Abstract
BACKGROUND
Influenza viruses can generate novel reassortants in coinfected cells. The global circulation and occasional introductions of pandemic H1N1/2009 virus in humans and in pigs, respectively, may allow this virus to reassort with other influenza viruses. These possible reassortment events might alter virulence and/or transmissibility of the new reassortants. Investigations to detect such possible reassortants should be included as a part of pandemic influenza surveillance plans.
METHODS
We established a real-time reverse-transcription (RT)-PCR–based strategy for the detection of reassortment of pandemic H1N1/2009 virus. Singleplex SYBR green–based RT-PCR assays specific for each gene segment of pandemic H1N1/2009 were developed. These assays were evaluated with influenza viruses of various genetic backgrounds.
RESULTS
All human pandemic H1N1 (n = 27) and all seasonal human (n = 58) isolates were positive and negative, respectively, for all 8 segments. Of 48 swine influenza viruses isolated from our ongoing surveillance program of influenza viruses in swine, 10 were positive in all reactions. All 8 viral segments of these 10 samples were confirmed to be of pandemic H1N1 origin, indicating that these were caused by zoonotic transmissions from human to pigs. The 38 swine viruses that were nonpandemic H1N1/2009 had 1–6 gene segments positive in the tests. Further characterization of these nonpandemic H1N1/2009 swine viruses indicated that these PCR-positive genes were the precursor genes of the pandemic H1N1/2009 virus.
CONCLUSIONS
Our results demonstrated that these assays can detect reintroductions of pandemic H1N1/2009 virus in pigs. These assays might be useful screening tools for identifying viral reassortants derived from pandemic H1N1/2009 or its precursors.
Influenza A virus belongs to the family Orthomyxoviridae. Its genome contains 8 RNA segments of negative polarity [polymerase basic protein 2 (PB2),3 PB1, polymerase acidic protein (PA), and hemagglutinin (HA), nucleoprotein (NP), neuraminidase (NA), matrix (M), and nonstructure (NS) gene]. These viruses can exchange their gene segments in coinfected cells and produce progeny viruses with new genotypes. The unpredictability of the virus can be demonstrated by the recent emergence of the pandemic H1N1/2009 virus with genes originally derived from avian, swine, and human viruses (1, 2). Such gene reassortment events play key roles in influenza virus evolution and have direct impacts on human health. The global cocirculation of pandemic H1N1/2009 and seasonal influenza viruses has raised concerns that these human viruses might reassort, thereby generating novel viral genotypes with altered virulence and/or transmissibility (3, 4). Furthermore, surveillance has repeatedly detected pandemic H1N1/2009 virus in pigs (5, 6). This provides the opportunity for the pandemic influenza H1N1 virus to reassort with other swine or avian influenza viruses that are endemic or transiently circulating in swine herds. Such reassortment events will have unpredictable public health consequences. Hence, there is an urgent need for tests that can rapidly identify each influenza virus gene segment to be of pandemic H1N1/2009 origin.
To differentiate pandemic H1N1/2009 from seasonal influenza viruses, we designed primers that target pandemic H1N1/2009, but not seasonal human H1N1 and H3N2, viral segments (Table 1). These singleplex real-time reverse-transcription (RT)-PCR assays were specifically designed to cross-react with the precursor genes of pandemic H1N1/2009 (2) (see below). RNA from viral culture or clinical samples was extracted as described (7, 8). In this study we adapted a 2-step RT-PCR approach, which allowed us to reduce the cost of running the assays and to use the screened cDNA samples for other molecular analyses (e.g., sequencing). All the specimens were tested with blinding. For each specimen, cDNA molecules generated from a universal RT-PCR were used as DNA inputs in all the segment-specific PCRs. For a typical RT reaction, 10 μL reaction containing 5.5 μL of purified RNA, 100 U of Superscript II reverse transcriptase (Invitrogen), 2 μL of 5× FS (first-strand) buffer (Invitrogen), 0.1 μg of Uni12 (5′-AGCAAAAGCAGG-3′) (9), 10 mmol/L of dithiothreitol, and 0.5 mmol/L of deoxynucleoside triphosphates was incubated at 42 °C for 50 min, followed by a heat inactivation step (72 °C for 15 min). We amplified 1 μL of a 10-fold diluted cDNA sample in a 20-μL reaction containing 10 μL of Fast SYBR Green Master Mix (Applied Biosystems) and 0.5 μmol/L of the corresponding forward and reverse primers. All 8 segment-specific PCRs were optimized and performed simultaneously in a 7500 Sequence Detection System (Applied Biosystems) with the following conditions: 20 s at 95 °C, followed by 30 cycles of 95 °C for 3 s and 62 °C for 30 s. To determine the specificity of the assay, PCR products were subjected to a melting-curve analysis at the end of the amplification step (62–95 °C; temperature increment: 0.1 °C/s). We used 10-fold diluted plasmid DNA (pHW2000) samples (9) containing the corresponding genes of pandemic H1N1/2009 virus (A/California/4/09) as standards to generate standard curves over a range of 102 to 108 copies per reaction (see Fig. 1 in the Data Supplement that accompanies the online version of this article at http://www.clinchem.org/content/vol56/issue8). cDNA of A/California/4/09 virus was used as a positive control.
