Abstract
1,2,3,4-Diepoxybutane (DEB) is a strongly genotoxic diepoxide hypothesized to be the ultimate carcinogenic metabolite of the common industrial chemical and environmental carcinogen 1,3-butadiene. DEB is a bis-electrophile capable of cross-linking cellular biomolecules to form DNA-DNA and DNA-protein cross-links (DPCs), which are thought to play a central role in its biological activity. Previous studies with recombinant proteins have shown that the biological outcomes of DEB-induced DPCs are strongly influenced by protein identities. The present work combines affinity capture methodology with mass spectrometry-based proteomics and immunological detection to identify the proteins which form DPCs in nuclear extracts from human cervical carcinoma (HeLa) cells. We identified 39 human proteins that form covalent DPCs in the presence of DEB. DNA-protein cross-linking efficiency following treatment with 25 mM DEB was 2–12%, depending on protein identity. HPLC-ESI+-MS/MS analysis of the total proteolytic digests of cross-linked proteins revealed the presence of 1-(S-cysteinyl)-4-(guan-7-yl)-2,3-butanediol conjugates, suggesting that DEB forms DPCs between cysteine thiols within proteins and the N-7 guanine positions within DNA.
Keywords: mass spectrometry, DNA-Protein cross-links, diepoxybutane, affinity capture, Western blot, DNA repair
Introduction
DNA-protein cross-links (DPCs) are bulky, helix-distorting lesions that are hypothesized to block the binding and progression of protein complexes, interfering with DNA replication, transcription, DNA repair, recombination, and chromatin remodeling.1 DPCs can form endogenously a result of oxidative stress and lipid peroxidation,2,3 or can be induced by exposure to ionizing radiation,4 metals,5 or common chemotherapeutic agents such as nitrogen mustards,6,7 platinum drugs,8 and alkylnitrosoureas.9 Although their biological relevance is poorly understood, certain types of DPCs persist through several cycles of DNA replication,1,10 potentially leading to cytotoxic and mutagenic outcomes such as sister chromatid exchanges and large deletions.
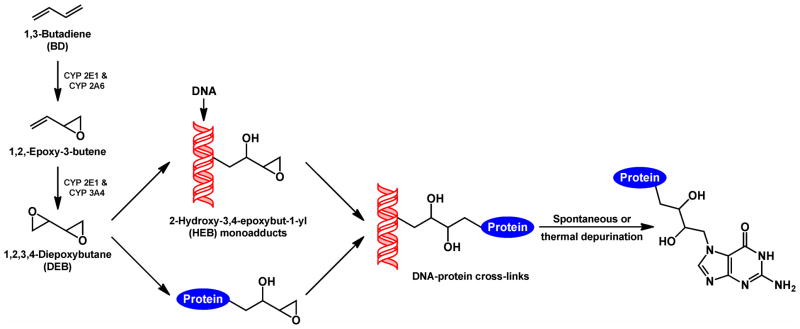
One prominent bis-electrophile capable of inducing DPCs is 1,2,3,4-diepoxybutane (DEB), the proposed ultimate carcinogenic metabolite of 1,3-butadiene (BD).11 BD is a known animal and human carcinogen present in automobile exhaust and in cigarette smoke.12,13 The adverse biological effects of DEB have been attributed to its ability to cross-link cellular biomolecules. Initial alkylation of adenine and guanine bases in DNA by DEB produces 2-hydroxy-3,4-epoxybut-1-yl (HEB) lesions, which contain an inherently reactive oxirane group that can alkylate neighboring nucleobases within the DNA duplex to form DNA-DNA cross-links.14 Alternatively, the 3,4-epoxy ring can be subject to nucleophilic attack by amino acid side chains of neighboring proteins, giving rise to DPCs (Scheme 1).15,16
Scheme 1.
Metabolic activation of BD to DEB and DEB-mediated formation of DNA-protein cross-links.
DNA-protein cross-linking by DEB was first observed by Jelitto et al. who employed alkaline elution methodology to detect DPC formation in liver tissue of B6C3F1 mice following exposure to BD.15 This observation was subsequently confirmed by other groups that utilized similar biophysical methods for detecting DEB-induced DPCs.15,17 Although these authors have established the ability of DEB to covalently cross-link proteins to DNA, they did not identify specific proteins which participate in DPC formation, nor did they provide information regarding the chemical structure of the resulting amino acid-nucleobase conjugates.
In recent years, mass spectrometry has become an increasingly valuable tool in the study of DNA-protein interactions. 18–21 Recent in vitro studies with purified recombinant proteins have identified three proteins that can form covalent cross-links to DNA in the presence of DEB: O6-alkylguanine DNA alkyltransferase (AGT),16 glyceraldehyde 3-phosphate dehydrogenase (GAPDH),22 and histone H3.23 Our laboratory has described DEB-mediated cross-linking between the DNA repair protein AGT and DNA. 16 Mass spectrometric analysis of tryptic peptides identified two cross-linking sites within the AGT protein: the catalytic alkyl acceptor site (Cys145), and a neighboring active site residue (Cys150). Loecken and colleagues have reported that DEB also forms DPCs involving GAPDH22 and histones H2b and H3.23 Like AGT, DPC formation by GAPDH involves a cysteine residue (Cys246).22 Alkylation of Cys246 in vitro results in the inhibition of GAPDH activity.22 Similarly, DEB is capable of cross-linking the Cys111 residue of histone H3 to DNA.23 In vitro DPC formation by all three proteins required treatment with relatively high concentrations of DEB (20 mM for GAPDH and histone proteins and 15 mM for AGT).16,22,23 Interestingly, the over-expression of human AGT in bacteria enhanced the cytotoxic and mutagenic effects of DEB, presumably through the formation of toxic DPC lesions.24,25 In contrast, no enhanced mutagenesis was observed in cells over-expressing GAPDH22 or histone H2b.23 These results suggest that the identity of the cross-linked protein influences the biological effects of DPCs. However, the identities of other nuclear proteins that participate in cross-linking to DNA in the presence of DEB, as well as the abundance with which DNA-protein lesions are produced in mammalian cells following DPC exposure, have not been established, limiting our understanding of the role of DNA-protein cross-linking in the genotoxicity and cytotoxicity of DEB.
The purpose of the current study was to identify human proteins participating in DEB-mediated DPC formation. We have recently developed an affinity capture technique which can be coupled with mass spectrometry-based proteomics and immunological detection to identify nuclear extract proteins involved in DPC formation in the presence of bis-electrophiles.26 In the present study, the new methodology was used to investigate DEB-mediated DNA-protein cross-linking. The identities of nuclear proteins which become covalently cross-linked to DNA in the presence of DEB were established by mass spectrometry-based proteomics, and immunological approaches were employed to estimate the cross-linking efficiency for specific protein targets. These results are significant because the identification of specific nuclear proteins that become cross-linked to DNA in the presence of DEB will allow a better understanding of their role in DEB-mediated cytotoxicity.
Materials And Methods
Safety Statement
DEB is a known human carcinogen and should be handled with caution in a well-ventilated fume hood with appropriate personal protective equipment.
Chemicals and Reagents
DEB, phenylmethanesulfonyl fluoride (PMSF), pepstatin, leupeptin, aprotinin, dithiothreitol (DTT), and iodoacetamide were purchased from Sigma-Aldrich (St. Louis, MO). Mass spectrometry grade Trypsin Gold was purchased from Promega (Madison, WI). Proteinase K was obtained from Worthington Biochemical Corp. (Lakewood, NJ). Primary polyclonal antibodies to human actin, DNA-(apurinic or apyrimidinic site) lyase (Ref-1), GAPDH, and poly(ADP-ribose) polymerase (PARP) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). The primary polyclonal antibody to the ATP-dependent DNA helicase subunit 2 (Ku) was purchased from Lab Vision/NeoMarkers (Fremont, CA). The primary monoclonal antibody to AGT was purchased from Millipore (Temecula, CA). Alkaline phosphatase-conjugated anti-mouse and anti-rabbit IgG secondary antibodies were obtained from Sigma-Aldrich (St. Louis, MO). Synthetic DNA oligodeoxynucleotides were prepared at the University of Minnesota’s Biomedical Genomics Center (Minneapolis, MN). 1-(S-cysteinyl)-4-(guan-7-yl)-2,3-butanediol (Cys-N7G-BD) was prepared as described previously. 16
Cell Culture
Human cervical carcinoma (HeLa) cells were generously provided by Dr. Jonathan Marchant (University of Minnesota). The cells were maintained as exponentially growing monolayer cultures in Dulbecco’s Modified Eagle’s Medium supplemented with 9% fetal bovine serum, in a humidified incubator at 37°C with 5% CO2.
