, USA). This assay proposes to detect amplification of S. aureus-specific phages in the presence of methicillin. A multicenter, preclinical study that evaluated the effectiveness of MicroPhage’s S. aureus/MSSA/MRSA test directly from leftover positive blood culture specimens has recently been completed [101]. The MicroPhage test will be compared with site standards (gold standard) and market-available tests with similar indications (comparators). The results of this trial have not been reported. Identification of bacteria and fungi using proteomic profiles by matrix-assisted laser desorption ionization time of flight mass spectrometry (MALDI-TOF MS) has also been evaluated [73–75]. This technique, although rapid, requires specialized equipment and preliminary growth in blood cultures. The combination of PCR amplification followed by MALDI-TOF MS provides the advantage of increased sensitivity, as well as the high-throughput nature of mass spectrometry [76]. This method consists of uracil-DNA-glycosylase-mediated base-specific fragmentation of 16S rDNA PCR products followed by MALDI-TOF MS. Raman and Fourier transform infrared spectroscopy have been used to detect and identify microorganisms, including yeast, after growth in blood cultures. Identification is based on the property of specificity of the spectroscopic fingerprint of the organisms. Identification accuracy was 92.2% for Raman and 98.3% for infrared spectroscopic methods, respectively [77].
Expert commentary
We reviewed studies that have evaluated molecular microbiological methods in the diagnoses of neonatal sepsis. These studies show that molecular assays, including PCR and hybridization methods, are feasible in neonates and have rapid detection times compared with blood cultures. In neonatal studies (Table 1), the sensitivity of molecular methods used to diagnose sepsis ranged from 41.1 to 100%, and specificity from 77.2 to 100%. These variations can be attributed to differences in methodology and study design, including DNA extraction methods, as well as the characteristics of the population studied (term vs preterm infants). In addition, prevalence rates of sepsis in the population studied impacts PPVs and NPVs of the diagnostic test. The most widely studied method was broad-range PCR targeting sequences within the 16S rRNA gene. The sensitivity of PCR improved with preamplification culture of samples for 5 h [33,38]. The sensitivity was low (50%) in the only study that evaluated multiplex PCR targeting eight pathogens [51]. Combination of PCR with hybridization to specific probes or microarrays may improve the diagnostic characteristics of the assays. FISH and PNA FISH techniques enable rapid detection of the organisms after blood cultures are determined to be positive. These assays may impact clinical decisions regarding antibiotic therapy and have the potential to improve clinical outcomes. Organism proteomics and spectroscopic methods used for organism identification require specialized equipment and technical expertise. Cost–effectiveness and the ability of molecular assays to impact clinical outcomes should be evaluated before widespread use in clinical practice. Molecular assays might eventually replace blood cultures, but will continue as a supplement to blood cultures until they are adequately evaluated.
Five-year view
Molecular assays may have a significant impact on the diagnosis and management of neonatal sepsis. Advances in molecular microbiology will lead to more reliable and efficient techniques that can be cost effective. Microarray hybridization and next-generation sequencing techniques can lead not only to rapid identification of organisms, but also to evaluation of organism characteristics such as virulence and antibiotic susceptibility. Another exciting prospect is the ability to quantify bacterial loads (analogous to viral loads), which can then be followed during therapy to assess response. In one study, quantitative assessment of meningococcal burden by real-time PCR correlated with the severity of meningococcal disease and predicted mortality [78]. Similarly, it is possible to estimate bacterial load in S. pneumoniae infection using real-time PCR targeting the pneumolysin (ply) gene and follow subsequent response to therapy [79]. Advances in mass spectrometry and spectroscopy may also identify specific microorganism signatures that can identify pathogens and their characteristics (e.g., virulence).
Key issues.
Molecular methods may offer advantages over blood cultures in the diagnosis of neonatal sepsis.
Molecular assays are rapid and require small sample volumes.
Molecular assays may be automated, enabling high throughput, and reduce microbiological workload compared with blood cultures.
