Abstract
Segregation of chromosomes during meiosis I is triggered by separase cleavage of the cohesin's Rec8 subunit along chromosome arms. Centromeric cohesin is protected from separase cleavage during meiosis I by Sgo1/MEI-S332 proteins in complex with protein phosphatase 2A (PP2A). This retention of centromeric sister chromatid cohesion is essential for faithful segregation of chromatids during the second meiotic division. While Sgo1/PP2A complex is required for protecting centromeric sister chromatid cohesion during meiosis I, it is not known what renders the centromeric cohesion sensitive to separase cleavage during meiosis II. Our data suggest that the absence of Sgo1 and PP2A from meiosis II centromeres is not sufficient to render centromeric cohesion sensitive to cleavage by separase and additional factors are required to ensure the removal of centromeric cohesion during meiosis II.
Keywords: S. pombe, shugoshin, protein phosphatase 2A, meiosis, chromosome segregation
Introduction
Step-wise loss of sister chromatid cohesion, mediated by the cohesin protein complex, is essential for proper segregation of chromosomes during meiosis.1-4 In meiosis I, cohesin located on chromosome arms is cleaved by a protease called separase, thus allowing segregation of homologous chromosomes. Centromeric cohesin is refractory to separase cleavage during meiosis I and is only cleaved during meiosis II, hence allowing segregation of sister chromatids. The protection of centromeric cohesion during meiosis I requires the conserved Sgo1/MEI-S332 proteins that recruit the protein phosphatase 2A (PP2A) to centromeric regions.5-11 Centromeric PP2A then blocks cohesin cleavage at centromeres, possibly by dephosphorylating the cohesin subunit Rec8. In the absence of Sgo1, the amount of PP2A associated with centromeric regions is severely reduced and centromeric cohesin is cleaved together with arm cohesin during meiosis I.7 The fission yeast Sgo1 protein is expressed specifically during meiosis I and is downregulated at the onset of anaphase I.9,10 Although PP2A is present during both meiotic divisions, it is not associated with centromeres during meiosis II.7 In fission yeast, Sgo1 and PP2A are, therefore, associated with centromeres during meiosis I but not during meiosis II. While Sgo1 in complex with PP2A is required to protect the cleavage of centromeric cohesin by separase during meiosis I, it is not known what renders the centromeric cohesion sensitive to separase cleavage during meiosis II. We asked whether the absence of Sgo1 and PP2A from meiosis II centromeres renders centromeric cohesin sensitive to separase cleavage. If the absence of Sgo1 and PP2A from the centromeres were the sole reason for the dissolution of centromeric cohesion, then artificially targeting Sgo1 and PP2A to meiosis II centromeres should inhibit cleavage of cohesin at this location and, therby, hinder the second meiotic division.
Results and Discussion
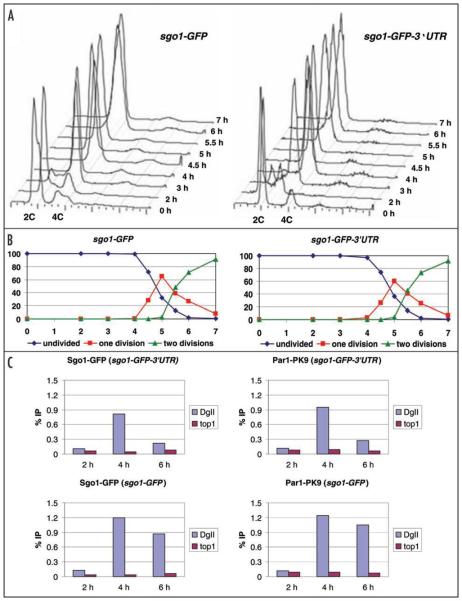
We have recently shown that expressing S. pombe Sgo1 in meiosis II is inhibited by sgo1's 3′UTR.10 Nevertheless, we artificially expressed Sgo1-GFP in meiosis II cells by removing its 3′UTR and analyzed progression through meiosis in pat1-synchronized cells. Similarly to the wild-type situation, cells lacking sgo1's 3′UTR underwent DNA replication and both meiotic divisions without any significant delay (Fig. 1A and 1B). This is consistent with our previous observation that spore viability is not significantly reduced when removal of sgo1's 3′UTR causes Sgo1-GFP to accumulate in meiosis II.10 Our observation is also consistent with data from other organisms where cohesion between sister chromatids can be released at the metaphase II/anaphase II transition even if Sgo1/MEI-S332 remains localized at centromeres.12-15
Figure 1.