Preparation of Nuclear Extracts
Nuclear protein extracts were prepared from HeLa cells as described in the literature.27 Briefly, ~108 cells were harvested, washed three times with ice cold phosphate-buffered saline, and suspended in hypotonic buffer (10 mM Tris-HCl – pH 7.4/10 mM MgCl2/10 mM KCl/1 mM DTT) containing 1 mM PMSF. After incubating for 5 min on ice, cells were broken by 20 strokes in a Dounce homogenizer and centrifuged at 2000g for 10 min. The sedimented nuclei were re-suspended in hypotonic buffer (see above) containing 350 mM NaCl and a protease inhibitor cocktail (1 μg/mL pepstatin; 0.5 μg/mL leupeptin; 0.75 μg/mL aprotinin; 1 mM PMSF), and incubated on ice for 1 h. The resulting nuclear lysate was centrifuged at 160,000g at 4 °C for 30 min, and the nuclear proteins were isolated in a clear supernatant. Extracts were dialyzed for 2 h at 4°C against 10 mM Tris-HCl – pH 7.4/10 mM KCl/10 mM MgCl2 buffer using Slide-A-Lyzer dialysis cassettes with a 3.5 kDa molecular weight cut-off (Pierce Biotechnology, Rockford, IL). Protein concentrations were determined using the Bio-Rad Protein Assay (Bio-Rad, Hercules, CA).
DNA-Protein Cross-linking and Biotin Capture
5′-Biotinylated double-stranded oligodeoxynucleotides (5′-GGA GCT GGT GGC GTA GGC-3′, (+) strand, 3.12 nmol) were combined with nuclear protein extracts from HeLa cells (500 μg total protein) in the absence or in the presence of DEB (5–500 mM, 1 mL total volume) and incubated at 37°C for 3 h to induce cross-linking. Biotinylated DNA, along with DNA-protein cross-links, was captured by incubation with Streptavidin Sepharose High Performance beads (750 μL slurry, GE Healthcare, Piscataway, NJ) overnight at 4°C, with rotation. To remove any non-covalently bound proteins, the beads were washed twice with 1% SDS (1 h followed by 30 min, with rotation, room temperature), twice with each 4 M urea and 1 M NaCl (both 30 min followed by 15 min, with rotation, room temperature), and twice with phosphate-buffered saline (no incubation). Following each washing step, the beads were centrifuged at 2,000g for 1 min, and the supernatant was discarded. DNA containing covalently bound proteins was released from the beads by adding 110 μL NuPage 4X LDS Sample Buffer (Invitrogen, Carlsbad, CA) and heating to 90°C for 15 min. Due to the thermal instability of DEB-induced cross-links, which form specifically at the N7 of guanine, 16 these conditions enabled a quantitative release of the proteins from DNA in the form of protein-guanine conjugates. The proteins were subsequently analyzed by HPLC-ESI+-MS/MS and western blotting as described below.
Protein Identification by Mass Spectrometry
For proteomic analyses, the cross-linked proteins were separated using 12% Tris-HCl Ready Gels (Bio-Rad, Hercules, CA) and stained with SimplyBlue SafeStain (Invitrogen, Carlsbad, CA). Gel lanes were cut into slices comprising the entire molecular weight range, washed with 100 mM ammonium bicarbonate, and subjected to reduction with dithiothreitol and iodoacetamide treatment as previously described.28 Gel pieces were dehydrated with acetonitrile, dried under vacuum, and reconstituted in 25 mM ammonium bicarbonate containing 10 μg of mass spectrometry grade trypsin. The samples were digested overnight at 37°C. Tryptic peptides were extracted with 1% aqueous formic acid/60% acetonitrile, evaporated to dryness, and re-suspended in 0.1% aqueous formic acid for mass spectrometric analysis.
Tryptic peptides were subjected to HPLC-ESI+-MS/MS analysis with a ThermoFinnigan LTQ ion trap mass spectrometer equipped with a Thermo MicroAS autosampler and a Thermo Surveyor HPLC pump, a nanospray source, and an Xcalibur 1.4 instrument control. Peptides were resolved on a 100 μm × 11 cm fused silica capillary column (Polymicro Technologies, LLC, Phoenix, AZ) packed with 5 μm, 300 Å Jupiter C18 packing (Phenomenex, Torrence, CA). The column was eluted at 0.6 μL/min with a gradient of 0.1% formic acid in water (A) and 0.1% formic acid in acetonitrile (B). The solvent composition was initially held at 2% B for 15 min, followed by a linear increase to 25% B over 25 min, and further to 90% B in 15 min. Liquid chromatography was carried out at an ambient temperature. Centroided MS-MS scans were acquired using an isolation width of 2 m/z, an activation time of 30 ms, an activation Q of 0.250 and 30% normal collision energy using 1 microscan with a max ion time of 100 ms for each MS/MS scan. The mass spectrometer was tuned prior to analysis using the synthetic peptide TpepK (AVAGKAGAR), so that some parameters may have varied slightly from experiment to experiment. Typically, the tune parameters were as follows: spray voltage of 2 kV, a capillary temperature of 150°C, a capillary voltage of 50 V, and tube lens of 120 V. The MS/MS spectra of the peptides were collected using data-dependent scanning, in which one full MS spectrum was followed by four MS/MS spectra. MS/MS spectra were recorded using dynamic exclusion of previously analyzed precursors for 60 s.
The “ScanSifter” algorithm v0.1, an in-house developed software, read tandem mass spectra stored as centroided peak lists from Thermo RAW files and transcoded them to DTA files. Spectra that contained fewer than 6 peaks or that had less than 20 measured total ion current (TIC) did not result in DTAs. If 90% of the intensity of a tandem mass spectrum appeared at a lower m/z than the precursor ion, a single precursor charge was assumed; otherwise, the spectrum was processed under both double and triple precursor charge assumptions. Proteins were identified using the SEQUEST v.27 algorithm29,30 on a high speed, multiprocessor Linux cluster in the Advanced Computing Center for Research & Education at Vanderbilt University using the human subset of the IPI human protein database, version 331, created 7/20/07. To estimate false discovery rates (FDRs), each sequence of the database was reversed and concatenated to the database, for a total of 135,168 entries for the human database. The database search encompassed tryptic peptides with a maximum of 5 missed cleavage sites for enzyme search and with a maximum number of 10 internal cleavage sites. Cysteines were expected to undergo carboxamidomethylation (+57 Da), and methionine oxidation (+16 Da) was permitted. DEB-induced alkylation at cysteine, arginine, lysine, histidine, and the N-terminus (hydrolyzed monoadduct: +104 Da; cross-link to guanine: +237 Da) were specified as dynamic modifications to identify spectra of adducted peptides. Precursor ions were required to fall within 1.25 m/z of the position expected from their average masses, and fragment ions were required to fall within 0.5 m/z of their monoisotopic positions. The database searches produced raw identifications in SQT format.31
Peptide identification, filtering, and protein assembly were done with IDPicker software 42, which filtered raw peptide identifications to a target FDR of 5%. The peptide filtering employed reversed sequence database match information to determine thresholds that yielded an estimated 5% FDR for the identifications of each charge state by the formula FDR = (2R)/(R+F), where R is the number of passing reversed peptide identifications and F is the number of passing forward (normal orientation) peptide identifications. A second round of filtering removed proteins supported by less than two distinct peptide identifications in the analyses. Indistinguishable proteins were recognized and grouped. Parsimony rules were applied to generate a minimal list of proteins that explained all of the peptides that passed our entry criteria.32 Finally, to be considered a positive identification, all proteins were required to have a minimum of three unique peptide spectra and 10 total spectral counts. Any protein identified in the DEB-treated samples that displayed comparable MS spectral counts to the untreated controls was disregarded.
Western Blot Analysis of Identified Proteins
Nuclear protein extracts from HeLa cells were exposed to 0–25 mM DEB in the presence of 5′-biotinylated oligonucleotide duplexes as described above, and the resulting DNA-protein cross-links were captured on streptavidin beads. The cross-linked proteins were released from the DNA backbone by thermal hydrolysis, separated by SDS-PAGE, and transferred to Trans-Blot nitrocellulose membranes (Bio-Rad, Hercules, CA). Membranes were immediately blocked in Tris-buffered saline (TBS) containing 5% (w/v) bovine serum albumin. Following a 1–2 h incubation with the primary antibody at room temperature, the blots were washed three times with TBS and incubated overnight at 4°C with the corresponding alkaline phosphatase-conjugated secondary antibody. Following three additional washes with TBS, the blots were developed using SIGMA Fast BCIP/NBT (Sigma, St. Louis, MO). The developed blots were scanned as image files, and the optical densities of the bands were then quantified using ImageJ software available free of charge from the NIH website (www.ncbi.nlm.nih.gov). The extent of DNA-protein cross-linking was estimated by comparing the band intensities in cross-linked samples to that of a known amount of nuclear proteins extract loaded on the gel as a positive control.