Molecular methods may evaluate virulence and antibiotic resistance markers that may inform antibiotic therapy.
Positive results of more sensitive molecular methods (detection of pathogen DNA) in the face of a negative blood culture (absence of viable organisms) need to be interpreted carefully in a clinical setting. Sample contamination may have to be excluded.
False-negative results from molecular assays may be due to inefficient DNA extraction, presence of low levels of pathogen DNA or the presence of inhibitors.
High negative predictive values of a diagnostic test may be clinically useful in ruling out sepsis and avoiding unnecessary antibiotics.
Costs, availability of equipment and technical skills in the microbiological laboratory are important considerations.
Cost–effectiveness of the newer molecular assays should be established before widespread acceptance in clinical practice.
Acknowledgments
The authors sincerely thank their colleagues in the neonatal intensive care unit at Texas Children’s Hospital (TX, USA), Michael Speer and Caraciolo Fernandes, for their critique and insight.
Footnotes
For reprint orders, please contact reprints@expert-reviews.com
Financial & competing interests disclosure
Funding for color printing of the figures was provided by AdvanDx (MA, USA). Mohan Pammi Venkatesh is funded by CHRCDA grant (NIH K12 HD41648). James Versalovic is supported by R01 AT004326-01A1 NIH/NCCAM, R01 DK065075-01 NIH/NIDDK and 1UH2HG083990-01 NIH/NHGRI. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
• of interest
•• of considerable interest
- 1.Stoll BJ, Hansen N, Fanaroff AA, et al. Late-onset sepsis in very low birth weight neonates: the experience of the NICHD Neonatal Research Network. Pediatrics. 2002;110(2 Pt 1):285–291. doi: 10.1542/peds.110.2.285. [DOI] [PubMed] [Google Scholar]
- 2.Adams-Chapman I, Stoll BJ. Neonatal infection and long-term neurodevelopmental outcome in the preterm infant. Curr Opin Infect Dis. 2006;19(3):290–297. doi: 10.1097/01.qco.0000224825.57976.87. [DOI] [PubMed] [Google Scholar]
- 3.Stoll BJ, Hansen NI, Adams-Chapman I, et al. Neurodevelopmental and growth impairment among extremely low-birth-weight infants with neonatal infection. JAMA. 2004;292(19):2357–2365. doi: 10.1001/jama.292.19.2357. [DOI] [PubMed] [Google Scholar]
- 4.Rodriguez-Creixems M, Alcala L, Munoz P, Cercenado E, Vicente T, Bouza E. Bloodstream infections: evolution and trends in the microbiology workload, incidence, and etiology, 1985–2006. Medicine. 2008;87(4):234–249. doi: 10.1097/MD.0b013e318182119b. [DOI] [PubMed] [Google Scholar]
- 5.Mirrett S, Hanson KE, Reller LB. Controlled clinical comparison of VersaTREK and BacT/ALERT blood culture systems. J Clin Microbiol. 2007;45(2):299–302. doi: 10.1128/JCM.01697-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Isaacman DJ, Karasic RB, Reynolds EA, Kost SI. Effect of number of blood cultures and volume of blood on detection of bacteremia in children. J Pediatr. 1996;128(2):190–195. doi: 10.1016/s0022-3476(96)70388-8. [DOI] [PubMed] [Google Scholar]
- 7.Kaditis AG, O’Marcaigh AS, Rhodes KH, Weaver AL, Henry NK. Yield of positive blood cultures in pediatric oncology patients by a new method of blood culture collection. Pediatr Infect Dis J. 1996;15(7):615–620. doi: 10.1097/00006454-199607000-00011. [DOI] [PubMed] [Google Scholar]
