Abstract
Diffusion-weighted magnetic resonance imaging (DWI) is a sensitive imaging biomarker for tumor cellularity. Functional diffusion maps (fDMs), which examine voxel-by-voxel changes in the apparent diffusion coefficient (ADC) calculated from serial DWIs, have previously been applied to regions of contrast-enhancement; however, application of fDMs to non-enhancing brain tumors has not been pursued. In this case study we demonstrate the utility of applying fDMs to regions of abnormal FLAIR signal intensity in a patient diagnosed with gliomatosis cerebri: a relatively rare, infiltrative, non-enhancing brain tumor. The absolute volume of hyper-cellularity extracted from fDMs was useful in tracking tumor growth, which correlated in time with a progressive decline in neurological status despite no change in traditional magnetic resonance images. Results of this study demonstrate the value of fDMs, applied to regions of FLAIR abnormal signal intensity, for localizing regions of hypercellularity and for monitoring overall tumor status.
Keywords: Gliomatosis cerebri, Functional diffusion maps, Diffusion MRI, Non-enhancing glioma, Cancer biomarkers
Introduction
Gliomatosis cerebri (GC) is a specific type of brain tumor classified by the WHO (World Health Organization) as having diffuse neoplastic glial cell infiltration with preservation of the architecture in normal-appearing surrounding tissue and absence of any identifiable tumor mass [1, 2]. In vivo diagnosis is often made solely on the basis of T2-weighted MRI or fluid-attenuated inversion recovery (FLAIR) abnormalities, since these tumors typically lack contrast-enhancement on T1-weighted images [3] and biopsy specimens often underestimate the degree of aggressive tumor infiltration and prognosis [4]. We propose the use of functional diffusion maps applied to regions of abnormal FLAIR signal intensity as a novel and sensitive method for clinically monitoring patients with GC and other non-enhancing brain tumors.
Apparent diffusion coefficients (ADCs) calculated from diffusion MRI data provide useful information about the suspected tumor microenvironment, exhibiting a marked decrease in regions with an increase in cellularity (i.e. hypercellularity) [5–7]. Functional diffusion maps (fDMs) were developed to examine voxel-by-voxel changes in ADC [8–11], thus providing spatial localization of hyper-cellular regions. Previous studies using the fDM technique have focused on contrast-enhanced regions within gliomas for analysis and visualization [8–11]; therefore, this technique had to be modified slightly for use in GC patients since regions of contrast-enhancement are often lacking [3]. In the current case study we demonstrate the potential of fDMs, applied to regions of abnormal FLAIR signal intensity, for tumor localization and clinical monitoring of a patient diagnosed with GC.
Case report
A 37-year-old, right-handed, white female with no past history of significant medical problems was admitted to the Neurology Service at our hospital after increasing confusion, fatigue, and lethargy for several weeks. Upon initial neurological examination (i.e. day 0) slow, wide-based gait was noted with a mild leftward drift. Slight left extremity weakness was also observed, along with left hand numbness and neglect. Standard clinical magnetic resonance images (see Clinical MRI methodology section for technical details) obtained on day 0 showed extensive, non-enhancing, abnormal T2-weighted signal involving the posterior right frontal cortex, and right temporoparietal white matter with extension into the splenium of the corpus callosum (see Fig. 1a–b). Serial conventional MR imaging showed no evidence of progressive cerebral swelling or other mass effect that would imply an increasing intracranial pressure. Single and multiple-voxel proton MR spectroscopy was performed at 1.5T on days 1 and 7 with the point resolved spectroscopy (PRESS) technique and a short TE value (35 ms). Increased choline peak amplitude (3.22 ppm), with increased myo-inositol (3.54 ppm) and decreased N-acetylaspartate (NAA) (2.01 ppm) were noted, with no definitive evidence of lipids or lactate in the spectra (0.8–1.5 ppm). STEALTH™-guided right temporal craniotomy, large open biopsy, and pathology performed on day 8 revealed an infiltrative, anaplastic mixed glioma (majority astrocytoma, minority oligodendroglioma) with no distinct tumor boundary (Fig. 1f). Twentyfive slides were sent to the Mayo Clinic in Rochester, MN, for consultation. Their evaluation confirmed our diagnosis of gliomatosis cerebri WHO grade 3. The patient was then started on a standard treatment regimen consisting of concurrent daily low-dose temozolomide (120 mg orally per day) and external beam radiation treatment, along with a continued dexamethasone dose of 4 mg QID from day 33 through day 75. From day 75 through the end of the observation period the dose of dexamethasone was reduced to 4 mg TID and no further chemotherapy or radiation therapy was implemented due to lack of evidence for progression on traditional MR images (Fig. 1e).