HPLC-ESI+-MS/MS Analysis of 1-(S-cysteinyl)-4-(guan-7-yl)-2,3-butanediol (Cys-N7G-BD)
Cross-linking reactions and biotin capture were carried out as described above, with the following exceptions: 1.3 mg total protein was incubated with 8 nmol of double-stranded 5′-bionylated oligodeoxynucleotides (5′-GGA GCT GGT GGC GTA GGC-3′, (+) strand) in the presence of 50 mM DEB, and the DPCs were eluted from the streptavidin beads by three incubations with 1 mL 70% acetonitrile/5% aqueous formic acid (overnight at 4°C, 30 min at room temperature, and 30 min at 90°C). Following each incubation, the beads were centrifuged for 1 min at 2,000g, and the supernatants containing DPCs were decanted and pooled. Cross-linked proteins were released from the DNA backbone by neutral thermal hydrolysis (1 h at 70°C) to produce protein-guanine conjugates. Samples were dried under vacuum, reconstituted in 25 mM ammonium bicarbonate, and digested to peptides with trypsin (20 μg, 0.86 nmol, 37°C overnight). To achieve complete hydrolysis of the peptides to amino acids, tryptic peptides were dried under vacuum, reconstituted in water, and digested with proteinase K (20 μg, 0.70 nmol) for 48 h at room temperature. To enrich for Cys-N7G-BD, the digest mixtures were purified by solid phase extraction. ExtractClean™ Carbo cartridges (150 mg, 4mL, Grace Davison Discovery Sciences, Deerfield, IL) were equilibrated with acetonitrile (3 mL), methanol (3 mL), and water (2 × 3 mL) prior to loading samples, which were adjusted to pH 9 by the addition of 1M ammonium hydroxide (1 mL total load volume). Samples were washed with water (3 mL), 50% methanol (3 mL), and 100% methanol (3 mL) prior to elution with 2:1 acetonitrile:water containing 1% formic acid (3 mL). The eluates were dried under vacuum and re-suspended in 15 mM ammonium acetate, pH 5.0 (25 μL) prior to HPLC-ESI+-MS/MS analysis (injection volume, 8 μL).
An Agilent 1100 capillary HPLC system interfaced to a Thermo-Finnigan TSQ Quantum Discovery mass spectrometer was utilized for MS detection of Cys-N7G-BD conjugates. Chromatographic separation was accomplished with a Phenomonex Synergi C18 column (250 mm × 0.5 mm, 4 μm) eluted with 15 mM ammonium acetate, pH 5.0 (A) and 3:1 methanol:acetonitrile (B) at a flow rate of 10 μL/min. The gradient program began at 2% B, followed by a linear increase to 8% B in 10 min, further to 11% B in 17 min, and finally to 30% B in 2 min. Using this gradient, Cys-N7G-BD eluted at ~19 min. ESI was achieved at a spray voltage of 3.2 kV and a capillary temperature of 250°C. CID was performed with Ar as a collision gas (1.0 mTorr) at a collision energy of 25V. The MS parameters were optimized for maximum response during infusion of a standard solution of Cys-N7G-BD. HPLC-ESI+-MS/MS analyses were performed in the selected reaction monitoring mode using the transition corresponding to a major fragment ion observed upon CID fragmentation of Cys-N7G-BD in a triple quadrupole mass spectrometer (m/z 359.7 [M + H]+ → 87.9 [M + H − Gua − C4H8O2S]+.
Results
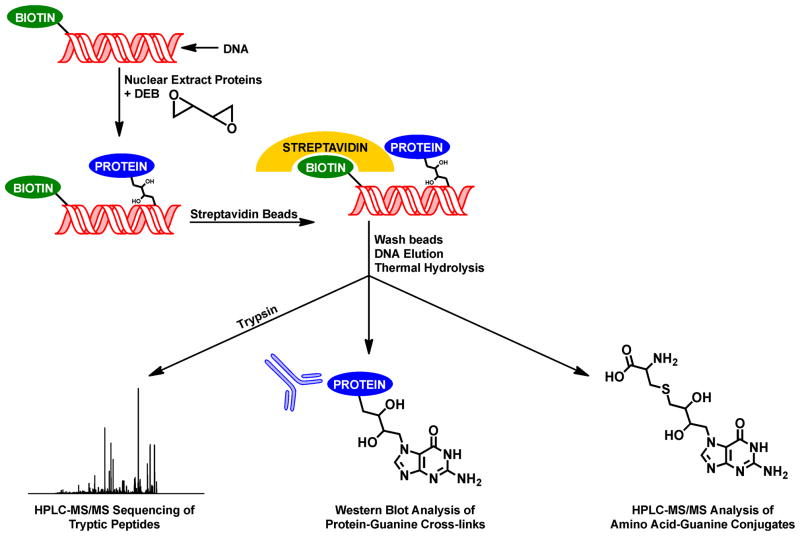
Strategy for Affinity Purification of Proteins Cross-linked to DNA by DEB
Our laboratory has recently developed an affinity capture-based methodology to investigate the in vitro formation of DPCs between nuclear extract proteins and biotinylated oligodeoxynucleotides.26 In this approach (Scheme 2), nuclear extract proteins are incubated with synthetic DNA duplexes containing a 5′-biotin tag (5′-GGA GCT GGT GGC GTA GGC-3′, derived from codons 10–15 of the K-ras protooncogene) in the absence or the presence of a bis-electrophile, such as DEB. Following affinity capture of biotinylated DNA and any covalently cross-linked proteins on streptavidin beads, the beads are subjected to stringent washing to remove any non-covalently bound proteins. Finally, the biotinylated DNA and the cross-linked proteins are eluted by heating the beads in SDS-containing gel loading buffer. Because the N7-alkylguanine adducts induced by DEB destabilize the glycosidic bond of DNA and are thus thermally labile, these elution conditions also release the cross-linked proteins from the DNA backbone, yielding protein-butanediol-guanine conjugates (Scheme 2). Following elution, the cross-linked proteins are separated by SDS-PAGE and are subjected to in-gel tryptic digestion followed by mass-spectrometry based proteomic analysis to identify the cross-linked proteins. Alternatively, the protein mixtures are analyzed by western blotting using commercial antibodies to target specific candidate proteins.
Scheme 2.
Experimental scheme for biotin capture enrichment of DPCs from nuclear protein extracts incubated with DEB in the presence of double-stranded DNA.
Mass Spectrometric Identification of Cross-Linked Proteins
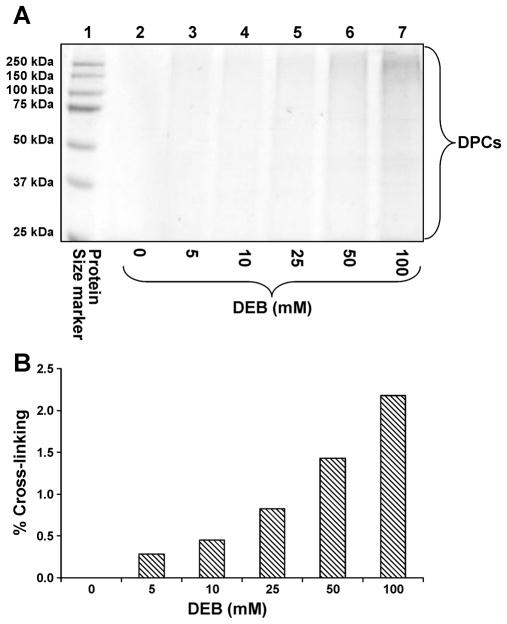
Previous studies have demonstrated the ability of AGT, GAPDH, and histone proteins to become cross-linked to DNA in vitro in the presence of DEB16,22,23 However, these studies were conducted using purified recombinant proteins in the absence of other cellular proteins which could also form DPCs or interfere with DNA-protein cross-linking. To determine whether DEB-induced DPC formation occurs in a more complex biological system, we employed the affinity capture approach developed in our laboratory26 (Scheme 2) to capture nuclear extract proteins from human cervical carcinoma (HeLa) cells which became cross-linked to biotinylated DNA duplexes in the presence of DEB. Following SDS-PAGE separation of the cross-linked proteins (Figure 1A), densitometric analysis of the stained protein bands over the 25 – 250 kDa molecular weight range showed a concentration-dependent increase in protein signal in samples treated with 5, 10, 25, 50, and 100 mM DEB, indicating the formation of DPCs (Figure 1B). Minimal DPC background was present in control samples prepared in the absence of DEB, signifying that the majority of non-covalently bounds proteins are successfully removed by the washing procedures (Figure 1A, lane 2). A DEB concentration of 100 mM was required to achieve cross-linking of 2% of total protein to DNA. By comparison, 100-fold lower concentrations were required to obtain similar levels of cross-linking with the antitumor nitrogen mustard, mechlorethamine,26 indicating that DEB is a less effective DPC-inducing agent than mechlorethamine.
Figure 1.