- 8.Connell TG, Rele M, Cowley D, Buttery JP, Curtis N. How reliable is a negative blood culture result? Volume of blood submitted for culture in routine practice in a children’s hospital. Pediatrics. 2007;119(5):891–896. doi: 10.1542/peds.2006-0440. [DOI] [PubMed] [Google Scholar]
- 9.Schelonka RL, Chai MK, Yoder BA, Hensley D, Brockett RM, Ascher DP. Volume of blood required to detect common neonatal pathogens. J Pediatr. 1996;129(2):275–278. doi: 10.1016/s0022-3476(96)70254-8. [DOI] [PubMed] [Google Scholar]
- 10••.Mancini N, Carletti S, Ghidoli N, Cichero P, Burioni R, Clementi M. The era of molecular and other non-culture-based methods in diagnosis of sepsis. Clin Microbiol Rev. 2010;23(1):235–251. doi: 10.1128/CMR.00043-09. Comprehensive review of available molecular methods in the diagnosis of sepsis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Metwally L, Fairley DJ, Coyle PV, et al. Improving molecular detection of Candida DNA in whole blood: comparison of seven fungal DNA extraction protocols using real-time PCR. J Med Microbiol. 2008;57(Pt 3):296–303. doi: 10.1099/jmm.0.47617-0. [DOI] [PubMed] [Google Scholar]
- 12.Gescher DM, Kovacevic D, Schmiedel D, et al. Fluorescence in situ hybridisation (FISH) accelerates identification of Gram-positive cocci in positive blood cultures. Int J Antimicrob Agents. 2008;32(Suppl 1):S51–S59. doi: 10.1016/j.ijantimicag.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 13.Kudo M, Matsuo Y, Nakasendo A, et al. Potential clinical benefit of the in situ hybridization method for the diagnosis of sepsis. J Infect Chemother. 2009;15(1):23–26. doi: 10.1007/s10156-008-0655-7. [DOI] [PubMed] [Google Scholar]
- 14.Pellestor F, Paulasova P, Hamamah S. Peptide nucleic acids (PNAs) as diagnostic devices for genetic and cytogenetic analysis. Curr Pharm Des. 2008;14(24):2439–2444. doi: 10.2174/138161208785777405. [DOI] [PubMed] [Google Scholar]
- 15.Forrest GN, Roghmann MC, Toombs LS, et al. Peptide nucleic acid fluorescent in situ hybridization for hospital-acquired enterococcal bacteremia: delivering earlier effective antimicrobial therapy. Antimicrob Agents Chemother. 2008;52(10):3558–3563. doi: 10.1128/AAC.00283-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forrest GN, Mankes K, Jabra-Rizk MA, et al. Peptide nucleic acid fluorescence in situ hybridization-based identification of Candida albicans and its impact on mortality and antifungal therapy costs. J Clin Microbiol. 2006;44(9):3381–3383. doi: 10.1128/JCM.00751-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Della-Latta P, Whittier S, Wu F. Impact of rapid identification of C. albicans and C. glabrata directly from blood cultures using PNA FISH technology on selection of antifungal therapy. Presented at: 18th European Congress of Clinical Microbiology and Infectious Diseases; Barcelona, Spain. 19–22 April 2008. [Google Scholar]
- 18.Ly T, Gulia J, Pyrgos V, Waga M, Shoham S. Impact upon clinical outcomes of translation of PNA FISH-generated laboratory data from the clinical microbiology bench to bedside in real time. Ther Clin Risk Manag. 2008;4(3):637–640. doi: 10.2147/tcrm.s2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Forrest GN, Mehta S, Weekes E, Lincalis DP, Johnson JK, Venezia RA. Impact of rapid in situ hybridization testing on coagulase-negative staphylococci positive blood cultures. J Antimicrob Chemother. 2006;58(1):154–158. doi: 10.1093/jac/dkl146. [DOI] [PubMed] [Google Scholar]
- 20.Woese CR. Bacterial evolution. Microbiol Rev. 1987;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Relman DA. The search for unrecognized pathogens. Science. 1999;284(5418):1308–1310. doi: 10.1126/science.284.5418.1308. [DOI] [PubMed] [Google Scholar]