Table 1. Primers used for amplifying pandemic H1N1/2009 viral segments.
| Segment | Primera | Sequence |
|---|---|---|
| PB2 | PB2-1877F | 5′-AACTTCTCCCCTTTGCTGCT-3′ |
| PB2-2062R | 5′-GATCTTCAGTCAATGCACCTG-3′ | |
| PB1 | PB1-825F | 5′-ACAGTCTGGGCTCCCAGTA-3′ |
| PB1-1138R | 5′-TTTCTGCTGGTATTTGTGTTCGAA-3′ | |
| PA | PA-821F | 5′-GCCCCCTCAGATTGCCTG-3′ |
| PA-1239R | 5′-GCTTGCTAGAGATCTGGGC-3′ | |
| HA | HA-398F | 5′-GAGCTCAGTGTCATCATTTGAA-3′ |
| HA-570R | 5′-TGCTGAGCTTTGGGTATGAA-3′ | |
| NP | NP-593F | 5′-TGAAAGGAGTTGGAACAATAGCAA-3′ |
| NP-942R | 5′-GACCAGTGAGTACCCTTCCC-3′ | |
| NA | NA-163F | 5′-CATGCAATCAAAGCGTCATT-3′ |
| NA-268R | 5′-ACGGAAACCACTGACTGTCC-3′ | |
| M | M-504F | 5′-GGTCTCACAGACAGATGGCT-3′ |
| M-818R | 5′-GATCCCAATGATATTTGCTGCAATG-3′ | |
| NS | NS-252F | 5′-ACACTTAGAATGACAATTGCATCTGT-3′ |
| NS-691R | 5′-ACTTTTCATTTCTGCTCTGGAGGT-3′ |
Number indicates the nucleotide position of the first base in the target sequence (cRNA sense).
All 8 segment-specific PCRs achieved robust and highly specific DNA amplifications (see online Supplementary Fig. 1). These reactions were found to have a linear dynamic detection range from 102 to 108 copies/reaction. All the positive reactions yielded unique melting-curve patterns not observed in negative and water controls (Fig. 1 and online Supplementary Fig. 1). The melting temperature (Tm) of these PCR products was determined (see online Supplementary Fig. 1) and reactions with a Tm value within 2 SDs of the mean were considered to be positive. All serologically confirmed pandemic H1N1/2009 (n = 27) were positive in all 8 assays, as expected. Fifty-seven human seasonal viruses (H1N1 = 33, H3N2 = 25) were negative in all assays (Fig. 1A). These results also indicated that the tested human isolates are not reassortants between seasonal and pandemic H1N1 viruses.
Fig. 1. Real-time quantitative PCR assays for influenza viruses.
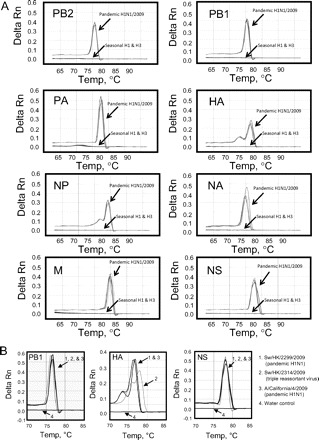
(A), Melting-curve analysis of PCR products generated in influenza segment-specific real-time PCR assays. The melting-curve signals of human pandemic H1N1/2009 (n = 4), seasonal H1 (n = 3), seasonal H3 (n = 3) are indicated as shown. (B), Melting-curve analysis of PCR products amplified in the PB1-, HA-, and NS-specific assays. The melting-curve signals of swine virus of pandemic H1N1/2009 origin (Sw/HK/2299/2009), triple reassortant swine virus (Swine/HK/2314/2009), human pandemic H1N1/2009(A/California/4/2009), and water control are indicated as shown. Delta Rn, normalized fluorescence signal.