Concentration-dependent formation of DPCs following DEB exposure of nuclear protein extracts prepared from HeLa cells in the presence of DNA. (A) Nuclear protein extracts from HeLa cells (500 μg) and 5′-biotinylated double-stranded oligodeoxynucleotides (3.12 nmol) were incubated in the presence of 0 (lane 2), 5 (lane 3), 10 (lane 4), 25 (lane 5), 50 (lane 6), or 100 mM DEB (lane 7). The resulting DPCs were subjected to biotin capture enrichment, hydrolyzed to release protein-guanine conjugates, and resolved by 12% SDS-PAGE. Gels were stained with SimplyBlue SafeStain to visualize the cross-linked proteins. (B) Densitometric analysis of protein bands in the 25 – 250 kDa molecular weight range to estimate the extent of total protein cross-linking to DNA in the presence of DEB. Band intensity was compared to staining of a known amount of nuclear protein extract analyzed as a control to estimate the cross-linking efficiency.
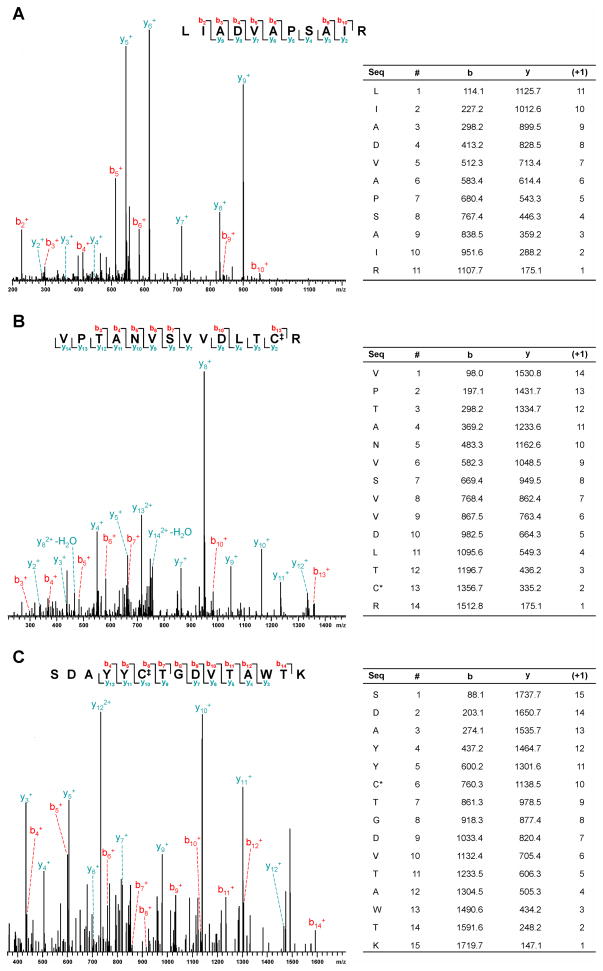
In order to identify human nuclear proteins which participate in DEB-induced DPC formation, nuclear protein extracts from HeLa cells were incubated in triplicate with biotinylated DNA in the absence or in the presence of DEB (500 mM). This high drug concentration was selected in order to maximize the number of low-abundance proteins identified in the proteomic analysis, due to the low numbers of DPCs observed in preliminary experiments (Figure 1). Following biotin capture enrichment (Scheme 2), the cross-linked proteins were separated by SDS-PAGE (Figure S1), and the gel lanes were separated into five fractions encompassing the entire molecular weight range and excised. Proteins in these gel pieces were subjected to in-gel tryptic digestion,28 and the resulting tryptic peptides were extracted and analyzed by HPLC-ESI+-MS/MS analysis to identify the cross-linked proteins. Protein identifications were based on the MS/MS spectra of unmodified tryptic peptides, as shown for representative peptides in Figure 2. Spectral data were subjected to parsimony analysis, resulting in the identification of a total of 39 HeLa nuclear proteins which participated in cross-linking to DNA in the presence of DEB (Table 1, Supplement S-3).
Figure 2.
Examples of HPLC-ESI+-MS/MS spectra of tryptic peptides used for protein identification: Fen-1 (A), GAPDH (B), and PARP (C) present in affinity-captured DPCs. ‡Cysteine carboxamidomethylation (+57).
Table 1.
HeLa nuclear extract proteins that form DNA-protein cross-links in the presence of 1,2,3,4-diepoxybutane (DEB)*
| Swiss-Prot-ID | Protein | Peptide Sequences | Total Spectra | % Coverage | Functional Classification | Protein MW (Da) | No. of Cysteines | No. of Lysines | Forms DPCs by Mechlorethamine? |
|---|---|---|---|---|---|---|---|---|---|
| P60709 | Actin | 16 | 225 | 47 | Architectural/Structural | 41737 | 6 | 19 | Yes |
| P68363 | Tubulin α-1B chain | 12 | 57 | 39 | 50152 | 12 | 19 | No | |
| P07437 | Tubulin β chain | 12 | 54 | 36 | 49671 | 8 | 15 | No | |
| P11586 | C-1-tetrahydrofolate synthase | 12 | 30 | 15 | Cellular Homeostasis/Cell Cycle | 101559 | 12 | 62 | No |
| O43175 | D-3-phosphoglycerate dehydrogenase | 13 | 55 | 31 | 56651 | 13 | 26 | No | |
| P04406 | Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) | 5 | 20 | 21 | 36053 | 3 | 26 | Yes | |
| P08107 | Heat shock 70 kDa protein 1 | 14 | 34 | 31 | 70052 | 5 | 50 | No | |
| P52789 | Hexokinase-2 | 14 | 32 | 20 | 102380 | 25 | 55 | No | |
| P12081 | Histidyl-tRNA synthetase | 6 | 18 | 15 | 57411 | 10 | 43 | No | |
| P22102 | Isoform Long of Trifunctional purine biosynthetic protein adenosine-3 | 7 | 15 | 10 | 107767 | 22 | 69 | No | |
| P14618 | Isoform M2 of Pyruvate kinase isozymes M1/M2 | 29 | 223 | 60 | 57937 | 10 | 37 | No | |
| P17987 | T-complex protein 1 subunit α | 8 | 16 | 17 | 60344 | 9 | 40 | No | |
| P50991 | T-complex protein 1 subunit δ | 5 | 17 | 11 | 57924 | 9 | 39 | No | |
| Q99832 | T-complex protein 1 subunit η | 6 | 13 | 14 | 59367 | 9 | 41 | No | |
| P40227 | T-complex protein 1 subunit ζ | 5 | 12 | 13 | 58024 | 8 | 45 | No | |
| Q00341 | Vigilin | 12 | 28 | 13 | 141456 | 11 | 109 | No | |
| Q9NTJ3 | Isoform 2 of Structural maintenance of chromosomes protein 4 | 6 | 10 | 5 | Chromatin Regulation | 147182 | 12 | 148 | No |
| Q16269 | Regulator of chromosome condensation 1 isoform a | 9 | 27 | 32 | 4915 | 2 | 5 | No | |
| P43246 | DNA mismatch repair protein Msh2 | 15 | 35 | 21 | DNA Repilication/Repair | 104743 | 13 | 70 | Yes |
| P28340 | DNA polymerase δ catalytic subunit | 6 | 13 | 6 | 12363 | 26 | 46 | No | |
| P39748 | Flap endonuclease 1 (Fen-1) | 4 | 13 | 16 | 42593 | 6 | 38 | Yes | |
| P33993 | Isoform 1 of DNA replication licensing factor MCM7 | 6 | 11 | 10 | 81308 | 11 | 32 | No | |
| P52701 | Isoform GTBP-N of DNA mismatch repair protein Msh6 | 13 | 24 | 10 | 152786 | 32 | 101 | No | |
| P09874 | Poly [ADP-ribose] polymerase 1 (PARP) | 10 | 20 | 13 | 113084 | 14 | 126 | Yes | |
| O00148 | ATP-dependent RNA helicase DDX39 | 13 | 63 | 33 | Transcription/Translation/RNA splicing | 49130 | 9 | 28 | No |
| P68104 | Elongation factor 1-α 1 (EF-1α1) | 14 | 147 | 45 | 50141 | 6 | 47 | Yes | |
| P26641 | Elongation factor 1-γ | 7 | 21 | 15 | 50119 | 6 | 32 | No | |
| P13639 | Elongation factor 2 | 12 | 24 | 17 | 95338 | 17 | 65 | No | |
| P60842 | Eukaryotic initiation factor 4A-I | 8 | 29 | 25 | 46154 | 4 | 20 | Yes | |
| P78347 | Isoform 1 of General transcription factor II-I | 11 | 17 | 14 | 112416 | 9 | 85 | No | |
| P61978 | Isoform 1 of Heterogeneous nuclear ribonucleoprotein K | 7 | 18 | 23 | 50976 | 5 | 22 | Yes | |
| P52272 | Isoform 1 of Heterogeneous nuclear ribonucleoprotein M | 5 | 11 | 9 | 77516 | 5 | 40 | Yes | |
| P26599 | Isoform 1 of Polypyrimidine tract-binding protein 1 | 7 | 27 | 21 | 57221 | 3 | 33 | Yes | |
| Q9Y265 | Isoform 1 of RuvB-like 1 | 6 | 14 | 17 | 50228 | 6 | 41 | No | |
| Q7L1Q6 | Isoform 2 of Basic leucine zipper and W2 domain-containing protein 1 | 3 | 13 | 12 | 48043 | 3 | 43 | No | |
| Q00839 | Isoform Short of Heterogeneous nuclear ribonucleoprotein U (hnRNP-U) | 5 | 12 | 8 | 90584 | 13 | 70 | No | |
| Q7KZF4 | Staphylococcal nuclease domain-containing protein 1 | 14 | 25 | 21 | 101997 | 12 | 58 | No | |
| A8K7X6 | poly(rC)-binding protein 2 isoform β | 4 | 11 | 14 | 38222 | 7 | 17 | No | |
| Q68DF1 | 91 kDa protein (putatuve uncharacterized protein DKFZP779B0247) | 7 | 12 | 11 | 90555 | 12 | 7 | No | |
500 mM for 3h
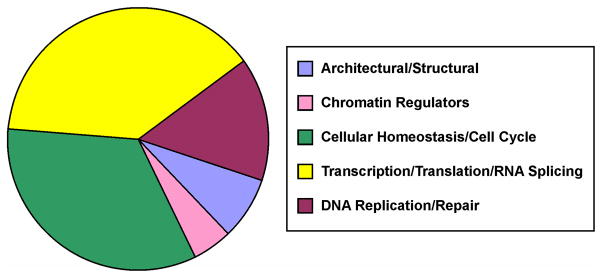