The presence of Sgo1 and Par1 at centromeres during meiosis II does not interfere with chromosome segregation. Schizosaccharomyces pombe pat1-114 homozygous diploid cells expressing Sgo1-GFP from sgo1-GFP and Par1-PK9 (JG14859) and cells expressing Sgo1-GFP from sgo1-GFP-3′UTR and Par1-PK9 (JG14857) were arrested by nitrogen starvation and released into meiosis at 34°C by inactivation of pat1. Cells were harvested at the indicated time points (hours) after meiosis induction. (A) Flow cytometry profiles showing the DNA content at the indicated timepoints. (B) Cells were stained with DAPI and nuclei were counted in 100 cells per time point. Shown are the fraction of cells that contained one nucleus (diamonds), two nuclei (squares) or more than two nuclei (triangles) at the indicated time points. (C) Chromatin binding (DgII—outer centromere, top1—chromosome arm) of epitope-tagged proteins was analyzed by chromatin immunoprecipitation (ChIP) followed by quantitative real-time PCR.7
The fact that the Sgo1 expressed in meiosis II did not block the second meiotic division may be due to Sgo1 mislocalization or the inability of Sgo1 to recruit PP2A to centromeres. We therefore analyzed the centromeric localization of the Sgo1-GFP and PP2A subunit Par1-PK9 by chromatin immunoprecipitation (ChIP) followed by real-time PCR quantification. Both Sgo1-GFP and Par1-PK9 were enriched at centromeres during both meiosis I and II (Fig. 1C). Although we cannot exclude the possibility that the Sgo1 and PP2A levels at meiosis II centromeres are slightly different from those of meiosis I centromeres, it is likely that the presence of Sgo1 and PP2A at centromeres during meiosis II does not interfere with the dissolution of centromeric cohesion. While Sgo1 and PP2A are essential for the protection of centromeric cohesion during meiosis I, our data suggest that they are not able to perform this function during meiosis II. Therefore, additional factors are required to ensure the removal of centromeric cohesion during the second meiotic division.
What makes the centromeric cohesion resistant to separase cleavage during meiosis I but not during meiosis II? The difference is likely to be an intrinsic property of meiotic chromosomes since chromosomes placed on spindles of a different meiotic division behave as they would have done on their native spindle; i.e. meiosis I chromosomes moved to a meiosis II spindle segregate sister chromatids to the same spindle pole and meiosis II chromosomes moved to a meiosis I spindle segregate sister chromatids to the opposite spindle poles.16 The regulation of sister centromeric cohesion may be linked to differences in the arrangement of kinetochores in the two meiotic divisions.14,16 It has been suggested that the protection of centromeric cohesion does not operate when sister kinetochores attach to opposite spindle poles.17 The generated tension may displace Sgo1 from cohesin-enriched regions and de-protect cohesin molecules.18,19 It is possible that such a mechanism prevents the protection of centromeric cohesion during meiosis II. Alternatively, phosphorylation of the cohesin subunit Rec8, required for its efficient cleavage by separase during meiosis I,20-22 may not be a prerequisite for its removal during meiosis II.22 The insensitivity of meiosis II to Rec8's phosphorylation status may explain our observation that the presence of Sgo1 and PP2A at meiosis II centromeres does not interfere with the segregation of chromosomes.
Acknowledgements
This work was supported by Austrian Science Fund (P18955-B03). L.C. was a recipient of EMBO short-term fellowship. Z.L was a recipient of EP-Uninet fellowship. We thank Franz Klein, Yoshinori Watanabe, Kim Nasmyth and members of his lab for helpful discussions and Maria Siomos for critically reading the manuscript.
References
- 1.Page SL, Hawley RS. Chromosome choreography: the meiotic ballet. Science. 2003;301:785–9. doi: 10.1126/science.1086605. [DOI] [PubMed] [Google Scholar]
- 2.Petronczki M, Siomos MF, Nasmyth K. Un menage a quatre: the molecular biology of chromosome segregation in meiosis. Cell. 2003;112:423–40. doi: 10.1016/s0092-8674(03)00083-7. [DOI] [PubMed] [Google Scholar]
- 3.Ishiguro K, Watanabe Y. Chromosome cohesion in mitosis and meiosis. J Cell Sci. 2007;120:367–9. doi: 10.1242/jcs.03324. [DOI] [PubMed] [Google Scholar]
- 4.Lee J, Okada K, Ogushi S, Miyano T, Miyake M, Yamashita M. Loss of Rec8 from chromosome arm and centromere region is required for homologous chromosome separation and sister chromatid separation, respectively, in mammalian meiosis. Cell Cycle. 2006;5:1448–55. doi: 10.4161/cc.5.13.2903. [DOI] [PubMed] [Google Scholar]