We applied this method to the rapid identification and characterization of influenza viruses isolated in the course of our ongoing influenza virus surveillance in swine. Nasal and tracheal swab samples collected at an abattoir in Hong Kong were cultured in Madin Darby canine kidney cells or embryonated eggs as described previously (10). Viral cultures found to be positive by hemagglutination assays were tested by use of the established real-time PCR assays. Forty-eight hemagglutination-positive samples collected from January 2009 to January 2010 were tested with these assays. Ten viral isolates were identified as pandemic H1N1/2009 in all 8 segments. Results of full-genome sequencing confirmed that these 10 viruses were of pandemic H1N1/2009 origin (see online Supplemental Fig. 2, clade pH1N1), indicating that there were interspecies transmissions of H1N1/2009 from humans to pigs (11).
The 38 swine viruses that are nonpandemic H1N1/2009 had 1–6 (but not all 8) gene segments positive in the established PCR tests. Based on the genotyping data generated from the above assays, we further selected 30 representative viral samples for full genome analysis. These viruses were all confirmed to be swine H1 viruses and their gene segments were derived either from the triple reassortant (TR) or Eurasian avianlike (EA) swine influenza virus lineage (2). It should be noted that all of the PCR-positive viral segments fall into the sister group of pandemic H1N1 (see online Supplemental Fig. 2), which demonstrates the feasibility of using these real-time RT-PCR assays to detect genes from contemporary TR (PB2, PB1, PA, HA, NP, and NS) and EA (NA and M) swine viruses (2). All of these PCR-positive reactions, except those for the HA gene, had melting curves that are similar to those derived from the pandemic H1N1/2009 virus (Fig. 1B and data not shown). In contrast, melting-curve signals of the HA gene derived from the TR-H1 swine viruses were found to be different from that of the pandemic H1N1/2009 virus (Fig. 1B, Sw/HK/2314/2009, Tm = 77.8 °C). These results demonstrated that this HA-specific PCR assay can differentiate the pandemic (H1N1) 2009 virus from other contemporary swine viruses with the same HA lineage. Nonetheless, full viral genome sequencing is still required for identifying all the genetic variations in the viruses of interest.
The sequence similarity and diversity of influenza viruses were the major hurdles for the primer design of this study. We tried to test degenerated primers that can cross-react with both pandemic H1N1/2009 and its precursor genes, but these primers were found to be highly nonspecific. Nondegenerated primers were therefore deliberately used in this work. These nondegenerated primers might cross-react with genes from TR (PB2, PB1, PA, HA, NP, and NS) and EA (NA and M) swine viruses, with some minor sequence mismatches. It should be noted that the NP of Sw/HK/2314/09 (TR swine lineage) and some NA in the EA swine lineage (e.g., Sw/HK/1105) were negative in the corresponding PCR (see online Supplemental Fig. 2). These results were due to several major mismatched base pairs between the primers and targets. The primer mismatches observed in the NP of Sw/HK/2314/09 could not be identified in other swine NP sequences available from the Genbank. We also used these real-time PCR assays to test 3 avian (H5N1, H7N7, and H9N2) and 1 classical swine (H1N1) influenza viruses. None of these animal viruses produced positive results in these tests, except for the NS of the classical swine virus.
Our work demonstrates that these assays might help to identify swine virus reassortants of interesting genotypes. Indeed, in the course of this study, we successfully used these assays to identify a novel reassortant between swine and pandemic H1N1 viruses (11). Our real-time PCR results revealed that this novel reassortant contains a previously unidentified viral gene combination. The detailed characterization of this novel reassortant is described elsewhere (12).
By use of the 8 established real-time PCR assays we tested RNA from 2 randomly selected samples of the original swine swab specimens that contained the pandemic H1N1/2009 virus. Both samples were found to be positive in all the PCR assays, demonstrating that these assays are sensitive enough to detect the pandemic H1N1 virus from the original specimen. We also tested the 8 remaining original swine samples positive for the pandemic H1N1/2009 virus with the HA-specific assay, and 6 of them were positive for the HA gene. Because the performance of these tests is strongly influenced by the amount of virus in the original sample, negative PCR results from reactions with extremely low viral RNA inputs (e.g., RNA extracted from original specimens) should be interpreted cautiously.
We developed 8 real-time RT-PCR tests to detect genes derived from the pandemic H1N1/2009 virus or viral gene segments of the same lineage. We are currently using this approach to screen viruses isolated from our surveillance work in humans and other animals. It is expected that these assays might help us to identify reassortant viruses between human seasonal and pandemic viruses as well as those generated from coinfections of pandemic H1N1/2009 and swine influenza viruses in pigs. It should be noted that none of these assays can detect viral genes derived from seasonal human influenza viruses. For testing human specimens containing influenza virus, it would be beneficial to include the PCR testing algorithm recommended by the WHO (13).