The proteins identified in affinity captured DPCs that were induced by DEB exposure encompass a variety of cellular functions (Figure 3). Over half of the identified proteins are known nucleic acid binding proteins known to participate in transcriptional regulation (e.g. PARP, heterogeneous nuclear ribonucleoprotein U (hnRNP-U), GAPDH, Ref-1, actin),33–37 chromatin remodeling (e.g. actin),37 DNA replication (e.g. DNA polymerase δ),38 and DNA repair (e.g. Ref-1, AGT, PARP, flap endonuclease-1 (Fen-1), Msh2).36,39–42 We also observed proteins involved in cell motility (e.g. actin),37 protein folding (e.g. T complex proteins),43 and biosynthesis (e.g. histidyl tRNA synthetase).44 Ten of the proteins identified in biotin capture fractions of DEB-induced DPCs, including actin, GAPDH, EF-1α1, PARP, and Fen-1, have been previously shown to form cross-links to DNA in the presence of the antitumor nitrogen mustard mechlorethamine (Table 1).26 Additionally, several of the identified proteins were closely related to proteins that form DPCs in the presence of mechlorethamine (e.g. different isoforms of tubulin). Altogether, approximately 25% the 39 proteins identified in the present work also formed DPCs in the presence of mechlorethamine.26 The remaining proteins were not common between the two lists, which is not surprising given the distinct mechanisms of cross-link formation by the two bis-electrophiles.
Figure 3.
Cellular functions of human proteins that form DPCs in the presence of DEB, as identified by affinity capture in combination with mass spectrometry-based proteomics.
The proteins identified in our proteomics screen were additionally categorized according to the molecular weight region of the gel in which they were present prior to in gel tryptic digestion (Figure S1). Peptides from all identified proteins were found in the correct molecular weight region as predicted by the protein’s molecular weight (Table 1). In addition, many proteins were also present in higher molecular weight fractions, suggesting that DEB can form ternary DNA-protein-protein complexes.
Identification and Quantitation of Individual Proteins that Cross-Link DNA by Western Blot Analysis
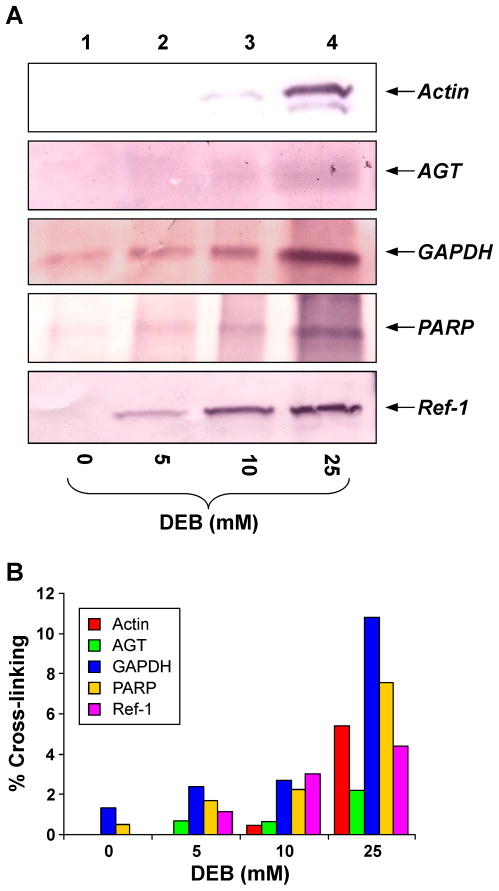
The identities of a subset of proteins which were detected by mass spectrometry-based proteomics were further confirmed by western blot analysis using commercial antibodies. Nuclear protein extracts derived from HeLa cells were incubated with biotinylated DNA in the presence of 0– 25 mM DEB to induce DPC formation. Following biotin capture (Scheme 2), thermal hydrolysis, and SDS-PAGE separation, protein-guanine conjugates were transferred to nitrocellulose membranes and subjected to western blot analysis using commercial antibodies against actin, AGT, GAPDH, PARP, Ref-1, and Ku (Figure 4). These proteins were selected based on either their identification from the proteomics screen (actin, PARP, GAPDH) or their previously demonstrated ability to form DPCs in the presence of other bis-electrophiles, such as antitumor nitrogen mustards (AGT, Ref-1, Ku).26 Densitometric analysis of protein signals observed in the western blots of biotin capture mixtures and total nuclear extracts was used to estimate the efficiency of DPC formation for specific proteins in the presence of DEB.
Figure 4.
Western blot analysis of DEB-induded DPCs in nuclear protein extracts from HeLa cells. (A) Nuclear extract proteins were incubated with 0 (lane 1), 5 (lane 2), 10 (lane 3), or 25 mM DEB (lane 4) in the presence of 5′-biotinylated double-stranded oligodeoxynucleotides (5′-GGA GCT GGT GGC GTA GGC-3′ (+) strand). Following biotin capture enrichment and hydrolysis to release protein-guanine conjugates, the cross-linked proteins were resolved by SDS-PAGE and transferred to nitrocellulose membranes. Western blots were performed using primary antibodies against actin, AGT, GAPDH, PARP, and Ref-1. (B) Densitometric analysis of western blots to estimate the extent of protein cross-linking to DNA in the presence of DEB.
Western blot analysis confirmed the identities of three proteins detected in mass spectrometric analysis of DEB-induced DPCs: actin, GAPDH, and PARP (Figure 4A). In addition, AGT and Ref-1 were found to form DPCs in the presence of DEB (Figure 4A). These proteins were not detected in the mass spectrometry-based proteomics screen (Table 1), likely due to their low abundance in the nucleus.45 The intensity of antigen-specific staining of streptavidin-captured fractions increased as DEB concentration was increased from 5 to 25 mM (Figure 4A, lanes 2–4). Densitometric analysis revealed that between 2 and 12% of total protein became cross-linked to DNA in the presence of 25 mM DEB (Figure 4B), with GAPDH forming the largest number of DPCs (~12% of total protein). Interestingly, the overall participation of all proteins in cross-linking to DNA under these conditions was less than 1% (Figure 1), suggesting that actin, AGT, GAPDH, PARP, and Ref-1 have a higher propensity to form DPCs in the presence of DEB as compared to other nuclear proteins. The DNA helicase protein Ku, which is present in the nuclear protein extract at high abundance, did not form detectable levels of DPCs in the presence of DEB (results not shown).
The requirement for high concentrations of DEB (5–25 mM) in order to induce DPC formation in our experiments (Figure 4) is consistent with previous studies investigating DEB-induced cross-linking to recombinant proteins.16,22,23 However, these concentrations are significantly higher than those required to obtain similar levels of cross-linking with antitumor nitrogen mustards. For example, about 3% of total Ref-1 in HeLa nuclear protein extracts became cross-linked to DNA in the presence of 10 mM DEB, but a similar cross-linking efficiency can be achieved in the presence of 0.5 mM mechlorethamine.26 Our recent data indicate that even lower drug concentrations are required for detection of DPCs in analogous experiments with cis-diamminedichloroplatinum (cisplatin), with approximately 3% of total Ref-1 proteins forming DPCs in the presence of 1 μM cisplatin (Ming et al, manuscript in preparation). These results indicate a 104-fold difference between the efficiency of DPC formation by different bifunctional alkylating agents. These pronounced differences may be attributable to variations in chemical reactivity, distinct mechanisms for cross-link formation, and different levels of hydrolytic stability of the resulting DPC lesions.