- 22.Lee JH, Park Y, Choi JR, Lee EK, Kim HS. Comparisons of three automated systems for genomic DNA extraction in a clinical diagnostic laboratory. Yonsei Med J. 2010;51(1):104–110. doi: 10.3349/ymj.2010.51.1.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fredricks DN, Relman DA. Application of polymerase chain reaction to the diagnosis of infectious diseases. Clin Infect Dis. 1999;29(3):475–486. doi: 10.1086/598618. quiz 487–478. [DOI] [PubMed] [Google Scholar]
- 24.Shrestha NK, Tuohy MJ, Hall GS, Isada CM, Procop GW. Rapid identification of Staphylococcus aureus and the mecA gene from BacT/ALERT blood culture bottles by using the LightCycler system. J Clin Microbiol. 2002;40(7):2659–2661. doi: 10.1128/JCM.40.7.2659-2661.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Millar BC, Xu J, Moore JE. Risk assessment models and contamination management: implications for broad-range ribosomal DNA PCR as a diagnostic tool in medical bacteriology. J Clin Microbiol. 2002;40(5):1575–1580. doi: 10.1128/JCM.40.5.1575-1580.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hogg GM, McKenna JP, Ong G. Rapid detection of methicillin-susceptible and methicillin-resistant Staphylococcus aureus directly from positive BacT/Alert blood culture bottles using real-time polymerase chain reaction: evaluation and comparison of 4 DNA extraction methods. Diagn Microbiol Infect Dis. 2008;61(4):446–452. doi: 10.1016/j.diagmicrobio.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 27.Riemann K, Adamzik M, Frauenrath S, et al. Comparison of manual and automated nucleic acid extraction from whole-blood samples. J Clin Lab Anal. 2007;21(4):244–248. doi: 10.1002/jcla.20174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Evertsson U, Monstein HJ, Johansson AG. Detection and identification of fungi in blood using broad-range 28S rDNA PCR amplification and species-specific hybridisation. APMIS. 2000;108(5):385–392. doi: 10.1034/j.1600-0463.2000.d01-73.x. [DOI] [PubMed] [Google Scholar]
- 29.Schabereiter-Gurtner C, Nehr M, Apfalter P, Makristathis A, Rotter ML, Hirschl AM. Evaluation of a protocol for molecular broad-range diagnosis of culture-negative bacterial infections in clinical routine diagnosis. J Appl Microbiol. 2008;104(4):1228–1237. doi: 10.1111/j.1365-2672.2007.03648.x. [DOI] [PubMed] [Google Scholar]
- 30•.Reier-Nilsen T, Farstad T, Nakstad B, Lauvrak V, Steinbakk M. Comparison of broad range 16S rDNA PCR and conventional blood culture for diagnosis of sepsis in the newborn: a case control study. BMC Pediatr. 2009;9:5. doi: 10.1186/1471-2431-9-5. Neonatal study evaluating broad-range PCR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Burik JA, Myerson D, Schreckhise RW, Bowden RA. Panfungal PCR assay for detection of fungal infection in human blood specimens. J Clin Microbiol. 1998;36(5):1169–1175. doi: 10.1128/jcm.36.5.1169-1175.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Imhof A, Schaer C, Schoedon G, et al. Rapid detection of pathogenic fungi from clinical specimens using LightCycler real-time fluorescence PCR. Eur J Clin Microbiol Infect Dis. 2003;22(9):558–560. doi: 10.1007/s10096-003-0989-0. [DOI] [PubMed] [Google Scholar]