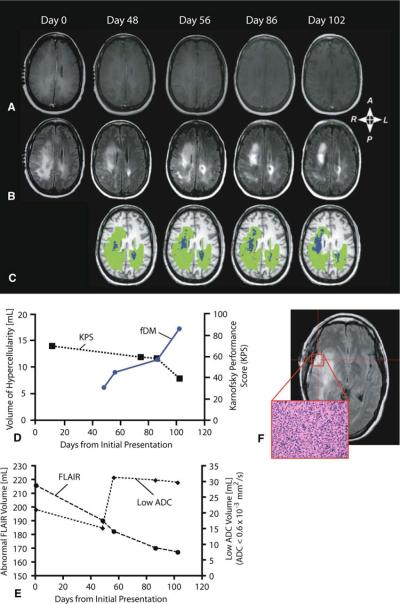
Fig. 1.
A 37-year-old woman diagnosed with gliomatosis cerebri. a Post-contrast T1-weighted images, b T2-weighted FLAIR images, and c functional diffusion maps (fDMs) at 0, 48, 56, 86, and 102 days from initial MRI scan upon admission. d Temporal changes in the volume of hypercelluarity (blue plot) and Karnofsky Performance Score (KPS; dashed black plot). e Temporal changes in the volume of abnormal FLAIR signal intensity and volume of tissue with abnormally low ADC (ADC < 0.6 × 10−3 mm2/s). f Hematoxylin and eosin (×200 magnification) slide of tissue from the biopsy site shows high cellularity and numerous mitotic figures. These results suggest fDMs can detect steady progressive disease in non-enhancing gliomas, even in the absence of progressive abnormalities on traditional MRI
Neurological status, as defined by the Karnofsky Performance Score (KPS), was approximately 70% during the first 11 days after admission according to experienced neuro-oncologists and neurologists working in our clinic. At day 48, fDMs (based on voxel-wise differences in ADC from day 0 to day 48) revealed spatially localized clusters of hypercellularity (blue regions in Fig. 1c), despite lack of changes on post-contrast T1-weighted or FLAIR images (Fig. 1a, b, e). By day 56, the volume of hypercellularity had increased approximately 43% relative to day 48 (Fig. 1d). The substantial increase in hypercellularity preceded a decrease in KPS from 70% (cares for self, but unable to carry on normal activity) to 60% (requires occasional assistance, but can care for most needs) by approximately 20 days. Clinical MRI showed a decrease in abnormal volume between day 56 and 86 (Fig. 1e); however, a 25% increase in the volume of hypercellularity with respect to the penultimate scan was observed (Fig. 1d). Despite this increase in hypercellular volume, neurological status did not change substantially at this time. The last set of fDMs, obtained on day 102, illustrated an additional 52% increase in hypercellular volume compared with day 86, contrary to the stable clinical MR images but consistent with the drastic decrease in KPS from 60 to 40% (disabled and requiring special assistance) at this time.
To verify that fDMs provide additional value beyond a simple ADC threshold, the volume of voxels having an ADC lower than 0.6 × 10−3 mm2/s were quantified (Fig. 1e). Results revealed an increase in the volume of low ADC voxels between days 48 and 56, during radiotherapy and concurrent chemotherapy. The volume of low ADC then appeared relatively stable through day 102, despite neurological progression. These results suggest that examining voxel-wise changes in ADC over time (i.e. fDMs) provide more information than solely examining the volume of tissue having a low ADC.