Identification of Protein Side Chains Participating in DPC Formation
The identities of protein amino acid side chains which participate in DEB-induced DPC formation were investigated by considering the MS/MS spectra of peptides containing DEB-induced adducts and by analyzing amino acid-nucleobase conjugates in total enzymatic digests of affinity captured DPCs. HPLC-MS/MS analysis of tryptic digests detected a peptide representing residues 207–238 of actin containing a DEB-mediated cysteine cross-link to guanine (Figure S2), and two peptides originating from actin and hnRNP-U implicated the involvement of lysine residues in DPC formation by DEB (results not shown). Furthermore, two peptides from elongation factor 1-α1 (EF-1α1) contained the hydrolyzed trihydroxybutyl monoadducts at cysteine and lysine residues. These results suggest that cysteine and lysine side chains of proteins can participate in DEB-mediated DPC formation.
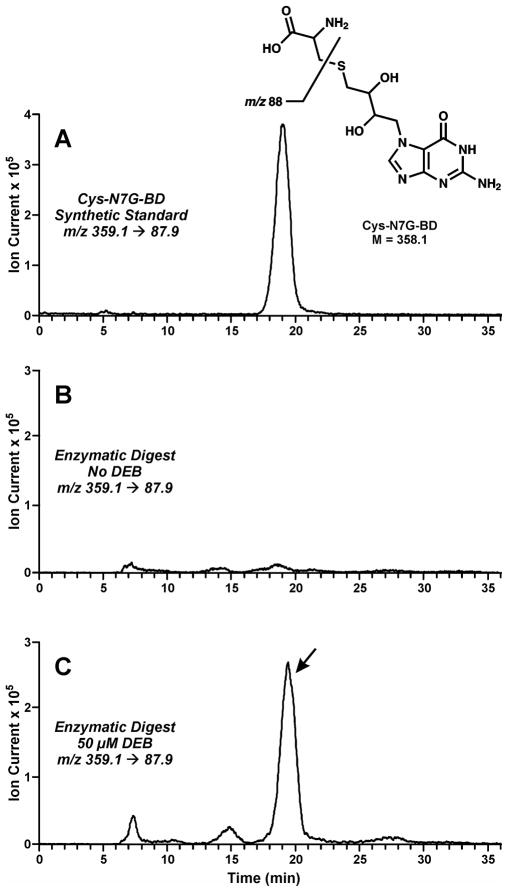
To determine the exact chemical structure of the DEB-induced amino acid-nucleobase conjugates, DNA and cross-linked proteins eluted from streptavidin beads were subjected to thermal hydrolysis to release protein-nucleobase conjugates, followed by digestion with trypsin and proteinase K to achieve complete proteolytic digestion of proteins to amino acids (Scheme 2, right). The resulting amino acid-nucleobase conjugates were enriched by solid phase extraction and analyzed by capillary HPLC-ESI+-MS/MS. Our previous studies conducted with recombinant AGT protein indicated that DEB-induced cross-linking takes place primarily between the cysteine sulfhydryl side chain within proteins and the N7-position to form 1-(S-cysteinyl)-4-(guan-7-yl)-2,3-butanediol (Cys-N7G-BD) conjugates. 16
Our selected reaction monitoring method for Cys-N7G-BD was based on a characteristic mass transition (m/z 359.1→ 87.9) corresponding to C-S bond cleavage within the cysteine moiety of Cys-N7G-BD (Figure 5A). Cys-N7G-BD was detected only in the total digests of DEB-treated nuclear extracts (Figure 5C), but not in the untreated control (Figure 5B). The presence of the Cys-N7G-BD conjugate in treated samples indicates that DEB forms covalent cross-links between the N7 position of guanine in duplex DNA and the side-chain sulfhydryls of cysteine residues in proteins. These results are consistent with our MS/MS data for alkylated peptides, which reveal DEB-mediated cross-links to guanine involving cysteine residues (Figure S2). Consistent with this interpretation, all of the identified proteins contain multiple cysteine residues (Table 1).
Figure 5.
HPLC-ESI+-MS/MS analysis of 1-(S-cysteinyl)-4-(guan-7-yl)-2,3-butanediol (Cys-N7G-BD) conjugates in total proteolytic digests of DEB-induced DPCs. Nuclear protein extracts from HeLa cells were exposed to DEB in the presence of biotinylated DNA duplexes. Following biotin capture enrichment, DPCs were subjected to thermal hydrolysis and enzymatic digestion of proteins to amino acids to release amino acid-nucleobase conjugates: (A) Synthetic Cys-N7G-BD; (B) enzymatic digests of HeLa nuclear protein extracts following incubation with DNA in the absence of DEB (negative control); (C) enzymatic digests of HeLa nuclear protein extracts following incubation with 50 mM DEB in the presence of DNA.
Discussion
We have developed a comprehensive approach for identification of the proteins that participate in DNA-protein cross-linking in the presence of bis-electrophiles in cell-free extracts (Scheme 2). This approach enabled us to identify 39 HeLa nuclear proteins which became cross-linked to DNA following exposure to 1,2,3,4-diepoxybutane (DEB). Among these, approximately 25% of proteins are also targeted for cross-linking to DNA in the presence of nitrogen mustards,26 while the remaining proteins appear unique to DEB-mediated cross-linking.
The ability of specific proteins to form DPCs is likely to be influenced by a number of factors, including protein abundance, structure, and cellular and nuclear localization. For example, DNA-binding proteins are in close proximity to DNA and readily available to form DPCs in the presence of cross-linking agents such as DEB. Several of the proteins found to form DEB-mediated DPCs, such as those involved in DNA replication and repair, are known to associate with DNA in the nucleus. For example, the DNA repair protein poly [ADP-ribose] polymerase I (PARP) is involved in the DNA damage signaling response, and has been previously shown to form cross-links to DNA as a result of exposure to nitrogen mustards,26 ionizing radiation,4 and formaldehyde.21 No DPC lesions involving histone proteins were detected, which could be a result of their low concentrations in our nuclear protein extracts prepared according to Jessberger et al.,27 their inefficient cross-linking to DNA, or their disrupted association with DNA. Interestingly, some other proteins identified in our screen (e.g. GAPDH, actin) (Table 1) are involved in cellular processes which do not involve association with DNA (e.g. glycolysis, cell motility). However, additional cellular roles have been proposed for many of these proteins, which would explain their ability to associate with DNA and form DPCs. For example, actin is primarily known for its role in cell motility, but is also believed to have regulatory roles in chromatin remodeling, DNA replication, and transcription.46,47
The biotinylated 18-mer DNA sequence used in the present study is derived from the frequently mutated region of the K-ras protooncogene (codons 10–15, 5′-GGA GCT GGT GGC GTA GGC-3′). The same sequence has been previously utilized for biotin capture of DPCs induced by other cross-linking agents, e.g. mechlorethamine48 and cispaltin (Ming and Tretyakova, manuscript in preparation), to allow for a direct comparison between different electrophiles. While it is possible that the selection of DNA sequence affects the types of proteins captured, systematic evaluation of the effects of DNA sequence on DPC formation was beyond the scope of the present study.
The biological consequences of DNA-protein cross-linking are not fully understood. It is hypothesized that the presence of bulky, helix-distorting DPC lesions interrupts critical cellular metabolic processes and results in damaging cytotoxic and genotoxic effects.1 Previous studies have shown that certain types of DPCs can persist through several cycles of DNA replication, resulting in permanent DNA alterations.1,10,49 Possible mechanisms for DPC repair include nucleotide excision repair (NER), homologous recombination (HR), and proteolytic degradation.50,51 More than one repair mechanism may be required.1,50,51 For example, de Graaf et. al. have reported that HR-deficient yeast strains showed the greatest sensitivity to chronic, low-exposure doses of formaldehyde, whereas NER-deficient yeast strains showed increased sensitivity to acute formaldehyde exposure.52 Nakano and colleagues have reported that the DPC repair mechanism is dependent on the size of the cross-linked protein, with NER repairing cross-links involving peptides and small proteins, and HR repairing cross-links involving larger proteins.50,51 The upper size limit of cross-linked proteins for NER repair of DPCs in bacteria was 12–14 kDa,50 whereas in mammalian cells it was 8–10 kDa.51 This would suggest that the majority of DPCs induced by DEB are expected to be repaired by HR, due to their significant size (Table 1).