- 33•.Jordan JA, Durso MB. Comparison of 16S rRNA gene PCR and BACTEC 9240 for detection of neonatal bacteremia. J Clin Microbiol. 2000;38(7):2574–2578. doi: 10.1128/jcm.38.7.2574-2578.2000. Neonatal study evaluating broad-range PCR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34•.Jordan JA, Durso MB, Butchko AR, Jones JG, Brozanski BS. Evaluating the near-term infant for early onset sepsis: progress and challenges to consider with 16S rDNA polymerase chain reaction testing. J Mol Diagn. 2006;8(3):357–363. doi: 10.2353/jmoldx.2006.050138. Neonatal study evaluating broad-range PCR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35•.Laforgia N, Coppola B, Carbone R, Grassi A, Mautone A, Iolascon A. Rapid detection of neonatal sepsis using polymerase chain reaction. Acta Paediatr. 1997;86(10):1097–1099. doi: 10.1111/j.1651-2227.1997.tb14815.x. Neonatal study evaluating broad-range PCR. [DOI] [PubMed] [Google Scholar]
- 36•.Yadav AK, Wilson CG, Prasad PL, Menon PK. Polymerase chain reaction in rapid diagnosis of neonatal sepsis. Indian Pediatr. 2005;42(7):681–685. Neonatal study evaluating broad-range PCR. [PubMed] [Google Scholar]
- 37•.Wu YD, Shang SQ, Li JP, et al. A broad-range 16S rRNA gene real-time PCR assay for the diagnosis of neonatal septicemia. Zhonghua Er Ke Za Zhi. 2007;45(6):446–449. Neonatal study evaluating broad-range PCR. [PubMed] [Google Scholar]
- 38••.Dutta S, Narang A, Chakraborty A, Ray P. Diagnosis of neonatal sepsis using universal primer polymerase chain reaction before and after starting antibiotic drug therapy. Arch Pediatr Adolesc Med. 2009;163(1):6–11. doi: 10.1001/archpediatrics.2008.513. Elegant study testing broad-range PCR in neonates, and the only neonatal study evaluating the effect of antibiotics on PCR. [DOI] [PubMed] [Google Scholar]
- 39.Chen LH, Duan QJ, Cai MT, Wu YD, Shang SQ. Rapid diagnosis of sepsis and bacterial meningitis in children with real-time fluorescent quantitative polymerase chain reaction amplification in the bacterial 16S rRNA gene. Clin Pediatr. 2009;48(6):641–647. doi: 10.1177/0009922809333972. [DOI] [PubMed] [Google Scholar]
- 40.Villanueva-Uy ME, Briones CR, Uy HG. Application of polymerase chain reaction in late-onset neonatal sepsis. Pediatr Res. 2003;53:313A. [Google Scholar]
- 41•.Paolucci M, Capretti MG, Dal Monte P, et al. Laboratory diagnosis of late-onset sepsis in newborns by multiplex real-time PCR. J Med Microbiol. 2009;58(Pt 4):533–534. doi: 10.1099/jmm.0.003848-0. Neonatal study evaluating a commercially available PCR assay for the diagnosis of neonatal sepsis. [DOI] [PubMed] [Google Scholar]
- 42.Briones CR, Villanueva-Uy ME, Uy HG. The use of polymerase chain reaction in neonatal candidemia. Pediatr Res. 2003;53:396A. [Google Scholar]
- 43.Fenollar F, Raoult D. Molecular diagnosis of bloodstream infections caused by non-cultivable bacteria. Int J Antimicrob Agents. 2007;30(Suppl 1):S7–S15. doi: 10.1016/j.ijantimicag.2007.06.024. [DOI] [PubMed] [Google Scholar]
- 44.Klingspor L, Jalal S. Molecular detection and identification of Candida and Aspergillus spp. from clinical samples using real-time PCR. Clin Microbiol Infect. 2006;12(8):745–753. doi: 10.1111/j.1469-0691.2006.01498.x. [DOI] [PubMed] [Google Scholar]
- 45.Klingspor L, Loeffler J. Aspergillus PCR formidable challenges and progress. Med Mycol. 2009;47(Suppl 1):S241–S247. doi: 10.1080/13693780802616823. [DOI] [PubMed] [Google Scholar]
- 46•.Bergseng H, Bevanger L, Rygg M, Bergh K. Real-time PCR targeting the sip gene for detection of group B Streptococcus colonization in pregnant women at delivery. J Med Microbiol. 2007;56(Pt 2):223–228. doi: 10.1099/jmm.0.46731-0. PCR method for evaluating intrapartum group B Streptococcus colonization. [DOI] [PubMed] [Google Scholar]