Supplementary Material
3 Nonstandard abbreviations:
- PB2
polymerase basic protein 2
- PA
polymerase acidic protein
- HA
hemagglutinin
- NP
nucleoprotein
- NA
neuraminidase
- M
matrix
- NS
nonstructure
- RT
reverse transcription
- Tm
melting temperature
- TR
triple reassortant
- EA
Eurasian avianlike.
Footnotes
Author Contributions: All authors confirmed they have contributed to the intellectual content of this paper and have met the following 3 requirements: (a) significant contributions to the conception and design, acquisition of data, or analysis and interpretation of data; (b) drafting or revising the article for intellectual content; and (c) final approval of the published article.
Authors' Disclosures of Potential Conflicts of Interest: Upon manuscript submission, all authors completed the Disclosures of Potential Conflict of Interest form. Potential conflicts of interest:
Employment or Leadership: None declared.
Consultant or Advisory Role: None declared.
Stock Ownership: None declared.
Honoraria: None declared.
Research Funding: L.L.M. Poon, Research Grant Council of Hong Kong (HKU 773408M). This work was supported by the Area of Excellence Scheme of the University Grants Committee Hong Kong (grant AoE/M-12/06), the Research Fund for the Control of Infectious Disease Commissioned Project Food and Health Bureau, and the NIH (NIAID contract HHSN266200700005C).
Expert Testimony: None declared.
Role of Sponsor: The funding organizations played no role in the design of study, choice of enrolled patients, review and interpretation of data, or preparation or approval of manuscript.
References
- 1. Dawood FS, Jain S, Finelli L, Shaw MW, Lindstrom S, Garten RJ et al. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N Engl J Med 2009;360:2605–15. [DOI] [PubMed] [Google Scholar]
- 2. Smith GJ, Vijaykrishna D, Bahl J, Lycett SJ, Worobey M, Pybus OG et al. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 2009;459:1122–5. [DOI] [PubMed] [Google Scholar]
- 3. Belshe RB. Implications of the emergence of a novel H1 influenza virus. N Engl J Med 2009;360:2667–8. [DOI] [PubMed] [Google Scholar]
- 4. Peiris JS, Poon LL, Guan Y. Emergence of a novel swine-origin influenza A virus (S-OIV) H1N1 virus in humans. J Clin Virol 2009;45:169–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hofshagen M, Gjerset B, Er C, Tarpai A, Brun E, Dannevig B et al. Pandemic influenza A(H1N1)v: human to pig transmission in Norway? Euro Surveill 2009;14:19406. [DOI] [PubMed] [Google Scholar]
- 6. Howden KJ, Brockhoff EJ, Caya FD, McLeod LJ, Lavoie M, Ing JD et al. An investigation into human pandemic influenza virus (H1N1) 2009 on an Alberta swine farm. Can Vet J 2009;50:1153–61. [PMC free article] [PubMed] [Google Scholar]
- 7. Poon LL, Wong OK, Chan KH, Luk W, Yuen KY, Peiris JS, Guan Y. Rapid diagnosis of a coronavirus associated with severe acute respiratory syndrome (SARS). Clin Chem 2003;49:953–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Poon LL, Chan KH, Smith GJ, Leung CS, Guan Y, Yuen KY, Peiris JS. Molecular detection of a novel human influenza (H1N1) of pandemic potential by conventional and real-time quantitative RT-PCR assays. Clin Chem 2009;55:1555–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hoffmann E, Stech J, Guan Y, Webster RG, Perez DR. Universal primer set for the full-length amplification of all influenza A viruses. Arch Virol 2001;146:2275–89. [DOI] [PubMed] [Google Scholar]
- 10. Peiris JS, Guan Y, Markwell D, Ghose P, Webster RG, Shortridge KF. Cocirculation of avian H9N2 and contemporary “human” H3N2 influenza A viruses in pigs in southeastern China: potential for genetic reassortment? J Virol 2001;75:9679–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cohen J. Hog in the limelight: swine flu's got new genes on. Science Insider. http://news.sciencemag.org/scienceinsider/2010/03/hog-in-the-limelight-swine-flus.html (Accessed June 2010).
- 12. Vijaykrishna D, Poon LL, Zhu HC, Ma SK, Li OT, Cheung CL et al. Reassortment of pandemic H1N1/2009 influenza A virus in swine. Science 2010;328:1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. WHO. WHO information for laboratory diagnosis of pandemic (H1N1) 2009 virus in humans—revised. http://www.who.int/csr/resources/publications/swineflu/WHO_Diagnostic_RecommendationsH1N1_20090521.pdf (Accessed May 2010).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