In conclusion, our study demonstrates the ability DEB to covalently cross-link a number of nuclear proteins to DNA. Although the DEB concentrations employed in this work are, for practical reasons, much higher than typical levels of human exposure, these results may still hold biological relevance due to the likelihood that DPCs are also formed in cells exposed to low levels of DEB. However, it should be noted that the induction of DPCs in vitro requires much higher concentrations of DEB relative to mechlorethamine and cisplatin. This is potentially of interest, considering that these agents are nearly equi-toxic in human fibrosarcoma HT1080 cells (Kurtz and Campbell, unpublished results). Taken together, these findings suggest that DPCs may contribute less to the cytotoxicity associated with DEB exposure as compared to other bis-electrophiles investigated previously (e.g. cisplatin, mechlorethamine). Regardless, the formation of these bulky, helix-distorting DPC lesions would have considerable potential to interfere with critical cellular processes such as replication and transcription, ultimately triggering programmed cell death or genotoxic outcomes.
Supplementary Material
Acknowledgments
We thank Prof. Jonathan Marchant (University of Minnesota) for generously providing HeLa cells, Brock Matter and Melissa Goggin (University of Minnesota Masonic Cancer Center) for assistance with MS experiments, Cate Tsufis for preparation of Cys-N7G-BD samples, Jamie Kurtz for help with cell culture, and Bob Carlson for preparing figures for this manuscript. Funding for this research was provided by the National Cancer Institute (R01-CA-100670), the American Chemistry Council, and a faculty development grant from the University of Minnesota Academic Health Center. E.M.R. and R.L. were partially supported by the NIH Chemistry-Biology Interface Training Grant (T32-GM08700), University of Minnesota Masonic Cancer Center, and University of Minnesota Graduate School. D.C.L. and S.G.C. acknowledge support from NIH grants ES010056 and ES013125.
Abbreviations
- AGT
O6-alkylguanine DNA alkyltransferase
- BD
1,3-butadiene
- cisplatin
cis-diamminodichloroplatinum
- Cys-N7G-BD
1-(S-cysteinyl)-4-(guan-7-yl)-2,3-butanediol
- DEB
1,2,3,4-diepoxybutane
- DPC
DNA-protein cross-link
- DTT
dithiothreitol
- EF-1α1
elongation factor 1-α1
- FDR
false discovery rate
- Fen-1
flap endonuclease 1
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- hnRNP-U
heterogeneous nuclear ribonucleoprotein U
- HPLC-ESI+-MS/MS
high-performance liquid chromatography-electrospray ionization-tandem mass spectrometry
- HR
homologous recombination
- Ku
ATP-dependent DNA helicase subunit 2
- NER
nucleotide excision repair
- PARP
poly(ADP-ribose) polymerase
- PBS
phosphate-buffered saline
- PMSF
phenylmethanesulfonyl fluoride
- Ref-1
DNA-(apurinic- or apyrimidinic-site) lyase
- TBS
Tris-buffered saline
- TIC
total ion current
Footnotes
Supporting Information Available: SDS-PAGE analysis of DPCs in nuclear protein extracts prepared from human cervical carcinoma HeLa cells following exposure to DEB, HPLC-ESI+-MS/MS analysis of a tryptic peptide from actin containing DEB-induced cross-link to guanine, and a list of peptides used for protein identification, database search scores etc. This information is available free of charge via the Internet at http://pubs.acs.org/.
Reference List
- 1.Barker S, Weinfeld M, Murray D. DNA-protein crosslinks: their induction, repair, and biological consequences. Mutat Res. 2005;589:111–135. doi: 10.1016/j.mrrev.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 2.Dizdaroglu M, Gajewski E. Structure and mechanism of hydroxyl radical-induced formation of a DNA-protein cross-link involving thymine and lysine in nucleohistone. Cancer Res. 1989;49:3463–3467. [PubMed] [Google Scholar]
- 3.Dizdaroglu M, Gajewski E, Reddy P, Margolis SA. Structure of a hydroxyl radical induced DNA-protein cross-link involving thymine and tyrosine in nucleohistone. Biochemistry. 1989;28:3625–3628. doi: 10.1021/bi00434a071. [DOI] [PubMed] [Google Scholar]
- 4.Barker S, Weinfeld M, Zheng J, Li L, Murray D. Identification of mammalian proteins cross-linked to DNA by ionizing radiation. J Biol Chem. 2005;280:33826–33838. doi: 10.1074/jbc.M502477200. [DOI] [PubMed] [Google Scholar]
- 5.Zhitkovich A, Voitkun V, Kluz T, Costa M. Utilization of DNA-protein cross-links as a biomarker of chromium exposure. Environ Health Perspect. 1998;106(Suppl 4):969–974. doi: 10.1289/ehp.98106s4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomas CB, Kohn KW, Bonner WM. Characterization of DNA-protein cross-links formed by treatment of L1210 cells and nuclei with bis(2-chloroethyl)methylamine (nitrogen mustard) Biochemistry. 1978;17:3954–3958. doi: 10.1021/bi00612a012. [DOI] [PubMed] [Google Scholar]
- 7.Baker JM, Parish JH, Curtis JP. DNA-DNA and DNA-protein crosslinking and repair in Neurospora crassa following exposure to nitrogen mustard. Mutat Res. 1984;132:171–179. doi: 10.1016/0167-8817(84)90035-x. [DOI] [PubMed] [Google Scholar]
- 8.Kloster M, Kostrhunova H, Zaludova R, Malina J, Kasparkova J, Brabec V, Farrell N. Trifunctional dinuclear platinum complexes as DNA-protein cross-linking agent. Biochemistry. 2004;43:7776–7786. doi: 10.1021/bi030243e. [DOI] [PubMed] [Google Scholar]
- 9.Ewig RA, Kohn KW. DNA-protein cross-linking and DNA interstrand cross-linking by haloethylnitrosoureas in L1210 cells. Cancer Res. 1978;38:3197–3203. [PubMed] [Google Scholar]
- 10.Oleinick NL, Chiu SM, Ramakrishnan N, Xue LY. The formation, identification, and significance of DNA-protein cross-links in mammalian cells. Br J Cancer Suppl. 1987;8:135–140. [PMC free article] [PubMed] [Google Scholar]
- 11.Swenberg JA, Boysen G, Georgieva N, Bird MG, Lewis RJ. Future directions in butadiene risk assessment and the role of cross-species internal dosimetry. Chem Biol Interact. 2007;166:78–83. doi: 10.1016/j.cbi.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 12.Morrow NL. The industrial production and use of 1,3-butadiene. Environ Health Perspect. 1990;86:7–8. doi: 10.1289/ehp.90867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hecht SS. Tobacco smoke carcinogens and lung cancer. J Natl Cancer Inst. 1999;91:1194–1210. doi: 10.1093/jnci/91.14.1194. [DOI] [PubMed] [Google Scholar]
- 14.Lawley PD, Brookes P. Interstrand cross-linking of DNA by difunctional alkylating agents. J Mol Biol. 1967;25:143–160. doi: 10.1016/0022-2836(67)90285-9. [DOI] [PubMed] [Google Scholar]
- 15.Jelitto B, Vangala RR, Laib RJ. Species differences in DNA damage by butadiene: role of diepoxybutane. Arch Toxicol Suppl. 1989;13:246–249. doi: 10.1007/978-3-642-74117-3_42. [DOI] [PubMed] [Google Scholar]
- 16.Loeber R, Rajesh M, Fang Q, Pegg AE, Tretyakova N. Cross-linking of the human DNA repair protein O6-alkylguanine DNA alkyltransferase in the presence of 1,2,3,4-diepoxybutane. Chem Res Toxicol. 2006;19:645–654. doi: 10.1021/tx0600088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Costa M, Zhitkovich A, Harris M, Paustenbach D, Gargas M. DNA-protein cross-links produced by various chemicals in cultured human lymphoma cells. J Toxicol Environ Health. 1997;50:433–449. doi: 10.1080/00984109708984000. [DOI] [PubMed] [Google Scholar]
- 18.Smith JC, Lambert JP, Elisma F, Figeys D. Proteomics in 2005/2006: developments, applications and challenges. Anal Chem. 2007;79:4325–4343. doi: 10.1021/ac070741j. [DOI] [PubMed] [Google Scholar]
- 19.Wong DL, Pavlovich JG, Reich NO. Electrospray ionization mass spectrometric characterization of photocrosslinked DNA-EcoRI DNA methyltransferase complexes. Nucleic Acids Res. 1998;26:645–649. doi: 10.1093/nar/26.2.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sperry JB, Shi X, Rempel DL, Nishimura Y, Akashi S, Gross ML. A mass spectrometric approach to the study of DNA-binding proteins: interaction of human TRF2 with telomeric DNA. Biochemistry. 2008;47:1797–1807. doi: 10.1021/bi702037p. [DOI] [PubMed] [Google Scholar]