- 47.Thomas LC, Gidding HF, Ginn AN, Olma T, Iredell J. Development of a real-time Staphylococcus aureus and MRSA (SAM-) PCR for routine blood culture. J Microbiol Methods. 2007;68(2):296–302. doi: 10.1016/j.mimet.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 48•.Makhoul IR, Smolkin T, Sujov P, et al. PCR-based diagnosis of neonatal staphylococcal bacteremias. J Clin Microbiol. 2005;43(9):4823–4825. doi: 10.1128/JCM.43.9.4823-4825.2005. Neonatal study evaluating pathogen-specific PCR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49•.Makhoul IR, Sprecher H, Smolkin T, et al. Approach to term neonates born after maternal intrapartum fever and unknown maternal group B Streptococcus status: value of serum C-reactive protein and 16S rRNA gene PCR amplification. Pediatr Infect Dis J. 2007;26(11):1064–1066. doi: 10.1097/INF.0b013e31812f5701. Neonatal study evaluating pathogen-specific PCR. [DOI] [PubMed] [Google Scholar]
- 50.Makhoul IR, Yacoub A, Smolkin T, Sujov P, Kassis I, Sprecher H. Values of C-reactive protein, procalcitonin, and Staphylococcus-specific PCR in neonatal late-onset sepsis. Acta Paediatr. 2006;95(10):1218–1223. doi: 10.1080/08035250600554250. [DOI] [PubMed] [Google Scholar]
- 51•.Enomoto M, Morioka I, Morisawa T, Yokoyama N, Matsuo M. A novel diagnostic tool for detecting neonatal infections using multiplex polymerase chain reaction. Neonatology. 2009;96(2):102–108. doi: 10.1159/000208791. Neonatal study evaluating multiplex PCR. [DOI] [PubMed] [Google Scholar]
- 52.Carroll NM, Jaeger EE, Choudhury S, et al. Detection of and discrimination between Gram-positive and Gram-negative bacteria in intraocular samples by using nested PCR. J Clin Microbiol. 2000;38(5):1753–1757. doi: 10.1128/jcm.38.5.1753-1757.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chan KY, Lam HS, Cheung HM, et al. Rapid identification and differentiation of Gram-negative and Gram-positive bacterial bloodstream infections by quantitative polymerase chain reaction in preterm infants. Crit Care Med. 2009;37(8):2441–2447. doi: 10.1097/CCM.0b013e3181a554de. [DOI] [PubMed] [Google Scholar]
- 54.Pryce TM, Palladino S, Kay ID, Coombs GW. Rapid identification of fungi by sequencing the ITS1 and ITS2 regions using an automated capillary electrophoresis system. Med Mycol. 2003;41(5):369–381. doi: 10.1080/13693780310001600435. [DOI] [PubMed] [Google Scholar]
- 55.Mancini N, Perotti M, Ossi CM, et al. Rapid molecular identification of fungal pathogens in corneal samples from suspected keratomycosis cases. J Med Microbiol. 2006;55(Pt 11):1505–1509. doi: 10.1099/jmm.0.46638-0. [DOI] [PubMed] [Google Scholar]
- 56.Mancini N, Carletti S, Ghidoli N, et al. Molecular diagnosis of polymicrobial sepsis. J Clin Microbiol. 2009;47(4):1274–1275. doi: 10.1128/JCM.00011-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Westh H, Lisby G, Breysse F, et al. Multiplex real-time PCR and blood culture for identification of bloodstream pathogens in patients with suspected sepsis. Clin Microbiol Infect. 2009;15(6):544–551. doi: 10.1111/j.1469-0691.2009.02736.x. [DOI] [PubMed] [Google Scholar]