Clinical MRI methodology
Clinical MRI scans consisted of a spoiled gradient recalled (SPGR) anatomical scan, pre-contrast T1-weighted (T1) scan, post-contrast T1-weighted (T1 + C) scan, and a fluid-attenuated inversion recovery scan (FLAIR) collected on a 1.5-T MR scanner (Signal Excite or CVi; GE Medical Systems, Milwaukee, WI). The 3D SPGR images were acquired using an echo-time (TE)/repetition time (TR) = 3.16 ms/8.39 ms, number of averages (NEX) = 2, slice thickness = 1.3 mm (contiguous), flip angle = 10°, field-of-view (FOV) = 240 mm, and a matrix size of 256 × 192 (reconstructed images were zero-padded and interpolated to 256 × 256) resulting in a total of 123 to 128 images. The axial pre- and post-contrast T1-weighted images were acquired before and after administration of 10 cc/kg body weight of gadobenate dimeglumine (Multihance; Bracco Diagnostics Inc., Princeton, NJ) or 20 cc/kg body weight of gadodiamine (Omniscan; GE Healthcare Systems) contrast agent using a spin-echo (SE) sequence, TE/TR = 24.16 ms/666.7 ms, NEX = 1, slice thickness of 5 mm with 6.5 mm interslice gap, flip angle = 90°, FOV = 240 mm, and a matrix size of 256 × 192 (reconstructed images were zero-padded and interpolated to 256 × 256) resulting in a total of 22 to 24 images. The axial FLAIR images were collected using a SE readout, inversion time (TI) = 1,250 ms, TE/TR = 125.2 ms/10,000 ms, NEX = 1, slice thickness of 5 mm with 6.5 mm interslice gap, flip angle = 90°, FOV = 240 mm, and a matrix size of 256 × 224 (reconstructed images were zero-padded and interpolated to 256 × 256) resulting in a total of 22 to 24 images.
Functional diffusion maps (fDMs) methodology
Diffusion-weighted images (DWIs) were collected using an echo planar imaging (EPI), spin-echo (SE) pulse sequence at b = 0 s/mm2 and b = 1,000 s/mm2. The sequence parameters used were TE/TR = 102.2 ms/8,000 ms, NEX = 1, slice thickness of 5 mm with 6.5 mm interslice gap, flip angle = 90°, FOV = 240 mm, and a matrix size of 128 × 128 (reconstructed images were zero-padded and interpolated to 256 × 256). After collecting the images, ADC maps were calculated from the b = 1,000 and b = 0 images.
All images were registered to a baseline, post-surgical, SPGR anatomical image using a mutual information algorithm and a 9-degree of freedom transformation using FSL (FMRIB, Oxford, UK; http://www.fmrib.ox.ac.uk/fsl/). Fine registration (1–2° and 1–2 voxels) was then performed using a Fourier transform-based, 6-degree of freedom, rigid body registration algorithm [12] followed by visual inspection to ensure adequate alignment. Total computation time for image registration (4 scan days registered to baseline images, with 4 image datasets per scan session) was 28 min, 34 s using a 32-bit MacBook Pro with an Intel Core 2 Duo (2.4 GHz), 4 GB of RAM, and an 800 MHz speed data bus. After proper registration was verified, voxel-by-voxel subtraction was performed between baseline DWIs on day 0 and DWIs acquired at subsequent time points. As per the standard fDM analysis previously applied to post-contrast-enhancing lesions [8–11], individual voxels were stratified into three categories based on the change in ADC from the baseline ADC. Red voxels represented areas within the ROI where ADC increased (>0.55 × 10−3 mm2/s), blue voxels represented areas where ADC decreased (<0.55 × 10−3 mm2/s), and green voxels represented no change. We chose to use these particular ADC thresholds based on previous fDM investigations in post-contrast-enhancing lesions [8, 10, 11], which suggested this was the 95% confidence interval for reproducibility of the ADC measurement within normal-appearing white and gray matter. The absolute volume (in milliliters, ml) of hypercellularity (blue, decreasing ADC) was calculated based on the physical resolution of the diffusion images.