- 21.Qiu H, Wang Y. Exploring DNA-binding proteins with in vivo chemical cross-linking and mass spectrometry. J Proteome Res. 2009;8:1983–1991. doi: 10.1021/pr8009319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loecken EM, Guengerich FP. Reactions of glyceraldehyde 3-phosphate dehydrogenase sulfhydryl groups with bis-electrophiles produce DNA-protein cross-links but not mutations. Chem Res Toxicol. 2007;21:453–458. doi: 10.1021/tx7003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loecken EM, Dasari S, Hill S, Tabb DL, Guengerich FP. The bis-electrophile diepoxybutane cross-links DNA to human histones but does not result in enhanced mutagenesis in recombinant systems. Chem Res Toxicol. 2009;22:1069–1076. doi: 10.1021/tx900037u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Valadez JG, Liu L, Loktionova NA, Pegg AE, Guengerich FP. Activation of bis-electrophiles to mutagenic conjugates by human O6-alkylguanine-DNA alkyltransferase. Chem Res Toxicol. 2004;17:972–982. doi: 10.1021/tx049897u. [DOI] [PubMed] [Google Scholar]
- 25.Guengerich FP. Principles of covalent binding of reactive metabolites and examples of activation of bis-electrophiles by conjugation. Arch Biochem Biophys. 2005;433:369–378. doi: 10.1016/j.abb.2004.07.035. [DOI] [PubMed] [Google Scholar]
- 26.Loeber RL, Michaelson-Richie ED, Codreanu SG, Liebler DC, Campbell CR, Tretyakova NY. Proteomic analysis of DNA-protein cross-linking by antitumor nitrogen mustards. Chem Res Toxicol. 2009;22:1151–1162. doi: 10.1021/tx900078y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jessberger R, Berg P. Repair of deletions and double-strand gaps by homologous recombination in a mammalian in vitro system. Mol Cell Biol. 1991;11:445–457. doi: 10.1128/mcb.11.1.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shin NY, Liu Q, Stamer SL, Liebler DC. Protein targets of reactive electrophiles in human liver microsomes. Chem Res Toxicol. 2007;20:859–867. doi: 10.1021/tx700031r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yates JR, III, Eng JK, McCormack AL. Mining genomes: correlating tandem mass spectra of modified and unmodified peptides to sequences in nucleotide databases. Anal Chem. 1995;67:3202–3210. doi: 10.1021/ac00114a016. [DOI] [PubMed] [Google Scholar]
- 30.Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. Journal of the American Society for Mass Spectrometry. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 31.McDonald WH, Tabb DL, Sadygov RG, MacCoss MJ, Venable J, Graumann J, Johnson JR, Cociorva D, Yates JR., III MS1, MS2, and SQT-three unified, compact, and easily parsed file formats for the storage of shotgun proteomic spectra and identifications. Rapid Commun Mass Spectrom. 2004;18:2162–2168. doi: 10.1002/rcm.1603. [DOI] [PubMed] [Google Scholar]
- 32.Zhang B, Chambers MC, Tabb DL. Proteomic parsimony through bipartite graph analysis improves accuracy and transparency. J Proteome Res. 2007;6:3549–3557. doi: 10.1021/pr070230d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jagtap P, Szabo C. Poly(ADP-ribose) polymerase and the therapeutic effects of its inhibitors. Nat Rev Drug Discov. 2005;4:421–440. doi: 10.1038/nrd1718. [DOI] [PubMed] [Google Scholar]
- 34.Gao C, Guo H, Mi Z, Wai PY, Kuo PC. Transcriptional regulatory functions of heterogeneous nuclear ribonucleoprotein-U and -A/B in endotoxin-mediated macrophage expression of osteopontin. J Immunol. 2005;175:523–530. doi: 10.4049/jimmunol.175.1.523. [DOI] [PubMed] [Google Scholar]
- 35.Sirover MA. New nuclear functions of the glycolytic protein, glyceraldehyde-3-phosphate dehydrogenase, in mammalian cells. J Cell Biochem. 2005;95:45–52. doi: 10.1002/jcb.20399. [DOI] [PubMed] [Google Scholar]
- 36.Xanthoudakis S, Curran T. Redox regulation of AP-1: a link between transcription factor signaling and DNA repair. Adv Exp Med Biol. 1996;387:69–75. [PubMed] [Google Scholar]
- 37.Coulombe RA, Jr, Drew GL, Stermitz FR. Pyrrolizidine alkaloids crosslink DNA with actin. Toxicol Appl Pharmacol. 1999;154:198–202. doi: 10.1006/taap.1998.8552. [DOI] [PubMed] [Google Scholar]
- 38.Burgers PM. Polymerase dynamics at the eukaryotic DNA replication fork. J Biol Chem. 2009;284:4041–4045. doi: 10.1074/jbc.R800062200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pegg AE. Repair of O6-alkylguanine by alkyltransferases. Mutat Res. 2000;462:83–100. doi: 10.1016/s1383-5742(00)00017-x. [DOI] [PubMed] [Google Scholar]
- 40.Nguewa PA, Fuertes MA, Valladares B, Alonso C, Perez JM. Poly(ADP-ribose) polymerases: homology, structural domains and functions. Novel therapeutical applications. Prog Biophys Mol Biol. 2005;88:143–172. doi: 10.1016/j.pbiomolbio.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 41.Finger LD, Blanchard MS, Theimer CA, Sengerova B, Singh P, Chavez V, Liu F, Grasby JA, Shen B. The 3′-flap pocket of human flap endonuclease 1 is critical for substrate binding and catalysis. J Biol Chem. 2009;284:22184–22194. doi: 10.1074/jbc.M109.015065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mukherjee S, Law SM, Feig M. Deciphering the mismatch recognition cycle in MutS and MSH2-MSH6 using normal-mode analysis. Biophys J. 2009;96:1707–1720. doi: 10.1016/j.bpj.2008.10.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Coghlin C, Carpenter B, Dundas SR, Lawrie LC, Telfer C, Murray GI. Characterization and over-expression of chaperonin t-complex proteins in colorectal cancer. J Pathol. 2006;210:351–357. doi: 10.1002/path.2056. [DOI] [PubMed] [Google Scholar]
- 44.McGinnis E, Williams LS. Role of histidine transfer ribonucleic acid in regulation of synthesis of histidyl-transfer ribonucleic acid synthetase of Salmonella typhimurium. J Bacteriol. 1972;109:505–511. doi: 10.1128/jb.109.2.505-511.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fountoulakis M, Tsangaris G, Oh J, Maris A, Lubec G. Protein profile of the HeLa cell line. J Chromatogr A. 2004;1038:247–265. doi: 10.1016/j.chroma.2004.03.032. [DOI] [PubMed] [Google Scholar]
- 46.Zhao K, Wang W, Rando OJ, Xue Y, Swiderek K, Kuo A, Crabtree GR. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell. 1998;95:625–636. doi: 10.1016/s0092-8674(00)81633-5. [DOI] [PubMed] [Google Scholar]
- 47.Mitsuzawa H, Kimura M, Kanda E, Ishihama A. Glyceraldehyde-3-phosphate dehydrogenase and actin associate with RNA polymerase II and interact with its Rpb7 subunit. FEBS Lett. 2005;579:48–52. doi: 10.1016/j.febslet.2004.11.045. [DOI] [PubMed] [Google Scholar]
- 48.Loeber R, Michaelson E, Fang Q, Campbell C, Pegg AE, Tretyakova N. Cross-linking of the DNA repair protein O6-alkylguanine DNA alkyltransferase to DNA in the presence of antitumor nitrogen mustards. Chem Res Toxicol. 2008;21:787–795. doi: 10.1021/tx7004508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cupo DY, Wetterhahn KE. Binding of chromium to chromatin and DNA from liver and kidney of rats treated with sodium dichromate and chromium(III) chloride in vivo. Cancer Res. 1985;45:1146–1151. [PubMed] [Google Scholar]
- 50.Nakano T, Morishita S, Katafuchi A, Matsubara M, Horikawa Y, Terato H, Salem AM, Izumi S, Pack SP, Makino K, Ide H. Nucleotide excision repair and homologous recombination systems commit differentially to the repair of DNA-protein crosslinks. Mol Cell. 2007;28:147–158. doi: 10.1016/j.molcel.2007.07.029. [DOI] [PubMed] [Google Scholar]
- 51.Nakano T, Katafuchi A, Matsubara M, Terato H, Tsuboi T, Masuda T, Tatsumoto T, Pack SP, Makino K, Croteau DL, Van Houten B, Iijima K, Tauchi H, Ide H. Homologous recombination but not nucleotide excision repair plays a pivotal role in tolerance of DNA-protein cross-links in mammalian cells. J Biol Chem. 2009;284:27065–27076. doi: 10.1074/jbc.M109.019174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.de Graaf B, Clore A, McCullough AK. Cellular pathways for DNA repair and damage tolerance of formaldehyde-induced DNA-protein crosslinks. DNA Repair (Amst) 2009;8:1207–1214. doi: 10.1016/j.dnarep.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.