- 58.Dierkes C, Ehrenstein B, Siebig S, Linde HJ, Reischl U, Salzberger B. Clinical impact of a commercially available multiplex PCR system for rapid detection of pathogens in patients with presumed sepsis. BMC Infect Dis. 2009;9:126. doi: 10.1186/1471-2334-9-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59•.Shang S, Chen Z, Yu X. Detection of bacterial DNA by PCR and reverse hybridization in the 16S rRNA gene with particular reference to neonatal septicemia. Acta Paediatr. 2001;90(2):179–183. doi: 10.1080/080352501300049389. Neonatal study evaluating PCR followed by reverse hybridization. [DOI] [PubMed] [Google Scholar]
- 60.Tong MQ, Shang SQ, Wu YD, Zhao ZY. Rapid diagosis of neonatal sepsis by 16SrRNA genes PCR amplification and genechip hybridization. Zhonghua Er Ke Za Zhi. 2004;42(9):663–667. [PubMed] [Google Scholar]
- 61•.Jordan JA, Butchko AR, Durso MB. Use of pyrosequencing of 16S rRNA fragments to differentiate between bacteria responsible for neonatal sepsis. J Mol Diagn. 2005;7(1):105–110. doi: 10.1016/s1525-1578(10)60015-3. Neonatal study evaluating PCR followed by pyrosequencing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ecker DJ, Massire C, Blyn LB, et al. Molecular genotyping of microbes by multilocus PCR and mass spectrometry: a new tool for hospital infection control and public health surveillance. Methods Mol Biol. 2009;551:71–87. doi: 10.1007/978-1-60327-999-4_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ecker DJ, Sampath R, Massire C, et al. Ibis T5000: a universal biosensor approach for microbiology. Nat Rev Microbiol. 2008;6(7):553–558. doi: 10.1038/nrmicro1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chakravorty S, Aladegbami B, Burday M, et al. Rapid universal identification of bacterial pathogens from clinical cultures by using a novel sloppy molecular beacon melting temperature signature technique. J Clin Microbiol. 2010;48(1):258–267. doi: 10.1128/JCM.01725-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Luna RA, Fasciano LR, Jones SC, Boyanton BL, Jr, Ton TT, Versalovic J. DNA pyrosequencing-based bacterial pathogen identification in a pediatric hospital setting. J Clin Microbiol. 2007;45(9):2985–2992. doi: 10.1128/JCM.00630-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cleven BE, Palka-Santini M, Gielen J, Meembor S, Kronke M, Krut O. Identification and characterization of bacterial pathogens causing bloodstream infections by DNA microarray. J Clin Microbiol. 2006;44(7):2389–2397. doi: 10.1128/JCM.02291-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Palka-Santini M, Putzfeld S, Cleven BE, Kronke M, Krut O. Rapid identification, virulence analysis and resistance profiling of Staphylococcus aureus by gene segment-based DNA microarrays: application to blood culture post-processing. J Microbiol Methods. 2007;68(3):468–477. doi: 10.1016/j.mimet.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 68•.Shang S, Chen G, Wu Y, Du L, Zhao Z. Rapid diagnosis of bacterial sepsis with PCR amplification and microarray hybridization in 16S rRNA gene. Pediatr Res. 2005;58(1):143–148. doi: 10.1203/01.PDR.0000169580.64191.8B. Neonatal study evaluating PCR followed by microarray hybridization. [DOI] [PubMed] [Google Scholar]
- 69.Palka-Santini M, Cleven BE, Eichinger L, Kronke M, Krut O. Large scale multiplex PCR improves pathogen detection by DNA microarrays. BMC Microbiol. 2009;9:1. doi: 10.1186/1471-2180-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tissari P, Zumla A, Tarkka E, et al. Accurate and rapid identification of bacterial species from positive blood cultures with a DNA-based microarray platform: an observational study. Lancet. 2010;375(9710):224–230. doi: 10.1016/S0140-6736(09)61569-5. [DOI] [PubMed] [Google Scholar]