Discussion
This case study demonstrates the potential of fDMs to spatially localize and quantitatively monitor regions of hypercellularity in non-enhancing, infiltrative brain tumors. In this example, the information provided by diffusion mapping could not be obtained with traditional MRIs and is thus offers complementary information. In particular, application of fDMs to this high-grade infiltrative tumor type (GC) demonstrated a better correlation with or prediction of changing neurological status while traditional MRIs did not. In addition, this study further supports the use of fDMs applied to regions of abnormal FLAIR signal intensity [13, 14], compared to the traditional method of applying fDMs to contrast-enhancing regions exclusively. Since 14–45% of high-grade gliomas [15–18] and more than 50% of low-grade gliomas [18, 19] present with little or no contrast-enhancement, the results from this case report suggest that the fDM technique applied to regions of abnormal FLAIR signal intensity may prove to be a powerful clinical tool for localizing and monitoring infiltrating tumor behavior not observable with standard anatomic MRI.
The possibility of changes in edema, localized infection, subacute stroke, substantial gliosis, tissue swelling from seizure activity, and changes in steroid use must be considered during interpretation of fDMs, since many factors can change measurements of ADC. For example, the possibility of progressive ischemia from radiation therapy could have caused the ADC to decrease in regions we termed “hypercellular” on fDMs; however, progressive ischemia was ruled out through the use of serial monitoring with dynamic-susceptibility contrast perfusion imaging (DSC-MRI). As such, the terms “hypercellular” and “hypocellular” instead of “increasing ADC” and “decreasing ADC” may be misleading if these factors have not been ruled out during differential diagnosis. At our institution, for example, clinical translation of the fDM technique involves integration of interpretations from board certified neurologists and radiologists, as well as biophysicists, to rule out the possibility of these confounding factors.
In addition, the KPS scale used in the current study may not be the ideal neurological assessment, since it is not a linear scale. Depending on the functioning level of the patient, neurological status may change substantially between even 10-point declines. We are currently exploring more comprehensive and specific neurological tests to address these issues.
Acknowledgments
This study was funded by a grant from the National Institutes of Health and National Cancer Institute (NIH/NCI R21 CA109820), the MCW Translational Brain Tumor Program, Advancing a Healthier Wisconsin, and a MCW Cancer Center Fellowship.
References
- 1.Nevin S. Gliomatosis cerebri. Brain. 1938;61:170–191. [Google Scholar]
- 2.Lantos P, Bruner J. Gliomatosis cerebri. In: Kleihues P, Cavanee W, editors. Pathology and genetics of tumors of the nervous system. IARC Press; Lyon, France: pp. 92–93. [Google Scholar]
- 3.Taillibert S, Chodkiewicz C, Laigle-Donadey F, Napolitano M, Cartalat-Carel S, Sanson M. Gliomatosis cerebri: a review of 296 cases from the ANOCEF database and the literature. J Neurooncol. 2006;76:201–205. doi: 10.1007/s11060-005-5263-0. [DOI] [PubMed] [Google Scholar]
- 4.Galanaud D, Chinot O, Nicoli F, Confort-Gouny S, Le Fur Y, Barrie-Attarian M, Ranjeva JP, Fuetès S, Viout P, Figarella-Branger D, Cozzone PJ. Use of proton magnetic resonance spectroscopy of the brain to differentiate gliomatosis cerebri from low-grade glioma. J Neurosurg. 2003;98:269–276. doi: 10.3171/jns.2003.98.2.0269. [DOI] [PubMed] [Google Scholar]