- 5.Gregan J, Rabitsch PK, Sakem B, Csutak O, Latypov V, Lehmann E, Kohli J, Nasmyth K. Novel genes required for meiotic chromosome segregation are identified by a high-throughput knockout screen in fission yeast. Curr Biol. 2005;15:1663–9. doi: 10.1016/j.cub.2005.07.059. [DOI] [PubMed] [Google Scholar]
- 6.Kitajima TS, Sakuno T, Ishiguro K, Iemura S, Natsume T, Kawashima SA, Watanabe Y. Shugoshin collaborates with protein phosphatase 2A to protect cohesin. Nature. 2006;441:46–52. doi: 10.1038/nature04663. [DOI] [PubMed] [Google Scholar]
- 7.Riedel CG, Katis VL, Katou Y, Mori S, Itoh T, Helmhart W, Galova M, Petronczki M, Gregan J, Cetin B, Mudrak I, Ogris E, Mechtler K, Pelletier L, Buchholz F, Shirahige K, Nasmyth K. Protein phosphatase 2A protects centromeric sister chromatid cohesion during meiosis I. Nature. 2006;441:53–61. doi: 10.1038/nature04664. [DOI] [PubMed] [Google Scholar]
- 8.Marston AL, Tham WH, Shah H, Amon A. A genome-wide screen identifies genes required for centromeric cohesion. Science. 2004;303:1367–70. doi: 10.1126/science.1094220. [DOI] [PubMed] [Google Scholar]
- 9.Kitajima TS, Kawashima SA, Watanabe Y. The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis. Nature. 2004;427:510–7. doi: 10.1038/nature02312. [DOI] [PubMed] [Google Scholar]
- 10.Rabitsch KP, Gregan J, Schleiffer A, Javerzat JP, Eisenhaber F, Nasmyth K. Two fission yeast homologs of Drosophila Mei-S332 are required for chromosome segregation during meiosis I and II. Curr Biol. 2004;14:287–301. doi: 10.1016/j.cub.2004.01.051. [DOI] [PubMed] [Google Scholar]
- 11.Kerrebrock AW, Moore DP, Wu JS, Orr-Weaver TL. Mei-S332, a Drosophila protein required for sister-chromatid cohesion, can localize to meiotic centromere regions. Cell. 1995;83:247–56. doi: 10.1016/0092-8674(95)90166-3. [DOI] [PubMed] [Google Scholar]
- 12.Clarke AS, Tang TT, Ooi DL, Orr-Weaver TL. POLO kinase regulates the Drosophila centromere cohesion protein MEI-S332. Dev Cell. 2005;8:53–64. doi: 10.1016/j.devcel.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 13.Katis VL, Galova M, Rabitsch KP, Gregan J, Nasmyth K. Maintenance of cohesin at centromeres after meiosis I in budding yeast requires a kinetochore-associated protein related to MEI-S332. Curr Biol. 2004;14:560–72. doi: 10.1016/j.cub.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 14.Watanabe Y. Unified mode of centromeric protection by shugoshin in mammalian oocytes and somatic cells. FEBS Journal. 2007:19. doi: 10.1038/ncb1667. [DOI] [PubMed] [Google Scholar]
- 15.Penkner AM, Prinz S, Ferscha S, Klein F. Mnd2, an essential antagonist of the anaphase-promoting complex during meiotic prophase. Cell. 2005;120:789–801. doi: 10.1016/j.cell.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 16.Paliulis LV, Nicklas RB. The reduction of chromosome number in meiosis is determined by properties built into the chromosomes. J Cell Biol. 2000;150:1223–32. doi: 10.1083/jcb.150.6.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vaur S, Cubizolles F, Plane G, Genier S, Rabitsch PK, Gregan J, Nasmyth K, Vanoosthuyse V, Hardwick KG, Javerzat JP. Control of Shugoshin function during fission-yeast meiosis. Curr Biol. 2005;15:2263–70. doi: 10.1016/j.cub.2005.11.034. [DOI] [PubMed] [Google Scholar]
- 18.Gomez R, Valdeolmillos A, Parra MT, Viera A, Carreiro C, Roncal F, Rufas JS, Barbero JL, Suja JA. Mammalian SGO2 appears at the inner centromere domain and redistributes depending on tension across centromeres during meiosis II and mitosis. EMBO Rep. 2007;8:173–80. doi: 10.1038/sj.embor.7400877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang H, Feng J, Famulski J, Rattner JB, Liu ST, Kao GD, Muschel R, Chan GK, Yen TJ. Tripin/hSgo2 recruits MCAK to the inner centromere to correct defective kinetochore attachments. J Cell Biol. 2007;177:413–24. doi: 10.1083/jcb.200701122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee BH, Amon A. Role of Polo-like kinase CDC5 in programming meiosis I chromosome segregation. Science. 2003;300:482–6. doi: 10.1126/science.1081846. [DOI] [PubMed] [Google Scholar]
- 21.Clyne RK, Katis VL, Jessop L, Benjamin KR, Herskowitz I, Lichten M, Nasmyth K. Polo-like kinase Cdc5 promotes chiasmata formation and cosegregation of sister centromeres at meiosis I. Nat Cell Biol. 2003;5:480–5. doi: 10.1038/ncb977. [DOI] [PubMed] [Google Scholar]
- 22.Brar GA, Kiburz BM, Zhang Y, Kim JE, White F, Amon A. Rec8 phosphorylation and recombination promote the step-wise loss of cohesins in meiosis. Nature. 2006;441:532–6. doi: 10.1038/nature04794. [DOI] [PubMed] [Google Scholar]