- 71.Inacio J, Flores O, Spencer-Martins I. Efficient identification of clinically relevant Candida yeast species by use of an assay combining panfungal loop-mediated isothermal DNA amplification with hybridization to species-specific oligonucleotide probes. J Clin Microbiol. 2008;46(2):713–720. doi: 10.1128/JCM.00514-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Marlowe EM, Hogan JJ, Hindler JF, Andruszkiewicz I, Gordon P, Bruckner DA. Application of an rRNA probe matrix for rapid identification of bacteria and fungi from routine blood cultures. J Clin Microbiol. 2003;41(11):5127–5133. doi: 10.1128/JCM.41.11.5127-5133.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Marvin LF, Delatour T, Tavazzi I, Fay LB, Cupp C, Guy PA. Quantification of o,o′-dityrosine, o-nitrotyrosine, and o-tyrosine in cat urine samples by LC/electrospray ionization-MS/MS using isotope dilution. Anal Chem. 2003;75(2):261–267. doi: 10.1021/ac020309w. [DOI] [PubMed] [Google Scholar]
- 74.Marvin LF, Roberts MA, Fay LB. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry in clinical chemistry. Clin Chim Acta. 2003;337(1–2):11–21. doi: 10.1016/j.cccn.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 75.van Baar BL. Characterisation of bacteria by matrix-assisted laser desorption/ionisation and electrospray mass spectrometry. FEMS Microbiol Rev. 2000;24(2):193–219. doi: 10.1016/S0168-6445(99)00036-4. [DOI] [PubMed] [Google Scholar]
- 76.von Wintzingerode F, Bocker S, Schlotelburg C, et al. Base-specific fragmentation of amplified 16S rRNA genes analyzed by mass spectrometry: a tool for rapid bacterial identification. Proc Natl Acad Sci USA. 2002;99(10):7039–7044. doi: 10.1073/pnas.102165899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Maquelin K, Kirschner C, Choo-Smith LP, et al. Prospective study of the performance of vibrational spectroscopies for rapid identification of bacterial and fungal pathogens recovered from blood cultures. J Clin Microbiol. 2003;41(1):324–329. doi: 10.1128/JCM.41.1.324-329.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hackett SJ, Guiver M, Marsh J, et al. Meningococcal bacterial DNA load at presentation correlates with disease severity. Arch Dis Child. 2002;86(1):44–46. doi: 10.1136/adc.86.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.van Haeften R, Palladino S, Kay I, Keil T, Heath C, Waterer GW. A quantitative LightCycler PCR to detect Streptococcus pneumoniae in blood and CSF. Diagn Microbiol Infect Dis. 2003;47(2):407–414. doi: 10.1016/s0732-8893(03)00129-9. [DOI] [PubMed] [Google Scholar]
- 80.Jordan JA, Durso MB. Real-time polymerase chain reaction for detecting bacterial DNA directly from blood of neonates being evaluated for sepsis. J Mol Diagn. 2005;7(5):575–581. doi: 10.1016/S1525-1578(10)60590-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wu YD, Chen LH, Wu XJ, et al. Gram stain-specific-probe-based real-time PCR for diagnosis and discrimination of bacterial neonatal sepsis. J Clin Microbiol. 2008;46(8):2613–2619. doi: 10.1128/JCM.02237-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tirodker UH, Nataro JP, Smith S, LasCasas L, Fairchild KD. Detection of fungemia by polymerase chain reaction in critically ill neonates and children. J Perinatol. 2003;23(2):117–122. doi: 10.1038/sj.jp.7210868. [DOI] [PubMed] [Google Scholar]
Website
- 101.Clinical trials.gov. MicroPhage S. aureus/MSSA/MRSA Blood Culture Beta Trial. http://clinicaltrials.gov/ct2/show/NCT00814151.