- 5.Sugahara T, Korogi Y, Kochi M, Ikushima I, Shigematu Y, Hirai T, Okudo T, Liang L, Ge Y, Komohara Y, Ushio Y, Takahashi M. Usefulness of diffusion-weighted MRI with echo-planar technique in the evaluation of cellularity in gliomas. J Magn Reson Imaging. 1999;9:53–60. doi: 10.1002/(sici)1522-2586(199901)9:1<53::aid-jmri7>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 6.Lyng H, Haraldseth O, Rofstad EK. Measurements of cell density and necrotic fraction in human melanoma xenografts by diffusion weighted magnetic resonance imaging. Magn Reson Med. 2000;43:828–836. doi: 10.1002/1522-2594(200006)43:6<828::aid-mrm8>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 7.Chenevert TL, Stegman LD, Taylor JM, Robertson PL, Greenberg HS, Rehemtulla A, Ross BD. Diffusion magnetic resonance imaging: an early surrogate marker for therapeutic efficacy in brain tumors. J Natl Cancer Inst. 2000;92:2029–2036. doi: 10.1093/jnci/92.24.2029. [DOI] [PubMed] [Google Scholar]
- 8.Moffat BA, Chenevert TL, Lawrence TS, Meyer CR, Johnson TD, Dong Q, Tsien CI, Mukherji S, Quint DJ, Gebarski SS, Robertson PL, Junck L, Rehemtulla A, Ross BD. Functional diffusion map: a noninvasive MRI biomarker for early stratification of clinical brain tumor response. Proc Natl Acad Sci USA. 2005;102:5524–5529. doi: 10.1073/pnas.0501532102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moffat BA, Chenevert TL, Meyer CR, McKeever PE, Hall DE, Hoff BA, Johnson TD, Rehemtulla A, Ross BD. The functional diffusion map: an imaging biomarker for the early prediction of cancer treatment outcome. Neoplasia. 2006;8:259–267. doi: 10.1593/neo.05844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamstra DA, Galbán CJ, Meyer CR, Johnson TD, Sundgren PC, Tsien CI, Lawrence TS, Junck L, Ross DJ, Rehemtulla A, Ross BD, Chenevert TL. Functional diffusion map as an early imaging biomarker for high-grade glioma: correlation with conventional radiologic response and overall survival. J Clin Oncol. 2008;26:3387–3394. doi: 10.1200/JCO.2007.15.2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamstra DA, Chenevert TL, Moffat BA, Johnson TD, Meyer CR, Mukherji S, Quint DJ, Gebarski SS, Fan X, Tsien CI, Lawrence TS, Junck L, Rehemtulla A, Ross BD. Evaluation of the functional diffusion map as an early biomarker of time-to-progression and overall survival in high-grade glioma. Proc Natl Acad Sci USA. 2005;102:16759–16764. doi: 10.1073/pnas.0508347102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cox RW, Jesmanowicz A. Real-time 3D image registration for functional MRI. Magn Reson Med. 1999;42:1014–1018. doi: 10.1002/(sici)1522-2594(199912)42:6<1014::aid-mrm4>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 13.Ellingson BM, Malkin MG, Rand SD, Hoyt A, Connelly J, Bedekar DP, Kurpad SN, Schmainda KM. Comparison of cytotoxic and anti-angiogenic treatment responses using functional diffusion maps in FLAIR abnormal regions. Proc Intl Soc Magn Reson Med. 2009;17:1010. [Google Scholar]
- 14.Ellingson BM, Malkin MG, Rand SD, Bedekar DP, Schmainda KM. Functional diffusion maps applied to FLAIR abnormal areas are valuable for the clinical monitoring of recurrent brain tumors. Proc Intl Soc Magn Reson Med. 2009;17:285. [Google Scholar]
- 15.Chamberlain MC, Murovic JA, Levin VA. Absence of contrast enhancement on CT brain scans of patients with supratentorial malignant gliomas. Neurology. 1988;38:1371–1374. doi: 10.1212/wnl.38.9.1371. [DOI] [PubMed] [Google Scholar]
- 16.Barker FG, II, Chang SM, Huhn SL, Davis RL, Gutin PH, McDermott MW, Wilson CB, Prados MD. Age and the risk of anaplasia in magnetic resonance-nonenhancing supratentorial cerebral tumors. Cancer. 1997;80:936–941. [PubMed] [Google Scholar]
- 17.Ginsberg LE, Fuller GN, Hashmi M, Leeds NE, Schomer DF. The significance of lack of MR contrast enhancement of supratentorial brain tumors in adults: histopathological evaluation of a series. Surg Neurol. 1998;49:436–440. doi: 10.1016/s0090-3019(97)00360-1. [DOI] [PubMed] [Google Scholar]
- 18.Scott JN, Brasher PM, Sevick RJ, Rewcastle NB, Forsyth PA. How often are nonenhancing supratentorial gliomas malignant? A population study. Neurology. 2002;59:947–949. doi: 10.1212/wnl.59.6.947. [DOI] [PubMed] [Google Scholar]
- 19.Kondziolka D, Lunsford LD, Martinez AJ. Unreliability of contemporary neurodiagnostic imaging in evaluating suspected adult supratentorial (low-grade) astrocytoma. J Neurosurg. 1993;79:533–536. doi: 10.3171/jns.1993.79.4.0533. [DOI] [PubMed] [Google Scholar]